Abstract
Apoptosis or programmed cell death (PCD) was initially described in metazoans as a genetically controlled process leading to intracellular breakdown and engulfment by a neighboring cell . This process was distinguished from other forms of cell death like necrosis by maintenance of plasma membrane integrity prior to engulfment and the well-defined genetic system controlling this process. Apoptosis was originally described as a mechanism to reshape tissues during development. Given this context, the assumption was made that this process would not be found in simpler eukaryotes such as budding yeast. Although basic components of the apoptotic pathway were identified in yeast, initial observations suggested that it was devoid of prosurvival and prodeath regulatory proteins identified in mammalian cells. However, as apoptosis became extensively linked to the elimination of damaged cells, key PCD regulatory proteins were identified in yeast that play similar roles in mammals. This review highlights recent discoveries that have permitted information regarding PCD regulation in yeast to now inform experiments in animals.
Keywords: cyclin C, apoptosis, oxidative stress, mitochondria, signal transduction
TWO types of regulated cell death, necrosis and programmed cell death, have been described in budding yeast (Lin and Austriaco 2014). Necrotic cell death was originally characterized as a simple collapse of the cell leading to cell wall breakdown and ultimately lysis. However, more recent studies report the existence of a regulatory network governing necrotic cell death (Eisenberg et al. 2010). This review concentrates on programmed cell death (PCD) in yeast, which closely resembles the intrinsic or mitochondrial-derived apoptosis in multicellular organisms (Perrone et al. 2008). Mammalian apoptosis is initiated by accumulation of Bcl2 homology 3 (BH3) containing proteins such as Bax on the mitochondrial outer membrane. Bax induces pore formation leading to the release of cytochrome c, which stimulates a cascade of proteases termed cysteine-dependent aspartate-specific proteases or caspases (Danial and Korsmeyer 2004). Plants and fungi possess a related protease family called metacaspases (Uren et al. 2000). Metacaspases share sequence and functional similarities but differ with respect to substrate recognition sites (asparagine/lysine rather than aspartic acid). Budding yeast possesses a single metacaspase (Yca1) and BH3 domain protein (Ybh3), which are both required for oxidative stress-induced PCD. Standard assays for PCD, such as double strand breaks or phophatidylserine externalization (Annexin V staining), routinely used to monitor apoptosis in metazoans, are also employed to assay PCD in yeast (Madeo et al. 1997). However, following excessive damage, these PCD hallmarks may be joined by necrotic markers (e.g., propidium iodide permeability) (Yamaki et al. 2001). Therefore, it is important to note that these different cell-death modes can be observed simultaneously within a population and care should be used when judging the contribution that each death pathway has on overall cell viability.
Oxidative Stress, a Common Denominator for PCD Initiation
There are many stimuli, either externally or internally derived, able to induce PCD in yeast. For example, aging (Corte-Real and Madeo 2013), extreme pH environment (Ludovico et al. 2001), plant toxins (Narasimhan et al. 2001), defects in actin function (Gourlay and Ayscough 2006), osmotic stress (Silva et al. 2005), acetic acid (Ludovico et al. 2002), the presence of lipid hydroperoxides (Alic et al. 2003), and prolonged mating-factor exposure (Severin and Hyman 2002) (although the exact nature of this cell death is in question) (Zhang et al. 2006) all stimulate PCD. Although these stressors appear different, many have in common the ability to generate internal reactive oxygen species (ROS). For example, a specific mutation in Cdc48 induces PCD in yeast (Madeo et al. 1997) due to elevated ROS (Madeo et al. 1999) produced from defective mitochondria (Braun et al. 2006; Braun and Zischka 2008). Similarly, defects in endoplasmic reticulum (ER)-dependent protein folding also produces ROS (Tu and Weissman 2004) to levels sufficient to induce PCD (Haynes et al. 2004). In addition, defects in the electron transport chain (ETC) lead to ER-produced ROS through hyperactivation of the ER NADPH oxidase Yno1 (Leadsham et al. 2013). These findings demonstrate the intricate relationships that have evolved between organelles that produce and respond to ROS-induced damage. The transcriptional response to, and the macromolecular damage caused by, oxidative stress in yeast are the subject of several excellent reviews (Avery 2011; Farrugia and Balzan 2012; Morano et al. 2012) and will not be detailed here. Rather, given the universal nature of the oxidative stress response from yeast to humans, this review focuses on recent insights into the signaling systems that transduce the ROS signal and the effector proteins that coordinate the response between organelles in budding yeast.
External origins of ROS
The cell maintains redox homeostasis by balancing low-level ROS produced by organelles or exogenous sources with an arsenal of antioxidant enzymes that neutralize reactive oxygen (e.g., superoxide dismutase, catalase) or repair oxidative damage (e.g., chaperones, DNA repair enzymes) once it occurs (Perrone et al. 2008). However, increased internal ROS concentrations above a certain threshold lead to an accumulation of oxidized lipids, proteins, and DNA, collectively termed oxidative stress (Tsuzi et al. 2004; Drakulic et al. 2005; Temple et al. 2005). Exogenous sources of ROS occur in many forms including prooxidants such as H2O2 (Veal et al. 2007), exposure to heavy metals that stimulate superoxide production through the Fenton reaction (Liang and Zhou 2007; Nargund et al. 2008), or treatment with certain anticancer drugs (Almeida et al. 2008) (Figure 1). Exogenous ROS can alter plasma membrane characteristics that trigger sensors able to induce signal transduction pathways such as the cell-wall integrity pathway (Levin 2011) or the osmolarity-sensing pathway (Singh 2000; Bilsland et al. 2004) resulting in dramatic changes in the transcriptome. In addition, direct oxidation of transcription factors (e.g., Yap1) promotes stress-responsive gene transcription (Delaunay et al. 2000; Kuge et al. 2001).
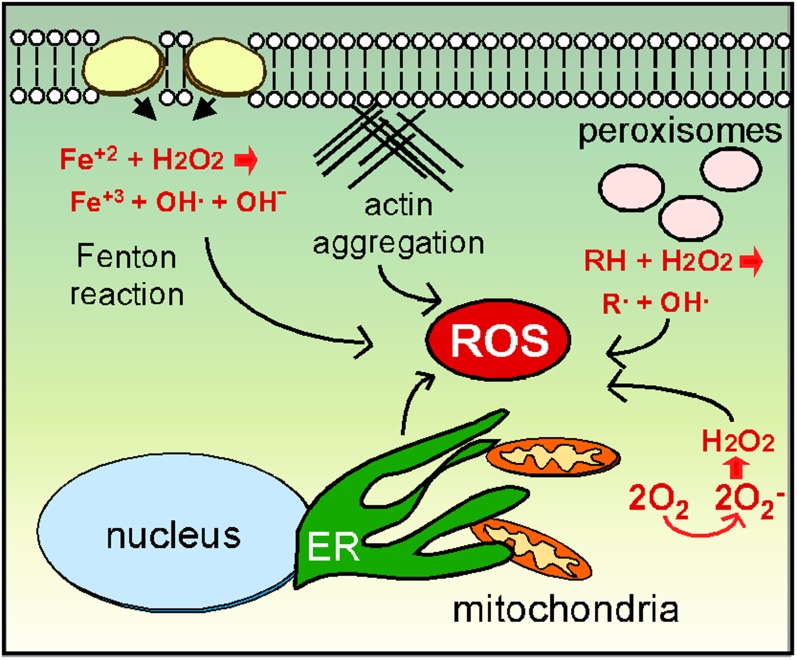
Figure 1.
Sources of reactive oxygen species (ROS). External sources of ROS can be derived from prooxidants like H2O2 or heavy metals that form reactive oxygen as a byproduct of the Fenton reaction. Internal sources of ROS sufficient to induce an oxidative stress response are the result of organelle dysfunction including the peroxisomes, mitochondria, and the ER. Fatty acid oxidation in the peroxisome leads to reactive oxygen intermediates. In the mitochondria, NADPH oxidase converts molecular oxygen to reactive species that are converted to H2O2 by super oxide dismutase. Actin aggregation indirectly induces ROS through elevated RAS signaling and regulation of mitochondrial dynamics.
Internal sources of ROS
Internal ROS is mostly derived from organelles performing their normal functions. The best studied and perhaps most important of these are the mitochondria (Figure 1). The mitochondrial function of ATP synthesis inherently produces reactive oxygen through the leakage of electrons from the ETC. This amount of ROS is limited and thought to represent a signaling molecule affecting many cellular processes (Guaragnella et al. 2012). However, mitochondrial dysfunction via mutations in ETC components, compounds that inhibit ETC function, or loss of mitochondrial inner membrane integrity, can generate sufficient ROS concentrations to induce the oxidative stress response (Eisenberg et al. 2007). For example, cytochrome c mutants display ETC defects that generate H2O2 (Barros et al. 2003). In addition, stimulating Ras signaling induces high protein kinase A (PKA) activity, leading to loss of mitochondrial integrity and elevated internal ROS (Hlavata et al. 2003; Hlavata et al. 2008; reviewed in Perrone et al. 2008).
In addition to defects in internal processes, mitochondrial-derived ROS can be caused via indirect mechanisms as well. For example, mutations or drugs that reduce actin dynamics cause elevated mitochondrially derived ROS (Gourlay et al. 2004). Interestingly, enhancing actin dynamics by deleting a gene (SCP1) encoding a bundling protein reduces ROS (Gourlay et al. 2004). A second connection between actin and mitochondrial fitness is observed during partitioning of this organelle to daughter cells. Myosin motors direct mitochondria toward the bud along F-actin cables to facilitate organelle partitioning (Mishra et al. 2014). In addition, a retrograde actin cable force is present that directs cargo toward the mother. Healthy mitochondria can bind the motors with sufficient strength to navigate to the bud despite the retrograde force moving in the opposite direction (McFaline-Figueroa et al. 2011). Pon and coworkers have likened this phenomenon to salmon swimming upstream against the river current (Higuchi et al. 2013). This process assures healthy mitochondria migrate to the bud while defective and ROS leaking mitochondria remain in the mother. As described later, this phenomenon may have consequences in aging-induced PCD.
In addition to the mitochondria, the ER is also a source of reactive oxygen in the cell. The ER provides the critical function of folding newly synthesized proteins and then sorting them for various cellular addresses (Chen et al. 2013). ER protein folding utilizes specialized chaperones (protein disulfide isomerases and Ero1) and an oxidative environment (Pollard et al. 1998) resulting in conversion of oxygen to H2O2 (Zito 2015). Defects in protein folding trigger the well-studied unfolded protein response (UPR) that induces ERO1 transcription. Prolonged Ero1p expression elevates ROS concentrations, ultimately leading to cell death (Haynes et al. 2004). Interestingly, the UPR leads to ROS generation by both the ER and the mitochondria. For example, Yno1/Aim14, a NADPH-oxidase found in the ER, generates ROS and promotes PCD (Rinnerthaler et al. 2012). Normally, Yno1-generated ROS concentrations are low and considered a signaling molecule in other fungi (Malagnac et al. 2004). However, yeast strains overexpressing Yno1 produce sufficient ROS to induce PCD. Although Yno1 is not part of the ER stress response, cytochrome oxidase c-defective mitochondria also raise Yno1 activity by preventing its normal turnover (Leadsham et al. 2013). Similar to the engineered overexpression studies, elevated Yno1 levels produce sufficient ROS to induce cell death. These studies highlight the intimate relationship between the ER and mitochondria with respect to ROS homeostasis.
Other organelles also contribute to oxidative stress. The peroxisome is important for β-oxidation of fatty acids that produce oxygen radicals and hydroperoxides (Manivannan et al. 2012). In addition, ROS are generated from peroxisomes that are defective in either form or function. For example, loss of Pex6 activity, a protein involved in peroxisome import, results in cells accumulating ROS to levels sufficient to induce cell death (Jungwirth et al. 2008). However, these cells show hallmarks of necrosis rather than PCD, indicating that internally produced ROS can induce multiple types of cell death. As discussed below, signaling systems that transduce the ROS signal have been identified. It will be interesting to determine if ROS generated from the mitochondria, ER, or peroxisomes activate similar or different pathways to trigger the oxidative stress response.
Aging and PCD
Two types of aging, chronological and replicative, are studied in yeast. Chronological aging examines how long cells can remain alive in stationary phase and is thought to be analogous to quiescent, postdifferentiated mammalian cells (Braun and Westermann 2011; Corte-Real and Madeo 2013). Conversely, replicative aging determines the number of cell divisions an individual mother cell can undergo and has been proposed to serve as a model for stem cell-like divisions. Both aging types are controlled by genetic factors as well as nutritional conditions, many of which impact mitochondrial function (Kaeberlein 2010; Corte-Real and Madeo 2013). Both replicative and chronological aging processes in budding yeast are driven by ROS accumulation that ultimately results in PCD (Laun et al. 2001; Fabrizio et al. 2004; Herker et al. 2004). For example, mother cells age through accumulation of oxidatively damaged proteins or mitochondria that are not passed on to their daughters (Aguilaniu et al. 2003; McFaline-Figueroa et al. 2011). In addition, protein aggregates are retained in aging mothers (Rujano et al. 2006; Spokoini et al. 2012) thus allowing daughter cells to start with a clean aging slate. The cell has multiple avenues to counteract these aging hallmarks. For example, protein aggregates are recognized as cellular damage and are degraded through the activity of protein chaperones and the metacaspase Mca1/Yca1 (Hill et al. 2014). Similarly, chronologically aged cells induce the NADP-dependent glutamate dehydrogenase Gdh3 that detoxifies ROS and prevents PCD initiation (Lee et al. 2012). These studies, as well as many others, provide a direct link between ROS accumulation, PCD initiation, and longevity. In mammals, this question is more complex as oxidative stress-induced pathology is influenced by the presence of cellular damage, and by other confounding factors including tissue type, stage in development, and the subcellular compartment in which the ROS originated (Cunningham et al. 2015). Therefore, the utility of yeast or mammalian tissue culture as a model to investigate some aspects of the free radical theory of aging may be limited.
Role of the mitochondrial dynamics in PCD execution
Mitochondria are dynamic organelles undergoing constant fusion and fission during normal cell division. The proper balance between these activities is required for normal mitochondrial function and to minimize ROS leakage (Ishihara et al. 2009; Wakabayashi et al. 2009). Under normal growth conditions, the mitochondria are elongated and interconnected. The high surface-to-volume ratio of this structure supports maximum ATP generation capacity and allows repair of damaged organelles through mixing of membrane components and recombination between nucleoids (Braun and Westermann 2011). Conversely, fission enhances the removal of damaged mitochondria via a specialized form of autophagy (mitophagy) and distribution of the organelle to daughter cells (Muller and Reichert 2011; Kurihara et al. 2012). The equilibrium between fission and fusion is controlled by the activity of conserved molecular machines driven by dynamin-like GTPases (see Westermann 2010 for review). In budding yeast, the fusion of the inner and outer mitochondrial membranes requires the Mgm1 and Fzo1 GTPases, respectively (Rapaport et al. 1998; Meeusen et al. 2006). Mitochondrial fission requires the GTPase Dnm1, which forms atypical helical filaments that first encircle, then constrict, mitochondria until scission is achieved (Otsuga et al. 1998; Bleazard et al. 1999; Sesaki and Jensen 1999). Recruitment of Dnm1 to the mitochondrial outer membrane (MOM) requires the receptor Fis1 (Mozdy et al. 2000; Tieu et al. 2002) and one of two adaptors, Mdv1 (Mozdy et al. 2000; Tieu et al. 2002; Cerveny and Jensen 2003) or Caf4 (Schauss et al. 2006; Motley et al. 2008). Interestingly, peroxisome fission also requires Fis1 and one of two dynamin-like proteins, Vps1 or Dnm1; the latter seems only important in cultures grown on oleic acid (Hoepfner et al. 2001; Kuravi et al. 2006). For mitochondria, fission often occurs at sites of interaction with the ER (Friedman et al. 2011). Many roles have been described for these junctions including sites of lipid transfer and Ca++ signaling (Michel and Kornmann 2012). Therefore, mitochondria–ER communication appears to be important for mitochondrial fission as well.
Of particular interest for this review, extensive mitochondrial fragmentation is a common feature following exposure to many types of damage including oxidative stress. Stress-induced mitochondrial hyperfission is conserved from yeast to mammals and represents an early morphological adaptation of the stress response (Youle and van der Bliek 2012). Mitochondrial hyperfission has been associated with the release of sequestered apoptotic factors (Frank et al. 2001; Breckenridge et al. 2003) while preventing fission protects cells from PCD. For example, mutants lacking Dnm1 or Fis1 are resistant to ROS-induced PCD (Fannjiang et al. 2004).
Although the basic fission machinery is required for stress-induced hyperfission, how their activity is enhanced occurs through an unlikely mechanism. In all eukaryotes examined, cyclin C (Ssn8) and Cdk8 (Ssn3) form a protein kinase that associates with the RNA polymerase II holoenzyme to control transcription (Bourbon 2008) (Figure 2). In budding yeast, this kinase represses ∼100 genes that are induced in response to environmental stress (Cooper et al. 1997; Holstege et al. 1998; van de Peppel et al. 2005). To relieve cyclin C–Cdk8 repression, stressed cells translocate cyclin C from the nucleus to the cytoplasm where it is ultimately destroyed through activity of the Not4 ubiquitin ligase (Cooper et al. 2012). However, cyclin C has a second function independent of Cdk8. Prior to its destruction in the cytoplasm, cyclin C associates with Mdv1 to induce extensive mitochondrial fragmentation (Cooper et al. 2014; reviewed in Strich and Cooper 2014). Deletion of its nuclear anchor, MED13, allows aberrant entry of cyclin C into the cytoplasm where it can induce fission in the absence of stress (Khakhina et al. 2014). These results indicate that cyclin C is both necessary and sufficient for hyperfission. Cyclin C-dependent hyperfission is directly related to the ability of the cell to induce PCD. Mutants lacking cyclin C are protected from ROS-induced PCD, whereas med13∆ mutants, in which the mitochondria are continuously fragmented, are hypersensitive to oxidative stress (Khakhina et al. 2014). It is important to note that continuous mitochondrial fission on its own is insufficient to induce cell death, although the health of this organelle suffers under these conditions through loss of mtDNA (Khakhina et al. 2014). These observations indicate that mitochondrial fragmentation potentiates the cell toward PCD initiation, but another stress signal is required to initiate this process. The role cyclin C plays in mitochondrial fission and PCD is remarkably well conserved. Mammalian cyclin C also translocates from the nucleus to the mitochondria in response to stress (Wang et al. 2015). Knockout mouse embryonic fibroblast (MEF) cells revealed that cyclin C is required for stress-induced mitochondrial fission and apoptotic cell death. Finally, the yeast cyclin C is able to induce complete mitochondrial fragmentation when purified protein is added to permeabilized MEF cultures. In the other direction, the human cyclin C complements the cell-death-resistance phenotype in cnc1∆ yeast mutants but not the transcriptional repression defect (Krasley et al. 2006). This analysis represents an example in which the direction of information understanding apoptotic control flowed from yeast to mammalian studies.
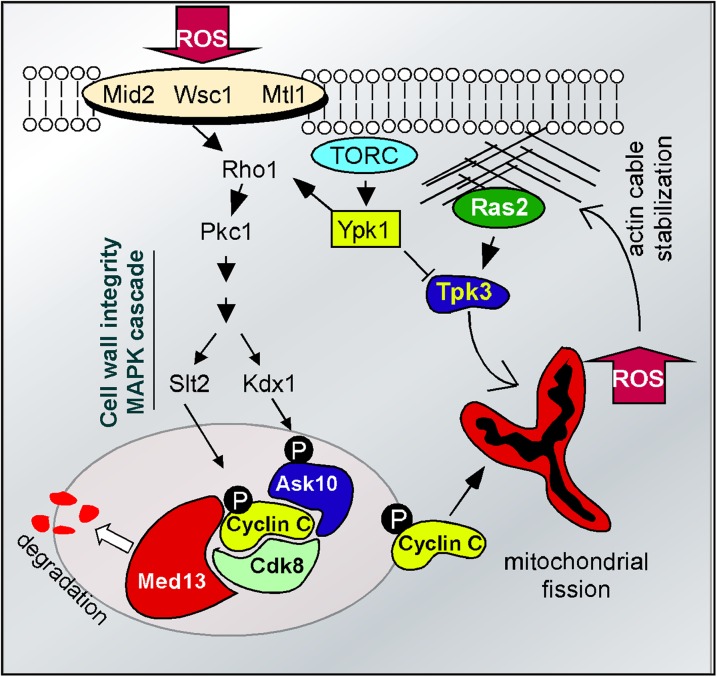
Figure 2.
Signal transduction pathways and the oxidative stress response. Exposure to ROS-generating compounds activates cell wall sensors (Mid2, Wsc1, Mtl1) connected to the cell wall integrity MAP kinase pathway. Activation of the MAP kinase (Slt2) and the pseudokinase (Kdx1) result in cyclin C and Ask10p phosphorylation, respectively. Phosphorylation of cyclin C initiates its relocation to the mitochondria. The role of Ask10 modification is currently unknown. Cyclin C relocalization also requires destruction of Med13, the anchor protein that tethers cyclin C to the RNA polymerase II holoenzyme. TORC signaling restricts ROS accumulation by regulating cyclin C levels through the CWI pathway and inhibiting the protein kinase A subunit Tpk3. When activated by Ras due to actin aggregation, Tpk3 causes accumulation of mitochondrial-dependent ROS that can result in more actin filament aggregation due to oxidation of conserved cysteine residues.
Signaling Pathways Directing ROS-Induced PCD: The Cell-Wall Integrity Pathway Controlling Cyclin C Nuclear Release
As indicated above, the failure to translocate cyclin C into the cytoplasm protects the cell from H2O2-induced PCD, while its aberrant release from the nucleus causes hypersensitivity to oxidative damage. Given the importance of this decision, it is not surprising that the switch controlling cyclin C release is complex and appears to be composed of at least two arms. First, the nuclear anchor, Med13 is destroyed in response to oxidative stress with kinetics similar to cyclin C release (Khakhina et al. 2014). This destruction is dependent on the 26S proteasome maturation factor Ump1, suggesting the involvement of ubiquitin-mediated proteolysis. Consistent with this model, the SCF ubiquitin ligase is required for ROS-induced Med13 destruction manner (K. F. Cooper, unpublished results). This result parallels a previous study in mammalian cells revealing a role for the SCF ligase in normal Med13 turnover (Davis et al. 2013). Currently, it is not known whether the yeast Med13 degradation is essential for cyclin C nuclear release or whether its proteolysis serves to prevent retention of the cyclin if it reenters the nucleus.
The second arm of the cyclin C control pathway is mediated by the cell-wall integrity MAP kinase pathway and includes a bifurcation at the MAP kinase step (Figure 2, see Table 1). The cell-wall integrity pathway transduces the oxidative stress signal from the cell wall to the nucleus to affect changes in transcription (Alic et al. 2003; Staleva et al. 2004; Vilella et al. 2005; Krasley et al. 2006; Petkova et al. 2010) and actin remodeling (Pujol-Carrion et al. 2013). Treating cells with H2O2 activates two cell-wall receptor groups containing Wsc1 and either Mid2 or Mtl1 (Vilella et al. 2005; Petkova et al. 2010; Jin et al. 2013). The receptors in turn stimulate the Rho1p GTPase, which activates Pkc1 and the cell-wall integrity MAP kinase pathway (Levin 2011). Recent studies revealed that the Slt2 MAPK directly phosphorylates cyclin C at Ser266 (Jin et al. 2014). Eliminating this phosphorylation site prevents cyclin C cytoplasmic translocation, while a phosphomimetic mutation enhances its translocation (Strich and Cooper 2014). The other branch contains the pseudokinase Kdx1 that associates with Ask10 (Jin et al. 2014), a previously identified cyclin C-associating factor (Cohen et al. 2003). Ask10 is required for cyclin C cytoplasmic translocation (Jin et al. 2014) and is phosphorylated in response to oxidative stress (Cohen et al. 2003). Surprisingly, Ask10p phosphorylation requires the two cell-wall-integrity pathway MAPKKs, Mkk1 and Mkk2, and the pseudokinase Kdx1, but not Slt2 (Cohen et al. 2003; Jin et al. 2014), suggesting the presence of yet another signaling system controlling cyclin C release.
Table 1. Signaling molecules.
| Yeast proteins | Mammalian orthologs | Function | Reference |
|---|---|---|---|
| Cdc48 | VCP/p97 | Protein turnover, ER stress response. | Madeo et al. (1997) |
| Mtl2, Mid2, Wsc1 | Unknown | Transmembrane receptors sensing oxidative stress. Required for cyclin C nuclear release. | Jin et al. (2013) |
| Ras2 | Ras | Relays plant antifungal and aging signals to stimulate PCD. | Narasimhan et al. (2005); Gourlay and Ayscough (2006) |
| Ste20 | Mst | Signals excessive mating pheromone response. Ca++ mobilization, phosphorylates histone H2B. | Ahn et al. (2005); Severin (2002) |
| Slt2/Mpk1 | MAP kinase | Downstream effector of cell-wall integrity. MAP kinase pathway. Required for ROS-induced cyclin C nuclear release. | Jin et al. (2014) |
| Kdx1 | MAP kinase pseudokinase | Downstream effector of cell-wall integrity. MAP kinase pathway. Required for ROS-induced cyclin C nuclear release. | Jin et al. (2014) |
Signaling Systems Directing ROS-Induced PCD: The Ras Signaling Pathway
Ras2 signaling also contributes to PCD initiation but in a manner different from the cell-wall integrity pathway. Rather than sensing and transducing the presence of ROS-induced damage, aberrant Ras signaling causes loss of mitochondrial integrity and subsequent ROS release (Smethurst et al. 2014). For example, aberrant actin aggregation, caused by specific actin monomer mutations or drugs that promote filament bundling, stimulates Ras2 signaling leading to activation of protein kinase A subunit Tpk3 (Gourlay et al. 2004; Gourlay and Ayscough 2006; Leadsham and Gourlay 2010). Tpk3 activation leads to elevated ROS levels in the cell (Figure 2). This system sets up a potential feedback loop in which the mitochondrial-derived ROS drives more actin aggregation through increased disulfide linkage of actin monomers (Haarer and Amberg 2004). Similarly, stationary-phase cells exposed to continuous Ras2 activation display elevated ROS levels and undergo PCD (Gourlay and Ayscough 2005). In both situations, constitutively active Ras results in a Tpk3-dependent loss of mitochondrial integrity and elevated ROS. These findings are similar to results obtained in mammalian cell culture in which prolonged RAS/RAF/ERK signaling also induces apoptosis (Cagnol and Chambard 2010).
The retrograde signaling pathway transduces information about mitochondrial activity and integrity to the nucleus to affect changes in gene expression (Jazwinski 2013). One component of this pathway is the target of rapamycin complex 2 (TORC2), which responds to cellular redox conditions through activation of Ypk1 protein kinase (Niles et al. 2014). Ypk1 stimulation activates the cell-wall integrity pathway through maintenance of sphingolipid levels required for proper localization of Rom2, the GAP activator of Rho1 (Niles et al. 2014) (Figure 2). This pathway keeps internal ROS levels in check, thus preventing cyclin C cytoplasmic relocalization and destruction (Niles and Powers 2014). Ypk1 also inhibits Tpk3 activity thereby maintaining normal mitochondrial function and reducing excessive ROS production (Niles et al. 2014). Thus, internal reactive oxygen levels are constantly monitored and adjusted to allow ROS to serve as a signaling molecule under certain situations.
Chromatin Modification and PCD Execution
Other, less-well-understood signaling pathways also play a conserved role in PCD execution. Ste20 is the founding member of the PAK (p21 activated protein kinase) protein family (Dan et al. 2001). A mammalian homolog of Ste20 (Mst1) (Creasy et al. 1996) is activated by caspase cleavage and phosphorylates histone H2B on serine 14 (Cheung et al. 2003). This modification promotes chromatin condensation and apoptotic cell death. Similarly, in response to oxidative stress, yeast Ste20 also phosphorylates the analogous H2B residue (Ser10) (Ahn et al. 2005) (Figure 3). Mutating H2B Ser10 to alanine protects the cell from H2O2-induced PCD, indicating an important role for this modification in the stress response. However, Ser10 phosphorylation occurs following H2O2 exposure only when the adjacent lysine (Lys11) is deacetylated by the Hos3 deacetylase (Ahn et al. 2006). Therefore, the stress signal must integrate both Hos3 and Ste20 activity. Conversely, monoubiquitylation of Lys123 on histone H2B by the Bre1 ligase prevents Yca1-dependent H2O2-induced PCD induced by chronological aging (Walter et al. 2010). In addition to H2B, histone H3 is also a target of posttranslational modifications that control PCD execution. Methylation of H3 Lys4 (H3K4) by Set1 reduces aging-dependent PCD (Walter et al. 2014). Consistent with this finding, deleting SET1 renders cells more sensitive to oxidative stress, whereas mutating the demethylase, JHD2, makes cells more resistant. These results reveal a chromatin-based tug-of-war between opposing signals that promote or inhibit PCD execution.
Figure 3.
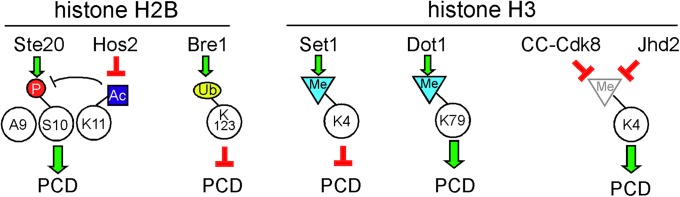
Chromatin modification and PCD regulation. H2K11 acetylation prevents H2S10 phosphorylation and chromatin condensation. Activation of Hos2 deacetylase removes the acetylation mark, allowing Ste20-dependent phosphorylation of H2S10 and PCD completion. Separate methylation marks at H3K4 and H3K79 have opposite impacts on PCD progression. Independent cyclin C–Cdk8 and Jhd2 demethylase activities reduce H3K4 methylation antagonizing Set1 activity and promoting cell death.
Regulators of mitochondrial outer membrane permeability
Similarly to mammalian cells, mitochondrial outer membrane permeabilization represents the commitment step to PCD execution (Green and Kroemer 2004; Antignani and Youle 2006). The loss of inner and outer mitochondrial membrane integrity is required for release of proapoptotic factors such as cytochrome c and two nucleases, Nuc1 and Aif1 (see Table 2). In mammalian cells, mitochondrial permeability is regulated through the competing activities of prosurvival (e.g., Bcl-2) and proapoptotic (e.g., Bax, Bak) Bcl-2 homology (BH) family members (Green and Kroemer 2004). In response to proapoptotic stimuli, Bax is recruited to the mitochondrial outer membrane, where it, in conjunction with Bak, forms pores that permeabilize the outer membrane. Therefore, the proper control of Bax and Bcl-2 activity is critical for the correct response to cellular damage. In yeast, loss of the BH-homology protein Ybh3 function reduces PCD efficiency in response to oxidative stress or aging, whereas its overexpression increases H2O2 sensitivity (Buttner et al. 2011). In addition, Ybh3 function requires the proposed BH3 domain and its activity is suppressed by expression of the prosurvival human Bcl-XL (Buttner et al. 2011). Finally, similarly to mammalian Bax, which relocalizes from the cytoplasm to the mitochondria (Lovell et al. 2008; reviewed in Renault et al. 2013), Ybh3 translocates from the vacuole to the mitochondria in response to stress (Buttner et al. 2011). Interestingly, Ybh3 function requires two associated proteins, Cor1 and Mir1, a ubiquinol–cytochrome c oxidoreductase subunit, and a mitochondrial phosphate carrier, respectively (Buttner et al. 2011). A similar function was confirmed for the mammalian orthologs of these proteins, QCR1 and PHC (Buttner et al. 2011). These results illustrate that, as with the cyclin C studies, the information obtained in analyzing yeast PCD is helping to instruct similar studies in mammalian cells.
Table 2. Executioner molecules.
| Yeast protein | Mammalian orthologs | Function | Reference |
|---|---|---|---|
| Yca1/Mca1 | Metacaspase | Cleave proteins | Madeo et al. (2002) |
| Nma111 | HtrA2/Omi | Nuclear serine protease required for ROS-induced PCD, cleaves Bir1. | Fahrenkrog et al. (2004) |
| Bir1 | IAP | Inhibitor of apoptosis. Substrate of Nma111. | Walter et al. (2006) |
| Aif1, Ndi1 | Aif/AMID | Mitochondrial nuclease released following permeability. Required for chromatin destruction. | Wissing et al. (2004) |
| Esp1 | Separin | Caspase-like protease, cleaves the cohesion Mcd1. | Yang et al. (2008) |
| Nuc1 | EndoG | Mitochondrial nuclease released following permeability. Required for chromatin destruction. | Buttner et al. (2007) |
| Kex1 | Caspase-like | Required for PCD in response to glycosylation defects, acetic acid, aging. | Hauptmann and Lehle (2008) |
| Cyclin C/Ssn3p | Cyclin C | Translocates to mitochondria following stress. Associates with mitochondrial fission machinery, required for mitochondrial fragmentation and permeability. | Cooper et al. (2012, 2014) |
| Ybh3 | Bax | Translocate to the mitochondria following stress. Induce mitochondrial outer membrane permeability. | Buttner et al. (2011) |
Executioners of the programmed cell death pathway
The ultimate goal of mitochondrial outer membrane permeability is the release of proapoptotic proteins including cytochrome c and two nucleases (Aif1 and Nuc1, see Table 2). Genetic studies have verified their role in PCD. Deleting these nucleases increases resistance to ROS-induced cell death, whereas their overexpression causes hypersensitivity (Wissing et al. 2004; Buttner et al. 2007). Similar to their mammalian counterparts Aif1 and EndoG, the yeast Aif1 and Nuc1 enter the nucleus and fragment chromatin. In mammalian cells, Aif1 activity requires association with the peptidylprolyl cis-trans isomerase cyclophilin A (Cande et al. 2004). Likewise, yeast Aif1 function is dependent on the yeast homolog of cyclophilin A (Cpr1) but not cyclophilin B (Cpr2). Taken together with the chromatin modification similarities, the nuclear changes in response to PCD execution are remarkably similar in yeast and mammals.
In mammalian cells, release of cytochrome c from mitochondria activates the caspase 9 initiator protease, which resides in the Apaf-1 apoptosome complex (Riedl and Salvesen 2007). Yeast genetic evidence indicates that cytochrome c is partially required for efficient PCD (Ludovico et al. 2002; Giannattasio et al. 2008), although no Apaf-1 ortholog has been identified. Genetic studies have identified several proteases that are required for PCD execution. Similarly to the caspase cascade in mammalian cells, Yca1 is activated by proteolysis and required for H2O2 and acetic acid-induced PCD (Madeo et al. 2002). Esp1 cleaves the cohesin Mcd1 in response to H2O2 treatment (Yang et al. 2008). Nma111, an ortholog of the human HtrA protease (Belanger et al. 2009), cleaves Bir1, the yeast ortholog of the mammalian inhibitor of apoptosis factor (Walter et al. 2006). Interestingly, these proteases exhibit full, partial, or no role in PCD execution depending on the stress (Liang et al. 2008; Madeo et al. 2009). These results suggest that different stimuli utilize specific caspases to execute the cell-death pathway. Alternatively, these proteases may perform overlapping activities masking their roles. The genetic analyses possible in yeast will be able to address whether functional overlap exists between proteases, or whether additional proteases exist that have not been ascribed a role in PCD control. In support of the latter possibility, protease activities that do not correspond to known caspase-like enzymes have been identified that are able to cleave fluorescent substrates with specificities similar to mammalian caspases (Wilkinson and Ramsdale 2011).
Coordinating the oxidative stress response throughout the cell
The oxidative-stress response is a culmination of changes in gene expression, organelle structure/function, and the cytoskeleton (reviewed in Smethurst et al. 2014). The organellar communication between the nucleus and mitochondria has been well studied (Hill and Van Remmen 2014; Shaughnessy et al. 2014). One example of this coordination, and insight into the complexity of the regulatory system governing this process, is demonstrated by examining cyclin C–Cdk8 activity in stressed and nonstressed cells. Several studies indicate both a prosurvival and prodeath role for cyclin C translocation from the nucleus to the cytoplasm. As described above, transcriptome analysis and more directed studies indicate that cyclin C-Cdk8 represses genes involved in the stress response (Cooper et al. 1997; Holstege et al. 1998). Therefore, the stress-induced nuclear release of cyclin C inactivates Cdk8, which remains nuclear (Cooper et al. 2012). The inactivation of Cdk8 allows complete and timely induction of meiotic (Cooper et al. 1997) or stress-responsive (Cooper et al. 2012) genes. In addition, cyclin C–Cdk8 restricts H3K4 methylation (Law and Ciccaglione 2015), a chromatin mark that prevents PCD-induced chromatin condensation (Walter et al. 2014). As H3K4 methylation is associated with transcriptional activation, these processes may well be related. Finally, cyclin C translocation to the cytoplasm induces extensive mitochondrial fragmentation, which may aid in the removal of damaged, ROS-leaking organelles (Youle and van der Bliek 2012). Therefore, derepressing stress response genes, enhancing H3K4 methylation, and removing damaged mitochondria all protect cells from PCD-inducing insults. These findings would explain why cnc1∆ mutants are more resistant to oxidative stress than either fis1∆ or dnm1∆ mutants (Cooper et al. 2014). However, the tipping point toward PCD is not mitochondrial fission. Therefore, the cell requires an additional signal, perhaps mitochondrial recruitment of Ybh3, to initiate the cell death pathway. In this process, the cell has utilized cyclin C relocalization to affect changes in gene expression, chromatin remodeling, and mitochondrial dynamics.
Physiological role for PCD in a single-celled organism
Due to the lack of obvious counterparts (e.g., p53, Bcl-2), many early studies considered yeast an in vivo test tube in which to analyze mammalian apoptotic regulators free of complications from yeast-based PCD (Manon et al. 1997; Ligr et al. 1998; Lisa-Santamaria et al. 2009; Greenwood and Ludovico 2010; reviewed in Silva et al. 2011; Clapp et al. 2012). However, extensive studies in budding yeast, fission yeast, and other single-cell eukaryotes challenge this view (Shemarova 2010). The identification of conserved regulatory proteins such as cyclin C, Ybh3, and Yca1 in budding yeast argues that PCD regulation is an ancient process. Several models have been put forth to explain the early evolutionary appearance of both pro- and antiapoptotic proteins (Taylor-Brown and Hurd 2013). Given the prominent role of the mitochondria in PCD regulation, it is not surprising that many models start at the conception of the eukaryotic cell with a bacterial parasite that eventually became endosymbiotic with its host. As cellular stress is a universal PCD initiator, one possibility is that ancient parasites recognized that their host was compromised and elicited cell death. This provided the bacteria a last gasp of nutrients as well as a free path to find another host (Nedelcu et al. 2011). As this relationship evolved to be less selfish and more mutually beneficial, the health of the newly identified mitochondria became coordinated with the rest of the cell.
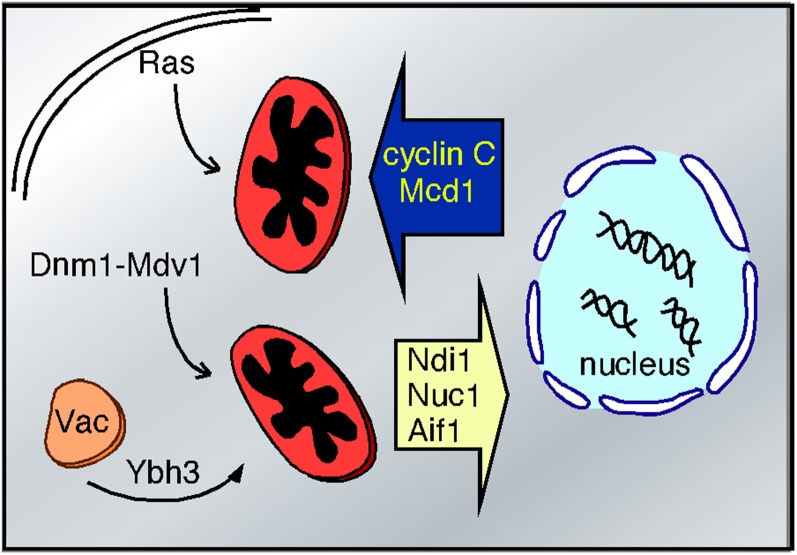
As the eukaryotic cell and its symbiont became more intertwined, regulatory systems evolved to take advantage of this unique situation (Ameisen 2002). For example, proteins that regulate the newly evolving PCD would be predicted to have “day jobs” required for normal cellular growth and development (Kroemer 1997). As described earlier, the yeast metacaspase Yca1 helps resolve protein aggregates. Nuc1 and Aif1 are important for mitochondrial RNA processing (Zassenhaus and Denniger 1994), while cyclin C regulates transcription in unstressed cells. However, it would be important for the cell to prevent the precocious activation of the PCD pathway until the proper stress signal occurs. To separate their cell death functions from their important day jobs, the cell utilizes regulated subcellular relocalization. For example, Ybh3 is found on the vacuole in unstressed cells but relocalizes to the mitochondria following stress (Figure 4). Likewise, cyclin C translocates from the nucleus to the mitochondria upon stress. Conversely, Nuc1 (Buttner et al. 2007), Aif1 (Wissing et al. 2004), and the AMID ortholog Ndi1 (Li et al. 2006) leave the mitochondria and are targeted to the nucleus where they fragment chromatin. In addition, Ras2 translocates from the plasma membrane in response to loss of mitochondria activity (Amigoni et al. 2013) or defects in ETC function (Leadsham et al. 2013). Mitochondrial Ras2 accumulation increases ROS production and sensitizes cells to PCD (Amigoni et al. 2013; Leadsham et al. 2013). In addition, H2O2 treatment induces the Esp1-dependent cleavage of the chromosomal cohesion Mcd1, resulting in a carboxyl terminal fragment that relocalizes to the mitochondria to drive loss of mitochondrial integrity (Yang et al. 2008). Therefore, the increased compartmentalization of the eukaryotic cell stages proteins at one address but allows their translocation to a different location in response to stress.
Figure 4.
Stress-induced relocalization of PCD regulators. The direction of protein translocation is depicted in stressed cells. Ras2 and Ybh3 transit to the mitochondria from the plasma membrane and vacuole (Vac), respectively. Mdv1 and Dnm1 relocalize from the cytoplasm to the mitochondria to induce fission. Cyclin C and Mcd1 relocalization from the nucleus to the mitochondria induces fission and loss of mitochondrial outer membrane integrity. Translocation of mitochondrial proteins Aif1, Nuc1, and Ndi1 stimulate chromatin breakdown in the nucleus.
Why would yeast maintain a cell death pathway? Altruism has been argued to provide a selective pressure to maintain PCD based on the normal colony mode for yeast growth. For example, colonies contain regions of young and old cells (Vachova and Palkova 2005; reviewed in Gourlay et al. 2006) with the death of older cells no longer capable of cell division providing metabolites for the younger cells. Sporulating colonies also provide evidence for more complex architecture in that zones of sporulating cells are separated by vegetative layers (Piccirillo and Honigberg 2010). This patterning is consistent with cells possessing different “identities” based on their age, location within the colony, and environmental signals. Therefore, recycling the components of severely damaged or nonreplicative cells within a colony would maximize growth chances for younger, reproductive cells.
Future challenges for the singled-cell model community
As the single-celled eukaryotic community moves past the “if” and “why” questions concerning PCD, attention can now be focused on “how.” It seems clear that as eukaryotes became more complex, additional layers of regulation were required to fulfill the requirements for tissue sculpting and removal of unwanted immune cells and damaged cells. Although some of these regulatory systems may be missing in single-celled organisms, the basic switches that recognize damage, transmit the signals, and coordinate the responses between the different organelles appear well conserved. Therefore, understanding how organelle-to-organelle communication coordinates both the stress response and ultimately PCD initiation represents a key challenge for the community in the near future.
Acknowledgments
I thank Scott Moye-Rowley, Campbell Gourlay, and Katrina Cooper for helpful comments and Katrina Cooper for permission to include unpublished results. This work was supported by a grant from the National Institutes of Health (GM113052).
Footnotes
Communicating editor: J. Rine
Literature Cited
- Aguilaniu H., Gustafsson L., Rigoulet M., Nystrom T., 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299: 1751–1753. [DOI] [PubMed] [Google Scholar]
- Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., et al. , 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120: 25–36. [DOI] [PubMed] [Google Scholar]
- Ahn S. H., Diaz R. L., Grunstein M., Allis C. D., 2006. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol. Cell 24: 211–220. [DOI] [PubMed] [Google Scholar]
- Alic N., Higgins V. J., Pichova A., Breitenbach M., Dawes I. W., 2003. Lipid hydroperoxides activate the mitogen-activated protein kinase Mpk1p in Saccharomyces cerevisiae. J. Biol. Chem. 278: 41849–41855. [DOI] [PubMed] [Google Scholar]
- Almeida B., Silva A., Mesquita A., Sampaio-Marques B., Rodrigues F., et al. , 2008. Drug-induced apoptosis in yeast. Biochim. Biophys. Acta 1783: 1436–1448. [DOI] [PubMed] [Google Scholar]
- Ameisen J. C., 2002. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 9: 367–393. [DOI] [PubMed] [Google Scholar]
- Amigoni L., Martegani E., Colombo S., 2013. Lack of HXK2 induces localization of active Ras in mitochondria and triggers apoptosis in the yeast Saccharomyces cerevisiae. Oxid. Med. Cell. Longev. 2013: 678473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignani A., Youle R. J., 2006. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr. Opin. Cell Biol. 18: 685–689. [DOI] [PubMed] [Google Scholar]
- Avery S. V., 2011. Molecular targets of oxidative stress. Biochem. J. 434: 201–210. [DOI] [PubMed] [Google Scholar]
- Barros M. H., Netto L. E., Kowaltowski A. J., 2003. H(2)O(2) generation in Saccharomyces cerevisiae respiratory pet mutants: effect of cytochrome c. Free Radic. Biol. Med. 35: 179–188. [DOI] [PubMed] [Google Scholar]
- Belanger K. D., Walter D., Henderson T. A., Yelton A. L., O’Brien T. G., et al. , 2009. Nuclear localisation is crucial for the proapoptotic activity of the HtrA-like serine protease Nma111p. J. Cell Sci. 122: 3931–3941. [DOI] [PubMed] [Google Scholar]
- Bilsland E., Molin C., Swaminathan S., Ramne A., Sunnerhagen P., 2004. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 53: 1743–1756. [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J. M., King E. J., Bale S., Mozdy A., et al. , 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H. M., 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36: 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R. J., Zischka H., 2008. Mechanisms of Cdc48/VCP-mediated cell death: from yeast apoptosis to human disease. Biochim. Biophys. Acta 1783: 1418–1435. [DOI] [PubMed] [Google Scholar]
- Braun R. J., Westermann B., 2011. Mitochondrial dynamics in yeast cell death and aging. Biochem. Soc. Trans. 39: 1520–1526. [DOI] [PubMed] [Google Scholar]
- Braun R. J., Zischka H., Madeo F., Eisenberg T., Wissing S., et al. , 2006. Crucial mitochondrial impairment upon CDC48 mutation in apoptotic yeast. J. Biol. Chem. 281: 25757–25767. [DOI] [PubMed] [Google Scholar]
- Breckenridge D. G., Stojanovic M., Marcellus R. C., Shore G. C., 2003. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 160: 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H., et al. , 2007. Endonuclease G regulates budding yeast life and death. Mol. Cell 25: 233–246. [DOI] [PubMed] [Google Scholar]
- Buttner S., Ruli D., Vogtle F. N., Galluzzi L., Moitzi B., et al. , 2011. A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 30: 2779–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnol S., Chambard J. C., 2010. ERK and cell death: mechanisms of ERK-induced cell death–apoptosis, autophagy and senescence. FEBS J. 277: 2–21. [DOI] [PubMed] [Google Scholar]
- Cande C., Vahsen N., Kouranti I., Schmitt E., Daugas E., et al. , 2004. AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene 23: 1514–1521. [DOI] [PubMed] [Google Scholar]
- Cerveny K. L., Jensen R. E., 2003. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol. Biol. Cell 14: 4126–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Novick P., Ferro-Novick S., 2013. ER structure and function. Curr. Opin. Cell Biol. 25: 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. L., Ajiro K., Samejima K., Kloc M., Cheung P., et al. , 2003. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113: 507–517. [DOI] [PubMed] [Google Scholar]
- Clapp C., Portt L., Khoury C., Sheibani S., Eid R., et al. , 2012. Untangling the roles of anti-apoptosis in regulating programmed cell death using humanized yeast cells. Front Oncol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T. J., Lee K., Rutkowski L. H., Strich R., 2003. Ask10p mediates the oxidative stress-induced destruction of the Saccharomyces cerevisiae C-type cyclin Ume3p/Srb11p. Eukaryot. Cell 2: 962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. F., Mallory M. J., Smith J. B., Strich R., 1997. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). EMBO J. 16: 4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. F., Scarnati M. S., Krasley E., Mallory M. J., Jin C., et al. , 2012. Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction. J. Cell Sci. 125: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. F., Khakhina S., Kim S. K., Strich R., 2014. Stress-induced nuclear-to-cytoplasmic translocation of cyclin C promotes mitochondrial fission in yeast. Dev. Cell 28: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corte-Real M., Madeo F., 2013. Yeast programed cell death and aging. Front Oncol 3: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasy C. L., Ambrose D. M., Chernoff J., 1996. The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J. Biol. Chem. 271: 21049–21053. [DOI] [PubMed] [Google Scholar]
- Cunningham G. M., Roman M. G., Flores L. C., Hubbard G. B., Salmon A. B., et al. , 2015. The paradoxical role of thioredoxin on oxidative stress and aging. Arch. Biochem. Biophys. 576: 32–38. [DOI] [PubMed] [Google Scholar]
- Dan I., Watanabe N. M., Kusumi A., 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11: 220–230. [DOI] [PubMed] [Google Scholar]
- Danial N. N., Korsmeyer S. J., 2004. Cell death: critical control points. Cell 116: 205–219. [DOI] [PubMed] [Google Scholar]
- Davis M. A., Larimore E. A., Fissel B. M., Swanger J., Taatjes D. J., et al. , 2013. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 27: 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay A., Isnard A. D., Toledano M. B., 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19: 5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakulic T., Temple M. D., Guido R., Jarolim S., Breitenbach M., et al. , 2005. Involvement of oxidative stress response genes in redox homeostasis, the level of reactive oxygen species, and ageing in Saccharomyces cerevisiae. FEMS Yeast Res. 5: 1215–1228. [DOI] [PubMed] [Google Scholar]
- Eisenberg T., Buttner S., Kroemer G., Madeo F., 2007. The mitochondrial pathway in yeast apoptosis. Apoptosis 12: 1011–1023. [DOI] [PubMed] [Google Scholar]
- Eisenberg T., Carmona-Gutierrez D., Buttner S., Tavernarakis N., Madeo F., 2010. Necrosis in yeast. Apoptosis 15: 257–268. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Battistella L., Vardavas R., Gattazzo C., Liou L. L., et al. , 2004. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166: 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B., Sauder U., Aebi U., 2004. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell Sci. 117: 115–126. [DOI] [PubMed] [Google Scholar]
- Fannjiang Y., Cheng W. C., Lee S. J., Qi B., Pevsner J., et al. , 2004. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 18: 2785–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia G., Balzan R., 2012. Oxidative stress and programmed cell death in yeast. Front Oncol 2: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E. S., Leitner W. W., Robert E. G., et al. , 2001. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1: 515–525. [DOI] [PubMed] [Google Scholar]
- Friedman J. R., Lackner L. L., West M., DiBenedetto J. R., Nunnari J., et al. , 2011. ER tubules mark sites of mitochondrial division. Science 334: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio S., Atlante A., Antonacci L., Guaragnella N., Lattanzio P., et al. , 2008. Cytochrome c is released from coupled mitochondria of yeast en route to acetic acid-induced programmed cell death and can work as an electron donor and a ROS scavenger. FEBS Lett. 582: 1519–1525. [DOI] [PubMed] [Google Scholar]
- Gourlay C. W., Ayscough K. R., 2005. Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J. Cell Sci. 118: 2119–2132. [DOI] [PubMed] [Google Scholar]
- Gourlay C. W., Ayscough K. R., 2006. Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 6487–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C. W., Carpp L. N., Timpson P., Winder S. J., Ayscough K. R., 2004. A role for the actin cytoskeleton in cell death and aging in yeast. J. Cell Biol. 164: 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C. W., Du W., Ayscough K. R., 2006. Apoptosis in yeast–mechanisms and benefits to a unicellular organism. Mol. Microbiol. 62: 1515–1521. [DOI] [PubMed] [Google Scholar]
- Green D. R., Kroemer G., 2004. The pathophysiology of mitochondrial cell death. Science 305: 626–629. [DOI] [PubMed] [Google Scholar]
- Greenwood M. T., Ludovico P., 2010. Expressing and functional analysis of mammalian apoptotic regulators in yeast. Cell Death Differ. 17: 737–745. [DOI] [PubMed] [Google Scholar]
- Guaragnella N., Zdralevic M., Antonacci L., Passarella S., Marra E., et al. , 2012. The role of mitochondria in yeast programmed cell death. Front Oncol 2: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B. K., Amberg D. C., 2004. Old yellow enzyme protects the actin cytoskeleton from oxidative stress. Mol. Biol. Cell 15: 4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hauptmann P., Lehle L., 2008. Kex1 protease is involved in yeast cell death induced by defective N-glycosylation, acetic acid, and chronological aging. J. Biol. Chem. 283: 19151–19163. [DOI] [PubMed] [Google Scholar]
- Haynes C. M., Titus E. A., Cooper A. A., 2004. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell 15: 767–776. [DOI] [PubMed] [Google Scholar]
- Herker E., Jungwirth H., Lehmann K. A., Maldener C., Frohlich K. U., et al. , 2004. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164: 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Vevea J. D., Swayne T. C., Chojnowski R., Hill V., et al. , 2013. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr. Biol. 23: 2417–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S., Van Remmen H., 2014. Mitochondrial stress signaling in longevity: a new role for mitochondrial function in aging. Redox Biol 2: 936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. M., Hao X., Liu B., Nystrom T., 2014. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science 344: 1389–1392. [DOI] [PubMed] [Google Scholar]
- Hlavata L., Aguilaniu H., Pichova A., Nystrom T., 2003. The oncogenic RAS2(val19) mutation locks respiration, independently of PKA, in a mode prone to generate ROS. EMBO J. 22: 3337–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavata L., Nachin L., Jezek P., Nystrom T., 2008. Elevated Ras/protein kinase A activity in Saccharomyces cerevisiae reduces proliferation rate and lifespan by two different reactive oxygen species-dependent routes. Aging Cell 7: 148–157. [DOI] [PubMed] [Google Scholar]
- Hoepfner D., van den Berg M., Philippsen P., Tabak H. F., Hettema E. H., 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155: 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., et al. , 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S. O., et al. , 2009. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11: 958–966. [DOI] [PubMed] [Google Scholar]
- Jazwinski S. M., 2013. The retrograde response: when mitochondrial quality control is not enough. Biochim. Biophys. Acta 1833: 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Parshin A. V., Daly I., Strich R., Cooper K. F., 2013. The cell wall sensors Mtl1, Wsc1, and Mid2 are required for stress-induced nuclear to cytoplasmic translocation of cyclin C and programmed cell death in yeast. Oxid. Med. Cell. Longev. 2013: 320823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Strich R., Cooper K. F., 2014. Slt2p phosphorylation induces cyclin C nuclear-to-cytoplasmic translocation in response to oxidative stress. Mol. Biol. Cell 25: 1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth H., Ring J., Mayer T., Schauer A., Buttner S., et al. , 2008. Loss of peroxisome function triggers necrosis. FEBS Lett. 582: 2882–2886. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., 2010. Lessons on longevity from budding yeast. Nature 464: 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhina S., Cooper K. F., Strich R., 2014. Med13p prevents mitochondrial fission and programmed cell death in yeast through nuclear retention of cyclin C. Mol. Biol. Cell 25: 2807–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasley E., Cooper K. F., Mallory M. J., Dunbrack R., Strich R., 2006. Regulation of the oxidative stress response through Slt2p-dependent destruction of cyclin C in Saccharomyces cerevisiae. Genetics 172: 1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., 1997. Mitochondrial implication in apoptosis. Towards an endosymbiont hypothesis of apoptosis evolution. Cell Death Differ. 4: 443–456. [DOI] [PubMed] [Google Scholar]
- Kuge S., Arita M., Murayama A., Maeta K., Izawa S., et al. , 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21: 6139–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi K., Nagotu S., Krikken A. M., Sjollema K., Deckers M., et al. , 2006. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119: 3994–4001. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Kanki T., Aoki Y., Hirota Y., Saigusa T., et al. , 2012. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 287: 3265–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laun P., Pichova A., Madeo F., Fuchs J., Ellinger A., et al. , 2001. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39: 1166–1173. [PubMed] [Google Scholar]
- Law M. J., Ciccaglione K., 2015. Fine-tuning of histone H3 Lys4 methylation during pseudohyphal differentiation by the CDK submodule of RNA Polymerase II. Genetics 199: 435–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadsham J. E., Gourlay C. W., 2010. cAMP/PKA signaling balances respiratory activity with mitochondria dependent apoptosis via transcriptional regulation. BMC Cell Biol. 11: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadsham J. E., Sanders G., Giannaki S., Bastow E. L., Hutton R., et al. , 2013. Loss of cytochrome c oxidase promotes RAS-dependent ROS production from the ER resident NADPH oxidase, Yno1p, in yeast. Cell Metab. 18: 279–286. [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Kim K. J., Kang H. Y., Kim H. R., Maeng P. J., 2012. Involvement of GDH3-encoded NADP+-dependent glutamate dehydrogenase in yeast cell resistance to stress-induced apoptosis in stationary phase cells. J. Biol. Chem. 287: 44221–44233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189: 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sun L., Liang Q., Wang J., Mo W., et al. , 2006. Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Mol. Biol. Cell 17: 1802–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Zhou B., 2007. Copper and manganese induce yeast apoptosis via different pathways. Mol. Biol. Cell 18: 4741–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Li W., Zhou B., 2008. Caspase-independent apoptosis in yeast. Biochim. Biophys. Acta 1783: 1311–1319. [DOI] [PubMed] [Google Scholar]
- Ligr M., Madeo F., Frohlich E., Hilt W., Frohlich K. U., et al. , 1998. Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett. 438: 61–65. [DOI] [PubMed] [Google Scholar]
- Lin S. J., Austriaco N., 2014. Aging and cell death in the other yeasts, Schizosaccharomyces pombe and Candida albicans. FEMS Yeast Res. 14: 119–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisa-Santamaria P., Neiman A. M., Cuesta-Marban A., Mollinedo F., Revuelta J. L., et al. , 2009. Human initiator caspases trigger apoptotic and autophagic phenotypes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., et al. , 2008. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135: 1074–1084. [DOI] [PubMed] [Google Scholar]
- Ludovico P., Sousa M. J., Silva M. T., Leao C., Corte-Real M., 2001. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 147: 2409–2415. [DOI] [PubMed] [Google Scholar]
- Ludovico P., Rodrigues F., Almeida A., Silva M. T., Barrientos A., et al. , 2002. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Frohlich E., Frohlich K. U., 1997. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139: 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Frohlich E., Ligr M., Grey M., Sigrist S. J., et al. , 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145: 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Herker E., Maldener C., Wissing S., Lachelt S., et al. , 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9: 911–917. [DOI] [PubMed] [Google Scholar]
- Madeo F., Carmona-Gutierrez D., Ring J., Buttner S., Eisenberg T., et al. , 2009. Caspase-dependent and caspase-independent cell death pathways in yeast. Biochem. Biophys. Res. Commun. 382: 227–231. [DOI] [PubMed] [Google Scholar]
- Malagnac F., Lalucque H., Lepere G., Silar P., 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41: 982–997. [DOI] [PubMed] [Google Scholar]
- Manivannan S., Scheckhuber C. Q., Veenhuis M., van der Klei I. J., 2012. The impact of peroxisomes on cellular aging and death. Front Oncol 2: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manon S., Chaudhuri B., Guerin M., 1997. Release of cytochrome c and decrease of cytochrome c oxidase in Bax-expressing yeast cells, and prevention of these effects by coexpression of Bcl-xL. FEBS Lett. 415: 29–32. [DOI] [PubMed] [Google Scholar]
- McFaline-Figueroa J. R., Vevea J., Swayne T. C., Zhou C., Liu C., et al. , 2011. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 10: 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., et al. , 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 127: 383–395. [DOI] [PubMed] [Google Scholar]
- Michel A. H., Kornmann B., 2012. The ERMES complex and ER-mitochondria connections. Biochem. Soc. Trans. 40: 445–450. [DOI] [PubMed] [Google Scholar]
- Mishra M., Huang J., Balasubramanian M. K., 2014. The yeast actin cytoskeleton. FEMS Microbiol. Rev. 38: 213–227. [DOI] [PubMed] [Google Scholar]
- Morano K. A., Grant C. M., Moye-Rowley W. S., 2012. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190: 1157–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A. M., Ward G. P., Hettema E. H., 2008. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 121: 1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A. D., McCaffery J. M., Shaw J. M., 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151: 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Reichert A. S., 2011. Mitophagy, mitochondrial dynamics and the general stress response in yeast. Biochem. Soc. Trans. 39: 1514–1519. [DOI] [PubMed] [Google Scholar]
- Narasimhan M. L., Damsz B., Coca M. A., Ibeas J. I., Yun D. J., et al. , 2001. A plant defense response effector induces microbial apoptosis. Mol. Cell 8: 921–930. [DOI] [PubMed] [Google Scholar]
- Narasimhan M. L., Coca M. A., Jin J., Yamauchi T., Ito Y., et al. , 2005. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol. Cell 17: 171–180. [DOI] [PubMed] [Google Scholar]
- Nargund A. M., Avery S. V., Houghton J. E., 2008. Cadmium induces a heterogeneous and caspase-dependent apoptotic response in Saccharomyces cerevisiae. Apoptosis 13: 811–821. [DOI] [PubMed] [Google Scholar]
- Nedelcu A. M., Driscoll W. W., Durand P. M., Herron M. D., Rashidi A., 2011. On the paradigm of altruistic suicide in the unicellular world. Evolution 65: 3–20. [DOI] [PubMed] [Google Scholar]
- Niles B. J., Powers T., 2014. TOR complex 2-Ypk1 signaling regulates actin polarization via reactive oxygen species. Mol. Biol. Cell 25: 3962–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles B. J., Joslin A. C., Fresques T., Powers T., 2014. TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Reports 6: 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., Keegan B. R., Brisch E., Thatcher J. W., Hermann G. J., et al. , 1998. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143: 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone G. G., Tan S. X., Dawes I. W., 2008. Reactive oxygen species and yeast apoptosis. Biochim. Biophys. Acta 1783: 1354–1368. [DOI] [PubMed] [Google Scholar]
- Petkova M. I., Pujol-Carrion N., Arroyo J., Garcia-Cantalejo J., Angeles de la Torre-Ruiz M., 2010. Mtl1 is required to activate general stress response through Tor1 and Ras2 inhibition under conditions of glucose starvation and oxidative stress. J. Biol. Chem. 285: 19521–19531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., Honigberg S. M., 2010. Sporulation patterning and invasive growth in wild and domesticated yeast colonies. Res. Microbiol. 161: 390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M. G., Travers K. J., Weissman J. S., 1998. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1: 171–182. [DOI] [PubMed] [Google Scholar]
- Pujol-Carrion N., Petkova M. I., Serrano L., de la Torre-Ruiz M. A., 2013. The MAP kinase Slt2 is involved in vacuolar function and actin remodeling in Saccharomyces cerevisiae mutants affected by endogenous oxidative stress. Appl. Environ. Microbiol. 79: 6459–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B., 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273: 20150–20155. [DOI] [PubMed] [Google Scholar]
- Renault T. T., Teijido O., Antonsson B., Dejean L. M., Manon S., 2013. Regulation of Bax mitochondrial localization by Bcl-2 and Bcl-x(L): keep your friends close but your enemies closer. Int. J. Biochem. Cell Biol. 45: 64–67. [DOI] [PubMed] [Google Scholar]
- Riedl S. J., Salvesen G. S., 2007. The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8: 405–413. [DOI] [PubMed] [Google Scholar]
- Rinnerthaler M., Buttner S., Laun P., Heeren G., Felder T. K., et al. , 2012. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc. Natl. Acad. Sci. USA 109: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano M. A., Bosveld F., Salomons F. A., Dijk F., van Waarde M. A., et al. , 2006. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 4: e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauss A. C., Bewersdorf J., Jakobs S., 2006. Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface. J. Cell Sci. 119: 3098–3106. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E., 1999. Division vs. fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147: 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F. F., Hyman A. A., 2002. Pheromone induces programmed cell death in S. cerevisiae. Curr. Biol. 12: R233–R235. [DOI] [PubMed] [Google Scholar]
- Shaughnessy D. T., McAllister K., Worth L., Haugen A. C., Meyer J. N., et al. , 2014. Mitochondria, energetics, epigenetics, and cellular responses to stress. Environ. Health Perspect. 122: 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemarova I. V., 2010. Signaling mechanisms of apoptosis-like programmed cell death in unicellular eukaryotes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155: 341–353. [DOI] [PubMed] [Google Scholar]
- Silva R. D., Sotoca R., Johansson B., Ludovico P., Sansonetty F., et al. , 2005. Hyperosmotic stress induces metacaspase- and mitochondria-dependent apoptosis in Saccharomyces cerevisiae. Mol. Microbiol. 58: 824–834. [DOI] [PubMed] [Google Scholar]
- Silva R. D., Manon S., Goncalves J., Saraiva L., Corte-Real M., 2011. The importance of humanized yeast to better understand the role of bcl-2 family in apoptosis: finding of novel therapeutic opportunities. Curr. Pharm. Des. 17: 246–255. [DOI] [PubMed] [Google Scholar]
- Singh K. K., 2000. The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic. Biol. Med. 29: 1043–1050. [DOI] [PubMed] [Google Scholar]
- Smethurst D. G., Dawes I. W., Gourlay C. W., 2014. Actin: a biosensor that determines cell fate in yeasts. FEMS Yeast Res. 14: 89–95. [DOI] [PubMed] [Google Scholar]
- Spokoini R., Moldavski O., Nahmias Y., England J. L., Schuldiner M., et al. , 2012. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Reports 2: 738–747. [DOI] [PubMed] [Google Scholar]
- Staleva L., Hall A., Orlow S. J., 2004. Oxidative stress activates FUS1 and RLM1 transcription in the yeast Saccharomyces cerevisiae in an oxidant-dependent Manner. Mol. Biol. Cell 15: 5574–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R., Cooper K. F., 2014. The dual role of cyclin C connects stress regulated gene expression to mitochondrial dynamics. Microbial Cell 1: 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Brown E., Hurd H., 2013. The first suicides: a legacy inherited by parasitic protozoans from prokaryote ancestors. Parasit. Vectors 6: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple M. D., Perrone G. G., Dawes I. W., 2005. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 15: 319–326. [DOI] [PubMed] [Google Scholar]
- Tieu Q., Okreglak V., Naylor K., Nunnari J., 2002. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158: 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzi D., Maeta K., Takatsume Y., Izawa S., Inoue Y., 2004. Regulation of the yeast phospholipid hydroperoxide glutathione peroxidase GPX2 by oxidative stress is mediated by Yap1 and Skn7. FEBS Lett. 565: 148–154. [DOI] [PubMed] [Google Scholar]
- Tu B. P., Weissman J. S., 2004. Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 164: 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren A. G., O’Rourke K., Aravind L. A., Pisabarro M. T., Seshagiri S., et al. , 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6: 961–967. [DOI] [PubMed] [Google Scholar]
- Vachova L., Palkova Z., 2005. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 169: 711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T. T., van Leenen D., et al. , 2005. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19: 511–522. [DOI] [PubMed] [Google Scholar]
- Veal E. A., Day A. M., Morgan B. A., 2007. Hydrogen peroxide sensing and signaling. Mol. Cell 26: 1–14. [DOI] [PubMed] [Google Scholar]
- Vilella F., Herrero E., Torres J., de la Torre-Ruiz M. A., 2005. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J. Biol. Chem. 280: 9149–9159. [DOI] [PubMed] [Google Scholar]
- Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., et al. , 2009. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186: 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D., Wissing S., Madeo F., Fahrenkrog B., 2006. The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J. Cell Sci. 119: 1843–1851. [DOI] [PubMed] [Google Scholar]
- Walter D., Matter A., Fahrenkrog B., 2010. Bre1p-mediated histone H2B ubiquitylation regulates apoptosis in Saccharomyces cerevisiae. J. Cell Sci. 123: 1931–1939. [DOI] [PubMed] [Google Scholar]
- Walter D., Matter A., Fahrenkrog B., 2014. Loss of histone H3 methylation at lysine 4 triggers apoptosis in Saccharomyces cerevisiae. PLoS Genet. 10: e1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Yan R., Cooper K. F., Strich R., 2015. Cyclin C mediates stress-induced mitochondrial fission and apoptosis. Mol. Biol. Cell 26: 1030–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., 2010. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11: 872–884. [DOI] [PubMed] [Google Scholar]
- Wilkinson D., Ramsdale M., 2011. Proteases and caspase-like activity in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 39: 1502–1508. [DOI] [PubMed] [Google Scholar]
- Wissing S., Ludovico P., Herker E., Buttner S., Engelhardt S. M., et al. , 2004. An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 166: 969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki M., Umehara T., Chimura T., Horikoshi M., 2001. Cell death with predominant apoptotic features in Saccharomyces cerevisiae mediated by deletion of the histone chaperone ASF1/CIA1. Genes Cells 6: 1043–1054. [DOI] [PubMed] [Google Scholar]
- Yang H., Ren Q., Zhang Z., 2008. Cleavage of Mcd1 by caspase-like protease Esp1 promotes apoptosis in budding yeast. Mol. Biol. Cell 19: 2127–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J., van der Bliek A. M., 2012. Mitochondrial fission, fusion, and stress. Science 337: 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zassenhaus H. P., Denniger G., 1994. Analysis of the role of the NUC1 endo/exonuclease in yeast mitochondrial DNA recombination. Curr. Genet. 25: 142–149. [DOI] [PubMed] [Google Scholar]
- Zhang N. N., Dudgeon D. D., Paliwal S., Levchenko A., Grote E., et al. , 2006. Multiple signaling pathways regulate yeast cell death during the response to mating pheromones. Mol. Biol. Cell 17: 3409–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E., 2015. ERO1: a protein disulfide oxidase and H2O2 producer. Free Radic. Biol. Med. 83: 299–304. [DOI] [PubMed] [Google Scholar]