Abstract
A central goal in the development of genome engineering technology is to reduce the time and labor required to produce custom genome modifications. Here we describe a new selection strategy for producing fluorescent protein (FP) knock-ins using CRISPR/Cas9-triggered homologous recombination. We have tested our approach in Caenorhabditis elegans. This approach has been designed to minimize hands-on labor at each step of the procedure. Central to our strategy is a newly developed self-excising cassette (SEC) for drug selection. SEC consists of three parts: a drug-resistance gene, a visible phenotypic marker, and an inducible Cre recombinase. SEC is flanked by LoxP sites and placed within a synthetic intron of a fluorescent protein tag, resulting in an FP–SEC module that can be inserted into any C. elegans gene. Upon heat shock, SEC excises itself from the genome, leaving no exogenous sequences outside the fluorescent protein tag. With our approach, one can generate knock-in alleles in any genetic background, with no PCR screening required and without the need for a second injection step to remove the selectable marker. Moreover, this strategy makes it possible to produce a fluorescent protein fusion, a transcriptional reporter and a strong loss-of-function allele for any gene of interest in a single injection step.
Keywords: CRISPR/Cas9, homologous recombination, gene tagging, Caenorhabditis elegans, self-excising cassette
A common goal in biological and biomedical research is to visualize the localization of a protein of interest within a cell or organism. This is often accomplished by fusing GFP or another fluorescent protein (FP) to the protein of interest. In the nematode Caenorhabditis elegans, GFP fusions were historically generated by injecting plasmids into the gonad of the adult hermaphrodite worm, resulting in the formation of extrachromosomal arrays (Mello et al. 1991). However, the resulting fusion proteins were typically strongly overexpressed in somatic tissues and silenced in the germline. Microparticle bombardment allowed the generation of low-copy transgenes that in some cases more closely recapitulated endogenous expression levels (Praitis et al. 2001; Sarov et al. 2012), but this technique is inefficient, time consuming, and difficult and requires expensive equipment and materials. More recently, we and others have reported CRISPR/Cas9-based approaches that together can be used to make essentially any desired change to the C. elegans genome, including insertion of GFP into endogenous loci (Friedland et al. 2013; Dickinson et al. 2013; Lo et al. 2013; Chiu et al. 2013; Cho et al. 2013; Katic and Großhans 2013; Tzur et al. 2013; Waaijers et al. 2013; Chen et al. 2013; Zhao et al. 2014; Kim et al. 2014; Arribere et al. 2014; Paix et al. 2014; Ward 2015; Farboud and Meyer 2015). The resulting GFP knock-in strains express 100% labeled protein under the control of all native regulatory elements, resulting in endogenous levels and patterns of expression in all cases reported to date (Dickinson et al. 2013; Kim et al. 2014).
Our published approach for generating GFP knock-ins (Dickinson et al. 2013) made use of a selection strategy that was originally developed for single-copy transgene construction (Frøkjaer-Jensen et al. 2008, 2012). This strategy is based on rescue of an unc-119 mutant phenotype. unc-119(ed3) animals are nearly paralyzed, and when a functional copy of unc-119 is integrated into the genome along with GFP or another modification, knock-in animals are easily identified by their wild-type movement. Although this strategy is extremely robust, it suffers from several important limitations. First, knock-ins must be generated in an unc-119 mutant background. unc-119 mutant animals are sick, difficult to inject, and likely carry additional undesired background mutations besides the mutation in unc-119. Second, the unc-119 rescue strategy requires integration of unc-119(+) into the genome along with the desired genome modification. unc-119(+) can be flanked by LoxP sites, allowing it to be removed from the genome after knock-in isolation by injecting a plasmid encoding Cre recombinase (Dickinson et al. 2013); however, this requires an extra injection step, followed by outcrossing to remove the unc-119(ed3) mutant allele and still leaves behind a 34-bp LoxP scar. Finally, our published approach required construction of a complex homologous repair template plasmid for each new knock-in, which represents a significant investment of time and materials. These limitations have motivated other groups to develop alternative screening strategies that do not involve inserting a selectable marker along with the desired modification (Kim et al. 2014; Arribere et al. 2014; Paix et al. 2014; Ward 2015). Although valuable, these alternative strategies rely on PCR screening to identify knock-in animals, which is much more labor intensive than unc-119 selection. In addition, selectable markers allow one to interrogate all progeny of injected animals (∼10,000 in a typical experiment), whereas it is impractical to screen more than a few hundred animals by PCR. As a result, selection-based strategies are less sensitive to the choice of sgRNA and can more robustly identify genome editing events that occur at low frequency.
Here, we present a novel selection strategy especially tailored for inserting fluorescent proteins into the genome of C. elegans. Our approach does not require PCR screening or a second injection step; it inserts fluorescent proteins into the genome cleanly, leaving no exogenous sequences outside the fluorescent protein tag; and it can be used in wild-type animals and in most other genetic backgrounds. Moreover, this strategy can be used to produce a fluorescent protein fusion, a transcriptional reporter, and a strong loss-of-function allele for any gene of interest in a single injection step. Central to our approach is a new self-excising drug selection cassette that affords the benefits of positive selection without the limitations associated with unc-119 selection. The total amount of hands-on labor required to tag a new gene is less than 1 day, which makes our new protocol the least labor-intensive approach reported to date for construction of new fluorescent protein fusions in C. elegans, to our knowledge.
Materials and Methods
Availability of protocols and reagents
A complete protocol for gene tagging using the self-excising cassette (SEC) is included in the Supporting Information. A continuously updated version will also be available on our website (http://wormcas9hr.weebly.com). FP–SEC vectors carrying worm codon-optimized GFP, the yellow FP YPET and the red FPs mKate2 and TagRFP-T will be deposited at Addgene. The mNeonGreen (mNG) vector is available from the authors upon completion of a license agreement.
Strains and culture conditions
Supporting Information, Table S1 lists the strains constructed for this study. Bristol N2 was used as wild type and is the parent strain of all new strains reported here. Worms were raised on standard NGM plates, fed Escherichea coli OP50, and kept at 20° except where otherwise noted.
Gene tagging using SEC
To generate knock-in strains using long homology arms, we followed exactly the protocol presented in the Supporting Information. In brief, 500–700 bp homology arms for each target were PCR amplified from N2 genomic DNA and inserted into the mNG^SEC^3xFlag vector pDD268 using Gibson assembly (New England BioLabs) (throughout this article, we use the ^ symbol to denote a synthetic intron containing a LoxP site). The complete sequences of all repair templates used in this study are available upon request. Cas9 target sites were chosen using the MIT CRISPR design tool (http://crispr.mit.edu) and inserted into the Cas9–sgRNA vector pDD162 as previously described (Dickinson et al. 2013). Table S2 lists the sgRNA sequences used in this study. For each tagging experiment, a mixture of 50 ng/µl Cas9–sgRNA plasmid, 10 ng/µl repair template, and red fluorescent co-injection markers (Frøkjaer-Jensen et al. 2008; Dickinson et al. 2013) was injected into the gonads of young adults. Injected animals were transferred to new OP50 plates (three animals per plate) and allowed to lay eggs for 2–3 days at 25° in the absence of selection. Then, hygromycin was added to a final concentration of 250 µg/ml and the plates were returned to 25° for an additional 3–4 days. Candidate knock-in animals (those that survived hygromycin selection, were Rollers (Rol) and lacked the red fluorescent extrachromosomal array markers) were singled to establish lines. Only one line from each injection plate was kept.
To generate knock-in strains using short homology arms, we designed primers to amplify mNG^SEC^3xFlag and add 35–40 bp homology arms, following the recommendations in Paix et al. (2014). PCR products were purified using a MinElute spin column (Qiagen) and injected at 50 ng/µl, along with a Cas9–sgRNA plasmid and red fluorescent co-injection markers.
To excise SEC, L1/L2 larvae from insertion strains were heat shocked for 4–5 hr at 32°, then placed at 20° for 5–7 days. Wild-type F1 progeny of the heat-shocked animals were singled to establish marker-excised strains. Initial insertion strains for ebp-2, his-72, oma-2, and rap-1 were homozygous viable and segregated 100% Rol progeny prior to heat shock. Therefore, wild-type animals picked after heat shocking these strains had lost both copies of SEC. gex-3, mex-5, and nmy-2 are essential genes (Guo and Kemphues 1996; Schubert et al. 2000; Soto et al. 2002) and so insertions at these loci were isolated and maintained as heterozygotes. The resulting animals segregated ∼1/4 wild-type progeny at each generation, so a wild-type phenotype alone was not a reliable indicator of SEC loss. For mNG^SEC^3xFlag::mex-5 and 6/9 mNG^SEC^3xFlag::nmy-2 strains, mNG fluorescence was easily visible on a dissecting microscope; therefore, to excise SEC from these strains, heterozygous insertion strains were heat shocked, and wild-type animals that had excised SEC were identified by mNG fluorescence. Some of these worms segregated 100% mNG-positive progeny, indicating homozygosity for the marker-excised alleles. For mNG^SEC^3xFlag::gex-3, which is located on chromosome IV, fluorescence was too dim to see by eye. Therefore, to excise SEC from this strain, heterozygous insertion animals were first mated to males carrying the nT1 [qIs51] (IV;V) balancer chromosome. The resulting mNG^SEC^3xFlag::gex-3 IV / nT1 [qIs51] (IV;V) animals segregated 100% Rol progeny, as expected (because nT1 [qIs51] (IV;V) is homozygous lethal). These animals were heat shocked, and wild-type progeny were picked to establish marker-excised lines. Marker-excised animals that had lost the nT1 [qIs51] balancer (that is, mNG^3xFlag::gex-3 homozygotes) were viable and fertile, indicating that gex-3 function was restored after SEC excision. A similar approach (using the hT2 [bli-4(e937) let-?(q782) qIs48] (I;III) balancer) was used to excise SEC from the 3/9 mNG^SEC^3xFlag::nmy-2 I strains that displayed fainter mNG fluorescence (we attribute the faint fluorescence in these strains to germline silencing (Lee et al. 2012; Shirayama et al. 2012; Leopold et al. 2015)).
Microscopy
Embryos were dissected in egg buffer, transferred to poly-L-lysine-coated coverslips using a mouth pipet, and gently flattened using 2.5% agar pads. Whole worms were mounted on 2.5% agar pads containing 10 mM sodium azide as a paralytic. Immunostaining was done using a tube fixation protocol (Finney and Ruvkun 1990). Mixed stage worms were washed in deionized water, resuspended in fixative (30 mM PIPES pH 7.4, 160 mM KCl, 40 mM NaCl, 20 mM EGTA, 10 mM spermidine, 50% methanol, 1% paraformaldehyde) and frozen in liquid nitrogen. Worms were frozen and thawed three times to crack the cuticle, then incubated for 1 hr at 4° with gentle mixing on a turning wheel. After washing twice with Tris/Triton (TT) buffer (100 mM Tris pH 7.4, 1% Triton X-100, 1 mM EGTA), the worms were incubated for 2 hr at 37° in TT containing 1% β-mercaptoethanol. Worms were next washed with borate buffer (BB; 50 mM HBO3, 25 mM NaOH, 0.01% Triton X-100); incubated for 15 min at 37° in BB containing 10 mM dithiothreitol; washed with BB; incubated for 15 min at room temperature in BB containing 0.3% hydrogen peroxide; washed with BB; and finally washed with antibody buffer (PBS containing 1 mM EDTA, 0.5% Triton X-100, 1 mg/ml BSA and 0.03% sodium azide). The worms were blocked with 10% goat serum in antibody buffer and then stained with anti-Flag (Sigma) followed by Cy5-anti-mouse (Jackson Laboratories). The stained specimens were mounted in Slowfade Diamond mounting medium with DAPI (Life Technologies).
mNG::his-72 animals (Figure 2B, Figure 3C, and Figure S1B) were imaged using a Zeiss LSM710 laser scanning confocal microscope equipped with a 40×, 1.2 NA water immersion objective. mNG was excited using the 514-nm line of an Argon ion laser and detected using emission filters for YFP. DAPI was excited with a 405-nm laser, and Cy5 was excited with a 633-nm laser; the DAPI and Cy5 signals were collected simultaneously. mNG::ebp-2 embryos (Figure 5) were imaged using a Nikon Ti-E microscope equipped with a 100×, 1.49 NA objective, a Yokogawa CSU-X1 spinning disk head, and a Hamamatsu C9100-13 camera operated in non-EM mode. mNG was excited with a 488-nm diode laser and detected using emission filters for GFP. mNG::mex-5 embryos (Figure 5) were imaged using the same Ti-E microscope, but using a 60×, 1.4 N objective, a 514-nm diode laser for excitation, and YFP emission filters for detection. mNG::gex-3 and mNG::nmy-2 worms and embryos (Figure 5) were imaged using a Nikon TE2000 microscope equipped with a 60×, 1.4 NA objective, a Yokogawa CSU-10 spinning disk head, and a Hamamatsu Orca ER camera. mNG was excited with a 514-nm diode laser and detected using emission filters for YFP. mNG::rap-1 worms (Figure 5) were imaged with the same TE2000 microscope but using a 20×, 0.5 NA objective. mNG::oma-2 worms (Figure 5) were imaged using a Nikon Eclipse Ti microscope equipped with epifluorescence illumination and a 20×, 0.5 NA objective. A FITC filter cube was used to excite and detect mNG.
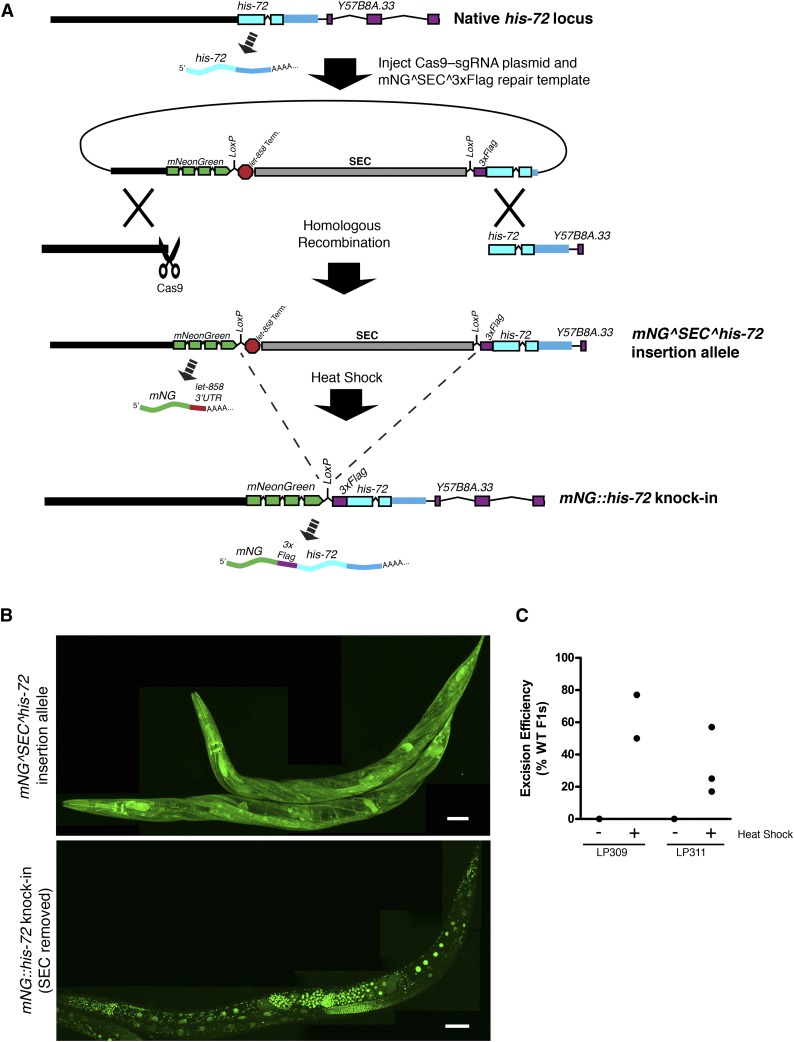
Figure 2.
Tagging of his-72 with mNG^3xFlag. (A) Illustration of the organization of the his-72 locus and the predicted transcripts from this gene before editing (top), after homologous recombination (middle), and after SEC removal (bottom). (B) Images of adult mNG::his-72 worms before (strain LP309, top) and after (strain LP310, bottom) SEC removal. Shown are maximum intensity projections of a confocal Z series through entire worms. Scale bars, 50 µm. (C) Efficiency of SEC excision following heat shock for two independent mNG^SEC^3xFlag::his-72 insertion strains. For each experiment, L1/L2 larvae were heat shocked, and the number of wild-type (WT) and Roller progeny were counted. Each data point represents an independent experiment in which all F1’s present were counted (n = 26–394 animals counted per experiment).
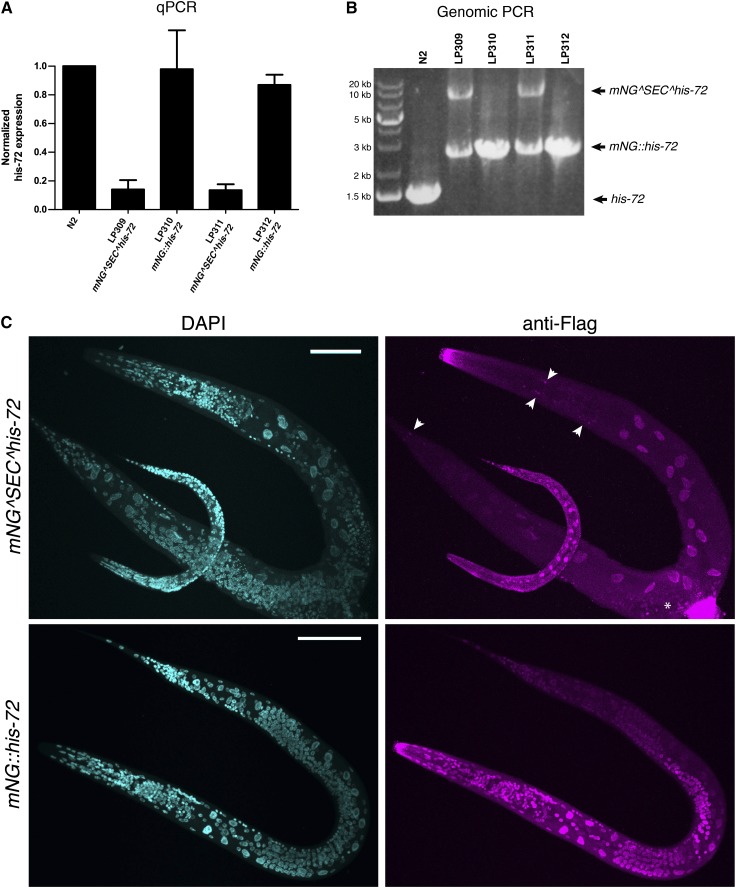
Figure 3.
his-72 expression levels and spontaneous loss of SEC from a subset of cells. (A) Expression of the his-72 ORF measured by qPCR in wild-type (N2), initial insertion (LP309 and LP311), and marker excised (LP310 and LP312) strains. Results are the means of three independent experiments, and error bars indicate 95% confidence intervals. (B) Genotyping of the strains in A. The lower band in LP309 and LP311 indicates spontaneous self-excision of SEC in a population of cells in the absence of heat shock. (C) Images of L4 worms of the indicated genotypes stained with anti-Flag antibodies to label cells that have excised SEC (spontaneously in the mNG^SEC^his-72 strain LP309, or after heat shock in the mNG::his-72 strain LP310). Shown are maximum intensity projections of a confocal Z series through entire worms. Scale bars, 50 µm. Arrowheads indicate stained cells in the head and tail, and the asterisk indicates staining near the developing vulva that may be nonspecific (see text for details).
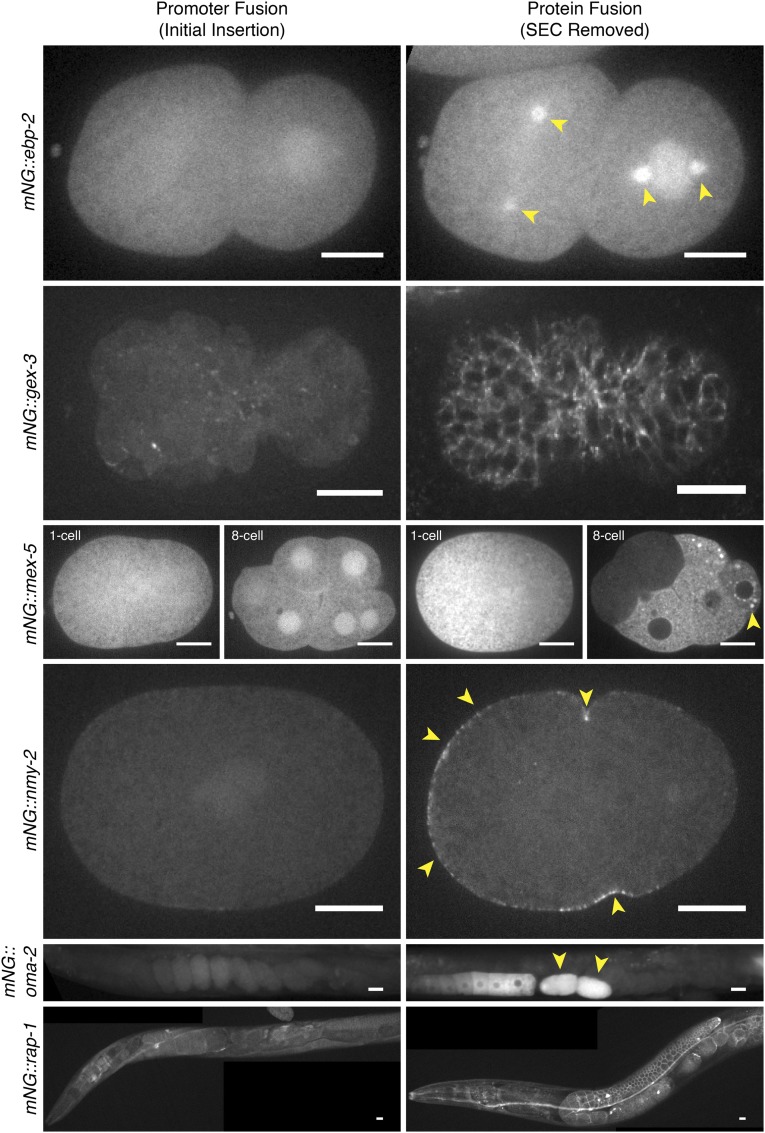
Figure 5.
Images of knock-in strains generated using SEC. mNG fluorescence was imaged in the indicated strains. Left: initial insertions, which are predicted to behave as transcriptional reporters. Right: marker-excised strains, which express mNG^3xFlag fused to the protein of interest. The follow strains are shown: mNG^SEC^3xFlag::ebp-2, LP345; mNG^3xFlag::ebp-2, LP346; mNG^SEC^3xFlag::gex-3, LP361; mNG^3xFlag::gex-3, LP362; mNG^SEC^3xFlag::mex-5, LP366; mNG^3xFlag::mex-5, LP367; mNG^SEC^3xFlag::nmy-2, LP388; mNG^3xFlag::nmy-2, LP389; mNG^SEC^3xFlag::oma-2, LP390; mNG^3xFlag::oma-2, LP391; mNG^SEC^3xFlag::rap-1, LP394; mNG^3xFlag::rap-1, LP395. Arrowheads indicate expected localization of the fusion proteins (see text for details). Scale bars, 10 µm.
To prepare figures for publication, images were cropped and rotated, brightness and contrast were adjusted, and maximum intensity projections (where applicable) were performed using FIJI. In addition, the “despeckle” function was applied to the mNG::oma-2 images to remove hot pixels that are a feature of the camera. No other image manipulations were performed.
RNA isolation and qRT–PCR
For RNA isolation, mixed stage worms were washed with M9 and dissolved in Trizol (Life Technologies). After addition of chloroform to separate phases, RNA was isolated from the upper aqueous phase using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. Genomic DNA contamination was removed using an on-column DNase digestion kit (Qiagen). Poly(T)-primed cDNA was then prepared using the Superscript III reverse transcriptase kit (Life Technologies). qRT–PCR was performed using a Viia7 real-time PCR instrument and SYBR Green master mix (Life Technologies). Transcripts containing the his-72 ORF were detected with forward primer 5′-TCGTTCGTGAGATTGCCCAG-3′ and reverse primer 5′-GAGTCCGACGAGGTATGCTT-3′. Y45F10D.4 was used for normalization (Hoogewijs et al. 2008; Zhang et al. 2012). The data were analyzed using Viia7 software, with the default settings for a relative standard curve experiment.
Genomic DNA isolation and PCR
Genomic DNA was isolated from the organic phase of Trizol extracts (see above) after phase separation, according to the manufacturer’s instructions. For genotyping of the his-72 locus (Figure 3B) we used forward primer 5′-GACCCCACAAAATCGATACG-3′ and reverse primer 5′-GAGTCCGACGAGGTATGCTT-3′. Genotyping reactions were run using LongAmp Taq DNA polymerase (New England BioLabs). For qPCR of genomic DNA, we used the same instrument, reagents, and analysis settings as above. Marker-excised mNG^3xFlag was detected with forward primer 5′-GAGAATCTGTACTTTCAATCCGGA-3′ and reverse primer 5′-TCTCTTGTCATCGTCATCCT-3′. The his-72 ORF, detected using the primers above, was used for normalization.
RNAi
Depletion of hsf-1 was done by feeding using clone I-6C09 from the Ahringer library (Kamath and Ahringer 2003). We sequenced our clone to verify that the correct gene was targeted. Young adults that had not yet begun to produce embryos were placed on feeding plates, and their progeny were collected for imaging and qPCR when they reached adulthood (4 days later).
Results
Design of a new gene tagging strategy
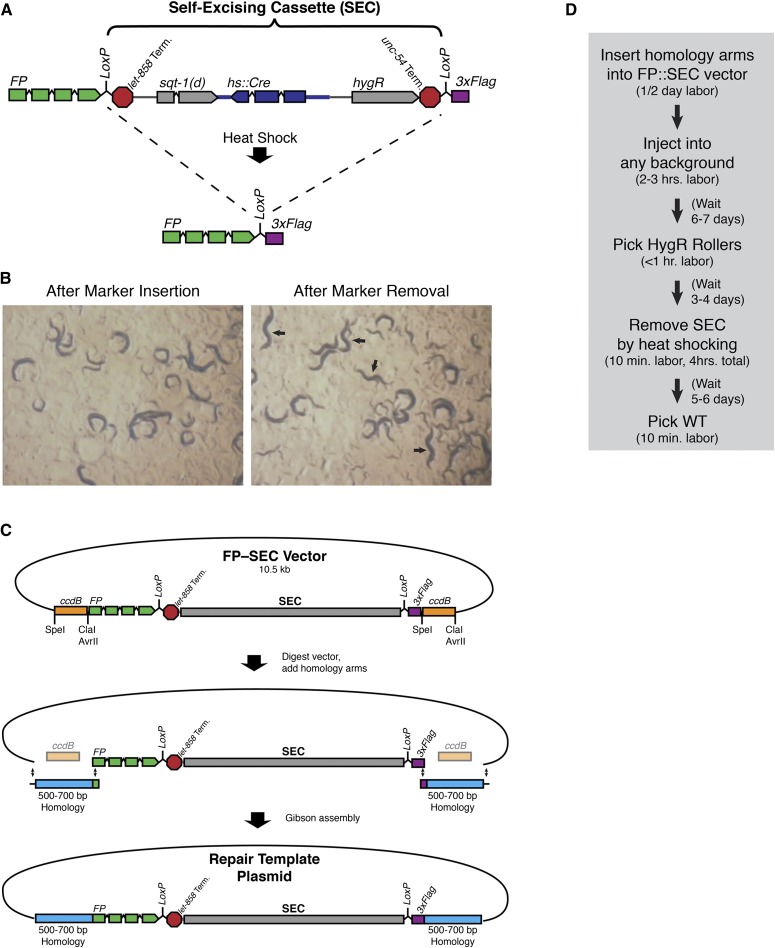
Because of the limitations of our published unc-119 selection approach (Dickinson et al. 2013) (see Introduction), we set out to develop a new selection approach for fluorescent protein tagging of endogenous genes via Cas9-triggered homologous recombination. Our goal was to minimize the hands-on labor required at every step of the procedure. We first sought a positive selectable marker that could be used in a wild-type background, as an alternative to unc-119 selection that requires working in an unc-119 mutant background. Hygromycin resistance has been reported to be an effective selectable marker in C. elegans (Greiss and Chin 2011; Radman et al. 2013; Chen et al. 2013). We confirmed that hygromycin killed 100% of nontransformed worms within 2–3 days, and a single heterozygous copy of the hygromycin phosphotransferase gene (hygR) was sufficient to confer resistance. Hygromycin selection can in principle be used in any genetic background, and it is also slightly faster than unc-119 selection (6 days for hygromycin vs. 8–10 days for unc-119).
The only disadvantage of hygromycin selection compared to unc-119 selection for knock-in experiments is that hygR does not confer a visible plate-level phenotype (in contrast, unc-119(+) worms are wild type, while their nontransformed siblings are Uncoordinated, Unc). Our published protocol (Dickinson et al. 2013) relied on the visible phenotype conferred by unc-119(+) at two stages: first, to identify animals that were homozygous for an insertion, and second, to identify animals that had excised the unc-119(+) marker after Cre injection. We therefore sought a dominant marker that we could integrate along with hygR in order to confer a visible plate-level phenotype. We tested several classical dominant alleles and found that sqt-1(e1350) confers a strong and 100% penetrant Rol phenotype when expressed transgenically from a single-copy insertion (Figure 1B, left). We therefore included sqt-1(e1350) as a dominant phenotypic marker along with hygR within our selection cassette (Figure 1A). Compared to the more widely used rol-6(su1006), sqt-1(e1350) had a stronger and more penetrant phenotype. Note that sqt-1(e1350) has also been used as a marker in “co-conversion” approaches (Arribere et al. 2014), but co-conversion of the endogenous sqt-1 gene in that application differs from the use of transgenically expressed sqt-1(d) as a dominant marker here.
Figure 1.
Design of an improved gene tagging workflow. (A) Design of a self-excising cassette for drug selection. SEC consists of a hygromycin resistance gene (hygR), a visible marker [sqt-1(d)], and an inducible Cre recombinase (hs::Cre). SEC is flanked by LoxP sites and placed within a synthetic intron in an FP::3xFlag tag, so that the LoxP site that remains after marker excision is within an intron. (B) Plate phenotype of animals homozygous for a sqt-1(d)::hygR selection marker (left) and appearance of wild-type animals after marker excision (right). Arrows indicate wild-type animals. See also File S2. (C) Schematic of an expedited cloning procedure for insertion of homology arms into an FP–SEC vector. The FP–SEC vector is first digested with restriction enzymes to release the ccdB markers, and 500–700 bp homology arms are inserted by Gibson assembly to generate the repair template plasmid. (D) Workflow for generation of new FP knock-ins using our strategy. The time required for each step is listed in parentheses.
Next, we sought to eliminate the second injection step that was required to remove a LoxP-flanked selectable marker in our original protocol (Dickinson et al. 2013). We constructed a mini-gene (hs::Cre) composed of Cre recombinase under the control of the hsp-16.41 heat-shock-inducible promoter and inserted it between the sqt-1(d) and hygR genes in our selection cassette (Figure 1A). The entire sqt-1(d)::hsCre::hygR construct is flanked by LoxP sites (Figure 1A). Thus, upon heat shock, expression of Cre recombinase should excise the selection cassette from the genome. We therefore refer to the selection cassette composed of sqt-1(d), hs::Cre, and hygR as a self-excising cassette (SEC). In pilot experiments, we found that animals carrying SEC were 100% Rol, whereas after heat shock, wild-type animals appeared and were easy to distinguish from their Rol siblings (Figure 1B and File S2; see Figure 2C and Figure 4B, below, for measurements of self-excision efficiency).
Figure 4.
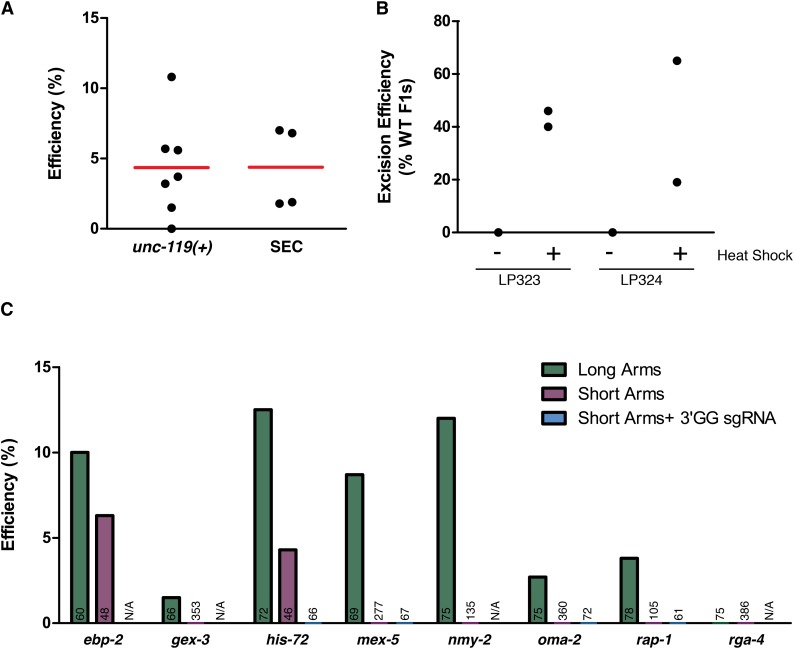
Efficiency of genome engineering using SEC. (A) Comparison of single-copy transgene insertion at the ttTi5605 locus using either unc-119(+) or SEC selection. Each data point represents a single experiment, and red lines show the means across experiments; 45–90 animals were injected for each experiment. Efficiency is defined as the fraction of injected animals yielding insertions. (B) Efficiency of SEC excision following heat shock, measured as in Figure 2C, for two transgenes at the ttTi5605 locus. Each data point represents an independent experiment in which all F1’s present were counted (n = 299–913 animals counted per experiment). (C) Efficiency of precise mNG^SEC^3xFlag insertion into eight different endogenous loci using long homology arms (500–700 bp; green bars), short homology arms (35–40 bp; purple bars) or short homology arms and 3′GG sgRNAs (blue bars). Numbers on each bar indicate the number of animals injected, and efficiency is defined as the fraction of injected animals yielding precise insertions. N/A, not applicable (3′GG sgRNAs were tested only for the genes were a 3′GG target was present near the site of insertion). See also Table 1.
Excision of a LoxP-flanked selectable marker from the genome leaves behind a 34-bp LoxP site. In principle, this residual LoxP site could interfere with gene regulation by, for example, disrupting a transcription factor binding site. To minimize the potential impact of the residual LoxP site, we placed SEC within a synthetic intron between the FP and 3xFlag sequences of an FP::3xFlag tag (Figure 1A, top). After SEC removal, the location of the residual LoxP site is within this synthetic intron (Figure 1A, bottom), where it is unlikely to interfere with gene regulation. This approach allows clean insertion of FP::3xFlag into the genome, with no exogenous sequences left outside the fluorescent protein tag.
Finally, we addressed the labor-intensive cloning step that was required in our previous unc-119(+)-based approach (Dickinson et al. 2013). Importantly, the inclusion of SEC within the FP tag generates a 1-piece FP^SEC^3xFlag module that can be inserted anywhere in the genome when appropriate homology arms are added. To facilitate addition of homology arms, we generated constructs in which the FP^SEC^3xFlag module is flanked by ccdB negative selection markers (Figure 1C, top). ccdB is toxic to E. coli, and ccdB negative selection is one of the key features that accounts for the high efficiency of Gateway cloning. In our constructs, each ccdB marker is flanked by unique restriction sites, which were chosen because they do not leave any residual sequence after digestion, allowing seamless fusion of FP^SEC^3xFlag to the homology arms. Homology arms can be cloned into these constructs using the following simple procedure (Figure 1C). First, the vector is digested with restriction enzymes to liberate the ccdB markers. Individual vector fragments are not purified; the entire digested vector is kept in one tube. A batch of digested vector may be stored and used to make multiple repair templates. Second, homology arms are PCR amplified using primers that add 20–30 bp of sequence overlapping the vector, to allow Gibson assembly. Third, the homology arms are mixed with the digested vector. The four repair template fragments (two homology arms plus the FP^SEC^3xFlag and backbone vector fragments) are joined together by Gibson assembly, and the resulting plasmids are transformed into competent E. coli. Since the parent vector (containing ccdB markers) does not transform, only clones that have correctly inserted both homology arms should grow. Therefore, correct clones can be identified by sequencing alone, without screening clones for inserts.
To test this cloning strategy, we generated 8 different homologous repair templates (targeting 8 different genes) in parallel, which took approximately half of a day. We directly sequenced 6 random clones of each of the 8 constructs (48 clones total). For 7/8 constructs, a total of >90% of clones (39 of 42 clones total) contained correctly inserted homology arms. For the eighth construct, one of the homology arm PCR products was obtained at low yield, and so the assembly was less efficient. This reaction still yielded a correct clone, although 12 clones had to be sequenced to identify one that was correct. We conclude that this cloning strategy allows robust insertion of homology arms into FP–SEC vectors with minimal labor.
With these technical and methodological innovations, our workflow for gene tagging consists of four steps (Figure 1D): (1) insertion of homology arms into an FP^SEC^3xFlag module; (2) injection; (3) picking Rollers that survive hygromycin selection; and (4) heat shocking to remove SEC. Importantly, the same workflow can be used to place FP::3xFlag at the N terminus, C terminus, or internally for any gene of interest, or to generate whole-gene deletions by replacing an entire coding region with FP^SEC^3xFlag. This workflow eliminates the most labor-intensive steps that were required in previous protocols (Dickinson et al. 2013; Paix et al. 2014).
Generation of a loss-of-function allele, promoter fusion, and protein fusion in a single injection step
As a first test of our approach, we inserted fluorescent protein at the N terminus of the his-72 gene, which encodes a broadly expressed Histone H3.3 (Ooi et al. 2006). We chose to target the N terminus of his-72 rather than the C terminus to take advantage of an additional feature of the FP^SEC^3xFlag design. We predicted that because SEC contains transcriptional terminators, insertion of SEC at the 5′ end of his-72 should disrupt the gene by separating the promoter and coding region, resulting in a loss-of-function allele that expresses mNG in place of HIS-72 (Figure 2A). This allele should then convert to an N-terminal protein tag after SEC excision. Thus, N-terminal insertion of mNG^SEC^3xFlag into a gene of interest should produce a loss-of-function allele, a promoter fusion, and a protein fusion in a single injection step (Figure 2A).
We generated a homologous repair template for mNG^SEC^3xFlag::his-72 using the procedure outlined in Figure 1C and injected it into the germlines of wild-type animals along with an appropriate Cas9–sgRNA plasmid. Our injection mix also contained mCherry markers that we used to distinguish extrachromosomal arrays from insertions, as has been done previously (Frøkjaer-Jensen et al. 2008, 2012; Dickinson et al. 2013). From 72 injected animals, we obtained 9 independent strains carrying an insertion of mNG^SEC^3xFlag at the 5′ end of his-72. Because his-72 is nonessential (Ooi et al. 2006), we were able to readily isolate homozygous insertion strains by choosing animals that segregated 100% Rol progeny. These animals showed cytosolic mNG fluorescence in most tissues, consistent with the known expression pattern of his-72 (Figure 2B, top).
Next, we heat shocked animals from these strains to remove SEC. After heat shocking L1/L2 animals, we readily identified wild-type animals in the F1 progeny of the heat-shocked animals for 9/9 strains. Strikingly, these animals exhibited strictly nuclear mNG fluorescence (Figure 2B), which is the expected pattern for a HIS-72 fusion protein (Ooi et al. 2006; Dickinson et al. 2013). These data indicate that N-terminal insertions of mNG^SEC^3xFlag behave as expected: the insertion allele expresses mNG from the gene’s promoter, and an N-terminal protein fusion is produced after SEC removal by heat shock.
We quantified the frequency of SEC excision for two mNG^SEC^3xFlag::his-72 strains. In five independent experiments, the F1 progeny of heat-shocked animals ranged from 17 to 77% wild type, with a mean of 45% (Figure 2C) (note that animals must excise both copies of SEC in order to display a wild-type phenotype in the F1). Although the frequency of excision varied between individual experiments, wild-type animals that had excised SEC were easily identified even on the plates that showed the lowest excision frequency. These results indicate that SEC excision by heat shock is highly efficient. By picking wild-type animals to new plates, we established strains with mNG^3xFlag fused cleanly to his-72. For clarity, we refer to mNG^SEC^3xFlag knock-in strains (prior to heat shock) as “initial insertion” strains, and mNG^3xFlag protein fusion strains (after heat shock) as “marker-excised” strains.
Spontaneous SEC excision can occur in certain tissues
We sought to test more quantitatively our prediction that expression of a gene of interest should be abolished upon N-terminal SEC insertion and restored following SEC excision. To do this, we measured the expression of the his-72 ORF in two pairs of strains before and after SEC removal (strains LP310 and LP312 are marker-excised derivatives of LP309 and LP311, respectively). As expected, his-72 expression was strongly reduced in the mNG^SEC^3xFlag insertion strains, and expression was restored to wild-type levels after SEC removal (Figure 3A). However, we were surprised to find that his-72 was still expressed at detectable levels (∼15% of wild type) in the initial insertion strains. The reason for this became clear when we genotyped these strains with PCR primers that flank the insertion site. As expected, we detected a 1.5-kb band in N2 lysates, corresponding to the unmodified locus; an 8 kb band in initial insertion strains, indicating correct single-copy insertion of mNG^SEC^3xFlag; and a 2.5-kb band in marker-excised strains following SEC removal (Figure 3B). However, we also observed the 2.5-kb band, indicating excision of SEC, in the initial insertion strains that had not been heat shocked (Figure 3B, lanes 2 and 4). These data indicate that either a fraction of worms in each sample, or a fraction of cells within each worm, spontaneously excised SEC in the absence of heat shock. We were able to rule out the former possibility because the initial insertion strains continued to produce 100% Rol progeny over multiple generations. We therefore inferred that some cells in each worm spontaneously excise SEC in the absence of heat shock.
To estimate the prevalence of spontaneous self-excision, we performed qPCR on genomic DNA samples. We used primers flanking the LoxP-containing intron left behind after SEC excision, which amplified a single product corresponding to marker-excised mNG^3xFlag. The abundance of mNG^3xFlag in each sample is a measure of the fraction of haploid genomes in that sample that have undergone SEC excision. Using qPCR, we determined that the initial insertion strains LP309 and LP311 had 2.5 and 2.3% as much genomic mNG^3xFlag, respectively, as their marker-excised derivatives LP310 and LP312. These data suggest that spontaneous self-excision occurs in a relatively small fraction of cells.
To identify the cells that spontaneously excise SEC, we stained whole worms with anti-Flag antibodies. Because the 3xFlag tag is located downstream of SEC and does not carry its own start codon, only cells that have excised SEC should be stained. As a control, anti-Flag staining was detected in all, or almost all, nuclei in marker-excised animals (Figure 3C, bottom). In the initial insertion strains, reproducible anti-Flag staining was observed in the nuclei of the intestine and in a small number of nuclei in the head and tail (Figure 3C, top). Intestinal anti-Flag staining was observed in every animal examined, while the staining in head and tail nuclei was fainter and more variable. The stained nuclei in the head and tail were concentrated in the nerve ring between the two pharyngeal bulbs and postanally in a bilaterally symmetric pattern in the tail, respectively, suggesting that these cells are likely to be neurons. Consistent with a tendency for neurons to spontaneously excise SEC, we also saw stained cells in the ventral nerve cord in a minority of specimens (D.J.D., unpublished results). Additionally, in a minority of specimens we observed nuclear staining in cells in the center of the animal, near the developing vulva (Figure 3C, asterisk), but this staining may be nonspecific as it was also observed in one N2 animal.
Because Cre recombinase is under the control of a heat shock promoter in our strains, we tested whether spontaneous self-excision required a functional heat-shock response. Treatment of animals with RNAi against hsf-1, which is known to be required for the transcriptional response to heat shock (Hajdu-Cronin et al. 2004), did not prevent intestinal nuclei from spontaneously excising SEC (Figure S1). To verify the effectiveness of RNAi, we heat shocked L1/L2 larvae from control or hsf-1 RNAi plates. Control animals produced abundant wild-type progeny after heat shock, but hsf-1 RNAi animals produced 100% Rol progeny, indicating that hsf-1 RNAi prevented heat-shock-induced activation of Cre recombinase. Growth of worms at 15° also had little to no effect on expression levels of the his-72 ORF in initial insertion strains (Figure S1), suggesting that SEC excision in intestinal cells is not due to low-level heat stress in worms maintained at 20° or 25°. Spontaneous SEC excision may therefore reflect a normal physiological activation of the hsp-16.41 promoter during intestinal development. In summary, these data are consistent with our prediction that N-terminal insertions of FP^SEC^3xFlag behave as strong loss-of-function alleles, with the caveat that spontaneous self-excision—resulting in expression of the protein of interest—can occur in the absence of heat shock in the intestine and in certain neurons.
SEC selection is robust across a wide range of loci
Having established that our tagging strategy performed as designed for the his-72 locus, we next tested whether it would be generally applicable to a wider range of genes. First, we sought to quantitatively compare the efficiency of SEC selection to unc-119(+) selection. To do this, we generated a derivative of the MosSCI targeting vector pCFJ150 (Frøkjaer-Jensen et al. 2008) that carried SEC in place of unc-119(+). We previously showed that Cas9 could be used in place of Mos1 to generate the DNA double-strand break that allows insertion of transgenes cloned into pCFJ150 at the ttTi5605 locus on chromosome II (Dickinson et al. 2013). We compared the efficiency of Cas9-mediated transgene insertion using either unc-119(+) or SEC selection. Although the insertion frequency varied between individual experiments, as previously reported (Dickinson et al. 2013), overall the efficiency of SEC selection was indistinguishable from unc-119(+) (Figure 4A). To ensure that the ability to excise SEC following heat shock was not specific to any one locus, we selected two transgenic strains from this experiment and measured the frequency of SEC excision following heat shock. Similar to our observations at the his-72 locus (Figure 2C), SEC excision frequency at the ttTi5605 locus varied from plate to plate but was always high enough that it was easy to identify marker-excised animals (Figure 4B).
Next, we designed and cloned homologous repair templates to generate N-terminal mNG^3xFlag tags on seven additional genes. We injected 60–78 animals per construct and obtained mNG^SEC^3xFlag insertions on the first attempt for six of seven targets, five of which yielded multiple independent lines (Figure 4C and Table 1). Plasmid construction and injections for these genes were done in parallel, requiring a total of ∼4 days of hands-on labor for all seven targets (∼1/2 day total per target), including all cloning steps. Of the strains generated in these experiments, 91% (32/35) yielded marker-excised derivatives following heat shock. Interestingly, a few strains (6/35) spontaneously excised SEC in the germline and produced marker-excised progeny without needing to be heat shocked. These events were sufficiently rare that they did not undermine the efficacy of SEC selection, nor did they prevent us from using the Rol phenotype to derive or maintain homozygous insertion lines. Overall, these results demonstrate that our procedure is robust and efficient and can be used to tag a wide range of C. elegans genes.
Table 1. Efficiency of mNG^SEC^3xFlag insertion into 8 different endogenous loci.
| Target | Homology arms | sgRNA | Doench et al. sgRNA score | Distance to cut (bp) | No. injected | Precise insertions | Imprecise insertions | False positives | Efficiency (%) |
|---|---|---|---|---|---|---|---|---|---|
| ebp-2 | Long | Non-3′GG | 0.555 | 4 | 60 | 6 | 0 | 0 | 10.0 |
| ebp-2 | Short | Non-3′GG | 0.555 | 4 | 48 | 3 | 0 | 2 | 6.3 |
| gex-3 | Long | Non-3′GG | 0.039 | 19 | 66 | 1 | 0 | 0 | 1.5 |
| gex-3 | Short | Non-3′GG | 0.039 | 19 | 353 | 0 | 0 | 1 | 0.0 |
| his-72 | Long | Non-3′GG | 0.027 | 5 | 72 | 9 | 0 | 0 | 12.5 |
| his-72 | Short | Non-3′GG | 0.027 | 5 | 46 | 2 | 0 | 0 | 4.3 |
| his-72 | Short | 3′gg | 0.281 | 40 | 66 | 0 | 2 | 0 | 0.0 |
| mex-5 | Long | Non-3′GG | 0.208 | 25 | 69 | 6 | 0 | 0 | 8.7 |
| mex-5 | Short | Non-3′GG | 0.208 | 25 | 277 | 0 | 2 | 4 | 0.0 |
| mex-5 | Short | 3′gg | 0.427 | 29 | 67 | 0 | 2 | 1 | 0.0 |
| nmy-2 | Long | Non-3′GG | 0.382 | 6 | 75 | 9 | 0 | 0 | 12.0 |
| nmy-2 | Short | Non-3′GG | 0.382 | 6 | 135 | 0 | 1 | 2 | 0.0 |
| oma-2 | Long | Non-3′GG | 0.112 | 2 | 75 | 2 | 0 | 0 | 2.7 |
| oma-2 | Short | Non-3′GG | 0.112 | 2 | 360 | 0 | 0 | 10 | 0.0 |
| oma-2 | Short | 3′gg | 0.392 | 26 | 72 | 0 | 1 | 3 | 0.0 |
| rap-1 | Long | Non-3′GG | 0.136 | 23 | 78 | 3 | 0 | 0 | 3.8 |
| rap-1 | Short | Non-3′GG | 0.136 | 23 | 105 | 0 | 1 | 3 | 0.0 |
| rap-1 | Short | 3′gg | 0.282 | 32 | 61 | 0 | 0 | 1 | 0.0 |
| rga-4 | Long | Non-3′GG | 0.150 | 4 | 75 | 0 | 0 | 0 | 0.0 |
| rga-4 | Short | Non-3′GG | 0.150 | 4 | 386 | 0 | 3 | 14 | 0.0 |
Precise insertions are the desired single-copy integration events confirmed by PCR and/or the correct pattern of mNG fluorescence. Imprecise insertions are insertion events at correct locus, but carry rearrangements within the insertion. False positives are animals that lacked the red fluorescent extrachromosomal array markers but did not carry insertions at the desired locus. Efficiency is defined as the fraction of injected animals yielding precise insertions. See also Figure 4C.
In C. elegans and other systems, Cas9 cleavage efficiency can vary substantially depending on the choice of sgRNA (Friedland et al. 2013; Waaijers et al. 2013; Shalem et al. 2014; Wang et al. 2014; Kim et al. 2014; Doench et al. 2014; Farboud and Meyer 2015), and sgRNA efficiency has been widely considered to be an important factor for the success of genome editing experiments in C. elegans (Kim et al. 2014; Arribere et al. 2014; Paix et al. 2014; Farboud and Meyer 2015). Interestingly, we designed our sgRNAs without regard to predicted efficiency and tested only a single sgRNA per target, and yet still obtained insertions on the first attempt at 7/8 loci, suggesting that high levels of Cas9 activity may not be a prerequisite for SEC-based homologous recombination. To explore further the relationship between sgRNA choice and recombination efficiency, we scored all of our sgRNAs using an activity prediction algorithm (Doench et al. 2014). This analysis revealed no statistically significant correlation between sgRNA score and recombination efficiency (Spearmann’s r = 0.18; P = 0.6; Figure S2 and Table 1). Although we would need to test many more targets to rule out a relationship between sgRNA activity and recombination efficiency, these data indicate that our selection strategy can yield knock-ins without the need for extensive sgRNA optimization.
Short homology arms can support insertion of mNG^SEC^3xFlag, but with lower efficiency
While our experiments were in progress, Paix et al. (2014) reported that short (30–70 bp) homology arms are in many cases sufficient to mediate precise repair of Cas9-induced double-strand breaks, supporting edits ranging in size from single base pairs to insertion of GFP. We reasoned that in principle, this finding could allow us to streamline our protocol even further, by entirely eliminating the need for homologous repair template cloning. To test this idea, we designed PCR primers to amplify mNG^SEC^3xFlag, adding 35–40 bp of homology for insertion at each of the 8 loci we targeted above. We repeated the injections, substituting the appropriate mNG^SEC^3xFlag PCR product (with short homology arms) for the homologous repair template plasmid (with long homology arms) in each case.
We confirmed the finding (Paix et al. 2014) that short homology arms are sufficient to mediate site-specific homologous recombination at several different loci (Figure 4C and Table 1). However, the efficiency and robustness of recombination with 35–40 bp homology arms were much lower than with 500–700 bp arms. Using short homology arms, we observed correct insertion of mNG^SEC^3xFlag for only 2/8 targets (his-72 and ebp-2). The remaining targets failed to yield precise knock-ins, despite hundreds of animals injected (Figure 4C and Table 1). In an attempt to improve the efficiency of recombination with short homology arms, we tested several different PCR purification kits, used PAGE-purified primers for PCR, and titrated the repair template concentration in the injection mix over a 100-fold range. However, none of these measures resulted in improved efficiency (D.J.D., unpublished results).
Besides low efficiency, we also encountered other challenges with the use of short homology arms to insert mNG^SEC^3xFlag into the genome. First, we observed a higher false-positive rate (that is, a larger proportion of animals that lacked extrachromosomal array markers but did not carry insertions) when injecting PCR products compared to plasmids. This may be due to the fact that PCR products were injected at a higher concentration (50 ng/µl; Paix et al. 2014) compared to plasmids (10 ng/µl; Dickinson et al. 2013). Second, even among strains that carried bona fide insertions at the desired locus, a high proportion (12/17) of the strains generated using short homology arms had rearrangements within the insertion (“Imprecise Insertions” in Table 1), whereas no rearrangements were observed in this study when long homology arms were used (although other studies have shown that rearrangements can occur, at low frequency, when using long homology arms with various means of transgene insertion; Berezikov et al. 2004; Frøkjaer-Jensen et al. 2008; Dickinson et al. 2013). The majority (11/12) of the rearranged strains generated using short homology arms failed to show detectable mNG fluorescence. Finally, we found that in practice, generating mNG^SEC^3xFlag PCR products with 35–40 bp homology arms often required a similar amount of effort compared to cloning 500–700 bp homology arms with our ccdB-based approach (Figure 1C), because amplification of mNG^SEC^3xFlag (∼6.5 kb) with long primers was more difficult than amplification and insertion of 500–700 bp homology arms into an FP–SEC vector. We conclude that, in our hands, the enhanced efficiency and robustness conferred by longer homology arms justifies any extra effort spent cloning repair template constructs.
Our results using long homology arms suggested that a high-efficiency sgRNA is not necessary for homologous recombination. Nevertheless, we wondered whether higher-activity sgRNAs could improve the low efficiency we observed for mNG^SEC^3xFlag insertion using short homology arms. Farboud and Meyer (2015) recently reported that sgRNAs whose target sequences end in a GG dinucleotide (3′GG sgRNAs) support much higher levels of Cas9 activity. Therefore, we generated 3′GG sgRNAs for the four loci on our targets list for which it was possible to design a 3′GG sgRNA that would cleave within 30–40 bp of the desired insertion site, and we repeated the injections targeting these loci. We obtained a modestly higher frequency of insertions using a 3′GG sgRNA for some of the loci tested (2/4 loci), but 5/5 of the resulting insertions carried rearrangements (Table 1). Even when these imprecise insertion events are included in the total, 3′GG sgRNAs with short homology arms yielded a lower frequency of insertions than non-3′GG sgRNAs with long homology arms. We conclude that the use of 3′GG sgRNAs does not increase short homology arm insertion efficiency to the level seen with long homology arms and does not overcome the propensity of short homology arm inserts to rearrange.
mNG^SEC^3xFlag knock-in strains behave as expected
Finally, we characterized the knock-in strains that we generated using our new tagging strategy. Of the eight genes we tagged N terminally with mNG^SEC^3xFlag, three (gex-3, mex-5, and nmy-2) are essential genes (Guo and Kemphues 1996; Schubert et al. 2000; Soto et al. 2002). As expected, the initial insertion strains for these loci were viable and fertile only as heterozygotes. Homozygous mNG^SEC^3xFlag::gex-3 and mNG^SEC^3xFlag::mex-5 insertions were maternal-effect lethal, as are previously described loss-of-function mutations in these genes (Schubert et al. 2000; Soto et al. 2002). Animals homozygous for an mNG^SEC^3xFlag::nmy-2 insertion survived to adulthood, likely due to perdurance of maternal protein, but were sterile. In each of these cases, viability and fertility were fully restored upon SEC excision. These results support our conclusion that N-terminal insertion of FP^SEC^3xFlag generates a loss-of-function allele and that gene function is restored upon SEC removal.
We examined mNG localization in all of our strains. In every case, we observed localization of the marker-excised fusion proteins that was consistent with our prior knowledge of the tagged proteins (Figure 5, right). mNG::EBP-2 localized prominently around centrosomes (Figure 5, arrowheads), especially during mitosis, similar to the localization of endogenous EBP-2 revealed by immunostaining (Srayko et al. 2005). mNG::GEX-3 localized to cell boundaries throughout embryogenesis (Figure 5) (Soto et al. 2002). mNG::MEX-5 was concentrated in the anterior cytoplasm of polarized one-cell embryos and localized to punctate structures (P granules) in the cytoplasm of germline precursor cells (Figure 5, arrowheads), and its expression became confined to the posterior blastomeres in older embryos (Schubert et al. 2000; Tenlen et al. 2008). mNG::NMY-2 was enriched at the anterior cell cortex and in the cleavage furrow in the one-cell embryo (Figure 5, arrowheads) (Munro et al. 2004). mNG::OMA-2 localized in the cytoplasm, and its expression was restricted to oocytes and one-cell embryos (Figure 5, arrowheads) (Detwiler et al. 2001). The localization of RAP-1 has not been previously reported, but we found that mNG::RAP-1 localized to the plasma membrane in a wide range of tissues including neurons, intestine, and the germline (Figure 5), consistent with the fact that RAP-1 is a Ras-family small GTPase with a stereotypical C-terminal membrane-targeting sequence. In each of these cases, the expression of mNG in the initial insertion strain broadly mimicked that of the protein fusion, but lacked subcellular localization and, in the cases of MEX-5 and OMA-2, regulation of localized expression that is known to be mediated at the protein level (Detwiler et al. 2001; Tenlen et al. 2008). These observations are consistent with our prediction that the initial insertion alleles behave as transcriptional reporters and are converted into protein fusions after SEC excision.
Discussion
We have developed a gene-tagging strategy that, to our knowledge, is the least labor-intensive method currently available for insertion of fluorescent protein tags into the C. elegans genome. Central to our approach is a novel self-excising drug selection cassette (SEC) that enables robust selection in any genetic background without PCR screening, following insertion alleles based on plate phenotype, and marker excision without a second injection step. SEC-based homologous recombination is also relatively insensitive to the choice of sgRNA, which simplifies experimental design. We anticipate that this tool will greatly facilitate tagging of C. elegans proteins for cell biological assays. Indeed, given the low amount of hands-on labor required to tag a new gene, it may now be feasible to generate a collection of fluorescent protein knock-ins for every gene in the C. elegans genome. In addition, the ability to generate a loss-of-function mutation, transcriptional reporter, and tagged protein using a single workflow and injection step will simplify the initial characterization of unstudied or little-studied genes. Although we focused here on gene tagging, we anticipate that SEC will also facilitate the construction of other kinds of genome modifications.
For this study, we exclusively generated N-terminal mNG^SEC^3xFlag knock-ins, because we wanted a large panel of N-terminal mNG^SEC^3xFlag insertions in order to test our predictions about their behavior. However, it is important to emphasize that our workflow does not require placing a tag at the N terminus of a protein of interest. By choosing an appropriate sgRNA and homology arms, it is feasible to insert FP^SEC^3xFlag at any location in the genome, including at the C terminus, internally, or in place of any gene of interest. A C-terminal insertion of FP^SEC^3xFlag generates a protein fusion immediately, but with the let-858 3′UTR that is part of SEC in place of the endogenous 3′UTR; the native 3′UTR is restored upon SEC excision (A.M.P and J.K.H., unpublished results). In addition, it should be possible to use SEC to produce other modifications besides simple FP insertions. For example, by cloning mutant sequences into FP–SEC vectors along with the homology arms, one could in principle tag a protein and at the same time introduce targeted mutations. More generally, we anticipate that SEC can be used in place of unc-119(+) for any application that relies on a selectable marker.
One potential limitation of SEC is the tendency for the marker to spontaneously self-excise in the absence of heat shock. Spontaneous self-excision occurs in a fraction of cells in each worm, particularly in the intestine, and also occurs on rare occasions in the germline leading to permanent loss of SEC in an animal’s progeny. This phenomenon does not affect the utility of SEC for gene tagging or other applications that involve positive selection, but it does prevent N-terminal FP^SEC^3xFlag insertions from being treated as true null alleles. Spontaneous self-excision is almost certainly due to low-level expression of Cre recombinase in the absence of heat shock. Therefore, it might be possible to prevent spontaneous self-excision by treating animals with RNAi against Cre. However, given the small amount of time and labor involved, deleting the entire coding region of a gene of interest and replacing it with FP^SEC^3xFlag might be a better strategy if a true null allele is required.
In the course of our experiments, we performed a direct comparison of homologous recombination efficiency mediated by short (35–40 bp) vs. long (500–700 bp) homology arms. Although we confirmed the finding that short homology arms can mediate homologous recombination (Paix et al. 2014), we found substantially higher efficiencies using longer arms. One important difference between our study and that of Paix et al. (2014) is the size of the DNA insertions we generated: the largest sequence inserted by Paix et al. was GFP, which is ∼1 kb in size, whereas mNG^SEC^3xFlag is ∼6.5 kb. Thus, our results may simply reflect a limit to the size of insertion that can be made using short homology arms. We also found that short homology arms yielded a much higher frequency of rearranged insertions. This result might not reflect a property of short homology arms per se, but instead may be due to the fact that our short homology arm repair templates were PCR products, while the long homology arm repair templates were plasmids. We suspect that linear DNA repair templates might be more prone to rearrangements because of their free ends. Although the need to clone longer homology arms might at first be viewed as a disadvantage of the SEC-based strategy, we emphasize that in our experience, the amount of labor required to clone homology arms using our expedited cloning strategy was very similar to that required to generate PCR repair templates using primers that include the homology arms. Moreover, any added effort spent cloning homology arms into an FP–SEC construct is made up for by higher recombination efficiency (Figure 4C) and by elimination of labor-intensive single worm PCR screening steps that are required for other protocols (Kim et al. 2014; Arribere et al. 2014; Paix et al. 2014; Ward 2015).
Finally, we note that, although we have demonstrated the utility of SEC for gene tagging in C. elegans, the design principles behind a self-excising selection cassette are not specific to this organism. The use of drug selection, which does not require preexisting mutations, should allow straightforward extension of this approach to nonmodel nematodes, tissue culture cells, and possibly other invertebrates. Although the sqt-1(d) phenotypic marker is worm specific, it could easily be replaced by GFP or another marker. Finally, the placement of SEC within a synthetic intron of an FP::epitope tag should be applicable to any system where splicing rules are sufficiently well understood. Thus, our strategy may facilitate seamless insertion of fluorescent proteins via genome editing in a variety of organisms.
Supplementary Material
Acknowledgments
We thank Amy Maddox, David Reiner, Geraldine Seydoux, Jordan Ward, and members of the Goldstein lab for helpful discussions and comments on the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the U.S. National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported by NIH T32 CA009156 (D.J.D. and A.M.P.); a Howard Hughes postdoctoral fellowship from the Helen Hay Whitney Foundation (D.J.D.); a U.S. National Science Foundation (NSF) graduate research fellowship (J.K.H.); and NIH R01 GM083071 and NSF IOS 0917726 (B.G.).
Footnotes
Communicating editor: O. Hobert
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.178335/-/DC1.
Literature Cited
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Bargmann C. I., Plasterk R. H. A., 2004. Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res. 32: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Fenk L. A., de Bono M., 2013. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 41: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195: 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J.-S., Lee J., 2013. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E., Lin R., 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1: 187–199. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., et al. , 2014. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B., Meyer B. J., 2015. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63: 895–905. [DOI] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S., Chin J. W., 2011. Expanding the genetic code of an animal. J. Am. Chem. Soc. 133: 14196–14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Kemphues K. J., 1996. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature 382: 455–458. [DOI] [PubMed] [Google Scholar]
- Hajdu-Cronin Y. M., Chen W. J., Sternberg P. W., 2004. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics 168: 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J., Vanfleteren J. R., 2008. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Katic I., Großhans H., 2013. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics 195: 1173–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., et al. , 2014. A Co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-C., Gu W., Shirayama M., Youngman E., Conte D., et al. , 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold L. E., Heestand B. N., Seong S., Shtessel L., Ahmed S., 2015. Lack of pairing during meiosis triggers multigenerational transgene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 112(20): E2667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T.-W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Heritable genome editing using TALENs and CRISPR/Cas9 to engineer precise insertions and deletions in evolutionarily diverse nematode species. Genetics 195: 331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E., Nance J., Priess J. R., 2004. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell 7: 413–424. [DOI] [PubMed] [Google Scholar]
- Ooi S. L., Priess J. R., Henikoff S., 2006. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2: e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C.-Y. S., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman I., Greiss S., Chin J. W., 2013. Efficient and rapid C. elegans transgenesis by bombardment and hygromycin B selection. PLoS ONE 8: e76019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., Murray J. I., Schanze K., Pozniakovski A., Niu W., et al. , 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert C. M., Lin R., de Vries C. J., Plasterk R. H., Priess J. R., 2000. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol. Cell 5: 671–682. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., et al. , 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.-C., Gu W., Ishidate T., et al. , 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto M. C., Qadota H., Kasuya K., Inoue M., Tsuboi D., et al. , 2002. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 16: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M., Kaya A., Stamford J., Hyman A. A., 2005. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell 9: 223–236. [DOI] [PubMed] [Google Scholar]
- Tenlen J. R., Molk J. N., London N., Page B. D., Priess J. R., 2008. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development 135: 3665–3675. [DOI] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers S., Portegijs V., Kerver J., Lemmens B. B. L. G., Tijsterman M., et al. , 2013. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M., Lander E. S., 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D., 2015. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen D., Smith M. A., Zhang B., Pan X., 2012. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS ONE 7: e31849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Ke H., Yue Y., Xue D., 2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.