Abstract
Sex chromosomes in plants and animals and fungal mating-type chromosomes often show exceptional genome features, with extensive suppression of homologous recombination and cytological differentiation between members of the diploid chromosome pair. Despite strong interest in the genetics of these chromosomes, their large regions of suppressed recombination often are enriched in transposable elements and therefore can be challenging to assemble. Here we show that the latest improvements of the PacBio sequencing yield assembly of the whole genome of the anther-smut fungus, Microbotryum lychnidis-dioicae (the pathogenic fungus causing anther-smut disease of Silene latifolia), into finished chromosomes or chromosome arms, even for the repeat-rich mating-type chromosomes and centromeres. Suppressed recombination of the mating-type chromosomes is revealed to span nearly 90% of their lengths, with extreme levels of rearrangements, transposable element accumulation, and differentiation between the two mating types. We observed no correlation between allelic divergence and physical position in the nonrecombining regions of the mating-type chromosomes. This may result from gene conversion or from rearrangements of ancient evolutionary strata, i.e., successive steps of suppressed recombination. Centromeres were found to be composed mainly of copia-like transposable elements and to possess specific minisatellite repeats identical between the different chromosomes. We also identified subtelomeric motifs. In addition, extensive signs of degeneration were detected in the nonrecombining regions in the form of transposable element accumulation and of hundreds of gene losses on each mating-type chromosome. Furthermore, our study highlights the potential of the latest breakthrough PacBio chemistry to resolve complex genome architectures.
Keywords: Microbotryum violaceum, finished genome assembly, intratetrad mating, MAT, basidiomycete, selfing, bipolarity
SEX chromosomes often show exceptional genome features with extensive suppression of homologous recombination and cytological differentiation between members of the diploid chromosome pair (Bergero and Charlesworth 2009). Determination of male vs. female development by such nonrecombining sex chromosomes has evolved independently in diverse lineages, including mammals, birds, fishes insects, vascular plants, bryophytes, and macroalgae (Ohno 1967; Charlesworth 1991; Itoh et al. 2006; Yamato et al. 2007; Marais et al. 2008; Bergero and Charlesworth 2009; Cock et al. 2010; Kaiser and Bachtrog 2010; Page et al. 2010; Bachtrog et al. 2011; McDaniel et al. 2013), where incorporation of multiple genes responsible for sexually antagonistic traits is a common explanation for expansion of recombination suppression (Rice 1984; Lahn and Page 1999). Despite the fundamental roles that sex chromosomes play, their genic content is particularly prone to degenerative influences of recombination suppression and enforced heterozygosity (Bergero and Charlesworth 2009).
In fungi, mating-type chromosomes contain the genes for molecular mechanisms of mating compatibility (e.g., via pheromones and receptors), and they can display recombination suppression and size dimorphism analogous to sex chromosomes (Bakkeren and Kronstad 1994; Hood 2002; Fraser et al. 2004; Fraser and Heitman 2004; Hood et al. 2004, 2013; Fraser and Heitman 2005; Menkis et al. 2008; Ellison et al. 2011; Grognet et al. 2014), as well as degeneration (Hood et al. 2004; Whittle and Johannesson 2011; Whittle et al. 2011; Fontanillas et al. 2015). Grouped together, heteromorphic sex chromosomes and mating-type chromosomes can therefore be considered allosome pairs (Montgomery 1911), which are similarly maintained and influenced by their central role in regulating gamete fusion, contrasted by the homomorphic autosomes. Nonrecombining fungal mating-type chromosomes can also show footprints of degeneration (Hood et al. 2004; Whittle and Johannesson 2011; Whittle et al. 2011; Fontanillas et al. 2015). However, important differences exist between sex and mating-type chromosomes: because mating types are expressed as non-self-recognition at the haploid stage, they are found only in the heterogametic condition in diploids (similar to macroalgae and bryophytes), and thus with symmetrical roles, and mating types are not associated with male/female functions in fungi (Billiard et al. 2011). These common features and differences help make fungal mating-type chromosomes valuable models for the broader phenomenon of allosome evolution, with some shared and some distinguishing expectations compared to sex chromosomes, e.g., on the asymmetry of allosome degeneration (Bull 1978) and on the role of sexually antagonistic selection in driving suppressed recombination (Bergero and Charlesworth 2009).
Examples of nonrecombining fungal allosomes include the mating-type chromosomes of Microbotryum lychnidis-dioicae (Hood 2002) and the fungus-causing anther-smut disease on Silene latifolia (Votintseva and Filatov 2009; Hood et al. 2013; Fontanillas et al. 2015; Perlin et al. 2015; Whittle et al. 2015). These nonrecombining fungal mating-type chromosomes have been extensively studied, and degeneration has been reported in the form of transposable element (TE) accumulation and high rates of nonsynonymous substitutions (Hood et al. 2004; Fontanillas et al. 2015). The buildup of TEs in regions of suppressed recombination has, however, prevented genome assembly for these mating-type chromosomes. Therefore, the exact content in genes and transposable elements, the extent of the nonrecombining regions (NRRs), and the existence of evolutionary strata of recombination suppression have remained elusive (Votintseva and Filatov 2009; Hood et al. 2013; Fontanillas et al. 2015; Perlin et al. 2015; Whittle et al. 2015).
The spread of recombination suppression outward from key compatibility-determining loci to form evolutionary strata of differentiation is described for plant and animal sex chromosomes (Bergero and Charlesworth 2009) and may also occur on fungal mating-type chromosomes (Fraser et al. 2004; Fraser and Heitman 2004, 2005; Menkis et al. 2008; Votintseva and Filatov 2009). Suppressed recombination has been shown to link the two mating-type-determining loci (i.e., encoding mating pheromone/receptor and homeodomain proteins, respectively), including basidiomycete fungal pathogens of plants and humans (Ustilago hordei and Cryptococcus neoformans, respectively) (Bakkeren and Kronstad 1994; Fraser et al. 2004). This linkage of two mating-type-determining loci may be advantageous under selfing mating systems, where it increases the compatibility of gamete combinations (Billiard et al. 2011; Nieuwenhuis et al. 2013). In other fungi, suppressed recombination links mating-type loci to the centromere, causing segregation of mating types in the first meiotic division. This is associated with automictic reproduction (mating within the meiotic tetrad), e.g., in M. lychnidis-dioicae and Neurospora tetrasperma (Zakharov 1987; Hood and Antonovics 2000; Giraud et al. 2008; Menkis et al. 2008). Such intratetrad mating may favor successive linkage to mating type for a suite of genes that experience deleterious, recessive mutations, as shown by theoretical models (Antonovics and Abrams 2004; Johnson et al. 2005) (Supporting Information, Figure S1). Indeed, deleterious mutation occurring at the margin of the nonrecombining regions may be partially sheltered in a heterozygous state due to less frequent recombination, and extension of the region of suppressed recombination would then be selected for permanent sheltering of these deleterious mutations (Ironside 2010). This could lead to the formation of evolutionary strata, reflected in increasing allelic differentiation at physical positions along the chromosome when coming closer to the mating-type genes. Although in plants and in animals evolutionary strata are generally thought to be due to sexually antagonistic selection, the definitive linkage of partially linked deleterious alleles to the sex-determining region has also been proposed as an alternative explanation (Ironside 2010). Elucidating the existence of evolutionary strata in fungal mating-type chromosomes may thus generally provide insights on the shared evolutionary processes acting on allosomes.
In this research, we aimed to produce a finished assembly of the mating-type chromosomes of the anther-smut fungus M. lychnidis-dioicae for elucidating the extent of suppressed recombination, transposable element accumulation, gene content, and rearrangements and whether evolutionary strata are present. For this goal, we took advantage of the recent developments of the PacBio sequencing technology, which allowed full-length physical mapping of the mating-type chromosomes and the resolution of these important genomic properties.
Materials and Methods
DNA extraction and sequencing
DNA was extracted with the Qiagen Kit 10243 (Courtaboeuf, France) following manufacturer instructions and using a Carver hydraulic press (reference 3968, Wabash, IN) for breaking cell walls. Haploid a1 and a2 genomes of the Lamole reference strain of M. lychnidis-dioicae were sequenced separately using the Pacific Bioscience (PacBio) method (Institute for Genomic Medicine, University of California, San Diego, La Jolla, CA).
Assembly and annotation
The PacBio P5/C3 sequencing of haploid a1 and a2 genomes produced 658,501 and 648,664 reads, respectively (mean read length: 7.925/7.569; coverage: 180×/160×). Independent assemblies of the a1 and a2 genomes were generated with the wgs-8.2beta version of the PBcR assembler (Koren et al. 2012), with the following parameters: -pbCNS -length 500 -partitions 200 genomeSize = 27000000 -maxCoverage 150. Contigs were aligned with optical maps of the two mating-type chromosomes (Hood et al. 2013) with MapSolver software (OpGen). The alignment of the contig corresponding to the a2 mating-type chromosome with the corresponding optical map was highly significant. The a1 optical map was used to create an oriented a1 pseudomolecule composed of two contigs for each chromosome arm, aligned on the optical map, plus a 50-kb unanchored contig in the centromeric region. The two contigs corresponding to the two a1 chromosome arms produced full-length alignments that matched only with the a1 mating-type chromosome optical map; alignment scores of the contigs were 46 and 143, exceeding the default MapSolver threshold range for significance of 3 to 6.
Mauve was used to compare the two assemblies (Darling et al. 2010) (Figure S6). In this genome comparison, a contig belonging to autosomes in one assembly was found to be a subsequence of a contig in the second assembly (and vice versa). Manual curation was performed to select the largest contigs and remove the few short degenerate contigs, probably resulting from the incorrect editing of reads (Figure S6). Comparison with the reference sequence (GenBank no. NC_020353) revealed that the mitochondrial genome was not correctly solved by the assembler, and this genome sequence was therefore removed from the assembly. The PacBio assembly was compared with the Illumina-based assembly available from the Broad Institute and the Illumina reads generated in a previous study (Perlin et al. 2015). The PacBio assembly was edited at positions for which both the genome alignment obtained with Mugsy (Angiuoli and Salzberg 2011) and a standard variant-calling method based on read mapping (samtools, bcftools, and VarScan) (Li et al. 2009; Koboldt et al. 2012) gave concordant findings for the polymorphism. This conservative approach led to the editing of 1332 SNPs, 456 insertions, and 19,528 deletions. The 11,057 protein-coding gene models were predicted with EuGene (Foissac et al. 2008), trained for Microbotryum. Available Microbotryum EST data (Aguileta et al. 2010) and similarities to the fungi subset of the uniprot database (Consortium 2011) were integrated into EuGene for gene prediction. InterPro (Mitchell et al. 2014) was used to identify protein domains and families and to infer an automatic functional annotation from protein domain content.
Analysis of genome structure
Repetitive DNA content was analyzed with RepeatMasker (Smit 2013), using REPBASE v19.11 (Jurka 1998). We searched for subtelomeric motifs with Meme (Bailey et al. 2006) (Figure S2). Figure 1, Figure 2, and Figure S5 were prepared with Circos (Krzywinski et al. 2009). We identified syntenic blocks from the alignment of the two chromosomes with Mauve, using standard parameters (Darling et al. 2010). We analyzed gene order after filtering out transposable elements (TEs) to identify larger blocks of synteny. Alleles were assigned between the two mating-type chromosomes by applying orthomcl (Li et al. 2003) to the protein data sets for unique a1 and a2 orthologs. The signed permutations were used to calculate minimal reversal distances with Grimm (Tesler 2002). The sequences of TE-like copias and minisatellites specific for the centromeres are provided in File S1.
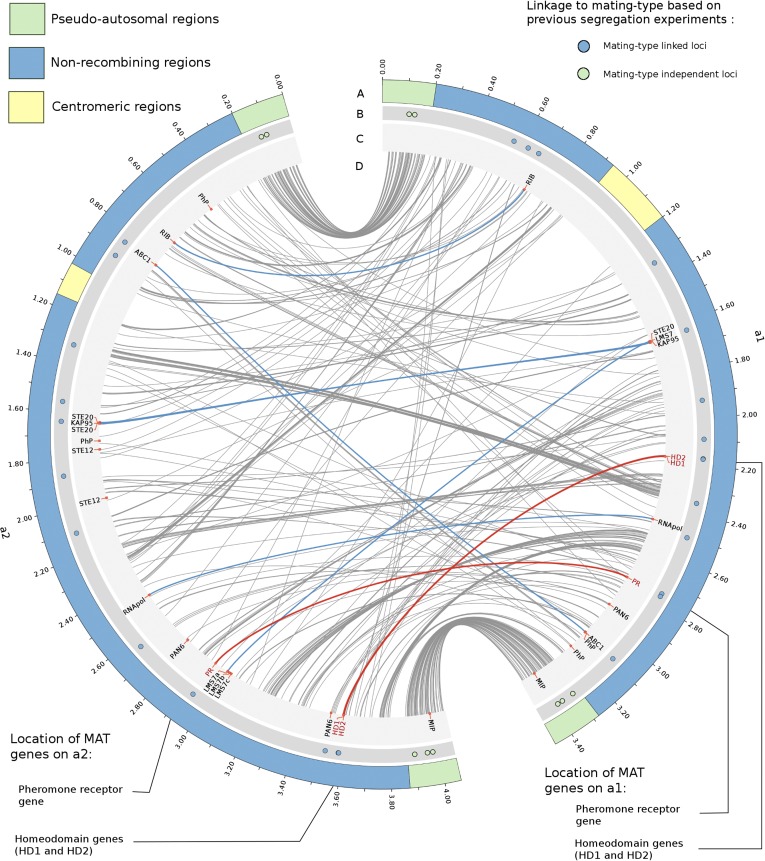
Figure 1.
Finished assembly of the mating-type chromosomes of the anther-smut fungus M. lychnidis-dioicae. Tracks A–D show the location of different genomic elements. A: Structure of the chromosomes, with the PARs in green, the NRRs in blue, and the centromeres in yellow. B: Location of loci that have been shown to be linked (blue circles) or unlinked (green circles) to mating type by previous segregation analyses (Votintseva and Filatov 2009; Abbate and Hood 2010; Petit et al. 2012). C: Location of the genes related to the mating-type function—pheromone receptor (PR) and HD homeodomain genes (in red), of the other genes likely involved in mating (STE12, STE20, and the precursors of pheromones, PhP), and of the genes located around the pheromone receptor gene in the closely related S. salmonicolor (Coelho et al. 2010) (KAP95, RNAPol, RIB, and ABC1). D: Links between orthologous alleles of a1 and a2, in red for HD and PR and in blue for KAP95, RNAPol, RIB, and ABC1.
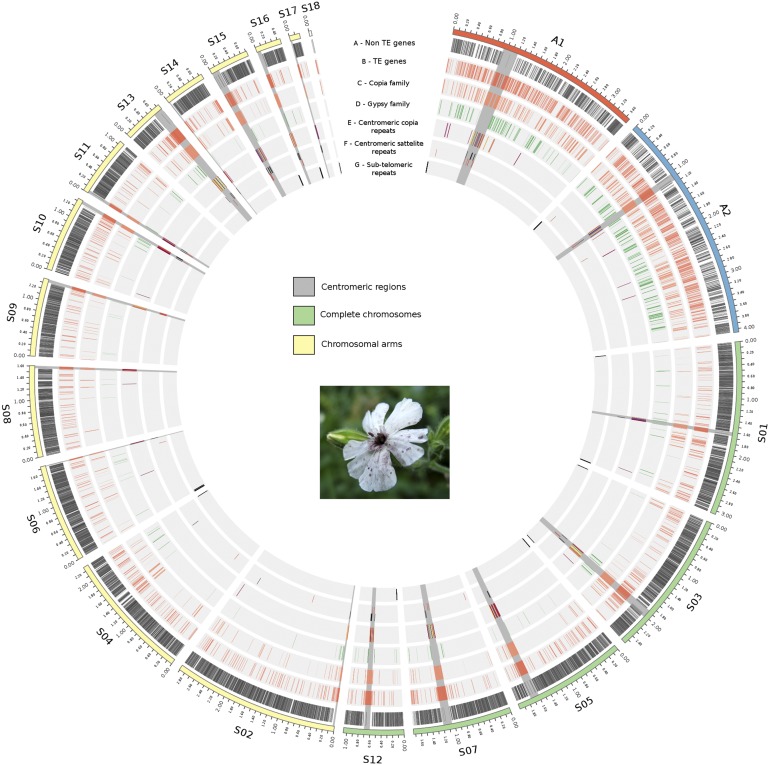
Figure 2.
PacBio assembly of the nuclear M. lychnidis-dioicae genome. The picture shows the fungal dark spores in the anthers of a S. latifolia flower, replacing the pollen. The mating-type chromosomes (blue) and finished autosomes (green) or autosome arms (yellow) are represented. The identified centromeric regions are shaded. Tracks A–G show the location of different genomic elements. A: Genes after filtering out TEs. B: Genes encoding TEs. C: TEs from the copia family. D: TEs from the gypsy family. E: TEs from the copia subfamily particularly abundant at centromeres (Figure 3). F: Satellite repeat of the type particularly abundant at centromeres (Figure 3). G: Subtelomeric motifs (Figure S2).
Divergence between a1 and a2 alleles
Alleles were identified as indicated above, and MACSE (Ranwez et al. 2011) was used to align DNA-coding sequences, respecting phase. Synonymous divergence and its standard error were estimated with the yn00 program of the PAML package (Yang 2007). The correlation between dS and physical distance was assessed with JMP (SAS Institute). As we have here a single individual genome sequence, some of the substitutions between a1 and a2 identified here could theoretically include within-species polymorphism rather than fixed differences between the a1 and a2 alleles. However, this should not be a problem for identifying the nonrecombining regions from the pseudo-autosomal regions (PARs). Indeed, given its highly selfing mating system, M. lychnidis-dioicae is largely homozygous in regions unlinked to mating type (Giraud et al. 2008; Vercken et al. 2010), so that high levels of synonymous divergence between two a1 and a2 alleles from a given diploid individual reliably indicate suppressed recombination.
Data access
The assembly is available at EMBL-ENA (accession no. PRJEB7910).
Results and Discussion
Finished genome assembly and new insights about centromere and telomere structures
The recent PacBio P5/C3 chemistry produced long reads, averaging 8 kb. The imperfect quality of PacBio reads was fully compensated by >100× sequencing depth, as illustrated by the almost complete assembly of the M. lychnidis-dioicae genome and including the mating-type chromosomes (Figure 1 and Figure 2), while several previous efforts and chemistries had been unsuccessful (Votintseva and Filatov 2009; Hood et al. 2013; Fontanillas et al. 2015). The PacBio genome assembly indeed yielded 22 contigs, including 18 autosomal contigs, 1 contig for the a2 mating-type chromosome, and 3 contigs for the a1 mating-type chromosome that could be assembled in a scaffold (see below) (Figure 2, Table 1). The total size of the assembly was 33.3 Mb, i.e., almost 7 Mb larger than that of the very same a1 haploid M. lychnidis-dioicae strain sequenced at the Broad Institute using Illumina technology (Perlin et al. 2015), as the long reads of the PacBio chemistry allowed assembly by more complete incorporation of repetitive DNA elements and included both mating-type chromosomes. In particular, putative centromeric regions could be assembled and were identified as the most repeat-rich regions, where we found two copia-like TEs and two minisatellite repeats specifically clustering in these regions at one location per contig (tracks E and F in Figure 2). Both copia-like TEs possessed two open reading frames, coding for an integrase and for a reverse transcriptase, but no long terminal repeats, and many copies were truncated. The minisatellites consisted of a 66-bp repeat motif of GGCCCA and a 110-bp repeat motif of CGACGG; the 66-bp repeat was specific for centromeric regions and identical in all iterations. The structures of the finished centromeres are shown in Figure 3. Eleven of the contigs had these specific repeats at one end, suggesting that some autosomes were split at their centromeres in the assembly (Figure 2). On finished chromosomes, the centromeric regions spanned 65–155 kb. Centromeric regions have been elucidated in a single other basidiomycete fungus so far, Cryptococcus neoformans var. grubii, in which they were instead enriched in Tcn transposons (Janbon et al. 2014).
Table 1. Assembly statistics of the M. lychnidis-dioicae genome.
| No. of contigs | 22 |
| Minimum contig size | 44,493 |
| Maximum contig size | 4,061,474 |
| N50 (bp) | 2,275,168 |
| N50 (no. of contigs) | 6 |
| N90 (bp) | 1,030,812 |
| N90 (no. of contigs) | 15 |
| Mean contig size | 1,505,578.59 |
| Median contig size | 1,345,833 |
| Total length of the assembly (bp) | 33,122,729 |
The PacBio genome assembly yielded 22 contigs—18 for autosomes, 1 for the a2 mating-type chromosome, and 3 for the a1 mating-type chromosome—that could be assembled in a single scaffold.
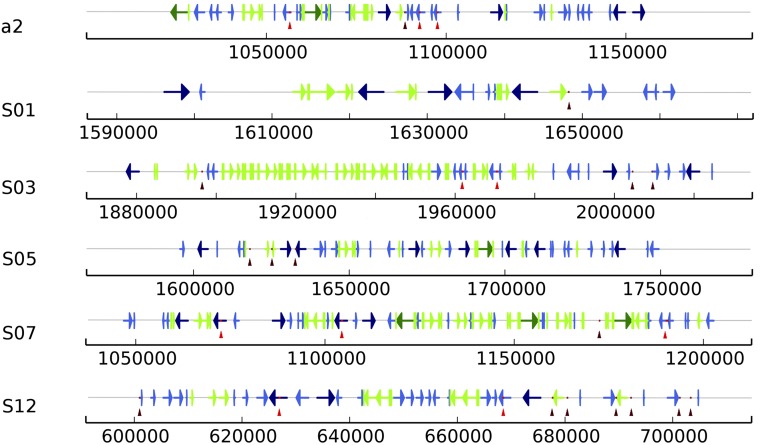
Figure 3.
Structure of the centromeres. Localization of copia-like transposable elements and specific mini-satellite repeats in the finished centromeric regions (scaffolds names are indicated as in Figure 1). Full-length copies of two types of copia-like transposable elements are shown in blue and green. Lighter shades indicate incomplete copies of these two types. Arrows indicate the orientation of copies. Dots and vertical arrows indicate repeats of the 66-bp motif minisatellite (GGCCCA)n and of the 110-bp motif minisatellite (CGACGG)n in brown and red, respectively.
We found no typical telomeric repeats (TTAGGG) while they were identified at the edge of five scaffolds in the Broad Institute M. lychnidis-dioicae genome (Perlin et al. 2015). Here, we identified instead a specific 50-bp motif repeated at some, but not all, chromosome ends (Figure 2, Figure S2), which thus likely represent a subtelomeric motif. This motif was in fact found near the TTAGGG telomeric repeats (80–1500 bp apart) in two of the five contigs from the Broad Institute containing TTAGGG repeats (the three other contigs being only 500–850 bp long). This suggests that PacBio technology does not sequence the very edges of telomeres well and that other motifs may be useful for detecting ends of chromosomes.
Previous pulse-field gel electrophoresis estimated 11 autosomes for the haploid stage (Perlin et al. 2015), corresponding precisely to the 5 complete autosomes plus the 6 autosomes as two contigs split in their centromeric regions (the S18 contig corresponded only to a telomeric region). The autosomes were thus assembled into full or two-arm contigs (Figure 2). The a2 mating-type chromosome was completely assembled from PacBio sequences, including the submetacentric centromeric region. Available optical maps (Hood et al. 2013), i.e., ordered, genome-wide restriction maps obtained from single DNA molecules, allowed orienting the two contigs covering the arms of the a1 mating-type chromosome and attributing a 50-kb contig composed of repeated DNA to its centromeric region. Collinearity between the optical maps and the contigs confirmed the high quality of the assembly (Figure S3). The combination of PacBio chemistry and optical maps thus revealed the right approach to finally resolve the mating-type chromosome structure and content.
Identification of pseudo-autosomal regions and extensive nonrecombining regions on the mating-type chromosomes
Assemblies of the dimorphic M. lychnidis-dioicae mating-type chromosomes (a1, 3.5 Mbp; a2, 4.0 Mbp) encompassed 614 and 683 predicted genes, respectively, filtering out TEs. Among these genes, only a few had putative functions known to be involved in mating (Petit et al. 2012; Fontanillas et al. 2015), i.e., the PR genes encoding the pheromone receptors, the PhP genes encoding the precursors of pheromones, the HD1 and HD2 homeodomain genes, STE20 encoding a protein kinase regulating mating (Smith et al. 2004), and STE12 encoding a transcription factor regulating mating and invasive hyphal growth in fungal pathogens (Hoi and Dumas 2010) (Figure 1). Actually, two homologs of STE12 were found, both present only on the a2 mating-type chromosome, while absent from the a1 mating-type chromosome; this seems consistent with previous observations that initiation of the conjugation tubes is earlier and to a greater extent from cells of the a2 mating type in M. lychnidis-dioicae (Day 1976; Xu et al. 2015). The PhP pheromone genes were recently characterized as encoding proteins with the typical C terminus CAAX motif and producing functional pheromone peptides triggering the mating response in several Microbotryum species (Xu et al. 2015).
We identified 305 predicted genes shared between the a1 and a2 mating-type chromosomes. Synonymous divergence between a1 and a2 alleles of these shared genes, when plotted along their physical positions on both mating-type chromosomes, clearly identified the nonrecombining regions; suppressed recombination can lead to divergence between alleles while recombination homogenizes allele sequences. Synonymous divergence values between alleles confirmed the existence of recombining regions at both ends of the mating-type chromosomes, called PARs (Figure 4) (Votintseva and Filatov 2009; Hood et al. 2013). The PARs displayed almost no substitutions between a1 and a2 alleles (Figure 4) and included loci that have previously been shown to be unlinked to the mating type (Votintseva and Filatov 2009) (Figure 1). PARs on the short arm (pPAR) and long arm (qPAR) of the mating-type chromosomes spanned 0.20 and 0.19 Mb, respectively, and contained 102 genes. PAR boundaries corresponded precisely to the two distal regions of collinearity between a1 and a2 mating-type chromosomes on the restriction digest optical maps (Hood et al. 2013).
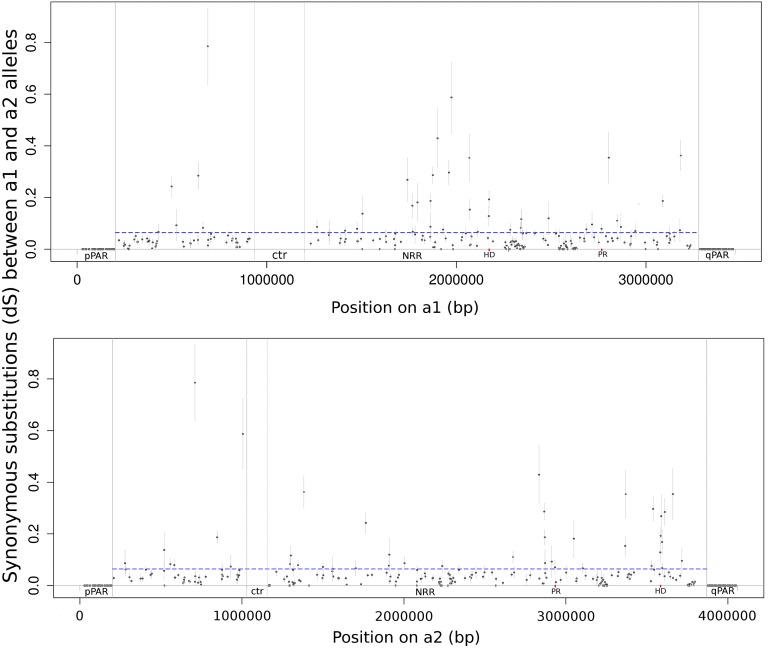
Figure 4.
Divergence between a1 and a2 mating-type chromosomes in M. lychnidis-dioicae. (A) Synonymous divergence dS ± SE is plotted against the genomic coordinates of the a1 (A) and a2 (B) mating-type chromosomes for all nontransposable element genes shared by the mating-type chromosomes. The boundaries between the PARs and the NRRs are indicated, as well as the locations of the mating-type loci (PR: pheromone receptor gene; HD1 and HD2: homeodomain genes). The mean value of dS in the NRRs is shown as a blue dotted line. The pheromone receptor could not be plotted as its a1–a2 divergence was too extensive to be aligned in nucleotides.
The NRRs were characterized by high divergence between the a1 and a2 alleles of the single-copy genes (Figure 4 and Figure S4). Fourteen widely distributed loci known to cosegregate with mating-type (Votintseva and Filatov 2009; Abbate and Hood 2010; Petit et al. 2012) were found within these divergent regions, validating the NRRs (Figure 1). The a1 and a2 NRRs spanned the majority of the mating-type chromosomes (>88%), covering 3.08 Mb with 506 genes and 3.67 Mb with 580 genes for the a1 and a2 mating-type chromosomes, respectively (Figure 1), confirming conclusions based on optical maps and marker segregation (Hood et al. 2004, 2013; Fontanillas et al. 2015) and contrasting with reports of smaller regions of suppressed recombination (Votintseva and Filatov 2009; Whittle et al. 2015).
Extensive rearrangements between mating-type chromosomes and degeneration
Comparison of aligned sequences between the a1 and a2 NRRs revealed extensive rearrangement (Figure 1 and Figure S5), probably reflecting and contributing to the current degree of suppressed recombination. We identified 40 syntenic blocks of shared genes in the NRRs, encompassing 2–18 genes. At least 210 inversion events were inferred to account for these rearrangements (Figure 1 and Figure S5). In contrast, all autosomes were collinear (Figure S6). Suppressed recombination on fungal mating-type chromosomes has been shown to be associated with rearrangements in N. tetrasperma (Ellison et al. 2011) and C. neoformans (Fraser et al. 2004; Fraser and Heitman 2004, 2005) or to occur despite conserved collinearity in Podospora anserina (Grognet et al. 2014). The lack of collinearity across several megabases and patterns of major inversions or translocations in M. lychnidis-dioicae is, however, to an exceptional degree among fungal mating-type chromosomes.
The two mating-type loci typical of basidiomycete fungi (i.e., the mating pheromone receptor determining premating recognition and the homeodomain proteins determining postmating compatibility) were 0.60 Mb apart on the a1 and a2 mating-type chromosomes, while inverted in orientation (Figure 1 and Figure S5). This distance is much greater than the one estimated recently based on Illumina sequencing (56 kb) (Whittle et al. 2015), again showing the great advantage of PacBio sequencing for chromosome assembly. These mating-type loci were not near the edges of the NRRs, unlike other basidiomycete fungi with linkage suppression in their mating-type regions, i.e., U. hordei (Bakkeren and Kronstad 1994) and C. neoformans (Fraser et al. 2004). The mating-type loci were instead on the same side of the centromere, on the long arm of the chromosomes. The pheromone precursor loci were linked to the receptor loci in the NRR, but were up to 1.4 Mb away, on opposite sides of the centromere on the a2 mating-type chromosome and the same side in the a1 mating-type chromosome. This distance between pheromone receptor and pheromone is also a novel observation among fungi. Despite rearrangements in NRRs, several genes close to the pheromone receptor genes in the closely related fungus Sporidiobolus salmonicolor (Microbotryomycetes) (Coelho et al. 2010) were also found on the mating-type chromosomes of M. lychnidis-dioicae (KAP95, RNAPol, RIB, and ABC1, Figure 1). In contrast, the homologs of the 40 predicted genes in the scaffold containing the HD locus in S. salmonicolor with blast hits in M. lychnidis-dioicae were all found on autosomes but one, and 34 of them on a single autosome arm (S09). Interestingly, we also found on the S09 contig a sequence with similarity to one of the two homeodomain genes (HD1). The possibility that the HD locus may have duplicated to a location in linkage with the PhP/PR locus warrants further study as a potential transition to mating-type bipolarity within the Microbotryomycetes.
The high degree of rearrangements between the two mating-type chromosomes in M. lychnidis-dioicae prevents us from obtaining definitive evidence for the evolutionary cause of the expansive nature of recombination suppression; nevertheless, the linkage of mating-type loci and the centromere within NRRs indicates that first-division segregation of both mating-type loci during meiosis may have been a primary evolutionary factor in recombination suppression, as suggested for other automictic fungi (Zakharov 2005).
As expected for nonrecombining regions, the NRRs had a higher frequency of repetitive elements than the PARs and autosomes (Figure 2). Repetitive elements accounted for 16.0 and 18.7% of the a1 and a2 NRRs, respectively, explaining why they have remained unassembled until now. Most of the repetitive elements were long terminal repeat retro-elements of the copia and gypsy families, as found previously (Hood et al. 2005; Fontanillas et al. 2015). In addition, gene density was lower in the NRRs (25.8% were coding sequences) than in the autosomes (62.7%) or PARs (51.2%).
The almost finished assembly of the genome allowed detection of 245 and 289 genes in a hemizygous state on the mating-type chromosomes a1 and a2, respectively, suggesting that functional copies have been lost from a single one of the mating-type chromosomes or have degenerated to a great extent. None of these hemizygous genes had putative functions known to be involved in mating except the STE12 homologs on the a2 mating-type chromosome. This degree of gene losses on mating-type chromosomes appears unprecedented among fungi. Together with the accumulation of TEs in NRRs, low gene density and hemizygous loss of genes constitute signs of advanced mating-type chromosome degeneration, as expected for long-term, nonrecombining regions (Bergero and Charlesworth 2009). Degeneration has also been reported for the mating-type chromosome NRRs of M. lychnidis-dioicae in terms of higher rates of nonsynonymous substitutions than for recombining regions, and these degenerative characteristics were shown to occur in homologous sequences from several other Microbotryum species (Fontanillas et al. 2015).
The a2 mating-type chromosome was 590 kb larger than the a1 mating-type chromosome, where TEs accounted for 26.4% (154 kb) and non-TE genes for 6.8% (40 kb) of the difference. In particular, gypsy copies were more abundant on a2 than a1 mating-type chromosome (129 vs. 93 copies, Figure 2). However, intergenic, nonrepetitive DNA accounted for the majority of the difference in size (59%, or 0.347 kb). This finding supports previous conclusions that TE accumulation and degeneration are not asymmetrical between the mating-type chromosomes (Fontanillas et al. 2015), as expected from their symmetrical role in mating (Bull 1978).
Ancient recombination suppression without clear evolutionary strata despite heterogeneity in a1–a2 divergence
Divergence between a1 and a2 alleles of predicted genes in the NRRs was substantial (mean synonymous substitution rates: dS = 0.064, Figure 4), indicating relatively ancient recombination suppression. However, this divergence was lower than for the pheromone receptor gene, which was previously shown to constitute the oldest trans-specific polymorphism in any organism so far (Devier et al. 2009).
In the evaluation of evolutionary strata within NRRs, we quantified synonymous divergence (dS) along the mating-type chromosomes, and there was no correlation with physical distance to the PAR boundary or to the mating-type loci on either the a1 or a2 chromosomes (Figure 3). In particular, the correlations between dS and gene order from PAR were nonsignificant (on a1, r = 0.06, P = 0.36; on a2, r = 0.02, P = 0.77), as were correlations between dS and gene order from the nearest mating-type gene (on a1 r = −0.04, P = 0.58; on a2 r = 0.02, P = 0.77). There was a particularly sharp boundary between the pPARs, with zero synonymous divergence between a1 and a2 alleles, and the adjacent NRRs, with extensive divergence (mean dS = 0.064). Thus, there was no evidence of evolutionary strata.
However, given that there is no homogametic diploid in heterothallic fungi (i.e., no a1a1 or a2a2 genotypes), which tends to retain chromosome structure in homogametic sex chromosomes of plants and animals (X or Z chromosomes), the mating-type chromosomes may more rapidly undergo rearrangements that obscure any history of evolutionary strata, had they existed. Actually, in XY or ZW systems, evolutionary strata can be inferred only by using as reference the less rearranged homogametic sex chromosome (Lahn and Page 1999).
It is possible that we may have detected one recent mini-stratum as a very small region (23 kb) at the edge of the NRR close to the qPAR that was collinear between a1 and a2 mating-type chromosomes but with a high rate of synonymous substitutions between mating types. This region was separated from the qPAR by a rearranged region rich with TEs (Figure S7). This pattern is consistent with a recent incorporation of a PAR fragment into the NRR, which would support the scenario of an extension of the NRR, and thus evolutionary strata.
Allele divergence between mating types in NRRs was characterized by heterogeneous values (Figure 4 and Figure S4), which may result from a continuing process of translocations between the chromosomes or the remnants of ancient evolutionary strata that have been scrambled by rearrangements. Alternatively, gene conversion may have played a role by recurrently resetting divergence to zero in some stretches of DNA.
Conclusions
The nearly finished assembly of the M. lychnidis-dioicae genome allowed the resolution of controversies over the extent of the nonrecombining regions of the mating-type chromosomes, the degree of divergence between the a1 and a2 mating-type chromosomes, the patterns of evolutionary strata, and the mechanisms of evolution to bipolarity (Votintseva and Filatov 2009; Hood et al. 2013; Fontanillas et al. 2015; Whittle et al. 2015). The great extent to which the mating-type chromosomes are consumed by the NRR and the chaos of rearrangements within this region indicate that these dimorphic chromosomes are the result of ancient and impactful evolutionary processes, approaching an ultimate state of genomic entropy, and thus showing striking convergence with some animals (Repping et al. 2006; Bachtrog 2013). Our study allows the broadening of evolutionary biology for nonrecombining chromosomes by contributing to evidence of convergence in genomic patterns between fungal mating-type chromosomes and sex chromosomes, as previously highlighted (Hood 2002; Fraser et al. 2004; Menkis et al. 2008). These distant systems share extensive suppressed recombination, a high level of rearrangements, numerous losses of genes, and accumulation of a high repetitive elements content. The high level of rearrangements and the possibility of gene conversion still make it difficult to reconstitute the full history of the recombination suppression of the mating-type chromosomes. Comparative genomics between closely related Microbotryum species and other members of the Microbotryomycetes with different degrees and/or ages of suppression of recombination between their mating-type chromosomes should help in resolving these questions in future studies.
Next generation sequencing is increasingly making it possible to obtain sequence data at low cost for nonmodel organisms. However, the short length of reads remains a major limitation, potentially preventing large-scale assembly, particularly for genomic regions rich in transposable elements (Treangen and Salzberg 2012). The lack of a complete physical assembly precludes investigation of some of the most fundamental evolutionary inferences. PacBio chemistry now makes it possible to obtain a complete and affordable assembly, even for notoriously challenging regions, such as highly repetitive centromeres and nonrecombining chromosomes (Treangen and Salzberg 2012). This breakthrough technology will now facilitate studies of the dynamics and evolutionary role of genome structures with unprecedented resolution and power.
Supplementary Material
Acknowledgments
We thank Stéphanie Le Prieur, Alodie Snirc, and Gilles Deparis for help with DNA extraction. PacBio sequencing was conducted at the Institute for Genomic Medicine, University of California, San Diego, La Jolla, CA. This work was supported by a European Research Council starting grant, GenomeFun 309403 and ANR-12-ADAP-0009 (Gandalf project) (to T.G.); an Institut Diversité et Evolution du Vivant grant (to T.G. and C.F.); National Science Foundation (NSF) grant DEB-1115765 to (M.E.H.); and NSF award #0947963 (to M.H.P. and C.A.C.). We declare no conflict of interest.
Footnotes
Communicating editor: J. Heitman
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177709/-/DC1.
Sequence data from this article have been deposited with EMBL-ENA under accession no. PRJEB7910.
Literature Cited
- Abbate J. L., Hood M. E., 2010. Dynamic linkage relationships to the mating-type locus in automictic fungi of the genus Microbotryum. J. Evol. Biol. 23: 1800–1805. [DOI] [PubMed] [Google Scholar]
- Aguileta G., Lengelle J., Marthey S., Chiapello H., Rodolphe F., et al. , 2010. Finding candidate genes under positive selection in non-model species: examples of genes involved in host specialization in pathogens. Mol. Ecol. 19: 292–306. [DOI] [PubMed] [Google Scholar]
- Angiuoli S., Salzberg S., 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics J., Abrams J. Y., 2004. Intratetrad mating and the evolution of linkage relationships. Evolution 58: 702–709. [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Kirkpatrick M., Mank J. E., McDaniel S. F., Pires J. C., et al. , 2011. Are all sex chromosomes created equal? Trends Genet. 27: 350–357. [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W., 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkeren, G., and J. W. Kronstad, 1994 Linkage of mating type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc. Natl. Acad. Sci. USA 91: 7085–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R., Charlesworth D., 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 24: 94–102. [DOI] [PubMed] [Google Scholar]
- Billiard S., Lopez-Villavicencio M., Devier B., Hood M., Fairhead C., et al. , 2011. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol. Rev. Camb. Philos. Soc. 86: 421–442. [DOI] [PubMed] [Google Scholar]
- Bull J. J., 1978. Sex chromosomes in haploid dioecy: unique contrast to Mullers theory for diploid dioecy. Am. Nat. 112: 245–250. [Google Scholar]
- Charlesworth B., 1991. The evolution of sex chromosomes. Science 251: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Cock J. M., Sterck L., Rouze P., Scornet D., Allen A. E., et al. , 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621. [DOI] [PubMed] [Google Scholar]
- Coelho M. A., Sampaio J. P., Goncalves P., 2010. A deviation from the bipolar-tetrapolar mating paradigm in an early diverged Basidiomycete. PLoS Genet. 6: e1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T. U., 2011. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39: D214–D219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T., 2010. ProgressiveMauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS ONE 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A., 1976. Communication through fimbriae during conjugation in a fungus. Nature 262: 583–584. [DOI] [PubMed] [Google Scholar]
- Devier B., Aguileta G., Hood M., Giraud T., 2009. Ancient trans-specific polymorphism at pheromone receptor genes in basidiomycetes. Genetics 181: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison C. E., Stajich J. E., Jacobson D. J., Natvig D. O., Lapidus A., et al. , 2011. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics 189: 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac S., Gouzy J., Rombauts S., Mathé C., Amselem J., et al. , 2008. Genome annotation in plants and fungi: EuGène as a model platform. Current Bioinformatics 3: 87–97. [Google Scholar]
- Fontanillas E., Hood M., Badouin H., Petit E., Barbe V., et al. , 2015. Degeneration of the non-recombining regions in the mating type chromosomes of the anther smut fungi. Mol. Biol. Evol. 32: 928–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. A., Heitman J., 2004. Evolution of fungal sex chromosomes. Mol. Microbiol. 51: 299–306. [DOI] [PubMed] [Google Scholar]
- Fraser J., Heitman J., 2005. Chromosomal sex-determining regions in animals, plants and fungi. Curr. Opin. Genet. Dev. 15: 645–651. [DOI] [PubMed] [Google Scholar]
- Fraser J. A., Diezmann S., Subaran R. L., Allen A., Lengeler K. B., et al. , 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2: 2243–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T., Yockteng R., Lopez-Villavicencio M., Refrégier G., Hood M. E., 2008. The mating system of the anther smut fungus, Microbotryum violaceum: selfing under heterothallism. Eukaryot. Cell 7: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grognet P., Bidard F., Kuchl C., Chan Ho Tong L., Coppin E., et al. , 2014. Maintaining two mating types: structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics 197: 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi J. W. S., Dumas B., 2010. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell 9: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. E., 2002. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics 160: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. E., Antonovics J., 2000. Intratetrad mating, heterozygosity, and the maintenance of deleterious alleles in Microbotryum violaceum (=Ustilago violacea). Heredity 85: 231–241. [DOI] [PubMed] [Google Scholar]
- Hood M. E., Antonovics J., Koskella B., 2004. Shared forces of sex chromosome evolution in haploids and diploids. Genetics 168: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. E., Katawezik M., Giraud T., 2005. Repeat-induced point mutation and the population structure of transposable elements in Microbotryum violaceum. Genetics 170: 1081–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. E., Petit E., Giraud T., 2013. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics 193: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ironside J. E., 2010. No amicable divorce? Challenging the notion that sexual antagonism drives sex chromosome evolution. BioEssays 32: 718–726. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Kampf K., Arnold A. P., 2006. Comparison of the chicken and zebra finch Z chromosomes shows evolutionary rearrangements. Chromosome Res. 14: 805–815. [DOI] [PubMed] [Google Scholar]
- Janbon G., Ormerod K. L., Paulet D., Byrnes E. J., III, Yadav V., et al. , 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10: e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. J., Antonovics J., Hood M. E., 2005. The evolution of intratetrad mating rates. Evolution 59: 2525–2532. [PubMed] [Google Scholar]
- Jurka J., 1998. Repeats in genomic DNA: mining and meaning. Curr. Opin. Struct. Biol. 8: 333–337. [DOI] [PubMed] [Google Scholar]
- Kaiser V. B., Bachtrog D., 2010. Evolution of sex chromosomes in insects. Annu. Rev. Genet. 44: 91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D., Zhang Q., Larson D., Shen D., McLellan M., et al. , 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22: 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Schatz M., Walenz B., Martin J., Howard J., et al. , 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30: 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. I., Schein J. E., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn B. T., Page D. C., 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., J. Ruan, et al, 2009. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Roos D., 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G. A. B., Nicolas M., Bergero R., Chambrier P., Kejnovsky E., et al. , 2008. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr. Biol. 18: 545–549. [DOI] [PubMed] [Google Scholar]
- McDaniel S. F., Neubig K. M., Payton A. C., Quatrano R. S., Cove D. J., 2013. Recent capture on the UV sex chromosomes of the moss Ceratodon purpureus. Evolution 67: 2811–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis A., Jacobson D. J., Gustafsson T., Johannesson H., 2008. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 4: e1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A., Chang H., Daugherty L., Fraser M., Hunter S., et al. , 2014. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43: D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T., 1911. Are particular chromosomes sex determinants? Biol. Bull. 19: 1–17. [Google Scholar]
- Nieuwenhuis B. P. S., Billiard S., Vuilleumier S., Petit E., Hood M. E., et al. , 2013. Evolution of uni- and bifactorial sexual compatibility systems in fungi. Heredity 111: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1967. Sex Chromosomes and Sex-Linked Genes. Springer-Verlag, Berlin. [Google Scholar]
- Page D. C., Hughes J. F., Bellott D. W., Mueller J. L., Gill M. E., et al. , 2010. Reconstructing sex chromosome evolution. Genome Biol. 11: I21. [Google Scholar]
- Perlin M., Amselem J., Fontanillas E., Toh S., Chen Z., et al. , 2015. Sex and parasites: genomic and transcriptomic analysis of Microbotryum lychnidis-dioicae, the biotrophic and plant-castrating anther smut fungus. BMC Genomics 16: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit E., Giraud T., de Vienne D. M., Coelho M., Aguileta G., et al. , 2012. Linkage to the mating-type locus across the genus Microbotryum: insights into non-recombining chromosomes. Evolution 66: 3519–3533. [DOI] [PubMed] [Google Scholar]
- Ranwez V., Harispe S., Delsuc F., Douzery E., 2011. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS ONE 6: e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S., van Daalen S. K. M., Brown L. G., Korver C. M., Lange J., et al. , 2006. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 38: 463–467. [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Smit, A. F. A, R. Hubley, and P. Green 2013 RepeatMasker Open-4.0. Available at: http://www.repeatmasker.org.
- Smith D. G., Garcia-Pedrajas M. D., Hong W., Yu Z. Y., Gold S. E., et al. , 2004. An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot. Cell 3: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesler G., 2002. GRIMM: genome rearrangements web server. Bioinformatics 18: 492–493. [DOI] [PubMed] [Google Scholar]
- Treangen T. J., Salzberg S. L., 2012. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercken E., Fontaine M., Gladieux P., Hood M., Jonot O., et al. , 2010. Glacial refugia in pathogens: European genetic structure of anther smut pathogens on Silene latifolia and S. dioica. PLoS Pathog. 6: e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votintseva A. A., Filatov D. A., 2009. Evolutionary strata in a small mating-type-specific region of the smut fungus Microbotryum violaceum. Genetics 182: 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Johannesson H., 2011. Evidence of the accumulation of allele-specific non-synonymous substitutions in the young region of recombination suppression within the mating-type chromosomes of Neurospora tetrasperma. Heredity 107: 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Sun Y., Johannesson H., 2011. Degeneration in codon usage within the region of suppressed recombination in the mating-type chromosomes of Neurospora tetrasperma. Eukaryot. Cell 10: 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Votintseva A., Ridout K., Filatov D. A., 2015. Recent and massive expansion of the mating-type specific region in the smut fungus Microbotryum. Genetics 199: 809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Petit E., Hood M. E., 2015. Variation in mate-recognition pheromones of the fungal genus Microbotryum. Heredity (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamato K. T., Ishizaki K., Fujisawa M., Okada S., Nakayama S., et al. , 2007. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc. Natl. Acad. Sci. USA 104: 6472–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zakharov I. A., 1987. Some principles of the gene localization in eukaryotic chromosomes. Formation of the problem and analysis of nonrandom localization of the mating-type loci in some fungi. Genetika 22: 2620–2624. [PubMed] [Google Scholar]
- Zakharov I. A., 2005. Intratetrad mating and its genetic and evolutionary consequences. Russ. J. Genet. 41: 508–519. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.