Abstract
The transcription factor NFκB is a central regulator of inflammation and genome-wide association studies in subjects with autoimmune disease have identified a number of variants within the NFκB signaling cascade. In addition, causal variant fine-mapping has demonstrated that autoimmune disease susceptibility variants for multiple sclerosis (MS) and ulcerative colitis are strongly enriched within binding sites for NFkB. Here, we report that MS-associated variants proximal to NFκB1 and in an intron of TNFRSF1A (TNFR1) are associated with increased NFκB signaling after TNFα stimulation. Both variants result in increased degradation of IκBα, a negative regulator of NFκB, and nuclear translocation of p65 NFκB. The variant proximal to NFκB1 controls signaling responses by altering expression of NFκB itself, with the GG risk genotype expressing 20-fold more p50 NFκB and diminished expression of the negative regulators of the NFκB pathway TNFAIP3, BCL3, and CIAP1. Finally naïve CD4 T cells from patients with MS express enhanced activation of p65 NFκB. These results demonstrate that genetic variants associated with risk of developing MS alter NFκB signaling pathways, resulting in enhanced NFκB activation and greater responsiveness to inflammatory stimuli. As such, this suggests that rapid genetic screening for variants associated with NFκB signaling may identify individuals amenable to NFκB or cytokine blockade.
Introduction
NFκB was one of the first transcription factors identified and is a central regulator of inflammation (1). The canonical p50/p65 NFκB signaling cascade is critical for activation of immune responses downstream of T and B-cell receptors, toll like receptors, and cytokines, including TNFα and IL-1β. Moreover, alterations in NFκB have been associated with both autoimmune disease and malignancies (2, 3). Inflammatory autoimmune diseases, which reflect complex interactions between genetic variation and environment, are important systems for genetic investigation of human disease. These diseases share a substantial degree of immunopathology, with increased activity of auto-reactive CD4+ T-cells secreting inflammatory cytokines and loss of regulatory T-cell (Treg) function (4–7). Multiple sclerosis (MS) is one such autoimmune disease where there is chronic inflammation in the central nervous system (CNS) with infiltration of activated mononuclear cells into the CNS that damage both myelin and axons. This complex genetic disease is associated with environmental factors that appear to drive a predominantly T cell autoimmune response against CNS antigens (8, 9).
Genome wide association studies (GWAS) and subsequent targeted genomic studies have identified 97 variants associated with MS susceptibility (10–12). While each of these variants contributes only a small increase in the complex phenotype of disease risk, the biologic function associated with individual allelic variants has been striking (13–17). Many of these variants fall within specific signaling cascades, suggesting alterations in pathways, rather than individual genes, may be the key to understanding how individual variants with small odds ratios result in disease susceptibility (18–20). Approximately 17% (17/97) of MS susceptibility variants identified by GWAS fall either within or proximal to NFκB signaling genes, including variants proximal to NFκB1 itself and within TNFR1 (10, 12, 21).
We recently integrated genetic and epigenetic fine-mapping to identify potentially causal variants in autoimmune disease-associated loci and explore their functions by generating cis-regulatory element maps for a spectrum of immune cell types. Approximately 60% of likely causal variants map to enhancer-like elements, with preferential enrichment in stimulus-dependent CD4+ T-cell enhancers. When overlapping causal SNPs with 31 TF binding maps generated by ENCODE, SNPs were strongly enriched within binding sites for immune-related TFs and variants associated with different diseases correlate to different combinations of TFs that control immune cell identity and response to stimulation. In patients with MS, SNPs preferentially coincide with NFκB, EBF1 and MEF2A-bound regions (22).
Previous studies have shown that total PBMCs from relapsing remitting MS (RRMS) patients exhibit increased levels of active NFκB (23). We similarly observe that naïve CD4 T cells from RRMS patients exhibit increased activation of the canonical p65 NFκB pathway compared to healthy controls, suggesting that this difference is not due to the activation status of the cells. Thus, we hypothesized that the alterations in NFκB signaling seen in patients with MS are a result of SNPs in the NFκB signaling cascade associated with MS susceptibility. As NFκB signaling is triggered by many environmental stimuli through toll-like receptors, this may represent a critical intersection between genetic and environmental factors resulting in MS development. However, the impact of autoimmunity-associated genetic variants on the biologic function of this major transcription factor is unknown.
It has been shown that mice with a constitutively active form of p65 NFκB exhibit multi-organ inflammation and die within three weeks of birth. However, when these mice with constitutively active p65 NFκB were bred back to mice lacking the Type I TNF receptor (TNFR1), they were protected from inflammation and exhibited only a mild Sjogren’s like ocular keratitis, indicating a critical role of the NFκB/TNF pathway in regulating autoimmune disease (24). Variants within or proximal to both TNFRSF1A (TNFR1) and NFκB1 have been identified by GWAS as being associated with risk of developing MS. Therefore, we chose to focus our studies on these SNPs to determine whether they alter NFκB responses.
Here, we investigated the effect of MS risk haplotypes on the function of the NFκB signaling cascade. We demonstrate that both the haplotype with the variant proximal to NFκB1 (rs228614-G) and within TNFR1 (rs1800693-C) result in enhanced NFκB responses to TNFα. The rs228614-G risk allele also enhances responses to PMA, suggesting that this variant strongly controls NFκB signaling. Mechanistically, we find that rs228614-G is associated with a 20-fold increase in p50 NFκB expression and decreased negative regulators of NFκB signaling. These findings elucidate a genetic mechanism controlling NFκB responses that results in predisposition to developing MS.
Results
Patients with MS have increased activation of the canonical NFκB cascade
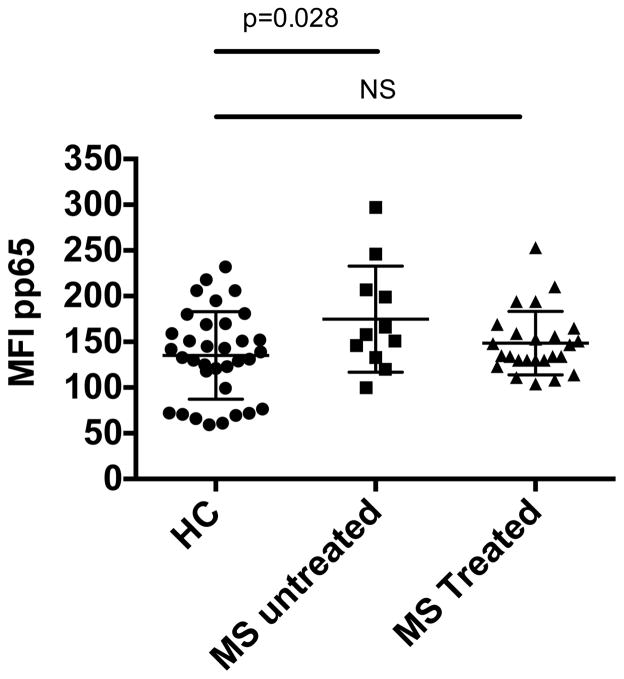
Previous reports suggest that total PBMCs from patients with MS exhibit increased activity of NFκB, consistent with excessive immune activation (23, 25). However, it was not possible to determine if this increased NFκB activation is due to upregulation of NFκB or to the increased frequency of activated and memory CD4 T cells in patients with MS. To resolve this question, we isolated total PBMCs from patients with MS and healthy control subjects ex vivo and directly stained for phospho-p65 NFκB. We found that naïve CD4 cells from patients with MS exhibit significantly higher phospho-p65 NFκB than those from age-matched healthy control donors and this increased activation of p65 NFκB was mitigated by treatment (Figure 1, Subject demographics listed in supplemental table 1). This increased constitutive expression of phospho-p65 NFκB was repeated in a second cohort of MS patients and healthy controls (Supplemental figure 1). The presence of enhanced activation of NFκB in naïve CD4 cells demonstrates that this is not due to an increase in the number of activated or memory cells, but rather a hyper-activated state of CD4 cells.
Figure 1. Naïve CD4 cells from patients with MS exhibit increased phospho-p65 NFκB.
Flow cytometry of PBMCs from age-matched healthy control (HC) and relapsing-remitting MS (RRMS) patients stained for CD4, CD45RA, CD45RO, and pS529 p65 NFκB. MFI of p65 results are shown gated on naïve CD4+CD45RA+CD45RO− T-cells. Healthy control n= 34, untreated MS n=11, MS treated n=25. (p-value, unpaired t-test)
The MS risk variant rs228614 near NFκB1 is associated with increased NFκB signaling
To determine whether the increased NFκB activation seen in patients with MS may be due to genetic variation in the NFκB signaling cascade associated with disease susceptibility, we next assessed whether the MS risk variant proximal to NFκB1 increases NFκB signaling. The SNP rs228614 on chromosome 4 is associated to MS susceptibility (OR 1.09 per G allele carried, p=1 × 10−8) and lies near the NFκB1 gene (12). It is part of a haplotype of over 90 variants in tight linkage disequilibrium that span the region encoding both NFκB1 and MANBA (Figure 2a). While this strong LD precludes identifying this specific SNP as the causal variant for disease, we stratified donors into those carrying one or two copies of the G risk allele (rs228614-G) and correlated NFκB signaling to genotype in total PBMCs. In addition, we also investigated a second variant, rs7665090, in linkage disequilibrium to rs228614 (r2>0.8) that was identified by fine-mapping as the most associated variant in the region (10). This allowed us to examine two variants within the same region to determine the impact on NFκB signaling, and thus address the difficulty of identifying the causal variant in genetic mapping studies. We used healthy control subjects carrying the risk or protective variants to avoid the confounding factors of on-going inflammation and therapeutic intervention seen in patients with MS.
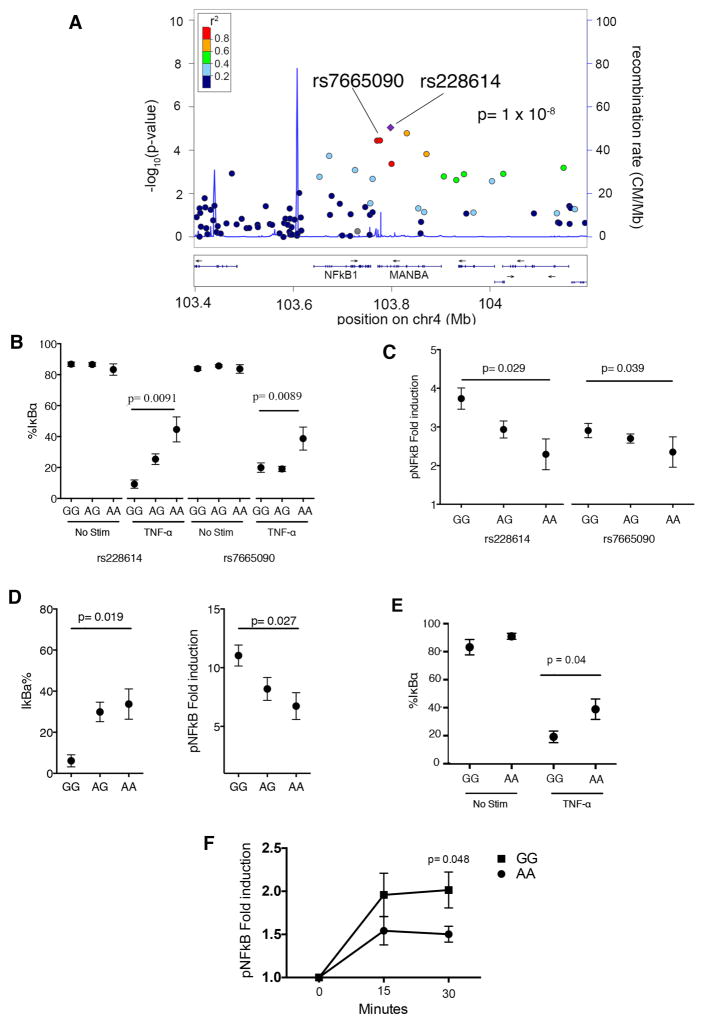
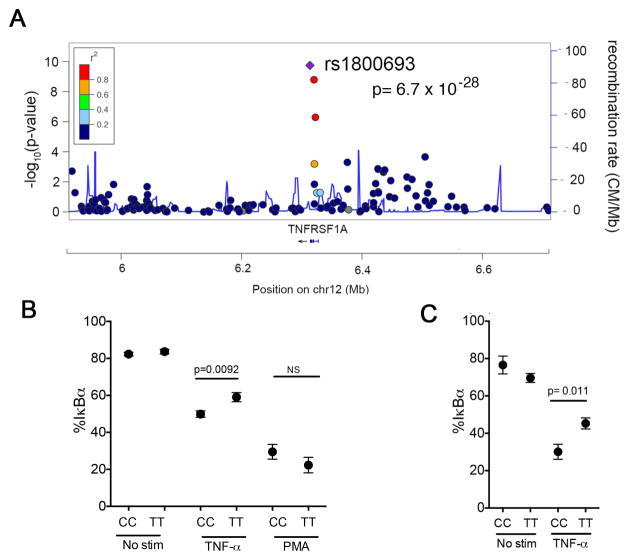
Figure 2. MS-associated allelic variants proximal to NFκB1 results in increased TNFα and PMA signaling.
A) Association to MS risk in the region surrounding NFκB. Y axis shows the GWAS –log(p value) for the allelic test of association as reported in (12). We have highlighted rs228614 and rs7665090, which are the most associated variants in the region. B) Degradation of IκBα in CD4+CD45RA+CD45RO− T cells after 15 minute TNFα stimulation by flow cytometry in healthy controls. C) phospho-p65 NFκB in CD4+CD45RA+CD45RO− naïve T cells after 15 minute TNFα stimulation by flow cytometry. Results shown normalized to unstimulated T cells. D) rs228614 CD4+CD45RA+CD45RO− T cell degradation of IκBα and phosphorylation of p65 NFκB after 15 minutes of PMA stimulation. E and F) Degradation of IκBα after 15 minutes (E) and phosphorylation of p65 NFκB at 15 and 30 minutes (F), in CD4+CD45RA+CD45RO− T cells after TNFα stimulation by flow cytometry in MS patients. For A–D, rs228614 GG n=4, AG n=18, AA n=6: rs7665090, AA n=8, AG n=49, GG n=23. For E and F, GG n=6, AA n=6.(p value shown for homozygous unpaired t-test)
We collected blood samples from healthy donors in our previously genotyped biorepository of recallable subjects (Yale Phenogenetic Project) (Demographics, supplemental table 2, 3). We found that the MS risk alleles of rs228614-G and rs7665090-G are significantly associated with an increase in IκBα degradation after TNFα stimulation (allelic logistic regression: rs228614 p = 0.00078; rs7665090 p = 0.01, Figure 2b). This difference is also significant when comparing homozygous risk and protective genotypes (Student’s t test between GG and AA carriers, rs228614 p = 0.0091; rs7665090 p=0.0089 Figure 2b). Risk allele homozygotes showed consistently increased phosphorylation of p65 NFκB (rs228614 Student’s t test between GG and AA carriers p = 0.029; rs7665090 p = 0.039 Figure 2c, representative histogram and gating strategy, supplemental figure 2). The trend to increased signaling from both rs228614 and rs7665090 was also observed at 15, 30, and 45 minutes (supplemental figure 3). Thus the region containing the rs228614 and rs7665090 SNPs modulates TNF-α signaling through NFκB.
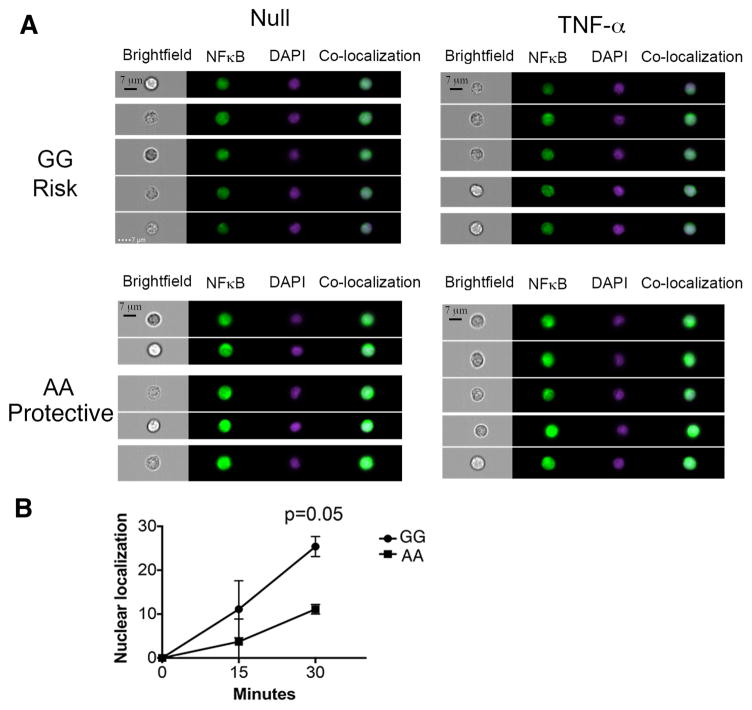
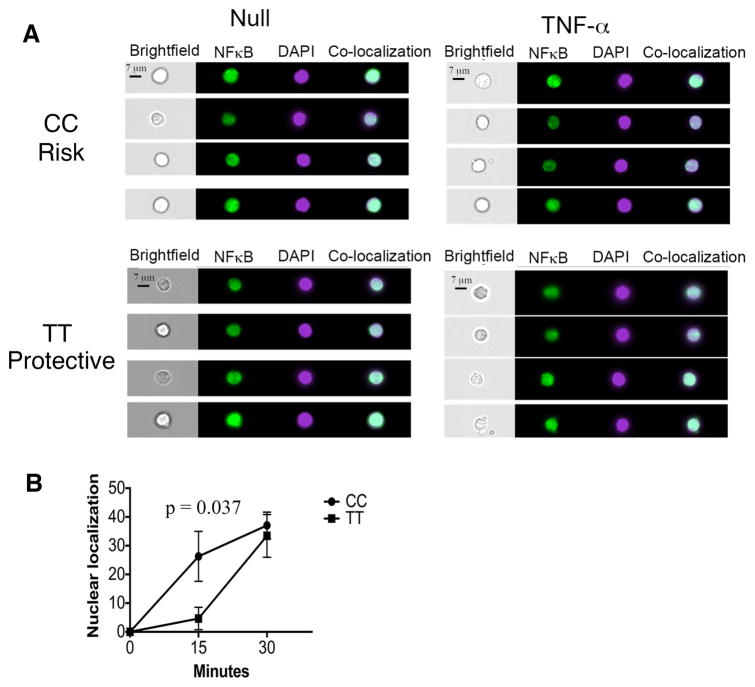
To determine if this represents a global impact on NFκB signaling or a specific alteration in TNF-α signaling, we stimulated total PBMCs from subjects with the different genotypes at rs228614 with Phorbol-12 myristate 13-acetate (PMA), an activator of NFκB independent of TNF-α. The risk genotype (GG) results in greater degradation of IκBα and phosphorylation of p65 NFκB in naïve CD4 cells after stimulation with PMA, suggesting a modulation of global NFκB responses rather than to a specific stimulus (Figure 2d, IκBα p = 0.019, pNFκB p = 0.027). To determine if this region was also impacting NFκB signaling in MS patients, we stimulated total PBMCs from rs7665090 homozygous risk (GG) and protective (AA) subjects with TNF-α and determined degradation of IκBα and phosphorylation of p65 NFκB by flow cytometry. Similar to the changes in NFκB signaling seen in healthy controls, the risk variant also resulted in increased IκBα degradation (Figure 2e) and phosphorylation of p65 NFκB (Figure 2f) in naïve CD4 cells in MS patients. We hypothesized that these divergent NFκB responses should result in differential rates of nuclear localization of p65 NFκB. We tested this by comparing healthy donors with AA (protective) and GG (risk) genotype at rs228614. We stimulated CD4 cells with TNFα for 15 or 30 minutes, stained for p65 NFκB and DAPI, and determined nuclear localization of p65. We find that GG carriers demonstrate increased p65 nuclear localization after 30 minutes, confirming stronger signaling responses (Figure 3a,b). Taken together, these data demonstrate that the MS risk haplotype captured by rs228614 and rs7665090 strongly modulates NFκB responses, with the GG risk genotype resulting in enhanced NFκB signaling.
Figure 3. The rs228614 GG risk genotype results in rapid nuclear localization of p65 NFκB.
A) Representative Imagestream data showing nuclear localization of p65 NFκB after 30 minute TNFα stimulation by Amnis Imagestreamx in CD4+ T cells. Green, p65 NFκB. Purple, DAPI. co-localization, overlap of p65 and DAPI by Pearson co-efficient. B) Compilation of rs228614 Nuclear localization of p65 NFκB after TNFα stimulation in CD4+ T cells(GG, n=5, AA n=4). Nuclear localization is shown normalized to unstimulated cells. (p-value shown for unpaired t test).
rs228614 is associated with changes in NFκB1 expression and negative regulators of the NFκB pathway
We did not find differences in IκBα and phospho-p65 NFκB expression in resting CD4 cells, suggesting that the effect of the rs228614 risk variant is only evident after signaling induction (Suppl. Figure 4). This suggests that the underlying causal variant perturbs regulatory elements controlling stimulus-dependent activation of the NFκB gene, consistent with the observation that most GWAS risk variants alter gene regulation rather than structure (26). The haplotype block containing rs228614 spans the coding regions of NFκB1 and MANBA and the intergenic space between them, which contains at least 62 transcription factor binding sites, 12 regions of DNA hypersensitivity, three insulator regions and multiple putative enhancer regions active in both T cell and myeloid cell lineages (27–33) suggestive of important regulatory functions. We therefore looked for changes in expression of p50 NFκB in rs228614 GG and AA genotype carriers by western blot.
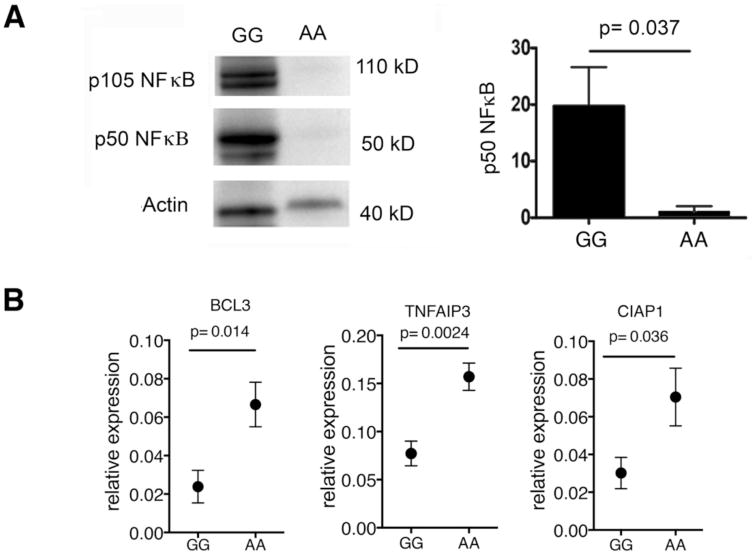
Remarkably, we found that subjects with the GG risk genotype express p50 NFκB at a 20-fold higher level than the AA genotype (Figure 4a, uncut blot supplemental figure 5). We also found a commensurate decrease of mRNA levels in total PBMCs of three key negative regulators of NFκB (TNFAIP3, BCL3, and CIAP1) in the GG homozygotes (Fig 4b). Taken together, these results suggest that an increase in total p50 NFκB and a loss of negative regulation of this pathway is responsible for enhanced signaling. While IκBα degradation is upstream of NFκB, signaling through NFκB regulates expression of NFκB1 itself, as well as the negative regulators of NFκB. Thus, we demonstrate alterations in both NFκB1 expression and the negative regulators of this pathway, suggesting that the MS risk allele causes a fundamental shift in the regulation of NFκB signaling.
Figure 4. rs228614 allelic variant results in increased NFκB1 expression and decreased negative regulators of NFκB.
A) Representative western blot of p105 and p50 NFκB in total PBMCs and densitometry of total p50 NFκB by western (GG n=7; AA, n=7). B) mRNA expression by Q-PCR of BCL3, TNFAIP3, and CIAP-1 in total PBMCs (GG n=6; AA n=7) by q-PCR. (p-value shown for unpaired t test)
NFκB responses are stable over time and are not associated with age or gender
If the strength of NFκB signaling is primarily mediated by genetic variability between individuals and not changes in environmental or external stimuli, we would expect consistent strength of signal across multiple time points. We redrew 15 subjects on average six months after the first blood draw. We found no statistically significant difference between draws (Suppl. figure 6). While there is significant variability between individuals in the strength of NFκB signaling, the stability in this variability across multiple draws is consistent with a genetically-mediated threshold for NFκB signaling.
It has been shown that individuals over 65 years old have an increase in constitutive activation of p65 NFκB (34). The phenogenetic project enrolled subjects up to age 56 at the time of enrollment and subjects were consented for redraws for up to 4 years. To determine if NFκB signaling changes with age, we compared age at blood draw to NFκB signaling after TNF-α stimulation. We found no correlation between age and strength of signal by either IκBα degradation or phosphorylation of p65 prior to age 60 (supplemental figure 7a,b). This suggests that changes to NFκB signaling associated with aging only occur after 60 years of age. We also found no differences in gender or ethnicity in NFκB signaling after TNF-α stimulation (supplemental figure 7c–f).
rs1800693 in TNFR1 is associated with enhanced NFκB responses to TNFα
As both NFκB and TNFR1 are important in autoimmune inflammation, and variants near both NFκB1 (rs266814) and TNFRSF1A (rs1800693) are associated with risk of developing MS (12), we investigated the impact of the TNFRSF1A variant on NFκB signaling. Unlike the NFκB1 locus where tight linkage disequilibrium precludes identifying the likely causal SNP, rs1800693 alone best explains the association signal in the TNFRSF1A region and is thus likely causal (10). The variant falls 10 base pairs upstream of TNFRSF1A exon 6 in a splice acceptor site (Figure 5a). We and others have previously shown that homozygous carriers of the rs1800693-C risk allele show loss of exon 6 and a premature stop codon in ~10% of TNFR1 mRNAs (35, 36). Moreover, this variant results in an exaggerated cytokine production response from monocytes after stimulation with TNF-α (35) suggesting this transcriptional change results in changes to overall signaling responses. We evaluated this hypothesis by stimulating PBMCs and purified monocytes with TNF-α and assessing resultant IκBα degradation (Demographics, supplemental Table 4. We found that carriers of the CC risk genotype show increased degradation of IκBα after stimulation with TNF-α in both naïve CD4 cells (p = 0.0092, figure 5b) and monocytes (p = 0.011, figure 5c), compared to TT genotype carriers. Unlike the variant proximal to NFκB1, there were no differences in signaling after PMA stimulation between the CC and TT genotypes (figure 5b). These data suggests that rs1800693 is associated with changes in signaling only when TNF-α binds to TNFR1.
Figure 5. The TNFR1 variant rs1800693 results in increased TNFα responses.
A) Association to MS risk in the region surrounding TNFRSF1A. Y axis shows the GWAS –log(p value) for the allelic test of association as reported in (10). We have highlighted rs1800693, the most associated variant in the region. With further replication data in independent samples, rs228614 meets the GWAS significance threshold of p < 5 × 10−8. B) Degradation of IκBα after 30 minutes TNFα stimulation in CD4+CD45RA+CD45RO− T cells. (CC n=14; TT n=20). C) Degradation of IκBα after 30 minutes TNFα stimulation in CD14+ monocytes. (CC n=5; TT n=12). (p-value shown for unpaired t test).
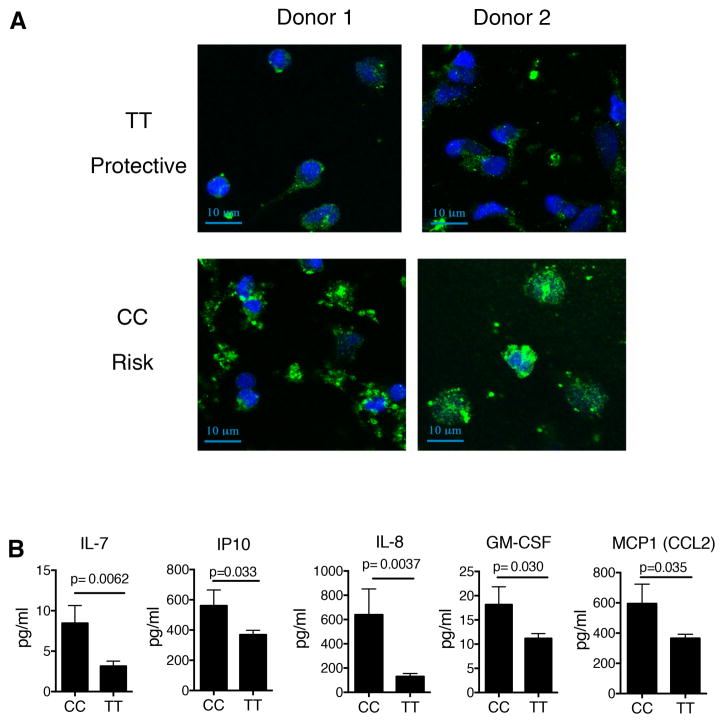
TNFα can signal through two receptors, TNFR1 and TNFR2. Naïve CD4 cells primarily express TNFR1 with TNFR2 being upregulated after activation (37). Our results suggest that altered TNFα-induced signaling is due to rs1800693-C exerting regulatory effects on TNFR1. To validate this model, we blocked TNFR1 and found that TNFα signaling in naïve CD4 cells occurs exclusively through TNFR1 with no contribution of TNFR2 (Supplemental. figure 8). As such, the changes in signaling observed in naïve CD4 cells can be attributed directly to alterations in TNFR1. To confirm these changes in NFκB signaling in CD4 cells, we purified total CD4 cells, stimulated them with TNFα for 15 or 30 minutes, and determined the amount of p65 NFκB nuclear localization. We observed an increase in nuclear localization 15 minutes after stimulation in carriers of the CC genotype compared to the TT genotype (Figure 6a,b). The CC genotype did not result in changes in the cell surface expression of TNFR1 on naïve CD4 cells, either as a percentage of cells expressing TNFR1, or the expression level of TNFR1 on the surface (Suppl Figure 9). This suggests that the change in signaling is not due to changes in overall TNFR1 levels on the cell surface. We and others have shown that expression of the prematurely terminated transcript missing exon 6 caused by the rs1800693-C allele results in altered intracellular accumulation of TNFR1 in HEK293 or HELA cells (35, 36). We investigated if this is true in primary immune cells directly ex vivo and found that CD14+ monocytes from CC risk genotype carriers exhibit altered intracellular accumulation of TNFR1 into punctate structures within the cytoplasm (Figure 7a, single stained control, supplemental figure 10). Taken together, these results demonstrate that the variant in TNFR1 results in increased signaling to TNFα in both naïve CD4 cells and monocytes and that this altered signaling may be due to altered localization of TNFR1 within the cell.
Figure 6. TNFR1 rs1800693 CC risk genotype results in increased NFκB nuclear localization.
A) Representative nuclear localization images from TT protective and CC risk genotypes in CD4+ T cells by Amnis Imagstreamx. Green, p65 NFκB. Purple, DAPI. co-localization, overlap of p65 and DAPI by Pearson co-efficient. B) Composite nuclear localization of p65 NFκB after TNFα stimulation in CD4+ T cells (CC n=9; TT n=9). Nuclear localization is shown normalized to unstimulated cells. (p-value shown for unpaired t test).
Figure 7. The TNFR1 variant rs1800693 results in altered intracellular accumulation of TNFR1 and plasma cytokines.
A) Confocal microscopy of CD14+ monocytes from the TT (protective) or CC (risk) variant in the TNFR1 region. (blue, DAPI; Green TNFR1). Two of four subjects from each genotype shown. B) Concentration of IL-7, IL-8, GM-CSF, MCP1 and IP10 in plasma samples from healthy control subjects with the CC (risk) or TT (protective) genotypes. (CC n=25; TT n=40). (p value shown for unpaired t test).
rs1800693 in TNFR1 is associated with increased plasma cytokines in healthy controls consistent with levels seen in MS patients
As TNFα signaling drives the expression of inflammatory cytokines, we examined whether changes to TNFR1-mediated signaling associated with the rs1800693-C risk allele results in altered plasma cytokines levels. We measured plasma levels for 29 cytokines in healthy subjects, and found that CC genotype carriers have increased plasma levels of IL-7, IL-8, GM-CSF, IP10, and MCP1 (Figure 7b and supplemental table 5). This is consistent with our previous findings that IP10 is overexpressed in monocytes after TNFα stimulation in CC genotype carriers (35). It has been reported that IL-7, GM-CSF, IP10, IL-8, and MCP1 are increased in serum of patients with MS (38–42). In addition, recent twin studies have shown that serum concentrations of all five of these cytokines are highly heritable (43). This demonstrates that the CC homozygous risk genotype alters levels of inflammation in healthy control subjects consistent with that seen in MS.
Discussion
NFκB is a central regulator of inflammation, controlling the activation, proliferation, and cytokine production of immune responses. We observed that naïve CD4 T cells from patients with the autoimmune disease MS exhibit increased activation of p65 NFκB, prompting us to investigate the genetic control of NFκB signaling in the disease. We found that variants near genes in the NFκB signaling cascade have large effects on NFκB signaling. Specifically, the allelic variant proximal to NFκB1 controls signaling responses by altering expression of NFκB itself with the GG risk genotype expressing 20-fold more p50 NFκB. This genotype is associated with altered NFκB responses both to TNFα and PMA, suggesting a global control of NFκB signaling. As causal SNPs are strongly enriched within binding sites and signaling molecules for NFkB, these results demonstrate a central role for NFκB in driving the pathophysiology of MS.
The region spanned by the MS risk haplotype near NFκB1 contains a large number of gene regulatory elements, as mapped by the ENCODE project (27–33). This suggested the hypothesis that the MS risk allele affects NFκB1 expression and thus alters NFκB signaling. To investigate this potential mechanism, we examined how allelic variation in this region influenced expression of p50 NFκB and found that MS risk variants are strongly associated with large increases in expression. Consistent with increased NFκB signaling, there was a decrease in expression of the negative regulators of NFκB, suggesting a global disruption in the NFκB cascade. The central role of NFκB in immune activation would suggest that this MS haplotype broadly increases inflammatory responses thereby predisposing individuals to autoimmunity.
We recently investigated the overlap of potentially causative SNPs with 31 transcription factor binding maps generated by ENCODE and observed they were strongly enriched within binding sites for immune-related transcription factors (22). Moreover, variants associated with different autoimmune diseases correlate with different combinations of transcription factors that control immune cell identity and response to stimulation. We found that MS SNPs preferentially coincide with NFkB, EBF1 and MEF2A-bound regions, whereas rheumatoid arthritis and celiac disease SNPs preferentially coincide with IRF4 regions (22). Thus, both GWAS and epigenetic mapping strongly implicates NFκB as a critical pathway associated with MS. That is, variants associated with MS tend to fall in or near genes involved in NFκB signaling and secondly, variants associated with MS frequently fall near known NFκB response elements. We propose a model in which variants associated with NFκB signaling and binding interact to form a cumulative burden on this critical pathway. The enhanced NFκB activity we observe in naïve CD4 cells suggests that these cells may have a decreased threshold of activation, thereby making them more susceptible to low levels of stimuli and ultimately autoimmunity.
We also established that an MS risk variant intronic to TNFRSF1A (TNFR1) is associated with increased NFκB signaling in response to TNFα. We confirmed previous reports that this risk variant results in an altered intracellular accumulation of TNFR1, and extended these findings to show that monocytes isolated from peripheral blood show this altered phenotype (35, 36). Of note, an R92Q mutation in TNFR1 is associated with TNF receptor associated periodic syndrome (TRAPS), a relapsing remitting peripheral inflammatory disorder. The R92Q mutation also results in an altered intracellular accumulation of TNFR1 and increased responses to inflammatory stimuli (44). This may suggest a common mechanism for altered NFκB mediated signaling between MS and TRAPS.
We have previously reported that CD4 cells from MS patients exhibit increased proliferation at low doses of anti-CD3 stimulation compared to healthy controls (45). These data suggest the presence of a decreased threshold for activation of CD4 cells in patients with MS, consistent with genetic risk variants perturbing NFκB pathway genes to create a net increase in signaling through this inflammatory cascade.
Treatments such as steroids that suppress NFκB signaling are routinely used in MS, though their prolonged use is ineffective, perhaps due to the many mechanisms associated with chronic steroid usage. While these studies might support the use of NFκB inhibitors in MS, they also suggest that it may be possible to determine the specific stimuli and downstream signaling cascades that may identify novel disease treatments. Of note, TNFα blockade by monoclonal antibodies or soluble receptors is highly effective in many autoimmune diseases. In contrast, TNFα blockade in patients with MS both worsens disease while inducing new onset MS in some patients with rheumatoid arthritis, Crohn’s disease, and ulcerative colitis (46). Moreover, TNFα blockade can also initiate psoriasis and induces the development of anti-nuclear or dsDNA antibodies in up to 70% of other autoimmune patients, with some progressing to anti-Rho/La antibodies and clinical lupus (46, 47). Blocking TNFα in patients with mutations in TNFR1 associated with TRAPS also results in increased NFκB signaling and inflammation similar to that seen in MS (48). These data emphasize the clinical importance in understanding the NFκB pathway in MS and other autoimmune diseases and implicates genetic variants in the TNF signaling pathway as controlling the altered responses to anti-TNFα therapy.
The haplotype block surrounding the intergenic region downstream of NFκB1 contains over 90 variants in strong LD and we were unable to identify a single most-likely causal variant by genetic and epigenetic fine mapping. In the future, the use of probabilistic identification of causative SNPs in larger cohorts may allow further fine-mapping of this region that may better identify the most-likely causative SNP (22). In spite of this lack of resolution in the discovery process, we are able to show that the underlying MS variant in this region tagged by the rs228614-G allele, is strongly associated to NFκB responses to TNFα.
We demonstrated significant inter-individual variation exists in NFκB responses and this variability is partially mediated by genetic variability in the NFκB signaling cascade. As many autoimmune diseases and cancers have genetic variants associated with NFκB, it is likely that genetically mediated variability in this pathway is central to disease risk. In addition, as the variants associated with each disorder are different, variation in the NFκB pathway may result in changes in signaling that are specific to each disease. Finally, the increased NFκB signaling we observed is consistent with the enhanced activation of the canonical NFκB pathway observed in naïve CD4 cells from patients with MS. Hyper-activation of NFκB in mice results in rapid post-natal death from massive, multi-organ inflammation not seen in human disease (24, 49). As such, it is likely that dysregulation of NFκB represents a necessary factor in the development of autoimmunity, but that other factors are also required to determine the location and nature of the inflammatory insult. Current GWAS studies have identified variants associated with disease susceptibility, but do not determine how these variants are impacting disease severity or progression. Given that NFκB signaling is critical for both inflammation and neuronal degeneration (50), this pathway may represent a node that both predisposes to disease and contributes to disease progression.
In conclusion, naïve CD4 T cells from patients with MS exhibit increased p65 activation, consistent with changes to NFκB signaling. We observe equivalent changes in healthy controls carrying MS risk variants, which alter NFκB responses to TNFα, suggesting disease risk is mediated by gene regulatory changes resulting in altered NFκB signaling. As SNPs causal for MS are enriched in binding sites and signaling molecules for NFκB, these data demonstrate a central role for NFκB in driving the pathophysiology of MS. Identifying these critical nodes may suggest novel therapeutics aimed at specifically targeting the underlying causes of disease while leaving systemic immune responses primarily intact.
Materials and Methods
Study Design
Healthy subjects between the age of 18 and 56 y.o. at the time of initial assessment were enrolled in the Yale phenogenetic project and could be recalled up to 6 times a year for up to 4 years. Subjects with autoimmune disorders were excluded from the repository. In addition, any subjects exhibiting illness or fever at the time of draw were excluded. Investigation of the impact of rs228614 and rs7665090 on phospho p65-NFκB and total IκBα was performed blinded with the strength of signal determined prior to genotyping the samples. Samples were un-blinded and comparison of strength of signal to genotype performed after un-blinding. For blinded studies, power calculations determined that a singleton quantitative trait locus, with a variance of 10% and an allele frequency of 20%, could generate significance of 0.05 for overall association with 50 samples and 0.01 with 90 samples. As rs228614, rs7665090, and rs1800693 all have minor allele frequencies of >30%, these studies were sufficiently powered to generate significance. Nuclear localization, western blotting, and Q-PCR studies were performed on recalled samples with specific genotypes. All TNFRSF1A studies were performed on samples of risk and protective genotypes recalled from the Brigham and Women’s Hospital (BWH) PhenoGenetic Project. All studies were performed to at least 3 biological replicates.
Blood Repositories
Samples were acquired from the Yale phenogenetic project or the Brigham and Women’s Hospital (BWH) PhenoGenetic Project. These samples have been genotyped for autoimmunity associated SNPs by Illumina Chip. Blood was drawn into Heparin tubes. Samples from Yale were prepared immediately after blood draw. TNFR1 samples from BWH were shipped overnight with room temperature gel packs (SAF-T-PAK) and prepared the following morning. RRMS from age and gender matched blood was drawn at the Yale MS Clinic and prepared the same as the healthy control samples. All samples were compared to samples from the same source (Yale or Brigham and Women’s Hospital).
PBMC preparation and Cell purification
PBMCs were prepared from whole blood by Ficoll-Hypaque density gradient centrifugation. For some experiments, magnetic negative selection for total CD4 Cells or CD14+ monocytes (containing both CD16+ and CD16−) cells were purified using Easysep magnetic separation kits (Stem Cell Technologies)
Cytokine stimulation
Total PBMCs were rested for 1 hour in RPMI media without FBS after isolation. Monocytes were plated overnight in RPMI media containing 5% FBS, L-glutamine, Non-essential amino acids, Sodium Pyruvate, and HEPES. Monocytes were washed 2x with RPMI and rested for 1 hour with RPMI without FBS prior to stimulation. Stimulation was performed with 50 ng/ml TNFα (R&D Systems) or phorbol 12-myristate 13-acetate (PMA, Sigma Aldrich, 500 ng/ml). Cells were fixed at 15, 30, or 45 minutes with BD Fixation Buffer (BD Biosciences), washed 2x with PBS, and permeabilized with ice-cold BD Perm Buffer III (BD Biosciences). Cells were permeabilized overnight at −80°C. After washing, the cells were stained for CD4 PE, CD45RA AF700, CD45RO PE-Cy7, phospho p65 NFκB (pS529) (BD Biosciences), and IκBα A488 (Cell Signaling Technologies). Flow cytometry was performed on a BD LSRII Fortessa (BD Biosciences). Analysis was performed using Flowjo software (TreeStar).
Nuclear Localization
Total CD4 cells were stimulated with 50 ng/ml TNFα (R&D Systems) for 15 or 30 minutes. Cells were fixed with 3% Formalin for 10 minutes, washed, and permeabilized with 0.1% Triton X-100 + 2% FBS in PBS (no Ca/mg). Cells were blocked with Fc Block (E-bioscience). p65 NFκB was stained with anti p65 NFκB (Santa Cruz) and FITC labeled donkey Fab2 anti-rabbit IgG (Jackson, West Grove, PA). Cells were stained with DAPI (4′,6-diamidino-2-phenylindole, Sigma Aldrich) nuclear stain for 10 minutes and washed twice with PBS. Nuclear localization was performed on an amnis Imagestreamx or Amnis Imagestreamx Mark 2 at 40x or 60x magnification. Nuclear localization was determined using Amnis IDEAS software (Amnis) by Pearson co-efficient co-localization of DAPI and p65 NFκB.
Western Blot
Total PBMCs were Lysed with RIPA buffer (Pierce) containing HALT protease inhibitors (Pierce). Total protein was determined by BCA assay (Pierce) and 10 ug total protein run/lane. Samples were prepared with Loading buffer (Life technologies), and heated for 10 minutes at 72°C prior to running on Western. Samples were run on a 10% Bis-Tris gel and transferred to nitrocellulose by XCELL western blot transfer apparatus (Life Technologies). Blots were blocked with 1X TBS, 0.1% Tween-20 with 5% w/v nonfat dry milk overnight. P105/p50 NFκB and actin (Cell Signaling Technology) were stained overnight, washed 3 times with 1x TBS and 0.1% Tween-20, and labeled with HRP linked anti-mouse IgG (Cell Signaling Technologies). Blots were developed using ECL Prime (GE Healthcare). Densitometry was performed using ImageJ analysis software (NIH)
Confocal microscopy
200,000 monocytes were plated overnight in 24 well plates with poly-l-lysine coated coverslips. Cells were incubated O/N, fixed with 3% Formalin, and permeabilized with 0.1% Triton X-100 + 2% FBS in PBS (no Ca/mg). Cells were blocked with Fc Block (e-bioscience) and a cocktail of 1% FBS, 1% donkey serum, and 1% human serum. TNFR1 was stained with polyclonal TNFR1 antibody (R&D Systems), labeled with FITC labeled anti Rabbit IgG (Jackson). Images were captured on a Leica microscope (Leica Biosystems) and analyzed using ImageJ software (NIH).
Luminex
Plasma was prepared by centrifuging total blood at 5000 rpm for 10 minutes and removing the plasma fraction from the red cell fraction. Plasma was immediately frozen at −80°C until run by Luminex. Samples were run by 30-plex luminex (Millipore).
Quantitative PCR
RNA was extracted from whole PBMCs using Qiagen RNeasy Plus Micro Kit, according to the manufacturer’s instructions. Sample concentration and purity were determined by spectrophotometer analysis on NanoDrop 2000. cDNA was prepared from RNA through RT-PCR with RT kits from Life Technologies. QPCR was performed using primers for TNFAIP3 (hs00234713_m1), BCL3 (hs00180403_m1), and cIAP1 (BIRC2, Hs01112284_m1) and the housekeeping genes HPRT (hs01003267_m1) and β2M (H500984230_m1) and Taqman Fast Universal PCR Master Mix (No AmpErase UNG) (Life Technologies). Samples were run on an ABI Prism quantitative PCR machine (Life technologies).
Statistics
We calculated genotype-phenotype associations in two ways: an allelic test as implemented in PLINK (51), and comparing opposite homozygote groups by unpaired T test, as implemented in PRISM (Graphpad). In both cases, we report uncorrected p values.
Study approval
The study was conducted in compliance with the Declaration of Helsinki. Before study initiation, approval was obtained from the ethics committee of Yale-New Haven Hospital (New Haven, Connecticut, USA). Informed consent was received from all subjects prior to inclusion in the study.
Supplementary Material
Supplementary Figure 1. Confirmation of increased pNFκB in multiple sclerosis patients.
Supplemental Figure 2: Gating strategy and representative histogram for rs228614 GG (risk) and AA (protective) variants.
Supplemental Figure 3: Rs228614 and rs7665090 proximal to NFκB1 result in increased IκBα degradation after TNFα treatment.
Supplemental Figure 4: Constitutive total IκBα and phospho-p65 NFκB are similar in rs228614 risk and protective variants.
Supplemental Figure 5: Full uncut gel for figure 4
Supplemental Figure 6: TNFα and PMA responses are stable across multiple draws.
Supplemental Figure 7: Responses to TNFα do not change with age, gender, or ethnicity.
Supplemental Figure 8: TNFα signals through TNFR1 on naïve CD4 cells.
Supplemental Figure 9: The rs1800693 CC variant does not change the surface expression of TNFR1.
Supplemental Figure 10: Representative single-stained control.
Supplemental Table 1. Demographics of healthy subjects and MS samples for figure 1 and suppl. figure 1.
Supplemental Table 2. Demographics of rs228614 results from figure 2.
Supplemental Table 3. Demographics of rs7665090 from figure 2.
Supplemental Table 4. Demographics of rs1800693 from figure 5.
Supplemental Table 5. Luminex data from rs1800693 subjects.
Supplemental Table 6. Raw Data
Acknowledgments
We thank Stephanie Eisenbarth, MD, PhD, Jonathan Cruz, and Arpita Singh, PhD for their assistance with sample acquisition. We thank Ewa Menet and Lesley Devine for their assistance with cell sorting and Amnis imaging.
Funding: This research was supported by grants from the National Institute of Allergy and Infectious Disease (AI045757, AI046130, AI070352, AI039671), the National Institute of Neurological Disorders and Stroke (NS24247, NS067305), the National Institute of General Medical Sciences (GM093080), the National Multiple Sclerosis Society (CA1061-A-18), the Penates Foundation, and the Nancy Taylor Foundation for chronic disease. WJH was supported by a training grant from the NIH (to David Schatz, 5T32AI007019-36) and is the Pfizer Fellow of the Life Sciences Research Foundation (to WJH).
Footnotes
Author Contribution: WJH designed, performed, and analyzed experiments and wrote the manuscript. SDF, KV, and SRM performed experiments, NC, LG, and PLD assisted in sample acquisition. MM and CC analyzed data. JG contributed to the confocal microscopy experiments. DAH oversaw the experiments and wrote the paper. CC performed statistical analyses.
Competing interests: The authors declare no competing interests.
References
- 1.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes & development. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng J, Montecalvo A, Kane LP. Regulation of NF-kappaB induction by TCR/CD28. Immunologic research. 2011;50:113–117. doi: 10.1007/s12026-011-8216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viglietta V, Baecher-Allan C, Weiner HL, Hafler DA. Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. The Journal of experimental medicine. 2004;199:971–979. doi: 10.1084/jem.20031579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nature medicine. 2011;17:673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, Kodama T, Tanaka S, Bluestone JA, Takayanagi H. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nature medicine. 2014;20:62–68. doi: 10.1038/nm.3432. [DOI] [PubMed] [Google Scholar]
- 7.McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, Hulme MA, Hoffmuller U, Baron U, Olek S, Bluestone JA, Brusko TM. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol. 2011;186:3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nylander A, Hafler DA. Multiple sclerosis. The Journal of clinical investigation. 2012;122:1180–1188. doi: 10.1172/JCI58649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han MH, Steinman L. Systems biology for identification of molecular networks in multiple sclerosis. Multiple sclerosis. 2009;15:529–530. doi: 10.1177/1352458509103318. [DOI] [PubMed] [Google Scholar]
- 10.C. International Multiple Sclerosis Genetics; Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D, Goris A, Oturai A, Saarela J, Fontaine B, Hemmer B, Martin C, Zipp F, D’Alfonso S, Martinelli-Boneschi F, Taylor B, Harbo HF, Kockum I, Hillert J, Olsson T, Ban M, Oksenberg JR, Hintzen R, Barcellos LF, Agliardi C, Alfredsson L, Alizadeh M, Anderson C, Andrews R, Sondergaard HB, Baker A, Band G, Baranzini SE, Barizzone N, Barrett J, Bellenguez C, Bergamaschi L, Bernardinelli L, Berthele A, Biberacher V, Binder TM, Blackburn H, Bomfim IL, Brambilla P, Broadley S, Brochet B, Brundin L, Buck D, Butzkueven H, Caillier SJ, Camu W, Carpentier W, Cavalla P, Celius EG, Coman I, Comi G, Corrado L, Cosemans L, Cournu-Rebeix I, Cree BA, Cusi D, Damotte V, Defer G, Delgado SR, Deloukas P, di Sapio A, Dilthey AT, Donnelly P, Dubois B, Duddy M, Edkins S, Elovaara I, Esposito F, Evangelou N, Fiddes B, Field J, Franke A, Freeman C, Frohlich IY, Galimberti D, Gieger C, Gourraud PA, Graetz C, Graham A, Grummel V, Guaschino C, Hadjixenofontos A, Hakonarson H, Halfpenny C, Hall G, Hall P, Hamsten A, Harley J, Harrower T, Hawkins C, Hellenthal G, Hillier C, Hobart J, Hoshi M, Hunt SE, Jagodic M, Jelcic I, Jochim A, Kendall B, Kermode A, Kilpatrick T, Koivisto K, Konidari I, Korn T, Kronsbein H, Langford C, Larsson M, Lathrop M, Lebrun-Frenay C, Lechner-Scott J, Lee MH, Leone MA, Leppa V, Liberatore G, Lie BA, Lill CM, Linden M, Link J, Luessi F, Lycke J, Macciardi F, Mannisto S, Manrique CP, Martin R, Martinelli V, Mason D, Mazibrada G, McCabe C, Mero IL, Mescheriakova J, Moutsianas L, Myhr KM, Nagels G, Nicholas R, Nilsson P, Piehl F, Pirinen M, Price SE, Quach H, Reunanen M, Robberecht W, Robertson NP, Rodegher M, Rog D, Salvetti M, Schnetz-Boutaud NC, Sellebjerg F, Selter RC, Schaefer C, Shaunak S, Shen L, Shields S, Siffrin V, Slee M, Sorensen PS, Sorosina M, Sospedra M, Spurkland A, Strange A, Sundqvist E, Thijs V, Thorpe J, Ticca A, Tienari P, van Duijn C, Visser EM, Vucic S, Westerlind H, Wiley JS, Wilkins A, Wilson JF, Winkelmann J, Zajicek J, Zindler E, Haines JL, Pericak-Vance MA, Ivinson AJ, Stewart G, Hafler D, Hauser SL, Compston A, McVean G, De Jager P, Sawcer SJ, McCauley JL C. Wellcome Trust Case Control, I. B. D. G. C. International. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nature genetics. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patsopoulos NA, Esposito F, Reischl J, Lehr S, Bauer D, Heubach J, Sandbrink R, Pohl C, Edan G, Kappos L, Miller D, Montalban J, Polman CH, Freedman MS, Hartung HP, Arnason BG, Comi G, Cook S, Filippi M, Goodin DS, Jeffery D, O’Connor P, Ebers GC, Langdon D, Reder AT, Traboulsee A, Zipp F, Schimrigk S, Hillert J, Bahlo M, Booth DR, Broadley S, Brown MA, Browning BL, Browning SR, Butzkueven H, Carroll WM, Chapman C, Foote SJ, Griffiths L, Kermode AG, Kilpatrick TJ, Lechner-Scott J, Marriott M, Mason D, Moscato P, Heard RN, Pender MP, Perreau VM, Perera D, Rubio JP, Scott RJ, Slee M, Stankovich J, Stewart GJ, Taylor BV, Tubridy N, Willoughby E, Wiley J, Matthews P, Boneschi FM, Compston A, Haines J, Hauser SL, McCauley J, Ivinson A, Oksenberg JR, Pericak-Vance M, Sawcer SJ, De Jager PL, Hafler DA, de Bakker PI M. S. G. W. G. Bayer Pharma, I.-b. Steering Committees of Studies Evaluating, C. C. R. A. a, A. N. Consortium, GeneMsa, C. International Multiple Sclerosis Genetics. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Annals of neurology. 2011;70:897–912. doi: 10.1002/ana.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, Freeman C, Hunt SE, Edkins S, Gray E, Booth DR, Potter SC, Goris A, Band G, Oturai AB, Strange A, Saarela J, Bellenguez C, Fontaine B, Gillman M, Hemmer B, Gwilliam R, Zipp F, Jayakumar A, Martin R, Leslie S, Hawkins S, Giannoulatou E, D’Alfonso S, Blackburn H, Martinelli Boneschi F, Liddle J, Harbo HF, Perez ML, Spurkland A, Waller MJ, Mycko MP, Ricketts M, Comabella M, Hammond N, Kockum I, McCann OT, Ban M, Whittaker P, Kemppinen A, Weston P, Hawkins C, Widaa S, Zajicek J, Dronov S, Robertson N, Bumpstead SJ, Barcellos LF, Ravindrarajah R, Abraham R, Alfredsson L, Ardlie K, Aubin C, Baker A, Baker K, Baranzini SE, Bergamaschi L, Bergamaschi R, Bernstein A, Berthele A, Boggild M, Bradfield JP, Brassat D, Broadley SA, Buck D, Butzkueven H, Capra R, Carroll WM, Cavalla P, Celius EG, Cepok S, Chiavacci R, Clerget-Darpoux F, Clysters K, Comi G, Cossburn M, Cournu-Rebeix I, Cox MB, Cozen W, Cree BA, Cross AH, Cusi D, Daly MJ, Davis E, de Bakker PI, Debouverie M, D’Hooghe BM, Dixon K, Dobosi R, Dubois B, Ellinghaus D, Elovaara I, Esposito F, Fontenille C, Foote S, Franke A, Galimberti D, Ghezzi A, Glessner J, Gomez R, Gout O, Graham C, Grant SF, Guerini FR, Hakonarson H, Hall P, Hamsten A, Hartung HP, Heard RN, Heath S, Hobart J, Hoshi M, Infante-Duarte C, Ingram G, Ingram W, Islam T, Jagodic M, Kabesch M, Kermode AG, Kilpatrick TJ, Kim C, Klopp N, Koivisto K, Larsson M, Lathrop M, Lechner-Scott JS, Leone MA, Leppa V, Liljedahl U, Bomfim IL, Lincoln RR, Link J, Liu J, Lorentzen AR, Lupoli S, Macciardi F, Mack T, Marriott M, Martinelli V, Mason D, McCauley JL, Mentch F, Mero IL, Mihalova T, Montalban X, Mottershead J, Myhr KM, Naldi P, Ollier W, Page A, Palotie A, Pelletier J, Piccio L, Pickersgill T, Piehl F, Pobywajlo S, Quach HL, Ramsay PP, Reunanen M, Reynolds R, Rioux JD, Rodegher M, Roesner S, Rubio JP, Ruckert IM, Salvetti M, Salvi E, Santaniello A, Schaefer CA, Schreiber S, Schulze C, Scott RJ, Sellebjerg F, Selmaj KW, Sexton D, Shen L, Simms-Acuna B, Skidmore S, Sleiman PM, Smestad C, Sorensen PS, Sondergaard HB, Stankovich J, Strange RC, Sulonen AM, Sundqvist E, Syvanen AC, Taddeo F, Taylor B, Blackwell JM, Tienari P, Bramon E, Tourbah A, Brown MA, Tronczynska E, Casas JP, Tubridy N, Corvin A, Vickery J, Jankowski J, Villoslada P, Markus HS, Wang K, Mathew CG, Wason J, Palmer CN, Wichmann HE, Plomin R, Willoughby E, Rautanen A, Winkelmann J, Wittig M, Trembath RC, Yaouanq J, Viswanathan AC, Zhang H, Wood NW, Zuvich R, Deloukas P, Langford C, Duncanson A, Oksenberg JR, Pericak-Vance MA, Haines JL, Olsson T, Hillert J, Ivinson AJ, De Jager PL, Peltonen L, Stewart GJ, Hafler DA, Hauser SL, McVean G, Donnelly P, Compston A. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kofler DM, Severson CA, Mousissian N, De Jager PL, Hafler DA. The CD6 multiple sclerosis susceptibility allele is associated with alterations in CD4+ T cell proliferation. J Immunol. 2011;187:3286–3291. doi: 10.4049/jimmunol.1100626. [DOI] [PubMed] [Google Scholar]
- 14.Maier LM, Anderson DE, De Jager PL, Wicker LS, Hafler DA. Allelic variant in CTLA4 alters T cell phosphorylation patterns. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18607–18612. doi: 10.1073/pnas.0706409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couturier N, Bucciarelli F, Nurtdinov RN, Debouverie M, Lebrun-Frenay C, Defer G, Moreau T, Confavreux C, Vukusic S, Cournu-Rebeix I, Goertsches RH, Zettl UK, Comabella M, Montalban X, Rieckmann P, Weber F, Muller-Myhsok B, Edan G, Fontaine B, Mars LT, Saoudi A, Oksenberg JR, Clanet M, Liblau RS, Brassat D. Tyrosine kinase 2 variant influences T lymphocyte polarization and multiple sclerosis susceptibility. Brain: a journal of neurology. 2011;134:693–703. doi: 10.1093/brain/awr010. [DOI] [PubMed] [Google Scholar]
- 16.Cerosaletti K, Schneider A, Schwedhelm K, Frank I, Tatum M, Wei S, Whalen E, Greenbaum C, Kita M, Buckner J, Long SA. Multiple autoimmune-associated variants confer decreased IL-2R signaling in CD4+ CD25(hi) T cells of type 1 diabetic and multiple sclerosis patients. PloS one. 2013;8:e83811. doi: 10.1371/journal.pone.0083811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturner KH, Borgmeyer U, Schulze C, Pless O, Martin R. A Multiple Sclerosis-Associated Variant of CBLB Links Genetic Risk with Type I IFN Function. J Immunol. 2014;193:4439–4447. doi: 10.4049/jimmunol.1303077. [DOI] [PubMed] [Google Scholar]
- 18.Cotsapas C, Hafler DA. Immune-mediated disease genetics: the shared basis of pathogenesis. Trends in immunology. 2013;34:22–26. doi: 10.1016/j.it.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, Abecasis GR, Barrett JC, Behrens T, Cho J, De Jager PL, Elder JT, Graham RR, Gregersen P, Klareskog L, Siminovitch KA, van Heel DA, Wijmenga C, Worthington J, Todd JA, Hafler DA, Rich SS, Daly MJ. Pervasive sharing of genetic effects in autoimmune disease. PLoS genetics. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voight BF, Cotsapas C. Human genetics offers an emerging picture of common pathways and mechanisms in autoimmunity. Current opinion in immunology. 2012;24:552–557. doi: 10.1016/j.coi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 21.De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C, Briskin R, Romano S, International MSGC, Baranzini SE, McCauley JL, Pericak-Vance MA, Haines JL, Gibson RA, Naeglin Y, Uitdehaag B, Matthews PM, Kappos L, Polman C, McArdle WL, Strachan DP, Evans D, Cross AH, Daly MJ, Compston A, Sawcer SJ, Weiner HL, Hauser SL, Hafler DA, Oksenberg JR. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nature genetics. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2014 doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggert M, Goertsches R, Seeck U, Dilk S, Neeck G, Zettl UK. Changes in the activation level of NF-kappa B in lymphocytes of MS patients during glucocorticoid pulse therapy. Journal of the neurological sciences. 2008;264:145–150. doi: 10.1016/j.jns.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 24.Dong J, Jimi E, Zeiss C, Hayden MS, Ghosh S. Constitutively active NF-kappaB triggers systemic TNFalpha-dependent inflammation and localized TNFalpha-independent inflammatory disease. Genes & development. 2010;24:1709–1717. doi: 10.1101/gad.1958410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoh J, Misawa T, Tabunoki H, Yamamura T. Molecular network analysis of T-cell transcriptome suggests aberrant regulation of gene expression by NF-kappaB as a biomarker for relapse of multiple sclerosis. Disease markers. 2008;25:27–35. doi: 10.1155/2008/824640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein MB, Ruan Y, Snyder M. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome research. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudson ME, Snyder M. High-throughput methods of regulatory element discovery. BioTechniques. 2006;41:673–675. 677. doi: 10.2144/000112322. passim. [DOI] [PubMed] [Google Scholar]
- 29.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 30.Segal E, Widom J. What controls nucleosome positions? Trends in genetics: TIG. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C. Genomes Project. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian F, Wang X, Zhang L, Chen S, Piecychna M, Allore H, Bockenstedt L, Malawista S, Bucala R, Shaw AC, Fikrig E, Montgomery RR. Age-associated elevation in TLR5 leads to increased inflammatory responses in the elderly. Aging cell. 2012;11:104–110. doi: 10.1111/j.1474-9726.2011.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ottoboni L, Frohlich IY, Lee M, Healy BC, Keenan BT, Xia Z, Chitnis T, Guttmann CR, Khoury SJ, Weiner HL, Hafler DA, De Jager PL. Clinical relevance and functional consequences of the TNFRSF1A multiple sclerosis locus. Neurology. 2013;81:1891–1899. doi: 10.1212/01.wnl.0000436612.66328.8a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory AP, Dendrou CA, Attfield KE, Haghikia A, Xifara DK, Butter F, Poschmann G, Kaur G, Lambert L, Leach OA, Promel S, Punwani D, Felce JH, Davis SJ, Gold R, Nielsen FC, Siegel RM, Mann M, Bell JI, McVean G, Fugger L. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–511. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faustman D, Davis M. TNF receptor 2 pathway: drug target for autoimmune diseases. Nature reviews Drug discovery. 2010;9:482–493. doi: 10.1038/nrd3030. [DOI] [PubMed] [Google Scholar]
- 38.Lund BT, Ashikian N, Ta HQ, Chakryan Y, Manoukian K, Groshen S, Gilmore W, Cheema GS, Stohl W, Burnett ME, Ko D, Kachuck NJ, Weiner LP. Increased CXCL8 (IL-8) expression in Multiple Sclerosis. Journal of neuroimmunology. 2004;155:161–171. doi: 10.1016/j.jneuroim.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Carrieri PB, Provitera V, De Rosa T, Tartaglia G, Gorga F, Perrella O. Profile of cerebrospinal fluid and serum cytokines in patients with relapsing-remitting multiple sclerosis: a correlation with clinical activity. Immunopharmacology and immunotoxicology. 1998;20:373–382. doi: 10.3109/08923979809034820. [DOI] [PubMed] [Google Scholar]
- 40.Bartosik-Psujek H, Stelmasiak Z. The levels of chemokines CXCL8, CCL2 and CCL5 in multiple sclerosis patients are linked to the activity of the disease. European journal of neurology: the official journal of the European Federation of Neurological Societies. 2005;12:49–54. doi: 10.1111/j.1468-1331.2004.00951.x. [DOI] [PubMed] [Google Scholar]
- 41.Scarpini E, Galimberti D, Baron P, Clerici R, Ronzoni M, Conti G, Scarlato G. IP-10 and MCP-1 levels in CSF and serum from multiple sclerosis patients with different clinical subtypes of the disease. Journal of the neurological sciences. 2002;195:41–46. doi: 10.1016/s0022-510x(01)00680-3. [DOI] [PubMed] [Google Scholar]
- 42.Lee LF, Axtell R, Tu GH, Logronio K, Dilley J, Yu J, Rickert M, Han B, Evering W, Walker MG, Shi J, de Jong BA, Killestein J, Polman CH, Steinman L, Lin JC. IL-7 promotes T(H)1 development and serum IL-7 predicts clinical response to interferon-beta in multiple sclerosis. Science translational medicine. 2011;3:93ra68. doi: 10.1126/scitranslmed.3002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJ, Furman D, Shen-Orr S, Dekker CL, Swan GE, Butte AJ, Maecker HT, Davis MM. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulua AC, Simon A, Maddipati R, Pelletier M, Park H, Kim KY, Sack MN, Kastner DL, Siegel RM. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS) The Journal of experimental medicine. 2011;208:519–533. doi: 10.1084/jem.20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kofler DM, Marson A, Dominguez-Villar M, Xiao S, Kuchroo VK, Hafler DA. Decreased RORC-dependent silencing of prostaglandin receptor EP2 induces autoimmune Th17 cells. The Journal of clinical investigation. 2014;124:2513–2522. doi: 10.1172/JCI72973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sfikakis PP. The first decade of biologic TNF antagonists in clinical practice: lessons learned, unresolved issues and future directions. Current directions in autoimmunity. 2010;11:180–210. doi: 10.1159/000289205. [DOI] [PubMed] [Google Scholar]
- 47.Atzeni F, Talotta R, Salaffi F, Cassinotti A, Varisco V, Battellino M, Ardizzone S, Pace F, Sarzi-Puttini P. Immunogenicity and autoimmunity during anti-TNF therapy. Autoimmunity reviews. 2013;12:703–708. doi: 10.1016/j.autrev.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 48.Jacobelli S, Andre M, Alexandra JF, Dode C, Papo T. Failure of anti-TNF therapy in TNF Receptor 1-Associated Periodic Syndrome (TRAPS) Rheumatology (Oxford) 2007;46:1211–1212. doi: 10.1093/rheumatology/kel298. [DOI] [PubMed] [Google Scholar]
- 49.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarnico I, Lanzillotta A, Benarese M, Alghisi M, Baiguera C, Battistin L, Spano P, Pizzi M. NF-kappaB dimers in the regulation of neuronal survival. International review of neurobiology. 2009;85:351–362. doi: 10.1016/S0074-7742(09)85024-1. [DOI] [PubMed] [Google Scholar]
- 51.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Confirmation of increased pNFκB in multiple sclerosis patients.
Supplemental Figure 2: Gating strategy and representative histogram for rs228614 GG (risk) and AA (protective) variants.
Supplemental Figure 3: Rs228614 and rs7665090 proximal to NFκB1 result in increased IκBα degradation after TNFα treatment.
Supplemental Figure 4: Constitutive total IκBα and phospho-p65 NFκB are similar in rs228614 risk and protective variants.
Supplemental Figure 5: Full uncut gel for figure 4
Supplemental Figure 6: TNFα and PMA responses are stable across multiple draws.
Supplemental Figure 7: Responses to TNFα do not change with age, gender, or ethnicity.
Supplemental Figure 8: TNFα signals through TNFR1 on naïve CD4 cells.
Supplemental Figure 9: The rs1800693 CC variant does not change the surface expression of TNFR1.
Supplemental Figure 10: Representative single-stained control.
Supplemental Table 1. Demographics of healthy subjects and MS samples for figure 1 and suppl. figure 1.
Supplemental Table 2. Demographics of rs228614 results from figure 2.
Supplemental Table 3. Demographics of rs7665090 from figure 2.
Supplemental Table 4. Demographics of rs1800693 from figure 5.
Supplemental Table 5. Luminex data from rs1800693 subjects.
Supplemental Table 6. Raw Data