Abstract
Background
In West Africa, tick-borne relapsing fever is a neglected arthropod-borne infection caused by Borrelia crocidurae transmitted by the argasid tick Ornithodoros sonrai. From an epidemiological point of view, it is of interest to know whether some genotypes of the vector are specialized in carrying certain genotypes of the pathogen.
Findings
Thirty-five O. sonrai ticks collected in Mali, Senegal, Mauritania and Morocco confirmed to be B. crocidurae-infected, were genotyped by 16S rRNA gene sequencing. B. crocidurae was genotyped by Multispacer Sequence Typing. The 35 O. sonrai ticks grouped into 12 genotypes with strong geographical structuration. MST resolved the 35 B. crocidurae isolates into 29 genotypes with pairwise divergence of 0.09 - 1.56 % without strict geographical structuration as genotype ST22 was found in Mali, Senegal and Mauritania. There was no evidence of tick-borrelia specialization as one O. sonrai genotype carried several B. crocidurae genotypes and one B. crocidurae genotype was found in different O. sonrai genotypes.
Conclusions
This report illustrates a non-specialized circulation of B. crocidurae borreliae within O. sonrai ticks in West Africa.
Keywords: Argasidticks, Borrelia, Multiple spacer sequence typing
Background
Borrelia crocidurae is one of the spirochetes responsible for tick-borne relapsing fever in North and West Africa [1]. In these countries, borreliae are maintained between Ornithodoros sonrai argasid ticks and rodents while the humans get infected accidently [2, 3]. Accordingly, B. crocidurae has been detected in O. sonrai ticks collected in Tunisia, Morocco, Mauritania, Senegal [4] and Mali [3]. In Morocco, borreliae infection was found in 10.2 % of ticks and 8.6 % of tested rodents and insectivores [5]. In Mali, B. crocidurae was found in 17.3 % of O. sonrai ticks and its diversity was established and characterized in 15 samples by multi-locus sequence typing (MLST) [3]. In patients, an average incidence of 11 per 100 person-years was reported in humans at Senegal [2].
It has already been reported that multiple strains of B. crocidurae were probably transmitted by ticks collected from the same geographical area and their progeny [6]. This observation however, was made before tools were available for a fine discrimination between both ticks and borreliae groups. Recently, such sequencing-based tools allowed refining the diversity of O. sonrai ticks into nine subgroups [7]. Likewise, MLST [3] and multiple spacer sequence typing (MST) [8] discriminated groups among B. crocidurae. Genotyping found a strong geographical structuration of the O. sonrai tick population [7], however, it remains unknown whether certain O. sonrai genetic variants are specialized in certain genetic variants of B. crocidurae. The aim of the present work was to investigate this question using modern molecular tools for genotyping O. sonrai ticks infected by B. crocidurae.
Findings
The National Ethics Committee of Senegal approved the protocol used for human samples. The study protocol was also approved by the Steering Committee of the Institut de Recherche pour le Développement (IRD) Special Programme Evolution Climatique et Santé (IRD, Montpellier, France), reference project ATI-ECS-07-H/2002.
O. sonrai ticks here studied are not registered as endangered species. Ticks were collected using a flexible tube and a portable aspirator, inside houses and private lands after the owner of the house and land gave permission to conduct the study on the site. Collected ticks were immediately preserved in ethanol 70 %, before total DNA (tick and Borrelia) was extracted and purified by using the DNeasy Tissue extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and stored at −20 °C until used as a template for the PCR-based amplification of tick or Borrelia genes.
The presence of B. crocidurae was confirmed in ticks by flagellin gene PCR-sequencing as previously described [4]. Genotyping of B. crocidurae was then done by using MST as previously [8]. The nucleotide sequences were edited using ChromasPro software (www.technelysium.com.au/chromas.html). Similarities between MST types were determined after multiple alignments using the Muscle software implemented in MEGA5 package [9]. The five-spacer sequences analyzed herein were concatenated. Relationships among B. crocidurae genotypes detected in ticks plus previously published 7 spacer types (STs) of human B. crocidurae isolates [8] were inferred using the Bayesian phylogenetic analyses in MrBayes v3.1.2 using default parameters [10]. Each particular combination of the five spacer sequences was assigned to a spacer type (ST) number.
In parallel, tick genotyping was done by 16S rRNA gene sequencing as previously described [7]. Modeltest 3.4 [11] was used to select the appropriate model of molecular evolution. The best-fitting ML model was HKY85 [12] for 16S rRNA sequences. The highest-likelihood DNA and corresponding bootstrap support values were obtained by PhyML (freely available at http://mobyle.pasteur.fr/cgi-bin/portal.py#welcome) using NNI (Nearest Neighbor Interchange) branch swapping and 1000 bootstrap replicates.
A total of 35 ticks - Borrelia crocidurae complexes collected in four countries (14 sites) and 13 infected patients were used in this study. All the new spacer nucleotide sequences of Borrelia crocidurae have been deposited in the GenBank database under accession numbers KF843734-KF843765 and described previously, including sequences of B. crocidurae from patients (JQ398819-JQ398841) [1]. The 16S rRNA sequences of Ornithodoros sonrai were also deposited in GenBank KP644211-KP644222).
Results
In all PCR-based experiments, negative controls remained negative. The B. crocidurae-infected ticks here investigated comprised one tick from Morocco (1 site), three ticks from Mauritania (1 site), twenty-seven ticks from Senegal (8 sites) and four ticks from Mali (4 sites) (Table 1).
Table 1.
Geographical sites and 16 s rRNA genotypes for Ornithodoros sonrai ticks used in this study from Trape and colleagues [7]
| Country | Location | Coordinates | Code of ticks | Code of 16S rRNA group of ticks |
|---|---|---|---|---|
| Morocco | Boudnib | 31°59’N-03°59’W | BOUD1310 | Os01 |
| Mauritania | Amridjiel (Oum El Khez) | 17°00’N-10°59’W | OUM1047 | Os06 |
| OUM1044 | Os05 | |||
| OUM 1063 | Os01 | |||
| Senegal | Dielmo | 13°43’N-16°24’W | DL1646 DL1650 | Os11 |
| DL1669 DL1675 | ||||
| DL1676 DL1706 | ||||
| DL1657 DL1661 | Os12 | |||
| DL1662 DL1674 | ||||
| DL1677 DL1680 | ||||
| DL1709 | ||||
| Thianène (Darou Mousti) | 15°04’N-16°00’W | DM129 DM136 | Os03 | |
| DM148 DM150 | ||||
| DM155 DM160 | ||||
| Néma-Nding | 13°42’N-16°25’W | NNT919 | Os10 | |
| Keur Aliou Gueye | 13°47’N-16°24’W | KAG349 KAG351 | Os12 | |
| Kanène Khar | 15°30’N-16°01’W | KKH113 | Os03 | |
| Keur Lamine Socé | 13°44’N-16°25’W | KLS363 KLS364 | Os12 | |
| Kéniéba | 14°05’N-12°03’W | KNB305 | Os07 | |
| Taouey (Richard-Toll) | 16°27’N-15°42’W | RT12 | Os02 | |
| Mali | Makania (Diamou) | 14°04’N-11°09’W | DIA999 | Os03 |
| Dianguirdé (Diéma) | 14°30’N-09°01’W | DIEM1207 | Os04 | |
| Kolomina (Doubala) | 14°28’N-08°01’W | DOUB1237 | Os09 | |
| Sogoli (Ké-Macina) | 13°58’N-05°28’W | KEM1410 | Os08 |
As for tick genotyping, all positions containing gaps and missing data were eliminated from the 441-bp aligned fragment of 16S rRNA gene sequences of O. sonrai. There was a total of 435 positions with 36 variable sites and 20 parsimony-informative sites in the final dataset. The 35 O. sonrai ticks used for the genetic analysis yielded 12 different genotypes (Table 1, Fig. 1). All ticks in the same clade (i.e. Os) had identical 16S rRNA sequences (Fig. 1).
Fig. 1.
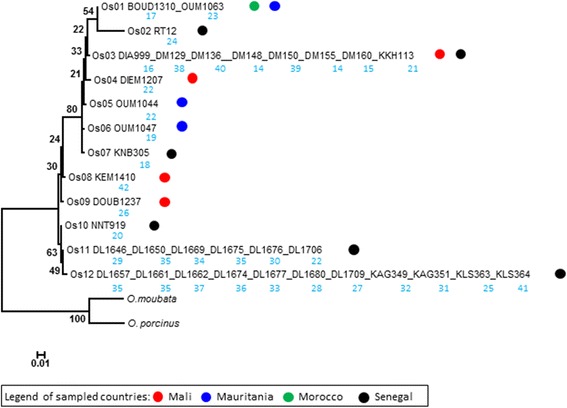
Phylogram (PhyML 1000 bootstraps) of partial 16S rRNA gene sequence data (441 nucleotides) of Ornithodoros sonrai sequences (35 sequences were grouped in 12 Os). Bootstrap values are shown in black (scale bar, 0.01 substitutions per site). Two 16S rRNA gene sequences of Ornithodoros moubata (GenBank accession number AB073679) and Ornithodoros porcinus (GenBank accession number AB105451), were treated as outgroups. Os: code of 16S rRNA groups. Number in blue: Spacer-Type of Borrelia crocidurae
One tick (Boud1310) collected in Morocco yielded one genotype (Os01) common with a tick (Oum1063) from Mauritania, the two other ticks collected in Mauritania yielded two genotypes. The twenty-seven ticks collected in Senegal (8 sites) yielded six genotypes and the four ticks collected in Mali yielded four genotypes (Fig. 1). More specifically, the Os04, Os09 and Os10 genotypes were found only in Mali whereas the Os03 genotype was found both in Mali and Senegal; the Os05 and Os06 genotypes were found only in Mauritania whereas the genotype Os01 was found both in Morocco and Mauritania; the genotypes Os02, Os07, Os10, Os11, Os12 were found only in Senegal (Fig. 1).
As for B. crocidurae genotyping, the analysis of the spacer sequences derived from the 35 B. crocidurae isolates from ticks yielded 29 spacer types (STs) named ST14–ST42 and 13 B. crocidurae isolates from patients yielded 7 spacer types (STs) previously named ST6–ST12 [8] (Fig. 2). Pairwise divergence between B. crocidurae ranged from 0.09 to 1.56 %. ST22 was detected in ticks collected in Mali, Senegal and Mauritania. The ST35 in Dielmo (Senegal) was found in tick genotype Os11 and Os12 and five STs (STs 14, 15, 38, 39 and 40) were found in Darou Mousti (Senegal). Three STs named ST38–ST40 were found in Senegal and carried by genotype Os03 (Figs. 1 and 2). Tick genotypes Os01, Os03, Os11 and Os12 carried more than one genotype of B. crocidurae. The phylogenetic tree constructed from 35 spacer sequences of B. crocidurae separated the strains into two clades. A first clade comprised of 32 B. crocidurae genotypes and a second clade comprised of three B. crocidurae genotypes (STs 38–40). A phylogenetic tree combining the STs found here in ticks, with the STs previously reported in patients [8], yielded a geographical clustering of B. crocidurae in patients and ticks from Dielmo, Senegal (Fig. 2).
Fig. 2.
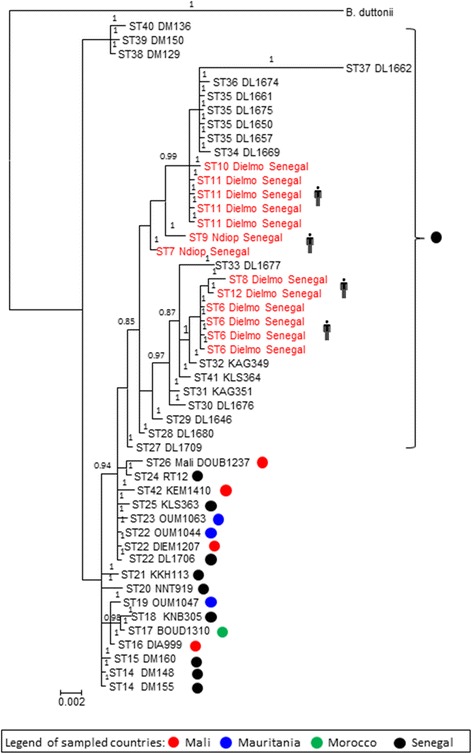
Bayesian MCMC analysis based on five intergenic spacers sequences for 48 Borrelia crocidurae strains in Ornithodoros sonrai ticks and human samples. The site of Dielmo (Senegal) is only indicated in the figure for comparison between STs B. crocidurae found in humans and ticks. Numbers represent posterior probabilities. Red colour indicates Borrelia from humans ( symbol) and black colour indicates Borrelia from ticks
symbol) and black colour indicates Borrelia from ticks
Discussion
The presence of several genetic variants of B. crocidurae has recently been reported in Mali [3]. In Mali, plasmidogram found six profiles among 15 isolates which yielded four MLST groups after sequencing the 16S rRNA, flaBand glpQ genes and the IGS [3]. Here, we further observed a high diversity of B. crocidurae in West Africa, in observing 27 genetic variants determined by MST, among 35 B. crocidurae samples. While plasmidogram and MLST required cultured borreliae [3], we applied MST directly to infected ticks as we previously did on the blood collected from infected patients in Senegal [1, 8].
Previous study showed differences in the infection rate of O. sonrai from different regions, compatible with differences in vector competence [4]. Our study does not verify this hypothesis. Indeed, the diversity of tick-borne B. crocidurae was not superimposed on that of O. sonrai. We found that one B. crocidurae ST was carried by different genotypes of O. sonrai. In addition, one O. sonrai genotype was carrying different B. crocidurae STs. Accordingly, we observed that one B. crocidurae ST22 was circulating in Mali, Mauritania and Senegal with different tick genotypes (Os04, Os05 and Os11). The ST35 (4 bacterial samples) was also circulating in two tick genotypes (Os11, Os12). Figure 2 also shows the B. crocidurae ST14 detected in two ticks of one sampled site (Darou Mousti, Senegal). These findings indicate that, within O. sonrai, there is no specialization of certain tick genetic variants for their capacity to host B. crocidurae.
Also, we were able to compare B. crocidurae MST genotypes in patients and ticks of the same village of Dielmo, rural Senegal. In this village, the B. crocidurae genotypes previously reported in patients unsurprisingly form a subset of the strains circulating in the tick vectors, confirming O. sonrai as the vector of B. crocidurae in this village.
This analysis indicates that MST is well adapted to study the diversity of borreliae on a small, kilometer geographical size; other tools may be more suitable on a larger scale.
Acknowledgments
This work is a part of an Agence Nationale de la Recherche program, France, and the IRD special programme “Evolution Climatique et Santé” (ref: IRD-ATI-ECS). Haitham Elbir and Aurélien Fotso Fotso benefited from grants by Méditerranée Infection Fondation, Marseille, France.
Footnotes
Haitham Elbir and Aurélien FotsoFotso contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
Conceived and Performed the experiments: EH AFF CA PD. Analyzed the data: AFF EH FR PD. Contributed reagents/materials/analysis tools: JFT GD CA. Wrote the manuscript: AFF EH FR PD. All authors read and approved the final version of the manuscript.
References
- 1.Elbir H, Raoult D, Drancourt M. Relapsing fever Borreliae in Africa. Am J Trop Med Hyg. 2013;89:288–92. doi: 10.4269/ajtmh.12-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trape JF, Duplantier JM, Bouganali H, Godeluck B, Legros F, Cornet JP, et al. Tick-borne borreliosis in West Africa. Lancet. 1991;337:473–5. doi: 10.1016/0140-6736(91)93404-W. [DOI] [PubMed] [Google Scholar]
- 3.Schwan TG, Anderson JM, Lopez JE, Fischer RJ, Raffel SJ, McCoy BN, et al. Endemic foci of the tick-borne relapsing fever spirochete Borrelia crocidurae in Mali, West Africa, and the potential for human infection. PLoSNegl Trop Dis. 2012;6:e1924. doi: 10.1371/journal.pntd.0001924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vial L, Durand P, Arnathau C, Halos L, Diatta G, Trape JF, et al. Molecular divergences of the Ornithodoros sonrai soft tick species, a vector of human relapsing fever in West Africa. Microbes Infect. 2006;8:2605–11. doi: 10.1016/j.micinf.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Diatta G, Souidi Y, Granjon L, Arnathau C, Durand P, Chauvancy G, et al. Epidemiology of tick-borne borreliosis in Morocco. PLoSNegl Trop Dis. 2012;6:e1810. doi: 10.1371/journal.pntd.0001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baltazard M. Sur le classement des spirochètes récurrents. Ann Parasitol Hum Comp. 1954;29:12–32. [PubMed] [Google Scholar]
- 7.Trape JF, Diatta G, Arnathau C, Bitam I, Sarih M, Belghyti D, et al. The epidemiology and geographic distribution of relapsing fever borreliosis in West and North Africa, with a review of the Ornithodoroserraticus Complex (Acari: Ixodida) PLoS One. 2013;8:e78473. doi: 10.1371/journal.pone.0078473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbir H, Gimenez G, Sokhna C, Bilcha K, Ali J, Barker SC, et al. Multispacer Sequence typing relapsing fever borreliae in Africa. PLoSNegl Trop Dis. 2012;6:e1652. doi: 10.1371/journal.pntd.0001652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 11.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa M, Kishino H, Yano T. Dating of human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]


