Abstract
The impetus for discovery and evaluation of protein biomarkers has been accelerated by recent development of advanced technologies for rapid and broad proteome analyses. Mass spectrometry (MS)-based protein assays hold great potential for in vitro biomarker studies. Described here is the development of a multiplex Mass Spectrometric Immunoassay (MSIA) for quantification of apolipoprotein C-I (apoC-I), apolipoprotein C-II (apoC-II), apolipoprotein C-III (apoC-III) and their proteoforms. The multiplex MSIA assay was fast (~40 min) and high-throughput (96 samples at a time). The assay was applied to a small cohort of human plasma samples, revealing the existence of multiple proteoforms for each apolipoprotein C. The quantitative aspect of the assay enabled determination of the concentration for each proteoform individually. Low-abundance proteoforms, such as fucosylated apoC-III, were detected in less than 20% of the samples. The distribution of apoC-III proteoforms varied among samples with similar total apoC-III concentrations. The multiplex analysis of the three apolipoproteins C and their proteoforms using quantitative MSIA represents a significant step forward toward better understanding of their physiological roles in health and disease.
Keywords: apolipoprotein C-I, C-II, C-III, proteoforms, biomarkers, mass spectrometric immunoassay, quantification
1. Introduction
As a result of alternative splicing, single-nucleotide polymorphisms (SNPs), and posttranslational modifications (PTMs), proteins can exist as multiple variants (or “proteoforms” - [1]) in vivo. These proteoforms may play important physiological roles, with possible implications in disease development. Thus, development of methods for detection and quantification of proteoforms is considered an important, albeit challenging task.
Mass spectrometry (MS)-based protein assays hold great potential for in vitro detection of protein biomarkers. Intrinsic to the MS-based methods is the measurement of molecular mass – a unique property of each protein. Several MS-based methodologies employ a combination of immuoaffinity enrichment and MS detection. Among them are MSIA [2, 3], SELDI [4], SISCAPA [5, 6], iMALDI [7] and SILAC [8]. Some of these methods are geared towards detection of proteolytic peptides as surrogate measures for protein quantification (e.g., SISCAPA, SILAC), leaving a large part of the protein sequence un-assessed. Hence, those approaches cannot detect proteoforms without a priori knowledge of their existence. Top-down MS-based approaches are better suited for proteoforms detection because they detect the mass of the intact proteins, thus covering putative modifications in the entire protein sequence [9, 10].
The mass spectrometric immunoassay (MSIA) is one such top-down proteomics method [2, 3, 11–14]. MSIA combines micro-scale immunoaffinity separation with mass spectrometric detection. Antibodies towards targeted proteins are attached to porous microcolumns fitted at the entrance of a pipette tip, and used for proteins affinity extraction directly from the biological sample. After the affinity capture, proteins are eluted either directly onto a target plate for MALDI-TOF MS analysis [15], or eluted with a small volume of elution solution for subsequent LC-ESI MS analysis [16]. When optimized, MSIA can provide detailed insights into the intrinsic protein properties, in both qualitative and quantitative manner [17–21]. The capability for multiplexing is another advantage of MSIA, because it enables simultaneous determination of several proteins in a single analysis [22, 23]. In relative quantification mode, the multiplex MSIA approach yields a protein profile consisting of all proteoforms, which can be used to assess the protein function. In fully quantitative mode (using internal reference standards), multiplex MSIA provides information about the concentration of the native (intact) protein and the various proteoforms that may exist alongside.
Apolipoprotein C-I (apoC-I), apolipoprotein C-II (apoC-II) and apolipoprotein C-III (apoC-III) are members of the same apolipoprotein family, and are synthesized in the liver. They are carried primarily in the very-low density lipoprotein (VLDL) fraction [24] although they become associated with other lipoproteins during normal lipid metabolism. They play important roles as co-factors and inhibitors in triglyceride metabolism. For example, apoC-II is required for efficient lipolysis of triglyceride (TG)-rich lipoproteins through lipoprotein lipase activation [25]. In contrast, apoC-III inhibits the lipolysis of TG-rich lipoproteins and is associated with elevated TG-rich lipoproteins and possibly CVD [26]. Interest in apoCs, particularly apoC-III, has grown considerably since recent studies indicate loss-of-function mutations in the apoC-III gene are associated with reduced triglyceride levels and cardiovascular risk [27–29]. In fact, there is intensive new drug development targeting apoC-III levels [30, 31]. Importantly, several proteoforms of apoCs exist [32] which reflect their function in vivo. MS-based methods provide a unique opportunity to study apoCs proteoforms [33–37], and correlate their structural modifications with function in physiological and pathological states.
Presented here is the development and application of a multiplex MSIA for detection and quantification of apoC-I, apoC-II, and apoC-III and their proteoforms. The main scope was to investigate the quantitative distribution and the heterogeneity of the apoCs in human plasma, and to provide insight into the potential role of these proteoforms as biomarkers.
2. Material and methods
2.1. Reagents
Polyclonal goat anti-human antibodies to apoC-I (Cat. No. 31A-G1b), apoC-II (32AG2b), apoC-III (33A-G2b), and ultra-pure human apolipoprotein C-I (31P-UP201) were obtained from Academy Bio-medical Co. (Houston, TX, USA). Native human apoC-II (Cat. No. MD-26-0012P), native human apoC-III (MD-26-0013P), and mouse anti-hen egg lysozyme (128-10094) were obtained from RayBiotech (Norcross, GA, USA). Protein calibration standard I (Cat. No. 206355) was purchased from Bruker (Billerica, MA). Phosphate buffered saline (PBS) buffer (Cat. No. 28372), MES buffered saline (28390), acetonitrile solution (ACN; A955-4), hydrochloric acid (HCl; A144-212), N-methylpyrrolidinone (NMP; BP1172-4, 1,1’ Carbonyldiimidazole (97%) (CDI, 530-62-1) and affinity pipettes fitted with porous microcolumns (991CUS01) were obtained from Thermo Scientific (Waltham, MA, USA). Tween20 (Cat. No. P7949), trifluoracetic acid (TFA, 299537), sinapic acid (85429-5G), sodium chloride (S7653), HEPES (H3375), ethanolamine (ETA; 398136), albumin from bovine serum (BSA; A7906) and lysozyme from chicken egg white (L7651) were obtained from Sigma Aldrich (St. Louis, MO, USA). Acetone (Cat. No. 0000017150) was obtained from Avantor Performance Materials (Center Valley, PA, USA).
2.2. Human samples
A small set of 82 human Na2EDTA plasma samples was used for the method development and validation. These samples were received from the National University of Singapore, with signed informed consent, without any identifiers, and labeled only with a barcode. The samples were centrifuged for 5 min at 3,000 rpm, and then aliquoted into 96-well microplates (V = 90 µL) and stored at −80°C. Prior to analysis, the samples were thawed and diluted by adding 3 µL of each sample aliquot into 117 µL PBS, 0.1%Tween (40× dilution).
2.3. Antibody immobilization
Activation of the microcolumns in the affinity pipettes was executed using a Multimek 96 channel pipettor (Beckman Coulter, Brea, CA). The affinity pipettes were first rinsed with 200 mM HCl (2 × 20 aspiration/dispense cycles, 100 µL volume), followed by acetone rinse (20 aspiration/dispense cycles, 100 µL). The pipettes were then immersed into a microplate containing 100 mg/mL 1,1’ Carbonyldiimidazole (in methyl-1pyrrolidone-2 (NMP)), and 450 cycles of 100 µL aspirations and dispenses through each affinity pipette were performed. Two rinses with NMP (10 cycles each, 150 µL) followed. The affinity pipettes were then immediately immersed into the wells of a 96-microwell microplate which contained the antibody mixture: 0.32 µg anti-apoC-I; 2.25 µg anti-apoC-II; 2.5 µg anti-apoC-II; and 0.40 µg anti-Lys; dissolved into 50 µL of 10 mM MES buffer (these amounts of antibodies were determined empirically to yield comparable signals of the targeted proteins in the mass spectra; they differed because of the unique affinities of each antibody toward the targeted protein). A total of 750 cycles of aspirations and dispenses of the antibody mixture were performed, followed by rinses with ethanolamine (50 cycles, 100 µL) and twice with HBS-N buffer (100 cycles, 100 µL). The derivatization process took 2 hours, for 96 pipettes. The antibody-derivatized pipettes were stored at +4°C until used (storage was good for > 6 months).
2.4. Sample preparation
For the standard curves, a mixture containing equal mass of the three purified human apolipoproteins was prepared to a final concentration of 10 mg/L (total apolipoproteins). The protein standards mixture was then serially diluted to 5 mg/L, 2.5 mg/L, 1.25 mg/L, 0.625 mg/L, 0.313 mg/L and 0.156 mg/L, in PBS buffer containing 3 g/L BSA. Analytical samples were prepared by combining 30 µL of the 40× diluted plasma (or the protein standards mixture) with 30 µL of a 0.1 mg/mL solution of lysozyme, and 60 µL PBS with 0.1% Tween, yielding a total of 120 µL analytical sample volume. Standards and analytical samples were run simultaneously on the same multi-channel robotic pipettor.
2.5. Assay execution
The antibody-derivatized affinity pipettes were mounted onto the head of the Multimek 96-channel pipettor. The MSIA started with an assay buffer rinse (PBS, 0.1% Tween, 10 aspirations and dispense cycles, 100 µL each). Next, the affinity pipettes were immersed into a microplate containing the analytical samples and 250 aspirations and dispense cycles were performed (100 µL each) allowing for affinity capture of all the proteins from the samples. The affinity pipettes were then rinsed with assay buffer (100 cycles, 100 µL volumes), and twice with water (10 cycles and 20 cycles, 100 µL) to remove non-specifically bound proteins from the microcolumns. For elution of the captured proteins, 6 µL aliquots of MALDI matrix (15 g/L sinapic acid in aqueous solution containing 33 % (v/v) ACN, and 0.4 % (v/v) TFA) were aspirated into each affinity pipette, and after a 10 second delay (to allow for the dissociation of the proteins from the capturing antibodies), the eluates were dispensed directly onto a 96-well formatted MALDI target. Following drying, linear mass spectra were acquired from each sample spot using Bruker’s Ultraflex III MALDI-TOF instrument (Bruker, Billerica, MA). The instrument was operated in positive ion mode, in the mass range from 5 to 30 kDa, with 200 ns delay, 20.00 kV and 18.45 kV ion source voltages, and signal suppression up to 500 Da. Approximately 5,000 laser-shot mass spectra were saved for each standard and sample.
2.6. Data analysis
Mass spectra were internally calibrated using the protein calibration standard I, and further processed with Flex Analysis 3.0 software (Bruker Daltonics). All peaks representing apolipoproteins and their proteoforms, along with the lysozyme IRS peak, were integrated baseline-to-baseline using Zebra 1.0 software (Intrinsic Bioprobes Inc), and the obtained peak area values were tabulated in a spreadsheet. To distinguish between noise and low intensity signals, the peak areas were corrected individually with baseline noise-bin signals (10 Da wide), picked from regions in the mass spectra in close proximity to the integrated protein signals. The corrected apolipoprotein peak areas were then normalized (divided) with the IRS peak area. For the plasma samples, the normalized peak areas for each apolipoprotein and its proteoforms were summed up, and the total apolipoprotein C-I, C-II, and C-III concentrations was determined using the corresponding apo C-I, C-II and C-III standard curves. The concentrations of the individual proteoforms were determined based on their percentage of the total concentrations for each individual apoC protein.
3. Results
3.1. Multiplex MSIA of human apolipoproteins C-I, C-II and C-III and their proteoforms
Multiplex assays have great allure in proteomics [38]. Currently, only a limited number of MS-based methodologies are amenable of multiplex protein analyses, including MSIA [39] and selected peptide-based proteomics approaches [40–42]. In this work we demonstrate a multiplex MSIA for the apoC-I, apoC-II, apoC-III and their proteoforms.
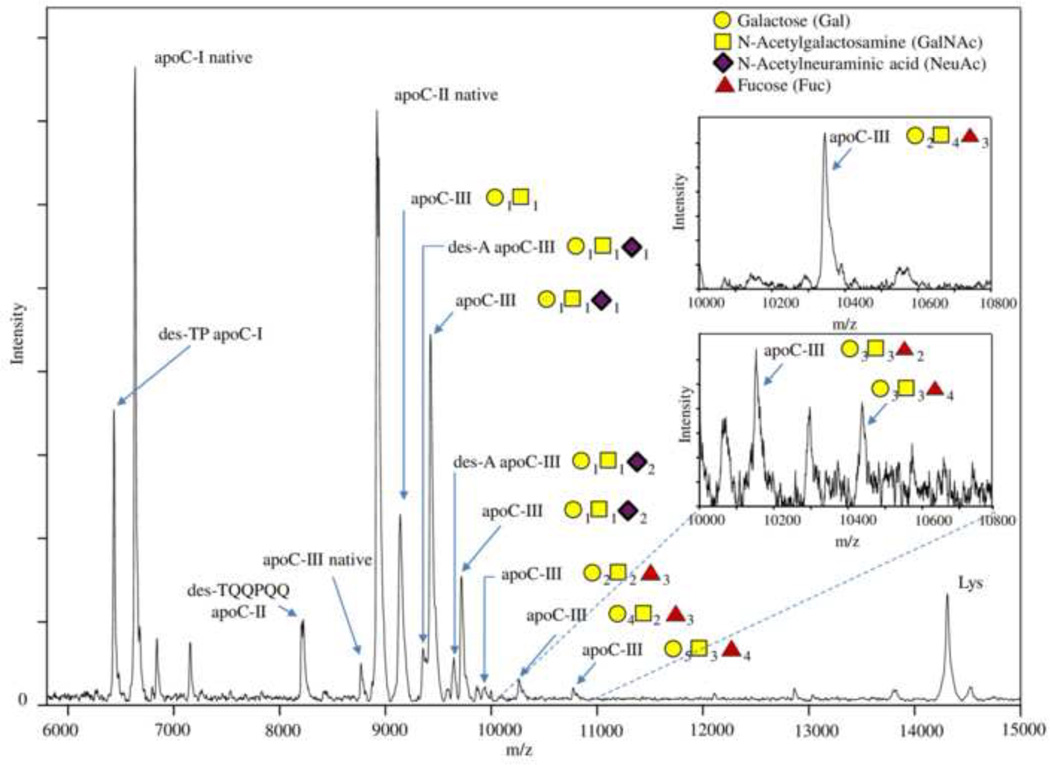
Presented in Fig. 1 are mass spectra resulting from the multiplex apoCs MSIA. Signals from native apoC-I (MW 6,630 Da), apoC-II (MW 8,915 Da), apoC-III (MW 8,765 Da) and the IRS, lysozyme (MW 14,305 Da), were clearly identified in all samples, along with several proteoforms. To identify the proteoforms, two internal calibrations were performed – first one using protein calibration standard I (Bruker, Billerica, MA), and second, using the IRS in the spectrum. Masses were calibrated up to 0.001 Da in m/z, in order to achieve accurate mass assignment for the peaks in the mass spectra. Using this approach, and accounting for the selectivity of the apoCs antibodies, we were able to identify several proteoforms, as listed in Table 1. The table shows the theoretical and observed mz/values of the proteoforms, their distribution (relative percent abundance, as calculated from the total for each apoC), and the frequency of the proteoforms in the cohort. Most of the proteoforms were detected in all samples, with the exception of the low abundance fucosylated apoC-III proteoforms.
Figure 1.
Mass spectra resulting from multiplex MSIA of apoC-I, apoC-II and apoC-III utilizing lysozyme as an IRS. Labeled on the mass spectra are the major C apolipoprotein proteoforms. The inlets represent mass spectra obtained from samples where the specific proteoforms were more prominent.
Table 1.
ApoC-I, apoC-II and apoC-III proteoforms, with their relative percent abundance (RPA) and frequency in a cohort of 82 human EDTA-plasma samples
| Proteoforms | Theoretical m/z value |
Observed m/z value |
RPA (%) Mean (range) |
Frequency (%) |
|---|---|---|---|---|
| des-TP apoC-I | 6432.344 | 6432.104 | 21.3 (13.6 – 27.0) | 100 |
| apoC-I native | 6630.565 | 6630.572 | 78.7 (72.9 – 86.4) | 100 |
| des-TQQPQQ apoC-II | 8204.162 | 8205.290 | 8.51 (4.09 – 13.7) | 100 |
| apoC-II native | 8914.906 | 8915.760 | 91.5 (86.3 – 95.9) | 100 |
| apoC-III native | 8764.652 | 8764.587 | 6.85 (1.06 – 19.0) | 100 |
| apoC-III-(Gal)1(GalNAc)1 | 9135.800 | 9137.925 | 29.9 (25.4 – 36.8) | 100 |
| des-A apoC-III-(Gal)1(GalNAc)1(NeuAc)1 | 9350.171 | 9351.781 | 1.17 (0.336 – 3.65) | 93.9 |
| apoC-III-(Gal)1(GalNAc)1(NeuAc)1 | 9422.249 | 9422.562 | 40.6 (29.1 – 48.3) | 100 |
| des-A apoC-III-(Gal)1(GalNAc)1(NeuAc)2 | 9641.429 | 9642.267 | 5.96 (4.32 – 9.26) | 100 |
| apoC-III-(Gal)1(GalNAc)1(NeuAc)2 | 9712.507 | 9713.492 | 12.1 (6.63 – 19.4) | 100 |
| apoC-III-(Gal)2(GalNAc)2(Fuc)3 | 9933.768 | 9933.941 | 2.61 (0.477 – 6.83) | 100 |
| apoC-III-(Gal)3(GalNAc)3(Fuc)2 | 10152.961 | 10153.559 | 0.949 (0.241 – 1.42) | 12.2 |
| apoC-III-(Gal)4(GalNAc)2(Fuc)3 | 10258.056 | 10259.700 | 1.74 (0.139 – 3.78) | 20.7 |
| apoC-III-(Gal)2(GalNAc)4(Fuc)3 | 10340.158 | 10341.098 | 1.40 (0.837 – 2.00) | 17.1 |
| apoC-III-(Gal)3(GalNAc)3(Fuc)4 | 10445.869 | 10446.206 | 0.507 (0.209 – 0.771) | 15.9 |
| apoC-III-(Gal)5(GalNAc)3(Fuc)4 | 10769.541 | 10770.404 | 0.924 (0.593 – 1.53) | 12.2 |
Gal – Galactose; GalNAc - N-acetylgalactosamine; NeuAc - N-acetyl neuraminic acid (sialic acid); Fuc – Fucose
For apoC-I, beside the native full-length form, one additional proteoform was detected and identified as a sequence lacking two N-terminal amino acids (des-TP apoC-I; MW 6,432 Da). This proteoform was identified in all samples and is a well-known product of dipeptidylpeptidase IV (DPP-4) enzymatic cleavage [43, 44].
For apoC-II, a proteoform lacking six N-terminal amino acids (des-TQQPQQ apoC-II; MW 8,204 Da) was identified in all samples. This proteoform is termed mature apoC-II [45], and it is of low abundance (accounting for <10 % of the total apoC-II). The full-length apoC-II, also called pro-apoC-II, represents ~ 90% of the total apoC-II.
For apoC-III, we detected a total of 12 proteoforms, including the full-length native apoC-III and numerous glycosylated proteoforms. ApoC-III is highly heterogeneous protein because of the extensive protein glycosylation [34]. The most abundant proteoform is apoC-III containing one N-acetyl neuraminic acid (NeuAc; sialic acid) attached to a glycan core of one galactose (Gal) and one N-acetylgalactosamine (GalNAc): apoC-III-(Gal)1(GalNAc)1(NeuAc)1. This proteoform is commonly referred to as apoC-III1 (one sialic acid) in the literature, with the other two most abundant forms being apoC-III-(Gal)1(GalNAc)1 and apoC-III-(Gal)1(GalNAc)1(NeuAc)2, commonly referred to as apoC-III0 (no sialic acid) and apoCIII2 (two sialic acids), respectively [32].
3.2 Quantitative MSIA analysis of apoC-I, apoC-II, apoC-III and their proteoforms
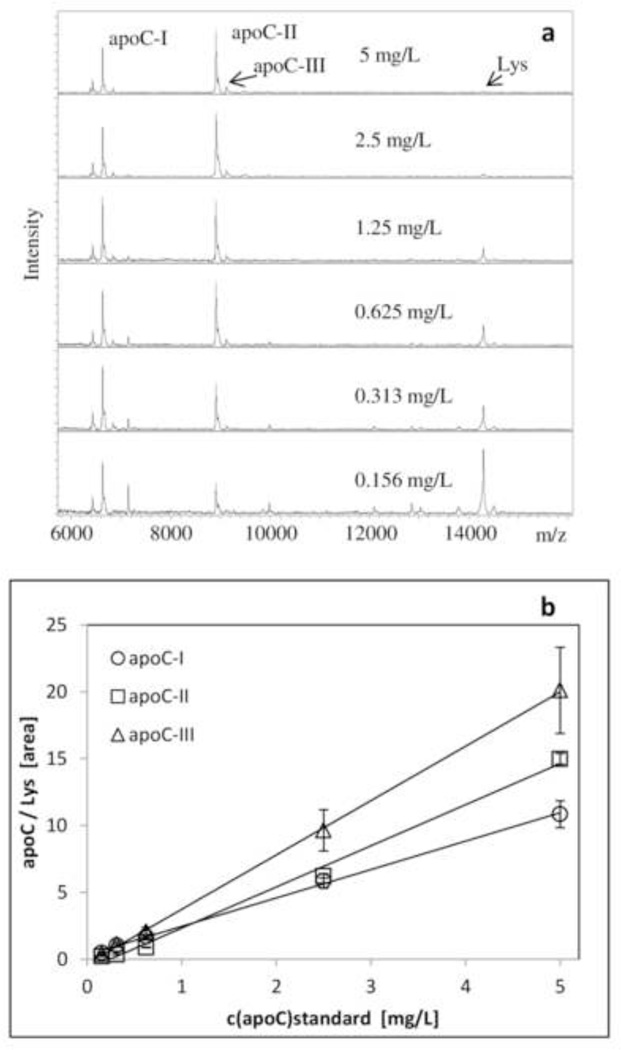
Quantification of the apoCs in this multiplex MSIA was achieved through the generation of three standard curves (one each for apoC-I, apoC-II and apoC-III), utilizing the corresponding protein standards. Because all three apolipoproteins were analyzed in the same assay, a single, exogenous protein (chicken-egg lysozyme) was added in the analytical samples as an internal reference standard (IRS) for quantification. The choice of IRS was critical step in the method development, because it had to satisfy all the necessary requirements in these “fit-for-purpose” approaches [46]. Chicken-egg lysozyme, a non-human protein, was a good choice for an IRS because it is not present in human plasma (therefore, avoiding cross-reactivity), and its signal in the mass spectrum (at m/z = 14,305 Da) is in close proximity to the signals from the apoCs (thus ensuring similar MS instrument settings use). The IRS was added in constant concentration in all analytical samples (standards and plasma), and produced a constant signal in the mass spectra.
The physiological concentrations of the three apoCs is in the mg/L range, with apoC-III having the highest range (80 – 150 mg/L), and apoC-I and apoC-II exhibiting similar concentration ranges (40 – 60 mg/L and 30 – 50 mg/L, respectively). The three standard curves, therefore were generated with an identical concentration range (0.156 – 5 mg/L) that was sufficient for quantitation of all the proteoforms (the plasma samples were diluted 40-fold). Examples of the standard curves, along with the corresponding mass spectra are presented in Fig. 2. The response for all standard curves is linear across the entire range (r2 = 0.9988, SEE = 0.0928 for apoC-I; r2 = 0.9940, SEE = 0.0262 for apoC-II; and r2 = 0.9996, SEE = 0.1605 for apoC-III). A control sample with known apoC-I, apoC-II and apoC-III concentrations (1 mg/L each) was analyzed in triplicate each day in order to assess the inter-day variability, and presented CVs of 8.82 % (apoC-I), 7.88 % (apoC-II) and 10.1 % (apoC-III).Linearity of the assay was evaluated by analyzing several admixtures of plasma samples with various concentrations of the apoCs, yielding a recovery of 89–124%.
Figure 2.
Quantitative apoC-I, apoC-II and apoC-III mass spectrometric immunoassay (MSIA). a) Representative MSIA mass spectra for the six standard apoCs samples containing lysozyme as an IRS; b) Representative standard curves generated with the multiplex apoCs MSIA, with an r2= 0.9988 and SEE = 0.0928 for apoC-I; r2 = 0.9940 and SEE = 0.0262 for apoC-II; and r2 = 0.9996 and SEE = 0.1605 for apoC-III.
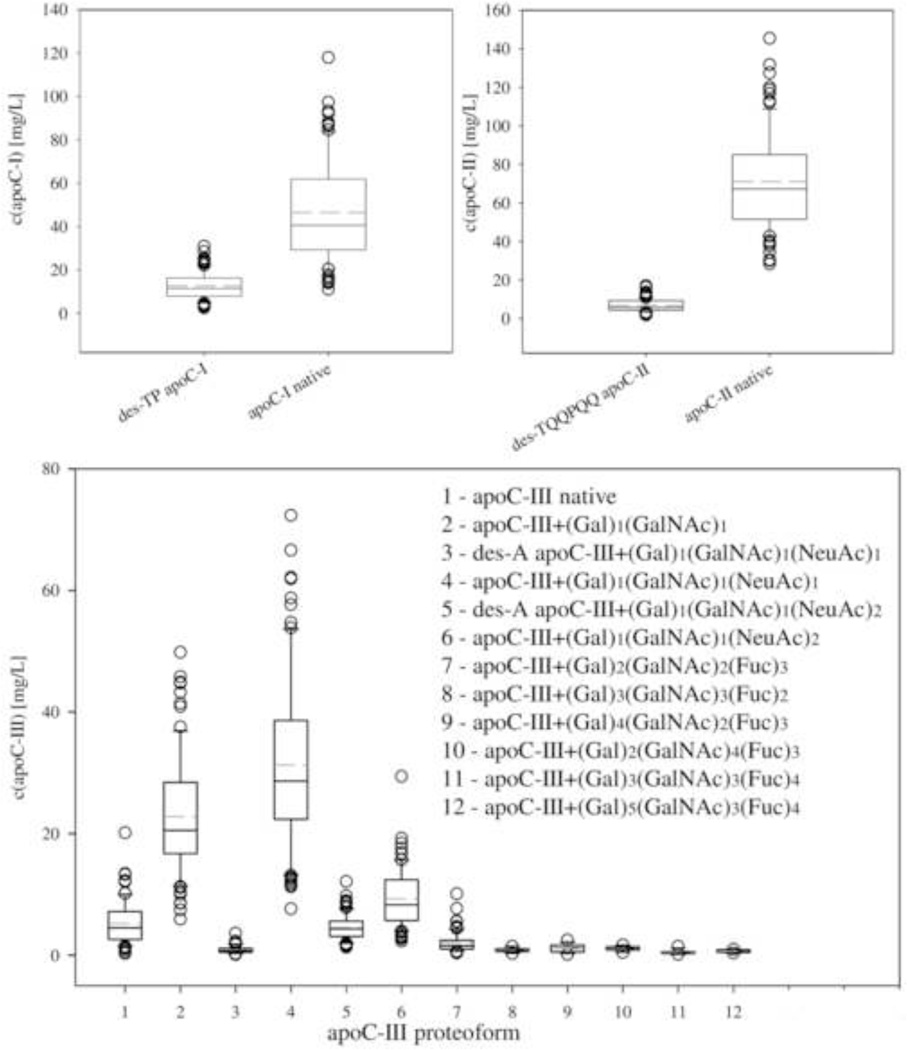
Using these standard curves we determined the concentrations of all apoCs and their proteoforms in the 82 human plasma samples (Fig. 3). We first determined the protein/lysozyme peak areas ratios for each proteoform, summed up the ratios of all proteoforms for an individual protein, determined the total individual protein concentration using the corresponding standard curve, and then determined the concentration of each proteoform based on its percentage of the total specific apoC concentration. This approach worked especially well for those proteoforms whose proteoform/lysozyme peak ratios were below those of the standard curve. The total concentrations were in the range of 13.8 to 148 mg/L for apoC-I (mean = 59.2 mg/L), 30.2 to 161 mg/L for apoC-II (77.8 mg/L) and 20.4 to 188 mg/L for apoC-III (76.7 mg/L), which is within the range of normal concentrations as determined by conventional immunoassays. The concentration ranges for the individual proteoforms are shown in Fig. 3.
Figure 3.
Individual concentrations of apolipoprotein C-I, C-II and C-III and their proteoforms in a cohort of 82 human EDTA-plasma samples. Box - 25–75th percentile; Solid line – median concentration; Short dash line – mean concentration; Error bars – 10th and 90th percentile; Symbols – outlying points.
4. Discussion
Delineating protein heterogeneity is an important undertaking because of its implications in physiological and pathological processes. The MSIA methodology can unambiguously distinguish between different proteoforms in a single assay. The approach is time efficient (~40 min) and high-throughput, allowing us to study the range of apoCs proteoforms from a small cohort of 82 human plasma samples in a single run on the automated 96-channel pipettor.
The apoC-I des-TP proteoform, a product of a DPP-4 enzymatic cleavage, was identified in all samples. Analyzing this proteoform is significant for several reasons. First, the des-TP apoC-I proteoform accounts for approximately a quarter of the total apoC-I in normal plasma (Table 1), making a significant contribution to the overall apoC-I concentration. Second, this proteoforms could potentially be used as a marker of DPP-4 inhibitor drugs activity. DPP-4 inhibitor drugs, such as vildagliptin, sitagliptin and sexagliptin, are prescribed for the treatment of type 2 diabetes (T2D) for controlling the HbA1C levels and preservation of the progressive loss of insulin-secreting capacity [47, 48]. DPP-4 cleaved apoC-I can therefore be a useful marker for the effect of DPP-4 inhibitor drugs and monitoring of therapy.
ApoC-II is a relatively homogenous protein exhibiting just two proteoforms: pro-apoC-II (the dominant proteoform), and mature apoC-II - a proteoform resulting from the removal of an N-terminal hexapeptide (TQQPQQ). ApoC-II has been primarily associated with activation of the lipoprotein lipase (LPL), and promoting triglyceride hydrolysis in lipid particles [24]. Deficiency of apoC-II was shown to cause hypertriglyceridemia, which can be reversed by infusion with normal plasma or synthetic apoC-II in affected individuals [49]. The MSIA approach measures both proteoforms - the pro- and mature apoC-II, and can be used for monitoring and predicting the kinetics of maturation of this apolipoprotein in vivo.
ApoC-III heterogeneity is a result of its glycosylation and presence of O-linked glycan motifs w/o and w/ one and/or two sialic acid molecules. These glycosylated forms (termed apoC-III0, apoC-III1 and apoC-III2) derive from glycosylation on threonine 74 (T74) in the amino acid sequence, and account for ~ 90 % of the total apoC-III concentration (Table 1). Our results indicate that apoC-III exhibits additional proteoforms: the low abundance C-terminal alanine cleaved proteoforms (present in majority of the samples, at 1 to 5 %), and also glycosylated proteoforms containing fucose in the glycan motif. The presence of fucosylated apoC-III has initially been detected by Balog et al., in relation to Schistosoma mansoni infection [50]. Nicolardi et al. confirmed the presence of fucosylated apoC-III proteoforms in their work [33], and went a step further in analyzing these glycan structures using MALDI-FTICR MS [51]. Their approach, however, is time consuming, and requires sample pretreatment using magnetic beads-based solid-phase extraction prior to MALDI and ESI-FTICR MS analysis. In our work we were able to apply the MSIA approach and detect these low abundance apoC-III proteoforms in a single run alongside the other apoCs, therefore significantly reducing the time and cost of the analysis. The concentrations of the fucosylated apoC-III proteoforms were determined to be in the very low mg/L range (Fig. 3). With the exception of apoC-III-(Gal)2(GalNAc)2(Fuc)3, they were detected in less than 20 % of samples. Finally, the distribution of the apoC-III proteoforms was noticeably different among the samples, even in samples with similar total apoC-III concentrations. This was especially evident for the fucosylated proteoforms of apoC-III, suggesting that fucosylation follows different pathways. Additional studies are needed in order to better understand these differences in the context of the physiological significance. Gaining an insight into the structural heterogeneity of apoC-III will certainly contribute to new findings and correlations, especially when assessed together with apoC-I and apoC-II, as done here with the multiplex MSIA.
5. Conclusion
The multiplex apoCs assay is based on the mass spectrometric immunoassay (MSIA) methodology. The assay’s two step approach is similar to that of well-established enzymatic immunoassays, with the secondary step of MALDI-MS detection an enabling factor in differentiating between multiple proteoforms. The quantification aspect further strengthens the methodology because the concentration of each proteoforms can be determined individually. The multiplex quantitative MSIA is a unique and high-throughput method for measuring the three apoproteins C and their proteoforms. The ability to detect and quantify all proteoforms in a single assay represents a significant step forward toward better understanding of their physiological roles in health and disease.
Highlights.
Proteoforms play important role in physiological and pathological processes
Top-down MS-based approaches are ideally suited for proteoforms detection
We developed multiplex MS- immunoassay for apoC-I, C-II and C-III and proteoforms
The multiplexed assay was fast (40 min) and high-throughput (96 samples at once)
The concentrations of each individual proteoform were determined and compared
ApoC-III proteoform distribution varied among samples with similar concentrations
Acknowledgements
The authors would like to acknowledge Dr. Tai E Shyong and Dr. Kao Shih Ling from National University Hospital, Singapore for providing us with the human plasma samples used in this analysis. This work was supported in part by Award Numbers R01DK082542 and R24DK090958 from the National Institute of Diabetes and Digestive and Kidney Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith LM, Kelleher NL C.f.T.D. Proteomics. Nat Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson RW, Krone JR, Bieber AL, Williams P. Anal Chem. 1995;67:1153–1158. doi: 10.1021/ac00103a003. [DOI] [PubMed] [Google Scholar]
- 3.Nelson RW, Borges CR. J Am Soc Mass Spectrom. 2011;22:960–968. doi: 10.1007/s13361-011-0094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Issaq HJ, Veenstra TD, Conrads TP, Felschow D. Biochem Biophys Res Commun. 2002;292:587–592. doi: 10.1006/bbrc.2002.6678. [DOI] [PubMed] [Google Scholar]
- 5.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 6.Razavi M, Frick LE, LaMarr WA, Pope ME, Miller CA, Anderson NL, Pearson TW. J Proteome Res. 2012;11:5642–5649. doi: 10.1021/pr300652v. [DOI] [PubMed] [Google Scholar]
- 7.Reid JD, Holmes DT, Mason DR, Shah B, Borchers CH. J Am Soc Mass Spectrom. 2010;21:1680–1686. doi: 10.1016/j.jasms.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 8.Mann M. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 9.Messana I, Cabras T, Iavarone F, Vincenzoni F, Urbani A, Castagnola M. J Sep Sci. 2013;36:128–139. doi: 10.1002/jssc.201200830. [DOI] [PubMed] [Google Scholar]
- 10.Weiß F, van den Berg BH, Planatscher H, Pynn CJ, Joos TO, Poetz O. Biochim Biophys Acta. 2014;1844:927–932. doi: 10.1016/j.bbapap.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Nedelkov D, Tubbs KA, Niederkofler EE, Kiernan UA, Nelson RW. Anal. Chem. 2004;76:1733–1737. doi: 10.1021/ac035105+. [DOI] [PubMed] [Google Scholar]
- 12.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Proc Natl Acad Sci U S A. 2005;102:10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nedelkov D, Phillips DA, Tubbs KA, Nelson RW. Mol Cell Proteomics. 2007;6:1183–1187. doi: 10.1074/mcp.M700023-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Trenchevska O, Phillips DA, Nelson RW, Nedelkov D. PLoS One. 2014;9:e100713. doi: 10.1371/journal.pone.0100713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oran PE, Trenchevska O, Nedelkov D, Borges CR, Schaab MR, Rehder DS, Jarvis JW, Sherma ND, Shen L, Krastins B, Lopez MF, Schwenke DC, Reaven PD, Nelson RW. PLoS One. 2014;9:e92801. doi: 10.1371/journal.pone.0092801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niederkofler EE, Phillips DA, Krastins B, Kulasingam V, Kiernan UA, Tubbs KA, Peterman SM, Prakash A, Diamandis EP, Lopez MF, Nedelkov D. PLoS One. 2013;8:e81125. doi: 10.1371/journal.pone.0081125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trenchevska O, Kamcheva E, Nedelkov D. J Proteome Res. 2010;9:5969–5973. doi: 10.1021/pr1007587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trenchevska O, Nedelkov D. Proteome Sci. 2011;9:19. doi: 10.1186/1477-5956-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trenchevska O, Kamcheva E, Nedelkov D. Proteomics. 2011;11:3633–3641. doi: 10.1002/pmic.201100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiernan UA, Phillips DA, Trenchevska O, Nedelkov D. PLoS One. 2011;6:e17282. doi: 10.1371/journal.pone.0017282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherma ND, Borges CR, Trenchevska O, Jarvis JW, Rehder DS, Oran PE, Nelson RW, Nedelkov D. Proteome Sci. 2014;12:52. doi: 10.1186/s12953-014-0052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiernan UA, Nedelkov D, Nelson RW. J Proteome Res. 2006;5:2928–2934. doi: 10.1021/pr060062+. [DOI] [PubMed] [Google Scholar]
- 23.Kiernan UA, Addobbati R, Nedelkov D, Nelson RW. J Proteome Res. 2006;5:1682–1687. doi: 10.1021/pr0601133. [DOI] [PubMed] [Google Scholar]
- 24.Jong MC, Hofker MH, Havekes LM. Arterioscler Thromb Vasc Biol. 1999;19:472–484. doi: 10.1161/01.atv.19.3.472. [DOI] [PubMed] [Google Scholar]
- 25.Kei AA, Filippatos TD, Tsimihodimos V, Elisaf MS. Metabolism. 2012;61:906–921. doi: 10.1016/j.metabol.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Zheng C, Khoo C, Furtado J, Sacks FM. Circulation. 2010;121:1722–1734. doi: 10.1161/CIRCULATIONAHA.109.875807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hegele RA, Connelly PW, Hanley AJG, Sun F, Harris SB, Zinman B. Arteriosclerosis, Thrombosis, and Vascular Biology. 1997;17:2753–2758. doi: 10.1161/01.atv.17.11.2753. [DOI] [PubMed] [Google Scholar]
- 28.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. New England Journal of Medicine. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 29.Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO, Lange LA, Lu Y, Tang ZZ, Zhang H, Hindy G, Masca N, Stirrups K, Kanoni S, Do R, Jun G, Hu Y, Kang HM, Xue C, Goel A, Farrall M, Duga S, Merlini PA, Asselta R, Girelli D, Olivieri O, Martinelli N, Yin W, Reilly D, Speliotes E, Fox CS, Hveem K, Holmen OL, Nikpay M, Farlow DN, Assimes TL, Franceschini N, Robinson J, North KE, Martin LW, DePristo M, Gupta N, Escher SA, Jansson JH, Van Zuydam N, Palmer CN, Wareham N, Koch W, Meitinger T, Peters A, Lieb W, Erbel R, Konig IR, Kruppa J, Degenhardt F, Gottesman O, Bottinger EP, O'Donnell CJ, Psaty BM, Ballantyne CM, Abecasis G, Ordovas JM, Melander O, Watkins H, Orho-Melander M, Ardissino D, Loos RJ, McPherson R, Willer CJ, Erdmann J, Hall AS, Samani NJ, Deloukas P, Schunkert H, Wilson JG, Kooperberg C, Rich SS, Tracy RP, Lin DY, Altshuler D, Gabriel S, Nickerson DA, Jarvik GP, Cupples LA, Reiner AP, Boerwinkle E, Kathiresan S N.H. TG and HDL Working Group of the Exome Sequencing Project, L.ng, and Blood Institute. N Engl J Med. 2014;371:22–31. [Google Scholar]
- 30.Huff MW, Hegele RA. Circ Res. 2013;112:1405–1408. doi: 10.1161/CIRCRESAHA.113.301464. [DOI] [PubMed] [Google Scholar]
- 31.Graham MJ, Lee RG, Bell TA, Fu W, Mullick AE, Alexander VJ, Singleton W, Viney N, Geary R, Su J, Baker BF, Burkey J, Crooke ST, Crooke RM. Circ Res. 2013;112:1479–1490. doi: 10.1161/CIRCRESAHA.111.300367. [DOI] [PubMed] [Google Scholar]
- 32.Bondarenko PV, Cockrill SL, Watkins LK, Cruzado ID, Macfarlane RD. J Lipid Res. 1999;40:543–555. [PubMed] [Google Scholar]
- 33.Nicolardi S, van der Burgt YE, Wuhrer M, Deelder AM. Proteomics. 2013;13:992–1001. doi: 10.1002/pmic.201200293. [DOI] [PubMed] [Google Scholar]
- 34.Wada Y, Kadoya M, Okamoto N. Glycobiology. 2012;22:1140–1144. doi: 10.1093/glycob/cws086. [DOI] [PubMed] [Google Scholar]
- 35.Flood-Nichols SK, Tinnemore D, Wingerd MA, Abu-Alya AI, Napolitano PG, Stallings JD, Ippolito DL. Mol Cell Proteomics. 2013;12:55–64. doi: 10.1074/mcp.M112.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezeli M, Végvári Á, Donnarumma F, Gidlöf O, Smith JG, Erlinge D, Marko-Varga G. J Proteomics. 2013;87:16–25. doi: 10.1016/j.jprot.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Jian W, Edom RW, Wang D, Weng N, Zhang SW. Anal Chem. 2013;85:2867–2874. doi: 10.1021/ac3034757. [DOI] [PubMed] [Google Scholar]
- 38.Kingsmore SF. Nat Rev Drug Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterman S, Niederkofler EE, Phillips DA, Krastins B, Kiernan UA, Tubbs KA, Nedelkov D, Prakash A, Vogelsang MS, Schoeder T, Couchman L, Taylor DR, Moniz CF, Vadali G, Byram G, Lopez MF. Proteomics. 2014;14:1445–1456. doi: 10.1002/pmic.201300300. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, Sabatine MS, Gerszten RE, Carr SA. Clin Chem. 2009;55:1108–1117. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chapman JD, Goodlett DR, Masselon CD. Mass Spectrom Rev. 2014;33:452–470. doi: 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- 43.Augustyns K, Bal G, Thonus G, Belyaev A, Zhang XM, Bollaert W, Lambeir AM, Durinx C, Goossens F, Haemers A. Curr Med Chem. 1999;6:311–327. [PubMed] [Google Scholar]
- 44.Lambeir AM, Durinx C, Scharpé S, De Meester I. Crit Rev Clin Lab Sci. 2003;40:209–294. doi: 10.1080/713609354. [DOI] [PubMed] [Google Scholar]
- 45.Fojo SS, Taam L, Fairwell T, Ronan R, Bishop C, Meng MS, Hoeg JM, Sprecher DL, Brewer HB. J Biol Chem. 1986;261:9591–9594. [PubMed] [Google Scholar]
- 46.Carr SA, Abbatiello SE, Ackermann BL, Borchers C, Domon B, Deutsch EW, Grant RP, Hoofnagle AN, Hüttenhain R, Koomen JM, Liebler DC, Liu T, MacLean B, Mani DR, Mansfield E, Neubert H, Paulovich AG, Reiter L, Vitek O, Aebersold R, Anderson L, Bethem R, Blonder J, Boja E, Botelho J, Boyne M, Bradshaw RA, Burlingame AL, Chan D, Keshishian H, Kuhn E, Kinsinger C, Lee JS, Lee SW, Moritz R, Oses-Prieto J, Rifai N, Ritchie J, Rodriguez H, Srinivas PR, Townsend RR, Van Eyk J, Whiteley G, Wiita A, Weintraub S. Mol Cell Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pratley RE, Salsali A. Curr Med Res Opin. 2007;23:919–931. doi: 10.1185/030079906x162746. [DOI] [PubMed] [Google Scholar]
- 48.Barnett A. Int J Clin Pract. 2006;60:1454–1470. doi: 10.1111/j.1742-1241.2006.01178.x. [DOI] [PubMed] [Google Scholar]
- 49.Goldberg IJ, Scheraldi CA, Yacoub LK, Saxena U, Bisgaier CL. J Biol Chem. 1990;265:4266–4272. [PubMed] [Google Scholar]
- 50.Balog CI, Mayboroda OA, Wuhrer M, Hokke CH, Deelder AM, Hensbergen PJ. Mol Cell Proteomics. 2010;9:667–681. doi: 10.1074/mcp.M900537-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicolardi S, van der Burgt YE, Dragan I, Hensbergen PJ, Deelder AM. J Proteome Res. 2013;12:2260–2268. doi: 10.1021/pr400136p. [DOI] [PubMed] [Google Scholar]