Abstract
In Staphylococcus aureus, a T-box riboswitch exists upstream of the glyS gene to regulate transcription of the sole glycyl-tRNA synthetase, which aminoacylates five tRNAGly isoacceptors bearing GCC or UCC anticodons. Subsequently, the glycylated tRNAs serve as substrates for decoding glycine codons during translation, and also as glycine donors for exoribosomal synthesis of pentaglycine peptides during cell wall formation. Probing of the predicted T-box structure revealed a long stem I, lacking features previously described for similar T-boxes. Moreover, the antiterminator stem includes a 42-nt long intervening sequence, which is staphylococci-specific. Finally, the terminator conformation adopts a rigid two-stem structure, where the intervening sequence forms the first stem followed by the second stem, which includes the more conserved residues. Interestingly, all five tRNAGly isoacceptors interact with S. aureus glyS T-box with different binding affinities and they all induce transcription readthrough at different levels. The ability of both GCC and UCC anticodons to interact with the specifier loop indicates ambiguity during the specifier triplet reading, similar to the unconventional reading of glycine codons during protein synthesis. The S. aureus glyS T-box structure is consistent with the recent crystallographic and NMR studies, despite apparent differences, and highlights the phylogenetic variability of T-boxes when studied in a genome-dependent context. Our data suggest that the S. aureus glyS T-box exhibits differential tRNA selectivity, which possibly contributes toward the regulation and synchronization of ribosomal and exoribosomal peptide synthesis, two essential but metabolically unrelated pathways.
Keywords: T-box riboswitch, tRNA, transcription regulation, glycyl-tRNA synthetase, Staphylococcus aureus
INTRODUCTION
Riboswitches are important bacterial regulatory RNAs that modulate either transcription elongation or translation initiation in bacteria (Winkler and Breaker 2005). Ιn order to act as modulators of gene expression, riboswitches exhibit a certain plasticity that depends on several factors. The majority of riboswitches bind rather small molecule ligands, such as amino acids (aa) or nucleotides (nt), through characteristic conserved motifs of their aptamer domain (Breaker 2011; Mellin and Cossart 2015). The aptamer domain in turn transmits the conformational changes induced by ligand binding to the expression platform, thus affecting gene expression (Deigan and Ferré-D'Amaré 2011). Due to its central role in bacterial metabolism, riboswitches are considered as a novel class of molecular targets for the development of specific antibacterial drugs (Blount and Breaker 2006; Breaker 2012; Jentzsch and Hines 2012).
T-box riboswitches represent a special class of riboswitches that modulate gene expression through binding of tRNAs. They control transcription of downstream genes that either encode proteins involved in biosynthesis and transport of aa or aminoacyl-tRNA synthetases (aaRSs) which supply ribosomal protein synthesis with the aminoacylated tRNAs (aa-tRNAs) substrates (Henkin 2008, 2014). Although their main regulatory mechanism is transcriptional attenuation, recently it was shown that the ileS T-box in Actinobacteria acts via sequestration of the Shine–Dalgarno sequence, thus modulating ribosome binding and translation initiation (Sherwood et al. 2015).
The stabilization of the T-box:tRNA complex depends on contacts between two distinct but variably distant domains that are important for the correct orientation of the tRNA (Vitreschak et al. 2008; Gutiérrez-Preciado et al. 2009). The first domain of a T-box (stem I) contains the specifier loop (SL) that varies in size and contains a codon-like nucleotide triplet. The SL is responsible for recognition of the bound tRNA through “scanning” of its anticodon sequence by Watson–Crick base-pairing with the tRNA anticodon (Grigg et al. 2013; Zhang and Ferré-D'Amaré 2013). At the beginning of the stem I duplex, a kink turn (or K-turn) is formed. This motif is a common structural feature for many functional RNA molecules (Winkler et al. 2001; Schroeder et al. 2010; Wang and Nikonowicz 2011). Upon binding of uncharged cognate tRNA, the T-box adopts a characteristic conformation (termed antiterminator) that allows transcription elongation of the downstream gene by RNA polymerase. Since their discovery, the few characterized T-boxes were considered of single-specificity for the tRNA ligand mainly because of the existence of a single codon-like triplet in the specifier loop (Grundy and Henkin 1993; Green et al. 2010). However, it was shown recently in Clostridium acetobutylicum that T-box riboswitches of dual specificity also exist in a genome-dependent context (Saad et al. 2012). Moreover, T-boxes displaying SLs with relaxed codon specificity are more widespread than previously thought. It has been proposed that the specificity of the T-box for tRNA recognition depends both on the number of the nucleotides forming the SL and most importantly, on the downstream genes, which can be organized in operons and may control more than one metabolic pathway (Saad et al. 2013).
The second essential domain that contains the conserved T-box signature sequence is the terminator/antiterminator stem. Due to its conservation, it is easily recognizable by the available bioinformatics tools and plays a role in the recognition and binding of tRNA's discriminator base and universal 3′CCA end (Vitreschak et al. 2008; Wels et al. 2008; Chang and Nikonowicz 2013). This interaction is equally critical for the correct positioning of tRNAs and moreover, it is important for sensing uncharged or charged tRNAs (Zhang and Ferré-D'Amaré 2014). Once bound to the SL, the uncharged tRNA is available to interact through its 3′CCA to the T-box bulge (Gerdeman et al. 2003; Yousef et al. 2005; Fauzi et al. 2009; Grigg and Ke 2013b). Given all the structural requirements, the mRNA adopts an overall rather simple conformation for completion of gene transcription (Grundy et al. 2002b; Grigg and Ke 2013a). Stem I and the antiterminator stem are connected through an inter-stem linker of variable length and role (Rollins et al. 1997; Grundy et al. 2002a; Grundy and Henkin 2004). Using this elegant system, bacteria (mainly gram positive) can control their metabolic rate under various environmental conditions (i.e., under amino acid starvation). On the other hand, stimulation of transcription of the gene under control is achieved only upon binding of cognate uncharged tRNAs to the specifier loop of the T-box, or via conformational changes which affect translational initiation (Sherwood et al. 2015). By using the actual carriers of amino acids as ligands, bacteria can modulate the levels of their growth depending on the availability of amino acids as essential growth ingredients (Smith et al. 2010; Raina and Ibba 2014).
Recent structural studies, focused mainly on stem I, have provided a more detailed illustration of the conformation changes that occur during the interaction of the T-box with the tRNA (Chang and Nikonowicz 2013; Grigg et al. 2013; Grigg and Ke 2013b; Zhang and Ferré-D'Amaré 2013). In addition, the important role of tRNA's elbow for efficient interaction also became evident (Lehmann et al. 2013). Finally, T-boxes can sense and discriminate the volume of charged and uncharged tRNAs in an EF-Tu-independent manner (Zhang and Ferré-D'Amaré 2014). It must be noted that all the above-mentioned interactions occur in the absence of protein factors (Grigg and Ke 2013a; Zhang and Ferré-D'Amaré 2014).
The first T-boxes were initially discovered and characterized, based on the observation that the mRNAs of many aminoacyl-tRNA synthetases (aaRSs) have a very long 5′ leader region, which exhibits conservation in the T-box region (Green et al. 2010). This domain is quite flexible and it has been proposed to facilitate interaction with tRNA via an “induced fit” model (Zhang and Ferré-D'Amaré 2013). Although bioinformatics tools are able to identify numerous T-boxes, the secondary structures from only a few species were experimentally verified (Vitreschak et al. 2008; Gutiérrez-Preciado et al. 2009; Chang and Nikonowicz 2013; Sun and Rodionov 2014). Despite the recent progress (Miao et al. 2015), it is even more difficult to reliably predict the tertiary structure of T-boxes. A more detailed look at the available genomes indicates that there is an extensive variability in T-boxes’ structural features among species that merits further investigation (Vitreschak et al. 2008; Wels et al. 2008; Gutiérrez-Preciado et al. 2009; Sun and Rodionov 2014). Most recently, the description of a riboswitch, which controls the tyrS paralog tyrZ, indicated that variability and differences among T-boxes might reflect evolutionary events which favored the conservation of structural elements that could vary among species for the same type of T-box (Williams-Wagner et al. 2015). Various detailed bioinformatics analyses have proposed that there is species-specific occurrence of various T-boxes, raising questions on the evolution and propagation of T-boxes as regulatory elements. Therefore, a closer and more detailed look on the relationship between specific structural features of T-boxes and the metabolic pathways that they control may exist and could provide further insights.
In Staphylococcus aureus, a sole glyS gene (encoding an α2 type glycyl-tRNA synthetase; GlyRS) and five tRNAGly genes exist bearing one GCC and four UCC anticodons (Giannouli et al. 2009). All genes serve the uninterrupted supply of glycine for incorporation into nascent polypeptides during ribosomal translation as well as during synthesis of the bacterial cell wall. The S. aureus cell wall contains characteristic pentaglycine bridges that stabilize peptidoglycan chain interconnection (Schneider et al. 2004). They also represent an example of exoribosomal peptide synthesis catalyzed by a family of three nonribosomal peptidyl-transferases known as Fem factors (Factors Essential for Methicillin Resistance; FemX, A and B) (Rohrer and Berger-Bachi 2003; Hübscher et al. 2007; Giannouli et al. 2010). As has been shown previously, the five tRNAGly isoacceptors might serve as either proteinogenic or nonproteinogenic carriers of glycine for the two distinct and metabolically unrelated pathways (Giannouli et al. 2009). This role of each tRNA could be assigned by the strength of binding affinity to EF-Tu. In addition, it is known that the tRNAs that participate as carriers of amino acids for cell wall synthesis exhibit idiosyncratic features (Villet et al. 2007; Shepherd and Ibba 2015). Previous bioinformatics analyses indicated that a T-box riboswitch most likely precedes and possibly regulates the sole glyS gene in S. aureus and other staphylococci (Vitreschak et al. 2008; Wels et al. 2008; Gutiérrez-Preciado et al. 2009). However, the sequence corresponding to the overall T-box riboswitch appeared longer and only the T-box sequence shared an adequate degree of conservation compared with previously characterized glyQS T-boxes from Bacillus subtilis, Geobacillus kaustophilus, and Oceanobacillus iheyensis.
To investigate the role of the S. aureus glyS T-box, we cloned it and determined its secondary structure. Our results revealed essential differences in stem I, as well as in the antiterminator domain structure, which is constituted of two stems instead of one. Despite the observed differences, a detailed analysis of the T-box:tRNA interaction showed that the S. aureus T-box binds its tRNA ligand using essentially the same mode that has been described previously, and satisfies both the proposed “two-ruler” and the induced fit models. Moreover, the S. aureus glyS T-box interacts with all five tRNAGly isoacceptors, albeit with different affinities, revealing an intriguing ambiguity for the reading of the SL codon by the tRNA anticodon. Overall, the S. aureus glyS T-box represents the longest riboswitch of each kind that has been studied so far, and we propose that it is able to regulate binding and transcription readthrough by utilizing all five glycine isoacceptors, independently of their anticodon (GCC or UCC). This is the first example of an SL codon-reading ambiguity of tRNAs that synchronizes the regulation of two different cellular processes: the protein and the cell wall synthesis.
RESULTS
Identification of the glyS leader sequence and transcription initiation point
In the current work, we investigated the existence of a functional T-box riboswitch in the 5′UTR of the mRNA encoding for the sole α2-type GlyRS in S. aureus. Although previous bioinformatics analyses predicted the existence of such a T-box, they were based only on the recognition of the highly conserved T-box bulge sequence (Vitreschak et al. 2008; Wels et al. 2008; Sherwood et al. 2015). In the case of the S. aureus glyS T-box, it was evident that stem I with the exception of the SL GGC codon did not exhibit the same conservation with other characterized T-boxes based on comparison with glyQS T-boxes from B. subtilis, O. iheyensis, and G. kaustophilus that have been structurally characterized (Fig. 1). Analysis using the RegPrecise 3.0 database indicated that the S. aureus glyS T-box exhibited a very low score of conservation (12.9) compared with the other T-boxes which exhibit significantly higher scores (above 60) (Novichkov et al. 2013). To clarify whether the ∼340-nt upstream of the glyS start codon region was part of its 5′UTR, we tested its expression under minimal growth conditions (M9 mineral salt medium supplemented with amino acids, see Materials and Methods). The primers that were used were designed to anneal at the predicted 5′UTR region upstream of the glyS gene. When glycine was omitted from the medium, the region upstream of glyS showed elevated expression (Fig. 2A, upper panel, left lane), which was attributed to glycine deprivation. When glycine concentration was restored in the medium (Fig. 2A, upper panel, right lane), the levels of expression were reduced. The sequence of the glyS 5′UTR was confirmed (Fig. 2A, lower panel) and the promoter region was identified (nucleotides −10 and −35, Fig. 2B). Both the bioinformatics prediction and the experimental verification of the T-box sequence revealed that although the SL loop/codon region and the conserved T-box sequence can be traced, the remaining predicted structure was significantly divergent (Fig. 1).
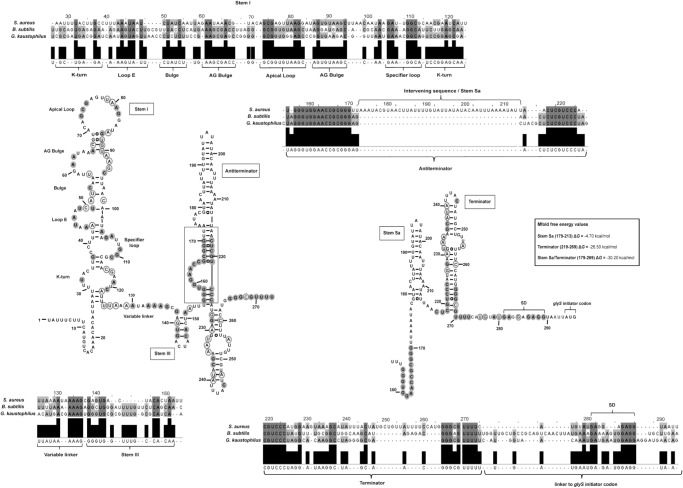
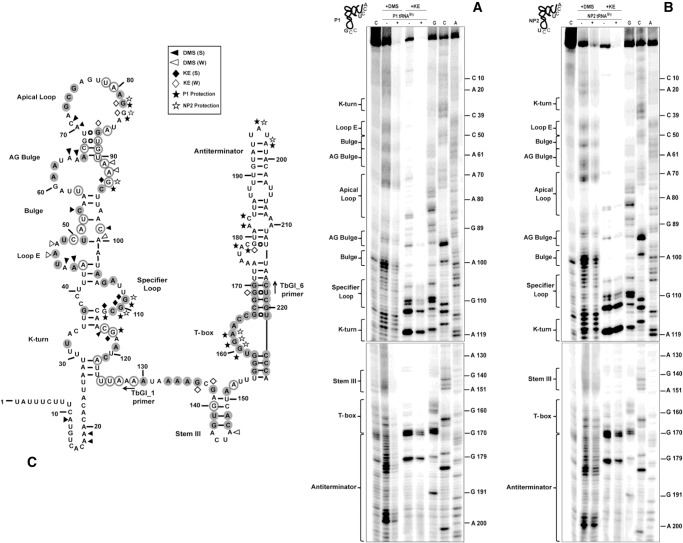
FIGURE 1.
Proposed S. aureus glyS T-box secondary structure based on in silico and biochemical analysis. For the alignment, highly conserved (100%) nucleotides are indicated in dark gray filled circles and moderately conserved (66%) appear in light gray open circles. Alignment numbering corresponds to S. aureus glyS leader sequence. Values corresponding to the thermodynamic stability of the terminator region were calculated according to Mfold free energy prediction tool and appear in a box for each structural element. Annotation numbers: SA1394 S. aureus N315; BSU25270 B. subtilis 168; GK3430 G. kaustophilus HTA426.
FIGURE 2.
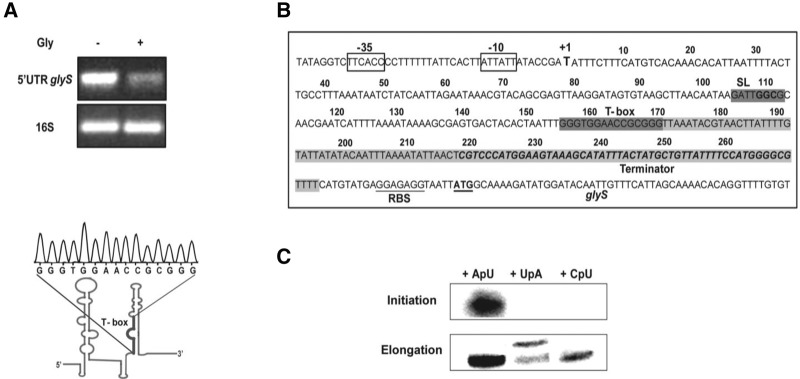
The S. aureus glyS mRNA leader includes a T-box transcription regulator. (A) (Upper panel) glyS leader mRNA in vivo transcription levels under different nutrition conditions during growth. 16S rRNA was used as control. (Lower panel) T-box sequence confirmation of the amplified bands. (B) The region upstream of glyS gene in S. aureus. Boxed regions indicate putative −35 and −10 promoter sites and the transcription start is indicated as +1. The predicted specifier loop (SL) and T-box regions have a dark gray background. In italics and gray is the potential Ρ-independent transcription terminator region; the ribosome binding site (RBS) and glyS start codon (ATG) are underlined. (C) Dinucleotide-primed initiation of transcription. The upper panel indicates the initiation reaction and the lower panel the elongation reaction. ApU, UpA, and CpU correspond to dinucleotides that were used for each reaction.
The transcription starting point (Fig. 2B, nucleotide +1) was determined based on bioinformatic tools that predicted as possible starting points positions U1, A2, and U7 (Tanabe and Kanehisa 2012). The transcription start was determined by dinucleotide-primed transcription initiation reactions using three different dinucleotides, ApU, UpA, and CpU (Samuels et al. 1984) (see Materials and Methods). Initiation reactions were performed by omitting the G nucleotide, which was used subsequently at the elongation phase of the reaction. Transcription initiation could be induced only in the presence of ApU (Fig. 2C) without excluding the possibility of transcription initiation at positions +2 and +3. Bioinformatics analysis using the BPROM online tool (Solovyev and Salamov 2011) in Softberry database was also used for the same prediction (data not shown). The length of the glyS upstream region is 294 nt (until the AUG initiator codon), and the full-length T-box leader according to sequence alignments is 225 nt.
Prediction of the glyS T-box secondary structure indicates unusual structural features
The putative secondary structure of the S. aureus glyS T-box was predicted based on bioinformatic analysis and subsequent thermodynamic analysis of the secondary structure stability using the Mfold web server (Zuker 2003). Prediction of the secondary structure was also based in a multiple sequence alignment of stem I, specifier loop, and terminator/antiterminator stem (Fig. 1). The predicted S. aureus glyS T-box structure forms a long riboswitch with the following unusual structural features: (i) the K-turn motif (also known as GA motif) exhibits very low conservation, (ii) the loop E is quite conserved but displaced and not opposite to the SL loop as in the case of B. subtilis, (iii) the apical (distal) loop of stem I, although adequately conserved, contains six additional nucleotides, and (iv) the SL loop belongs to the 8-nt-long class, and includes the GGC triplet that is recognized by the GCC anticodon of the sole tRNAGly(GCC) ligand. The SL codon is followed by the conserved purine which is a guanosine (instead of adenosine in B. subtilis and G. kaustophilus). As has been described for the B. subtilis glyQS T-box, the characteristic stem II and pseudoknot stem IIA/B are also absent from S. aureus glyS T-box (Henkin 2014). It has been proposed that the absence of stem II and pseudoknot stem IIA/B indicates that no additional trans-acting factors mediate the binding of the T-box with the tRNAGly (Grundy and Henkin 2004; Vitreschak et al. 2008). Similarly, it contains a very short variable linker and the stem III is predicted shorter compared with that from B. subtilis (4 bp instead of 7). Surprisingly, the S. aureus glyS T-box seems to form a very long antitermination stem that includes an intervening sequence of 42 additional nucleotides (nt 173–214, Fig. 1). This unusually long antiterminator conformation is similar to that of B. subtilis only in the bottom half (Fig. 1A, boxed region), and contains the 7-nt T-box bulge including the UGGA conserved sequence that interacts with the UCCA 3′ end of tRNAGly. This additional sequence forms almost exclusively the first stem of the terminator structure (Fig. 1B). This domain (termed stem Sa) is followed by the second and more conventional terminator stem that, despite a quite adequate conservation, is also 17-nt longer between positions 246 and 264. All the predictions for the possible folding of individual regions that we examined suggested an overall T-box riboswitch structure which is appropriate for tRNA binding and includes the unusual two-stem terminator structure (Fig. 1B) which is restricted in staphylococci (Supplemental Fig. 1). The two-stem terminator conformation facilitates sequestration of the conserved T-box sequence and possibly confers a stronger terminator signal during transcription. To verify the S. aureus glyS T-box predicted structure we prepared three constructs of different length. The first construct included the whole sequence upstream of the glyS gene (position +1 to position +275, termed T275), the second included essentially the full-length T-box (position +10 to position +225, termed T225), and the third construct was a truncated version of stem I without the K-turn (position +34 to position +115, termed T115). All the constructs were used for subsequent experimentation.
Structural analysis reveals a divergent stem I
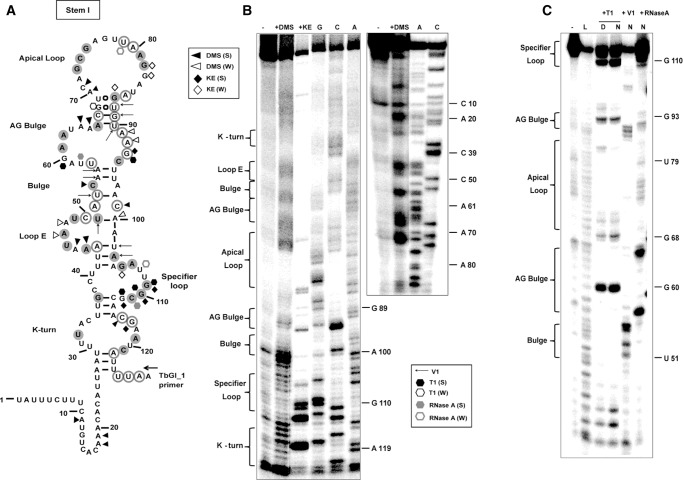
In order to get insights on the actual structural features of the individual regions from the predicted model, we performed chemical and enzymatic probing of stem I (Fig. 3). Stem I is well-structured in the absence of tRNA and therefore initial analysis was performed in the absence of the cognate tRNAGly(GCC) (Wang et al. 2010; Wang and Nikonowicz 2011). The construct T275, which includes the T-box leader and the long antiterminator stem with the intervening sequence, was treated with DMS (modifies A and C bases) and kethoxal (modifies G bases). Primer extension analysis was performed by using a specific primer for stem I (Fig. 3A; Supplemental Table 1). Analysis of the structural probing that we performed (Fig. 3B) confirmed the predicted structural features. More specifically, the T-box includes a broader K-turn region with only a few of the conserved nucleotides that have been reported previously for this motif (Figs. 1, 3A, filled gray circles). Although the K-turn region is not typical, it appears protected from chemical modification with the exception of C116 and G117 (Wang and Nikonowicz 2011; Zhang and Ferré-D'Amaré 2013). As a result, the necessary bend of K-turn into the helical path can be formed with the nucleotides that constitute the region (i.e., U32 and A33 with A119 and U120). In addition, the K-turn does not contain a typical GA motif as for the majority of the organisms belonging to the Bacillus–Clostridium group (Winkler et al. 2001).
FIGURE 3.
(A) Chemical and enzymatic probing analysis of stem I region. TbGl_1 primer was used for Stem I primer extension analysis. Highly conserved (100%) and moderately conserved (66%) nucleotides are indicated with gray filled or open circles, respectively. (B) Chemical modification; (S) corresponds to strong and (W) to weak DMS or KE base modification. (C) Enzymatic probing analysis; (D) indicates denaturant conditions and (N) native conditions; (L) corresponds to the ladder constructed by alkaline hydrolysis reaction. (S) indicates strong and (W) weak susceptibility to cleavage by T1 RNase or RNase A. Arrows correspond to V1 RNase cleavage sites.
Another striking difference is the displaced E-loop, starting at positions A44 and A45, which appear exposed and more modification-prone than A47 and A48. Loop E is an important structural element, responsible for the S-turn formation, which preserves the helical form of the riboswitch and exposes the SL for interaction with the tRNA's anticodon. In the case of S. aureus glyS T-box, the S-turn can be formed via interactions with A101 that was found protected by DMS modification. Most of the E-loop region includes conserved nucleotides (A44, U46, A47, and U49), followed by a short bulge, which contains a conserved C54. Stem I structure continues with a wider AG bulge, which contains a 9-nt loop instead of 8 nt in Bacilli. The AG bulge is resistant to endonucleolytic cleavage, something that we also observed. In addition, it contains the conserved nucleotides A61, A62, and A66. The A62, which in our case appears protected from either chemical modification or enzymatic digestion, could potentially interact with A64 or A65, since the conserved A66 was found to base pair with U90 in S. aureus glyS T-box, as reported before (Vitreschak et al. 2008; Zhang and Ferré-D'Amaré 2013). This interaction is essential in order to bring the AG bulge in proximity to the distal (apical) loop (A72–A76), as was also observed in a previous study, in the absence of tRNA.
The distal loop was also found broader, spanning from A70 till A86 (17 nt) compared with the 11-nt apical loop in Bacilli, and exhibits different arrangement in sequence although it includes the conserved nucleotides G73, C74, G75, and A81 and G82. Nucleotide G82 was the only one found susceptible to modification. This observation suggests that the apical loop is able to form long-range interactions with the AG bulge and eventually shape the necessary conformation to interact with tRNA's D- and T-loop. Moreover, both primer extension analysis and enzymatic probing using the T115 variant (Fig. 3C) indicated that (i) a possible interaction can be formed between G77 and A81, and (ii) the region A72–A81 is inaccessible to modifying agents and enzymatic cleavage, indicating that this region is protected more likely because of a possible interaction with the AG bulge. This observation is in agreement with the available structural data and in addition, both the observed differences in the AG bulge and the distal loop support the previously described ambiguity over the exact interactions that take place in the upper part of stem I with the tRNA ligand (Vitreschak et al. 2008; Grigg and Ke 2013b; Zhang and Ferré-D'Amaré 2013). Our analysis showed that the SL loop belongs to the 8-nt loop T-box class. This type of T-box has been described to exhibit dual-specificity in a genome-dependent context depending on the genes that regulate. In the case of the S. aureus glyS T-box, the downstream sequence encodes only a single gene and not an operon (as in the case of the NT-box from C. acetobutylicum), although a second codon sequence can be identified for tRNATrp(CCA) (U108, G109, G110). Therefore, the possibility of dual-specificity for the S. aureus glyS T-box cannot be excluded and remains to be investigated in future experiments. The specifier loop contains the conserved nucleotides G105 and A106 and follows the pattern of previously characterized T-boxes (Yousef et al. 2003). It also contains the G109, G110, and C111 codon sequence corresponding to the anticodon tRNAGly(GCC) isoacceptor (termed P1 tRNA, Supplemental Fig. 3). The codon sequence is followed by the conserved purine (G112), which is characteristic of all T-boxes, and is crucial for the interaction with the anticodon loop of the tRNA.
The terminator region forms an unusual two-stem structure
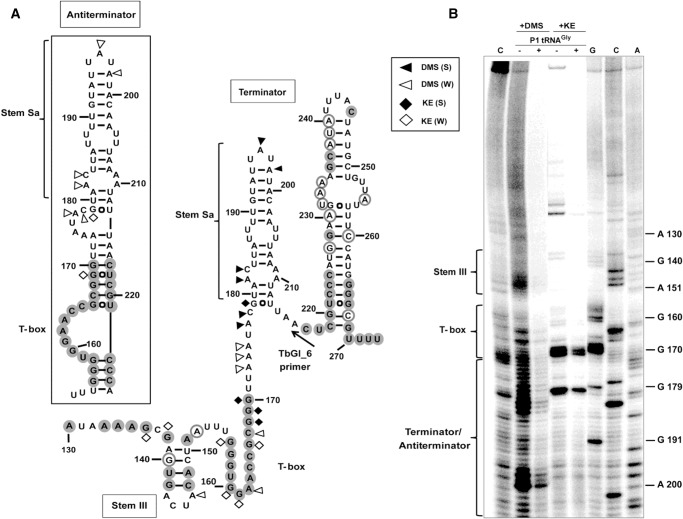
The S. aureus glyS T-box antiterminator region was predicted taking into account the unusual 42-nt intervening sequence that exists between the conserved T-box sequence and the well-conserved terminator sequence that follows. This intervening sequence forms an additional stem (termed stem Sa) in addition to the terminator stem (Fig. 4A). Attempts to successfully map both stems with one primer that was designed to anneal at the 3′ end of the terminator stem were unsuccessful, possibly due to the complexity of the region. After modification with DMS and kethoxal, followed by primer extension using a specific primer designed for the short inter-stem region between stem Sa and the terminator stem, we verified our model. In the absence of tRNA, the intervening sequence forms a very rigid stem which exhibits limited accessibility to extensive modification. Using the same line of experiments we also confirmed the secondary structure of part of the inter-stem sequence between stem I and the antiterminator region, including stem III (Fig. 4). A two-stem terminator region is reported for the first time for a glyS T-box and it seems to be specific for glyS T-boxes in staphylococci (Supplemental Fig. 1). Attempts to identify sequences exhibiting homology with stem Sa throughout bacterial genomes did not return any results, indicating the species-specific nature of this sequence.
FIGURE 4.
(A) Predicted secondary structure of terminator/antiterminator region. Highly conserved (100%) and moderately conserved (66%) nucleotides are indicated with gray filled or open circles, respectively. (B) Chemical modification; (S) and (W) indicate strong or weak base modifications by DMS or KE; (C) corresponds to control reaction performed in absence of modification reagent. Analysis of reaction products in the presence of P1 tRNAGly is representative of the antiterminator/terminator structure rearrangement. TbGl_6 primer was used for terminator/antiterminator primer extension analysis.
To verify the structure of the antiterminator conformation upon binding of the cognate tRNA, we performed chemical probing and primer extension analysis also in the presence of P1 tRNAGly(GCC). The predicted model was again confirmed by the structural probing showing that the antiterminator region shapes an unusually long stem, compared with previously characterized T-boxes (Fig. 4). Moreover, nucleotides between A173–C183 clearly participate in a conformational change. This observation is supportive of a more extensive involvement of the intervening sequence in interactions that stabilize the structure of the T-box upon tRNA binding. However, such interactions were difficult to detect because of the increased flexibility of the region.
Binding of all tRNAGly isoacceptors to the T-box induces transcription readthrough
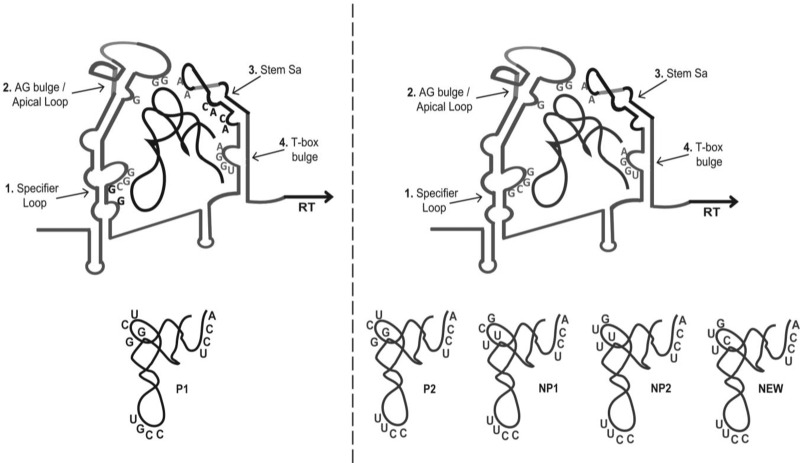
All the available S. aureus genomes encode for five tRNAGly isoacceptors. It has been previously proposed that the five tRNAGly molecules can potentially serve different roles during the S. aureus lifecycle based on their affinity for binding to the homologous EF-Tu. Two of them, termed P1 and P2 tRNAs (Supplemental Fig. 2; GCC and UCC anticodons respectively), can predominantly serve as glycine carriers for incorporation into nascent proteins during ribosomal protein synthesis. The remaining three molecules, termed NP1, NP2, and NEW (bearing UCC anticodons), potentially escape protein synthesis after glycylation as judged by their weak binding to EF-Tu. In turn, they probably serve as substrates for the formation of the characteristic pentaglycine interconnecting peptides that stabilize the staphylococcal cell wall, a process essential for cell viability (Rohrer and Berger-Bachi 2003; Schneider et al. 2004; Hübscher et al. 2007; Giannouli et al. 2009, 2010).
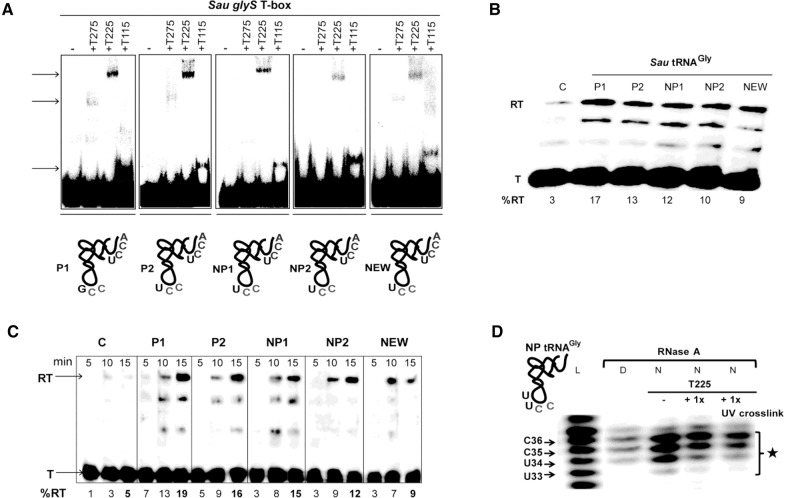
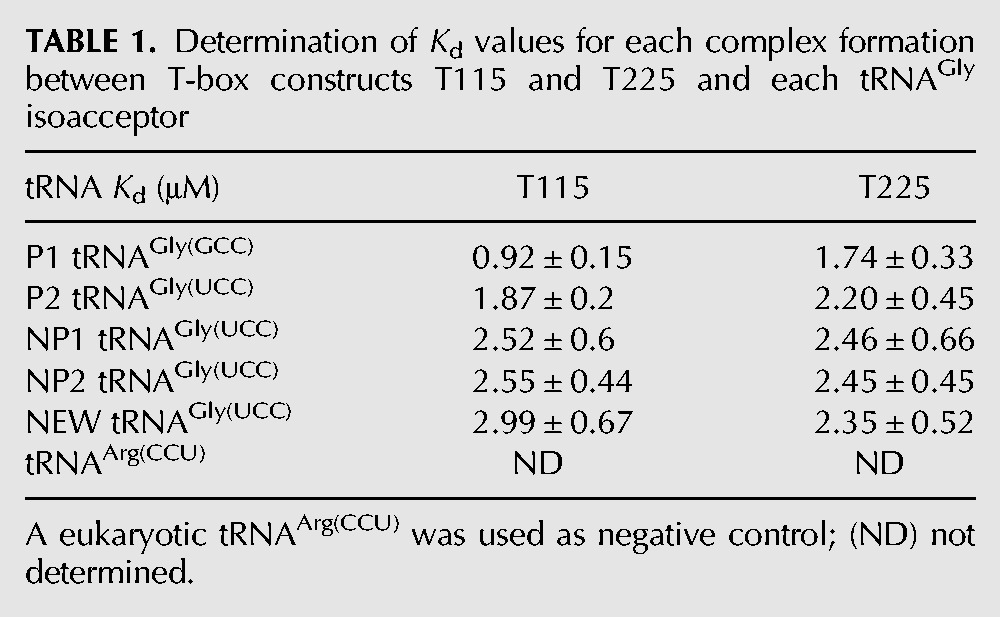
The only tRNAGly that fulfills the SL codon:tRNA anticodon-pairing requirements is P1, bearing a GCC anticodon that matches the SL GGC codon. So far, all the previous studies that have been performed to characterize the SL GGC codon:tRNA anticodon specificity examined interactions only with the cognate tRNAGly(GCC). Both proteinogenic tRNAGly include the D-loop nucleotides G18 and G19 and the T-loop nucleotides U55 and C56 that are considered important for the tRNAs’ tertiary folding (Supplemental Fig. 2). Previous reports have demonstrated that those nucleotides play a role in the interaction between the T-box and the tRNA ligand, thus affecting the RNA polymerase readthrough (Yousef et al. 2005). On the other hand, the three remaining nonproteinogenic tRNAs do not include these important nucleotides (Supplemental Fig. 2). Instead, they include either U18 and U19 or C18 and U19. Therefore, we decided to test all five tRNAs for binding to the T-box constructs, and on top we wanted to examine whether some of them (with the exception of P1) induce transcription readthrough. To our surprise, all five tRNAs could bind on the T-box constructs (T275, T225, and T115) albeit with different affinities. The T275 proved the less strong binder to probe this interaction, possibly due to the high complexity of its tertiary structure. The band shift corresponding to the T-box:tRNA complex was very weak and the corresponding Kd values could not be determined (Fig. 5A), despite the fact that several conditions were tested. However, when construct T225 (which includes essentially the full-length T-box leader) was used, the T-box:tRNA complex binding was enhanced and measured byEMSA gels. Similar results were observed also for construct T115, which includes stem I without the K-turn. The calculated Kd values (Table 1; Supplemental Fig. 3) indicate that the P1 isoacceptor was indeed the best ligand for stem I. However, the four remaining tRNAs also form complexes with T-box constructs. Interestingly, when we used the T225 to perform the same line of experiments, we noticed that the Kd for P1 tRNA was lower and comparable to those of nonproteinogenic tRNAs (Table 1; Supplemental Fig. 3). This observation indicates that in the presence of stem I, P1 tRNA binding appeared stronger, possibly due to the full complementarity of the SL:anticodon base-pairing. However, the binding of the full-length T-box with the tRNAs is probably stabilized by additional contact points beyond the anticodon reading that are most likely provided by stem Sa.
FIGURE 5.
(A) EMSA analysis of T-box:tRNA complex formation between T275, T225, and T115 constructs and either proteinogenic or nonproteinogenic tRNAGly isoacceptors. (B) In vitro tRNA-directed antitermination (readthrough) assay and (C) transcription elongation time plot using all five tRNAGly isoacceptors [P1(GCC), P2(UCC), NP1(UCC), NP2(UCC), NEW(UCC)]; lane C corresponds to transcription reaction in the presence of a eukaryotic tRNAArg(CCU) precursor used as negative control. (D) Enzymatic probing of the NEW tRNAGly:T225 complex. The region illustrated corresponds to anticodon loop sequence, and RNase A cleavage was performed either under denaturing (lane D) or native (lane N) conditions, with or without UV crosslink treatment; lane L indicates alkaline hydrolysis products (ladder).
TABLE 1.
Determination of Kd values for each complex formation between T-box constructs T115 and T225 and each tRNAGly isoacceptor
To further elucidate whether this kind of interaction could affect transcription readthrough, we used a 408-nt transcript that includes the T-box leader together with the endogenous promoter and part of the glyS encoded sequence (60 nt). Interestingly, all tRNAGly isoacceptors adequately induce time-dependent readthrough, independently of their anticodon sequence (Fig. 5B,C). In an attempt to probe whether the overall tRNA anticodon stacking is affected due to the interaction with the full-length T-box we performed enzymatic probing of the bound or unbound status of a nonproteinogenic tRNA, using also UV crosslinking to enhance the strength of binding. We noticed that upon formation of the complex, the protection of U34 but also U33 are enhanced possibly due to base stacking of the flexible anticodon loop. The flexibility of the anticodon is attributed to the absence of a typical U-turn in all the tRNAs under study (Fig. 5D; Chang and Nikonowicz 2012). The protection that we detected in combination with the lower affinity of P1 in the presence of the full-length T-box implies that a more extensive rearrangement must exist. As a result, both the anticodon and the upper part of the tRNA are affected and base stacking in the presence of the antiterminator occurred. Such an induced fit accommodates all the tRNAs (as we confirmed) and structural specificity can be achieved possibly through additional “sealing” by stem Sa (Supplemental Figs. 4B, 7). In the case where T115 (stem I) is used, the T-box achieves higher specificity for P1 exclusively through the SL:anticodon complementarity.
The very strong termination signal that was observed suggests that the S. aureus glyS T-box could behave in a different manner compared with its previously characterized counterpart from B. subtilis. This observation, in combination with the unusual and very rigid two-stem terminator conformation may further explain the lower readthrough bands that were observed. Finally, although it is known that K-turn contributes in general to the overall structure stabilization, tRNA orientation and transcription readthrough, it seems that its absence from T115 does not significantly affect tRNA binding. This observation is in agreement with data describing that K-turn mutations have milder effects for the B. subtilis glyQS T-box and do not affect tRNA binding capacity. Strong binding of all tRNAs to construct T115 despite the absence of one of the two crucial G–A pairs is in agreement with the suggestion that K-turn functions as an anchor of the stem I position relative to the antitermination stem (Winkler et al. 2001; Vitreschak et al. 2008; Wang and Nikonowicz 2011).
tRNAGly binding induces structural changes on S. aureus glyS T-box
The observation that all tRNAGly isoacceptors efficiently bind to the T-box and induce transcription readthrough, prompted us to examine possible differences in the structural level. Using the same methodology, we examined the changes in the pattern of the protected bases upon binding of P1 tRNAGly, which exhibits the highest binding affinity, and NP2 tRNAGly, which was chosen as representative of the nonproteinogenic tRNAs bearing the UCC anticodon. Although the mode of binding of both tRNAs is similar (Fig. 6A,B), a closer look revealed interesting differences. Upon binding of either P1 or NP2 tRNA it was obvious that only positions G109 and G110 of the SL codon were strongly protected. Instead, the third position of the SL codon (C111) remained accessible, at least in part, an observation that was quite intriguing and raised questions, given the specificity of the interaction between SL and both tRNAs. A previous study investigating the interaction of a tyrS mRNA leader in which the UAC specifier triplet was replaced by the glycyl GGC failed to detect binding when tRNAGly(UCC) was used (Chang and Nikonowicz 2013). This binding inability is very likely observed due to the nature of the hybrid T-box sequence that was tested and also possibly due to different conformational changes that were induced. Therefore, the observation that the native full-length S. aureus glyS T-box interacts mainly with the first two SL codon nucleotides (G109 and G110) with C35 and C36 of both tRNAs was intriguing. An equally interesting observation was the protection of position G112 in the SL loop. This position was protected by both tRNAs (albeit lower in NP2) and could indicate a wobble-pairing with U33. This interaction possibly facilitates the anticodon reading and also contributes to the orientation and binding of all tRNAs, as has been also suggested by the recent structures (Chang and Nikonowicz 2013; Grigg et al. 2013; Grigg and Ke 2013b; Zhang and Ferré-D'Amaré 2013). Since all the tRNAs contain anticodon loops with characteristic flexibility, it is possible that this interaction may be needed to stabilize the local structure, which in turn favors an induced fit (Chang and Nikonowicz 2013; Zhang and Ferré-D'Amaré 2015).
FIGURE 6.
Chemical probing of the T275:tRNAGly complex using a (A) proteinogenic (P1) and a (B) nonproteinogenic (NP2) tRNAGly; (S) indicates strong and (W) weak base modifications by DMS or KE; (C) indicates the unmodified control reaction. Primers TbGL_1 and TbGl_6 (Supplemental Table 1) were used for stem I and antiterminator region analysis, respectively. (C) Predicted antiterminator structure and base modification protection by tRNAGly binding. Filled black stars correspond to specific protection by P1 tRNAGly(GCC) and open stars to protection by NP2 tRNAGly(UCC). Highly conserved (100%) and moderately conserved (66%) nucleotides are indicated with gray filled or open circles, respectively.
Binding of both tRNAs protects positions G82 and G83 of the apical loop and G92 (Fig. 6C). The observed protections indicate possible interaction with the D- and T-loops of the tRNA as has been previously described, although with some differences (Vitreschak et al. 2008; Zhang and Ferré-D'Amaré 2013). However, it must be noted that G19 of the P1 tRNA is not present in the NP2 tRNA (replaced by U19) and therefore the protection is supportive of a nonspecific interaction due to the tRNA's induced fit. This observation is also in line with the recent structural analyses that showed extensive distortion of tRNA upon association with stem I, a bending of T-box and finally an induced fit of the tRNA. According to the two-checkpoint mechanism, the bound tRNA is first checked through the SL:anticodon interaction and in a next step the length of the anticodon arm is measured to secure the correct accommodation of the ligand. These extensive structural rearrangements seem to be very similar among the tRNAGly isoacceptors that exhibit almost identical antitermination strengths, suggesting that the discrete structural differences among tRNAs might mediate the interactions.
On the other side of the T-box leader, the antiterminator stem is also stabilized by both tRNAs. Extensive protection is observed as previously reported in the conserved G160, G161, and A162. A very interesting observation was the protection of A196 and A198 of the Sa loop (Fig. 6C). However, the interaction of the antiterminator stem with P1 tRNA revealed additional positions that are protected (A177 and C178 as well as A182 and C183). This observation implies that the higher affinity of P1 tRNA compared with the other tRNAs could be attributed in part to those additional contacts that facilitate tRNA binding stabilization and are probably part of the overall induced fit that occurs. The strength of the binding might be enhanced by an additional bend and turn of the stem Sa (positions A177–U213, Fig. 7). Such a conformational change facilitates a more stable fit and at the same time could waive the strong termination signal, due to the long intervening sequence. The observation of additional contact points between P1 tRNA and the stem Sa are further supported by the fact that P1 tRNA has been considered a molecule with enhanced flexibility under various conditions, a feature that has not been observed for the nonproteinogenic tRNAs (Chang and Nikonowicz 2013). However, the exact role of stem Sa and its role in the induced fit mechanism require further experimentation.
FIGURE 7.
Schematic representation of the proposed S. aureus glyS T-box:tRNAGly recognition model. (Left panel) S. aureus glyS T-box structural rearrangement upon P1 tRNAGly GCC isoacceptor binding. Nucleotides in black that are presented on T-box structure indicate additional interactions with P1 isoacceptor, according to the data presented herein. (Right panel) The corresponding T-box structural rearrangement upon binding of the remaining tRNAGly UCC isoacceptors. Nucleotides in gray represent common interaction with either GCC or UCC bearing tRNAs. Nucleotides on tRNAs correspond to major sequence differences among isoacceptors (Supplemental Fig. 2), namely, positions 18, 19, 33–36, 55, 56, 73–76.
DISCUSSION
T-box riboswitches represent a very elegant example of gene expression regulation at the transcriptional level. They give bacteria the ability to sense amino acid concentrations in various environmental niches (Winkler and Breaker 2005). Their regulatory function relies on specific structural features of the 5′UTR of mRNAs and also on several elements on tRNAs including the overall volumetric shape of the molecule (Grundy et al. 2005; Yousef et al. 2005; Zhang and Ferré-D'Amaré 2014). Due to the essential dynamics that are established during this interaction, it has been proposed that tRNA is an integral part of the transcription termination and through contact points as well as favorable thermodynamic associations, the T-box senses the aminoacylation status of the tRNA (Zhang and Ferré-D'Amaré 2015). The synchronization of pathways that are activated by cellular demands (i.e., for the utilization of a specific amino acid) must be precise and could involve additional reactants that have not yet been identified. All the recent biochemical and structural data suggest that species-specific variations of this theme must exist, and can probably serve specific metabolic needs. Most interestingly, it was recently shown that in Actinobacteria, the ileS T-boxes lack either partially or completely the stem I but can still specifically bind tRNAIle. However, those T-boxes act as sequesters of the Shine–Dalgarno sequence and thus regulate gene expression at the level of translation and not exclusively via transcriptional attenuation.
The few glyQS T-boxes that have been extensively characterized so far derive only from Bacilli species. Therefore, studies on the same type of regulons from different species could provide further essential information, at both the biochemical and phylogenetic levels (Supplemental Fig. 4). In the present study, we cloned and characterized the glyS T-box riboswitch from S. aureus. This specific T-box deviates from the usual structural pattern that has been described for other glyQS T-boxes. The glyS T-box presented herein precedes the sole α2 type GlyRS and shows remarkable conservation in all staphylococci (Supplemental Fig. 1). Stem I contains an atypical K-turn region with limited conservation. It should be mentioned that the K-turn region has been proposed to serve as a hub region for proteins that may facilitate the overall riboswitch conformation. However, although the K-turn is required in vivo, its distortion or absence does not essentially affect the tRNA binding ability of stem I in vitro, as we also observed in our EMSA experiments (Winkler et al. 2001; Grigg et al. 2013; Zhang and Ferré-D'Amaré 2013). The most striking differences are the lack of a characteristic AG bulge (forming a K-turn in many RNAs), the displaced E-loop, the wider distal loop (or apical loop), and an 8 nt SL, which includes the GGC triplet. Most interestingly, the domain that switches from the terminator to the antiterminator conformation contains a 42-nt long intervening sequence, which is glycine-specific in all known staphylococcal genomes. Extensive chemical and enzymatic probing analyses showed that this additional sequence (termed stem Sa) contributes to the formation of a two-stem rigid terminator conformation, which is unusual and possibly confers a more strict regulation of transcriptional readthrough by RNA polymerase. Moreover, it is involved in a local conformational rearrangement through a helix turn, which leads to interaction of specific nucleotides during antitermination upon tRNA binding (Fig. 7). This interaction is possibly used for discrimination, based on differences of the T-stem or the acceptor stem of the tRNAs. These differences may confer a way to enhance both sequence and structural specificity. Overall, the divergent structural features of the S. aureus glyS T-box may be the reason why the available bioinformatics tools failed to provide a high score for the putative regulon, although its presence had been previously predicted (Vitreschak et al. 2008; Gutiérrez-Preciado et al. 2009).
Based on the above observations and taking into account the recent notions that the observed riboswitch variation seems to be related to different metabolic profiles among bacterial species, we examined the ability of the T-box to induce transcriptional antitermination in vitro. In S. aureus, transcriptional regulation of the GlyRS affects both ribosomal and exoribosomal glycine incorporation into nascent polypeptides. This elegant regulation potentially involves all five tRNAGly isoacceptors and most probably depends on the synchronization of the two metabolically unrelated pathways, since both need glycine. Interestingly, all the tRNAGly molecules induced transcriptional readthrough, independently of the anticodon type (GCC or UCC). This intriguing observation was also supported by the probing data and the determination of the Kd values for tRNAGly binding. Both primer extension and kinetic analyses suggested that at least in vitro, the S. aureus glyS T-box interacts with all the available tRNAGly isoacceptors.
Among the most intriguing results was the fact that both proteinogenic and nonproteinogenic tRNAs interact with the T-box and induce antitermination. The interaction of the SL with the anticodon appears stronger between the two first bases of the SL codon (G109, G110) and the two last bases of the tRNA anticodon, indicating an ambiguity during the SL codon reading by the tRNA's anticodon. A similar observation has been reported for the lysK (encoding a Class I LysRS) T-box from B. cereus (SL, AAA), which responds to both tRNALys(UUU) and tRNAAsn(GUU) from the same organism (Foy et al. 2010). An additional interesting protection involves position G112, which represents a conserved purine, next to the cytidine that possibly interacts with G34 (in P1 and all the tRNAGly(GCC) isoacceptors, Supplemental Figs. 2, 7) as has been shown in the available structural analyses. In the case of the S. aureus T-box, the flexibility of the anticodon loop is possibly transduced and/or enhanced by additional contacts promoted most likely by stem Sa. Moreover, existing studies on the tRNA features that are important for interaction with the T-box were focused on tRNAGly bearing GCC but not UCC anticodons, which, in the case of S. aureus, correspond to all the nonproteinogenic tRNAs (Yousef et al. 2003, 2005; Grundy et al. 2005; Zhang and Ferré-D'Amaré 2015). In addition, the existence of GGA SL triplets in some glyQS T-boxes, as has been shown by bioinformatics prediction, suggests that in other organisms binding of tRNAGly(UCC) and possible contribution to the regulation of glyQS transcription attenuation must exist and cannot be excluded (Supplemental Figs. 2, 4; Chang and Nikonowicz 2012). Our study provides evidence that in the case of the S. aureus glyS T-box, this regulation occurs in the context of the GGC SL triplet. Moreover, an overall synchronization of gene regulation can involve interactions with additional features of the T-box, like the stem Sa.
The unconventional reading of the GGC codons for glycine is not unusual and has also been observed during protein synthesis. It is known that glycine is a member of a four-codon family with the first two nucleotides being guanosines matching the C35 and C36 of the anticodon. In bacteria, all four codons are read by up to three different tRNA anticodons (GCC, UCC, and CCC). In addition, some Bacillus and Staphylococcus species use two tRNAGly isoacceptors bearing UCC and GCC anticodons, where U is post-transcriptionally modified. Modifications of U34 can lead to opposite functional effects, such as enhancement of the ability of U to wobble pair or restriction of wobble pairing and enhancement of discrimination. Moreover, in Mycoplasma mycoides, as well as in mitochondria and chloroplasts, the tRNAGly(UCC) recognizes all four codons with equal efficiency. Therefore, the ambiguity that is observed in the S. aureus glyS T-box SL codon reading can be attributed to this intrinsic idiosyncrasy of the tRNAGly isoacceptors (Samuelsson et al. 1983; Chang and Nikonowicz 2012). However, it is still unknown whether this ambiguity plays a significant role in the cellular environment. Based on previous reports and using data from the MODOMICS database (http://modomics.genesilico.pl/sequences/list/), it is evident that the ability of the tRNAGly(UCC) to discriminate among the four codons might also be affected by the presence of a 5′-carboxymethylaminomethyluridine modification in the first position of the anticodon. However, although this modification has been identified in B. subtilis and increases codon discrimination, it is absent from S. epidermidis that also encodes nonproteinogenic tRNAGly molecules identical to those of S. aureus (Machnicka et al. 2013). Based on the above studies, it has been suggested that the ability of the tRNAs to discriminate codons (or in this case SL triplets) in the third position depends to a certain extent on the distribution of existing modifications at positions 34 and/or 37, the identity of positions 32 and 38, and the nature of positions 35 and 36 (Nelson et al. 2006). Therefore, it is still unclear whether yet unidentified modifications in specific positions, together with structural changes in the overall shape of the tRNA, could enhance or lower the SL:anticodon binding affinity and recognition in vitro or during the bacterial growth. When tRNA molecules were added during cotranscription reactions in order to achieve proper folding of the T-box, only minor alternative conformations were observed. This observation may also be relevant with the use of unmodified in vitro transcripts.
In conclusion, this study provides an additional example of the structural and functional diversity of T-box riboswitches. Our data, in line with the recent and earlier studies, suggest that T-box regulons adopt species-specific patterns that facilitate the overall metabolic strategy of each species. In the case of S. aureus (and most likely for other similar Staphylococcus species like S. epidermidis), the glyS T-box synchronizes two essential but metabolically unrelated pathways, namely the incorporation of glycine during ribosomal protein synthesis and for cell wall stabilization via the pentaglycine interconnecting peptides. All tRNAs participate in the regulation process and stabilize the transcriptional readthrough of GlyRS. This is probably achieved through induced fit and is enhanced by additional specificity constraints provided by additional contacts with the intervening sequence. Both pathways require all tRNAGly isoacceptors to be available and aminoacylated and moreover, all tRNAGly isoacceptors seem to synchronize this regulation through binding and interaction with the T-box riboswitch, thus exhibiting the same ambiguity as that which has been described for the glycine codon reading during protein synthesis.
MATERIALS AND METHODS
Chemicals, enzymes, plasmid vectors, and bacterial strains
All primers were synthesized by VBC biotech (Austria). All enzymes were purchased from New England Biolabs, except T7 RNA polymerase, SalI (Takara) and AMV Reverse Transcriptase (Promega). RNase T1, V1, and RNase A used for enzymatic probing were from Life Technologies. DMS and Kethoxal reagents for chemical modification experiments were from Sigma-Aldrich. Plasmid DNA was prepared using the NucleoSpin Plasmid Mini or Midi prep Kit and PCR products were purified by NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel). [γ-32P] ATP (6000 Ci/mmol) and [α-32P] UTP (800 Ci/mmol) were from PerkinElmer. Radiolabeled RNA elution was performed using mini Quick Spin RNA Columns (Roche). Gel Filtration chromatography column Superdex 200 10/300 GL was from GE Healthcare Life Sciences attached to an ÄKTA FPLC system (Amersham Biosciences). Heparin used for in vitro antitermination assays were from Sigma-Aldrich. RNA extraction and purification from S. aureus strains was performed using RiboPure-bacteria RNA Purification Kit (Life Technologies, USA). For analysis of RNA purity and quality, RNA 6000 Nano Kit was used (Agilent). RT-PCR was performed by using the Brilliant III SYBR Master Mix (Agilent). The pUC57 vector was used for T275 glyS T-box cloning (GenScript) and the pSC-A vector (StrataClone PCR Cloning Kit, Agilent) was used for cloning of T225 and T115 constructs. S. aureus strains were kindly provided by Prof. S. Pournaras (Department of Microbiology, School of Medicine, University of Athens, Greece).
RT-PCR validation of the endogenous S. aureus glyS T-box sequence
S. aureus strains were grown in M9 minimal salts medium supplemented by 12 amino acids (0.2 mg/mL L-proline; L-phenylalanine; L-tryptophan; L-histidine; L-tyrosine, 0.1 mg/mL L-glutamine; L-glutamic acid; L-cysteine; L-arginine; L-methionine; L-threonine; L-leucine) at 37°C for 16 h under shaking (250 rpm). Glycine starvation and nonstarvation conditions were performed by addition of 5 μg/mL and 100 μg/mL glycine, respectively. For lysis and total RNA extraction, 109 cells were used according to RiboPure-Bacteria Kit instruction manual (Protocol for Gram-positive bacteria). All total RNA samples were checked for their quality and possible genomic or plasmid DNA contamination (RNA 6000 Nano Kit) and were subsequently used as templates for the RT-PCR reactions using Brilliant III SYBR Master Mix. A specific primer pair was designed for both cDNA synthesis and amplification reactions (269-bp product, including the region from the transcription start to terminator region of glyS T-box leader sequence; Supplemental Table 1). An additional primer pair for Sau 16 S rRNA was used as internal control (Supplemental Table 1). Finally, RT-PCR-amplified fragments were extracted (NucleoSpin Gel and PCR Clean-up PCR Kit) and were sequenced (VBC biotech, Austria) for further validation.
In silico analysis
The region containing the T-box sequence [Score:12.9%, Position: −162] upstream of the S. aureus glyS coding region was identified using RegPrecise database prediction (Novichkov et al. 2013). The broader full-length region upstream of the S. aureus glyS gene (SA1394), which includes the T-box sequence, was further verified using the KEGG database tool (Tanabe and Kanehisa 2012) and “The T-box search pattern” (Vitreschak et al. 2008). Promoter sequence and the transcription start site were predicted using the BPROM online tool (Solovyev and Salamov 2011) in Softberry database. Thermodynamic analysis for the secondary structure stability of the antiterminator/terminator region was performed using the Mfold web server (Zuker 2003). All multiple sequence alignments were created using the T-Coffee server algorithm (Notredame et al. 2000), based on the full-length T-box riboswitch sequences or individually based on highly conserved regions of stem I, specifier loop, terminator stem, and antiterminator stem. T-box sequences used for multiple alignments were obtained from RegPrecise and were further verified using the KEGG database tools. The tRNAGly anticodon sequences in each species were obtained from the Genomic tRNA database (Schattner et al. 2005). Phylogenetic trees of T-boxes were constructed using the ClustalW2 algorithm (Larkin et al. 2007) and were visualized using Phylogeny.fr online tool (Dereeper et al. 2008).
Cloning and in vitro transcription of glyS T-box constructs and tRNAGly genes
The S. aureus glyS T-box T275 was synthesized and cloned into pUC57 vector by GenScript. Constructs T225 (10–225) and T115 (34–115) were PCR-amplified using specific primers and cloned into pSC-A vector (see Supplemental Table 1). All constructs were verified through sequencing to avoid random mutations (VBC, Austria). The tRNAGly genes encoding the five different isoacceptors (P1, P2, NP2, NP1, and NEW) were cloned in pUC18 vector (Giannouli et al. 2009). All plasmid constructs were designed with a T7 promoter leader sequence and a terminal restriction enzyme recognition site, BstNI for T275 and tRNAGly, SalI for T225 and T115 constructs. Digestion of plasmids with the appropriate restriction enzyme produced a linear template for subsequent in vitro transcription using T7 RNA polymerase. Run-off in vitro transcription reactions were carried out at 37°C, except T275, T225, and T115 (30°C), for 16 h in the presence of inorganic pyrophosphatase (8 U). After DNase I treatment and Phenol:Chloroform:Isoamyl alcohol (25:24:1) extraction, transcripts were purified on 8% PAGE 8 M urea, excised after visualization under a UV lamp and bands corresponding to the tRNAs were eluted. All the in vitro transcripts were further purified on a gel filtration column (Superdex 200 10/300 GL-ÄKTA FPLC system).
Identification of the glyS leader transcription starting point and transcription readthrough assay
In order to confirm the in silico predicted transcription starting point of T-box glyS leader RNA, we performed dinucleotide-primed initiation of transcription reactions using three different dinucleotides, ApU, UpA, and CpU (Samuels et al. 1984). Template for in vitro transcription reactions (10-ng per reaction) was a 408-bp PCR fragment, starting 48-nt upstream of the transcription start site, and extending to 354-nt downstream, which included the endogenous promoter, the full-length T-box sequence, and part of the glyS coding sequence (up to 60 nt) (Supplemental Table 1). The template was previously purified and was sequenced prior to use. Transcription termination site was predicted approximately at position +274. The assays were carried out essentially as previously described with minor modifications (Grundy et al. 2002b). Transcription initiation reactions were performed by omitting the G nucleotide, incubated at 37°C for 15 min. [α-32P] UTP (800 Ci/mmol) was used (0.25 μM) to visualize the size of T-box conformations products during transcription by 1 U of recombinant E. coli RNAP holoenzyme. The initiation step is paused with heparin (20 μg/mL) and elongation of transcription is allowed by the addition of MgCl2 (28 μΜ), KCl (86 μΜ), and all nucleotides in a final concentration of 5 μM each. Elongation reactions were carried out at 37°C for 15 min.
Each tRNAGly isoacceptor was tested for its antitermination capacity by using the same in vitro transcription protocol. Initiation reaction was carried out as described above in the presence of ApU dinucleotide, which is obtained by transcription initiation priming reactions. Before transcription elongation induction, tRNAGly transcripts were refolded (5 min at 65°C) in the presence of 1 mM MgCl2, and added in the reactions to a final concentration of 300 nM. Elongation reactions were stopped at different time points (5, 10, and 15 min). An additional elongation reaction was performed in the presence of a eukaryotic precursor tRNAArg(CCU) (from Dictyostelium discoideum), which was used as a negative control both in EMSA and readthrough assays. All transcription products were extracted and analyzed on 6% PAGE/8 M urea, after denaturation at 50°C for 5 min. Bands corresponding to termination (T) and readthrough (RT) transcripts were visualized by scanning on phosphoimager (Fujifilm FLA 3000 platform). The corresponding “band volumes” were quantified using AIDA image analyzer software (version 5.0). The (%) RT measurement was calculated according to the equation: (%) RT = [(volume RT − volume background)/(volume T + volume RT − 2 × volume background)] × 100.
Electrophoretic mobility shift assay (EMSA) and determination of dissociation constant values (Kd)
All tRNAGly molecules were radiolabeled at the 5′ end according to standard protocols. Of note, 5 pmol of each tRNA transcript was dephosphorylated at the 5′ end using 7.5 U Antarctic phosphatase and subsequently phosphorylated using 30 pmol of [γ-32P]-ATP in the presence of T4 polynucleotide kinase (10 U). Binding between each of five tRNAGly isoacceptors (P1, P2, NP2, NP1, and NEW) and glyS T-box constructs (T275, T225, T115) was analyzed using electrophoretic mobility shift assay (EMSA). The conditions for the binding assay were 10 mM Tris-HCl; pH 7.1, 100 mM KCl, and 10 mM MgCl2. Each radiolabeled tRNAGly transcript (100 nM) was simultaneously mixed with each glyS T-box construct (5 μΜ) in the reaction buffer and was denatured for 3 min at 65°C. After renaturation, reactions were incubated at 25°C for 1 h, and stopped with 8% v/v glycerol, remained on ice for at least 10 min and analyzed on a 6% v/v nondenaturing PAGE in the presence of 0.35× Tris-borate buffer supplemented with 5 mM MgCl2. Native PAGE electrophoretic conditions were 400 V for 15 min and 200 V for 3 h at 4°C. After electrophoresis, the gels were vacuum dried and autoradiographed on phosphoimager. Analysis of the complexes formation between T-box constructs T225 and T115 and each tRNAGly isoacceptor were performed using the same experimental conditions described above. Concentration of each tRNA transcript was 100 nM. T225 and T115 transcripts were used in increasing concentrations of 0.5, 1, 2, 5, 10 and 0.5, 1, 2, 3, 5 μΜ, respectively. Bound and free radiolabeled tRNAGly was visualized by phosphoimager scanning. Band intensity measurement was performed using AIDA image analyzer software (version 5.0). The measurement results were expressed as the percent integrated raw intensity of the bands corresponding to bound tRNAGly transcript (namely the amount of T225 or T115 transcripts that is complexed with each tRNAGly) as a result of one-site specific binding. GraphPad Prism Software (version 5.00) was used to plot and analyze binding affinities by nonlinear regression (amount of specific binding versus T225 or T115 increasing concentrations). The calculated values were used for the corresponding Scatchard plots (Bound/free versus Bound) to calculate binding constants (Kd) according to the equation: slope = −1/Kd. All experiments were performed in triplicates and each point value used in Kd determination was the mean of two independent measurements.
Chemical and enzymatic probing
Modifications by DMS and kethoxal (KE) were performed as previously described, with minor modifications (Saad et al. 2013). Chemical modifications were analyzed by primer extension. Two oligonucleotides were used: TbGl_1 that hybridizes at nucleotides 129–148 and TbGl_6 that hybridizes at nucleotides 216–238 of T275 glyS T-box construct (Supplemental Table 1). T275 transcript (20 pmol) alone or in combination with P1 or NP2 tRNAGly transcripts (100 and 200 pmol) were mixed in the presence of modification buffer [70 mM Hepes-KOH, pH 7.8, 10 mM Mg(OAc)2, and 270 mM KOAc] and denatured at 60°C for 10 min, followed by slow cooling in water bath until the mixture reaches up to 25°C. Reactions left for 30 min at 25°C and for 10 min on ice. After DTT addition (1 mM), the reaction mixtures were subjected to modification by addition of DMS (1:1 dilution) or Kethoxal (1:1 dilution), at 30°C for 30 min. DMS and Kethoxal modification reactions were stopped by adding 0.25 M Tris-acetate; pH 7.5, 0.25 M β-mercaptoethanol, 0.3 M sodium acetate; pH 9.2, 0.025 mM EDTA and 0.3 M sodium acetate; pH 6.0, 25 mM potassium borate; pH 6.0, respectively, followed by phenol extraction and ethanol precipitation. Modified transcripts were resuspended in 1×TE buffer (10 mM Tris-acetate; pH 7.5, 0.1 mM EDTA), except Kethoxal modified transcripts, which resuspended in 1×TE buffer supplemented with 50 mM potassium borate; pH 7.5. Furthermore, in primer extension reactions, each modified transcript (2 pmol) was hybridized with [γ-32P]-labeled TbGl_1 or TbGl_6 primer, which probe either T275 stem I or the terminator/antiterminator region, respectively. Extension reactions were performed in the presence of 20 mM Tris-acetate, pH 8.3, 10 mM Mg (OAc)2, 5 mM DTT, 1 mM of each dNTP, and 5 U AMV reverse transcriptase at 47°C for 1 h. Reaction products were ethanol precipitated and analyzed on 6% PAGE 8 M urea after denaturation at 80°C for 2 min. For the enzymatic probing, T115 transcript (20 pmol of unlabeled transcript mixed with 0.2 pmol of [γ-32P] ATP 5′-end-labeled transcript) was digested with RNase T1 (0.1 U), V1 (0.01U), and RNase A (0.2 pg) in denaturing (20 mM sodium citrate, pH 5.0, 1 mM EDTA, 7 M urea) or native conditions (10 mM Tris-HCl, pH 7.1, 100 mM KCl and 10 mM MgCl2). The same digestion protocol was used for analysis of NEW tRNAGly isoacceptor in complex with T225 transcript. Both transcripts were mixed simultaneously (1:1 stoichiometry), denatured at 65°C for 3 min, and incubated for 1 h at 25°C under native conditions. Subsequently, reactions were radiated for 5 min by UV (254 nm) in 8-cm distance and digested by 0.2 pg of RNase A. Alkaline hydrolysis conditions of either tRNA or T-box transcript (50 mM sodium carbonate, pH 9.2, and 1 mM EDTA) were used for ladder construction. Digestion reactions under both native and denaturing conditions were incubated at 25°C for 15 min. For the denaturing conditions we performed preincubation of the transcript at 50°C for 5 min. Alkaline hydrolysis reactions were performed at 95°C for 10 min. All reactions were stopped by placing reaction mixtures on ice and after ethanol precipitation were denaturized at 80°C for 5 min and analyzed on 10% PAGE 8 M urea. Finally, both primer extension and enzymatic probing analysis were visualized by phosphoimager scanning.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported in part and implemented under the ARISTEIA I Action of the Operational Programme “Education and Lifelong Learning” and is cofunded by the European Social Fund (ESF) and National Resources (MIS 1225, no. D608 to C.S.) This work was also supported in part by University of Patras Research Committee “K. Karatheodoris” grant D164 (to C.S.), the French National Program Investissement d'Avenir administered by the Agence National de la Recherche (ANR), MitoCross Laboratory of Excellence (LabEx), funded as ANR-10-IDEX-0002-02, the University of Strasbourg and the CNRS (to H.D.B.). H.D.B and N.Y.S. would like to thank Dr. T. Henkin for sharing a protocol for the readthrough assays. FEBS is gratefully acknowledged for granting a short-term fellowship to M.A. related to this study.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.052712.115.
REFERENCES
- Blount KF, Breaker RR. 2006. Riboswitches as antibacterial drug targets. Nat Biotechnol 24: 1558–1564. [DOI] [PubMed] [Google Scholar]
- Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol Cell 43: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2012. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AT, Nikonowicz EP. 2012. Solution nuclear magnetic resonance analyses of the anticodon arms of proteinogenic and nonproteinogenic tRNA(Gly). Biochemistry 51: 3662–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AT, Nikonowicz EP. 2013. Solution NMR determination of hydrogen bonding and base pairing between the glyQS T box riboswitch Specifier domain and the anticodon loop of tRNA(Gly). FEBS Lett 587: 3495–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Ferré-D'Amaré AR. 2011. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res 44: 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauzi H, Agyeman A, Hines JV. 2009. T box transcription antitermination riboswitch: influence of nucleotide sequence and orientation on tRNA binding by the antiterminator element. Biochim Biophys Acta 1789: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy N, Jester B, Conant GC, Devine KM. 2010. The T box regulatory element controlling expression of the class I lysyl-tRNA synthetase of Bacillus cereus strain 14579 is functional and can be partially induced by reduced charging of asparaginyl-tRNAAsn. BMC Microbiol 10: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdeman MS, Henkin TM, Hines JV. 2003. Solution structure of the Bacillus subtilis T-box antiterminator RNA: seven nucleotide bulge characterized by stacking and flexibility. J Mol Biol 326: 189–201. [DOI] [PubMed] [Google Scholar]
- Giannouli S, Kyritsis A, Malissovas N, Becker HD, Stathopoulos C. 2009. On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie 91: 344–351. [DOI] [PubMed] [Google Scholar]
- Giannouli S, Labrou M, Kyritsis A, Ikonomidis A, Pournaras S, Stathopoulos C, Tsakris A. 2010. Detection of mutations in the FemXAB protein family in oxacillin-susceptible mecA-positive Staphylococcus aureus clinical isolates. J Antimicrob Chemother 65: 626–633. [DOI] [PubMed] [Google Scholar]
- Green NJ, Grundy FJ, Henkin TM. 2010. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 584: 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg JC, Ke A. 2013a. Sequence, structure, and stacking: specifics of tRNA anchoring to the T box riboswitch. RNA Biol 10: 1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg JC, Ke A. 2013b. Structural determinants for geometry and information decoding of tRNA by T box leader RNA. Structure 21: 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg JC, Chen Y, Grundy FJ, Henkin TM, Pollack L, Ke A. 2013. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc Natl Acad Sci 110: 7240–7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. 1993. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74: 475–482. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. 2004. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J Bacteriol 186: 5392–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Moir TR, Haldeman MT, Henkin TM. 2002a. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res 30: 1646–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Winkler WC, Henkin TM. 2002b. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci 99: 11121–11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Yousef MR, Henkin TM. 2005. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J Mol Biol 346: 73–81. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. 2009. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev 73: 36–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM. 2008. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 22: 3383–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM. 2014. The T box riboswitch: A novel regulatory RNA that utilizes tRNA as its ligand. Biochim Biophys Acta 1839: 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübscher J, Jansen A, Kotte O, Schäfer J, Majcherczyk PA, Harris LG, Bierbaum G, Heinemann M, Berger-Bachi B. 2007. Living with an imperfect cell wall: compensation of femAB inactivation in Staphylococcus aureus. BMC Genomics 8: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentzsch F, Hines JV. 2012. Interfacing medicinal chemistry with structural bioinformatics: implications for T box riboswitch RNA drug discovery. BMC Bioinformatics 13: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lehmann J, Jossinet F, Gautheret D. 2013. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders. Nucleic Acids Res 41: 5494–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. 2013. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res 41: D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Cossart P. 2015. Unexpected versatility in bacterial riboswitches. Trends Genet 31: 150–156. [DOI] [PubMed] [Google Scholar]
- Miao Z, Adamiak RW, Blanchet MF, Boniecki M, Bujnicki JM, Chen SJ, Cheng C, Chojnowski G, Chou FC, Cordero P, et al. 2015. RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures. RNA 21: 1066–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AR, Henkin TM, Agris PF. 2006. tRNA regulation of gene expression: interactions of an mRNA 5′-UTR with a regulatory tRNA. RNA 12: 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, Kazanov MD, Riehl W, Arkin AP, Dubchak I, et al. 2013. RegPrecise 3.0—a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14: 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M, Ibba M. 2014. tRNAs as regulators of biological processes. Front Genet 5: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer S, Berger-Bachi B. 2003. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and β-lactam resistance in gram-positive cocci. Antimicrob Agents Chemother 47: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins SM, Grundy FJ, Henkin TM. 1997. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol Microbiol 25: 411–421. [DOI] [PubMed] [Google Scholar]
- Saad NY, Schiel B, Brayé M, Heap JT, Minton NP, Durre P, Becker HD. 2012. Riboswitch (T-box)-mediated control of tRNA-dependent amidation in Clostridium acetobutylicum rationalizes gene and pathway redundancy for asparagine and asparaginyl-trnaasn synthesis. J Biol Chem 287: 20382–20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad NY, Stamatopoulou V, Braye M, Drainas D, Stathopoulos C, Becker HD. 2013. Two-codon T-box riboswitch binding two tRNAs. Proc Natl Acad Sci 110: 12756–12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels M, Fire A, Sharp PA. 1984. Dinucleotide priming of transcription mediated by RNA polymerase II. J Biol Chem 259: 2517–2525. [PubMed] [Google Scholar]
- Samuelsson T, Axberg T, Borén T, Lagerkvist U. 1983. Unconventional reading of the glycine codons. J Biol Chem 258: 13178–13184. [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Senn MM, Berger-Bächi B, Tossi A, Sahl HG, Wiedemann I. 2004. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol Microbiol 53: 675–685. [DOI] [PubMed] [Google Scholar]
- Schroeder KT, McPhee SA, Ouellet J, Lilley DM. 2010. A structural database for k-turn motifs in RNA. RNA 16: 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J, Ibba M. 2015. Bacterial transfer RNAs. FEMS Microbiol Rev 39: 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood AV, Grundy FJ, Henkin TM. 2015. T box riboswitches in Actinobacteria: translational regulation via novel tRNA interactions. Proc Natl Acad Sci 112: 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Fuchs RT, Grundy FJ, Henkin TM. 2010. Riboswitch RNAs: regulation of gene expression by direct monitoring of a physiological signal. RNA Biol 7: 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and its applications in agriculture, biomedicine and environmental studies (ed. Li RW), pp. 61–78. Nova Science Publishers, NY. [Google Scholar]
- Sun EI, Rodionov DA. 2014. Computational analysis of riboswitch-based regulation. Biochim Biophys Acta 1839: 900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Kanehisa M. 2012. Using the KEGG database resource. Curr Protoc Bioinformatics 11: 1.12.1–1.12.54. [DOI] [PubMed] [Google Scholar]
- Villet R, Fonvielle M, Busca P, Chemama M, Maillard AP, Hugonnet JE, Dubost L, Marie A, Josseaume N, Mesnage S, et al. 2007. Idiosyncratic features in tRNAs participating in bacterial cell wall synthesis. Nucleic Acids Res 35: 6870–6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. 2008. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14: 717–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Nikonowicz EP. 2011. Solution structurez of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA. J Mol Biol 408: 99–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Henkin TM, Nikonowicz EP. 2010. NMR structure and dynamics of the Specifier Loop domain from the Bacillus subtilis tyrS T box leader RNA. Nucleic Acids Res 38: 3388–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wels M, Groot Kormelink T, Kleerebezem M, Siezen RJ, Francke C. 2008. An in silico analysis of T-box regulated genes and T-box evolution in prokaryotes, with emphasis on prediction of substrate specificity of transporters. BMC Genomics 9: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Wagner RN, Grundy FJ, Raina M, Ibba M, Henkin TM. 2015. The Bacillus subtilis tyrZ gene encodes a highly selective tyrosyl-tRNA synthetase and is regulated by a MarR regulator and T box riboswitch. J Bacteriol 197: 1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. 2005. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 59: 487–517. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Grundy FJ, Murphy BA, Henkin TM. 2001. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7: 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef MR, Grundy FJ, Henkin TM. 2003. tRNA requirements for glyQS antitermination: a new twist on tRNA. RNA 9: 1148–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef MR, Grundy FJ, Henkin TM. 2005. Structural transitions induced by the interaction between tRNA(Gly) and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol 349: 273–287. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ferré-D'Amaré AR. 2013. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500: 363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ferré-D'Amaré AR. 2014. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol Cell 55: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ferré-D'Amaré AR. 2015. Structure and mechanism of the T-box riboswitches. Wiley Interdiscip Rev RNA 6: 419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.