Abstract
Manganese plays a central role in cellular detoxification of reactive oxygen species (ROS). Therefore, manganese acquisition is considered to be important for bacterial pathogenesis by counteracting the oxidative burst of phagocytic cells during host infection. However, detailed analysis of the interplay between bacterial manganese acquisition and phagocytic cells and its impact on bacterial pathogenesis has remained elusive for Staphylococcus aureus, a major human pathogen. Here, we show that a mntC mutant, which lacks the functional manganese transporter MntABC, was more sensitive to killing by human neutrophils but not murine macrophages, unless the mntC mutant was pre-exposed to oxidative stress. Notably, the mntC mutant formed strikingly small colonies when recovered from both type of phagocytic cells. We show that this phenotype is a direct consequence of the inability of the mntC mutant to reinitiate growth after exposure to phagocytic oxidative burst. Transcript and quantitative proteomics analyses revealed that the manganese-dependent ribonucleotide reductase complex NrdEF, which is essential for DNA synthesis and repair, was highly induced in the mntC mutant under oxidative stress conditions including after phagocytosis. Since NrdEF proteins are essential for S. aureus viability we hypothesize that cells lacking MntABC might attempt to compensate for the impaired function of NrdEF by increasing their expression. Our data suggest that besides ROS detoxification, functional manganese acquisition is likely crucial for S. aureus pathogenesis by repairing oxidative damages, thereby ensuring efficient bacterial growth after phagocytic oxidative burst, which is an attribute critical for disseminating and establishing infection in the host.
Introduction
Staphylococcus aureus is a major cause of nosocomial and community-acquired infections leading high rates of hospitalization and mortality worldwide [1]. A hallmark of S. aureus as a major human pathogen is its large arsenal of virulence factors involved in colonizing diverse host niches, evading innate immunity, and resisting major antibacterial therapies [2]. Accordingly, S. aureus is able to infect a wide variety of organs and cause a multitude of diseases such as skin and soft tissue infections, pneumonia, endocarditis and sepsis [3]. As bacteria continue to develop resistance, it is important to define new anti-bacterial targets for the effective treatment of S. aureus infections. Currently available antibiotics target only a limited number of bacterial cellular functions such as biosynthesis of macromolecules like DNA and protein. Targeting bacterial nutrient uptake systems is an attractive alternative approach, since acquiring essential elements, such as iron and manganese, is critical for the survival and replication of pathogens [4]. In fact, one of the host defense systems to combat infection is to restrict the availability of essential elements from invading pathogens, a process termed “nutritional immunity” [4]. Therefore, a recently identified antibody fragment that interferes with manganese uptake of S. aureus might provide an alternative therapeutic intervention in treating bacterial infections [5].
While the most prominent example of nutritional immunity is the restriction of iron by the host, recent work has revealed that vertebrates also limit manganese availability during infection [6]. Vertebrates produce two proteins to sequester manganese: calprotectin, a member of the S100 class of EF-hand calcium binding proteins [7], and Nramp1, a multi-spanning integral membrane protein [8]. Both calprotectin and Nramp1 are highly expressed in phagocytic cells that act as a first defense against invading pathogens [6, 9].
The S. aureus genome encodes for two distinct Mn2+ uptake systems: MntABC and MntH [10, 11]. MntABC is an ATP-binding cassette (ABC)-type transporter, which consists of three proteins, the ATP-binding protein MntA, the permease MntB and the metal binding protein MntC, while MntH is a proton-dependent NRAMP transporter. Simultaneous inactivation of MntABC and MntH was shown to attenuate the virulence of the laboratory strain 8325–4 and the methicillin-sensitive clinical strain Newman in a murine skin abscess model and in a murine systemic infection model, respectively [10, 11]. In the methicillin-resistant S. aureus (MRSA) strain USA300, inactivation of MntABC alone was sufficient to attenuate its virulence in a murine systemic infection model [12]. The differences in the requirement of functional manganese acquisition systems for S. aureus pathogenesis might be strain- and infection model-dependent, but the functionality of the host Nramp1 locus is known to be important for analyzing manganese-dependent processes of pathogens within the host [13]. While the major laboratory mouse strains such as BALB/c and C57BL/6, which were used in the previous studies [10, 11], do not have a functional Nramp1 locus, the A/J strain that was used in the latest study does [12, 14]. Functional Mn2+ acquisition systems are also critical for full virulence of several other pathogens, including Neisseria gonorrhoeae [15], Salmonella species [16], and Streptococcus species [17], suggesting the universal requirement of manganese for bacterial pathogenesis.
During the oxidative burst (or respiratory burst) of phagocytic cells, a significant amount of reactive oxygen species (ROS) is released [18, 19]. Oxidative burst is initiated by the assembly of the NADPH oxidase complex, which catalyzes the reduction of oxygen (O2) to superoxide (O2 −), which is further reduced to hydroxyl radical (OH·) or dismutated to hydrogen peroxide (H2O2) [20]. Moreover, neutrophils (but not macrophages) contain high concentrations of myeloperoxidase (MPO), which together with H2O2 and Cl− produces the strong oxidant hypochlorous acid (HOCl) [18]. O2 −, OH·, H2O2, and HOCl are ROS and are capable of damaging a number of cellular structures including membranes and macromolecules in particular DNA [21, 22]. Manganese has been proposed to detoxify ROS in various ways: the detoxification of O2 − by Mn2+-dependent superoxide dismutases is the best-characterized Mn2+-dependent ROS detoxification mechanism [23]. Indeed, it was shown that concomitant inactivation of both MntC and MntH ablated S. aureus superoxide dismutase activity under manganese-restricting conditions [11]. In E. coli, it has been proposed that Mn2+ might be able to replace the more reactive Fe2+ in Fe2+-containing proteins, thereby reducing oxidative damage to these proteins [24]. Non-enzymatic detoxification of ROS by Mn2+ has been proposed for S. aureus and other bacteria [25, 26], but its relevance has been controversially discussed [27]. Since phagocytic cells are the major source of ROS during infection [18], functional manganese acquisition systems are thought to be important for bacterial survival in the host. To date, however, few studies have directly investigated the role functional manganese acquisition plays in the interaction of S. aureus with phagocytic cells. The majority of published work has focused on in vitro experiments where the response of S. aureus mutant strains towards oxidative stress was investigated using the ROS-generating agent methyl viologen whose mechanism of ROS generation has shown to be distinct from the oxidative burst of phagocytic cells [28–31]. Moreover, the in vitro experiments have been usually performed under non-physiological conditions, e.g. bacteria are incubated with high concentrations of methyl viologen for an extended time, often overnight. Therefore, the goal of our study was to directly investigate the role functional manganese acquisition plays in the interaction of S. aureus and phagocytic cells. Moreover, we aimed to elucidate the biological processes that are affected by manganese depletion during oxidative stress in order to better understand the molecular basis underlying the strong attenuation of the MRSA strain USA300 lacking MntABC during murine systemic infection [12].
Materials and Methods
Ethics Statement
All research involving human participants were approved by Western Institutional Review Board (WIRB). All individuals who donated blood for research use have signed an IRB approved consent form. All procedures involving mice were compliant with the ILAR guidelines, and were approved by the IACUC at Genentech, Inc.
Bacterial strains
Methicillin-resistant S. aureus (MRSA) SF8300, a clinical isolate representative of the epidemic strain USA300 [32], and its isogenic mntC mutant, in which the mntC gene was inactivated by changing its 8th and 12th codon to a stop codon, were obtained as described previously [12].
Bone marrow derived macrophage isolation
Bone marrow-derived macrophages (BMDMs) were isolated from the femurs of A/J mice purchased from the Jackson Laboratory. Bone marrow was eluted using Bone Marrow Macrophage medium (BMM), consisting of a 50:50 mix of Dulbecco's Modified Eagle Medium and F12 media with 20% heat-inactivated Fetal Bovine Serum (FBS), 100 ng/ml rM-CSF, 2 mM glutamine, 110 μg/ml sodium pyruvate, penicillin and streptomycin. Bone marrow cells were cultured for 6–7 days in a tissue culture incubator. Adherent cells were harvested by gentle scraping after washing with cold PBS (Mg2+- and Ca2+-free) plus 1 mM EDTA and frozen in FBS + 10% DMSO for storage in liquid nitrogen until use.
Neutrophil killing assay
Polymorphonuclear leukocytes (PMN) were isolated from heparin-treated whole human blood (from healthy donors) by the addition of an equal volume of 3% Dextran and 0.9% NaCl. After incubation at room temperature for 20 min, PMNs were pelleted from the upper layer by centrifugation at 200 × g for 5 min. Cells were washed and resuspended in Hanks Balanced Salt Solution containing 10 mM HEPES pH 7.4 and 1% BSA (HB media). S. aureus was grown to mid-log phase (OD600 = 0.5) in Roswell Park Memorial Institute (RPMI) media (RPMI 1640, Sigma-Aldrich) supplemented with 10 mM HEPES (RPMI-H). S. aureus cells were spun down and resuspended to an OD600 of 0.5 in 10 mL RPMI-H ± 1 μM methyl viologen, then incubated at 37°C with shaking for 1 hour. S. aureus cells were washed once in fresh RPMI-H, resuspended in 10 ml HB media, and pre-opsonized in 10% human serum at 37°C for 30 min. Bacteria were then added to 9 × 104 PMNs at an MOI of 10 and incubated at 37°C for 90 min in a tissue culture incubator. PMNs were lysed in PBS with 0.1% Triton X-100. Viable bacterial counts were determined by plating serial dilutions on Tryptic Soy Agar (TSA) containing 5% defibrinated sheep’s blood.
Macrophage killing assay
BMDMs were seeded from frozen stocks into 24-well tissue culture plates at a density of 2 × 105 cells/well in BMM. After 24 hours of culture in a tissue culture incubator, cells were given fresh media plus 10 ng/ml IFNγ (Peprotech) without antibiotics. Macrophages were then washed and infected with prepared S. aureus cells (±1 μM methyl viologen as described above without pre-opsonization) at an MOI of 10. After incubation at 37°C for 90 min, the inoculum was removed from BMDMs and replaced with BMM containing 50 μg/ml gentamycin to kill extracellular S. aureus cells. After 24 hours of incubation, bacteria were harvested from macrophages by adding PBS containing 0.1% Triton X-100 to lyse macrophages. Viable bacterial counts were determined by plating serial dilutions on TSA containing 5% defibrinated sheep’s blood.
CFSE proliferation assay
Bacteria were grown overnight in RPMI medium buffered with 10 mM HEPES pH 7.2, then diluted into fresh media and grown to mid-log phase. Bacteria were then harvested, washed, and resuspended in PBS buffer containing 0.1% Bovine Serum Albumin (PBS-BSA) to a concentration of 1 × 109 cfu/ml. Labeling reaction was carried out at 37°C for 30 minutes with 100 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes, Eugene, OR) diluted in PBS-BSA. Excess label was removed by washing 3 times in PBS-BSA. To confirm that the CFSE label was retained in the absence of cell division, bacterial cell division was inhibited by irradiation or co-culturing with high concentrations of unlabeled stationary phase bacteria. For irradiation, bacteria were diluted to 1 × 109 cfu/ml in PBS containing 0.1% BSA. 1 ml of the bacterial suspension was placed in each well of a 24-well tissue culture dish and irradiated 2 times at 400 mJoules using a UV stratalinker 2400 (Stratagene). For co-culturing, 100 μl CFSE-labeled bacteria (1 × 108 cfu/ml) were added directly to a 10 ml overnight culture of unlabeled bacteria (~3 × 109 cfu/ml). Macrophage infection was carried out with CFSE-labeled bacteria as described before with the exception that dH2O and agitation were utilized to lyse the macrophages. CFSE-labeled bacteria that were recovered from macrophages were resuspended in TSB media and allowed to grow aerobically at 37°C. Samples were taken at indicated time points and were fixed by the addition of 2% paraformaldehyde in PBS. Retention of CFSE label was determined by flow cytometry using a FACSAria cell sorter (BD Biosciences, San Jose, CA) and data analysis was performed using FlowJo software (Tree Star Inc, Ashland, OR). To distinguish bacteria from macrophage debris during FACS analysis, samples were immunolabeled with an anti-S. aureus Alexa647-conjugated antibody (Genentech) and gates were set with controls that were singly labeled with either the anti-S. aureus Alexa647 antibody or with CFSE but not exposed to phagocytosis or conditions that induced proliferation.
Growth after methyl viologen treatment
S. aureus was grown to mid-log phase (OD600 = 0.5) in RPMI-H. Cells were spun down and resuspended to an OD600 of 0.5 in 10 mL RPMI-H ± 1 μM methyl viologen, then incubated at 37°C with shaking for 1 hour. Cells were washed once in fresh RPMI-H, inoculated 1:100 in 10 ml fresh RPMI-H and incubated at 37°C with shaking. At the indicated time points, aliquots of culture were removed and assayed in triplicate for intracellular ATP concentrations using BacTiter-Glo (Promega) according to manufacturer’s instructions with the following modification: samples were incubated in the dark for 15 minutes to ensure lysis of the bacteria. Luminescence was measured on a Synergy 2 Multi-Mode Microplate Reader (BioTek). Bacterial growth was measured as the fold change in luminescence relative to the starting culture.
Recovery after phagocytosis
To measure the recovery of S. aureus after phagocytosis, 200 μl aliquots of recovered lysate, which contained comparable amount of wild-type and mntC mutant bacteria as determined by Bac-Titer-Glo luminescence, were added to 10 ml of RPMI-H. Bacterial growth was measured at the indicated time points as the fold change in the BacTiter-Glo luminescence relative to the starting culture as described above.
Harvesting of S. aureus RNA after methyl viologen treatment
S. aureus was grown to mid-log phase (OD600 = 0.5) in RPMI-H. Cells were spun down and resuspended to an OD600 of 0.5 in 10 mL RPMI-H ± 1 μM methyl viologen, then incubated at 37°C with shaking for 1 hour. Cells were washed once in fresh RPMI-H and pelleted by centrifugation at 2400 × g for 15 min at 4°C. Bacterial pellets were resuspended in PBS with 2 volumes RNAProtect Bacteria Reagent (Qiagen), and incubated at room temperature with gentle rotation for 5 minutes. Samples were then spun down at 8500 × g for 10 min at 4°C and bacterial pellets were stored at −80°C until RNA isolation. To harvest RNA, bacteria were first incubated with 80 μg lysostaphin (Sigma), 80,000 U RNasin (Promega) and 200 μg Proteinase K (Qiagen) for 30 min. Samples were then lysed via bead-beating using 0.1 mm glass beads (BioSpec) and RNA was subsequently isolated per manufacturer’s instructions using the RNeasy mini kit (Qiagen).
Harvesting of S. aureus RNA after phagocytosis
8 × 105 BMDMs or 3.6 × 105 PMNs were seeded in 6 well tissue culture plates and infected with S. aureus at an MOI of 20 as described above for killing assays. After infection for 45 minutes (PMNs) or 2 hours (macrophages), cells were lysed by incubating with PBS + 0.1% Triton X-100 for 2 min. Lysate from 3 wells was pooled and phagocytosed bacteria were recovered by centrifugation at 2400 × g for 20 min at 4°C. Bacterial pellets were resuspended in PBS with 2 volumes RNAProtect Bacteria Reagent (Qiagen) and incubated at room temperature with gentle rotation for 5 min. Samples were then spun down at 8500 × g for 10 min at 4°C and bacterial pellets were stored at −80°C until RNA was isolated as described above.
Analysis of S. aureus mRNA expression
cDNA was synthesized from 500−1000 ng of Turbo-DNase treated RNA using the Superscript III First-Strand Synthesis Supermix for qRT-PCR (Life Technologies). cDNA reactions were diluted 1:10 using nuclease-free water and analyzed by qRT-PCR. qRT-PCR was performed in triplicate using custom designed Taqman primers on a 7500 Real-Time PCR System (S1 Table) (Applied Biosciences). As an internal control, primers specific for the 16S rRNA were used.
Cell Culture and SILAC Labeling
Three independent cultures from independent clones of wild-type and mntC mutant strains were grown in RPMI 1640 (Thermo Scientific, 89984) media containing 1 μM of methyl viologen supplemented with either light arginine (Sigma, A5006) and lysine (Sigma, L5501) or heavy arginine (Sigma, 608033) and lysine (Sigma, 608041) to OD600 of 0.3, washed and resuspended in PBS. Both heavy and light SILAC media were supplemented with light proline (Sigma, P0380). Bacteria were lysed by mechanical disruption using a Mini-Beadbeater (Biospec Products). The lysates were centrifuged at 18,000 × g for 10 min. Bradford assays (BioRad) were performed on each lysate to determine the protein concentration.
SILAC Incorporation Analysis
To determine the incorporation rate of heavy amino acids, 5 μg of lysate of cells grown in heavy media was reduced with 10 mM dithioreitol (DTT) and loaded onto a NuPage 4–12% Bis-Tris gel (Invitrogen). A gel band in the region around 60 kDa from each lane was excised, destained with 50 mM ammonium bicarbonate/50% methanol, and digested with 0.02 μg/μl trypsin (Promega) in 50 mM ammonium bicarbonate overnight at 37°C. Digests were subjected to LC-MS/MS analysis using similar conditions as described earlier [11]. Tandem mass spectral data was submitted for Mascot version 2.3.02 (Matrix Science) database search against a concatenated target-decoy database consisting of S. aureus proteins from UniProtKB (version 2011_12) and common laboratory contaminants. Database search parameters included full tryptic specificity, variable modifications of heavy lysine (K8, +8.014Da) and heavy arginine (R10, +10.008Da), methionine oxidation (+15.995Da), 2 missed cleavages, and 30 ppm precursor ion mass and 0.5 Da fragment ion mass tolerance. Peptide spectral matches were post-filtered to a 10% peptide false discovery rate using a linear discriminant approach as previously described [33]. Quantification was performed using the VistaQuant [34] algorithm and only high quality peptides with VistaQuant scores of 95 and above were employed to verify SILAC incorporation rates.
SILAC Proteome Profiling
“Light” wild-type and “Heavy” mntC mutant lysates were mixed in a 1:1 ratio based on protein concentration. The reverse labeled lysates “Heavy” wild-type and “Light” mntC mutant were also similarly mixed. A total of 60 μg of protein for each mixed SILAC pair was reduced with DTT and loaded onto a 4–12%Bis-Tris gel (Invitrogen). The resulting gel lanes were excised into 15 slices and subjected to in-gel trypsin digestion as described above. Following digestion, each sample was analyzed via LC-MS/MS with duplicate injections using similar conditions as described previously [12], however with a 67 min gradient from 2−90% Buffer B (Buffer B comprises of 2% water/ 98% acetonitrile/ 0.1% formic acid) at 1.00 μl/min with a total analysis time of 90 min. Tandem mass spectral data was submitted for Mascot (Matrix Science) database search against a concatenated target-decoy database consisting of S. aureus proteins and common laboratory contaminants as described above. The data was searched with full trypsin enzyme specificity, variable modifications of heavy lysine (K8, +8.014Da) and heavy arginine (R10, +10.008Da), and methionine oxidation (+15.995Da), and 30 ppm precursor ion mass and 0.5 Da fragment.
Protein-level Quantification and Normalization
Quantification was performed using the VistaQuant algorithm [34]. Quantified peptide spectral matches (PSMs) with VistaQuant scores of 83 and above were combined across all gel fractions for a given condition and labeling set and normalized to the median relative abundance ratio of each combined dataset. Log2 protein relative abundance ratios were calculated as the log-transformed ratio of the summed area under curve measurements of the labeled and unlabeled peptide species. In instances where multiple quantification events were observed for the same species, as determined by overlapping ion masses and peak retention times, the event with the largest total area under the curve was chosen as representative and all others were discarded. Events for which either the light or heavy species was below the limit of detection of the instrument were set to the signal to noise ratio of the observed species and denoted by an inequality symbol in the results. Standard scores (z-scores) for each protein ratio were calculated against a moving standard deviation calculated as a function of overall abundance as previously described [34]. Proteins were stratified into two lists: first, a list for which all absolute protein standard scores when observed were ≥2, and a second slightly relaxed list where at least one observation had an absolute protein standard score ≥2 while requiring that other observations be consistent (that is, the sign of all standard scores for a given protein are the same).
Results
Lack of a functional MntABC system renders S. aureus more sensitive to killing by human neutrophils but not murine macrophages, unless S. aureus is pre-exposed to oxidative stress
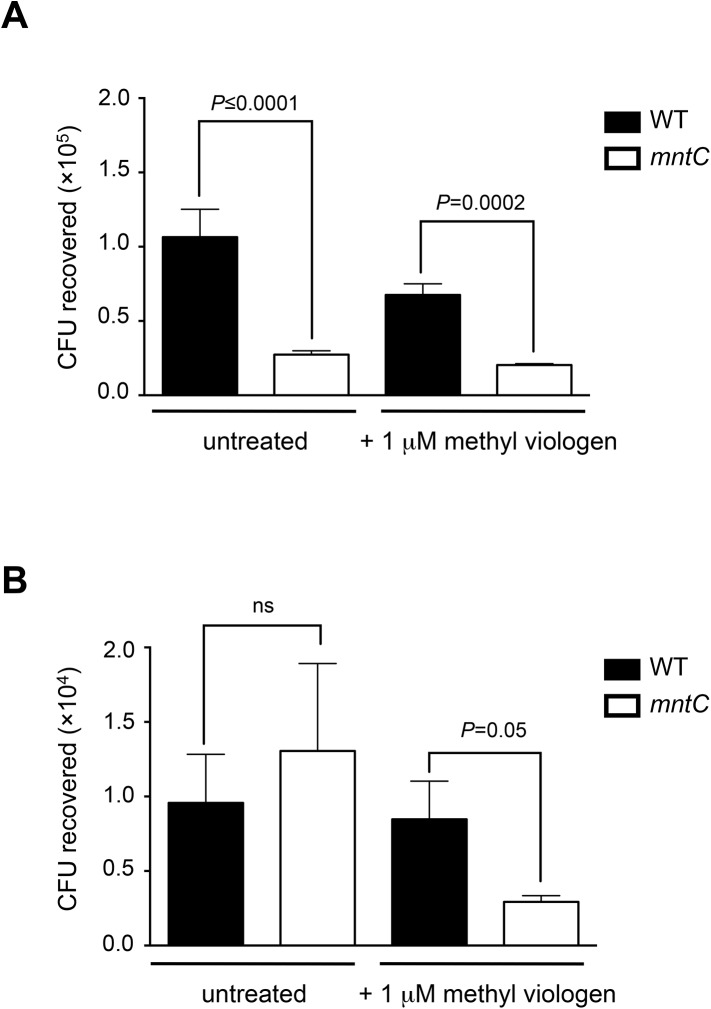
Previous studies showed that S. aureus cells lacking a functional MntABC system are more susceptible to killing by methyl viologen [10], a ROS-generating quaternary ammonium compound [28]. Since the mechanism of ROS generation by methyl viologen has shown to be distinct from the oxidative burst of phagocytic cells [28–31], we wondered whether the higher susceptibility of S. aureus cells lacking a functional MntABC system to methyl viologen in vitro is translated into an increased killing by phagocytic cells during host infection. To this end, we sought to compare the survival of wild-type and mntC mutant strains upon exposure to neutrophils and macrophages isolated from A/J mice ex vivo. Due to the limited volume of whole blood that can be recovered from mice, we decided to use neutrophils that were isolated from human donors instead of murine neutrophils for our experiment. Human neutrophils were infected with either the opsonized wild-type or mntC mutant strain at a multiplicity of infection (MOI) of 10:1 and the number of viable intracellular bacteria was determined 90 min later. Compared to the wild-type strain, the mntC mutant strain was statistically significantly more susceptible to killing by neutrophils (Fig 1A). Notably, we observed increased killing of the mntC mutant only when neutrophils were isolated from whole blood that was treated with heparin, but not with EDTA, as an anticoagulant (data not shown). Several studies have reported that treatment of whole blood with EDTA results in significantly reduced ROS production by neutrophils compared to heparinized whole blood [35, 36]. It is possible that the increased sensitivity of the mntC mutant to the phagocytic oxidative burst manifests only at a high amount of ROS, similar to sensitivity to killing by methyl viologen (S1 Fig). Next, we tested the survival of wild-type and mntC mutant strains within macrophages. Interferon-gamma (IFN-γ) activated bone-derived murine macrophages were infected with either wild-type or mntC mutant cells at a MOI of 10:1 and the number of viable intracellular bacteria was determined 24 hours later. In contrast to the survival within human neutrophils, there was no difference between wild-type and mntC mutant strains in killing by activated murine macrophages (Fig 1B).
Fig 1. Lack of a functional MntABC system renders S. aureus more sensitive to killing by methyl viologen and human neutrophils but not murine macrophages, unless S. aureus is pre-exposed to oxidative stress.
(A, B) Survival of wild-type and mntC mutant strains within neutrophils harvested from heparin-treated human blood (A) and INF-γ-activated murine macrophages (B). Bacteria were either untreated or pre-exposed to 1 μM methyl viologen for 1 hour. Neutrophils (A) and macrophages (B) were lysed after 90 min and 24 hours of infection, respectively, to enumerate CFU. Bars represent the mean value of triplicate samples and error bars are standard deviation. P-values were determined using one-way ANOVA with multiple comparisons between samples via Tukey’s post-test.
Several lines of evidence indicate that S. aureus is able to survive inside phagocytic cells and contribute to infection [37–39]. Moreover, it has been suggested that neutrophil destruction after phagocytosis of USA300 is in part a form of programmed necrosis [38]. Therefore, it is likely that surviving S. aureus cells experience repetitive cycles of oxidative burst during infection. We wondered whether S. aureus cells that were previously phagocytosed and have already experienced an oxidative burst become more susceptible to killing during a second round of phagocytosis. To pre-expose bacteria to oxidative stress before phagocytic infection we incubated bacteria with a sub-lethal concentration of methyl viologen in which the survival rate of both strains was equal (S2 Fig), since it was not feasible to recover a sufficient number of intracellular bacteria to perform a second round of infection. Pre-treatment with a sub-lethal concentration of methyl viologen rendered the mntC mutant strain significantly more susceptible to killing by murine macrophages, while it had a minor effect on the susceptibility of the mntC mutant strain towards neutrophil killing (Fig 1A and 1B). It is possible that the observed difference in the susceptibility of the mntC mutant to killing by neutrophils and macrophages is due to differences in the amount and species of ROS produced by these phagocytic cells (cf. Discussion for details).
S. aureus cells lacking MntABC have delayed resumption of growth following internalization by phagocytic cells
Notably, we observed that the mntC mutant strain formed significantly smaller colonies on agar plates compared to the wild-type strain after recovery from phagocytic cells (Fig 2A). After additional incubation at 37°C for 24 h, the colony size reached that of the wild-type strain (Fig 2A), suggesting that the nature of the small mntC mutant colonies is distinct from that of small colony variants [40]. Similarly, smaller-sized colonies were observed when the mntC strain was recovered from infected kidneys of immune-sufficient mice or cultures containing sub-lethal concentrations of methyl viologen (Figure A in S3 Fig). Since growth in manganese-restricted media alone did not result in smaller mntC mutant colonies (Figure B in S3 Fig), we hypothesized that the mntC mutant might be impaired in resuming growth after recovery from the oxidative burst of phagocytic cells. To this end, wild-type and mntC mutant cells recovered from murine macrophages were inoculated in fresh liquid media and growth was determined by measuring the amount of intracellular ATP, which is indicative of the presence of metabolically active cells [41]. Measurement of the total amount of ATP is significantly more sensitive than the measurement of optical density when analyzing bacterial growth at low cell density [41]. Both the wild-type and mntC mutant samples had comparable amounts of total ATP at the time of inoculation into fresh media (Fig 2B). Strikingly, while the number of metabolically active wild-type cells significantly increased after inoculation in liquid culture that of the mntC mutant cells recovered from macrophages increased only marginally (Fig 2B). Both wild-type and mntC mutant cells proliferated similarly when they were not pre-exposed to murine macrophages (Fig 2B). Similar results were obtained when the growth of wild-type and mntC mutant cells recovered from human neutrophils was analyzed (Fig 2C). These results clearly indicate that MntABC is crucial for the growth of S. aureus cells following phagocytosis.
Fig 2. Recovery of the mntC mutant is delayed after phagocytosis.
(A) Wild-type and mntC mutant strains that were harvested from murine macrophages were plated onto agar plates containing 5% defibrinated sheep blood. Images were taken after 16 and 40 hours of incubation at 37°C. (B, C) Growth after phagocytosis by macrophages (B) or neutrophils (C) was measured by BacTiter-Glo (Promega) 22 hours after recovery from phagocytic cells and inoculation into manganese-restricted media. Data are mean values of triplicate samples with standard deviation.
To confirm that MntABC is important for reinitiation of S. aureus replication following internalization by phagocytic cells we ascertained the proportion of wild-type and mntC bacteria that have started to divide by labeling the bacteria with the fluorescent dye carboxyfluorescein diacetate succinimidyl ester (CFSE) prior to infection of murine macrophages. CFSE covalently binds to intracellular amines and as cells divide CFSE segregates equally among daughter cells with each division; thus, diminishing signal intensity, as measured by flow cytometry, is indicative of cell division [42]. As a proof of concept, we monitored proliferation of CFSE-labeled wild-type S. aureus with or without irradiation exposure, which inhibits cell division. While non-irradiated cells started to lose CFSE labeling soon after inoculation into liquid culture in vitro, irradiated cells fully retained CFSE labeling for at least 24 hours indicating that the CFSE label is stably maintained in the absence of cell division (Figure A in S4 Fig). In addition, CFSE-labeled wild-type bacteria that were inoculated into a saturated overnight culture of unlabeled wild-type bacteria, which inhibit the growth of CFSE-labeled bacteria, retained the CFSE signal significantly longer compared to cells that were inoculated into a fresh growth media (S5 Fig) Moreover, flow cytometry analysis of CFSE-labeled wild-type and mntC mutant bacteria at hourly intervals yielded a series of overlapping, normally distributed curves reflecting uniform replication within the bacterial population and confirming that mntC mutant bacteria, which were not previously exposed to phagocytic cells, have a comparable growth rate with that of wild-type bacteria (Fig 3A, Figure B in S4 Fig). In contrast, when mntC mutant cells were recovered from macrophages, they had a significant growth defect as was evident by the significant retention of CFSE fluorescence, which is indicative of non-dividing cells (Fig 3A).
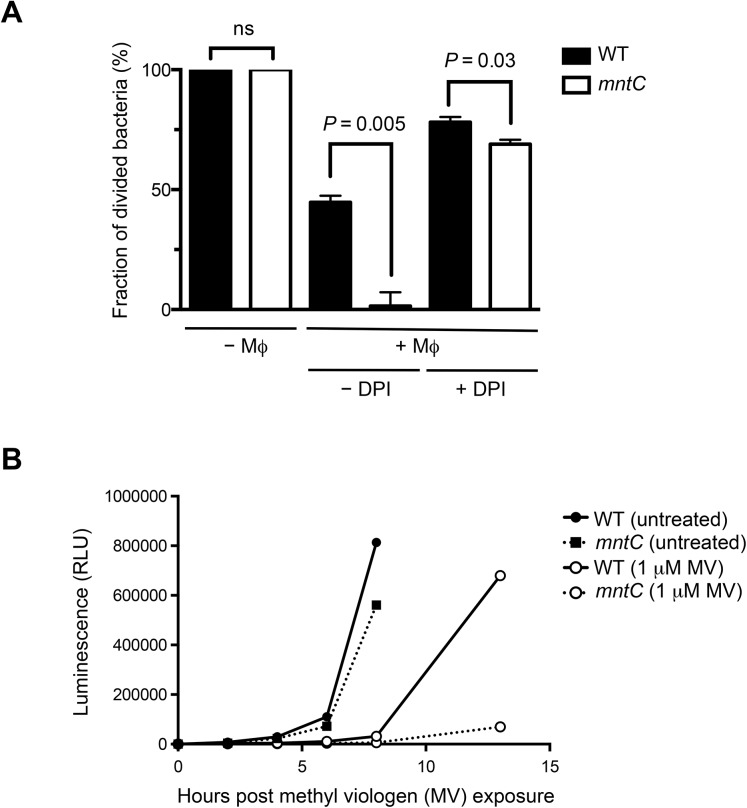
Fig 3. MntABC is important for efficient proliferation of S. aureus after oxidative stress.
(A) Percentage of wild-type and mntC mutant cells that lost CFSE fluorescence when inoculated into TSB media after recovery from IFN-γ activated murine macrophages with and without DPI treatment. Bacteria were identified by cell size and further differentiated from macrophage debris by co-staining with an anti-S. aureus antibody. Data are median values from triplicate samples with standard deviation. (B) Wild-type and mntC mutant cells were treated with 1 μM methyl viologen (MV) for 1 hour. Growth after treatment was measured at indicated time points for 12 hours post exposure, via an increase in BacTiter-Glo luminescence values. Once bacterial cultures reached stationary phase growth, growth was no longer measured. Data are mean values of triplicated samples with standard deviation.
Delay in replication of the mntC strain is associated with oxidative stress
To determine whether the delay in replication of the mntC mutant strain was a consequence of the oxidative stress experienced inside macrophages, we inhibited the oxidative burst of IFN-γ-activated macrophages with diphenylene iodonium (DPI), which is a well-known inhibitor of NADPH oxidase [43]. The DPI treatment of IFN-γ activated macrophages prior to infection diminished the previously observed growth defect of mntC mutant cells (Fig 3A) suggesting that the growth defect of the mntC mutant strain was primarily a consequence of the phagocytic oxidative burst.
We used methyl viologen to confirm that the growth defect of mntC mutant cells that suffered oxidative stress ex vivo can be reproduced in vitro. To this end, we monitored the growth of both wild-type and mntC mutant strains after exposure to a sub-lethal concentration of methyl viologen at which the survival rates of both strains are equivalent (S2 Fig). Without pre-exposure to methyl viologen, both strains had comparable growth profile (Fig 3B). Pre-treatment with methyl viologen substantially extended the lag-phase of both wild-type and mntC mutant strains (Fig 3B). Notably, while the wild-type strain had grown substantially by 13 hours after inoculation into fresh media, the growth of the mntC mutant strain was still significantly impaired (Fig 3B). Together, our results demonstrate that a functional MntABC system is crucial for S. aureus to recommence growth after enduring oxidative stress.
Genes involved in DNA damage repair are highly upregulated in S. aureus cells lacking MntABC after phagocytosis
In order to better understand the molecular mechanism underlying the severe growth defect of S. aureus cells lacking MntABC after the phagocytic oxidative burst, we sought to investigate the differences in the cellular response of the wild-type and mntC mutant strains to oxidative stress. Bacterial survival under oxidative stress conditions relies on a series of distinct defense mechanisms including detoxification of ROS through enzymes and repair of damaged macromolecules, in particular DNA, which was shown to be the most sensitive target of oxidative damage [22, 44]. Therefore, we aimed to study the differences in the transcription of genes involved in these two key processes between wild-type and mntC mutant strains under oxidative stress conditions (Table 1). First, we performed quantitative real-time reverse transcription PCR (qRT-PCR) analysis on bacteria that were exposed to a sub-lethal concentration of methyl viologen in vitro. The methyl viologen-exposed mntC mutant bacteria consistently upregulated genes involved in oxidative stress resistance and DNA repair/synthesis compared to those that were not treated with methyl viologen (Fig 4A and 4B). Moreover, the expression levels of genes involved in oxidative stress resistance and DNA repair/synthesis in the mntC mutant strain were significantly higher compared to those of the wild-type strain (Fig 4A and 4B). Addition of excess manganese repressed the induction of the majority of the genes involved in oxidative stress resistance and DNA repair/synthesis in the mntC mutant strain upon exposure to a sub-lethal concentration of methyl viologen (Fig 4A and 4B). The expression of srtA, which encodes for an enzyme involved in anchoring LPXTG-proteins to the cell wall and whose expression is unrelated to oxidative stress [45], was comparable between wild-type and mntC mutant strains and between methyl viologen exposed and non-exposed bacteria (Fig 4C).
Table 1. List of genes analyzed by qRT-PCR.
| Gene | Name | Locus tag | GeneID | Function | Mn 2+ dependency | Essential | Reference |
|---|---|---|---|---|---|---|---|
| sodM | superoxide dismutase | SAUS300_0135 | 3913957 | oxidative stress response | requires Mn2+ as a cofactor | no | 25, 47 |
| sodA | superoxide dismutase | SAUS300_1513 | 3915273 | oxidative stress response | requires Mn2+ as a cofactor | no | 25, 48 |
| katA | catalase A | SAUSA300_1232 | 3914723 | oxidative stress response | regulated through PerR regulon | no | 46, 47 |
| trxB | thioredoxin-disulfide reductase | SAUSA300_0747 | 3913567 | oxidative stress response | regulated through PerR regulon | no | 46 |
| ahpC | alkyl hydroperoxide reductase subunit C | SAUSA300_0380 | 3914843 | oxidative stress response | regulated through PerR regulon | no | 46, 47 |
| nrdE | ribonucleotide diphosphate reductase subunit αlpha | SAUSA300_0716 | 3913497 | DNA repair/synthesis | yes | 49 | |
| nrdF | ribonucleotide diphosphate reductase subunit beta | SAUSA300_0717 | 3913498 | DNA repair/synthesis | requires Mn2+ as a cofactor | yes | 49 |
| recA | recombinase A | SAUSA300_1178 | 3913385 | DNA repair/synthesis | no | 52 | |
| nth | endonuclease III | SAUSA300_1343 | 3912944 | DNA repair/synthesis | no | 51 | |
| uvrA | excinuclease ABC subunit A | SAUSA300_0742 | 3913512 | DNA repair/synthesis | no | 51 | |
| srtA | sortase A | SAUSA300_2467 | 3913526 | cell wall biosynthesis | no | 45 |
Fig 4. Genes involved in oxidative stress response and DNA repair are highly upregulated in the mntC mutant after exposure to a sub-lethal concentration of methyl viologen in vitro.
Expression of genes involved in oxidative stress response (A), DNA repair (B) and the srtA gene (C) was determined via qRT-PCR in wild-type (filled bars) and mntC (open bars) cells grown in RPMI-H media and treated with or without 1 μM methyl viologen (MV) for 1 hour. Bars represent the mean value of triplicate samples and error bars are standard deviation. P-values (* = <0.05, *** = <0.001, **** = <0.0001) were determined using student’s t-test.
Expression of the katA, trxB and ahpC genes is controlled by the peroxide resistance regulon repressor PerR, whose activity depends on intracellular metal concentrations, in particular those of iron and manganese [46, 47]. Since a significant increase in the expression of these three genes in the mntC mutant strain was only observed when bacteria were exposed to a sub-lethal concentration of methyl viologen, low intracellular manganese concentration alone likely is not sufficient to induce the expression of genes regulated by the PerR operon (Fig 4A and 4B). Among the genes tested, three genes encode manganese-dependent enzymes: SodM, SodA and NrdF [48, 49, 50]. Their enzymatic activity is expected to be impaired in the mntC mutant strain due to a non-functional MntABC system. Despite the fact that all the genes involved in oxidative stress response and DNA repair/synthesis tested were highly upregulated in the mntC mutant strain, the mntC mutant strain exposed to a sub-lethal concentration of methyl viologen had a significant growth defect when bacteria were inoculated into fresh media lacking methyl viologen (Fig 3B). This suggests that the proper function of manganese-dependent enzymes during oxidative stress is likely crucial for protecting S. aureus cells from oxidative damage and ensuring efficient growth after oxidative stress.
Next, we analyzed whether similar transcriptional differences between wild-type and mntC mutant strains are observed upon phagocytosis ex vivo. In contrast to bacteria exposed to a sub-lethal concentration of methyl viologen in vitro, nrdEF were the only genes that were induced 2-fold more in the mntC mutant strain than the wild-type strain after exposure to both murine macrophages and human neutrophils (Fig 5A and 5B). nth, which encodes for a DNA repair enzyme [51] was highly induced in murine macrophages, but not in human neutrophils. The gene encoding for the protein RecA [52] was induced neither in murine macrophages nor human neutrophils. The nrdEF genes, which are transcribed together [50], encode the NrdEF class Ib ribonucleotide reductase complex that catalyzes the conversion of nucleotides to deoxynucleotides, providing the monomeric building blocks required for DNA replication and repair [53].
Fig 5. The nrdEF genes are highly upregulated in the mntC mutant after exposure to murine macrophages and human neutrophils ex vivo.
Expression of genes involved in oxidative stress response (filled bars) and DNA repair (striped bars) in the mntC mutant strain relative to the wild-type strain that were phagocytosed by murine macrophages for 2 hours (A) or human neutrophils for 45 minutes (B). Expression levels were determined via qRT-PCR. Bars represent the mean value of triplicate samples and error bars are standard deviation. P-values (ns = non-significant, * = <0.05, *** = <0.001, **** = <0.0001) were determined using student’s t-test.
In order to determine whether the observed upregulation of the nrdEF genes directly translates to an increased protein level and if other proteins are specifically induced or repressed in the mntC mutant strain compared to the wild-type strain in response to oxidative stress, we took a non-biased proteomics approach to compare the protein abundance between the wild-type and mntC mutant strains under oxidative stress conditions using stable isotope labeling by amino acids in cell culture (SILAC) [54]. Proteomic analysis requires a large number of bacterial cells, thus limiting our analysis to bacteria grown in vitro. The wild-type and mntC strains were grown in the presence of a sub-inhibitory concentration of methyl viologen in liquid culture containing either natural (“light”) amino acids or non-radioactive, stable isotope containing (“heavy”) amino acids. When “light” and “heavy” cell populations are mixed, they remain distinguishable by mass spectrometry (MS) and protein abundances are determined from the relative MS signal intensities by LC-MS/MS analysis [54]. We selected proteins that were identified and quantified with a minimum of 2 distinct peptide observations, with the greatest overall mass spectrometric signal levels which had relative abundance ratios 2 or more standard deviations from the mean (Fig 6A and 6B, S2 and S3 Tables). Of those, we focused on proteins that were most upregulated or downregulated in both biological replicas (Fig 6A and 6B, S2 and S3 Tables). 5 proteins were more abundant in the mntC strain compared to the wild-type strain and 4 proteins were present at lower abundance in the mntC mutant strain compared to wild-type (Fig 6A and 6B). Strikingly, NrdE and NrdF were among the most highly upregulated proteins in the mntC mutant strain (Fig 6A and 6B, S2 and S3 Tables). PurH, which is involved in the biosynthesis of purines, the pyruvate kinase Pyk, and IsdB were also more abundant in the mntC strain compared to wild-type (Fig 6A and 6B, S2 and S3 Tables). IsdB is known to be the primary receptor for hemoglobin, but has also been reported to play a role in promoting resistance to hydrogen peroxide and neutrophil killing [55]. Among the proteins that were less abundant in the mntC mutant are SerS, which is involved in protein biosynthesis, as well as ButA and AtpD that function in general energy metabolism [32] (Fig 6A and 6B, S2 and S3 Tables). SerS is likely to be essential for S. aureus viability as no transposon mutants have been identified so far [56–59]. Further studies are needed to determine whether its down regulation has any effect on S. aureus fitness and whether the growth defect of the mntC mutant strain after oxidative stress is in part caused by the down regulation of this essential gene.
Fig 6. The NrdEF proteins are highly induced in the mntC mutant when exposed to oxidative stress in vitro.
(A) MA plot of the log2 transformed normalized ratio of “light” mntC mutant relative to the wild-type strain against the log2 transformed signal intensity arising from the “light” mntC mutant. (B) MA plot of the log2 transformed normalized ratio of “heavy” mntC mutant relative to the wild-type strain against the log2 transformed signal intensity arising from the “heavy” mntC mutant. Proteins with z-scores for ratios between the mntC mutant and wild-type strains in all replicates greater than 2 and smaller than −2 are shown in green and red dots, respectively. Proteins that were identified both in (A) and (B) that had the strongest mass spectrometric signals are shown in bold.
Overall, our transcript and proteomic analyses revealed that the manganese-dependent NrdEF protein complex, which is essential for DNA synthesis and repair, might play a critical role in maintaining S. aureus fitness under oxidative stress conditions. Since NrdEF requires manganese as a cofactor, its function is likely impaired in the mntC mutant strain in the absence of a functional uptake manganese uptake system. The observed induction of NrdEF, both on the mRNA and protein level, in the mntC mutant strain could be an attempt of bacteria to compensate for the impaired function of NrdEF by increasing the protein copy number. NrdEF proteins were shown to be essential for the viability of S. aureus [56–59]. Therefore, an impaired protein function during infection would be detrimental to the growth and survival of the pathogen inside the host.
Discussion
Increasing evidence suggests that the ability of S. aureus, in particular MRSA, to survive within and eventually escape from phagocytic cells contributes to its dissemination and pathogenesis within the host [37–39]. S. aureus readily causes lysis of both neutrophils and macrophages through the production of leukocidins, which allows the bacteria to not only survive inside phagocytes, but also to escape [60–62]. In order to thrive in the host, S. aureus cells that have survived the first oxidative burst of primary phagocytic cells need to rapidly recover from oxidative stress and prevent further oxidative damage by potential secondary phagocytosis events. In this study, we showed that MntABC plays a critical role in ensuring efficient growth of S. aureus cells after phagocytic oxidative burst.
The role of manganese in detoxifying ROS is well established and it has been anticipated that an important role of manganese acquisition in the pathogenesis of S. aureus is to protect bacteria from killing by phagocytic cells. Our analysis revealed that the mntC mutant strain was indeed highly susceptible to the ROS-generating reagent methyl viologen in vitro (S1 Fig); however, the mntC mutant was only marginally more susceptible to phagocytic killing ex vivo compared to the wild-type strain (Fig 1A and 1B). A possible explanation is that the mechanism of ROS generation by methyl viologen and oxidative burst of phagocytic cells is different. Methyl viologen causes redox cycling, i.e. a repetitive cycle of oxidation and reduction, so that ROS generation is continuous [28, 29]. In contrast, the oxidative burst of phagocytic cells represents a rapid, transient release of ROS [30, 31]. Therefore, the amount of ROS that bacteria experience in vitro in the presence of methyl viologen is expected to be significantly higher than in vivo and the greater susceptibility of the mntC mutant to ROS might become more obvious under conditions where high amounts of ROS accumulates. Interestingly, we showed that the mntC mutant strain was killed more rapidly by neutrophils than macrophages. This may be due to differences in the antimicrobial mechanisms of these two phagocytic cell types. For example, neutrophils kill S. aureus more rapidly than macrophages [63]. This is partly attributed to an increased intensity of oxidative burst in neutrophils compared to macrophages [63–68]. HOCl, which is a strong oxidant, is produced by neutrophils, but not by macrophages due to the lack of myeloperoxidase [18].
Although the presence of a functional MntABC system did not fully protect S. aureus cells from phagocytic killing (Fig 1A and 1B), it played a significant role in ensuring efficient bacterial growth after phagocytosis (Fig 2A–2C). It seems likely that in the absence of a functional MntABC system, S. aureus cells endure more oxidative damage due to an impaired ability to acquire a sufficient amount of manganese as evidenced by a profound delay in resuming growth after oxidative stress (Fig 3A and 3B). Although the exact physiological and molecular processes underlying bacterial lag phase are not fully understood, the repair of macromolecular damage [69] and the synthesis of cellular components necessary for growth [70] are thought to be critical. There are two possible mechanisms by which the uptake of manganese could influence the processes underlying the lag phase. The first involves the reduction of ROS-mediated cellular damage, as manganese plays a key role in detoxifying ROS [23, 71]. S. aureus possesses at least two different manganese-dependent superoxide dismutases, SodM and SodA, which play distinct roles in counteracting oxidative stress depending on bacterial growth phase and the nature of oxidative stress [48, 49]. A previous study has demonstrated that the loss of functional manganese acquisition system in S. aureus results in reduced superoxide dismutase activity when the availability of manganese is restricted [11]. The second mechanism involves the repair of cellular damage induced by ROS. Our transcript analysis revealed that among the genes involved in ROS detoxification and DNA repair tested, nrdEF are the highest upregulated genes in the mntC mutant strain relative to the wild-type strain upon phagocytosis (Fig 5A and 5B). Notably, proteomic analysis showed that the NrdEF proteins were the most highly induced proteins in the mntC mutant strain relative to the wild-type strain after exposure to a sub-lethal concentration of methyl viologen in vitro (Fig 6A and 6B). Since NrdEF requires manganese as a cofactor, its function is likely impaired in the mntC mutant strain. The observed induction of NrdEF, both on the mRNA and protein level, in the mntC mutant strain could be an attempt by the bacteria to compensate for the impaired function of NrdEF by increasing the protein copy number. NrdEF proteins have been shown to be essential for the viability of S. aureus [56–59]. Therefore, the potentially impaired function of the protein during infection would be detrimental to the growth and survival of the pathogen inside the host. Additionally, a recent study revealed that the NrdEF complex is important for the replication of E. coli under oxidative stress conditions when the iron concentration is low as found in the host [72].
Overall, our results suggest that a functional manganese acquisition system is not only important for ROS detoxification, as has been previously proposed, but also for repairing oxidative damage. The severe growth defect of the mntC mutant strain after exposure to oxidative stress is therefore likely a consequence of increased damage due to an impaired activity of manganese-dependent superoxide dismutases [11] and the decreased ability in repairing the damage due to an impaired function of the manganese-dependent NrdEF complex.
Supporting Information
Survival of wild-type and mntC mutant strains after overnight incubation with methyl viologen at different concentrations. In addition, survival of the mntC strain in which the stop-codon was repaired, and that of the mntC strain in the presence of 5 μM MgCl2 is shown.
(TIF)
Wild-type and mntC mutant strains were incubated in 1 μM methyl viologen in RPMI-H for 1 hour. The initial inoculum as well as the number of viable cells post-treatment was determined by plating samples onto agar plates containing 5% defibrinated sheep’s blood. Relative CFU/mL (survival) was determined by dividing cell count post-treatment by the cell count of the initial inoculum.
(TIF)
Wild-type and mntC mutant strains were exposed to 1 μM methyl viologen for 1 hour (Figure A) or grown in RPMI-H (Mn-low media) (Figure B) and plated onto agar plates containing 5% defibrinated sheep blood. Images were taken after 16 hours of inoculation at 37°C.
(TIF)
Histogram plots demonstrating the influence of irradiation on the loss of CFSE fluorescence in wild-type cells grown in TSB media (Figure A). Histogram plots showing the loss of CFSE fluorescence in wild-type or mntC mutant cells grown in TSB media (Figure B).
(TIF)
CFSE-labeled wild-type bacteria were diluted into the fresh TSB media (Figure A) or saturated overnight culture of unlabeled bacteria (Figure B). The cultures were shaken at 37°C and samples were taken at indicated time points for FACS analysis.
(TIF)
Survival of wild-type and mntC mutant strains after overnight incubation in 40 μM H2O2 and 5.5 mg/ml NaOCl. The number of viable bacteria was determined by BacTiter-Glo luminescence measurement (Figure A) or by CFU determination (Figure B). Data are from triplicate samples and error bars are standard deviation. * indicates statistical significance (P-values = <0.05) based on student’s t-test.
(TIF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Binh An Diep (California, USA) for providing the bacterial strains.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
All work was supported by internal funds at Genentech, Inc. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript The funder provided support in the form of salaries for authors CA, MX, QP, TKC, CB, MKA, SML, JK, SP, MWT, MN], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis. 2007;13: 1840–1846. 10.3201/eid1312.070629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barlett AH, Hulten KG. Staphylococcus aureus pathogenesis: secretion systems, adhesins, and invasins. Pediatr Infect Dis J. 2010;29: 860–861. 10.1097/INF.0b013e3181ef2477 [DOI] [PubMed] [Google Scholar]
- 3. Lowy FD. Staphylococcus aureus infections. New Engl J Med. 1998;339: 520–532. [DOI] [PubMed] [Google Scholar]
- 4. Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10: 525–537. 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahuja S, Rougé L, Swem DL, Sudhamsu J, Wu P, Russell SJ, et al. Structural analysis of bacterial ABC transporter inhibition by an antibody fragment. Structure. (2015;23: 713–723. 10.1016/j.str.2015.01.020 [DOI] [PubMed] [Google Scholar]
- 6. Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319: 962–965. 10.1126/science.1152449 [DOI] [PubMed] [Google Scholar]
- 7. Yui S, Nakatani Y, Mikami M. Calprotectin (S100A8/S100A9), an inflammatory protein complex from neutrophils with a broad apoptosis-inducing activity. Biol Pharma Bull. 2003;26: 753–760. [DOI] [PubMed] [Google Scholar]
- 8. Forbes JR, Gros P. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol. 2001;9: 397–403. [DOI] [PubMed] [Google Scholar]
- 9. Govoni G, Gauthier S, Billia F, Iscove NN, Gros P. Cell-specific and inducible Nramp1 gene expression in mouse macrophages in vitro and in vivo. J Leukoc Biol. 1997;62: 277–286. [DOI] [PubMed] [Google Scholar]
- 10. Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Paecock S, Foster SJ. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol. 2002;44: 1269–1286. [DOI] [PubMed] [Google Scholar]
- 11. Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, et al. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun. 2013;81: 3395–3405. 10.1128/IAI.00420-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diep BA, Phung Q, Date S, Arnott D, Bakalarski C, Xu M, et al. Identifying potential therapeutic targets of methicillin-resistant Staphylococcus aureus through in vivo proteomics analysis. J Infect Dis. 2014;209: 1533–1541. 10.1093/infdis/jit662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Papp-Wallace KM, Maguire ME. Manganese transport and the role of manganese in virulence. Annu Rev Microbiol. 2006;60: 187–209. [DOI] [PubMed] [Google Scholar]
- 14. Malo D, Vogan K, Vidak S, Hu J, Cellier M, Schurr E, et al. Halotype mapping and sequence analysis of the mouse Nramp gene predict susceptibility to infection with intracellular parasites. Genomics. 1994;23: 51–61. [DOI] [PubMed] [Google Scholar]
- 15. Lim KHL, Jones CE, vanden Hoven RN, Edwards JL, Falsetta ML, Apicella MA, et al. Metal binding specificity of the MntABC permease of Neisseria gonorrhoeae and its influence on bacterial growth and interaction with cervical epithelial cells. Infect Immun. 2008;76: 3569–3576. 10.1128/IAI.01725-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boyer E, Bergevin I, Malo D, Gros P, Cellier MFM. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2002;70: 6032–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McAllister LJ, Tseng HJ, Ogunniyi AD, Jennings MP, McEwan AG, Paton JC. Molecular analysis of the psa permease complex of Streptococcus pneumonia. Mol Microbiol. 2012;53: 215–235. [DOI] [PubMed] [Google Scholar]
- 18. Hampton MB, Kettle AJ, Winterbourn CC. Involvement of superoxide and myeloperoxidase in oxygen-dependent killing of Staphylococcus aureus by neutrophils. Infect Immun. 1996;64: 3512–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DeLeo FR, Diep BA, Otto M. Host defense and pathogenesis in Staphylococcus aureus infections. Infect Dis Clin North Am. 2009;23: 17–34. 10.1016/j.idc.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fang FC. Antimicrobial actions of reactive oxygen species. mBio. 2011;2: e00141–11. 10.1128/mBio.00141-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240: 1302–1309. [DOI] [PubMed] [Google Scholar]
- 22. Imlay JA. How oxygen damages microbes: Oxygen tolerance and obligate anaerobiosis. Adv Microb Physiol. 2002;46: 111–153. [DOI] [PubMed] [Google Scholar]
- 23. Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64: 97–112. [DOI] [PubMed] [Google Scholar]
- 24. Sobota JM, Imlay JA. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc Natl Acad Sci U.S.A. 2011;108: 5402–5407. 10.1073/pnas.1100410108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karavolos MH, Horsburgh MJ, Ingham E, Foster SJ. Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology. 2003;149: 2749–2758. [DOI] [PubMed] [Google Scholar]
- 26. Berlett BS, Chock PB, Yim MB, Stadtman ER. Manganese(II) catalyzes the bicarbonate-dependent oxidation of amino acids by hydrogen peroxide and the amino acid-facilitated dismutation of hydrogen peroxide. Proc Natl Acad Sci U.S.A. 1990;87: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77: 755–776. 10.1146/annurev.biochem.77.061606.161055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hassan HM, Fridovich I. Paraquat and Escherichia coli. Mechanism of production of exracellular superoxide radical. J Biol Chem. 1979;254: 10846–10852. [PubMed] [Google Scholar]
- 29. Gray JP, Heck DE, Mishin V, Smith PJ, Hong JY, Thiruchelvam M, et al. Paraquat increases cyanide-insensitive respiration in murine lung epithelial cells by activating an NAD(P)H:paraquat oxidoreductase: identification of the enzyme as thioredoxin reductase. J Biol Chem. 2007;282: 7939–7949. [DOI] [PubMed] [Google Scholar]
- 30. Steinbeck MJ, Hegg GG, Karnovsky MJ. Arachidonate activation of neutrophil NADPH-oxidase. J Biol Chem. 1991;266: 16366–16342. [PubMed] [Google Scholar]
- 31. Badwey JA, Karnovsky ML Active oxygen species and the functions of phagocytic leukocytes. Annu Rev Biochem. 1980;49: 16366–16342. [DOI] [PubMed] [Google Scholar]
- 32. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet. 2006;367: 731–739. [DOI] [PubMed] [Google Scholar]
- 33. Pham VC, Pitti R, Anania VG, Bakalariski CE, Bustos D, Jhunjhunwala S, et al. Complementary proteomic tools for the dissection of apoptotic proteolysis events. J Proteome Res. 2012;11: 2947–2954. 10.1021/pr300035k [DOI] [PubMed] [Google Scholar]
- 34. Bakalarski CE, Elias JE, Villén J, Haas W, Gerber SA, Everley PA, et al. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J Proteome Res. 2008;11: 4756–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Freitas M, Porto G, Lima JL, Fernandes E. Isolation and activation of human neutrophils in vitro. The importance of the anticoagulant used during blood collection. Clin Biochem. 2008;41: 570–575. 10.1016/j.clinbiochem.2007.12.021 [DOI] [PubMed] [Google Scholar]
- 36. Mohanty K, Mishra S, Pani J, Hasan T, Purohit A, Sharma S, et al. Heparin or EDTA; anticoagulant of choice in free radical estimation? Oxid Antioxid Med Sci. 2012;1: 21–24. [Google Scholar]
- 37. Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, Gajkowska B, et al. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. Plos One. 2008;3: e1409 10.1371/journal.pone.0001409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kobayashi SD, Braughton KR, Palazzolo-Balance AM, Kennedy AD, Sampaio E, Kristosturyan E, et al. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J Innate Immun. 2010;2: 560–575. 10.1159/000317134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gresham HD, Lowrance JH, Caver TE, Wilson BS, Cheung AL, Lindberg FP. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J Immunol. 2000;164: 3713–3722. [DOI] [PubMed] [Google Scholar]
- 40. Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Hermann M, et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4: 295–305. [DOI] [PubMed] [Google Scholar]
- 41. Stanley PE. A review of bioluminescent ATP techniques in rapid microbiology. J Biolumin Chemilumin. 1989;4: 375–380. [DOI] [PubMed] [Google Scholar]
- 42. Lyons AB, Parish CR. Determination of lymphocyte division by flow-cytometry. J Immunol Methods. 1994;171: 131–137. [DOI] [PubMed] [Google Scholar]
- 43. O’Donnell VB, Tew DG, Jones OTG, England PJ. Studies on the inhibitory mechanism of iodonium compounds with special reference to neutrophil NADPH oxidase. Biochem J. 1993;290: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hassett DJ, Cohen MS. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J. 1989;3: 2574–2582. [DOI] [PubMed] [Google Scholar]
- 45. Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285: 760–763. [DOI] [PubMed] [Google Scholar]
- 46. Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun. 2001;69: 3744–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, et al. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol. 2007;189: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Valderas MW, Hart ME. Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus. J Bacteriol. 2001;183: 3399–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clements MO, Watson SP, Foster SJ. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181: 3898–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Masalha M, Borovok R, Schreiber Y, Aharonowitz, Cohen G. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J Bacteriol. 2001;183: 7260–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang W, Small DA, Toghrol F, Bentley WE. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J Bacteriol. 2006;188: 1648–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bayles KW, Brunskill EW, Iandolo JJ, Hruska LL, Huang S, Pattee PA, et al. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Gene. 1994;147: 13–20. [DOI] [PubMed] [Google Scholar]
- 53. Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75: 681–706. [DOI] [PubMed] [Google Scholar]
- 54. Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;5: 376–386. [DOI] [PubMed] [Google Scholar]
- 55. Palazzolo-Balance AM, Reniere ML, Braughton KR, Sturdevant DE, Otto M, Kreiswirth BN, et al. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180: 500–509. [DOI] [PubMed] [Google Scholar]
- 56. Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, et al. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridization (TMDH). BMC Genomics. 2009;10: 291 10.1186/1471-2164-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, et al. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci U.S.A. 2004;101: 12312–12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4: e00537–12. 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chrisiansen MT, Kaas RS, Chaudhuri RR, Holmes MA, Hasman H, Aarestrup FM. (2014) Genome-wide high-throughput screening to investigate essential genes involved in methicillin-resistant Staphylococcus aureus sequence type 398 survival. PLoS One. 2013;9: e89018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. DuMont AL, Yoong P, Surewaard BGJ, Benson MA, Nijland R, van Strijp JAG, et al. Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect Immun. 2013;81: 1830–41. 10.1128/IAI.00095-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dumont AL, Nygaard TK, Watkins RL, Smith A, Kozhaya L, Kreiswirth BN, et al. Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol Microbiol. 2011;79: 814–25. 10.1111/j.1365-2958.2010.07490.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol. 2012;34: 237–59. 10.1007/s00281-011-0295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Silva MT. When two is better than one: macrophages and neutrophils work in concert in innate immunity as complementary and cooperative partners of a myeloid phagocyte system. J Leukoc Biol. 2009;87: 93–106. [DOI] [PubMed] [Google Scholar]
- 64. Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U.S.A. 2000;97: 8841–8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nordenfelt P, Tapper H. Phagosome dynamics during phagocytosis by neutrophils. J Leukoc Biol. 2011;90: 271–284. 10.1189/jlb.0810457 [DOI] [PubMed] [Google Scholar]
- 66. Johansson A, Jesaitis AJ, Lundqvist H, Magnusson KE, Sjölin C, Karlsson A, et al. Different subcellular localization of cytochrome b and the dormant NADPH-oxidase in neutrophils and macrophages: effect on the production of reactive oxygen species during phagocytosis. Cell Immunol. 1995;161: 61–71. [DOI] [PubMed] [Google Scholar]
- 67. VanderVen BC, Yates RM, Russell DG. Intraphagosomal measurement of the magnitude and duration of the oxidative burst. Traffic. 2009;10: 372–378. 10.1111/j.1600-0854.2009.00877.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Silva MT, Correia-Neves M. (2012) Neutrophils and macrophages: the main partners of phagocyte cell systems. Front Immunol. 3: 174 10.3389/fimmu.2012.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dukan S, Nystrom T. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 1998;12: 3431–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rolfe MD, Rice CJ, Lucchini S, Pin C, Thompson A, Cameron AD, et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J Bacteriol. 2012;194: 686–701. 10.1128/JB.06112-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jakubovics NS, Jenkinson HF. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology. 2001;147: 1709–1718. [DOI] [PubMed] [Google Scholar]
- 72. Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol. 2011;80: 319–334. 10.1111/j.1365-2958.2011.07593.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Survival of wild-type and mntC mutant strains after overnight incubation with methyl viologen at different concentrations. In addition, survival of the mntC strain in which the stop-codon was repaired, and that of the mntC strain in the presence of 5 μM MgCl2 is shown.
(TIF)
Wild-type and mntC mutant strains were incubated in 1 μM methyl viologen in RPMI-H for 1 hour. The initial inoculum as well as the number of viable cells post-treatment was determined by plating samples onto agar plates containing 5% defibrinated sheep’s blood. Relative CFU/mL (survival) was determined by dividing cell count post-treatment by the cell count of the initial inoculum.
(TIF)
Wild-type and mntC mutant strains were exposed to 1 μM methyl viologen for 1 hour (Figure A) or grown in RPMI-H (Mn-low media) (Figure B) and plated onto agar plates containing 5% defibrinated sheep blood. Images were taken after 16 hours of inoculation at 37°C.
(TIF)
Histogram plots demonstrating the influence of irradiation on the loss of CFSE fluorescence in wild-type cells grown in TSB media (Figure A). Histogram plots showing the loss of CFSE fluorescence in wild-type or mntC mutant cells grown in TSB media (Figure B).
(TIF)
CFSE-labeled wild-type bacteria were diluted into the fresh TSB media (Figure A) or saturated overnight culture of unlabeled bacteria (Figure B). The cultures were shaken at 37°C and samples were taken at indicated time points for FACS analysis.
(TIF)
Survival of wild-type and mntC mutant strains after overnight incubation in 40 μM H2O2 and 5.5 mg/ml NaOCl. The number of viable bacteria was determined by BacTiter-Glo luminescence measurement (Figure A) or by CFU determination (Figure B). Data are from triplicate samples and error bars are standard deviation. * indicates statistical significance (P-values = <0.05) based on student’s t-test.
(TIF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.