Abstract
Objective
Dementia with Lewy bodies is an α-synucleinopathy characterized by neocortical Lewy-related pathology (LRP). We carried out a genome-wide association study (GWAS) on neocortical LRP in a population-based sample of subjects aged 85 or over.
Methods
LRP was analyzed in 304 subjects in the Vantaa 85+ sample from Southern Finland. The GWAS included 41 cases with midbrain, hippocampal, and neocortical LRP and 177 controls without midbrain and hippocampal LRP. The Medical Research Council Cognitive Function and Ageing Study (CFAS) material was used for replication (51 cases and 131 controls).
Results
By analyzing 327,010 markers the top signal was obtained at the HLA-DPA1/DPB1 locus (P = 1.29 × 10−7); five other loci on chromosomes 15q14, 2p21, 2q31, 18p11, and 5q23 were associated with neocortical LRP at P < 10−5. Two loci were marked by multiple markers, 2p21 (P = 3.9 × 10−6, upstream of the SPTBN1 gene), and HLA-DPA1/DPB1; these were tested in the CFAS material. Single marker (P = 0.0035) and haplotype (P = 0.04) associations on 2p21 were replicated in CFAS, whereas HLA-DPA1/DPB1 association was not. Bioinformatic analyses suggest functional effects for the HLA-DPA1/DPB1 markers as well as the 15q14 marker rs8037309.
Interpretation
We identified suggestive novel risk factors for neocortical LRP. SPTBN1 is the candidate on 2p21, it encodes beta-spectrin, an α-synuclein binding protein and a component of Lewy bodies. The HLA-DPA1/DPB1 association suggests a role for antigen presentation or alternatively, cis-regulatory effects, one of the regulated neighboring genes identified here (vacuolar protein sorting 52) plays a role in vesicular trafficking and has been shown to interact with α-synuclein in a yeast model.
Introduction
Analysis of abnormal protein accumulation plays an important role in the neuropathological classification of neurodegenerative disorders. Alzheimer's disease (AD) is characterized by β-amyloid plaques and intracellular neurofibrillary tangles, composed of hyperphosphorylated tau protein. Parkinson's disease (PD) is characterized by intraneuronal Lewy bodies and Lewy neurites (Lewy-related pathology, LRP) in the brainstem. The main component of Lewy bodies is conformationally modified α-synuclein.1,2 Anatomical spreading of the LRP into neocortex often results in cognitive and behavioral symptoms.
Neocortical LRP is found in at least three clinically defined conditions: in PD with dementia, in dementia with Lewy bodies (DLB) and in Lewy body variant of AD. These disorders are considered to constitute a continuum with varying weighting of the symptoms and neuropathological features. Yoshimura suggested that an intermediate phenotype between AD and PD represents a disorder of its own, which he termed “Diffuse Lewy body disease”.3 However, clinical characterization of this disorder has been difficult and no specific biomarkers have been available. These ambiguities are reflected in the various terms that have been used, the most common of which is DLB. Neuropathological classification of Lewy body disorders has also been challenging, the criteria have been widely debated and subject to many revisions. Today both classical Lewy bodies and Lewy neurites are regarded as neuropathological hallmarks of DLB and termed as “LRP.” The most recent proposal classifies LRP as brainstem, limbic, or neocortical-predominant categories based on the anatomical spreading.4 Virtually all subjects with neocortical LRP have brainstem and limbic pathology, too.
There has been significant progress in deciphering the genetic background of AD and PD. However, the “intermediate phenotype” DLB, has remained genetically less well characterized. Most DLB patients are sporadic, but a few DLB families have been identified. Mutations in PD-related genes α-synuclein (SNCA), Leucine-rich repeat kinase-2 (LRRK2), and Glucocerebrosidase-A (GBA) have been described in DLB patients with onset before age 65.5–11 Overlap with AD is found, too, both pathologically and genetically. Cortical Lewy bodies are relatively commonly found in combination with AD pathology in patients diagnosed as AD. Amyloid precursor protein (APP) and Presenilin-2 (PSEN-2) mutations typically lead to early-onset AD, but the phenotypic spectrum may include features of DLB.12,13 In addition to the genetic findings overlapping with PD or AD, two different presumably pathogenic β-synuclein (SNCB) mutations have been found in two unrelated DLB patients14 and, in a Belgian family, linkage between DLB and chromosome 2q35–q36 has been reported.15 Genetic analyses of sporadic late-onset DLB cases have identified associations with both AD and PD genes, such as APOE, SNCA,and SCARB2.16–18
Despite these advances, the genetic background of the common late-onset sporadic form of DLB has remained unclear. Here, we have carried out a neuropathology-based genome-wide association study (GWAS) using the presence of neocortical LRP as the phenotypic trait in a population-based setting. Such analysis is free from ambiguities of clinical diagnostics (differentiation between PD-dementia, DLB, and Lewy body variant of AD) and from selection bias often involved in patient materials collected from referral-based institutions.
Subjects and Methods
Subjects in Vantaa 85+
The Vantaa 85+ study includes all 601 persons aged 85 years or over who were living in the city of Vantaa (Southern Finland), on 1 April 1991. The study design has been described in detail earlier.19,20 Autopsies were carried out in 304 subjects, median age at death was 92.2 years (females 83%). The study was approved by the Ethical review committee of the City of Vantaa. The use of the health and social work records and death certificates was approved by the Finnish Health and Social Ministry by the Finnish Ministry of Social Affairs and Health. The collection of the tissue samples at autopsy, and their use for research, was approved by the National Authority for Medicolegal Affairs and coordinating ethical committee of the Helsinki and Uusimaa Health care district (74/13/03/00/2014). Consent for participation in the study and autopsy was obtained from the subjects and/or their nearest relatives.
Pathology in Vantaa 85+
The brains of the autopsied subjects were fixed in phosphate-buffered 4% formaldehyde for at least 2 weeks before sampling. Tissue samples were obtained following recommendations of the first Consortium for DLB (CDLB) workshop for assessing LRP.21 The analysis of LRP has been described in detail earlier.19 Briefly, a two-step analysis was used. First, sections from the midbrain and hippocampus were stained with the hematoxylin and eosin method and with immunohistochemical method for α-synuclein (primary antibody from Transduction Laboratories, Lexington, KY, clone42, mouse monoclonal, diluted 1:800). Second, if any LRP was detected in the screened areas, immunohistochemical staining for α-synuclein was performed on samples from the temporal, frontal, and parietal neocortex and cingulate gyrus. Semiquantitative scoring of LRP (none, mild, moderate, severe, and very severe) and assignment of the type of LRP (none, brainstem-predominant, limbic, diffuse neocortical) was performed by a single investigator (M. Oinas) following the modified Third CDLB guidelines for diagnosis.21 There were 47 subjects (15%) with neocortical LRP in the 304 brains analyzed in the Vantaa 85+ study; 20 of these 47 had a Braak stage V–VI.19 Genotyping was possible in 41 subjects (cases) with diffuse neocortical LRP and in 177 subjects (controls) with no LRP in the brainstem and hippocampus.
CFAS study
The Medical Research Council Cognitive Function and Ageing Study (CFAS) is a longitudinal, prospective, population-based cohort study undertaken in six UK centers initiated in 1989 (www.cfas.ac.uk). It has been previously described.22 The study included a random sample of 18,226 people 65 years and over. A subsample of respondents was asked whether they, with family support, were willing to consent to brain donation after their death. Median age at death for CFAS brain donors was 87 years (females 41%, donations ongoing). The burden and anatomic distribution of α-synuclein was investigated in a subsample of donations (in two of the six centers) before July 2003 (n = 208). The method to assess LRP has been previously described.23 A hierarchical sampling strategy, based on evaluation of the midbrain (substantia nigra), medulla, and amygdala, was used to immunohistochemically detect α-synuclein in this cohort (primary monoclonal antibody LB509; Zymed Laboratories Inc., San Francisco, CA). If an α-synuclein immunoreactive profile was found in a screening area a further five areas recommended by the First CDLB 1996,21 the same as in the Vantaa 85+ study, were investigated. There were 54 subjects (cases) who showed LRP in at least one of the three regions brainstem, limbic, or neocortex (brainstem only n = 24, limbic n = 2, brainstem + limbic n = 9, neocortical n = 19) and 138 subjects (controls) who did not show LRP in the three aforementioned regions23 (Fig.2 therein). The controls included the brainstem-negative amygdala-predominant group (n = 22) because subjects with this type of pathology were classified as control in the Vantaa 85+ material (they were negative for brainstem and hippocampal LRP). Genotyping was successful in 51 cases with LRP and in 131 controls. Because of the lower sensitivity of the antibody used in the CFAS study,4 and lower number of subjects with neocortical LRP (19/208 [9%] overall; genotyping successful in 17 cases), we chose to pool the subjects with brainstem, limbic, and neocortical LRP for the genetic analyses. Thus, we increased the number of cases at the expense of the regional specificity of LRP.
Figure 2.
Regional association plot and linkage disequilibrium structure of the chromosome 2p21 markers at the C2ORF73 and SPTBN1 genes.
Genotyping
Infinium Human370 BeadChips (Illumina, San Diego CA), which assay 345,111 single-nucleotide polymorphisms (SNPs) across the genome, was used for genotyping the Vantaa 85+ samples. Standard quality control procedures were applied as follows: exclusion of samples with SNP call rates of less than 95%, cryptic relatedness, non-European ancestry, minor allele frequency (MAF) less than 0.01, and Hardy–Weinberg equilibrium P value of less than 0.001 as reported.24 Two-hundred and eighteen subjects with 327,010 SNPs, including sex-chromosomal SNPs, were analyzed. Bonferroni corrected threshold for genome-wide significance with this data would be 1.56 × 10−7 (α = 0.05/327,010 SNPs). Genotyping of the CFAS study was carried out by Sanger sequencing with the following forwards (F) and reverse (R) primers: rs9277685-rs9277682-F 5′-tct ggt ggt cca att tcc-3′; rs9277685-rs9277682-R 5′-cca ctg act cca agt atg-3′; rs2071349-F1 5′-gag gtg tgg cag aat tgg-3′, rs2071349-R1 5′-tct gtg acc ctg gga ttg-3′; rs2301226-F1 5′-ttg cag ggt tgct gga gat g-3′; rs2301226-R1 5′-cca agg aga cag ttg cca gaa g-3′; rs9277334-F1 5′-ata tgg gca tgg cgt gat gag-3′; rs9277334-R1 5′-tgg aag tgg gta cgt cac aac-3′; rs4671212-F1 5′-ttc aca gtg tgg agc aga ac-3′; rs4671212-R1 5′-agc ctc tgt ctc tac tca cta c-3′; rs4315567-F1 5′-cct cct atg tcc tcc ctt aac-3′; rs4315567-R1 5′-tag tct gtg ctg cca gat g-3′. HLA-DPB1 typing was carried out by sequence-specific oligonucleotide probes using OLERUP SSP DPB1 kit (www.olerup-ssp-com). SPTBN1 and C2ORF73 re-sequencing was carried out by Sanger sequencing of PCR products using the following primers. SPTBN1 promoter-exon 1: 5′-cgt gaa att ggc cct ctc cg-3′ and 5′-tcc cgc atc atc cgt ga tacc-3′; SPTBN1 exon 2: 5′- gat atc ggc tca cta caa cct-3′ and 5′-ttc agg cca gct caa gaa aga tc-3′; SPTBN1 exon 3: 5′- gca ggt gaa gac ggt cat tgc-3′ and 5′-cat gtg ctc tgg gag gat aca-3′; C2ORF73 promoter-exon 1: 5′-cca ctc ctt act cac caa ac-3′ and 5′-cgc tca gcc aac tgg aaa tta g-3′; C2ORF73 exon 2: 5′- aac aca aag ccc ttt atc g-3′ and 5′- cac tta gct cat tcc tag aac-3′; C2ORF73 exon 3: 5′- ggc tag gga cta aaa ctt c-3′ and 5′- tgg tgg caa caa caa tga g-3′; C2ORF73 exons 4–5: 5′-cac cat gcc tga cca tat tg-3′ and 5′- gcc tac tgc ctg gtt tta tc-3′; C2ORF73 exon 6: 5′- ctg gct ttg cct aat ttc-3′ and 5′- agt acc caa atg gta ctg-3′, C2ORF73 exons 7–8: 5′-cgg cgg aga tgg cag tat atg ac-3′ and 5′- gtc aga agg cag aca gcc aag ag-3′.
Statistical analyses and bioinformatics
Whole genome associations were calculated with PLINK (allelic chi-square test without covariates, and by logistic regression with age, sex, and AD-pathology as co-variates http://pngu.mgh.harvard.edu/purcell/plink/). Beadstudio was used in the first quality control to determine that a beadchip had worked and transferring data from beadchips to PLINK format. Haplotype association and linkage disequilibrium structures were calculated with the Haploview software. In silica quantitative trait locus (QTL) analysis methods was carried out in the North American Brain Expression Consortium (NABEC) and United Kingdom Brain Expression Consortium (UKBEC) data.25–27 Brain mRNA expression and DNA methylation have been assayed in brains without determinable neuropathological evidence of disease. Expression of mRNA was assayed using Illumina HumanHT-12 v3 Expression Beadchips, methylation was assayed on bisulfite converted DNA using the Illumina Infinium HumanMethylation27 BeadChips. Genotyping was performed using Illumina HumanHap550 v3, Human610-Quad v1 or Human660W-Quad v1 Infinium Beadchips. The combined annotation-dependent depletion (CADD) tool28 was also use to analyze possible functionality of the top SNPs.
Results
In the GWAS we compared the 41 cases with neocortical LRP to the 177 controls without midbrain and hippocampal LRP. Five association peaks with P < 10−5 were found (Fig.1, Table1). Two of these signals showed multiple flanking-associated SNPs, one on chromosome 2p21 between the C2ORF73 and beta-spectrin family gene (SPTBN1) (P = 3.86 × 10−6, allelic test), the other on chromosome 6p21 at the HLA-DPA1 and -DPB1 loci (P = 1.29 × 10−7, allelic test). Logistic regression using AD pathology as a covariate did not abolish the five association peaks, suggesting that these associations are largely driven by neocortical LRP (Table S1). The Q-Q plot indicates that the number of observations at P < 10−4 is higher than expected (Fig. S1). By imputation using MAF filter >0.02 and r2 > 0.30 we did not detect any association reaching genome-wide significance (threshold set at 5 × 10−8 for imputation-derived signals).
Figure 1.

Manhattan plot of Lewy-related pathology in the Vantaa 85+ study, showing −log10 P-values for the 327,010 markers ordered by their chromosomal position. The horizontal lines indicate the threshold for genome-wide significance (P = 1.56 × 10−7) and P = 10−5.
Table 1.
P-values, positions, nearest gene, frequencies and OR of all the SNPs associated with Lewy-related pathology at P < 10−5 in the Vantaa 85+ genome-wide association study
| Chr | SNP | Position | Gene | P | Risk allele | Risk allele frequency | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| 6 | rs9277685 | 33196062 | HLA-DPB1 | 1.29E-07 | A | 0.214485 | 5.31 (2.59 to 10.91) |
| 6 | rs9277334 | 33138090 | HLA-DPA1 | 9.65E-07 | C | 0.192308 | 5.27 (2.56 to 10.81) |
| 6 | rs2301226 | 33142574 | HLA-DPA1 | 1.16E-06 | T | 0.19346 | 3.75 (2.15 to 6.54) |
| 15 | rs8041665 | 35937471 | Intergenic | 1.39E-06 | A | 0.045726 | 7.41 (2.92 to 18.81) |
| 15 | rs8037309 | 35937730 | Intergenic | 1.39E-06 | T | 0.045726 | 7.41 (2.92 to 18.81) |
| 6 | rs4713610 | 33215933 | HLA-DPB1 | 1.51E-06 | G | 0.207756 | 3.51 (2.02 to 6.11) |
| 6 | rs2071349 | 33151498 | HLA-DPB1 | 2.08E-06 | G | 0.197802 | 3.63 (2.08 to 6.32) |
| 6 | rs9277656 | 33192126 | HLA-DPB1 | 2.50E-06 | T | 0.252174 | 3.41 (2.01 to 5.79) |
| 2 | rs7595929 | 54479744 | SPTBN1 | 3.86E-06 | T | 0.301493 | 3.23 (1.93 to 5.39) |
| 2 | rs4315567 | 54509448 | SPTBN1 | 4.86E-06 | T | 0.273256 | 3.21 (1.92 to 5.38) |
| 2 | rs3796058 | 172650694 | MAP10 | 4.97E-06 | C | 0.258621 | 3.23 (1.92 to 5.44) |
| 6 | rs2395349 | 33191112 | HLA-DPB1 | 5.01E-06 | A | 0.258621 | 3.27 (1.93 to 5.52) |
| 6 | rs9277682 | 33195662 | HLA-DPB1 | 5.01E-06 | C | 0.279805 | 3.27 (1.93 to 5.52) |
| 18 | rs1472194 | 1200675 | Intergenic | 5.19E-06 | G | 0.137662 | 8.06 (2.84 to 22.87) |
| 5 | rs6872138 | 116447410 | Intergenic | 6.40E-06 | G | 0.171123 | 3.82 (2.07 to 7.45) |
| 5 | rs1459086 | 116416478 | Intergenic | 7.15E-06 | T | 0.214485 | 3.54 (1.99 to 6.30) |
OR, odds ratios; SNP, single-nucleotide polymorphism.
A list of all SNPs with a P < 10−3 (n = 336) are shown in Table S2. The results at the previously implicated DLB-loci (GBA,LRRK2, SNCA,SNCB,2q35-q36, APP, PSEN2, APOE, SCARB2) are provided in Table S3, of these, the lowest P-value was observed with a SNP (rs12694814, P = 0.0011) within the delta/notch-like EGF repeat containing (DNER) gene on 2q36. APOE ε4 was nominally associated with neocortical LRP (P = 0.004, Table S3). However, when a logistic regression analysis was applied with AD pathology as a covariate, this association was lost (P = 0.5279, Table S1) suggesting that the APOE ε4 association is largely driven by concomitant AD pathology. A more thorough analysis on AD and PD loci in neocortical LRP and its pathological subtypes using other pathologies as covariates will be reported separately (L. Myllykangas et al., unpubl. ms.).
A more detailed view to the chromosome 2p21 peak is given in Figure2. Based on the haplotype block structure the associated block is between the C2ORF73 and SPTBN1 genes. A 9-SNP haplotype within this block was associated with neocortical LRP (P = 5.2 × 10−7). This haplotype was ∼48 kb wide and was located upstream of the SPTBN1 including its promoter. The whole C2ORF73 gene and the SPTBN1 promoter and exons 1–3 (located within ∼100 kb from the two top SNPs) were re-sequenced in three cases with this haplotype. One missense variation was found in the C2ORF73 gene (Asn29His, rs55714450). No sequence variations were found in the SPTBN1 promoter and exons 1–3. The rs5571450 allele A associated with neocortical LRP (allelic test χ2 = 8.18, 1 df, P = 4.2 × 10−3, recessive test χ2 = 12.7, 2 df, P = 3.6 × 10−4).
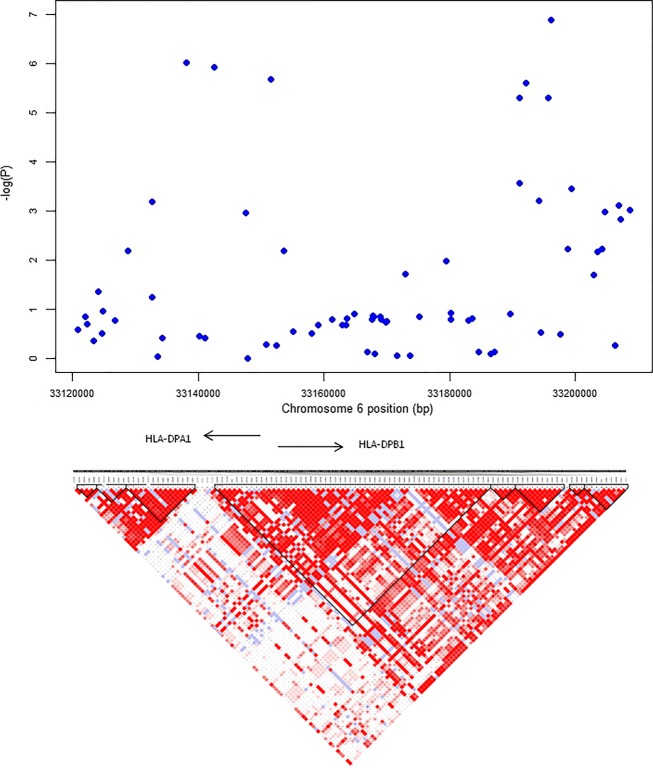
The associated haplotype block in the HLA region was ∼150 kb wide and included HLA-DPA1 and -DPB1 genes (Fig.3). A six-SNP haplotype was associated with LRP (P = 1.10 × 10−7, markers listed in Fig.3) and another haplotype defined by the same six SNPs was associated with protection against neocortical LRP (P = 0.005). Four individuals homozygous for the predisposing haplotype and three individuals homozygous for the putative protective haplotype were typed for HLA-DPB1. All carriers of the predisposing haplotype were HLA-DPB1*0201 homozygotes. All carriers of the protective haplotype were carriers of HLA-DPB1*0401, two homozygous, one heterozygous.
Figure 3.
Regional association plot and linkage disequilibrium structure of the chromosome 6 markers at the HLA-DPA1,HLA-DPB1, and COL11A2 genes. Haplotype analysis was performed with the following six markers: rs2395349, rs9277656, rs3117035, rs1883414, rs9277682, and rs9277685. The predisposing haplotype was defined by the alleles ATACCA and the putative protective haplotype by the alleles GGGCTG.
We analyzed two 2p21 SNPs and five HLA-DPA1/DPB1 SNPs in the CFAS material, three additional SNPs failed in genotyping by Sanger sequencing. One of the chromosome 2p21 SNPs (P = 0.0035) and haplotypes associated with either predisposition to (P = 0.044) or protection from LRP (P = 0.011) were replicated in the CFAS material (Table2). The joint analysis of the predisposing haplotype strengthened the association (P = 4.0 × 10−7). The HLA-DPA1/DPB1 SNPs did not show nominally significant associations with neocortical LRP in the CFAS material (Table2).
Table 2.
Chromosomes 2 and 6 associations in the CFAS materials
| Vantaa-85+ P-value | CFAS P-value | Combined P-value | |
|---|---|---|---|
| Chromosome 2 | |||
| rs4671212 | 0.012 | 0.0035 | 3.6 × 10−5 |
| rs43155671 | 2.6 × 10−6 | 0.12 | 3.1 × 10−6 |
| GT-haplotype | 9.5 × 10−7 | 0.044 | 4.0 × 10−7 |
| TG-haplotype | 0.016 | 0.011 | 1.2 × 10−4 |
| Chromosome 6 | |||
| rs9277334 | 1.4 × 10−6 | 0.48 | 9.7 × 10−5 |
| rs2301226 | 1.6 × 10−6 | 0.24 | 2.3 × 10−5 |
| rs2071349 | 2.8 × 10−6 | 0.075 | 5.4 × 10−6 |
| rs9277682 | 5.9 × 10−6 | 0.97 | 1.1 × 10−3 |
| rs9277685 | 1.1 × 10−7 | 0.97 | 2.2 × 10−4 |
The two SNPs that best separated the chromosome 2 locus haplotype (rs7595929, rs4315567) were selected for genotyping by sequencing in the CFAS material. One of the SNPs (rs7595929) failed in sequencing but another SNP rs4671212, was located in the sequenced area. The GT-haplotype was associated with predisposition to and the TG-haplotype with protection from Lewy-related pathology in the Vantaa 85+ and CFAS materials. Seven SNPs with a P-value under 10−5 were selected from the HLA-DPA1/DPB1 locus, two of the SNPs failed in sequencing in the CFAS material. CFAS, Cognitive Function and Ageing Study; SNPs, single-nucleotide polymorphisms.
To analyze possible functional effects of the 16 top SNPs in the GWAS (P < 10−5 shown in Table1), we analyzed possible association of these SNPs with chromosomal methylation and mRNA expression (cis QTLs) from the NABEC-UKBEC frontal cortex and cerebellum data.25–27 The mRNA expression analysis (data shown in Table S4) suggest that the HLA-DPA1/DPB1 locus risk alleles modify the expression of the Vacuolar protein sorting 52 (VPS52, downregulation), Beta 1,3 galactosyltransferase, polypeptide 4 (B3GALT4, upregulation) and Transporter associated with antigen processing binding protein (TAPBP, upregulation) genes, which are located 160–220 kb centromeric from HLA-DPB1. The methylation analysis indicates that the HLA-DPA1/DPB1 locus SNPs modify the methylation of VPS52. We also analyzed CADD scores28 of the same 16 top SNPs. The chromosome 15 rs8037309 showed a significant CADD-score 29.3 suggesting a possible functional role for this intergenic SNP (Table S4).
Discussion
Although DLB was first recognized as a disease entity already 30 years ago, understanding of its pathogenesis and genetic background is still very limited. The development of neocortical LRP is part of a spectrum of neurodegenerative mechanisms that overlaps with both AD and PD.21,29 Accordingly, many of the previous genetic findings implicate AD and PD genes.5–18 A GWAS meta-analysis was recently reported in which LRP as a trait was analyzed slightly differently from our study by dichotomy (absent vs. present in any brain region), three category endpoint (none, brainstem-predominant, and all other regions or not specified) or five category endpoint (none, brainstem-predominant, limbic, neocortical, and other regions or not specified).18 Using these endpoints APOE ε4 associated with LRP at the genome-wide significant level illustrating a strong link with a major AD gene.18 In our data the APOE association was driven by the subjects with concomitant AD pathology suggesting that a subgroup reminiscent of the “Lewy body variant of AD” would be responsible for the APOE signal in the Vantaa 85+ material.
Here, we report the results of a GWAS using “neocortical LRP versus none” as the endpoint in a population-based neuropathologically examined material of very elderly subjects (Vantaa 85+). At least two interesting loci were revealed: the chromosome 2p21 locus and the chromosome 6p21/HLA-DPA1/DPB1 locus. The top SNPs were not replicated in the CFAS material, but nominally significant associations were found with the chromosome 2p21 locus markers and haplotypes (Table2). The replication analysis of the HLA-DPA1/DPB1 locus did not yield nominally significant associations in the CFAS material. A few other potentially interesting loci were detected at P < 10−5 (Table1) and a larger list of other possible risk loci (P < 10−3) is provided in Table S2.
It is possible that the differences in the HLA-DPA1/DPB1 results reflect the differences in the study populations or neuropathological methods. First, the CFAS study population is somewhat younger than the Vantaa 85+ and with more males. The risk allele profile may vary as a function of age and sex. Second, the British population is genetically more heterogeneous than the Finns, thereby genetic association maybe harder to detect. Third, different methods were used when assessing the LRP, which may have affected the sensitivity of detecting LRP.4 The neuropathological phenotype of the cases was less purely neocortical in the CFAS material as in the Vantaa 85+.
The chromosome 2p21 peak is located between the C2ORF73 and SPTBN1 genes. The whole C2ORF73 gene and SPTBN1 promoter and exons 1–3 were re-sequenced. A common nonsynonymous (Asn29His) variant was found in the C2ORF73 gene, whereas no sequence variations were found in the SPTBN1. Although the Asn29His variant was associated with the disease in our sample, we consider SPTBN1 the more likely candidate in this region. First, SPTBN1 is known to be expressed in the brain and neurons, whereas C2ORF73 exhibits a restricted expression pattern; based on the expressed sequence tag and RNA sequencing data the highest expression levels is found in testis and fetus (https://www.ebi.ac.uk/gxa/experiments/E-MTAB-513). We did not detect any mRNA expression of C2ORF73 in RT-PCR experiments of frontal cortex specimen, whereas SPTBN1 mRNA expression was readily detected (data not shown). Second, SPTBN1 is functionally linked with Lewy bodies and α-synuclein. SPTBN1 has been identified as one of the constituents of neocortical Lewy bodies30 and it has been recently shown that SPTBN1 binds directly to α-synuclein.31 Furthermore, in dopaminergic neuronal cells SPTBN1 and α-synuclein are both functionally involved in the modulation of neurite outgrowth.31 Given the direct interaction between SPTBN1 and α-synuclein, SPTBN1 is an attractive candidate gene for modulating neocortical LRP. SPTBN1 is a 247-kDa cytoskeletal protein, which forms heterodimers with α-spectrins. These heterodimers have the capacity to bind to membranes at the cytoplasmic surfaces and they also bind to other cytoskeletal proteins such as actin and ankyrin. Presynaptic SPTBN1/α-spectrin heterodimers play an important physiological role in stabilization of synapses32 and are also involved with the regulation of exocytosis of neurotransmitters.33,34 We did not find any sequence variants that would lead to amino acid changes in the SPTBN1 exons 1–3 located within 100 kb of the top SNPs. Nor were such variants (with frequency of >2%) reported in the whole SPTBN1 gene in the Exome Aggregation Consortium database (http://exac.broadinstitute.org/). These data indicate that SPTBN1 exhibits very little common amino acid variation, and it is likely that the chromosome 2p21 risk locus regulates the expression of SPTBN1.
The HLA-DPA1/DPB1 region has previously been associated with allergic and immune-mediated disorders. Interestingly, recent studies have reported an association between PD and another HLA locus HLA-DRA/DRB1.35,36 The association of the HLA-DPA1/DPB1 locus with neocortical LRP and the association of the HLA-DRA/DRB1 locus with PD most likely represent two separate association signals. There was no linkage disequilibrium between the associated HLA-DPA1/DPB1 SNPs with HLA-DRA/DRB1 markers, and we did not find any association at P < 0.01 between neocortical LRP and the HLA-DRA/DRB1 locus (data not shown). The predisposing HLA-DPA1/DPB1 haplotype harbored the HLA-DPB1*0201 allele, whereas the putative protective haplotype harbored the DPB1*0401 allele. Similar pattern of predisposition (DPB1*0201) and protection (DPB1*0401) has been reported in chronic beryllium disease, which is a granulomatous lung disorder caused by hypersensitivity to beryllium and leads to the accumulation of beryllium-specific CD4 T lymphocytes in the lung upon exposure to beryllium metal.37 The role of metal exposure has been a subject of debate in the development of α-synuclein pathology since the discovery of increased amounts of iron, zinc, and aluminim in PD patients' substantia nigra.38 In addition to the immune-related functions, the HLA-DPA1/DPB1 locus SNPs may regulate expression of nearby genes. Based on the cis QTL analysis mRNA expression of VPS52,TAPBP, and B3GALT4 as well as methylation of VPS52 were modulated by these SNPs. This may be of interest because VPS52 yeast homologue has been shown to be part of a Golgi-associated retrograde protein (GARP) complex.39 Disruption of GARP-complex via VPS52 deletion has been shown to increase alpha-synuclein induced vesicle aggregation and toxicity in a yeast model.40
The GWAS in the Vantaa 85+ material is based on a small number of cases and controls (41 cases vs. 177 controls), which limits the statistical power. This limitation is, however, compensated by the precision of the neuropathological phenotype providing a good contrast of cases versus controls in the phenotypic axis (here spreading of LRP). Previous analysis on the association of neocortical beta-amyloid quantity with APOE ε4 has shown good statistical power in 282 subjects of the Vantaa 85+ (P = 4.9 × 10−17)20 illustrating the power gained by the phenotypic precision. It is clear that the present results, although hitting interesting genes, are preliminary and should be confirmed in similarly phenotyped elderly cases and controls.
Acknowledgments
This work has been supported by the Microsoft Research Foundation, the ALS Association, Helsinki University Central Hospital, the Parkinson Foundation of Finland, the Folkhälsan Research Foundation, the Finnish Academy (P. J. T. and L. M.) and by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services (project number Z01 AG000950-07). H. A. D. K. has been supported by a NHMRC Early Career Fellowship (568890). J. Z. has been supported by Medical Research Council Ph.D. and Newton European Research Studentships and a Wingate Foundation Scholarship in her work on LRP in the CFAS material. Dr. Marja Liisa Lokki at the HLA laboratory of Haartmaninstitute, University of Helsinki, is thanked for the HLA-DPB1 typing. The CFAS I study was supported by the National Health Service and the Medical Research Council grant (G9901400). The individual CFAS centers (Cambridge, Newcastle, Oxford, Nottingham, Liverpool, Sheffield) were supported as follows. The Cambridge Brain Bank was supported by the NIHR Cambridge Biomedical Research Centre; The Cambridgeshire and Peterborough NIHR CLAHRC Newcastle was supported by the UKNIHR Biomedical Research Centre for Ageing and Age-related Disease Award to the Newcastle upon Tyne Hospitals Foundation Trust; Nottingham was supported by Nottingham University Hospitals NHS Trust; the Thomas Willis Oxford Brain Collection was supported by the Oxford Biomedical Research Centre; Liverpool was supported by the Walton Centre NHS Foundation Trust, Liverpool; Sheffield was supported by the University of Sheffield and the Sheffield Teaching Hospitals NHS Foundation Trust. The NABEC was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, part of the U.S. Department of Health and Human Services; project number ZIA AG000932-04. In addition this work was supported by a Research Grant from the Department of Defense, W81XWH-09-2-0128. Portions of this study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD, U.S.A. (http://biowulf.nih.gov). UKBEC was supported by the MRC through the MRC Sudden Death Brain Bank, by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust. King's College London, AROS Applied Biotechnology A/S company laboratories and Affymetrix.
Conflict of Interest
Dr. Traynor reports other from Intramural Research Program, NIA, grants from Microsoft Research Foundation, ALS Association, during the conduct of the study. Dr. Brayne reports grants from The Medical Research Council UK, University of South Australia (FB NHMRC), during the conduct of the study. Dr. Zaccai reports grants from Medical Research Council PhD Studentsship (UK), Newton European Research Studentship (UK), Wingate Foundation Scholarship (UK), during the conduct of the study. Dr. Ince's project was undertaken as part of the MRC Cognitive function and Ageing Neuropathology Study. Dr. Tienari reports grants from The Finnish Academy, The Helsinki University Central Hospital, during the conduct of the study. Dr. Singleton reports grants from Michael J Fox Foundation, during the conduct of the study. Dr. Peuralinna reports grants from Finnish Parkinson Foundation, during the conduct of the study.
Supporting Information
Figure S1. QQ plot for the P-values observed in the genome-wide association study of neocortical Lewy-related pathology in Vantaa 85+. Observed P-values of the SNPs are plotted against the expected P-values.
Table S1. Top signals (P < 10−5) and APOE ε4 analyzed by logistic regression using age, sex, and Alzheimer pathology (Braak, CERAD) as covariates. Logistic regression results in loss of power as compared to pure allelic test shown in Table1.
Table S2. Chromosomal positions, nearest genes and P-values of all 336 SNPs associated with neocortical Lewy-related pathology at P < 10−3 in the Vantaa 85+ study.
Table S3. Genes, chromosomal regions, SNPs and P-values in the genome-wide association study of Lewy-related pathology in Vantaa 85+ in previously implicated genes and regions.
Table S4. Significant cis eQTL results of the top 16 SNPs in methylation and expression derived from the NABEC-UKBEC frontal cortex and cerebellum data. CADD results are shown in the second sheet of the table. Two separate sheets: eQTL and CADD.
References
- Braak H, Del Tredici K, Rub U, et al. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Alafuzoff I, Parkkinen L, Al-Sarraj S, et al. Assessment of α-synuclein pathology: a study of the BrainNet Europe Consortium. J Neuropathol Exp Neurol. 2008;67:125–143. doi: 10.1097/nen.0b013e3181633526. [DOI] [PubMed] [Google Scholar]
- Yoshimura M. Cortical changes in the parkinsonian brain: a contribution to the delineation of “diffuse Lewy body disease”. J Neurol. 1983;229:17–32. doi: 10.1007/BF00313493. [DOI] [PubMed] [Google Scholar]
- Alafuzoff I, Ince PG, Arzberger T, et al. Staging/typing of Lewy body related α-synuclein pathology: a study of the BrainNet Europe Consortium. Acta Neuropathol. 2009;117:635–652. doi: 10.1007/s00401-009-0523-2. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, et al. Alpha-synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Zarranz JJ, Alegre J, Gomez-Esteban JC, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Hayashi S, Farrer MJ, et al. Clinical heterogeneity of alpha-synuclein gene duplication in Parkinson's disease. Ann Neurol. 2006;59:298–309. doi: 10.1002/ana.20753. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Covy JP, Bonini NM, et al. Biochemical and pathological characterization of LRRK2. Ann Neurol. 2006;59:315–322. doi: 10.1002/ana.20791. [DOI] [PubMed] [Google Scholar]
- Goker-Alpan O, Giasson BI, Eblan MJ, et al. Glucocerebrosidase mutations are an important risk factor for Lewy body disorders. Neurology. 2006;67:908–910. doi: 10.1212/01.wnl.0000230215.41296.18. [DOI] [PubMed] [Google Scholar]
- Mata IF, Samii A, Schneer SH, et al. Glucocerebrosidase gene mutations: a risk factor for Lewy body disorders. Arch Neurol. 2008;65:379–382. doi: 10.1001/archneurol.2007.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang D, Leverenz JB, Lopez OL, et al. GBA mutations increase risk for Lewy body disease with and without Alzheimer disease pathology. Neurology. 2012;79:1944–1950. doi: 10.1212/WNL.0b013e3182735e9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscopo P, Marcon G, Piras MR, et al. A novel PSEN2 mutation associated with a peculiar phenotype. Neurology. 2008;70:1549–1554. doi: 10.1212/01.wnl.0000310643.53587.87. [DOI] [PubMed] [Google Scholar]
- Guyant-Marechal I, Berger E, Laquerrière A, et al. Intrafamilial diversity of phenotype associated with APP duplication. Neurology. 2008;71:1925–1926. doi: 10.1212/01.wnl.0000339400.64213.56. [DOI] [PubMed] [Google Scholar]
- Ohtake H, Limprasert P, Fan Y, et al. Beta-synuclein gene alterations in dementia with Lewy bodies. Neurology. 2004;63:805–811. doi: 10.1212/01.wnl.0000139870.14385.3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaerts V, Engelborghs S, Kumar-Singh S, et al. A novel locus for dementia with Lewy bodies: a clinically and genetically heterogeneous disorder. Brain. 2007;130:2277–2291. doi: 10.1093/brain/awm167. [DOI] [PubMed] [Google Scholar]
- Tsuang D, Leverenz JB, Lopez OL, et al. APOE ε4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 2013;70:223–228. doi: 10.1001/jamaneurol.2013.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bras J, Guerreiro R, Darwent L, et al. Genetic analysis implicates APOE, SNCA and suggests lysosomal dysfunction in the etiology of dementia with Lewy bodies. Hum Mol Genet. 2014;23:6139–6146. doi: 10.1093/hmg/ddu334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham GW, Hamilton K, Naj AC, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer's disease and related dementias. PLoS Genet. 2014;10:e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinas M, Polvikoski T, Sulkava R, et al. Neuropathologic findings of dementia with Lewy bodies (DLB) in a population-based Vantaa 85+ study. J Alzheimers Dis. 2009;18:677–689. doi: 10.3233/JAD-2009-1169. [DOI] [PubMed] [Google Scholar]
- Peuralinna T, Tanskanen M, Mäkelä M, et al. APOE and AβPP gene variation in cortical and cerebrovascular amyloid β pathology and Alzheimer's disease – a population-based analysis. J Alzheimers Dis. 2011;26:377–385. doi: 10.3233/JAD-2011-102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeith IG, Galasko D, Kosaka K, et al. Consensus guidelines for the clinical and pathologic diagnosis of dementia with Lewy bodies (DLB): report of the consortium on DLB international workshop. Neurology. 1996;47:1113–1124. doi: 10.1212/wnl.47.5.1113. [DOI] [PubMed] [Google Scholar]
- Brayne C, McCracken C, Matthews FE. Medical Research Council Cognitive Function and Ageing Study (CFAS) Int J Epidemiol. 2006;35:1140–1145. doi: 10.1093/ije/dyl199. [DOI] [PubMed] [Google Scholar]
- Zaccai J, Brayne C, McKeith I, et al. Patterns and stages of α-synucleinopathy: relevance in a population-based cohort. Neurology. 2008;70:1042–1048. doi: 10.1212/01.wnl.0000306697.48738.b6. [DOI] [PubMed] [Google Scholar]
- Laaksovirta H, Peuralinna T, Schymick JC, et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9:978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Wray S, Vandrovcova J, et al. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl DP, Olanow CW, Calne D. Alzheimer's disease and Parkinson's disease: distinct entities or extremes of a spectrum of neurodegeneration? Ann Neurol. 1998;44(3 Suppl 1):S19–S31. doi: 10.1002/ana.410440705. [DOI] [PubMed] [Google Scholar]
- Leverenz JB, Umar I, Wang Q, et al. Proteomic identification of novel proteins in cortical Lewy bodies. Brain Pathol. 2007;17:139–145. doi: 10.1111/j.1750-3639.2007.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Lee K, Im H. α-Synuclein modulates neurite outgrowth by interacting with SPTBN1. Biochem Biophys Res Commun. 2012;424:497–502. doi: 10.1016/j.bbrc.2012.06.143. [DOI] [PubMed] [Google Scholar]
- Pielage J, Fetter RD, Davis GW. Presynaptic spectrin is essential for synapse stabilization. Curr Biol. 2005;15:918–928. doi: 10.1016/j.cub.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Sikorski AF, Terlecki G, Zagon IS, Goodman SR. Synapsin I-mediated interaction of brain spectrin with synaptic vesicles. J Cell Biol. 1991;114:313–318. doi: 10.1083/jcb.114.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski AF, Sangerman J, Goodman SR, Critz SD. Spectrin (betaSpIIsigma1) is an essential component of synaptic transmission. Brain Res. 2000;852:161–166. doi: 10.1016/s0006-8993(99)02253-2. [DOI] [PubMed] [Google Scholar]
- Hamza TH, Zabetian CP, Tenesa A, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissemann WT, Hill-Burns EM, Zabetian CP, et al. Association of Parkinson disease with structural and regulatory variants in the HLA region. Am J Hum Genet. 2013;93:984–993. doi: 10.1016/j.ajhg.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Falta MT, Bowerman NA, et al. T cell recognition of beryllium. Curr Opin Immunol. 2013;25:775–780. doi: 10.1016/j.coi.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaglia M, Tessari I, Mammi S, Bubacco L. Interaction between alpha-synuclein and metal ions, still looking for a role in the pathogenesis of Parkinson's disease. Neuromolecular Med. 2009;11:239–251. doi: 10.1007/s12017-009-8082-1. [DOI] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell. 2003;14:1610–1623. doi: 10.1091/mbc.E02-10-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper JH, Kehm V, Burd CG, et al. Aggregation of α-synuclein in S. cerevisiae is associated with defects in endosomal trafficking and phospholipid biosynthesis. J Mol Neurosci. 2011;43:391–405. doi: 10.1007/s12031-010-9455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. QQ plot for the P-values observed in the genome-wide association study of neocortical Lewy-related pathology in Vantaa 85+. Observed P-values of the SNPs are plotted against the expected P-values.
Table S1. Top signals (P < 10−5) and APOE ε4 analyzed by logistic regression using age, sex, and Alzheimer pathology (Braak, CERAD) as covariates. Logistic regression results in loss of power as compared to pure allelic test shown in Table1.
Table S2. Chromosomal positions, nearest genes and P-values of all 336 SNPs associated with neocortical Lewy-related pathology at P < 10−3 in the Vantaa 85+ study.
Table S3. Genes, chromosomal regions, SNPs and P-values in the genome-wide association study of Lewy-related pathology in Vantaa 85+ in previously implicated genes and regions.
Table S4. Significant cis eQTL results of the top 16 SNPs in methylation and expression derived from the NABEC-UKBEC frontal cortex and cerebellum data. CADD results are shown in the second sheet of the table. Two separate sheets: eQTL and CADD.