Abstract
Aim
To investigate the expression of hepatoma-derived growth factor (HDGF) in osteosarcoma (OS) and the correlation with clinicopathologic factors, prognosis, and tumor progression.
Method
HDGF expression in OS tissues was detected by immunohistochemistry. The correlation between HDGF and clinicopathologic factors was analyzed by chi-square test, and the association between HDGF expression and the overall survival rates was evaluated by univariate analysis using Kaplan–Meier method. HDGF concentration in cell medium or cell lysates was detected by enzyme-linked immunosorbent assay method. The effect of extrinsic and intrinsic HDGF on OS cell proliferation was detected by MTT assay after recombinant HDGF stimulation or HDGF knockdown, respectively.
Results
Proportion of HDGF high expression was 18.69% (20/107) in OS. HDGF high expression was significantly associated with larger tumor size (P=0.004). With experiments in vitro, we demonstrated that human recombinant HDGF could activate AKT and MAPK signaling pathway, resulting in OS cell proliferation. By knocking down HDGF expression, we proved that intrinsic HDGF was required in OS proliferation.
Conclusion
High HDGF expression was significantly associated with larger OS tumor size and could promote OS cell proliferation, indicating that HDGF could be an effective biomarker and a potential drug target in OS treatment.
Keywords: hepatoma-derived growth factor, osteosarcoma, tumor size, proliferation, overall survival rate
Introduction
Osteosarcoma (OS) is a primary solid malignancy mainly originating from bone and rarely from soft tissue, with the highest incidence in bone malignancy.1 The incidence of OS is 2–3/million/year in the general population. In adolescence, the incidence of OS peaks at 15–19 years of age at approximately 8–11/million/year, which accounts for 15% of all solid extracranial cancers in adolescence.2 OS is featured with early metastasis and unfavorable outcome without treatment, with more than 90% patients dying from pulmonary metastasis before poly-chemotherapy treatment. The histologic types of OS include conventional, telangiectatic, parosteal, periosteal, low-grade central, and small cell subtypes. The conventional type is the most frequent and can be further classified as osteoblastic, chondroblastic, and fibroblastic. Thanks to the application of chemotherapy in OS treatment, the OS survival rate has significantly improved from less than 20% up to 70%–80% in patients without clinically evident metastatic disease at the time of presentation in high-grade OS,3 which accounts for 80%–90% of all OS.4 However, despite the important progresses made by chemotherapy implementation, the survival rates of OS continue to be unsatisfactory in metastatic and relapse cases and new chemotherapy targets are still urgently needed.5
HDGF is an acidic HBGF which was first purified from the medium of human hepatoma cell line Huh-7.6 It is a member of a new family of growth factors called HDGF-related proteins (HRPs), including HRP1, HRP2, HRP3, HRP4, and LEDGF p52/p75, which are characterized by the homology in N-terminal amino acids containing the DNA-binding PWWP domain.7 Based on this specific structure, HDGF can translocate to the nucleus and function as a transcription factor by binding DNA. Over the last 2 decades, HDGF has been proven to be involved in numerous biological processes including regeneration, remodeling, neurotrophic effect, wound repair, angiogenesis, radiosensitivity, chemotherapy resistance, mitogenic function, transcriptional regulation, apoptosis, growth and differentiation, epithelial-to-mesenchymal transition, and migration.8–13 The oncogenic role of HDGF overexpression has been revealed in many kinds of cancers and the underlying mechanisms include HDGF-involved proliferation, invasion, angiogenesis, and epithelial-to-mesenchymal transition, etc.14–17 In bone malignancy, HDGF was reported by Savola et al to be the most interesting candidate gene in Ewing’s sarcoma, and the following experiments by Yang et al demonstrated that HDGF could promote the proliferation and was an prognostic factor of Ewing’s sarcoma.18,19 Although the prognostic value and molecular mechanisms of HDGF in Ewing’s sarcoma have been revealed gradually, the clinical significance of HDGF in OS has still not been explored.10
In our study, we investigated the expression of HDGF in 107 OS tissue samples and different OS cell lines. Moreover, we evaluated the correlation between HDGF expression and clinicopathologic factors. With univariate analysis, we further estimated the prognostic value of HDGF. To verify the results obtained in clinical observation, we stimulated different OS cell lines with recombinant HDGF and studied the changes of signaling pathways and cell proliferation. Moreover, HDGF concentration in medium and cell lysates was detected, and cell proliferation was evaluated after HDGF knockdown to estimate HDGF’s role in OS proliferation.
Materials and methods
Patients and samples
From 2002 to 2012, 183 patients underwent surgical treatment and were pathologically diagnosed as having OS in multi-medical centers including People’s Hospital of Linyi City, Yishui Central Hospital, and Central Hospital of Jinan City. The study was approved by the Ethics Boards of all these medical centers, and the samples of OS were obtained from the Pathology Department with the prior consent of the patients. A total of 107 patients were selected for the validation cohort if immunohistochemical (IHC) specimens and follow-ups were available and patients underwent standard adjuvant therapy if possible. Clinical stage of OS was determined according to the criteria by Enneking et al.20 The final diagnosis was confirmed by two senior pathologists, and cases without consensus were identified by the third pathologist. In addition, in the validation cohort, 49 patients presented good response to the neoadjuvant chemotherapy (necrosis >90%), which was based on multi-agent chemotherapy including high-dose methotrexate with leucovorin rescue, adriamycin, cisplatin, and ifosfamide. The representing tumor areas of tumors for immunohistochemistry were usually the center of the tumor if there was no necrosis. If the tumor was fully necrotic and no tumor area could be selected for immunohistochemistry, the case was regarded as an unavailable sample and excluded from the validation cohort. The follow-ups ranged from 3 to 107 months. Cases with survival time less than 3 months were identified as perioperative death and excluded from the cohort. The overall survival rate was calculated from the operation date to the date of death or the last follow-up.
Cell lines and reagents
Human OS cell lines U2-OS, Saos-2, and MG-63 were all bought from Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). Roswell Park Memorial Institute-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) was used to culture cell line U2-OS, Dulbecco’s Modified Eagle’s Medium was used to culture MG-63, and McCoy’s 5A (Sigma-Aldrich Co., St Louis, MO, USA) was used to culture Saos-2. All media were supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) and 1% ampicillin/streptomycin in 5% CO2 resuscitation at 37°C with saturated humidity.
Recombinant HDGF protein was purchased from Pepro-Tech (Rocky Hill, NJ, USA). Anti-HDGF and anti-β-actin antibodies were from Santa Cruz Biotechnology Inc. (Dallas, TX, USA), anti-ERK, anti-pERK-Thr202/Tyr204, anti-AKT, anti-pAKT-Ser473, anti-p38, and anti-p-p38-Thr180/Tyr182 antibodies were from Cell Signaling Technology (Danvers, MA, USA). AKT inhibitor wortmannin was purchased from Sigma-Aldrich Co.
Immunohistochemistry and evaluation
IHC staining was performed by the streptavidin peroxidase complex method. After deparaffinization and rehydration with xylene and graded alcohol, samples were incubated in 3% hydrogen peroxide for 10 minutes to inactivate endogenous peroxidase, and then soaked in citrate buffer (pH =6.0) for 10 minutes for antigen retrieval. Unspecific binding was blocked by 5% bovine serum albumin, after which the specimens were incubated in primary antibody at 1:100 dilution at 4°C overnight. After recycling the primary antibodies and washing the slides with phosphate buffered saline three times, the slides were then incubated in secondary antibodies, streptavidin peroxidase complex reagent and 3,3′-diaminobenzidine solution orderly. Every stained section was blindly evaluated by two pathologists unaware of the clinical data. The standard of IHC score was determined according to the previous study.21 The score was quantified by the calculation of software Image-Pro-Plus, and the mean value of IHC scores of all samples was set as the cut-off, which divided the validation cohort into higher-expression and lower-expression groups.
Tissue specimens and RNA extraction
Twenty-five pairs of OS specimens and corresponding adjacent tissues were collected immediately after surgery and stored in liquid nitrogen until RNA extraction. Total mRNAs of the tumor and adjacent tissues were extracted with TRIzol reagent (Thermo Fisher Scientific) and RNeasy protect mini kit (Qiagen NV, Venlo, the Netherlands) according to the manual. The extracted RNAs were dissolved in diethyl pyrocarbonate-treated water at a concentration of 1.0 mg/mL and then stored at −80°C. Quantification of mRNAs was achieved by one-step real-time reverse transcription polymerase chain reaction (RT-PCR) method following the guideline of SYBR-Green PCR Master Mix (Thermo Fisher Scientific). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. Specific primers for HDGF were as follows: sense primers as CCGCCATGTCGCGATCCAACC; and antisense primers as ACATTGGTGGCTACAGGCTCT.
Immunoblotting assay
HDGF expression in OS cell lines was detected by immunoblotting. Total cellular protein, including the protein from both nuclear and cytoplasm, was first extracted by Total Cellular Protein Kit (Sangon Company, Shanghai, People’s Republic of China, Cat No BSP003). Concentration of protein was detected by BCA detection kit (P0010S; Beyotime Institute of Biotechnology, Jiangsu, People’s Republic of China) before electrophoresis. Equal amount of 20 µg total protein was electrophoresed with sodium dodecyl sulfate polyacrylamide gel electrophoresis gel, then transferred to polyvinyl difluoride (PVDF) membrane (Pall Corporation, Port Washington, NY, USA). After soaking in 5% defatted milk for 1 hour for blocking unspecific binding, the membranes were incubated in primary antibody (1:1000) overnight at 4°C, followed by incubation in corresponding secondary antibody, labeled with horseradish peroxidase for 2 hours at 37°C and visualized by enhanced chemiluminescence (ECL) (EMD Millipore, Billerica, MA, USA).
RNA knockdown and transfection
HDGF knockdown was achieved by transfection of oligo-siRNA designed during a previous study.22 The sequence of HDGF siRNA was 5′-GCCAUGUCUUCUCCCUGGA and 5′-UCCAGGGAGAAGACAUGGC. Control scrambled siRNA was also ordered from Thermo Fisher Scientific. Transfection of siRNA was performed by reagent Lipofectamine 2000 (Thermo Fisher Scientific) according to the manual.
HDGF detection by enzyme-linked immunosorbent assay (ELISA)
HDGF concentration was detected by ELISA kit from Antibodies-online (Aachen, Germany) according to the instructions. Tested samples were made by collecting the cell medium, centrifuging it at 2,000 rpm, and filtrating it with 0.22 µm filter. Samples were loaded into the wells and incubated for 2 hours, followed by addition with 100 µL detection reagent A and incubation for 1 hour. Wells were subsequently washed with phosphate buffered saline and incubated in substrate solution for 15 minutes. Finally, optical density (OD) at 450 nm was read after 50 µL stop solution was added and mixed.
Proliferation assay
Cell proliferation was detected and quantified by MTT assay. Briefly, ~4,000 OS cells were seeded into 96-well plate and subsequently starved in serum-free medium for 12 hours for similar cellular status. Recombinant HDGF was added into 96-well plate for stimulation at final dose of 10 µg/mL for 24 or 48 hours. Cell proliferation was terminated and visualized by adding 10 mg/mL MTT, then the crystals were dissolved in 100 µL dimethyl sulfoxide. Absorbance of OD 490 nm was read by microplate reader (SpectraMax; Molecular Devices LLC, Sunnyvale, CA, USA). The OD of cells in 0 ng HDGF was set as a base line and percentage of proliferation index was standardized by ratio to the base line. To detect the influence of cellular HDGF on proliferation, cells were transfected with HDGF siRNA 48 hours prior to MTT test, and the rest of the procedures were all the same. Cells with scrambled siRNA were set as the baseline. All analyzed data were from at least three independent experiments and P-value was generated from Student’s t-test.
Statistical analysis
SPSS17.0 software (SPSS Inc., Chicago, IL, USA) was used to analyze all the data. The correlation between HDGF expression and other clinicopathologic parameters was evaluated by chi-square test, and the correlation between HDGF and overall survival rate was analyzed by Kaplan–Meier method, with difference in survival curves evaluated with log-rank test. Significance of data from experiments in vitro was detected by Student’s t-test. It was considered statistically significant when P<0.05.
Results
HDGF expression in OS tissue and cell line
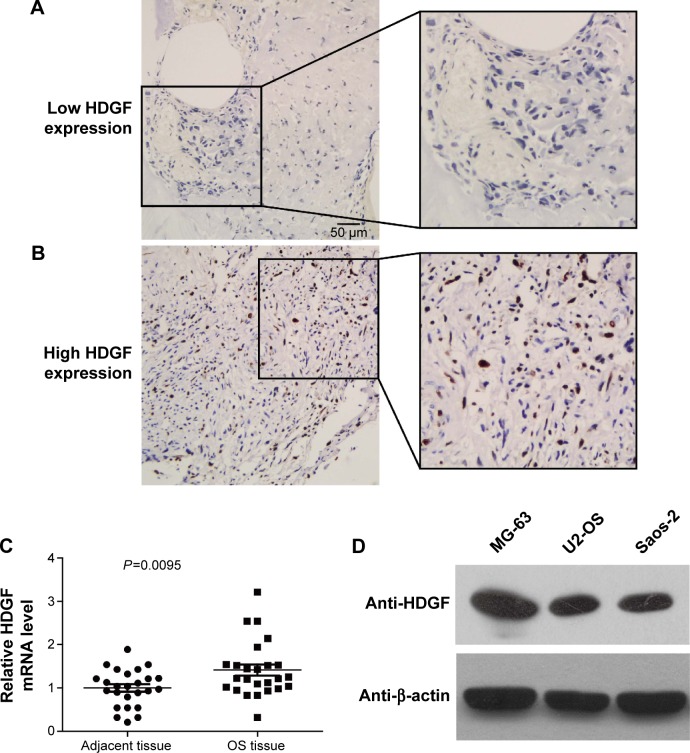
HDGF expression was detected in 107 cases of OS by immunohistochemistry. As a transcription factor, HDGF was mainly observed in nucleus but also detectable in cytoplasm in some cases (Figure 1A and B). The validation group was further divided into HDGF higher-expression group and HDGF lower-expression group according to the IHC score of HDGF as the criteria described in “Patients and Methods”. The proportion of HDGF higher-expression and lower-expression was 18.7% (20/107) and 81.3% (87/107), respectively.
Figure 1.
HDGF expression in OS tissues and cells.
Notes: Representative figures of HDGF with low expression (A) and high expression (B) in OS tissues detected by immunohistochemistry. (C) HDGF mRNA in 20 pairs of OS tissues and corresponding adjacent tissues was detected with real-time PCR and compared with paired Student’s t-test. (D) HDGF expression in OS cell line MG-63, U2-OS, and Saos-2 was detected by immunoblotting.
Abbreviations: OS, osteosarcoma; PCR, polymerase chain reaction; HDGF, hepatoma-derived growth factor.
HDGF mRNA levels were further detected by real-time PCR in 25 pairs of fresh OS tumor tissues and the corresponding adjacent tissues. Interestingly, HDGF mRNA levels in tumor tissues were significantly higher than those in adjacent tissues (P=0.0095), suggesting the oncogenic role of HDGF in OS tumorigenesis or progression (Figure 1C). Moreover, we performed immunoblotting to detect the HDGF expression in different OS cell lines. As the result, HDGF expression varied in these three cell lines. HDGF expression in MG-63 was the highest, and the left two cell lines had the similar HDGF expression (Figure 1D).
Correlation between HDGF and clinicopathologic parameters
The correlations between HDGF expression and the clinicopathologic factors were analyzed by chi-square test (Table 1). The clinicopathologic factors were obtained from medical and surgery records, including patients’ sex, age, tumor size, Enneking stage, tumor site, histopathology, metastasis, and response to chemotherapy. In our experiments, high HDGF expression was demonstrated to be significantly associated with larger tumor size (P=0.004), suggesting that HDGF may facilitate the proliferation of OS, which needed further experiments to verify.
Table 1.
Correlation between HDGF and clinicopathologic factors
| Characters | Number | HDGF
|
P-value* | |
|---|---|---|---|---|
| Low | High | |||
| Sex | ||||
| Female | 24 | 17 | 7 | 0.151 |
| Male | 83 | 70 | 13 | |
| Age | ||||
| <20 | 86 | 70 | 16 | 0.963 |
| ≥20 | 21 | 17 | 4 | |
| Tumor size (cm) | ||||
| <8 | 68 | 61 | 7 | 0.004 |
| ≥8 | 39 | 26 | 13 | |
| Enneking stage | ||||
| I | 11 | 10 | 1 | 0.619 |
| II | 68 | 54 | 14 | |
| III | 28 | 23 | 5 | |
| Site | ||||
| Femur | 49 | 41 | 8 | 0.637 |
| Tibia | 25 | 20 | 5 | |
| Humerus | 16 | 15 | 1 | |
| Fibula | 9 | 5 | 4 | |
| Others | 8 | 6 | 2 | |
| Histopathology | ||||
| Osteoblastic | 37 | 27 | 10 | 0.483 |
| Fibroblastic | 25 | 21 | 4 | |
| Chondroblastic | 13 | 11 | 2 | |
| Telangiectatic | 17 | 16 | 1 | |
| Others | 15 | 12 | 3 | |
| Metastasis | ||||
| No | 79 | 64 | 15 | 0.895 |
| Yes | 28 | 23 | 5 | |
| Response to chemotherapy | ||||
| Poor | 58 | 46 | 12 | 0.563 |
| Good | 49 | 41 | 8 | |
Note:
Chi-square test.
Abbreviation: HDGF, hepatoma-derived growth factor.
Prognostic value of HDGF
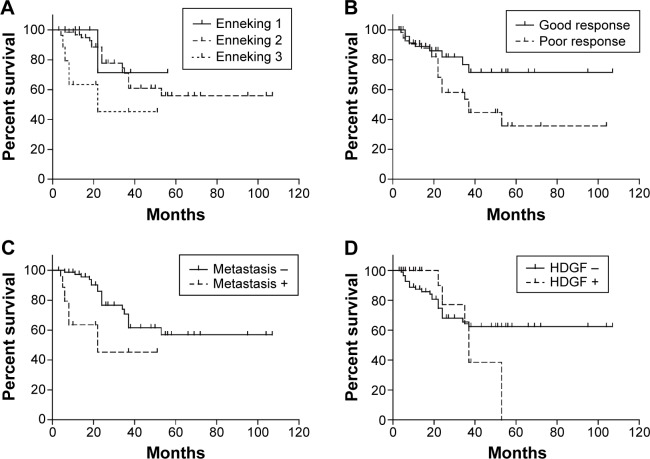
High HDGF expression was proven to be related to unfavorable prognosis in Ewing sarcoma in previous studies,23 and there was no relevant report about the prognostic value of HDGF in OS. In our study, we evaluated the correlation between HDGF expression and the overall survival rates in OS with Kaplan–Meier method (Table 2). As the result, advanced Enneking stage, positive metastasis, and poorer response to chemotherapy were proven to lead to poorer prognosis in OS (P=0.006, 0.001 and 0.043, respectively), which corresponded to previous studies (Figure 2A–C). However, the HDGF expression had no statistically significant correlation with prognosis (P=0.613) (Figure 2D).
Table 2.
Correlation between clinicopathologic factors and survival rate
| Characters | Average survival time (months) | 5-year survival rate | P-value* |
|---|---|---|---|
| Sex | |||
| Female | 64 | 58.6 | 0.342 |
| Male | 67.2 | 51.3 | |
| Age | |||
| <20 | 70.5 | 57.9 | 0.312 |
| ≥20 | 56.8 | 37.3 | |
| Tumor size (cm) | |||
| <8 | 72.8 | 61.2 | 0.968 |
| ≥8 | 59 | 43.7 | |
| Enneking stage | |||
| I | 46.3 | 71.4 | 0.006 |
| II | 72.9 | 55.9 | |
| III | 29.5 | 45.4 | |
| Site | |||
| Femur | 74.2 | 62.1 | 0.065 |
| Tibia | 50.9 | 82.8 | |
| Humerus | 45.9 | 31.9 | |
| Fibula | 39.6 | 40 | |
| Others | 35.1 | 0 | |
| Histopathology | |||
| Osteoblastic | 50 | 42.7 | 0.083 |
| Fibroblastic | 53.2 | 65 | |
| Chondroblastic | 29.2 | 23.4 | |
| Telangiectatic | 80 | 83.3 | |
| Others | 87.2 | 77.8 | |
| Metastasis | |||
| No | 73.4 | 56.8 | 0.001 |
| Yes | 29.5 | 45.4 | |
| Response to chemotherapy | |||
| Poor | 54.2 | 35.7 | 0.043 |
| Good | 82.6 | 71.6 | |
| HDGF | |||
| Low | 73.7 | 62.5 | 0.613 |
| High | 39.7 | 0 | |
Note:
Log-rank test.
Abbreviation: HDGF, hepatoma-derived growth factor.
Figure 2.
Correlation between clinicopathologic factors and survival rate.
Notes: The correlation between overall survival rate and Enneking stage (A), response to chemotherapy (B), metastasis (C), and HDGF expression (D). The survival curve was analyzed by Kaplan–Meier analysis; and the differences between the two groups were analyzed by the log-rank test.
Abbreviation: HDGF, hepatoma-derived growth factor.
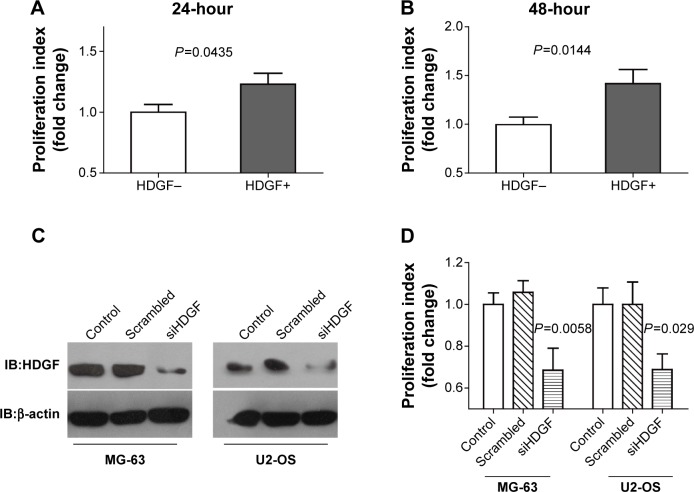
Both intrinsic and extrinsic HDGF could promote the proliferation of OS cells
In Table 1, we observed that high HDGF expression was significantly associated with larger tumor size and hypothesized that HDGF could facilitate the proliferation in OS, thus we performed the following experiments in vitro to identify our hypothesis. U2-OS cells were stimulated with/without 10 µg/mL human recombinant HDGF for 24 or 48 hours, followed by detection with MTT assay (Figure 3A and B). Human recombinant HDGF was demonstrated to promote the proliferation of U2-OS cells and this HDGF-induced proliferation at 48 hours was higher than that at 24 hours. To evaluate whether intrinsic HDGF could affect the proliferation of OS cells, we knocked down HDGF expression with transfection of HDGF siRNA and verified successful knockdown with immunoblotting with MG-63 and U2-OS cells. After siRNA transfection, HDGF expression was significantly decreased ~70% in both MG-63 and U2-OS cells (Figure 3C). Cell proliferation was subsequently detected with MTT assaŷ48 hours after successful HDGF siRNA transfection. Compared with control group and scrambled RNA group, the proliferation of both MG-63 and U2-OS cells decreased remarkably after HDGF knockdown, indicating that cellular HDGF was required in OS cell proliferation (Figure 3D).
Figure 3.
HDGF could promote the proliferation of OS cells.
Notes: (A and B) Recombinant HDGF effect on U2-OS proliferation. Human recombinant HDGF at a dose of 10 µg/mL was used to stimulate U2-OS for 24 (A) or 48 (B) hours and cell proliferation was then detected by MTT assay. (C) Successful HDGF knockdown was verified by immunoblotting of MG-63 and U2-OS cells after transfection with scrambled siRNA and HDGF siRNA. (D) Proliferation of MG-63 and U2-OS cells was detected by MTT assay 48 hours after HDGF knockdown. P-value was calculated by Student’s t-test between the HDGF siRNA group and scrambled siRNA group. Every tested group contained at least six parallel wells in each independent experiment, and the analyzed data were from three independent tests.
Abbreviations: IB, immunoblotting; HDGF, hepatoma-derived growth factor; OS, osteosarcoma.
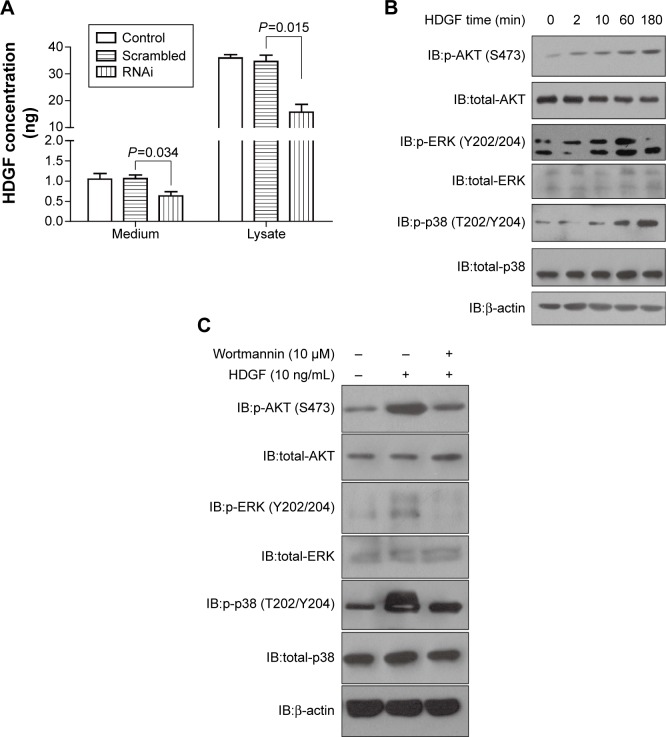
We detected the HDGF concentration in cell medium with ELISA method to evaluate whether HDGF could be released from cells. The HDGF concentration of MG-63 medium and MG-63 cell lysate was both detected (Figure 4A). HDGF concentration waŝ1 ng/mL in medium, although it was much lower than that in cell lysate. Additionally, HDGF in medium was significantly lower when intrinsic HDGF was knocked down, suggesting that HDGF was released from intracellular cytoplasm. Furthermore, we explored the signaling pathways which may be involved in HDGF-induced proliferation. Under extrinsic HDGF activation from 2 minutes to 180 minutes, the phosphorylation of AKT, ERK and p38 of U2-OS cells was remarkably increased, and this increase was associated with the stimulation time (Figure 4B), indicating that extrinsic HDGF could trigger AKT and MAPK signaling pathways which were essential for cell proliferation. Moreover, U2-OS cells were pre-incubated with AKT inhibitor wortmannin at a dose of 10 µM for 30 minutes and then stimulated in 10 ng/mL recombinant HDGF for 30 minutes, and subsequently lysed for detection of phosphorylation. As a result, we proved that wortmannin could significantly decrease the HDGF-induced AKT/ERK/p38 phosphorylation (Figure 4C).
Figure 4.
HDGF could stimulate AKT and MAPK signaling pathway.
Notes: (A) HDGF concentration in medium and cell lysate of U2-OS was detected with ELISA method 48 hours after transfection with scrambled RNA, HDGF siRNA, or Lipofectamine 2000. (B) Recombinant HDGF effect on phosphorylation of AKT, ERK, and p38 in U2-OS cells. U2-OS cells were stimulated with 10 µg/mL HDGF for 0 to 180 minutes, then lysed and detected by primary antibodies of AKT, p-AKT-Ser473, ERK, p-ERK-Thr202/Tyr204, p38, p-p38-Thr180/Tyr182, and β-actin. (C) AKT inhibitor wortmannin was able to decrease HDGF-induced phosphorylation of AKT/ERK/p38. U2-OS cells were pre-incubated in 10 µM wortmannin for 30 minutes and then stimulated in recombinant HDGF for 30 minutes. Immunoblotting was used to detect the phosphorylation of AKT/ERK/p38.
Abbreviations: ELISA, enzyme-linked immunosorbent assay; IB, immunoblotting; HDGF, hepatoma-derived growth factor; OS, osteosarcoma.
Discussion
OS is the most frequent primary cancer of bone, usually with dismal outcome because of the local and metastatic progression. Thanks to the major advances in chemotherapy and surgical resection, the prognosis and overall survival rate for OS has been improved dramatically over the past 50 years. The chemical regimen has improved the survival rate from less than 20% tô70% at 5 years for patients without known metastases,3 and 5-year overall survival rate for recurrent OS has been reported to be 23%~29%.24 However, despite the developing progress made in OS treatment, the prognosis of OS is still not optimistic and it is still a major threat to health, especially in teenagers. New approaches are still eagerly needed to improve the prognosis, and new potentially effective chemical drugs seem to be the most promising way to help improve the survival time of OS patients. However, the discovery of new drugs relies on the finding of new drug targets and exploration of more underlying signaling pathways. There is a high priority need in drug development for a new agent that can be administered as adjuvant to conventional chemotherapeutics and control the local or metastatic disease better. The molecular targeted drug may be a good addition to current chemical therapy but there is no available targeted drug for OS currently, even though the genomic complexity of OS has been confirmed by whole-genome sequencing.25
Among all the candidate genes which may affect the progression of OS, HDGF is so suspicious because it functions as a growth factor when existing extracellularly and a transcription factor when existing intracellularly. Additionally, the ectopic expression of HDGF was reported in many kinds of malignancies.26 Moreover, recent studies demonstrated that HDGF was significantly associated with aggressive biological behavior and prognosis in Ewing sarcoma.23 In our study, we proved that HDGF expression was significantly associated with tumor size in clinical observation and that HDGF could promote OS cell proliferation with experiments in vitro, which resulted in the hypothesis that HDGF could be a potential biomarker and may help find new targeted therapy for OS.
We verified our hypothesis that HDGF may promote the proliferation of OS cells, which explained why HDGF overexpression was more frequent in larger OS specimens. Interestingly, both intrinsic and extrinsic HDGF played an important role in OS proliferation in our experiment. We boldly suspected that HDGF may be released from OS cells and function as a growth factor in a paracrine or autocrine way, which resulted in OS proliferation and larger tumor size. However, the mechanism of how HDGF is released is still controversial. A previous study proved that HDGF can be regarded as a secreted factor in human glioblastoma,27 but more scientists considered that HDGF functions as a damage-associated molecular pattern. HDGF is passively released from dead, dying, or injured cells partially because it lacks the secretion signal sequence which is found in most secreted proteins. Regardless of how, it is acknowledged that HDGF could be released from tumor cells, which could subsequently activate downstream signaling pathways and influence cell proliferation. However, the underlying mechanisms why HDGF results in OS proliferation is still unclear, and experiments in vivo are urgently needed, especially experiments on whether HDGF functions as a transcription factor and which downstream gene is regulated in OS. We hope our finding of HDGF significance in OS can trigger more interest in the exploration of HDGF-stimulated signaling pathways and develop an available HDGF inhibitor, which may be used as a targeted drug for OS clinically.
In conclusion, we detected HDGF expression in 107 cases of OS with immunohistochemistry and analyzed the correlation between HDGF and clinicopathologic factors as well as overall survival rate. As a result, we demonstrated that high HDGF expression was significantly associated with larger OS size and both intrinsic and extrinsic HDGF could promote OS cell proliferation, indicating that HDGF may be a potential biomarker and a promising drug target in OS treatment.
Footnotes
Disclosure
The authors have no conflicts of interest to disclose.
References
- 1.Arndt V, Lacour B, Steliarova-Foucher E, et al. Up-to-date monitoring of childhood cancer long-term survival in Europe: tumours of the sympathetic nervous system, retinoblastoma, renal and bone tumours, and soft tissue sarcomas. Ann Oncol. 2007;18(10):1722–1733. doi: 10.1093/annonc/mdm189. [DOI] [PubMed] [Google Scholar]
- 2.Collins M, Wilhelm M, Conyers R, et al. Benefits and adverse events in younger versus older patients receiving neoadjuvant chemotherapy for osteosarcoma: Findings from a meta-analysis. J Clin Oncol. 2013;31(18):2303–2312. doi: 10.1200/JCO.2012.43.8598. [DOI] [PubMed] [Google Scholar]
- 3.Bernthal NM, Federman N, Eilber FR, et al. Long-term results (>25 years) of a randomized, prospective clinical trial evaluating chemotherapy in patients with high-grade, operable osteosarcoma. Cancer. 2012;118(23):5888–5893. doi: 10.1002/cncr.27651. [DOI] [PubMed] [Google Scholar]
- 4.Bielack S, Carrle D, Casali PG. Osteosarcoma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(Suppl 4):137–139. doi: 10.1093/annonc/mdp154. [DOI] [PubMed] [Google Scholar]
- 5.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment – where do we stand? A state of the art review. Cancer Treat Rev. 2014;40(4):523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura H, Kambe H, Egawa T, et al. Partial purification and characterization of human hepatoma-derived growth factor. Clin Chim Acta. 1989;183(3):273–284. doi: 10.1016/0009-8981(89)90361-6. [DOI] [PubMed] [Google Scholar]
- 7.Gijsbers R, Vets S, De Rijck J, et al. Role of the pwwp domain of lens epithelium-derived growth factor (ledgf)/p75 cofactor in lentiviral integration targeting. J Biol Chem. 2011;286(48):41812–41825. doi: 10.1074/jbc.M111.255711. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Clermont F, Gonzalez NS, Communi D, Franken S, Dumont JE, Robaye B. HDGF is dephosphorylated during the early steps of endothelial cell apoptosis in a caspase-dependent way. J Cell Biochem. 2008;104(4):1161–1171. doi: 10.1002/jcb.21788. [DOI] [PubMed] [Google Scholar]
- 9.Wang CH, Davamani F, Sue SC, et al. Cell surface heparan sulfates mediate internalization of the PWWP/HATH domain of HDGF via macropinocytosis to fine-tune cell signalling processes involved in fibroblast cell migration. Biochem J. 2011;433(1):127–138. doi: 10.1042/BJ20100589. [DOI] [PubMed] [Google Scholar]
- 10.Chen SC, Kung ML, Hu TH, et al. Hepatoma-derived growth factor regulates breast cancer cell invasion by modulating epithelial – mesenchymal transition. J Pathol. 2012;228(2):158–169. doi: 10.1002/path.3988. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Everett AD. Hepatoma-derived growth factor represses SET and MYND domain containing 1 gene expression through interaction with c-terminal binding protein. J Mol Biol. 2009;386(4):938–950. doi: 10.1016/j.jmb.2008.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enomoto H, Nakamura H, Liu W, et al. Hepatoma-derived growth factor is induced in liver regeneration. Hepatol Res. 2009;39(10):988–997. doi: 10.1111/j.1872-034X.2009.00532.x. [DOI] [PubMed] [Google Scholar]
- 13.Hollander A, D’Onofrio PM, Magharious MM, Lysko MD, Koeberle PD. Quantitative retinal protein analysis after optic nerve transection reveals a neuroprotective role for hepatoma-derived growth factor on injured retinal ganglion cells. Invest Ophthalmol Vis Sci. 2012;53(7):3973–3989. doi: 10.1167/iovs.11-8350. [DOI] [PubMed] [Google Scholar]
- 14.Lin YW, Li CF, Chen HY, et al. The expression and prognostic significance of hepatoma-derived growth factor in oral cancer. Oral Oncol. 2012;48(7):629–635. doi: 10.1016/j.oraloncology.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto S, Tomita Y, Hoshida Y, et al. Expression of hepatoma-derived growth factor is correlated with lymph node metastasis and prognosis of gastric carcinoma. Clin Cancer Res. 2006;12(1):117–122. doi: 10.1158/1078-0432.CCR-05-1347. [DOI] [PubMed] [Google Scholar]
- 16.Uyama H, Tomita Y, Nakamura H, et al. Hepatoma-derived growth factor is a novel prognostic factor for patients with pancreatic cancer. Clin Cancer Res. 2006;12(20 Pt 1):6043–6048. doi: 10.1158/1078-0432.CCR-06-1064. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto S, Tomita Y, Hoshida Y, et al. Expression level of hepatoma-derived growth factor correlates with tumor recurrence of esophageal carcinoma. Ann Surg Oncol. 2007;14(7):2141–2149. doi: 10.1245/s10434-007-9369-9. [DOI] [PubMed] [Google Scholar]
- 18.Savola S, Klami A, Tripathi A, et al. Combined use of expression and CGH arrays pinpoints novel candidate genes in Ewing sarcoma family of tumors. BMC Cancer. 2009;9:17. doi: 10.1186/1471-2407-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Li H, Zhang F, et al. Clinical and biological significance of hepatoma-derived growth factor in Ewing’s sarcoma. J Pathol. 2013;231(3):323–334. doi: 10.1002/path.4241. [DOI] [PubMed] [Google Scholar]
- 20.Enneking WF. A system of staging musculoskeletal neoplasms. Clin Orthop Relat Res. 1986;(204):9–24. [PubMed] [Google Scholar]
- 21.Liu YF, Zhao R, Guo S, et al. Expression and clinical significance of hepatoma-derived growth factor as a prognostic factor in human hilar cholangiocarcinoma. Ann Surg Oncol. 2011;18(3):872–879. doi: 10.1245/s10434-010-1303-x. [DOI] [PubMed] [Google Scholar]
- 22.Guo Z, He Y, Wang S, et al. Various effects of hepatoma-derived growth factor on cell growth, migration and invasion of breast cancer and prostate cancer cells. Oncol Rep. 2011;26(2):511–517. doi: 10.3892/or.2011.1295. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Zhen T, Zhang F, et al. P53 and hepatoma-derived growth factor expression and their clinicopathological association with Ewing family tumour. J Clin Pathol. 2014;67(3):235–242. doi: 10.1136/jclinpath-2013-201705. [DOI] [PubMed] [Google Scholar]
- 24.Carrle D, Bielack S. Osteosarcoma lung metastases detection and principles of multimodal therapy. Cancer Treat Res. 2009;152:165–184. doi: 10.1007/978-1-4419-0284-9_8. [DOI] [PubMed] [Google Scholar]
- 25.Hattinger CM, Fanelli M, Tavanti E, et al. Advances in emerging drugs for osteosarcoma. Expert Opin Emerg Drugs. 2015 May 28; doi: 10.1517/14728214.2015.1051965. Epub. [DOI] [PubMed] [Google Scholar]
- 26.Bao C, Wang J, Ma W, Wang X, Cheng Y. HDGF: A novel jack-of-all-trades in cancer. Future Oncol. 2014;10(16):2675–2685. doi: 10.2217/fon.14.194. [DOI] [PubMed] [Google Scholar]
- 27.Thirant C, Galan-Moya EM, Dubois LG, et al. Differential proteomic analysis of human glioblastoma and neural stem cells reveals HDGF as a novel angiogenic secreted factor. Stem Cells. 2012;30(5):845–853. doi: 10.1002/stem.1062. [DOI] [PubMed] [Google Scholar]