Abstract
Cryptococcus neoformans is the etiologic agent of cryptococcal meningitis that causes more than half a million deaths worldwide each year. This capsulated basidiomycetous yeast also serves as a model for micropathogenic studies. The ability to make stable mutants, either via ectopic integration or homologous recombination, has been accomplished using biolistic transformation. This technical advance has greatly facilitated the research on the basic biology and pathogenic mechanisms of this pathogen in the past two decades. However, biolistic transformation is costly, and its reproducibility varies widely. Here we found that stable ectopic integration or targeted gene deletion via homologous replacement could be accomplished through electroporative transformation. The stability of the transformants obtained through electroporation and the frequency of homologous replacement is highly dependent on the selective marker. A frequency of homologous recombination among the stable transformants obtained by electroporation is comparable to those obtained by biolistic transformation (~10%) when dominant drug selection markers are used, which is much higher than what has been previously reported for electroporation when auxotrophic markers were used (0.001% to 0.1%). Furthermore, disruption of the KU80 gene or generation of gene deletion constructs using the split marker strategy, two approaches known to increase homologous replacement among transformants obtained through biolistic transformation, also increase the frequency of homologous replacement among transformants obtained through electroporation. Therefore, electroporation provides a low cost alternative for mutagenesis in Cryptococcus.
Keywords: Cryptococcus neoformans, ectopic integration, dominant markers, electroporation, gene disruption, transformation
Introduction
Cryptococcus neoformans is the major etiologic agent of fungal meningitis [1-6]. This clinically important fungal pathogen also serves as a model for micropathogenic studies. There are complete genome sequences, several congenic pairs, robust mammalian and invertebrate host models, and various genetic tools available [6-12]. In particular, the introduction of biolistic transformation in early 1990s enables target mutagenesis (e.g., gene deletion) via homologous recombination, and it has been central to for fungal studies of Cryptococcus genes and their roles in pathogenicity [13-20].
Transformants obtained by biolistic introduction of DNA are generally stable (17.5% to 100%) [14,21]. The frequencies of homologous recombination vary widely depending on the gene and the strain background, but a frequency in the range of 1–10% is typical [13-19,22]. However, biolistic transformation is costly due to the requirement of the expensive Biolistic ® PDS-1000/He Particle Delivery System only available from BioRad. In addition, it requires a vacuum pump and a helium gas tank as well as other pricey consumables sold by BioRad (gold beads, macrocarriers, stopping screens, and the rupture disks). Repeated transformations are often necessary to obtain knockout mutants due to variability of this technique and frequencies of homologous recombination in this fungus. All these factors make the biolistic transformation inhibitory for resource-limited laboratories.
Other transformation systems have been explored, including protoplasting, Agrobacterium-mediated transformation (AMT), and electroporation. None of these attempts were successful for targeted mutagenesis [23-28].
Electroporation has been used in Cryptococcus research since early 1990s. All previous reports on electroporation were done in strains belonging to Cryptococcus neoformans var. neoformans (serotype D). Most of the electroporation reported earlier used nutritional auxotrophic markers such as URA5 to select for transformants. The vast majority of transformants obtained through electroporation were unstable, and half of them tended to lose the introduced DNA even when they were grown on selective minimum medium [27-29]. The introduced DNA often acquires telomeric sequences from Cryptococcus genome and replicates autonomously [23,27,28]. Consequently, the introduced DNA is often maintained episomally without being integrated into the genome. A minor proportion of the transformants were stable and these transformants usually contained ectopic integration of the URA5 sequences [23,27,28]. Homologous recombination by electroporation occurred at extremely low frequencies, in the range of 1/1000 to 1/100,000 [30-33]. Therefore, electroporation has not been used for targeted mutagenesis. Rather it is used to introduce DNA that is desirable to be episomally maintained, such as in a multicopy suppressor screen [34-36].
One reported study of Cryptococcus electroporation did not use auxotrophic markers [37]. Instead, a native Cryptococcus L41 ribosomal gene with a dominant mutation was used as a selective marker gene because the protein produced confers resistance to cycloheximide, an inhibitor of protein synthesis. The cycloheximide-resistant transformants obtained were stable, resulted from ectopic integration events [37]. However, whether homologous recombination occurred in these transformants were not investigated. Given that bacterium-originated genes whose products confer resistance to nourseothricin (NAT) [35], geneticin (G418) [38,39], and hygromycin B (HYB) [40,41] are currently preferably used in Cryptococcus studies because these marker genes lack homology to the Cryptococcus genome, it will be important to know whether these dominant markers could increase the stability of introduced DNA in these transformants obtained by electroporation.
Unfortunately, electroporation in var. grubii (serotype A) has been considered extremely inefficient, and successful transformation by electroporation were previously done in var. neoformans (serotype D). A previous study obtained no transformants at all by electroporation in four strains in the grubii H99 genetic background [14]. However, C. neoformans var. grubii causes the vast majority of all cryptococcosis cases globally [3,6,42] and is in general more virulent than var. neoformans [43,44]. This current study is designed to examine whether electroporation with dominant selective markers (NAT and G418) can generate stable transformants in the H99 background, whether decreasing nonhomologous end joining by deletion of the KU80 gene [45] or using a split marker strategy can increase the rate of homologous recombination, and whether electroporation can be practically used for targeted mutagenesis. We chose the ADE2 locus for the primary investigation as deletion of the ADE2 gene not only confers auxotrophy but also red pigmentation to the mutant colony that can be easily visualized [46]. We also included other genetic loci and a different genetic background in this study to demonstrate that this approach is not locus- or strain-specific.
Materials and methods
Strains and Growth Conditions
Strains used and generated in this study and their sources are listed in Table 1. Cells were maintained on YPD (1% yeast extract, 2% BactoPeptone, and 2% dextrose) agar medium at 30°C unless indicated otherwise.
Table 1.
Strains used in this study.
| Strain name | Genotype | Source and comments |
|---|---|---|
| H99 | Serotype A wild type | 65 |
| JLCN488 | MATα cku80∷NEOr | 45 |
| XL1681 | MATα DsRED-NEOr znf2∷NEOr ZNF2-NATr | this study |
| DSF51 | MATα znf1∷NAT, ste12∷URA5, ade2 | this study |
| YP22 | MATα cku80∷NEOr ade2∷NATr | this study |
| YP23 | MATα cku80∷NEOr ade2∷NATr | this study |
| YP24 | MATα cku80∷NEOr ade2∷NATr | this study |
| YP25 | MATα cku80∷NEOr ade2∷NATr | this study |
| YP26 | MATα cku80∷NEOr ade2∷NATr | this study |
| YP27 | MATα cku80∷NEOr ade2∷NATr | this study |
| YP28 | MATα cku80∷NEOr ade2∷NATr | this study |
| YP29 | MATα cku80∷NEOr ade2∷NATr | this study |
| YP30 | MATα ade2∷NEOr | this study |
| XL1685 | MATα cku80∷NEOr CNAG2526∷NATr | this study |
| XL1690 | MATa cku80∷NEOr CNAG2526∷NATr | this study |
| XL1687 | MATα CNAG2526∷NATr | this study |
| XL1688 | MATa CNAG2526∷NATr | this study |
| XL280# | Serotype D wildtype | 66 |
| NC01# | MATα rze1∷NATr | this study |
| NC12 | MATα rze1∷NATr | this study |
| LW516α # | PFAD1-FAD1-mCherry∷G418r | |
| LW728α # | PFAS1-FAS1-mCherry∷G418r | 58 |
strains in the XL280 genetic background. All others are in the H99 genetic background.
Genomic DNA Preparations
Strains were grown in 50 ml YPD liquid medium at 30°C overnight with shaking. The cells were washed with sterile water, harvested by centrifugation, and frozen at −80°C. The cells were then lyophilized overnight and then broken into fine powder by glass beads. DNA was then purified from the powder using the CTAB protocol as described previously [47].
Generation of gene deletion constructs
To generate deletion constructs by triple-joint polymerase chain reaction (TJ-PCR) with the intact NAT marker, 5′ and 3′ flanking sequences (~1 kb each) of the gene of interest were amplified using the genomic DNA extracted from the wild-type H99 strain. Meanwhile, the NAT marker (~1.8 kb) that contains the Cryptococcus ACT1 promoter and TRP1 terminator was amplified using the pPZP-NATcc as the template [48]. Similarly, the G418 marker (~2.1 kb) was generated based on the plasmid pPZP-NEO1 [48]. The three products from the first round of PCR were then joined together to generate the deletion construct with the NAT marker flanked by the 5′ and 3′ flanking sequences of the gene of interest at either side (Fig. 1, left panel) by a triple-joint overlap PCR. Primers used are listed in Supplemental Table 1.
Figure 1.
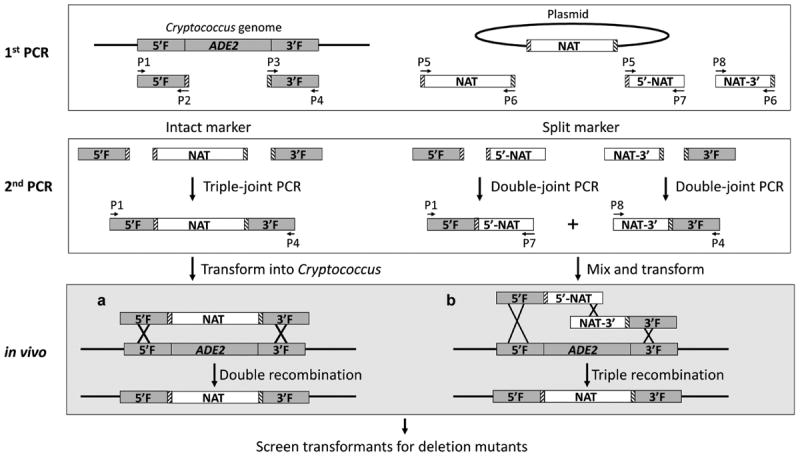
A schematic diagram showing the procedure of generating the ADE2 gene deletion constructs by a triple-joint polymerase chain reaction (PCR) or a double-joint PCR and transforming the constructs into Cryptococcus cells. During the first round of PCR, 5′ and 3′ flanking regions (5F and 3F) about ~1 kb each of the ADE2 gene were amplified using primers sets P1+P2 and P3+P8, respectively. The NAT marker was amplified using primers P5 and P6. The 5′ and 3′-NAT-split markers (5′-NAT and NAT-3′) were amplified using primer sets P5+ P7 and P8+P9, respectively. During the second round of PCR, three different overlap PCR products were generated. The first product contained the NAT marker flanked by the 5′and 3′-flanking regions of the ADE2 gene and was generated using primers P1 and P4 by a triple-joint PCR. The second product contained the 5′ flanking region of the ADE2 gene connected with the first 2/3 of the NAT marker and was generated using primers P1 and P7 by a double-joint PCR. The third product contains the last 2/3 of the NAT marker connected with the 3′ flanking region of the ADE2 gene and was amplified using primers P8+P4 by a double-joint PCR. The triple-joint PCR product or the mixed double-joint products were introduced into Cryptococcus cells by electroporation. Homologous recombination will lead to replacement of the ADE2 gene by the selective marker. The boxes with slashes bordering the NAT marker indicate the regions complement to the primers P2 and P3 that were used to amplify the flanking sequences of the ADE2 gene.
To generate deletion constructs by double-joint PCR (DJ-PCR) with NAT split markers, 5′ and 3′ flanking sequences (~1 kb each) of the gene of interest were amplified using the genomic DNA extracted from the wild type H99 strain. Meanwhile, the first 1.2 kb of the NAT marker (5′-NAT) and the last 1.2 kb of the NAT marker (NAT-3′) with ~600 bp overlap were amplified using the pPZP-NATcc as the template [48]. The product of the 5′ flanking sequence connected with 5′-NAT was produced by a double-joint overlap PCR. Similarly, the 3′ flanking sequence was connected with 5′-NAT (Fig. 1, right panel).
The DNA fragments (5F-NAT-3F, 5F-G418–3F, or 5F-5′NAT + NAT3′-3F) were then introduced into Cryptococcus yeast cells by electroporation as described below.
Electroporation of Cryptococcus
Electroporation of Cryptococcus was performed using previously described method with minor modifications [29,49,50]. Cryptococcus yeast cells were grown in 30 ml of YPD medium at 30°C with shaking overnight. Cells were then diluted with YPD medium to A600 of 0.3 to a final volume of 100 ml. The cells were cultured for additional three hours to reach a cell density between A600 of 0.6 to 1.0. Cells were harvested by centrifugation at 4,000 × g for 5 min. The cells were then washed twice in 50 ml EB buffer (10 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 270 mM Sucrose) and then resuspended in 50 ml of EB buffer with 1mM DTT. After 30–60 min of incubation on ice, cells were harvested, washed once with 50 ml of EB buffer, and then resuspended in 300 μl of EB buffer. The cell suspension (45 μl) was mixed with 5 μl of DNA (~100–400 ng), placed in a 0.2-cm electroporation cuvette, and transformed by electroporation using a BioRad gene pulser (0.45 kV, 125 μF, 400–600Ω) or an Eppendorf multiporator using the prokaryotic cell setting according to the manufacture’s instruction. The electroporated cells were then suspended with 1 ml of YPD medium and incubated at 30°C for 90 min before being plated to appropriate selective media (YPD supplemented with either NAT at 100 μg/ml or G418 at 200 μg/ml). Transformants became apparent after 2 days of incubation at 30°C.
Transformants stability testing
Transformants from selective media were picked and transferred to master YPD plates. The cells were incubated at 30°C for 2 days and then replicated on fresh YPD agar medium. After fifth passage, cells were replicated on YPD solid medium with the appropriate antibiotic to examine the stability of the transformants. Colonies that showed spotty growth on the selective medium compared to those on non-selective YPD medium were scored as unstable. Colonies that grew similarly robustly on selective and nonselective media were scored as stable transformants.
Genetic cross
Mating partners were co-cultured together on V8 medium and incubated at 22°C in the dark for 2 weeks. Spores were microdissected using a dissection microscope. The mating type of the progeny was confirmed by crossing with reference strains as previously described [51].
Results
The stability of transformants obtained by electroporation is highly dependent on the marker used and targeted gene deletion can be achieved through electroporation
To examine the stability of DNA fragments containing the dominant NAT or G418 selective markers introduced into H99 by electroporation, we first generated the ADE2 gene deletion constructs by triple-joint overlap PCR with the NAT or the G418 marker bordered by the 5′ and 3′ flanking sequences of the ADE2 gene as depicted in Figure 1 (left panel). These DNA fragments were then electroporated into H99, and the transformants were selected on YPD agar medium supplemented with either NAT or G418. After 2–3 days of additional incubation, transformants were transferred to nonselective YPD medium, passaged on YPD medium five times, and then replicated on selective medium to examine their stability. Colonies that showed spotty growth on the selective medium were considered unstable (Fig. 2), while transformants with robust growth of the whole colonies on the selective medium were considered stable (Fig. 2, pointed by the arrows). We found that the frequency of stable transformants using NAT or G418 dominant drug markers varies (Table 2) consistent with previous findings using electroporation (100% stability with the cycloheximide resistant marker in one study [37] and about 10% stability with the URA5 marker [29]).
Figure 2.
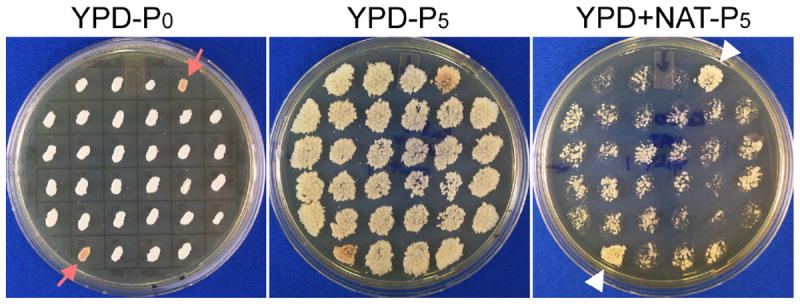
The stability test of transformants obtained by electroporation. Transformants were transferred to a gridded YPD agar plate (left panel, YPD-P0), passaged onto YPD solid medium for 5 consecutive times (YPD-P5), and examined for their resistance to the appropriate antibiotic on selective agar medium. After fifth passage, transformants with spotty growth on the selective medium (shown here YPD+NAT-P5 on the right panel) compared to that on nonselective YPD medium (YPD-P5 on the middle panel) were considered unstable. Transformants that grew robustly on the selective medium as they grew on nonselective medium were considered stable (pointed by the white triangles on the right image on YPD+NAT). This particular original master plate (YPD-P0) shown here have two colonies turned red, indicating of an ade2 mutation (pointed by red arrows in the left image). These red transformants grew robustly on the YPD+NAT medium, indicating that these transformants are stable and the construct likely has been integrated into the genome and disrupted the ADE2 gene.
Table 2.
Transformant stability and homologous replacement based on ADE2.
| Strain | Construct | Total transformants | Stable transformants | Homologous replacement |
|---|---|---|---|---|
| Wild type H99 | 5F-NAT-3F | 129 | 15 | 0 |
| 5F-5′-NAT + NAT-3′-3F | 49 | 11 | 1 | |
| 5F-G418-3F | 164 | 140 | 2 | |
| ku80 | 5F-NAT-3F | 102 | 8 | 6 |
| 5F-5′-NAT + NAT-3′-3F | 40 | 2 | 2 |
We then examined if any of the stable transformants were ade2Δ mutants resulting from the homologous replacement of the ADE2 gene by the marker. Two out of the 140 stable transformants obtained using the G418 marker turned red (Fig. 1), and the two transformants were also auxotrophic unable to grow on the YNB minimal medium, as expected for ade2 mutants (Fig. 3). As ectopic integration or episomal maintenance of the introduced DNA fragment will generate transformants capable of growing in the absence of adenine, the absence of growth of these transformants in the minimal medium further supports that the introduced DNA likely replaced the ADE2 gene in the genome. The replacement of the ADE2 gene by the marker in these transformants was further confirmed by diagnostic PCR (Fig. 4). Taken together, the data indicate that all these red transformants are indeed ADE2 deletion mutants as a result of homologous replacement of the ADE2 gene by the marker.
Figure 3.
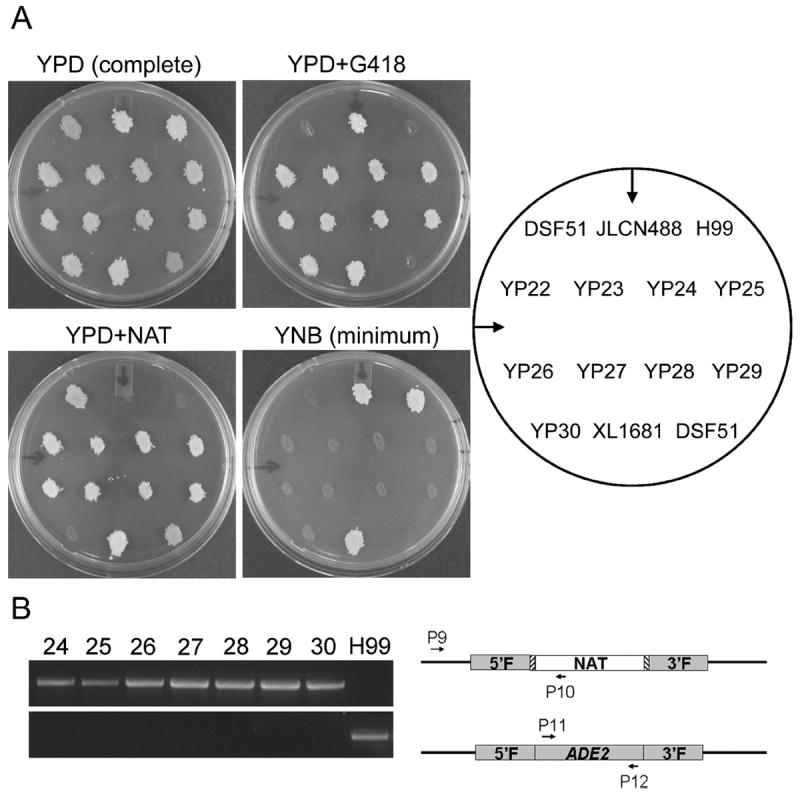
Phenotypical and polymerase chain reaction (PCR) confirmation of selected transformants for the replacement of the ADE2 gene by the marker. (A) The selected transformants YP22-YP29 and their parental strain JLCN488, transformant YP30 and its parental strain H99, and two control strains DSF51 (for ade2 and NAT resistant) and XL1681 (for NAT and G418 double resistant) were cultured on YPD agar medium at 30°C for 1 day and then replicated onto YPD medium for growth control. The colonies were also replicated onto YPD+G418 or YPD+NAT media to examine the presence of the G418 or the NAT marker. The colonies were replicated onto YNB minimum medium to examine auxotroph. Photographs were taken after 2 days of incubation at 30°C. (B) Genomic DNA extracted from red strains and the wild type strain H99 was used as the control. Only results of strains YP24-YP30 and H99 are shown here. The upper panel shows PCR amplicons using the primer P9 that localizes to the genomic region upstream of the 5′ flanking regions of the ADE2 gene and the primer P10 that localizes in the NAT marker of the introduced construct. Only correct integration of the construct will yield positive amplicons of 1.73 kb (DNA ladders not shown in the figure). The lower panel shows PCR amplicons using primers P11 and P12 that localize to the coding region of the ADE2 gene. Strains with the ADE2 gene present will yield positive amplicons of 1.70 kb.
Figure 4.
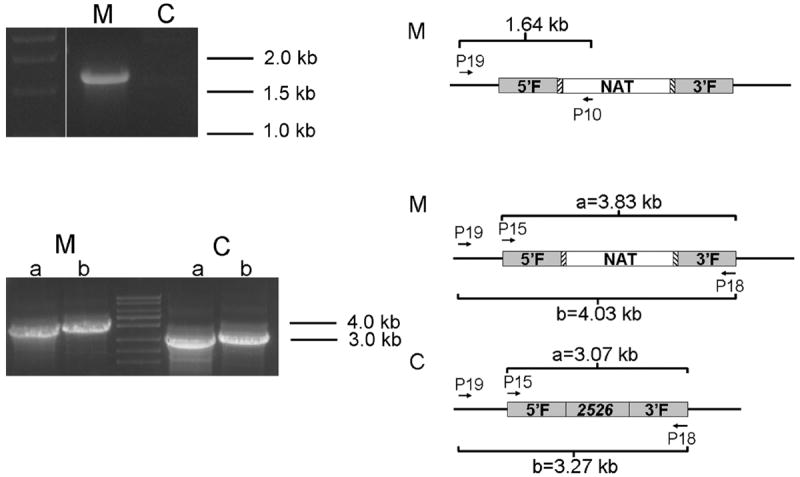
Polymerase chain reaction (PCR) confirmation of homologous replacement of the CNAG2526 gene in the stable transformant (XL1685). Genomic DNA extracted from XL1685 (labeled as M) and the control strain JLCN488 (labeled as C) was used as template. The upper panel shows PCR amplicons using the primer P19 that localizes to the genomic region upstream of the 5′ flanking regions of the CNAG2526 gene and the primer P10 that localizes in the NAT marker of the introduced construct. Only correct integration of the construct will yield positive amplicons of 1.64 kb. The lower panel shows PCR amplicons using primer sets P15+P18 and P19+P18 that localize to the flanking region of the CNAG2526 gene. Strains with the CNAG2526 gene replaced by the NAT marker will yield positive amplicons of 3.83 kb and 4.03 kb respectively, while the control with the wild type allele of the CNAG2526 gene will yield positive amplicons of 3.07 kb and 3.27 kb, respectively. (The expected sizes of the fragments were confirmed by longer electrophoresis.)
Disruption of Ku80 increases the frequency of homologous recombination among the transformants
Ku proteins are important for rejoining broken DNA ends by nonhomologous end-joining pathways [52]. Reducing or abolishing Ku protein activities lowers nonhomologous end joining events while promoting homologous recombination. Such changes occur naturally in meiotic cells in both yeasts and animals [53-55] and have been used to increase homologous recombination for target mutagenesis in Cryptococcus by biolistic transformation [45].
To examine if deletion of the KU80 gene also increases the frequency of homologous recombination in the transformants obtained through electroporation, we electroporatively transformed the ku80Δ mutant in the H99 background with the ADE2 deletion construct. Because the KU80Δ gene in that strain was replaced by the G418 marker [45], only the construct made with the NAT marker (5F-NAT-3F) was used in the electroporation. Out of eight stable transformants, six were ade2Δ mutants as confirmed by their phenotypes (red pigment and auxotrophy) and by PCR analysis as described earlier (Fig. 3; Table 2). Thus the frequency of homologous recombination among the stable transformants in the ku80 mutant background is high, similar to what is observed among transformants obtained with biolistic transformation.
The split marker strategy improves the rate of homologous replacement among stable transformants
The ADE2 gene deletion construct used earlier was generated with a triple-joint PCR and it is one DNA fragment containing an intact marker gene bordered by the ADE2 flanking regions (Fig. 1, left panel). Previous studies in other fungi and in Cryptococcus have demonstrated that a split-marker technique can increase homologous recombination events among transformants obtained by biolistic bombardment [21,56,57]. The split marker construct is generated by a double-joint PCR (Fig. 1, right panel) and is composed of two DNA fragments: one contains the 5′ flanking sequences of the ADE2 gene and the 5′ part of the NAT marker and the other contains the 3′ flanking sequences of the ADE2 gene and the remaining part of the NAT marker with some overlap sequences.
We introduced the split marker construct into both the wildtype H99 strain and the ku80 strain by electroporation. Out of 11 stable transformants obtained in the H99 strain, one is an ADE2 gene deletion mutant. Not surprisingly, two out of the two stable transformants obtained in the ku80 strain are the ade2Δ mutants. Our data indicate that split-marker strategy can indeed increase the frequency of homologous replacement. However, the gain would be minimal when the ku80 strain is used.
Deletion of other genes in both H99 background (var. grubii) and XL280 background (var. neoformans) can also be achieved by electroporation
We next examined whether this technique can be applied to other genetic loci or other strain background. We decided to test this in H99 ku80 mutant of an uncharacterized gene CNAG2526. Indeed, we successfully obtained a knockout mutant based on PCR screening (Fig. 4). Genetic crosses indicated a pattern of single-gene Mendelian segregation of the marker and also the creation of progeny with only CNAG2526 deletion in either mating type a or mating type α background (Table 1). To examine if this technique could also be applied to other strain background, we deleted the gene RZE1 in the H99 wildtype background (var. grubii) and in the XL280 wild-type background (var. neoformans). One out of 29 stable transformants screened in H99 background was a knockout mutant and one out of 19 stable transformants screened in XL280 background was a knockout mutant (Table 1). We also used this technique to generate ectopically integrated fluorescent protein fusions with Fad1 and Fas1 selected with the G418 resistant marker in the XL280 background [58] (Table 1). About 10% (n = 50) and 12% (n = 50) of the transformants are genetically stable.
Discussion
The frequency of homologous integration is largely predetermined by the particular fungal species, although it can be affected by the genetic locus. For example, high frequencies of homologous integration are observed in Saccha-romyces cerevisiae and Aspergillus nidulans [59-61]. However, many other fungi, including Neurospora crassa [62], Coprinus cinereus [63], Ustilago maydis [64], and Cryptococcus neoformans predominantly have ectopic integration events. In Cryptococcus, homologous recombination typically occurs at the frequency of 1–10% based on targeted gene deletion using biolistic transformation. It was proposed that electroporation cannot deliver sufficient DNA to the nucleus for efficient integration into the genome [22], let alone homologous recombination. Although more transformants obtained by electroporation were unstable compared to biolistic transformation, we found that the frequency of homologous replacement among the stable transformants obtained by electroporation is comparable with those obtained by biolistic transformation. In practice, elimination of unstable transformants after several passages on nonselective media will enable similar screening of stable transformants for correct integration of the introduced DNA. Furthermore, the disruption of the KU80 gene or the adoption of the split-marker strategy can increase the frequency of homologous integration among transformants obtained (Table 2). In cases where the ku80 mutant strain is used for electroporation, the ku80 mutation can be easily separated from the mutations of the gene of interest by backcrossing selected transformants with a wildtype strain (Table 1), as reported previously [45].
In summary, our study has demonstrated that electroporation can provide an alternative low cost approach for targeted mutagenesis in Cryptococcus neoformans.
Supplementary Material
Acknowledgments
We thank Dr. Jenny Lodge for providing strains. We gratefully acknowledge the financial support from the National Institute of Allergy and Infectious Diseases (grants R01AI097599 and R21AI107138 to X. L.). X. L. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Footnotes
Supplementary material
Supplementary material is available at Medical Mycology online (http://www.mmy.oxfordjournals.org/).
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
References
- 1.Park BJ, Wannemuehler KA, Marston BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8(4):515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall A, Perfect JR. Cryptococcus neoformans. Washington, DC: ASM Press; 1998. [Google Scholar]
- 4.Chen J, Varma A, Diaz MR, et al. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis. 2008;14(5):755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain N, Wickes BL, Keller SM, et al. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J Clin Microbiol. 2005;43(11):5733–5742. doi: 10.1128/JCM.43.11.5733-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon-Chung KJ, Fraser JA, Doering TL, et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 2014;4(7):e1–e27. doi: 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni M, Feretzaki M, Li W, et al. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol. 2013;11(9):e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janbon G, Ormerod KL, Paulet D, et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014;10(4):e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mylonakis E, Casadevall A, Ausubel FM. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 2007;3(7):e101. doi: 10.1371/journal.ppat.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaragoza O, Alvarez M, Telzak A, Rivera J, Casadevall A. The relative susceptibility of mouse strains to pulmonary Cryptococcus neoformans infection is associated with pleiotropic differences in the immune response. Infect Immun. 2007;75(6):2729–2739. doi: 10.1128/IAI.00094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loftus BJ, Fung E, Roncaglia P, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307(5713):1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Idnurm A, Bahn YS, Nielsen K, et al. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol. 2005;3(10):753–764. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- 13.Perfect JR, Toffaletti DL, Rude TH. The gene encoding phos-phoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61(10):4446–4451. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175(5):1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodge JK, Jackson-Machelski E, Toffaletti DL, Perfect JR, Gordon JI. Targeted gene replacement demonstrates that myristoyl-CoA: protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc Natl Acad Sci U S A. 1994;91(25):12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 1997;11(23):3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odom A, Muir S, Lim E, et al. Calcineurin is required for virulence of Cryptococcus neoformans. Embo J. 1997;16(10):2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz MC, Cavallo LM, Gorlach JM, et al. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19(6):4101–4112. doi: 10.1128/mcb.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Poeta M, Toffaletti DL, Rude TH, et al. Topoisomerase I is essential in Cryptococcus neoformans: role In pathobiology and as an antifungal target. Genetics. 1999;152(1):167–178. doi: 10.1093/genetics/152.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu OW, Chun CD, Chow ED, et al. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135(1):174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim MS, Kim SY, Yoon JK, Lee YW, Bahn YS. An efficient gene-disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochem Biophys Res Commun. 2009;390(3):983–988. doi: 10.1016/j.bbrc.2009.10.089. [DOI] [PubMed] [Google Scholar]
- 22.Davidson RC, Cruz MC, Sia RA, et al. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol. 2000;29(1):38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- 23.Kwon-Chung KJ, Goldman WE, Klein B, Szaniszlo PJ. Fate of transforming DNA in pathogenic fungi. Med Mycol. 1998;36(Suppl 1):38–44. [PubMed] [Google Scholar]
- 24.McClelland CM, Chang YC, Kwon-Chung KJ. High frequency transformation of Cryptococcus neoformans and Cryptococcus gattii by Agrobacterium tumefaciens. Fungal Genet Biol. 2005;42(11):904–913. doi: 10.1016/j.fgb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Idnurm A, Reedy JL, Nussbaum JC, Heitman J. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryotic Cell. 2004;3:420–429. doi: 10.1128/EC.3.2.420-429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 2010;6(5):e1000953. doi: 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varma A, Edman JC, Kwon-Chung KJ. Molecular and genetic analysis of URA5 transformants of Cryptococcus neoformans. Infect Immun. 1992;60(3):1101–1108. doi: 10.1128/iai.60.3.1101-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edman JC. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol Cell Biol. 1992;12(6):2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edman JC, Kwon-Chung KJ. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10(9):4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang YC, Kwon-Chung KJ. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14(7):4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang YC, Kwon-Chung KJ. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect Immun. 1998;66(5):2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang YC, Penoyer LA, Kwon-Chung KJ. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun. 1996;64(6):1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salas SD, Bennett JE, Kwon-Chung KJ, Perfect JR, Williamson PR. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184(2):377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mondon P, Chang YC, Varma A, Kwon-Chung KJ. A novel episomal shuttle vector for transformation of Cryptococcus neoformans with the ccdB gene as a positive selection marker in bacteria. FEMS Microbiol Lett. 2000;187(1):41–45. doi: 10.1111/j.1574-6968.2000.tb09134.x. [DOI] [PubMed] [Google Scholar]
- 35.McDade HC, Cox GM. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol. 2001;39(1):151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- 36.Fox DS, Cox GM, Heitman J. Phospholipid-binding protein Cts1 controls septation and functions coordinately with calcineurin in Cryptococcus neoformans. Eukaryot Cell. 2003;2(5):1025–1035. doi: 10.1128/EC.2.5.1025-1035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varma A, Kwon-Chung KJ. Characterization of the L41 gene in Cryptococcus neoformans: its application as a selectable transformation marker for cycloheximide resistance. Yeast. 2000;16(15):1397–1403. doi: 10.1002/1097-0061(200011)16:15<1397::AID-YEA636>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 38.Dolan CT. Optimal combination and concentration of antibiotics in media for isolation of pathogenic fungi and Nocardia asteroides. Appl Microbiol. 1971;21(2):195–197. doi: 10.1128/am.21.2.195-197.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua J, Meyer JD, Lodge JK. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin Diagn Lab Immunol. 2000;7(1):125–128. doi: 10.1128/cdli.7.1.125-128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox GM, Toffaletti DL, Perfect JR. Dominant selection system for use in Cryptococcus neoformans. J Med Vet Mycol. 1996;34(6):385–391. [PubMed] [Google Scholar]
- 41.Varma A, Kwon-Chung KJ. Characterization of the glyceraldehyde-3-phosphate dehydrogenase gene [correction of glyceraldehyde-3-phosphate gene] and the use of its promoter for heterologous expression in Cryptococcus neoformans, a human pathogen. Gene. 1999;232(2):155–163. doi: 10.1016/s0378-1119(99)00132-8. [DOI] [PubMed] [Google Scholar]
- 42.Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 43.Barchiesi F, Cogliati M, Esposto MC, et al. Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. J Infect. 2005;51(1):10–16. doi: 10.1016/j.jinf.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Lin X, Nielsen K, Patel S, Heitman J. Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans. Infect Immun. 2008;76(7):2923–2938. doi: 10.1128/IAI.00168-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goins CL, Gerik KJ, Lodge JK. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet Biol. 2006;43(8):531–544. doi: 10.1016/j.fgb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Dorfman BZ. The isolation of adenylosuccinate synthetase mutants in yeast by selection for constitutive behavior in pigmented strains. Genetics. 1969;61(2):377–389. doi: 10.1093/genetics/61.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitkin JW, Panaccione DG, Walton JD. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology. 1996;142:1557–1565. doi: 10.1099/13500872-142-6-1557. [DOI] [PubMed] [Google Scholar]
- 48.Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol. 2005;57(5):1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- 49.Cruz MC, Fox DS, Heitman J. Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. EMBO J. 2001;20(5):1020–1032. doi: 10.1093/emboj/20.5.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickes BL, Edman JC. Development of a transformation system for Cryptococcus neoformans. In: Maresca B, Kobayashi GS, editors. Molecular Biology of Pathogenic Fungi : A Laboratory Manual. New York: Telos Press; 1994. pp. 309–313. [Google Scholar]
- 51.Zhai B, Zhu P, Foyle D, et al. Congenic strains of the filamentous form of Cryptococcus neoformans for studies of fungal morphogenesis and virulence. Infect Immun. 2013;81(7):2626–2637. doi: 10.1128/IAI.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valencia M, Bentele M, Vaze MB, et al. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414(6864):666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- 53.Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet. 1999;23(2):194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- 54.Astrom SU, Okamura SM, Rine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397(6717):310. doi: 10.1038/16833. [DOI] [PubMed] [Google Scholar]
- 55.Lee SE, Paques F, Sylvan J, Haber JE. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol. 1999;9(14):767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- 56.Fu J, Hettler E, Wickes BL. Split marker transformation increases homologous integration frequency in Cryptococcus neoformans. Fungal Genet Biol. 2006;43(3):200–212. doi: 10.1016/j.fgb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 57.Fairhead C, Llorente B, Denis F, Soler M, Dujon B. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast. 1996;12(14):1439–1457. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1439::AID-YEA37%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 58.Wang L, Tian X, Gyawali R, et al. Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Pathog. 2014;10(6):e1004185. doi: 10.1371/journal.ppat.1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yelton MM, Hamer JE, Timberlake WE. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A. 1984;81(5):1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Struhl K. The new yeast genetics. Nature. 1983;305(5933):391–397. doi: 10.1038/305391a0. [DOI] [PubMed] [Google Scholar]
- 62.Case ME. Genetical and molecular analyses of qa-2 transformants in Neurospora crassa. Genetics. 1986;113(3):569–587. doi: 10.1093/genetics/113.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Binninger DM, Skrzynia C, Pukkila PJ, Casselton LA. DNA-mediated transformation of the basidiomycete Coprinus cinereus. Embo J. 1987;6(4):835–840. doi: 10.1002/j.1460-2075.1987.tb04828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Holden DW, Leong SA. Gene transfer system for the phytopathogenic fungus Ustilago maydis. Proc Natl Acad Sci U S A. 1988;85(3):865–869. doi: 10.1073/pnas.85.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101(1):177–194. [PMC free article] [PubMed] [Google Scholar]
- 66.Lin X, Huang JC, Mitchell TG, Heitman J. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genet. 2006;2(11):e187. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


