Abstract
The mode of succinate mediated repression of mineral phosphate solubilization and the role of repressor in suppressing phosphate solubilization phenotype of two free-living nitrogen fixing Klebsiella pneumoniae strains was studied. Organic acid mediated mineral phosphate solubilization phenotype of oxalic acid producing Klebsiella pneumoniae SM6 and SM11 were transcriptionally repressed by IclR in presence of succinate as carbon source. Oxalic acid production and expression of genes of the glyoxylate shunt (aceBAK) was found only in glucose but not in succinate- and glucose+succinate-grown cells. IclR, repressor of aceBAK operon, was inactivated using an allelic exchange system resulting in derepressed mineral phosphate solubilization phenotype through constitutive expression of the glyoxylate shunt. Insertional inactivation of iclR resulted in increased activity of the glyoxylate shunt enzymes even in succinate-grown cells. An augmented phosphate solubilization up to 54 and 59% soluble phosphate release was attained in glucose+succinate-grown SM6Δ and SM11Δ strains respectively, compared to glucose-grown cells, whereas phosphate solubilization was absent or negligible in wildtype cells grown in glucose+succinate. Both wildtype and iclR deletion strains showed similar indole-3-acetic acid production. Wheat seeds inoculated with wildtype SM6 and SM11 improved both root and shoot length by 1.2 fold. However, iclR deletion SM6Δ and SM11Δ strains increased root and shoot length by 1.5 and 1.4 folds, respectively, compared to uninoculated controls. The repressor inactivated phosphate solubilizers better served the purpose of constitutive phosphate solubilization in pot experiments, where presence of other carbon sources (e.g., succinate) might repress mineral phosphate solubilization phenotype of wildtype strains.
Introduction
Mineral phosphate solubilization (MPS) is one of the most desirable traits of plant growth promoting rhizobacteria (PGPR). Klebsiella is known for plant growth promoting (PGP) activities like phosphate (P) solubilization, indole-3-acetic acid (IAA) production, siderophore production and HCN production [1–3]. The existence of Klebsiella pneumoniae associated to nitrogen fixation in wheat (Triticum aestivum L.) has been reported [4], while Klebsiella strains endophytic to rice, maize, sugarcane and banana have also been reported [5–8].
The principal mode of MPS is organic acid secretion by rhizospheric microorganisms [9]. Among organic acids responsible for MPS phenotype, gluconic acid, 2-ketogluconic acid, oxalic acid, acetic acid, citric acid, succinic acid and glyoxylic acid are the major ones [10]. Organic acid production by bacteria depends on the carbon source utilized, as the organic acids produced are intermediates of the central carbon metabolism.
The central metabolic pathways in bacteria are controlled by a set of global and local regulators depending on the carbon sources available and the culture conditions [11]. Carbon catabolite repression (CCR) controls the uptake and metabolism of a preferred carbon source when bacteria are exposed to more than one carbon sources. CCR has been extensively studied in the Enterobacteriaceae and Firmicutes where it is mediated by components of the phosphoenolpyruvate (PEP): carbohydrate phosphotransferase system (PTS), but the mechanisms are different in these groups [12]. In E. coli, the preferential utilization of glucose over other carbon sources is controlled by catabolite repression protein (CRP) and PTS [13].
In our previous report, we have shown that Klebsiella pneumoniae SM6 and SM11 solubilized mineral phosphate by production of oxalic acid when grown in glucose as sole carbon source [14]. However, acid production and MPS phenotype of both these strains were repressed in the presence of succinate [14]. Oxalic acid was produced in these bacteria from glyoxylate by the action of glyoxylate oxidase (GO) [14]. Genes of the glyoxylate shunt in the Enterobacteriaceae family are clustered in the aceBAK operon under the control of an isocitrate lyase repressor (IclR) [15, 16]. The aceBAK operon codes for malate synthase (aceB), isocitrate lyase (aceA) and isocitrate dehydrogenase kinase/phosphatase (aceK) [15, 16]. Isocitrate lyase (ICL) cleaves isocitrate into glyoxylate and succinate, malate synthase (MS) catalyzes the condensation of acetyl-CoA and glyoxylate to form malate, while isocitrate dehydrogenase kinase/phosphatase inactivates/activates isocitrate dehydrogenase [15, 16].
The IclR class of transcriptional regulators was first reported in E. coli [17, 18]. Regulators of the IclR family have been shown to control diverse cell functions like carbon metabolism in the Enterobacteriaceae [19], catabolism of aromatic compounds in soil bacteria [20], quorum-sensing signals in Agrobacterium [21], and plant virulence in certain enterobacteria [22]. IclR (glyoxylate shunt repressor) is the best-characterized member of the IclR family in E. coli, which regulates the aceBAK operon encoding proteins for acetate utilization [15, 16].
In the present study, we investigated the role of iclR in catabolite repression of oxalic acid production in P solubilizing Klebsiella pneumoniae SM6 and SM11. To understand the molecular mechanism of succinate mediated repression of oxalic acid linked MPS phenotype, ΔiclR mutants Klebsiella pneumoniae SM6Δ and SM11Δ were generated and assessed for relieved repression of MPS phenotype. Although both strains are genetically similar and produce oxalic acid, they vary considerably in quantity of oxalic acid produced and activities of ICL and GO, as described previously [14]. Hence, both strains were considered for further studies. Based on expression profile, enzyme activities and physiological studies, we have identified iclR as putative repressor of oxalic acid linked MPS phenotype in our isolates. iclR inactivation showed derepression of the glyoxylate shunt enzymes and enhanced the MPS phenotype in presence of succinate and glucose. To check the effect of wildtype and ΔiclR strains on plant growth, pot experiments were conducted under controlled conditions. Indeed, the deletion of iclR improved plant growth, compared to the wildtype.
Materials and Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH5α, K. pneumoniae SM6, K. pneumoniae SM11 K. pneumoniae SM6Δ and K. pneumoniae SM11Δ were grown aerobically on LB medium (Himedia Laboratories, India) at 30°C. K. pneumoniae SM6 and K. pneumoniae SM11 were deposited at Microbial Culture Collection (MCC), Pune with accession numbers MCC2730 and MCC2716, respectively. For plate cultures, 1.5% agar was added to LB medium before autoclaving. Temperature sensitive mutants and temperature sensitive plasmid-carrying strains were grown at 28°C. Antibiotics ampicillin, erythromycin and kanamycin were added to the medium at a concentration of 50 μg ml-1, as and when required. Culture stocks were preserved in 50% (v/v) glycerol and stored at -20°C. Antibiotics, salts, reagents and carbon sources were acquired from Merck-Millipore, Germany.
Table 1. Bacterial strains and plasmids used in the study.
| Strains | Relevant Characteristics | Source or Reference |
|---|---|---|
| E. coli DH5α | Host, ΔlacU169 (ϕ80lacZΔM15) recA1 | [23] |
| K. pneumoniae SM6 | P solubilizing, free-living N2 fixer, Host, Wildtype (GenBank accession number JX139003) | [14] |
| K. pneumoniae SM11 | P solubilizing, free-living N2 fixer, Host, Wildtype (GenBank accession number JX139004) | [14] |
| K. pneumoniae SM6Δ | ΔiclR, P solubilizing, free-living N2 fixer, Host | This study |
| K. pneumoniae SM11Δ | ΔiclR, P solubilizing, free-living N2 fixer, Host | This study |
| Plasmids | ||
| pUC18 | AmpR, LacZα, pBR322 origin, cloning vector | [24] |
| pcDNA3.1 (+) | KanR, AmpR, T7 promoter, CMV promoter, Mammalian expression vector | Invitrogen, USA |
| pGRG36 | AmpR, Tn7 insertion vector, pSC101 origin of replication oriTS | [25] |
| pRV85 | EryR, gfp uv, pldhL promoter | [26] |
| pSMB1a | 386 bp SM6 iclR-N’ fragment at PstI:XbaI site of pUC18, AmpR, pBR322 origin | This study |
| pSMB2a | 363 bp SM6 iclR-C’ fragment at SmaI:SacI site of pSMB1a, AmpR, pBR322 origin | This study |
| pSMB3a | 890 bp kanR gene at BamHI site of pSMB2a, AmpR pBR322 origin, | This study |
| pSMB4a | pSC101 temperature sensitive origin of replication, cloned in NdeI:PciI digested pSMB3a, oriTS, iclR-N’, iclR-C’, KanR | This study |
| pSMB5a | pldhL:gfp uv fragment from pRV85 digested with EcoRI, cloned at EcoRI site of pSMB4a, pSC101 oriTS, iclR-N’, iclR-C’, KanR | This study |
| pSMB1b | 386 bp SM11 iclR-N’ fragment at PstI:XbaI site of pUC18, AmpR, pBR322 origin | This study |
| pSMB2b | 363 bp SM11 iclR-C’ fragment at SmaI:SacI site of pSMB1b, AmpR, pBR322 origin | This study |
| pSMB3b | 890 bp kanR gene at BamHI site of pSMB2b, AmpR pBR322 origin, | This study |
| pSMB4b | pSC101 temperature sensitive origin of replication, cloned in NdeI:PciI digested pSMB3b, oriTS, iclR-N’, iclR-C’, KanR | This study |
| pSMB5b | pldhL:gfp uv fragment from pRV85 digested with EcoRI, cloned at EcoRI site of pSMB4b, pSC101 oriTS, iclR-N’, iclR-C’, KanR | This study |
AmpR ampicillin resistance, KanR kanamycin resistance, EryR erythromycin resistance, oriTS temperature sensitive origin of replication, pldhL Lactobacillus lactate dehydrogenase promoter, gfp uv UV excitable green fluorescent protein.
Growth studies and enzyme assays
Effect of carbon source on growth of wildtype and mutant strains was studied as described previously [14]. To study mono- and diauxic growth of the cultures, they were grown in M9 minimal medium with 20 mM glucose, 20 mM succinate and 10 mM glucose+succinate as carbon source. Glucose utilization in liquid media was quantified by Glucose-SLR Reagent (Lab-Care Diagnostics, India) [14]. Activities of ICL and GO were determined in cell extracts of glucose-, succinate- and glucose+succinate-grown cells as described previously by Dixon and Kornberg [27] and Akamatsu and Shimada [28], respectively, with some modifications [14]. Cell extract was prepared as described by Buch et al. [29], with mid-log phase cells, harvested by centrifugation at 12,000 rpm for 5 min at 4°C. The cell pellet was washed with 80 mM phosphate buffer (pH 7.5) and resuspended in small volume of same buffer with 20% glycerol and 1 mM DTT. The cell suspension was sonicated using Ultrasonic homogenizer JY92-IIDN (Syclon, China) for 2 min at a pulse rate of 30 at 500 Hz on ice. Cell debris were removed by centrifugation at 12,000 rpm at 4°C for 30 min and the supernatant was used for enzyme assays.
Determination of MPS phenotype
Qualitative and quantitative estimation of phosphate solubilization was carried out to assess if iclR inactivation relieved succinate-mediated repression of MPS phenotype in SM6Δ and SM11Δ strains. Qualitative estimation of MPS phenotype of wildtype and mutant strains was performed on Pikovskaya agar [30] and Tris rock phosphate (TRP) agar, as described previously [14]. TRP agar contained 100 mM glucose, 100 mM succinate or equimolar mixture of glucose and succinate (50 mM each; repression medium) as carbon sources; Pikovskaya agar contained glucose (10 g L-1) or equimolar mixture of glucose and succinate (5 g L-1 each; repression medium) as carbon sources; Pikovskaya broth contained glucose (10 g L-1) or equimolar mixture of glucose and succinate (5 g L-1 each; repression medium) as carbon sources. Quantitative estimation of P solubilization was carried out in Pikovskaya broth, as described by Rajput et al [14], and phosphate released was quantified as described by Ames [31]. In this method, soluble phosphate reacts with ammonium molybdate to give phosphomolybdate, which is further reduced by ascorbic acid to give a blue colored complex that can be quantified at 820 nm [31].
Polymerase chain reaction (PCR) and product purification
PCR amplifications were carried out for expression studies and vector construction. Primers used for DNA amplification are listed in Table 2. Primers for K. pneumoniae SM6, SM11, SM6Δ and SM11Δ were designed using nucleotide sequences of closely related K. pneumoniae strains available at NCBI. Genomic DNA was isolated from overnight grown cultures using GeneJET genomic DNA purification kit (Thermo Scientific, USA) following the manufacturer’s instructions. Plasmid isolation from cells grown in LB medium with ampicillin, erythromycin and kanamycin was performed using a GeneJET plasmid miniprep kit (Thermo Scientific, USA). pcDNA3.1 was used as PCR template for amplification of kanamycin resistance gene, pGRG36 for pSC101 oriTS, and genomic DNA of K. pneumoniae SM6 and SM11 for iclR-N’ and iclR-C’ fragments. All PCR amplifications were carried out in a Nexus Gradient Mastercycler (Eppendorf, Germany). The reaction mixture for PCR amplification was as follows: 200 ng template DNA, 200 μM of each of the four dNTPs, 20 pmol of each forward and reverse primer, 2 U Maxima Hot Start Taq DNA polymerase (Thermo Scientific, USA) and 1X Maxima Hot Start Taq Buffer (Thermo Scientific, USA), in final volume of 20 μl. The PCR parameters were as follows: 4 min initial denaturation at 95°C, 35 X (1 min denaturation at 95°C, 1 min primer annealing at 50–60°C (depending on the Tm and G+C content of the primers, see Table 2), 0.5–2.5 min primer extension at 72°C (see Table 2)) and a final extension of 10 min at 72°C. PCR products were analysed on 1% agarose gel. PCR products used for cloning were purified using GeneJET gel extraction and DNA cleanup micro kit (Thermo Scientific, USA). Primers were procured from Integrated DNA Technologies, Inc. (Coralville, USA)
Table 2. Primers used in the study.
| Primers | Relevant Characteristics (5’→3’) | Annealing Temperature | Primer Extension time |
|---|---|---|---|
| IclRN’-F | AATCTGCAGTTCGCGATGGCCACTAC, with PstI underlined site, located 58 bp downstream of iclR start codon | 55°C | 0.5 min |
| IclRN’-R | CCGTCTAGAGAGCACCGCCAGATTCA, with XbaI underlined site, located 445 bp downstream of iclR start codon | 55°C | 0.5 min |
| IclRC’-F | TATCCCGGGGACCACCAGGCGATCATTA, with SmaI underlined site, located 454 bp downstream of iclR start codon | 58°C | 0.5 min |
| IclRC’-R | ACGGAGCTCCATCGGTCATACGCGAGAT, with SacI underlined site, located 818 bp downstream of iclR start codon | 58°C | 0.5 min |
| KanR-F | CCCGGATCCCCGGGAGCTTGTATATCCATTT, with BamHI underlined site, located 60bp upstream of kanR start codon | 57°C | 1 min |
| KanR-R | CCCGGATCCTCGCTTGGTCGGTCATTTC, with BamHI underlined site, located 17 bp downstream of kanR stop codon | 57°C | 1 min |
| OriTS-F | CCCACATGTCCCTGTAGCGGCGCATTAAG, with PciI underlined site, located 973 bp upstream of pSC101 oriTS. | 59°C | 2.5 min |
| OriTS-R | AAACATATGTGGTGAACAGCTTTAAATGCACC, with NdeI underlined site, located 1098 bp downstream of pSC101 oriTS. | 59°C | 2.5 min |
| AceB-F | CACCGATGAACTGGCCTTTA, binds 21 bp downstream of aceB start codon | 54°C | 0.5 min |
| AceB-R | GATTGGGCTGCAACTGATAGA, binds 478 bp downstream of aceB start codon | 54°C | 0.5 min |
| AceA-F | CCGCGCTTTATCGACTACTT, binds 433 bp downstream of aceA start codon | 54°C | 0.5 min |
| AceA-R | AGATCCGGTTTCGACGTTTC binds 865 bp downstream of aceA start codon | 54°C | 0.5 min |
| AceK-F | GTAAACGCCTTATCCGGTCTAC, binds 35 bp downstream of aceK start codon | 54°C | 0.5 min |
| AceK-R | CGTGATGCGGTAGAAGAGTATG, binds 462 bp downstream of aceK start codon | 54°C | 0.5 min |
| IclR-F | TCATCGGTCATACGCGAGAT binds 77 bp downstream of iclR start codon | 54°C | 0.5 min |
| IclR-R | CGCCTGCTGATGGAAGATT binds 480 bp downstream of iclR start codon | 54°C | 0.5 min |
| GyrA-F | GGAAATCAGGGCCAGGAATATG, binds 1983 bp downstream of gyrA start codon | 54°C | 0.5 min |
| GyrA-R | CGTTGGTGACGTAATCGGTAAA, binds 2407 bp downstream of gyrA start codon | 54°C | 0.5 min |
Restriction sites added at the 5’ end of primer are underlined.
Expression studies
Total RNA was isolated from glucose-, succinate- and glucose+succinate-grown K. pneumoniae SM6, SM11, SM6Δ and SM11Δ cells using GeneJET RNA purification kit (Thermo Scientific, USA) following manufacturer’s instructions, except that the cells were washed twice with sterile NaCl solution (0.8%) and resuspended in the same solution before RNA purification. Purified RNA was quantified by measuring absorbance at 260 nm, and the quality and purity was confirmed by electrophoresis on 1.5% agarose gel containing 1.9% (v/v) formaldehyde. cDNA was synthesized from isolated RNA using First Strand cDNA synthesis kit (Thermo Scientific, USA) following manufacturer’s instructions. cDNA was used as template for analysing expression of aceB, aceA, aceK, iclR and gyrA genes in glucose-, succinate- and glucose+succinate-grown cells. DNA Gyrase A (gyrA) was used as internal control to standardize expression and mRNA levels.
Construction of iclR mutants by insertion of a kanamycin cassette
The iclR gene was inactivated employing an allelic exchange system in SM6 and SM11 strains to generate SM6Δ and SM11Δ strains, respectively. To achieve this, pSMB5a and pSMB5b vectors were constructed with 386 bp and 363 bp iclR N’ and C’ gene fragments (of SM6 and SM11, respectively) flanking kanamycin resistance gene, pldhL:gfp uv, and pSC101 temperature controlled origin of replication to direct the integration and excision of vector. E. coli DH5α was used routinely as a host strain for cloning experiments. DNA was introduced into E. coli using Bacterial TransformAid kit (Thermo Scientific, USA). 386 bp iclR-N’ PstI-XbaI fragment was amplified from genomic DNA of K. pneumoniae SM6 and SM11 using primers IclRN’-F and IclRN’-R. The amplicon was cloned in pUC18 digested with PstI-XbaI to generate pSMB1a and pSMB1b, respectively. 363 bp iclR-C’ SmaI-SacI fragment was amplified with IclRC’-F and IclRC’-R primers and cloned in SmaI-SacI digested pSMB1a and pSMB1b, generating pSMB2a and pSMB2b, respectively. Kanamycin resistance gene from pcDNA3.1(+) was amplified using KanR-F and KanR-R primers with BamHI sites. 890 bp kanR gene was ligated at BamHI site of pSMB2a and pSMB2b between iclR-N’ and iclR-C’ fragments creating pSMB3a and pSMB3b, respectively. Successful insertion was confirmed by kanamycin resistant colonies on LB agar plate with kanamycin (50 μg ml-1), restriction digestion of isolated plasmids and amplification of kanamycin resistance gene from pSMB3a and pSMB3b. 2297 bp oriTS was amplified from pGRG36 using oriTS-F and oriTS-R primers with PciI and NdeI flanking sites. pSMB3a and pSMB3b were then digested with PciI and NdeI, and resultant 2288 bp fragment was ligated with 2297 bp PciI and NdeI digested oriTS. Thus, ampicillin resistance gene with pBR322 origin of replication in pSMB3a and pSMB3b was replaced by pSC101 temperature sensitive origin of replication to give pSMB4a and pSMB4b. pRV85 was digested with EcoRI to release pldhL:gfp uv fragment and was cloned at EcoRI site of pSMB4a and pSMB4b. Restriction enzymes were acquired from New England Biolabs (Ipswich, Mass.). S1 Fig shows diagrammatic representation of pSMB5a and pSMB5b construction. pSMB5a-transformed SM6 and pSMB5b-transformed SM11 cells were grow on LB medium with kanamycin and checked for fluorescence under UV light at 28°C (permissive temperature for replication of oriTS) and at 40°C (non-permissive temperature) for integration into genome. The ΔiclR strains were selected for kanamycin resistance and absence of green fluorescence. The integration of pSMB5a and pSMB5b in the genome of SM6 and SM11 strains, respectively, was verified by PCR, loss of green fluorescence and expression of iclR. For PCR verification, chromosomal DNA of transformants was amplified using specific forward primer annealing to regions upstream of iclR and reverse primer of kanamycin resistance gene (data not shown). The loss of green fluorescence was due to curing of pSMB5a and pSMB5b. iclR inactivation and resultant expression of aceBAK operon genes were also confirmed by expression studies.
Indole-3-acetic acid (IAA) production
K. pneumoniae wildtype and ΔiclR strains were screened for IAA production in the LB medium containing tryptophan (1 mg ml-1). The medium was inoculated with 100 μl of overnight-grown bacterial culture and incubated on orbital shaker at 150 rpm for 48 h at 28°C. Quantitative estimation of IAA production was performed, as described by Gordon and Weber [32], using Salkowski reagent containing 10 g L-1 (NH4)2MoO4, 10 ml L-1 H2SO4 and 5 g L-1 FeSO4.7H2O. Uninoculated medium was taken as control.
Pot experiments
Pot experiments were carried out with Wheat (var. Lokwan) (Triticum aestivum), as described by Sachdev et al [33]. Soil samples were collected, air dried, sieved (2 mm mesh) and sterilized by repeated cycles of autoclaving. pH and total P content of the soil was determined by AES-ICP (Model: 330RL, Perkin-Elmer). Soil used for pot experiments was brown, sandy and loamy, with pH 7.3 and total P content 0.12 g L-1. Approximately 1 kg of sterile soil was distributed per pot, and sterile seeds were inoculated with SM6, SM11, SM6Δ and SM11Δ strains (106−107 cfu ml-1) while seeds treated with sterile medium served as control. All pots were maintained in a greenhouse under natural sunlight (10 h day and 14 h night cycle), temperature (average 25°C) and supplied with 150–200 ml autoclaved water per pot per day. Plants were gently uprooted on 31st day and scored for root length (cm), shoot length (cm), root: shoot ratio and dry mass (g).
Statistical analysis
All the experiments were performed in triplicates, and the values in results are mean ± standard deviation, averaged for data analysis. The plant experiment data were expressed as mean ± standard deviation and an analysis of variance (ANOVA) to test the significance of differences was performed with the statistical program Prism® (GraphPad 5.0, USA). The differences between the parameters were evaluated using Tukey’s test (Prism® software). P values ≤ 0.05 were considered to be statistically significant.
Results
Expression analysis of iclR and aceBAK operon genes
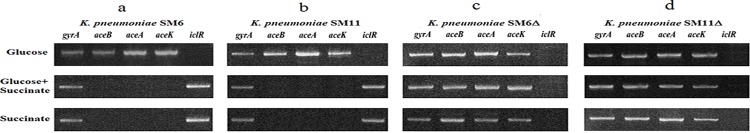
The differential operation of the glyoxylate shunt in glucose-, succinate- and glucose+succinate-grown K. pneumoniae SM6 and SM11 was studied through expression analysis of the glyoxylate shunt (i.e. aceBAK operon) genes and its transcriptional repressor iclR. Genes of the glyoxylate shunt aceB, aceA and aceK were expressed only when glucose was the sole carbon source, whereas iclR was expressed when succinate was the sole carbon source or when present along with glucose (Fig 1A and 1B).
Fig 1. Expression profile of aceBAK operon genes and iclR in glucose-, glucose+succinate-and succinate-grown K. pneumoniae strains (a) SM6, (b) SM11, (c) SM6Δ and (d) SM11Δ.
iclR inactivation and confirmation
iclR deletion strains SM6Δ and SM11Δ retained kanamycin resistance while gfp uv used as second selectable marker was lost after the second recombination. This was due to plasmid curing of pSMB5a and pSMB5b when transformant SM6 and SM11 were subjected to reinouclation in LB with kanamycin and temperature cycles, and kanR integration disrupts iclR (S2 Fig). Fig 1C and 1D show expression analysis of aceBAK operon genes and iclR of glucose-, succinate- and glucose+succinate-grown SM6Δ and SM11Δ. Expression of iclR was not observed in succinate- and glucose+succinate-grown iclR inactive SM6Δ and SM11Δ.
Physiological activities and enzyme activities of the glyoxylate shunt and GO
Fig 2A and 2B show monoauxic growth profile of SM6Δ and SM11Δ when grown on glucose and succinate. No significant difference in growth pattern was observed for both mutants in either of the carbon sources as compared to the wildtype cultures. Glucose favoured the maximum growth, and cells entered exponential phase within 2–3 h of inoculation, reaching maximum optical density (OD) with stationary phase in 4–5 h. Growth on succinate was observed after a lag phase and was slower than on glucose. This may be because succinate is a poor carbon source for enterobacteria like E. coli [34]. Similarly, no significant difference was observed in the diauxic growth profile and carbon utilization pattern of iclR mutants SM6Δ and SM11Δ, as compared to wildtype SM6 and SM11. Glucose was utilized preferentially over succinate in both the mutants and 90–100% of glucose was consumed within first 5–6 h of incubation, while succinate was utilized only when glucose was exhausted in the media (Fig 2C and 2D). The specific activities of ICL and GO in glucose- and succinate-grown SM6 and SM11 cells and their iclR inactivated counterparts SM6Δ and SM11Δ are depicted in Fig 3.
Fig 2. Monoauxic growth profile of (a) SM6Δ and (b) SM11Δ, and diauxic growth profile of (c) SM6Δ and (d) SM11Δ.
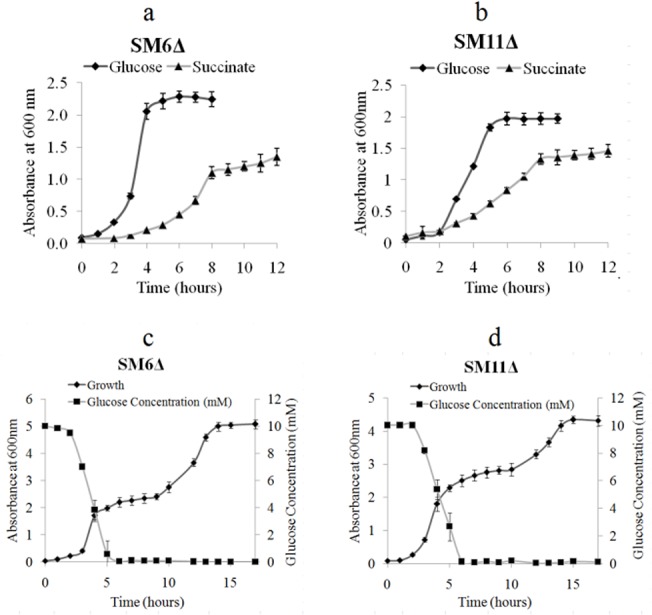
Fig 3. Enzyme activity of glucose- and succinate-grown SM6, SM11, SM6Δ and SM11Δ: (a) ICL (b) GO.
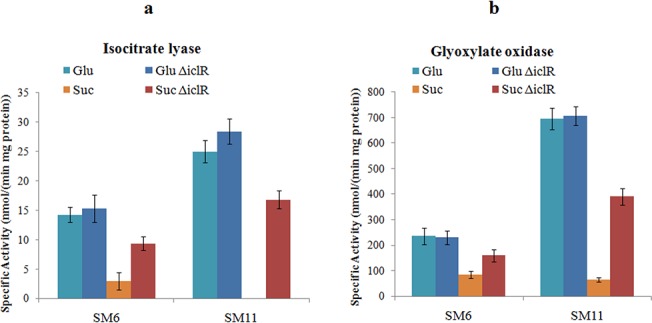
iclR inactivation resulted in 3.2 fold increase in ICL activity of succinate-grown SM6Δ, as compared to wildtype SM6, and in high activity in succinate-grown SM11Δ, whereas ICL was undetectable in SM11 strain. The ICL activity of succinate-grown SM6Δ and SM11Δ was 74.6% and 73.2% as compared to that of glucose-grown cells. The ICL activity in succinate-grown wildtype SM6 and SM11 cells was 20.7% and 0% respectively, when compared to the glucose-grown cells [14]. Similarly, an enhanced GO activity was observed in succinate-grown SM6Δ and SM11Δ (Fig 3A). Based on the published data [14] and present findings, we propose a metabolic scheme that explains the MPS phenotype in the wildtype SM6 and SM11 strains during grown on glucose or on succinate (or on glucose+succinate) (Fig 4).
Fig 4. Carbon metabolism linked P solubilization in (A) glucose and (B) succinate or glucose+succinate-grown Klebsiella pneumoniae SM6 and SM11.
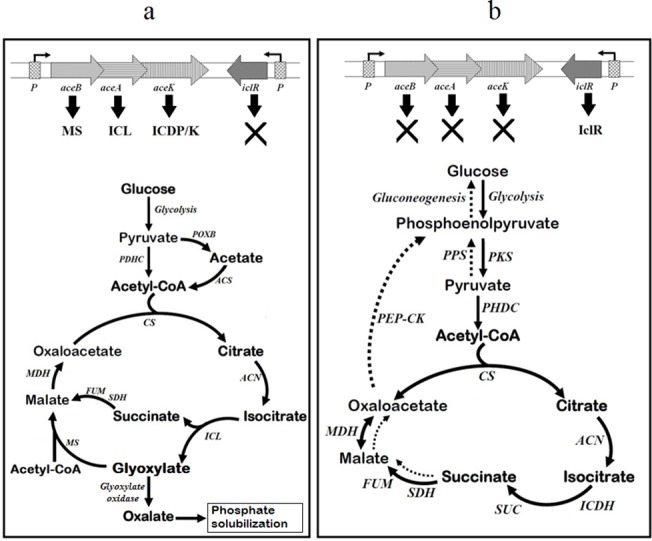
Glucose is metabolized by EMP pathway and acetate is overproduced, which can be then activated to acetyl-CoA. Acetyl-CoA is incorporated in the TCA and the glyoxylate shunt. During growth on succinate, gluconeogenesis is required for the formation of PEP and further intermediates of the EMP pathway, whereas the TCA cycle serves as the main catabolic process under these conditions. MS, malate synthase; ICL, isocitrate lyase; ICDP/K, isocitrate dehydrogenase kinase/phosphatase; IclR, isocitrate lyase repressor; P, promoter; POXB, pyruvate oxidase B; ACS, AMP-forming acetyl-CoA synthetase; PDHC, pyruvate dehydrogenase complex; PPS, phosphoenolpyruvate synthase; PKS, pyruvate kinase; PEP-CK, phosphoenolpyruvate carboxykinase; CS, citrate synthase; ACN, aconitase; SUC, succinyl-CoA synthetase complex; ICDH, isocitrate dehydrogenase; SDH, succinate dehydrogenase; FUM, fumarase; MDH, malate dehydrogenase.
P solubilization by iclR mutants
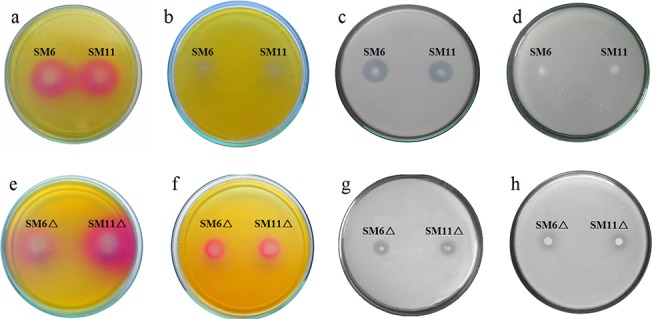
Fig 5 shows soluble P released in Pikovskaya broth with glucose and glucose+succinate. The maximum P solubilized by SM6 and SM11 strains in media with glucose+succinate was only 3.8 and 1.5%, respectively, as compared to P solubilized with glucose [14]. The amount of P solubilized in glucose media by the wildtype strains was almost equivalent to that of the corresponding ΔiclR strains. SM6Δ and SM11Δ were enhanced in P solubilization in glucose+succinate medium, reaching up to 54 and 59% in comparison to glucose medium. Fig 6 shows mineral phosphate solubilization in Pikovskaya agar and rock phosphate solubilization in TRP agar by SM6, SM11, SM6Δ and SM11Δ in presence of glucose and glucose+succinate.
Fig 5. P released by (a) SM6 and SM11, and (b) SM6Δ and SM11Δ in Pikovskaya broth with glucose (G) and glucose+succinate (GS).
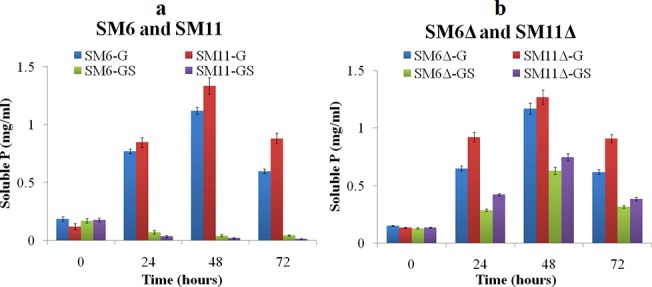
Fig 6. MPS phenotype of wildtype SM6 and SM11 on (a) TRP agar with glucose (b) TRP agar with glucose+succinate (c) Pikovskaya agar with glucose (d) Pikovskaya agar with glucose+succinate, and MPS phenotype of SM6Δ and SM11Δ on (e) TRP agar with glucose (f) TRP agar with glucose+succinate (g) Pikovskaya agar with glucose (h) Pikovskaya agar with glucose+succinate.
IAA Production
SM6 and SM11 produced 18.1 ± 0.9 μg ml-1 and 18.7 ± 1.1 μg ml-1 of IAA, respectively, while 17.6 ± 1.2 μg ml-1 and 19.5 ± 1.1 μg ml-1 IAA was produced by SM6Δ and SM11Δ respectively, in LB medium supplemented with tryptophan.
Effect of wildtype and iclR inactivated strains on plant growth
Wildtype and ΔiclR strains improved plant growth over the control (Table 3). Plants inoculated with the SM6 and SM11 strains increased root length and shoot length, while the growth improvement with SM6Δ and SM11Δ strains was even more prominent. SM6Δ and SM11Δ treated plants had significantly higher root length and shoot length compared to wildtype.
Table 3. Growth parameters of wheat plants in the presence of K. pneumoniae SM6, SM11, SM6Δ and SM11Δ strains.
| Root length (cm) * | Shoot length (cm) * | Root: Shoot ratio | Dry mass (mg) * | |
|---|---|---|---|---|
| Control | 19.2 ± 1.8 | 16.7 ± 1.7 | 1.14 | 241.1 ± 19.5 |
| K. pneumoniae SM6 | 25.7 ± 2.16 | 20.8 ± 1.8 | 1.23 | 290.4 ± 17.1 |
| K. pneumoniae SM11 | 24.3 ± 1.9 | 19.6 ± 1.5 | 1.23 | 282.9 ± 20.1 |
| K. pneumoniae SM6Δ | 30.2 ± 2.4 † | 23.5 ± 1.7 | 1.28 | 324.7 ± 22.8 † |
| K. pneumoniae SM11Δ | 29.9 ± 3.1 † | 23.0 ± 1.8 † | 1.3 | 330.2 ± 24.3 † |
* Significant as compared to control (P ≤ 0.05).
† Significant at P ≤ 0.05 as compared to their respective wildtype.
Values for root length, shoot length and dry mass are mean ± standard deviation of 12 plants (n = 12).
Discussion
iclR and aceBAK expression
Absence of expression of aceBAK operon genes was accompanied by the repression of phosphate solubilization and acid production in succinate or glucose+succinate-grown K. pneumoniae SM6 and SM11 [14]. In E. coli, IclR regulates the expression of the aceBAK operon coding for isocitrate lyase (AceA), malate synthase (AceB) and isocitrate dehydrogenase kinase/phosphatase (AceK) [17]. Isocitrate dehydrogenase kinase/phosphatase (AceK) phosphorylates isocitrate dehydrogenase (ICD), reducing the ICD activity and therefore the carbon flux through tricarboxylic acid (TCA) cycle [35]. Repression of the glyoxylate shunt genes is relieved and the pathway is activated, when IclR levels are low or when iclR is inactivated. This in particular occurs when cells are grown on acetate [36, 37], or in slow-growing glucose-utilizing cultures [38, 39].
The expression studies of aceBAK operon genes and iclR showed the repression of MPS phenotype by succinate in SM6 and SM11. ICL cleaves isocitrate to glyoxylate and succinate, and glyoxylate is oxidized by GO to produce oxalic acid [40]. Succinate in growth media induced the expression of iclR, which repressed aceBAK operon and oxalic acid-mediated MPS phenotype in SM6 and SM11. As iclR was not expressed in glucose-grown cells, the glyoxylate shunt and oxalic acid production were constitutive. Flow of carbon through the glyoxylate shunt can also be explained as a measure to utilize the acetate overproduced from pyruvate in glucose-grown cells. In E. coli, acetate production is usually credited to the “overflow metabolism” [40], and is believed to be a result of an imbalance between the energy requirement, biosynthesis and glucose uptake [41–43].
iclR inactivation
In the Enterobacteriaceae, genes of the glyoxylate shunt are encoded in aceBAK operon and are under the direct negative control of repressor IclR [15, 16]. The expression of aceBAK is induced, when organisms are grown on acetate or fatty acids [44]. The glyoxylate bypass is required for growth on fatty acids and acetate, as they enter intermediary metabolism as acetyl-CoA. When E. coli is grown in excess of glucose, the carbon flux through glycolysis surpasses the capacity of the TCA cycle, and acetate accumulation occurs [45]. Difference in carbon flux through the glyoxylate shunt and the TCA cycle is observed in low acetate producer E. coli K strain (JM109) and high acetate producer E. coli B strain (BL21). In glucose-grown E. coli B cell, acetate is produced from pyruvate by pyruvate oxidase (PoxB). The acetate accumulated in cell can be converted to acetyl-CoA by acetyl-CoA synthetase (Acs), or by reversing the phosphotransacetylase-acetate kinase (Pta-AckA) pathway [46]. Acetyl-CoA is metabolized through the TCA cycle and the glyoxylate shunt by ICL and MS [44, 47]. Under these conditions, activation of the glyoxylate shunt in E. coli B strains (BL21) has been proposed as a route for internal acetate utilization (acetyl-CoA consumption), in contrast to E. coli K strain (JM109) where excess acetate is secreted out [48]. Acetyl-CoA utilization with a higher ICL activity by activation of the glyoxylate shunt has been observed in BL21 only and not in JM109 in high glucose batch cultures [48]. K. pneumoniae SM6 and SM11 apparently resembled E. coli B strain, where operation of the glyoxylate shunt is constitutive when glucose is sole carbon source. Moreover, absence of acetic acid production by SM6 and SM11 [14] also related them to E. coli B strain, in contrast to E. coli K strain.
Physiological experiments and enzyme activities
The release of soluble P in the medium is the result of organic acid production [49]. The growth pattern of both iclR deletion strains was similar to that of parental wildtype strains, as reported earlier [14]. This showed that iclR inactivation did not interfere with glucose and succinate utilization. Unlike rhizobacteria Pseudomonas and Rhizobium, where C4 acids like succinate and malate are preferred over glucose, enterobacteria and firmicutes utilize glucose as the most preferred carbon source [50–52]. This is mainly because of the catabolite repression and carbon uptake PTS that controls the expression of carbon metabolism genes and carbon source uptake and subsequent utilization via EMP pathway. To further confirm that succinate mediated repression of MPS phenotype, activities of ICL and GO were analysed in glucose and succinate-grown cells of wildtype and SM6Δ and SM11Δ.
The ICL and GO activities measured in glucose- and succinate-grown SM6 and SM11 cells (Fig 3) were similar to those reported previously [14]. Interestingly, iclR inactivation did not alter ICL and GO activities of the glucose-grown cells. The possible explanation for these data is that the glucose-grown cells oxidise acetyl-CoA to glyoxylate via the glyoxylate cycle, and not via the classical TCA cycle, thus providing the substrate for oxalate synthesis.
MPS phenotype of iclR mutants
Although the release of P mediated by ΔiclR strains in glucose+succinate media was still not as high as in the media containing glucose only, the results are encouraging and reveal that regulators of carbon metabolism influence the MPS phenotype, depending on the nature of an available carbon source. Derepression of the MPS phenotype during growth on glucose+succinate observed in ΔiclR strains was due to enhanced organic acid production. This effect was accompanied by the acidification and rock phosphate solubilization in TRP media. Thus, the carbon source-mediated repression of the MPS phenotype may be one of the reasons of failure of several phosphate solubilizing microorganisms to improve plant growth in the field experiments. Indeed, the availability of multiple carbon sources in natural conditions may lead to repression of various functional genes, including those responsible for MPS phenotype. Identification and inactivation of such regulators may abolish the repression, thus resulting in engineering of microbes to constitutive P solubilization.
Plant experiments
Both, the wildtype and ΔiclR strains of K. pneumoniae SM6 and SM11 improved plant growth, whereas the mutant strains were significantly more efficient in terms of root and shoot length, root: shoot ratio and dry mass. This effect of iclR inactivation was not related to increase of IAA synthesis, but rather to the P release mediated by the oxalic acid production. Indeed, we showed that the synthesis of glyoxylate, oxalate precursor, was enhanced in the mutant. Thus, the repressor inactivation may result in better performance of PGP bacteria in the rhizosphere. Free-living N2 fixers with relieved constitutive MPS phenotype can serve dual purpose to plants.
Supporting Information
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Dr. SR received the financial grant from the Department of Science and Technology (Ministry of Science, India) (Project Sanction number SERB/F/1433/1014-15). The authors would like to acknowledge the Council of Scientific and Industrial Research (CSIR), Government of India, for providing senior research fellowship to Mahendrapal Singh Rajput (Ack No. 113827/2K12/1) and the Department of Science and Technology (DST), Government of India for providing INSPIRE fellowship to Bhagya Iyer (IF130895). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ahemad M, Khan MS. Effects of insecticides on plant growth-promoting activities of phosphate solubilizing rhizobacterium Klebsiella sp. strain PS19. Pestic Biochem Physiol. 2011;100: 51–56. [Google Scholar]
- 2. Ahemad M, Khan MS. Toxicological effects of selective herbicides on plant growth promoting activities of phosphate solubilizing Klebsiella sp. strain PS19. Curr Microbiol. 2011;62: 532–538. 10.1007/s00284-010-9740-0 [DOI] [PubMed] [Google Scholar]
- 3. Ahemad M, Khan MS. Biotoxic impact of fungicides on plant growth promoting activities of phosphate-solubilizing Klebsiella sp. isolated from mustard (Brassica campestris) rhizosphere. J Pest Sci. 2012;85: 29–36. [DOI] [PubMed] [Google Scholar]
- 4. Iniguez AL, Dong Y, Triplett EW. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol Plant Microbe Interact. 2004;17: 1078–1085. [DOI] [PubMed] [Google Scholar]
- 5. Chelius MK, Triplett EW. Immunolocalization of dinitrogenase reductase produced by Klebsiella pneumoniae in association with Zea mays L. Appl Environ Microbiol. 2000;66: 783–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. An QL, Yang XJ, Dong YM, Feng LJ, Kuang BJ, Li JD. Using confocal laser scanning microscope to visualize the infection of rice roots by GFP-labelled Klebsiella oxytoca SA2, an endophytic diazotroph. Acta Bot Sin. 2001;43: 558–564. [Google Scholar]
- 7. Martinez L, Caballero-Mellaod J, Orozco J, Martinez-Romero E. Diazotrophic bacteria associated with banana (Musa spp.). Plant Soil. 2003;257: 35–47. [Google Scholar]
- 8. Ando S, Goto M, Meunchang S, Thongraar P, Fujiwara T, Hayashi H, et al. Detection of nifH Sequences in sugarcane (Saccharum officinarum L.) and pineapple (Ananas comosus [L.] Merr.). Soil Sci Plant Nutr. 2005;51: 303–308. [Google Scholar]
- 9. Gyaneshwar P, Kumar GN, Parekh LJ, Poole PS. Role of soil microorganisms in improving P nutrition of plants. Plant Soil. 2002;245: 83–93. [Google Scholar]
- 10. Khan MS, Zaidi A, Wani PA. Role of phosphate-solubilizing microorganisms in sustainable agriculture-A review. Agron Sustain Dev. 2006;27: 29–43. [Google Scholar]
- 11. Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6: 613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- 12. Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2: 195–201. [DOI] [PubMed] [Google Scholar]
- 13. Saier MH Jr. Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Microbiol Rev. 1989;53: 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rajput MS, Naresh Kumar G, Rajkumar S. Repression of oxalic acid-mediated mineral phosphate solubilization in rhizospheric isolates of Klebsiella pneumoniae by succinate. Arch Microbiol. 2013;195: 81–88. 10.1007/s00203-012-0850-x [DOI] [PubMed] [Google Scholar]
- 15. Maloy SR, Nunn WD. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J Bacteriol. 1982;149: 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sunnarborg A, Klumpp D, Chung T, LaPorte DC. Regulation of the glyoxylate bypass operon: cloning and characterization of IclR. J Bacteriol. 1990;172: 2642–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamamoto K, Ishihama A. Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mol Microbiol. 2003;47: 183–194. [DOI] [PubMed] [Google Scholar]
- 18. Krell T, Molina-Henares AJ, Ramos JL. The IclR family of transcriptional activators and repressors can be defined by a single profile. Protein Sci. 2006;15: 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sarkar D, Siddiquee KA, Arauzo-Bravo MJ, Oba T, Shimizu K. Effect of cra gene knockout together with edd and iclR genes knockout on the metabolism in Escherichia coli . Arc Microbiol. 2008;190: 559–571. [DOI] [PubMed] [Google Scholar]
- 20. Brinkrolf K, Brune I, Tauch A. Transcriptional regulation of catabolic pathways for aromatic compounds in Corynebacterium glutamicum . Genet Mol Res. 2006;5: 773–789. [PubMed] [Google Scholar]
- 21. Zhang HB, Wang LH, Zhang LH. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens . Proc Natl Acad Sci USA. 2002;99: 4638–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Molina-Henares AJ, Krell T, Guazzaroni ME, Segura A, Ramos JL. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol Rev. 2006;30: 157–186. [DOI] [PubMed] [Google Scholar]
- 23. Seth GN, Gran T, Jesseet J, Bloomt FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants (bacterial restriction/ DNA methylation/ cloning mammalian DNA/ heterogeneous transgene expression/ insulin gene regulation). Proc Natl Acad Sci USA. 1990;87: 4645–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA-replication origins. Gene 1991;100: 189–194 [DOI] [PubMed] [Google Scholar]
- 25. McKenzie GJ, Craig NL. Fast, easy and efficient: site-specific insertion of transgenes into Enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 2006;6: 39 10.1186/1471-2180-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gory L, Montel MC, Zagorec M. Use of green fluorescent protein to monitor Lactobacillus sakei in fermented meat products. FEMS Microbiol Lett. 2001;194: 127–133. [DOI] [PubMed] [Google Scholar]
- 27. Dixon GH, Kornberg HL. Assay methods for key enzymes of the glyoxylate cycle. Proc Biochem Soc. 1959;72: 3P. [Google Scholar]
- 28. Akamatsu Y, Shimada M. Partial purification and characterization of glyoxylate oxidase from the brown-rot basidiomycete Tyromyces palustris . Phytochemistry. 1994;37: 649–653. [Google Scholar]
- 29. Buch AD, Archana G, Kumar G. Metabolic channeling of glucose towards gluconate in phosphate-solubilizing Pseudomonas aeruginosa P4 under phosphorus deficiency. Res Microbiol 2008;159:635–642. 10.1016/j.resmic.2008.09.012 [DOI] [PubMed] [Google Scholar]
- 30. Pikovskaya RI. Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiology. 1948;17: 362–370. [Google Scholar]
- 31. Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8: 115–118. [Google Scholar]
- 32. Gordon SA, Weber RP. Colorimetric estimation of indole acetic acid. Plant physiol. 1951;26: 192–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sachdev DP, Chaudhari HG, Kasture VM, Dhavale DD, Chopade BA. Isolation and characterization of indole acetic acid (IAA) producing Klebsiella pneumoniae strains from rhizosphere of wheat (Triticum aestivum) and their effect on plant growth. Indian J Exp Biol. 2009;47: 993–1000. [PubMed] [Google Scholar]
- 34. Chen G, Patten CL, Schellhorn HE. Positive selection for loss of RpoS function in Escherichia coli . Mutat. Res. 2004;554: 193–203. [DOI] [PubMed] [Google Scholar]
- 35. Rittinger K, Negre D, Divita G, Scarabel M, Bonod-Bidaud C, Goody RS, et al. Escherichia coli isocitrate dehydrogenase kinase/phosphatase. Eur J Biochem. 1996;237: 247–254. [DOI] [PubMed] [Google Scholar]
- 36. Cortay JC, Negre D, Galinier A, Duclos B, Perriere G, Cozzone AJ. Regulation of the acetate operon in Escherichia coli: purification and functional characterization of the IclR repressor. EMBO J. 1991;10: 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cozzone AJ. Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Ann Rev Microbiol. 1998;52: 127–164. [DOI] [PubMed] [Google Scholar]
- 38. Fischer E, Sauer U. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli . J Biol Chem. 2003;278: 46446–46451. [DOI] [PubMed] [Google Scholar]
- 39. Maharjan RP, Yu PL, Seeto S, Ferenci T. The role of isocitrate lyase and the glyoxylate cycle in Escherichia coli growing under glucose limitation. Res Microbiol. 2005;156: 178–183. [DOI] [PubMed] [Google Scholar]
- 40. Kayoko M, Takefumi H. Occurrence of enzyme systems for production and decomposition of oxalate in a white-rot fungus Coriolus versicolor and some characteristics of glyoxylate oxidase. Wood Res. 1996;83: 23–26. [Google Scholar]
- 41. Han K, Lim HC, Hong J. Acetic acid formation in Escherichia coli fermentation. Biotechnol Bioeng. 1992;39: 663–671. [DOI] [PubMed] [Google Scholar]
- 42. Kleman GL, Strohl WR. Acetate metabolism by Escherichia coli in high-cell-density fermentation. Appl Environ Microbiol. 1994;60: 3952–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aristidou AA, San KY, Bennett GN. Metabolic engineering of Escherichia coli to enhance recombinant protein production through acetate reduction. Biotechnol Prog. 1995;11: 475–478. [DOI] [PubMed] [Google Scholar]
- 44. Kornberg HL. The role and control of the glyoxylate cycle in Escherichia coli . Biochem J. 1966;99: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Majewski RA, Domach MM. Simple constrained optimization view of acetate overflow in E. coli . Biotechnol Bioeng. 1990;35: 732–738. [DOI] [PubMed] [Google Scholar]
- 46. Brown TDK, Jones-Mortimer MC, Kornberg HL. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli . J Gen Microbiol. 1977;102: 327–336. [DOI] [PubMed] [Google Scholar]
- 47. El-Mansi EM, Holms WH. Control of carbon flux to acetate excretion during overflow in Escherichia coli . Biotechnol Bioeng. 1989;35: 732–738. [DOI] [PubMed] [Google Scholar]
- 48. Van de Walle M, Shiloach J. Proposed mechanism of acetate accumulation in two recombinant Escherichia coli strains during high density fermentation. Biotechnol Bioeng. 1989;57: 71–78. [DOI] [PubMed] [Google Scholar]
- 49. Rodriguez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv. 1999;17: 319–339 [DOI] [PubMed] [Google Scholar]
- 50. Mandal NC, Chakrabartty PK. Succinate mediated catabolite repression of enzymes of glucose metabolism in root-nodule bacteria. Curr Microbiol. 1993;26: 247–251. [Google Scholar]
- 51. Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57: 543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collier DN, Hager PW, Phibbs PV Jr. Catabolite repression control in Pseudomonads. Res Microbiol. 1996;147: 551–561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


