Abstract
Axonal miRNAs locally regulate axonal growth by modulating local protein composition. Whether localized miRNAs in the axon mediate the inhibitory effect of CSPGs on the axon remains unknown. We showed that in cultured cortical neurons, axonal application of CSPGs inhibited axonal growth and altered axonal miRNA profiles, whereas elevation of axonal cGMP levels by axonal application of sildenafil reversed the effect of CSPGs on inhibition of axonal growth and on miRNA profiles. Specifically, CSPGs elevated and reduced axonal levels of miR-29c and ITGB1 proteins, respectively, while elevation of cGMP levels overcame these CSPG effects. Gain-of- and loss-of-function experiments demonstrated that miR-29c in the distal axon mediates axonal growth downstream of CSPGs and cGMP by regulating axonal protein levels of ITGB1, FAK, and RhoA. Together, our data demonstrate that axonal miRNAs play an import role in mediating the inhibitory action of CSPGs on axonal growth and that miR-29c at least partially mediates this process.
Introduction
The axon can function with a high degree of autonomy from the somata (Willis et al., 2007; Jung et al., 2012; Colak et al., 2013; Gomes et al., 2014). There is increasing knowledge of extracellular signals that can spatiotemporally regulate axonal growth and regeneration by mediating axonal levels of individual mRNAs and mRNA translation (Willis et al., 2007; Jung et al., 2012; Colak et al., 2013; Gomes et al., 2014). Chondroitin sulfate proteoglycans (CSPGs), extracellular matrix molecules that inhibit axonal growth, are generated by activated astrocytes and oligodendrocyte progenitor cells after injury (Hartmann and Maurer, 2001; Bradbury et al., 2002). Recent studies showed that CSPGs block axonal growth by locally activating axonal RhoA translation in dorsal root ganglia neurons (Walker et al., 2012). Activation of RhoA signaling leads to suppressed axonal growth, whereas inhibition of RhoA signaling increases axonal regeneration and improves neurological outcome in animal models of axonal injury (Monnier et al., 2003; Wu et al., 2005). Inactivation of integrins by CSPGs also inhibits axonal growth (Tan et al., 2011). In addition to mRNAs, axons contain abundant microRNAs (miRNAs) (Natera-Naranjo et al., 2010; Dajas-Bailador et al., 2012; Kaplan et al., 2013; Iyer et al., 2014; Sasaki et al., 2014). MiRNAs, noncoding RNAs, are post-transcriptional regulators of gene expression by binding the 3’ UTRs of mRNAs (Andreassi and Riccio, 2009). Axonal miRNAs regulate energy metabolism and axonal growth by locally modulating protein composition (Aschrafi et al., 2008; Dajas-Bailador et al., 2012; Kar et al., 2013; Wu and Murashov, 2013; Zhang et al., 2013; Hancock et al., 2014). However, whether localized miRNAs in the axon mediate CSPG-induced activation of RhoA and inactivation of integrins, has not been investigated.
Activation of cyclic guanosine monophosphate (cGMP) promotes neurite growth and axonal branching (Murray et al., 2009; Zhao et al., 2009; Xia et al., 2013). Inhibition of the RhoA signaling pathway by cGMP-activated protein kinase G is one of mechanisms that underscore cGMP-promoted axonal growth (Mandal et al., 2013; Xia et al., 2013). Thus, elevation of cGMP levels might overcome RhoA signaling activated by CSPGs and reverse the inhibitory effect of CSPGs on axonal growth. Hydrolysis of cGMP is mediated by phosphodiesterase type 5 (PDE5) enzyme and neurons express PDE5 (Corbin and Francis, 1999). Sildenafil is a highly specific inhibitor for PDE5 and increases cGMP (Corbin and Francis, 1999).
Using a microfluidic culture device, we investigated whether axonal miRNAs locally regulate the effect of CSPGs and cGMP on axonal growth of cortical neurons. Our studies indicate that localized miRNAs in axons mediate the CSPG-induced inhibitory effect on axonal growth and the cGMP-promotion of axonal growth. We also identified that miR-29c locally modulates integrin β1 (ITGB1) and its related signaling, including RhoA in axons. These data provide coherent mechanistic insight into the effect of CSPGs and cGMP on axonal growth.
Results
CSPGs alter distal axonal miRNA profile
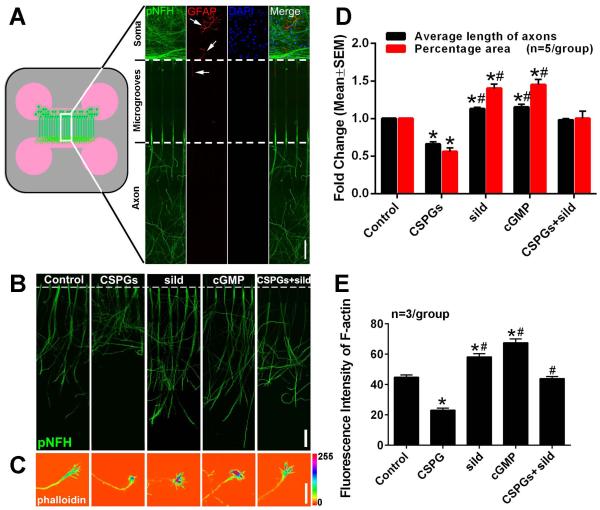
Selective exposure of distal axons to CSPGs occurs when axons approach gliotic regions after central nervous system (CNS) injury (Rudge and Silver, 1990; Monnier et al., 2003; Yiu and He, 2006). To mimic the in vivo situation, cortical neurons harvested from rat embryos were cultured in a microfluidic culture device that permits selective application of CSPGs to distal axons (Taylor et al., 2005; Zhang et al., 2013). To examine whether there are astrocytes in the culture, double immunofluorescent staining was performed. We found that cortical cells in the soma chamber were neurofilament H (NFH) positive neurons and that these neurons extended their axons into the axonal chamber (Fig. 1A). Only a few glial fibrillary acidic protein (GFAP) positive cells were detected in the soma chamber and no GFAP positive processes were detected in the axonal chamber (Fig. 1A). To examine the effect of CSPGs on axonal outgrowth, CSPGs were added into the axonal chamber and axonal growth was measured. After 5 days in vitro (DIV5), axonal application of CSPGs suppressed axonal outgrowth by 34% compared to non-CSPG treatment (Fig. 1B, D). CSPGs also significantly decreased F-actin fluorescent intensity in the axonal growth cone (Fig. 1C, E). These data indicate that axonal application of CSPGs inhibits distal axonal outgrowth, which is consistent with published studies (Yiu and He, 2006; Tan et al., 2011; Walker et al., 2012).
Figure 1.
The effect of axonal application of CSPGs and sildenafil on axonal outgrowth of cortical neurons. Panel A shows a schematic representation of the microfluidic device and double immunofluorescent images showing pNFH+ axons and dendrites (green) and GFAP+ astrocytes with their processes (red, arrows) in the cell body compartment and microgrooves, and the pNFH+ axons in the axonal compartment (green). Blue color indicates cell nuclei (DAPI). Representative images were taken from the boxed areas in the schematics. Panels B and C show representative pNFH+ axons and F-actin (phalloidin)+ growth cones, respectively, in the axonal chamber under control, CSPGs, sildenafil (sild), CSPGs+sildenafil (CSPGs+sild) and cGMP conditions on DIV5. Panel D shows quantitative data of average length of axons measured by the lengths of the 15 longest axons in one axonal compartment (black bar) and percentage of pNFH+ axonal area within the entire area of one axonal compartment (red bar). Panel E shows quantitative data of F-actin intensity within the growth cone. * and # p<0.05 vs control and CSPGs, respectively. sild=sildenafil, cGMP=8-Br-cGMP. Scale bar in A and B =100 μm; C =20 μm.
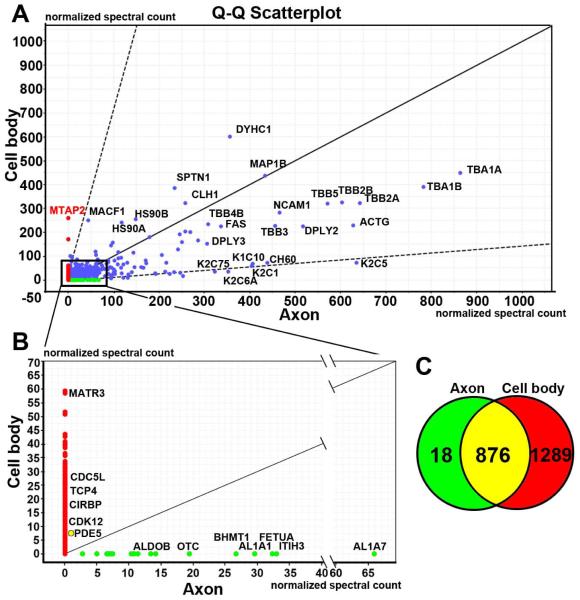
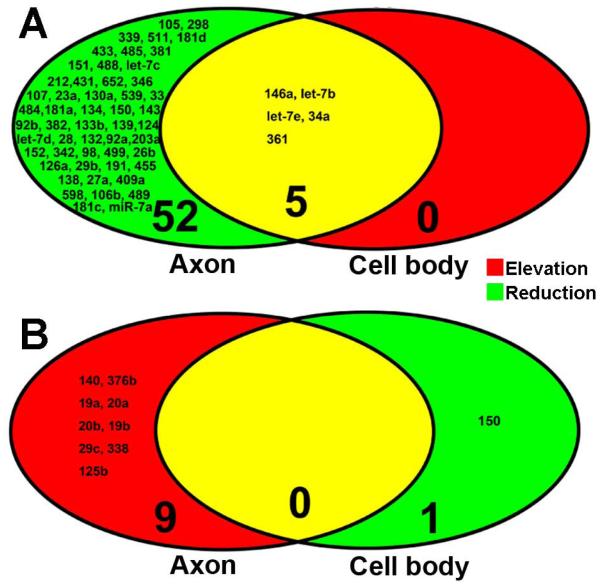
Molecular mechanisms underlying the inhibitory effect of CSPGs on axonal growth remain to be investigated. Enriched miRNAs in distal axons regulate energy metabolisms and axonal growth (Aschrafi et al., 2008; Dajas-Bailador et al., 2012; Kar et al., 2013; Wu and Murashov, 2013; Zhang et al., 2013; Hancock et al., 2014). We thus examined the effect of CSPGs on axonal miRNAs. To do so, we first tested whether biomaterials extracted from the axonal compartment are contaminated with extracts from the cell body compartments. Protein profiles were analyzed with proteomics for samples from the axonal or cell body compartment. Proteomic analysis revealed that the profile of proteins harvested from the axonal compartment was distinct from the cell body compartment (Fig. 2). Proteins such as MAP2 in the cell body samples are exclusively expressed in the soma and dendrites, which were not detected in distal axonal proteins. In addition, nuclear proteins such as MATR3 and CDC5L, were not detected in axonal proteins. To further examine purity of axonal samples, we measured mRNA levels of H1 histone (H1f0), calmodulin-dependent protein kinase II α (Camk2a), β-actin and synaptophysin (Syp) in axonal and cell body samples. Quantitative real-time RT-PCR analysis revealed that all four mRNAs were detected in samples prepared from the cell body compartment. However, in axonal samples, only β-actin and Syp mRNAs were detected, while mRNAs of H1f0 and Camk2a were not detected even after 55 cycles of PCR amplification, which is consistent with published data by others showing absence of H1f0 and Camk2a mRNAs in axons of cortical neurons (Taylor et al., 2005). These data indicate that there was no contamination of sample preparation between the cell body and axonal chambers and that we can isolate axonal proteins from the axonal compartment. (Taylor et al., 2005; Taylor et al., 2009). We then examined whether axonal application of CSPGs affects miRNA levels in the distal axons. After axonal application of CSPGs, total RNAs were harvested from the cell body compartment containing cell bodies and some axons and from the axonal compartment containing exclusively distal axons. miRNA profiles were analyzed by means of a miRNA PCR array kit including 84 miRNAs that are involved in neuronal development and neurological diseases. For miRNA array analysis, we used a fold change in levels of at least 1.5 relative to the non-CSPG treatment to select for CSPG-altered miRNAs, and found that in the distal axons 9 and 57 miRNAs were elevated and reduced, respectively (Table 1). Interestingly, axonal application of CSPGs also elevated and reduced 5 and 1 miRNAs, respectively, in the cell bodies (Fig. 3). These data indicate that the axonal application of CSPGs significantly changes the miRNA profile in distal axons and cell bodies, which may contribute to the inhibitory effect of CSPGs on axonal growth.
Figure 2.
Protein profiles in neuronal cell bodies and axons measured by proteomics. The quantified-quantified (Q-Q) Scatterplot of proteins (A) was generated by a Scaffold Viewer software based on protein extracts from cell body (Y axis) and axonal (X axis) compartments on DIV5. The numbers in X and Y axis are the normalized spectral counts which represent the relative content of proteins. The dots show the identified proteins exclusively in axon (green), cell body (red) and both (blue). The 45 degree line indicates the same abundance of proteins in cell body and axonal compartments, while the two dash lines show proteins with the two standard deviations above and below the 45 degree line. Red and green dots plotted outside the standard deviation lines likely represent that proteins were differentially expressed in cell body and axonal compartments, respectively. Panel B lists representative proteins exclusively detected in cell body (red) or axonal (green) compartment within the boxed area in panel A. Listed proteins exclusively in cell body compartment are nuclear proteins, while listed proteins in axonal compartment are involved in regulating axonal growth. As noted, PDE5 protein was detected in both cell body and axonal compartments (a yellow dot in B). A venn diagram of proteins (C) shows the total number of proteins identified in axon (green), cell body (red) and both (yellow).
Table 1.
Microarray data of axonal miRNAs altered by application of CSPGs.
| Down-regulated miRNAs |
Average CT (±SD) |
Fold changes vs control |
p-value |
|---|---|---|---|
| miR-105 | 32.4±0.1 | −26.0 | 0.017 |
| miR-298-5p | 29.0±0.2 | −11.7 | 0.007 |
| miR-339-5p | 31.4±0.2 | −8.8 | 0.055 |
| miR-34a-5p | 32.3±0.2 | −8.6 | 0.019 |
| miR-511-5p | 31.5±0.5 | −7.7 | 0.001 |
| let-7b-5p | 25.6±0.1 | −7.6 | 0.024 |
| miR-181d-5p | 28.1±0.1 | −7.3 | 0.020 |
| miR-433-3p | 28.9±0.2 | −6.8 | 0.005 |
| miR-485-5p | 32.1±0.4 | −6.7 | 0.009 |
| miR-381-3p | 30.9±0.3 | −6.2 | 0.014 |
| miR-151-3p | 29.3±0.2 | −5.8 | 0.053 |
| let-7e-5p | 25.6±0.1 | −5.7 | 0.008 |
| miR-488-3p | 32.2±0.5 | −5.5 | 0.044 |
| let-7c-5p | 23.9±0.1 | −5.5 | 0.008 |
| miR-212-3p | 31.4±0.1 | −5.4 | 0.025 |
| miR-361-5p | 31.1±0.4 | −5.3 | 0.005 |
| miR-431 | 30.4±0.6 | −5.2 | 0.017 |
| miR-652-3p | 29.0±0.1 | −5.0 | 0.005 |
| miR-346 | 30.3±0.0 | −4.5 | 0.032 |
| miR-107-3p | 31.4±0.2 | −4.5 | 0.001 |
| miR-146a-5p | 31.7±0.1 | −4.4 | 0.021 |
| miR-23a-3p | 27.8±0.0 | −4.1 | 0.065 |
| miR-130a-3p | 28.2±0.0 | −4.0 | 0.000 |
| miR-539-5p | 31.2±0.1 | −4.0 | 0.020 |
| miR-33-5p | 34.8±0.3 | −4.0 | 0.008 |
| miR-484 | 28.9±0.0 | −3.8 | 0.046 |
| miR-181a-5p | 27.5±0.1 | −3.7 | 0.021 |
| miR-134-5p | 29.3±0.1 | −3.6 | 0.001 |
| miR-150-5p | 33.2±1.8 | −3.6 | 0.182 |
| miR-143-3p | 31.7±0.3 | −3.3 | 0.051 |
| miR-92b-3p | 26.2±0.5 | −3.3 | 0.026 |
| miR-382-5p | 27.1±0.1 | −3.2 | 0.014 |
| miR-133b-3p | 34.7±0.4 | −3.1 | 0.801 |
| miR-139-5p | 28.9±0.1 | −3.1 | 0.072 |
| miR-124-3p | 25.3±0.0 | −3.0 | 0.043 |
| let-7d-5p | 26.1±0.0 | −2.9 | 0.040 |
| miR-28-5p | 31.6±0.4 | −2.7 | 0.018 |
| miR-132-3p | 27.5±0.1 | −2.7 | 0.029 |
| miR-92a-3p | 25.9±0.1 | −2.6 | 0.036 |
| miR-203a-3p | 33.7±0.6 | −2.3 | 0.093 |
| miR-152-3p | 29.4±0.4 | −2.3 | 0.133 |
| miR-342-3p | 25.8±0.2 | −2.3 | 0.126 |
| miR-98-5p | 28.2±0.0 | −2.2 | 0.005 |
| miR-499-5p | 34.2±1.1 | −2.2 | 0.597 |
| miR-26b-5p | 26.8±0.1 | −2.2 | 0.060 |
| miR-126a-5p | 30.3±0.3 | −2.1 | 0.171 |
| miR-29b-3p | 34.1±0.6 | −2.0 | 0.997 |
| miR-191a-5p | 26.3±0.0 | −2.0 | 0.101 |
| miR-455-5p | 33.4±0.7 | −1.9 | 0.443 |
| miR-138-5p | 29.8±0.2 | −1.8 | 0.339 |
| miR-27a-3p | 30.5±0.4 | −1.8 | 0.079 |
| miR-409a-3p | 29.0±0.1 | −1.7 | 0.005 |
| miR-598-3p | 29.1±0.1 | −1.6 | 0.223 |
| miR-106b-5p | 26.8±0.1 | −1.6 | 0.166 |
| miR-489-3p | 33.0±0.1 | −1.6 | 0.425 |
| miR-181c-5p | 28.0±0.1 | −1.5 | 0.232 |
| miR-7a-5p | 27.0±0.1 | −1.5 | 0.062 |
| Up-regulated miRNAs |
Average CT (±SD) |
Fold changes vs control |
p-value |
| miR-140-5p | 30.7±0.1 | 3.8 | 0.040 |
| miR-376b-3p | 29.2±0.0 | 2.9 | 0.012 |
| miR-19a-3p | 27.9±0.0 | 2.8 | 0.001 |
| miR-20a-5p | 25.4±0.1 | 1.9 | 0.005 |
| miR-20b-5p | 26.5±0.0 | 1.9 | 0.001 |
| miR-19b-3p | 28.2±0.0 | 1.8 | 0.002 |
| miR-29c-3p | 29.5±0.2 | 1.6 | 0.003 |
| miR-338-3p | 31.2±0.3 | 1.6 | 0.675 |
| miR-125b-5p | 22.2±0.1 | 1.5 | 0.095 |
Data are mean ± SD. n=4/group.
Figure 3.
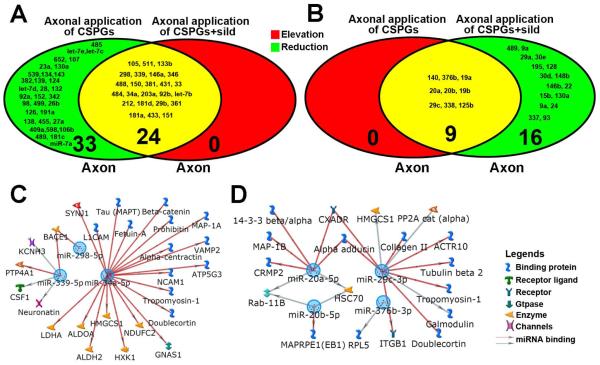
Profiles of miRNAs in RNAs extracted from the cell body and axonal compartments after axonal application of CSPGs. All miRNA data were derived from microarray analysis. Venn diagrams show reduced (A, green) and elevated (B, red) miRNAs in the axon (left) and elevated (A, red) and reduced (B, green) miRNAs in the cell body (right) after axonal application of CSPGs. Yellow color represents overlapped miRNAs. Numbers with small and large fonts in Venn diagrams represent the name of miRNAs and the total number of miRNAs, respectively.
Axonal application of sildenafil overcomes the inhibitory effect of CSPGs on axonal growth
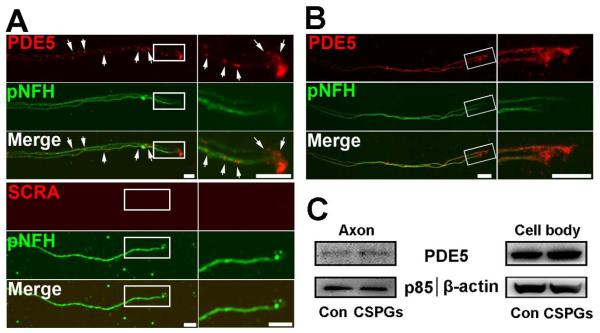
cGMP in neurons mediates neurite outgrowth (Ditlevsen et al., 2007; Murray et al., 2009; Zhao et al., 2009; Xia et al., 2013). PDE5 enzyme is highly specific for hydrolysis of cGMP (Corbin and Francis, 1999). We thus examined the effect of axonal application of sildenafil, a potent PDE5 inhibitor (Corbin and Francis, 1999), on axonal growth. Sildenafil was applied into the axonal chamber and axonal growth was measured on DIV5. Axonal application of sildenafil markedly promoted axonal outgrowth compared to the control group (Fig. 1B, D). In addition, sildenafil significantly elevated cGMP levels in distal axons (3.0±0.2 vs 1.5±0.1 pmol/μg in control, n=3/group. p<0.05), suggesting that increased cGMP levels regulate axonal growth. Indeed, axonal application of 8-Br-cGMP, a stable analog of cGMP, considerably increased axonal outgrowth (Fig. 1B, D). We then examined whether axons contain PDE5. FISH analysis showed the presence of PDE5 mRNA signals in axons with abundance in the growth cone (Fig. 4A). Double immunofluorescent analysis revealed that distal axons of the cortical neurons were PDE5 immunoreactive (Fig. 4B). Proteomic analysis showed the presence of PDE5 proteins in distal axons (Fig. 2B). Furthermore, the fluorescence polarization assay which detects PDE5 activity showed the presence of PDE5 activity in the axon (105.5±15.3 ΔmP, n=3) and the cell body (233.2±21.5 ΔmP, n=3) of neurons. Addition of 300 nM sildenafil into the axonal compartment decreased PDE5 activity by 59% (42.8±19.0 vs 105.5±15.3ΔmP, p<0.05, n=3). Collectively, these data indicate that distal axons of the cortical neurons contain mRNA and protein of PDE5 that is biologically active and that cGMP elevated by sildenafil likely via inhibition of PDE5 enhances axonal growth.
Figure 4.
The presence of mRNAs and proteins of PDE5 in distal axon of cortical neurons. Representative FISH in combination with immunofluorescent images (A) show the presence of punctate mRNA signals of PDE5 (A, PDE5, red, arrows) of pNFH+ axons with low (A, pNFH, green) and high (boxed area in A) magnifications. No signals were detected when FISH was performed with scramble probes (A, SCRA). Representative double immunofluorescent images (B, PDE5, red) show the presence of PDE5 immunoreactivity of pNFH+ axons with low (B, pNFH, green) and high (boxed area in B) magnifications (B, pNFH, green). Representative western blots (C) show the axonal application of CSPGs did not change the levels of PDE5 both in axons and their cell bodies. Scale bars in A and B =10 μm.
Next, we examined whether axonal application of sildenafil overcomes the inhibitory effect of CSPGs on axonal growth. Sildenafil and CSPGs were added into the axonal compartment. We found that axonal application of sildenafil significantly reversed the inhibitory effect of the CSPGs on axonal growth and on reduction of F-actin fluorescent intensity in the growth cone (Fig.1B-E). These data indicate that axonal application of sildenafil can block the local inhibitory effect of CSPGs on axonal growth.
To examine whether axonal application of CSPGs alters PED5 expression, levels of mRNA and protein of PDE5 were measured by quantitative RT-PCR and Western blot, respectively. Axonal application of CSPGs did not significantly alter axonal PDE5 mRNA (CT values, 32.3±0.5 vs 32.0±0.6 in scramble siRNA, n=3, p>0.05) and protein (Fig. 4C).
Sildenafil reverses CSPG-induced miRNA profile in axons
To examine whether axonal miRNAs are involved in the effect of axonal application of sildenafil on axonal growth, we assessed miRNA levels in axons by means of the miRNA PCR array. Axonal application of sildenafil elevated and reduced 3 and 46 of axonal miRNAs, respectively, compared to non-sildenafil treatment (Table 2). Moreover, the miRNA array analysis revealed that axonal application of sildenafil under CSPG condition induced elevation and reduction of 24 and 25 axonal miRNAs, respectively, compared to axonal application of CSPGs in the absence of sildenafil (Table 3). All 24 elevated miRNAs by axonal application of sildenafil under CSPG condition were originally reduced by axonal application of CSPGs (Fig. 5A), whereas 9 out 25 reduced-miRNAs by sildenafil were elevated by CSPGs (Fig. 5B). We speculated that these sildenafil-reversed miRNAs under axonal application of CSPGs may contribute to sildenafil-promoted axonal growth over CSPGs. Thus, these 33 (24 plus 9) miRNAs were ranked based on fold change in microarray analysis. Eleven out of the 33 miRNAs with a higher rank from the sildenafil-elevated or -reduced group were further analyzed by quantitative real-time RT-PCR (Table 4). Among these sildenafil-reversed miRNAs, we verified 3 sildenafil-elevated miRNAs, miR-34a-5p, miR-298-5p and miR-339-5p, and 4 sildenafil-reduced miRNAs, miR-20a-5p, miR-20b-5p, miR-29c-3p and miR-376b-3p (Table 4). Using the MetaCore® software, we then analyzed putative targets in the axon of these 7 miRNAs based on our axonal proteomic data including 894 proteins and on 308 mRNAs in axons published by Taylor et al (Taylor et al., 2009). We found that many genes and proteins in the axon targeted by these miRNA have critical roles in regulation of axonal growth, including beta site amyloid BACE1 (Farah et al., 2011), MAP1B (Bossel Ben-Moshe et al., 2012), CRMP2 (Kanzaki et al., 2011), and ITGB1 (Pasterkamp et al., 2003; Tan et al., 2011) (Fig. 5C, D). These results indicate that axonal application of sildenafil can reverse axonal miRNA profiles altered by axonal application of CSPGs.
Table 2.
Microarray data of axonal miRNAs altered by application of sildenafil.
| Down-regulated miRNAs |
Average CT (±SD) |
Fold changes vs control |
p-value |
|---|---|---|---|
| miR-23a-3p | 28.5±0.0 | −3.7 | 0.001 |
| miR-539-5p | 31.6±0.1 | −2.9 | 0.000 |
| miR-485-5p | 31.6±0.4 | −2.6 | 0.000 |
| miR-455-5p | 34.7±0.7 | −2.6 | 0.017 |
| miR-431 | 30.2±0.6 | −2.5 | 0.030 |
| miR-130a-3p | 28.3±0.0 | −2.5 | 0.013 |
| miR-151-3p | 28.9±0.2 | −2.3 | 0.006 |
| miR-34a-5p | 31.3±0.2 | −2.3 | 0.019 |
| miR-433-3p | 28.2±0.2 | −2.3 | 0.004 |
| miR-212-3p | 31.0±0.1 | −2.2 | 0.000 |
| miR-105 | 29.7±0.1 | −2.2 | 0.014 |
| miR-381-3p | 30.3±0.3 | −2.2 | 0.000 |
| miR-124-3p | 25.6±0.0 | −2.2 | 0.000 |
| let-7e-5p | 25.1±0.1 | −2.2 | 0.001 |
| miR-132-3p | 28.0±0.1 | −2.2 | 0.025 |
| miR-652-3p | 28.6±0.1 | −2.2 | 0.002 |
| miR-26b-5p | 27.6±0.1 | −2.1 | 0.003 |
| let-7c-5p | 23.3±0.1 | −2.1 | 0.000 |
| miR-298-5p | 27.4±0.2 | −2.0 | 0.030 |
| miR-98-5p | 28.9±0.0 | −2.0 | 0.009 |
| miR-382-5p | 27.3±0.1 | −2.0 | 0.005 |
| miR-181a-5p | 27.4±0.1 | −2.0 | 0.004 |
| miR-92a-3p | 26.3±0.1 | −2.0 | 0.000 |
| miR-33-5p | 34.6±0.3 | −1.9 | 0.009 |
| miR-152-3p | 30.0±0.4 | −1.9 | 0.028 |
| miR-511-5p | 30.3±0.5 | −1.9 | 0.020 |
| miR-409a-3p | 30.0±0.1 | −1.9 | 0.006 |
| let-7b-5p | 24.5±0.1 | −1.9 | 0.005 |
| miR-134-5p | 29.1±0.1 | −1.9 | 0.007 |
| miR-342-3p | 26.3±0.2 | −1.8 | 0.004 |
| miR-499-5p | 34.8±1.1 | −1.8 | 0.000 |
| miR-339-5p | 29.9±0.2 | −1.8 | 0.001 |
| miR-107-3p | 30.9±0.2 | −1.7 | 0.264 |
| miR-29a-3p | 32.2±0.1 | −1.7 | 0.005 |
| let-7d-5p | 26.2±0.0 | −1.7 | 0.004 |
| miR-28-5p | 31.8±0.4 | −1.7 | 0.006 |
| miR-181d-5p | 26.8±0.1 | −1.7 | 0.003 |
| miR-139-5p | 28.8±0.1 | −1.6 | 0.002 |
| miR-143-3p | 31.4±0.3 | −1.6 | 0.000 |
| miR-488-3p | 31.2±0.5 | −1.6 | 0.066 |
| miR-138-5p | 30.4±0.2 | −1.6 | 0.074 |
| miR-126a-5p | 30.7±0.3 | −1.6 | 0.006 |
| miR-27a-3p | 31.1±0.4 | −1.5 | 0.005 |
| miR-191a-5p | 26.7±0.0 | −1.5 | 0.035 |
| miR-146b-5p | 32.4±0.3 | −1.5 | 0.094 |
| miR-181c-5p | 28.8±0.1 | −1.5 | 0.026 |
| Up-regulated miRNAs |
Average CT (±SD) |
Fold changes vs control |
p-value |
| miR-376b-3p | 30.9±0.0 | 1.7 | 0.005 |
| miR-338-3p | 32.0±0.3 | 1.7 | 0.049 |
| miR-146a-5p | 29.2±0.1 | 2.3 | 0.000 |
Data are mean ± SD. n=4/group.
Table 3.
Microarray data of axonal miRNAs altered by axonal application of sildenafil and CSPGs.
| Down-regulated miRNAs |
Average CT (±SD) |
Fold changes vs CSPGs only |
p-value |
|---|---|---|---|
| miR-19a-3p | 29.4±0.1 | −3.0 | 0.001 |
| miR-29c-3p | 30.7±0.1 | −2.5 | 0.019 |
| miR-20b-5p | 27.7±0.0 | −2.4 | 0.001 |
| miR-489-3p | 34.2±0.2 | −2.4 | 0.013 |
| miR-9a-3p | 24.0±0.0 | −2.3 | 0.001 |
| miR-19b-3p | 29.3±0.0 | −2.3 | 0.001 |
| miR-20a-5p | 26.4±0.0 | −2.3 | 0.011 |
| miR-29a-3p | 31.4±0.2 | −2.2 | 0.024 |
| miR-376b-3p | 30.2±0.3 | −2.1 | 0.016 |
| miR-30e-5p | 26.8±0.0 | −2.1 | 0.001 |
| miR-195-5p | 27.9±0.2 | −2.1 | 0.009 |
| miR-128-3p | 24.9±0.1 | −2.0 | 0.000 |
| miR-125b-5p | 23.1±0.0 | −2.0 | 0.004 |
| miR-338-3p | 32.1±0.5 | −1.9 | 0.013 |
| miR-30d-5p | 26.8±0.0 | −1.8 | 0.000 |
| miR-30a-5p | 26.3±0.1 | −1.8 | 0.003 |
| miR-148b-3p | 29.5±0.2 | −1.7 | 0.020 |
| miR-146b-5p | 32.0±0.9 | −1.7 | 0.358 |
| miR-22-3p | 28.8±0.2 | −1.7 | 0.075 |
| miR-15b-5p | 27.9±0.1 | −1.7 | 0.072 |
| miR-140-5p | 31.3±0.2 | −1.6 | 0.042 |
| miR-130a-3p | 28.7±0.0 | −1.6 | 0.001 |
| miR-9a-5p | 21.8±0.1 | −1.5 | 0.011 |
| miR-24-3p | 27.5±0.1 | −1.5 | 0.004 |
| miR-337-3p | 33.1±0.5 | −1.5 | 0.322 |
| miR-93-5p | 27.9±0.2 | −1.5 | 0.063 |
| Up-regulated miRNAs |
Average CT (±SD) |
Fold changes vs CSPGs only |
p-value |
| miR-105 | 29.1±0.0 | 9.0 | 0.000 |
| miR-511-5p | 28.9±0.1 | 5.5 | 0.004 |
| miR-133b-3p | 32.5±0.2 | 4.5 | 0.027 |
| miR-298-5p | 26.8±0.0 | 4.3 | 0.002 |
| miR-339-5p | 29.4±0.3 | 3.7 | 0.042 |
| miR-146a-5p | 29.7±0.0 | 3.7 | 0.000 |
| miR-346 | 28.7±0.1 | 2.9 | 0.010 |
| miR-488-3p | 30.5±0.1 | 2.9 | 0.028 |
| miR-150-5p | 31.7±0.2 | 2.8 | 0.326 |
| miR-381-3p | 29.4±0.1 | 2.7 | 0.017 |
| miR-431 | 29.0±0.2 | 2.5 | 0.060 |
| miR-33-5p | 33.4±0.5 | 2.4 | 0.148 |
| miR-484 | 27.6±0.2 | 2.4 | 0.023 |
| miR-34a-5p | 31.0±0.1 | 2.4 | 0.011 |
| miR-203a-3p | 32.4±0.0 | 2.3 | 0.057 |
| miR-92b-3p | 24.9±0.0 | 2.3 | 0.043 |
| let-7b-5p | 24.5±0.0 | 2.1 | 0.004 |
| miR-212-3p | 30.3±0.4 | 2.1 | 0.125 |
| miR-181d-5p | 26.9±0.1 | 2.0 | 0.010 |
| miR-29b-3p | 33.0±0.4 | 2.0 | 0.202 |
| miR-361-5p | 30.1±0.1 | 1.9 | 0.063 |
| miR-181a-5p | 26.7±0.1 | 1.6 | 0.010 |
| miR-433-3p | 28.2±0.1 | 1.5 | 0.046 |
| miR-151-3p | 28.7±0.2 | 1.5 | 0.114 |
Data are mean ± SD. n=4/group.
Figure 5.
MiRNA profiles of RNA extracted from axon compartments and putative targets of axonal miRNAs. Venn diagrams of miRNA data derived from microarray analysis (A, B) show alteration of miRNA profiles in the distal axons after axonal application of CSPGs (left) and CSPGs+sildenafil (right). Colors in Venn diagrams represent that miRNAs were elevated (red), reduced (green), or overlapped (yellow). Numbers with small and large fonts in Venn diagrams represent the name of miRNAs and the total number of miRNAs, respectively. Metacore® analysis shows axonal gene networks that could be targeted by RT-PCR verified 3 elevated-(C) and 4 reduced-miRNAs (D) in the axon after axonal application of CSPGs and sildenafil.
Table 4.
Quantitative RT-PCR analysis with Taqman miRNA probes verified 11 miRNAs with a higher rank alteration in miRNA PCR array from the sildenafil-upregulated or -downregulated group.
| miRNA Name | miRNA PCR Array | qRT-PCR Taqman Assay | miRNA PCR Array | qRT-PCR Taqman Assay | ||||
|---|---|---|---|---|---|---|---|---|
| CSPGs vs control (n=4/group) |
p-value | CSPGs vs control (n=3/group) |
p-value | sildenafil +CSPGs vs CSPGs (n=4/group) |
p-value | sildenafil +CSPGs vs CSPGs (n=3/group) |
p-value | |
| miR-105 | −26.0 | 0.017 | −1.7 | 0.375 | 9.0 | 0.000 | −1.2 | 0.567 |
| miR-298-5p | −11.7 | 0.007 | −11.1 | 0.036 | 4.3 | 0.002 | 1.8 | 0.026 |
| miR-511-5p | −7.7 | 0.001 | − 1.1 | 0.919 | 5.5 | 0.004 | 1.4 | 0.512 |
| miR-339-5p | −8.8 | 0.055 | −5.9 | 0.026 | 3.7 | 0.042 | 1.5 | 0.047 |
| miR-34a-5p | −8.6 | 0.019 | −4.8 | 0.031 | 2.4 | 0.011 | 2.2 | 0.012 |
| miR-381-3p | −6.2 | 0.014 | Undetectable | N/A | 2.7 | 0.060 | Undetectable | N/A |
| miR-19a-3p | 2.8 | 0.001 | −2.9 | 0.004 | −3.0 | 0.001 | −1.5 | 0.111 |
| miR-376b-3p | 2.9 | 0.012 | 1.6 | 0.048 | −2.1 | 0.016 | −3.4 | 0.023 |
| miR-20b-5p | 1.9 | 0.001 | 5.9 | 0.012 | −2.4 | 0.001 | −12.5 | 0.013 |
| miR-20a-5p | 1.9 | 0.005 | 1.2 | 0.027 | −2.3 | 0.011 | −4.5 | 0.004 |
| miR-29c-3p | 1.6 | 0.003 | 2.6 | 0.011 | −2.5 | 0.019 | −14.3 | 0.001 |
Numbers are fold changes.
Axonal miR-29c induces sildenafil-enhanced axonal growth mitigating the CSPG inhibitory effect
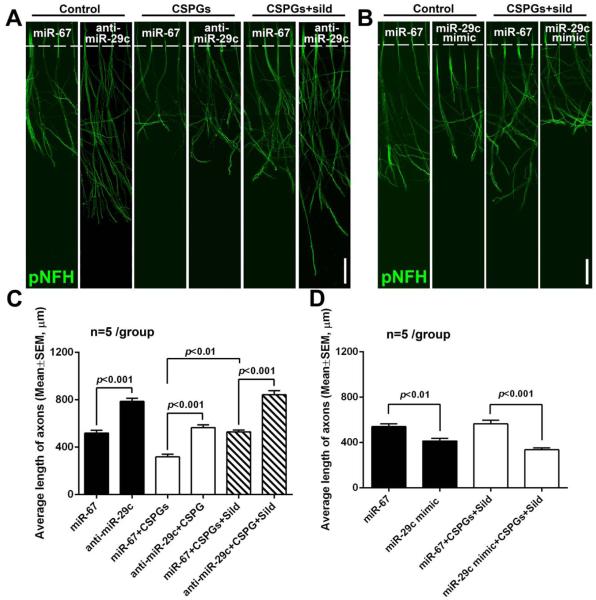
To test whether individual miRNAs in axons reversed by axonal application of sildenafil under axonal application of CSPGs contribute to axonal growth, we selected miR-29c, one of sildenafil-reduced miRNAs that were originally elevated by axonal application of CSPGs (Table 3). MetaCore data analysis (Fig. 5D) shows that miR-29c putatively targets ITGB1 that is known to regulate axonal function (Lei et al., 2012). A series of experiments were conducted to assay the cause-effect of axonal miR-29c on axonal growth. Initially, distal axons in the axonal compartment were locally transfected with the miR-29c hairpin inhibitor by a means of an N-TER Nanoparticle siRNA Transfection System. Quantitative RT-PCR analysis showed that endogenous miR-29c in the distal axons was reduced by the miR-29c hairpin inhibitor compared to miR-29c levels in axons transfected by the cel-miR-67 control inhibitor (0.3±0.1 vs 1 in cel-miR-67, p<0.05. n=3). On DIV5, reduction of axonal miR-29c enhanced axonal outgrowth of the cortical neurons cultured under the regular medium (Fig. 6A, C). Importantly, decrease of axonal miR-29c significantly mitigated the inhibitory effect of axonal application of CSPGs on axonal growth (Fig. 6A, C) and further enhanced axonal outgrowth induced by axonal application of sildenafil under axonal application of CSPGs compared to the cel-miR-67 control (Fig.6A, C). Axonal transfection of locked nucleic acid (LNA) anti-miR-29c oligonucleotides also blocked the effect of axonal application of CSPGs on axonal growth (521±18 vs 273±12 μm in CSPGs, p<0.05, n=3) and further elongated axons under axonal application of sildenafil and CSPGs (866±33 vs 561±20 μm in sildenafil+CSPGs, p<0.05, n=3). These data indicate the effect of reduced axonal miR-29c on axonal growth is specific. In contrast, axonal transfection of miR-29c mimics elevated miR-29c levels in the distal axons and inhibited the axonal outgrowth under the regular culture medium (Fig. 6B, D). Axonal transfection of miR-29c mimics also abolished axonal growth enhanced by axonal application of sildenafil when CSPGs were present in the axon (Fig. 6B, D). To examine whether other members of the miR-29 family, e.g. miR-29a and miR-29b, are involved in axonal growth regulated by axonal application of CSPGs and sildenafil, levels of miR-29a and miR-29b in distal axons were measured. Quantitative RT-PCR analysis of distal axons revealed that miR-29b was almost undetectable with a CT value of 40 and neither CSPGs (30.9±0.2, n=5) nor sildenafil (31.1±0.2, n=5) altered miR-29a levels compared to the controls (31.3±0.3, n=5). These data indicate that axonal miR-29c regulates axonal growth under basal, CSPG, and sildenafil conditions.
Figure 6.
The effect of miR-29c on axonal growth. Axons in the axonal compartment were transfected with miR-29c inhibitors (A, C) or mimics (B, D). Representative immunofluorescent images (A, B) and quantitative data (C, D) show axonal outgrowth (on DIV5) of distal axons locally transfected with the miR-29c hairpin inhibitor (A, C, anti-miR-29c) or miR-29c mimics (B, D) compared to axons transfected with cel-miR-67 control after axonal application of CSPG and CSPG+sildenafil (CSPGs+sild). Scale bar in A and B=100 μm.
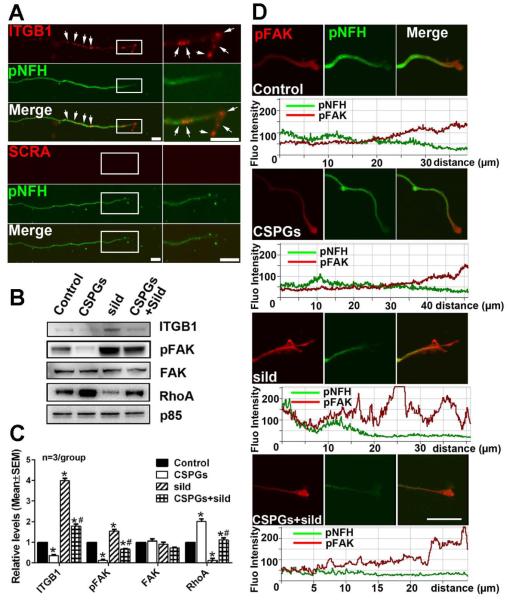
ITGB1 and its downstream focal adhesion kinase (FAK)/RhoA signals are involved in the CSPG inhibitory effect on axonal growth (Myers et al., 2011; Tan et al., 2011). ITGB1 mRNA signals were detected in pNFH positive axons (Fig. 7A). Axonal application of CSPGs substantially reduced protein levels of ITGB1 and phosphorylated FAK (pFAK), but increased RhoA (Fig. 7B, C). However, axonal application of sildenafil elevated levels of ITGB1 and pFAK and markedly reduced RhoA (Fig.7B, C). Furthermore, axonal application of sildenafil reversed the effect of CSPGs on axonal ITGB1 and pFAK and RhoA proteins (Fig. 7B, C). Sildenafil-elevated pFAK proteins in distal axons were further confirmed by double immunofluorescent staining, with the strongest pFAK signals in the ends of the distal axons (Fig. 7D), where integrin mediated axon outgrowth mainly takes place (Tan et al., 2011). These data indicate that axonal application of CSPGs and sildenafil locally modulates ITGB1/FAK/RhoA signals in the axon and that sildenafil suppresses the effect of CSPGs on these signals.
Figure 7.
The effect of axonal application of CSPGs and sildenafil on axonal ITGB1/pFAK/RhoA proteins. Representative FISH in combination with immunofluorescent images (A) show the presence of punctate mRNA signals of ITGB1 (A, red, arrows) in pNFH+ axons (A, green) with low (A, pNFH, green) and high (boxed area in A) magnifications. No signals were detected when FISH was performed with scramble probes (A, SCRA). Representative Western blots (B) and quantification data (C) show levels of ITGB1, pFAK, FAK and RhoA proteins in distal axons after axonal application of CSPGs, sildenafil (sild), or CSPGs+sildenafil (CSPGs+sild). Protein levels of p85 were used as internal controls for the distal axons. * and # p<0.05 vs control and CSPGs, respectively. Representative double immunofluorescent images and their relative fluorescent intensity along with the distal axons (D) show pFAK immunoreactivity (red) of pNFH+ (green) distal axons without any treatment (control) or after axonal application of CSPGs, sildenafil (sild), or CSPGs+sildenafil (CSPGs+sild). The distance (μm) under fluorescent intensity in panel D was measured from the distal axon tip. Scale bar in A=10 μm, in C=20 μm.
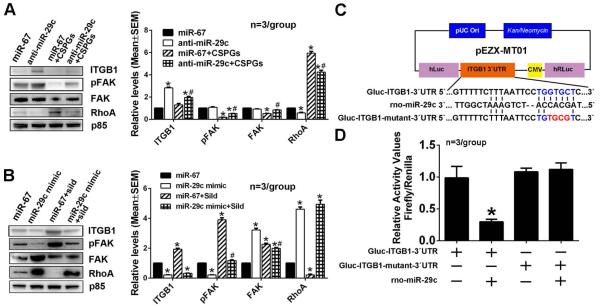
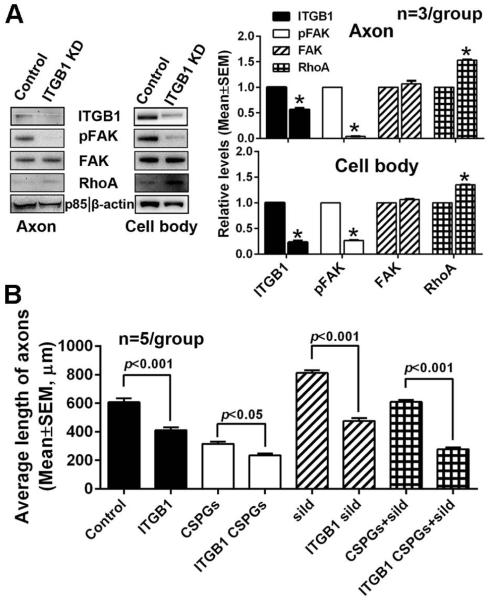
Based on the MetaCore and Targetscan analyses, ITGB1 is one of miR-29c putative target which have been validated by others (Wang et al., 2013). To examine whether axonal miR-29c is involved in regulation of ITGB1/FAK/RhoA signals, endogenous axonal miR-29c was reduced by axonal transfection of the miR-29c hairpin inhibitor. Axonal knockdown of miR-29c substantially increased ITGB1 protein levels, activated FAK, and decreased RhoA protein levels in axons compared to the cel-miR-67 control (Fig. 8A). In contrast, elevation of axonal miR-29c levels by axonal transfection of the miR-29c mimics reduced axonal ITGB1 protein levels, inactivated FAK and increased RhoA proteins in the axon (Fig. 8B). Moreover, reduction and elevation of axonal miR-29c levels abolished the effect of axonal application of CSPGs and sildenafil, respectively, on ITGB1/FAK/RhoA signals in axons (Fig. 8A, B). To examine whether miR-29c directly binds to 3’UTR of the ITGB1, we purchased a luciferase reporter plasmid containing 3’UTR of the ITGB1 gene encompassed the miR-29c binding site (Fig. 8C). Due to technical challenges to transfect primary cortical neurons with the plasmid, we failed to detect any luciferase activity. Thus, HEK293 cells were transfected with the plasmid and miR-29c mimics. The luminescence analysis revealed that co-transfection of the plasmid and miR-29c mimics reduced luminescence activity by ~70% compared to the cells transfected with the vector only or miR-29c mimics alone (Fig. 8D). However, mutation of miR-29c binding site in 3’UTR of ITGB1 abolished reduction of luminescence activity by miR-29c mimics (Fig. 8C, D). These data indicate that miR-29c regulates expression of ITGB1 by directly binding to 3’UTR of ITGB1gene. To examine the direct effect of ITGB1 on axonal growth, neurons were transfected with siRNA-ITGB1 and the transfected neurons were then cultured in a microfluidic culture device (Zhang et al., 2013). Western blot analysis revealed that neuronal transfection of siRNA-ITGB1 reduced ITGB1 levels in the distal axon and the cell body by approximately 75%, which was associated with a reduction of pFAK and an increase of RhoA proteins compared to neurons transfected with the scramble-siRNA control (Fig. 9A). Knockdown of ITGB1 in the neurons significantly decreased axonal growth in neurons cultured under the regular medium and further blocked axonal elongation with axonal application of CSPGs (Fig. 9B). Axonal application of sildenafil alone and with axonal application of CSPGs did not enhance axonal outgrowth in neurons transfected with siRNA-ITGB1 (Fig.9B). Knockdown of ITGB1 in neurons did not significantly affect the number of apoptotic neurons measured by TUNEL positive cells (9±2%, total 1,763 cells counted within 5 individual chambers) compared to the siRNA control (11±4%, total 1,805 cells counted within 5 individual chambers, p>0.05), suggesting that neuronal survival does not contribute to the effect of ITGB1 knock-down on suppression of axonal growth. Taken together, we conclude that miR-29c targets ITGB1 that activates FAK and inactivates RhoA signals in axons, which in-concert contribute to the effect on axonal growth induced by axonal application of CSPGs and sildenafil.
Figure 8.
The effect of miR-29c on on ITGB1/pFAK/RhoA proteins after axonal application of CSPGs or sildenafil in miR-29c reducted axons or miR-29c elevated axons, respectively. Representative Western blots and quantification data show levels of ITGB1, pFAK, FAK and RhoA proteins in distal axons transfected with the miR-29c hairpin inhibitor (A) and miR-29 mimics (B) after the axonal application of CSPGs and sildenafil, respectively. Protein levels of p85 were used as internal controls for the distal axons. * p<0.05 vs control; # p<0.05 vs anti-miR-29c (A) and miR-29c mimic (B), respectively. Panel C shows a schematic of the dual-luciferase reporter vector cloned with segments of the ITGB1 gene 3’UTR. Sequences with blue and red colors are the putative and mutant binding sites, respectively. Panel D shows luciferase activity data from HEK293 cells.* p<0.05 vs Gluc-ITGB1-3’UTR alone transfection group.
Figure 9.
The effect of ITGB1 knockdown on proteins of ITGB1/pFAK/RhoA and the axonal growth. Representative Western blots and quantification data (A) show levels of ITGB1, pFAK, FAK and RhoA proteins in distal axons (A, axon) and cell bodies (A, cell body) of cortical neurons transfected with siRNA against ITGB1. *, p<0.05 vs control. The quantitative results (B) show the effect of ITGB1 knockdown on distal axonal growth after axonal application of CSPGs, sildenafil (sild), or CSPGs+sildenafil (CSPGs+sild). Protein levels of p85 and β-actin were used as internal controls for the distal axons and cell bodies, respectively.
Discussion
Although previous studies have demonstrated the inhibitory effect of CSPGs on axonal growth, molecular mechanisms underlying the inhibitory effect are not well understood. By analyzing axonal miRNA content, the present study shows that axonal application of CSPGs substantially changed axonal miRNA profiles, which were associated with significant inhibition of axonal growth. Axonal application of the PDE5 inhibitor, sildenafil, significantly overcame the CSPG inhibitor effect on axonal outgrowth and reversed the majority of axonal miRNAs altered by CSPGs. Thus, we identified an axonal miRNA signature in the regulation of axonal growth by axonal application of CSPGs and sildenafil.
Extracellular signals regulate mRNAs and proteins in dendrites and axons of neurons (Steward and Schuman, 2003; Willis et al., 2007; Lin and Holt, 2008; Cohen et al., 2011; Colak et al., 2013). Axonal injury in the CNS leads to the generation of abundant CSPGs that inhibit axonal regeneration (Rudge and Silver, 1990; Hartmann and Maurer, 2001; Monnier et al., 2003; Yiu and He, 2006). Activation of cGMP promotes axonal growth (Murray et al., 2009; Zhao et al., 2009; Xia et al., 2013). However, whether extracellular matrix proteins modulate axonal miRNAs remains unknown. The axon contains numerous miRNAs and proteins that regulate miRNA biogenesis (Satkauskas and Bagnard, 2007; Natera-Naranjo et al., 2010; Deglincerti and Jaffrey, 2012; Kaplan et al., 2013; Wu and Murashov, 2013; Zhang et al., 2013; Hancock et al., 2014; Ji and Jaffrey, 2014). The present study demonstrated that local extracellular signals can alter axonal miRNA content. Alteration of axonal miRNAs in response to axonal applications of CSPGs and sildenafil is specific, because soma and axonal miRNA profiles exhibited distinct profiles in response to axonal application of CSPGs and sildenafil. The presence of the PDE5 enzyme in distal axons suggests that elevation of axonal cGMP by sildenafil likely occurs through inhibition of the PDE5 enzyme. Thus, our data indicate that axonal miRNA content can be altered by local extracellular signals, CSPGs and cGMP.
Our data indicate that endogenous axonal cGMP levels regulate alteration of axonal miRNAs. However, axonal application of CSPGs did not alter cGMP levels, suggesting that cGMP may not be involved in CSPGs-altered miRNAs. We speculate that downregulation of integrin signals by CSPGs may regulate miRNA expression, whereas elevated-cGMP by sildenafil upregulates integrins, leading to reversal of miRNA profiles induced by CSPGs, although it remains to be investigated how CSPGs and cGMP reciprocally regulate axonal miRNAs.
Previous studies suggest that the activation of RhoA in the axon by CSPGs contributes to impairment of integrin function, which is a general mechanism for inhibition of axon growth (Monnier et al., 2003; Tan et al., 2011; Walker et al., 2012). Studies in lung cancer cells showed that miR-29c acts as a tumor metastasis suppressor by targeting ITGB1 and matrix metalloproteinase 2 (Wang et al., 2013). The present study found that miR-29c, one of axonal miRNAs altered by axonal application of CSPGs and sildenafil, mediated the effect of CSPGs and sildenafil on axonal growth by regulating axonal ITGB1/FAK/RhoA signals. Local application of CSPGs in axons elevated miR-29c, which was associated with reduction of ITGB1, inactivation of FAK, and elevation of RhoA. In contrast, elevation of axonal cGMP by sildenafil suppressed CSPG-induced miR-29c and led to activation and inactivation of ITGB 1/FAK and RhoA, respectively. Loss-of- and gain-of-function experiments demonstrated that axonal miR-29c regulates local ITGB1/Akt and RhoA signaling. Thus, our data provide new insights into molecular mechanisms underscoring the inhibitory action of CSPGs on axonal growth. Future studies are required to determine whether axonal miRNAs-mediated activation of integrin signaling promote axonal regeneration after axonal injury.
Neurons transport mRNAs and proteins from their cell bodies to axons (Sabry et al., 1995; Willis et al., 2007; Jung et al., 2012) . Studies by others using the same microfluidic chambers have demonstrated that proteins such as tau and α-synuclein can anterogradely and retrogradely transport in the axon (Freundt et al., 2012). The present study found that axonal application of CSPGs and sildenafil altered miRNA profiles in the cell body, suggesting that axonal miRNAs may transport retrogradely upon axonal stimulation.
The present study only examined 84 miRNAs in the axon. It is likely that axonal miRNAs other than miR-29c can also locally regulate integrin signals. Studies have demonstrated that the elevation of miR-338 in the axon leads to reduction of local ATP levels and elevation of reactive oxygen species by targeting endogenous CoxIV and ATP5G1genes (Aschrafi et al., 2008). Thus, signals that regulate metabolism and microtubule proteins targeted by miRNAs in the axon could also mediate the effect of CSPGs and sildenafil on axonal growth. Much work is required to understand how axonal miRNAs locally regulate their target genes.
Materials and Methods
Primary cortical neurons cultured in a microfluidic culture platform
All experimental procedures were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Henry Ford Hospital. Cortical neurons were harvested from embryonic day-18 Wistar rats (Charles River Laboratories, Wilmington, MA). Cultures were prepared according to published studies with some modifications (Taylor et al., 2005; Zhang et al., 2013). Briefly, embryos were removed, and the cerebral cortex dissected, stripped of meninges, and dissociated by a combination of Ca2+ and Mg2+ free Hanks balance salt solution containing 0.125% trypsin digestion, then mechanically triturated. The triturated cells were passed through a 40 μm cell strainer to reach a cell density at 3×107cells/ml.
A microfluidic culture platform (Standard Neuron Device, Cat# SND450, Xona Microfluidics, Temecula, CA) was employed. The microfluidic device used in the present study contains 450 μm long microgrooves that connect cell body and axonal compartments and permit only distal axons to grow into the axonal compartment (Taylor et al., 2005). Briefly, cleaned, sterilized, and dried chambers were affixed to poly-D-lysine (Sigma-Aldrich) -coated dishes (35mm, Corning). The cortical neurons were plated at a density of 6×105cells/device in growth medium composed by neurobasal medium (Invitrogen), 2% B-27 supplement (Invitrogen), 1% GlutaMax (Invitrogen), and 1% antibiotic-antimycotic (Invitrogen). On DIV3, one-half of the medium was replaced with culture medium containing 20 μM 5-fluorodeoxyuridine. Thereafter, the growth media were replaced every other day.
Experimental groups
All the compounds used in the present study were added to the axonal compartment only on DIV2, when axons were starting to grow into the axonal compartment, and then daily during the experimental period. To examine the effect of CSPGs on axonal growth, CSPGs (2 μg/ml, Millipore, Cat# CC117) were applied to the axonal compartment. To examine the effect of sildenafil on axonal growth, sildenafil citrate (300 nM, Sigma) was added into the axons cultured under normal and CSPGs conditions. To examine the effect of cGMP on axons, a stable analog of cGMP, 8-Br-cGMP (20μM, Sigma) was added into axons cultured under normal and CSPGs conditions.
Isolation of total RNA
To analyze miRNAs in the axon or cell body, axons growing in the axonal compartment or neurons in the cell body compartment of microfluidic culture devices were lysed in Qiazol reagent, respectively. Then, total RNA was isolated using the miRNeasy Mini kit (Qiagen, Valencia, CA, USA). Briefly, medium in the axonal compartment was removed and washed with pre-warmed PBS, while medium in the cell body compartment was kept to minimize the contamination from the cell body. Then 20 μl of Qiazol reagent was applied to the axonal compartment for no more than 1 minute, and was totally collected. Then medium in the cell body compartment was removed and extracts were collected using the same procedures described above for harvesting axonal RNAs. Axonal or cell body miRNAs were extracted followed with a miRNeasy Mini kit according to the manufactor’s protocol.
miRNA PCR Array and Real-Time Reverse Transcriptase-PCR
To analyze miRNA profiles in axons or in cell body, a miRNA PCR array kit including 84 miRNAs (MIRN-107ZE-1, Qiagen) was employed. Briefly, the total RNAs at 1.5 to 2.0 μg were harvested from the axonal or cell body compartment of 20 microfluidic culture devices per experimental group. The reverse transcription was then performed using miScript HiSpec Buffer (miScript II RT Kit, Qiagen). cDNA prepared in a RT action was served as the template for the real-time PCR analysis using the miRNA PCR array kit (MIRN-107ZE-1, Qiagen) and the miScript SYBR Green kit on an ABI ViiATM 7 PCR instrument (Applied Biosystems). The data analysis was performed using an online data analysis software (http://pcrdataanalysis.sabiosciences.com/mirna). To verify miRNAs detected by the miRNA array, levels of selective miRNAs were measured by quantitative RT-PCR. Briefly, miRNAs were reversely transcribed with the miRNA Reverse Transcription kit (Applied Biosystems) and amplified with the TaqMan miRNA assay (Applied Biosystems).
Axonal transfection with miRNA hairpin inhibitors, locked nucleic acid (LNA) inhibitors and mimics
N-TER Nanoparticle siRNA Transfection System (Sigma-Aldrich, St. Louis, MO.) was employed to transfect axons with inhibitors and mimics. Briefly, on DIV3 when all of the microgrooves were fully filled with axons, miRNA hairpin inhibitors or LNA inhibitors against rat miR-29c (final concentration 20nM, mature sequence: UAGCACCAUUUGAAAUCGGUUA of hairpin inhibitor and ACCGATTTCAAATGGTGCT of LNA inhibitor) or miR-29c mimics along with nanoparticle packed in N-TER peptide in growth medium was applied to the axonal compartment for 72h. Caenorhabditis elegans miR-67 (cel-miR-67, mature sequence: UCACAACCUCCUAGAAAGAGUAGA), which lacks homolog in mammals, was used as a negative control (Dharmacon, Lafayette, CO, USA).
Knockdown of ITGB1 in cortical neurons
A pre-designed siRNA against ITGB1 gene (Life technologies, Silencer® Select Pre-Designed siRNA, siRNA ID 200619) was purchased to endogenously knockdown ITGB1 in cortical neurons. A Silencer® Select Negative Control (Life technologies, 4390843, USA) was used as the negative control. Briefly, the 200 pmol siRNA was mixed with 1×106 cortical neurons in 100 μl of Nucleofector solution (Mirus, USA), and cell-siRNA mixtures were transferred into a cuvette and electroporated using a program O-03 by Nucleofector Device (Lonza, USA). The transfected neurons were then seeded into microfluidic culture devices.
Dual-luciferase assay in HEK293 cells
A segment of the rat 3`UTR of ITGB1 gene encompassing the miR-29c binding site (1116 bases, Gluc-ITGB1-3‘UTR, RmiT049551-MT01, Genecopoeia, MD) was cloned into a pEZX-MT01 vector with Firefly/Renilla duo-Luciferase reporter driven by a CMV promoter (Genecopoeia, MD, Fig. 8C). Point mutation of 3‘UTR of ITGB1 gene for miR-29c, TGGTGCT to TGTGCGT, was made and confirmed by sequencing (Mutant ITGB1 clone, Gluc-ITGB1-mutant-3‘UTR, Genecopoeia, MD, Fig. 8C). The dual luciferase reporter system allows the normalization of luminescence to minimize variations of transfection efficiencies and cell viability. In our preliminary experiments, primary cortical neurons were transfected with these vectors. However, we failed to detect any luminescence signals in the neurons. Therefore, HEK293 cells were used. Briefly, each vector was transfected in HEK293 cells by lipofectamine (Life technologies) at a concentration of 2ug/106 cells. To assess the mRNA-miRNA interaction, each vector was co-transfected with miR-29c mimics (200pM/106 cells, Dharmacon, Lafayette, CO, USA) in HEK293 cells. Twenty-four hours later, the cells were lysised and treated with a Dual-luciferase assay kit (LucPair™ Duo-Luciferase Assay Kit, Genecopoeia, MD). Luciferase activity was detected using a multimode microplate reader (PerkinElmer/Fusion). The activity ratio of firefly to Renillain was normalized to 1 in the cells with single transfection of Gluc-ITGB1-3‘UTR.
Fluorescence in situ hybridization (FISH) and Immunocytochemistry
FISH was performed to detect mRNAs of PDE5 and ITGB1 in axons (Zhang et al., 2013) (Pena et al., 2009). Digoxigenin (DIG)-labeled (5’ and 3’ ends) LNA probes specifically against rat PDE5 and ITGB1 mRNA, and scramble probes (Exiqon) were used for hybridization according to a published protocol with some modifications (Pena et al., 2009; Dajas-Bailador et al., 2012). Briefly, cultured axons in microfluidic devices were fixed with 4% paraformaldehyde. LNA probes at 0.25 μM in hybridization buffer (50% Formamide, 5X SSC, 5X Denhardt’s solution, 500 μg/ml salmon sperm DNA, 2% Blocking reagent (Roche), 0.1% Chaps, 0.5% Tween-20) were applied to fixed axons for overnight in a humid chamber at 54°C for PDE5, 53°C for ITGB1, and 57°C for scramble. On the following day, endogenous peroxidase was inactivated by incubation with 3% hydrogen peroxide for 30min. Samples were further blocked with blocking solution (Roche) and were incubated with anti-DIG-HRP solution (Roche). Signals were detected using Fluorophore Amplification Reagent Working Solution (TSA PLUS Fluorescence Kits, PekinElmer). After in situ hybridization, immunofluorescent staining was performed with a monoclonal antibody against phosphorylated high molecular weight neurofilament (pNFH, SMI31, 1:500 Covance, Emeryville, CA).
Immunofluorescent staining was performed as previously described (Liu et al., 2009). Briefly, after washing with PBS twice and fixation with 4% formaldehyde, neurons in the microfluidic devices were incubated with the primary antibodies for one or two nights at 4°C, and then with Cy3 or FITC conjugated secondary antibodies for 2 hours at room temperature. Nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI, 1:10,000, Vector Laboratories, Burlingame, CA). The following primary antibodies were used: a monoclonal antibody against pNFH (SMI31, 1:500 Covance); a polyclonal antibody against phosphorylated focal adhesion kinase (pFAK, 1:200, Cell signaling) and a monoclonal antibody against Ago2 (eIF2C2, Abcam, ab57113). TUNEL staining was performed with ApopTag kit (Chemicon) according to the manufacturer’s instructions (Buller et al., 2010). For F-actin staining, Alexa Fluor® 568 Phalloidin kit (Invitrogen) was employed to stain F-actin in the growth cone according to the manufacturer’s instructions. The intensity of F-actin was converted into spectral images and the spectral signal was quantified using ImageJ (NIH).
Image acquisition and quantification
Distal axonal length was imaged under a 10x objective on DIV5. Five images per compartment were acquired, which encompass the majority of the compartment. The lengths of the 15 longest axons in each compartment were measured from the entrance of axonal compartment to their distal ends by tracing individual axon imageJ according to the published procedures (Meijering et al., 2004). The distal axons of the cortical neurons stained with immunocytochemistry were imaged under a 40x objective using a laser-scanning confocal microscope (Zeiss LSM 510 NLO, Carl Zeiss, Germany) (Zhang et al., 2009). Data of axonal length were normalized to the control group and are presented as fold change. Intensity of immunoreactive signals such as F-actin in growth cone area of distal axons was analyzed using the ImageJ, as previously described (Zhang et al., 2013).
To further verify whether the measurements of the 15 longest axons in the axonal compartment reflect overall axonal growth, all pNFH positive axons in the axonal compartment were imaged under a 10X objective (Axiophot2, ZEISS). Twelve images were acquired that encompass the entire axonal compartment, and they were tiled to form a single image. The number of fluorescent pixels within the compartment was measured with imageJ, divided by total pixels in the compartment and multiplied by 100. Axonal growth was calculated as percentage of pNFH fluorescent pixels within the axonal compartment area and was normalized to the control group. Data are presented as fold change. We found that data obtained from each experimental group using this method were comparable to data from the measurement of the longest length of the axon (Figure 1D). Thus, the measurement of the 15 longest lengths of the axons was performed for subsequent experiments.
Western blotting assay of axonal proteins
To measure the protein levels in the distal axons, proteins were extracted from distal axons cultured in the axonal compartment. Briefly, 10 μl of lysis buffer (RIPA with 1% protease inhibitor cocktails plus 1% phosphatase inhibitor cocktail, Sigma-Aldrich) was applied to the axonal compartment. Medium in the somal compartment was kept intact during axonal lysis to prevent lysis buffer in the axonal compartment flowing back to the cell body compartment, which minimizes cell body compartment contamination. During the entire period, the chamber was placed on the ice horizontally. Axonal lysis from 4 to 6 chambers was pooled for one Western blot. Protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce Biotechnology, Rockford, IL).
Western blots were performed according to published methods (Zhang et al., 2013). Briefly, equal amounts of total proteins (25~35μg) for each sample were loaded on 10% SDS-polyacrylamide gels. After electrophoresis, proteins were transferred to polyvinylidene fluoride membranes, and the blots were subsequently probed with the following primary antibodies: rabbit polyclonal anti-ITGB1 (1:1000, Abcam), rabbit polyclonal anti-pFAK (Tyr 397, 1:1000, Cell signaling), mouse monoclonal anti-FAK (1:1000, Santa Cruz), rabbit polyclonal anti-RhoA (1:1000, Santa Cruz), rabbit polyclonal anti-PI3 kinase p85 (1:1000, Millipore), mouse monoclonal anti β-actin (1:10000,Abcam). For detection, horseradish peroxidase-conjugated secondary antibodies were used (1:2000), and followed by enhanced chemiluminescence development (Pierce Biotechnology). Protein levels of β-actin and PI3K subunit p85 were used as the internal controls for somata and axons, respectively (Andreassi et al., 2010). Western blots were performed in at least 3 times in individual experiments.
Proteomics
Axonal and cell body proteins were measured using a Q-Exactive mass spectrometer (Thermo Fisher Scientific) (Lee et al., 2011). Briefly, the proteins from the axonal and cell body compartments were extracted as described above in the Western blot section. Briefly, protein extracts (35μg) from individual samples were loaded per lane on SDS-PAGE with a 4-12% gradient. After that, each lane was cut into 10 equal-sized pieces. Proteins in each gel piece were digested and peptides within the gel were extracted. Then, peptides were separated by reverse phase chromatography and analyzed with a Q-Exactive mass spectrometer (Thermo Fisher Scientific). Abundant species were fragmented in higher energy collision induced dissociation mode. Data analysis was performed using Proteome Discoverer 1.4 (Thermo) which incorporated the Mascot (Matrix Science) algorithm. The latest protein UniProtKB database was used for identification of rat protein sequences. Scaffold software (Proteome Software) was used to validate and compare MS/MS based peptide and protein identifications. Protein identifications were accepted if they could be established at greater than 99.0% probability with minimum 2 unique peptides detected. Proteins are presented by their official encoding gene names (Entrez Gene; NCBI). Spectral counts were calculated by normalizing the number of assigned spectra for each protein to the total number of assigned spectra for each sample.
ELISA analysis of axonal cGMP levels
The axonal protein lysates from microfluidic culture devices were collected with the same procedures for isolating axonal proteins for Western blot analysis. The concentration of axonal protein was determined and equaled before performing the cGMP Parameter Assay Kit (R&D systems), according to the manufacturer’s instruction. The cGMP levels in axons are presented as pmol cGMP per μg axonal proteins.
PDE5 activity
PDE5 activity was analyzed by Transcreener AMP2/GMP2 fluorescence polarization (FP) assay (Bell Brook) following manufacturer’s instruction. Enzyme activity is signaled by a decrease in FP as the bound tracer is displaced from the monoclonal AMP2/GMP2 antibody. Briefly, proteins extracted from axonal and cell body compartments were quantified by a bicinchoninic acid kit. Fluorescence polarization was measured in milliP (mP) at 590 nm excitation and 675 nm emission on a SpectraMax M5 microplate reader (Molecular Devices). For determination of optimal experimental settings for FP assay, the maximum polarization window (>175mP) was determined by minus the mP values of low polarization mixture (free tracer) from high polarization mixture (tracer plus antibody). A pilot experiment was performed on a gradient protein concentration from the axonal and cell body compartments, in order to verify the change in polarization of an optimal concentration that was located between EC50 to EC80. ATP (200μM) and cGMP (50 μM) were mixed with 1μg of axonal or cell body proteins in the presence or absence of sildenafil (300nM) for 20 min. The AMP/GMP detection mixture (1M Tris, 800nM AMP/GMP AlexaFluor 633 tracer, AMP2/GMP2 Antibody) was then added into the reaction component at room temperature for 90 min. The PDE5 activity is presented as a gap value, ΔmP, calculated by mP values from the control reaction and from axonal or cell body reactions.
Statistical analysis
All statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS, version 11.0; SPSS Inc, Chicago, IL). One-way ANOVA with post hoc Bonferroni tests was used when comparing more than two groups. Student’s t test was used when comparing two groups. Values presented in this study are expressed as mean ± standard error. A p-value < 0.05 was considered to be significant.
Table 5.
Quantitative RT-PCR data of selective axonal miRNAs altered by axonal application of CSPGs or CSPGs+sildenafil in neurons transfected by Ago2-siRNA or scramble probes.
| miRNA Name |
Scramble | Ago2 knockdown | Scramble | Ago2 knockdown | ||||
|---|---|---|---|---|---|---|---|---|
| CSPGs vs Control (n=4/group) |
p-value | CSPGs vs Control (n=4/group) |
p-value | sildenafil + CSPGs vs CSPGs (n=3/group) |
p-value | sildenafil + CSPGs vs CSPGs (n=3/group) |
p-value | |
| miR-298-5p | −11.1 | 0.036 | −1.2 | 0.588 | 1.8 | 0.026 | 1.3 | 0.336 |
| miR-339-5p | −5.9 | 0.026 | −1.4 | 0.506 | 1.5 | 0.272 | 3.9 | 0.155 |
| miR-34a | −4.8 | 0.031 | −1.2 | 0.412 | 2.2 | 0.012 | 2.0 | 0.051 |
| miR-376b-3p | 1.5 | 0.121 | 2.1 | 0.048 | −3.3 | 0.023 | 1.0 | 0.642 |
| miR-20b-5p | 5.9 | 0.012 | 1.8 | 0.045 | −12.5 | 0.013 | −2.2 | 0.100 |
| miR-20a-5p | 1.2 | 0.213 | 3.2 | 0.362 | −4.5 | 0.004 | 1.4 | 0.004 |
| miR-29c-3p | 2.6 | 0.011 | −1.4 | 0.265 | −14.3 | 0.001 | 1.7 | 0.136 |
Numbers are fold changes.
Acknowledgement
We thank Cynthia Roberts for technical assistance. This work was supported by the National Institutes of Health (RO1 NS75156 and RO1 AG037506). We thank Dr. Paul M. Stemmer at the Wayne State University Proteomic Core for analysis of proteomic data. The Wayne State University Proteomic Core is supported by the National Institutes of Health (P30 ES06639, P30 CA22453 and S10 OD 010700).
Abbreviations List
- Ago2
argonaute 2
- CSPGs
chondroitin sulfate proteoglycans
- cGMP
cyclic guanosine monophosphate
- CNS
central nervous system
- DIV
days in vitro
- FISH
Fluorescence in situ hybridization
- FP
fluorescence polarization
- FAK
focal adhesion kinase
- GFAP
glial fibrillary acidic protein
- ITGB1
integrin β1
- LNA
locked nucleic acid
- NFH
neurofilament H
- PDE5
phosphodiesterase type 5
Footnotes
Conflict of Interest: The authors declare no competing financial interests.
References
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends in cell biology. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossel Ben-Moshe N, Avraham R, Kedmi M, Zeisel A, Yitzhaky A, Yarden Y, Domany E. Context-specific microRNA analysis: identification of functional microRNAs and their mRNA targets. Nucleic Acids Res. 2012;40:10614–10627. doi: 10.1093/nar/gks841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- Buller B, Liu X, Wang X, Zhang RL, Zhang L, Hozeska-Solgot A, Chopp M, Zhang ZG. MicroRNA-21 protects neurons from ischemic death. FEBS J. 2010;277:4299–4307. doi: 10.1111/j.1742-4658.2010.07818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Bas Orth C, Kim HJ, Jeon NL, Jaffrey SR. Neurotrophin-mediated dendrite-to-nucleus signaling revealed by microfluidic compartmentalization of dendrites. Proc Natl Acad Sci U S A. 2011;108:11246–11251. doi: 10.1073/pnas.1012401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D, Ji SJ, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153:1252–1265. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin JD, Francis SH. Cyclic GMP phosphodiesterase-5: target of sildenafil. J Biol Chem. 1999;274:13729–13732. doi: 10.1074/jbc.274.20.13729. [DOI] [PubMed] [Google Scholar]
- Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nature Neuroscience. 2012;15:697–699. doi: 10.1038/nn.3082. [DOI] [PubMed] [Google Scholar]
- Deglincerti A, Jaffrey SR. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2012;2:120079. doi: 10.1098/rsob.120079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlevsen DK, Kohler LB, Berezin V, Bock E. Cyclic guanosine monophosphate signalling pathway plays a role in neural cell adhesion molecule-mediated neurite outgrowth and survival. J Neurosci Res. 2007;85:703–711. doi: 10.1002/jnr.21175. [DOI] [PubMed] [Google Scholar]
- Farah MH, Pan BH, Hoffman PN, Ferraris D, Tsukamoto T, Nguyen T, Wong PC, Price DL, Slusher BS, Griffin JW. Reduced BACE1 activity enhances clearance of myelin debris and regeneration of axons in the injured peripheral nervous system. J Neurosci. 2011;31:5744–5754. doi: 10.1523/JNEUROSCI.6810-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt EC, Maynard N, Clancy EK, Roy S, Bousset L, Sourigues Y, Covert M, Melki R, Kirkegaard K, Brahic M. Neuron-to-neuron transmission of alpha-synuclein fibrils through axonal transport. Ann Neurol. 2012;72:517–524. doi: 10.1002/ana.23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C, Merianda TT, Lee SJ, Yoo S, Twiss JL. Molecular determinants of the axonal mRNA transcriptome. Dev Neurobiol. 2014;74:218–232. doi: 10.1002/dneu.22123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock ML, Preitner N, Quan J, Flanagan JG. MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J Neurosci. 2014;34:66–78. doi: 10.1523/JNEUROSCI.3371-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Maurer P. Proteoglycans in the nervous system--the quest for functional roles in vivo. Matrix Biol. 2001;20:23–35. doi: 10.1016/s0945-053x(00)00137-2. [DOI] [PubMed] [Google Scholar]
- Iyer AN, Bellon A, Baudet ML. microRNAs in axon guidance. Front Cell Neurosci. 2014;8:78. doi: 10.3389/fncel.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji SJ, Jaffrey SR. Axonal transcription factors: novel regulators of growth cone-to-nucleus signaling. Dev Neurobiol. 2014;74:245–258. doi: 10.1002/dneu.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki H, Ito S, Hanafusa H, Jitsumori Y, Tamaru S, Shimizu K, Ouchida M. Identification of direct targets for the miR-17-92 cluster by proteomic analysis. Proteomics. 2011;11:3531–3539. doi: 10.1002/pmic.201000501. [DOI] [PubMed] [Google Scholar]
- Kaplan BB, Kar AN, Gioio AE, Aschrafi A. MicroRNAs in the axon and presynaptic nerve terminal. Front Cell Neurosci. 2013;7:126. doi: 10.3389/fncel.2013.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar AN, MacGibeny MA, Gervasi NM, Gioio AE, Kaplan BB. Intra-axonal synthesis of eukaryotic translation initiation factors regulates local protein synthesis and axon growth in rat sympathetic neurons. J Neurosci. 2013;33:7165–7174. doi: 10.1523/JNEUROSCI.2040-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DC, Hassan SS, Romero R, Tarca AL, Bhatti G, Gervasi MT, Caruso JA, Stemmer PM, Kim CJ, Hansen LK, Becher N, Uldbjerg N. Protein profiling underscores immunological functions of uterine cervical mucus plug in human pregnancy. J Proteomics. 2011;74:817–828. doi: 10.1016/j.jprot.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei WL, Xing SG, Deng CY, Ju XC, Jiang XY, Luo ZG. Laminin/beta1 integrin signal triggers axon formation by promoting microtubule assembly and stabilization. Cell Res. 2012;22:954–972. doi: 10.1038/cr.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Chopp M, Zhang RL, Hozeska-Solgot A, Gregg SC, Buller B, Lu M, Zhang ZG. Angiopoietin 2 mediates the differentiation and migration of neural progenitor cells in the subventricular zone after stroke. J Biol Chem. 2009;284:22680–22689. doi: 10.1074/jbc.M109.006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Stanco A, Buys ES, Enikolopov G, Rubenstein JL. Soluble guanylate cyclase generation of cGMP regulates migration of MGE neurons. J Neurosci. 2013;33:16897–16914. doi: 10.1523/JNEUROSCI.1871-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Monnier PP, Sierra A, Schwab JM, Henke-Fahle S, Mueller BK. The Rho/ROCK pathway mediates neurite growth-inhibitory activity associated with the chondroitin sulfate proteoglycans of the CNS glial scar. Mol Cell Neurosci. 2003;22:319–330. doi: 10.1016/s1044-7431(02)00035-0. [DOI] [PubMed] [Google Scholar]
- Murray AJ, Peace AG, Shewan DA. cGMP promotes neurite outgrowth and growth cone turning and improves axon regeneration on spinal cord tissue in combination with cAMP. Brain Res. 2009;1294:12–21. doi: 10.1016/j.brainres.2009.07.071. [DOI] [PubMed] [Google Scholar]
- Myers JP, Santiago-Medina M, Gomez TM. Regulation of axonal outgrowth and pathfinding by integrin-ECM interactions. Dev Neurobiol. 2011;71:901–923. doi: 10.1002/dneu.20931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasterkamp RJ, Peschon JJ, Spriggs MK, Kolodkin AL. Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature. 2003;424:398–405. doi: 10.1038/nature01790. [DOI] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge JS, Silver J. Inhibition of neurite outgrowth on astroglial scars in vitro. J Neurosci. 1990;10:3594–3603. doi: 10.1523/JNEUROSCI.10-11-03594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabry J, O'Connor TP, Kirschner MW. Axonal transport of tubulin in Ti1 pioneer neurons in situ. Neuron. 1995;14:1247–1256. doi: 10.1016/0896-6273(95)90271-6. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Gross C, Xing L, Goshima Y, Bassell GJ. Identification of axon-enriched microRNAs localized to growth cones of cortical neurons. Dev Neurobiol. 2014;74:397–406. doi: 10.1002/dneu.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satkauskas S, Bagnard D. Local protein synthesis in axonal growth cones: what is next? Cell Adh Migr. 2007;1:179–184. doi: 10.4161/cam.1.4.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–359. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- Tan CL, Kwok JCF, Patani R, ffrench-Constant C, Chandran S, Fawcett JW. Integrin Activation Promotes Axon Growth on Inhibitory Chondroitin Sulfate Proteoglycans by Enhancing Integrin Signaling. Journal of Neuroscience. 2011;31:6289–6295. doi: 10.1523/JNEUROSCI.0008-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nature methods. 2005;2:599. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. The Journal of Neuroscience. 2009;29:4697. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Ji SJ, Jaffrey SR. Intra-Axonal Translation of RhoA Promotes Axon Growth Inhibition by CSPG. Journal of Neuroscience. 2012;32:14442–14447a. doi: 10.1523/JNEUROSCI.0176-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang L, Zhang H, Chen X, Yang Y, Liu G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin beta1 and matrix metalloproteinase2 (MMP2) PLOS ONE. 2013;8:e70192. doi: 10.1371/journal.pone.0070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Murashov AK. MicroRNA-431 regulates axon regeneration in mature sensory neurons by targeting the Wnt antagonist Kremen1. Front Mol Neurosci. 2013;6:35. doi: 10.3389/fnmol.2013.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Nguyen M, Garrison AK, Zhao Z, Wang Z, Sutherland C, Ma L. CNP/cGMP signaling regulates axon branching and growth by modulating microtubule polymerization. Dev Neurobiol. 2013;73:673–687. doi: 10.1002/dneu.22078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RL, Chopp M, Gregg SR, Toh Y, Roberts C, Letourneau Y, Buller B, Jia L, S PND, Zhang ZG. Patterns and dynamics of subventricular zone neuroblast migration in the ischemic striatum of the adult mouse. J Cereb Blood Flow Metab. 2009;29:1240–1250. doi: 10.1038/jcbfm.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ueno Y, Liu XS, Buller B, Wang X, Chopp M, Zhang ZG. The MicroRNA-17-92 cluster enhances axonal outgrowth in embryonic cortical neurons. J Neurosci. 2013;33:6885–6894. doi: 10.1523/JNEUROSCI.5180-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Wang Z, Gu Y, Feil R, Hofmann F, Ma L. Regulate axon branching by the cyclic GMP pathway via inhibition of glycogen synthase kinase 3 in dorsal root ganglion sensory neurons. J Neurosci. 2009;29:1350–1360. doi: 10.1523/JNEUROSCI.3770-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]