Abstract
The brain circuitry that controls song learning and production undergoes marked changes in morphology and connectivity during the song learning period in juvenile zebra finches, in parallel to the acquisition, practice and refinement of song. Yet, the genetic programs and timing of regulatory change that establish the neuronal connectivity and plasticity during this critical learning period remain largely undetermined. To address this question, we used in situ hybridization (ISH) to compare the expression patterns of a set of thirty known robust molecular markers of HVC and/or area X, major telencephalic song nuclei, between adult and juvenile male zebra finches at different ages during development (20, 35, 50 days post-hatch, dph). We found that several of the genes examined undergo substantial changes in expression within HVC or its surrounds, and/or in other song nuclei. They fit into broad patterns of regulation, including those whose expression within HVC during this period increases (COL12A1, COL 21A1, MPZL1, PVALB, and CXCR7) or decreases (e.g. KCNT2, SAP30L), as well as some that show decreased expression in the surrounding tissue with little change within song nuclei (e.g. SV2B, TAC1). These results reveal a broad range of molecular changes that occur in the song system in concert with the song learning period. Some of the genes and pathways identified are potential modulators of the developmental changes associated with the emergence of the adult properties of the song control system, and/or the acquisition of learned vocalizations in songbirds.
Keywords: Zebra finch, Brain development, Gene expression, Song system, Vocal learning
INTRODUCTION
Songbirds are one of the few animal groups that learn their vocalizations by imitation of an adult tutor (Immelman 1969, Jarvis 2004, Nottebohm 1972). This form of learning requires the sensory acquisition of an adult song model, followed by a prolonged practice period by the pupil that is guided by auditory feedback (Marler 1981, Williams 2004). Accordingly, songbirds have a specialized neural circuitry for the acquisition and production of learned vocalizations (Bottjer et al. 1989, Nottebohm et al. 1982, Vates and Nottebohm 1995, Vates et al. 1997, Wild 2004). In adults, the nuclei within this song control system are structurally distinct from the surrounding tissues by cytoarchitectonic features (Nottebohm et al. 1976), reflecting their highly active and specialized function. Through detailed studies in zebra finches, much knowledge has been gained on the connectivity and functional organization of the song control system (Bottjer et al. 1989, Scharff and Nottebohm 1991, Sohrabji et al. 1990, Vates et al. 1997, Zeigler H. P. and Marler 2004, Zeigler H. Philip and Marler 2008), and more recently on molecular specializations of its component nuclei (Denisenko-Nehrbass et al. 2000, Kim et al. 2004, Li et al. 2007, Lovell et al. 2013, Lovell et al. 2008, Wada et al. 2006), and even on correspondences to expression patterns in human vocal areas (Pfenning et al. 2014). However, knowledge is still scarce about gene regulatory events that may occur in these song nuclei during the developmental period when song is learned.
Song learning in zebra finches occurs in three distinct but somewhat overlapping stages during juvenile growth (Williams 2004), bearing striking resemblance to human vocal learning (Doupe and Kuhl 1999, Jarvis 2004) and thus representing a highly informative model to understand the neural basis of that process. Starting at ~25 days post hatch (dph) and lasting to ~65 dph, juveniles are sensitive to memorizing a song heard from a tutor, typically the father, and form an auditory memory of that song (Eales 1985, Roper and Zann 2006). Juvenile subsong, which is imprecise and highly variable, is thought to start at ~30-35 dph (although some reports indicate an earlier onset for this behavior at or around 25 dph (Roper and Zann 2006). As birds mature, their songs increase in complexity during a lengthy period of sensorimotor practice that lasts several weeks. At sexual maturity (~90 dph) variability in the vocalizations declines markedly, resulting in a complex, yet highly stereotyped song that closely resembles that of the vocal tutor.
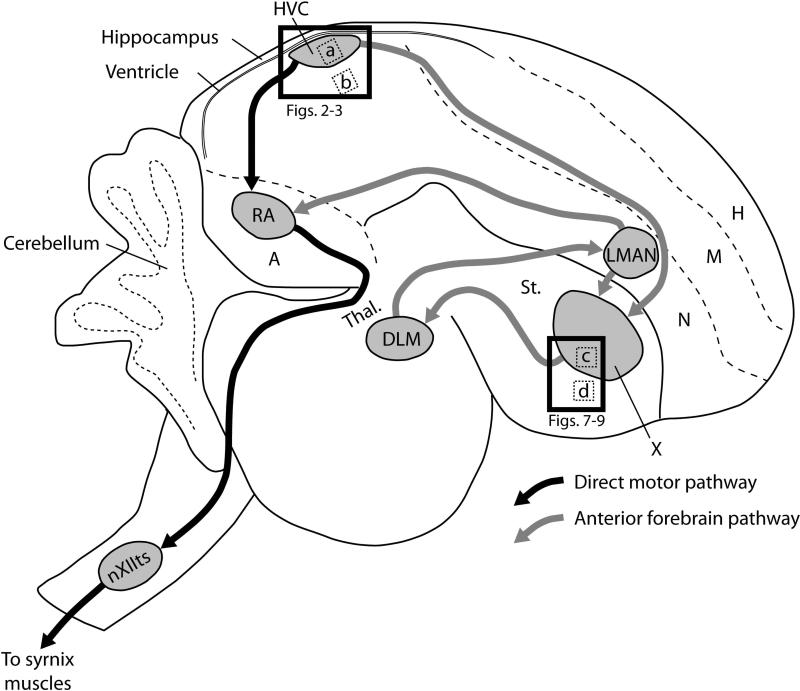
The song control system (Fig. 1) is composed of discrete interconnected pallial (cortical-like), basal ganglia, and thalamic nuclei that are solely dedicated to the learning and production of song. In the direct vocal-motor pathway (Fig. 1, black connections), the caudal nidopallial nucleus HVC (proper name) projects to the robust nucleus of the arcopallium (RA), and RA projections extend to brainstem nuclei that ultimately innervate the muscles of the syrinx and respiratory muscles necessary for song production. This portion of the circuit is largely responsible for the motor production and encoding of adult song (Hahnloser et al. 2002, Nottebohm et al. 1976, Yu and Margoliash 1996).
Figure 1.
Vocal pathways and major subdivisions of the zebra finch brain. Black arrows represent the direct motor pathway, and grey arrows represent the anterior forebrain pathway. HVC is used as a proper noun. Abbreviations: A, arcopallium; DLM, medial part of the dorsolateral nucleus of the anterior thalamus; H, hyperpallium; LMAN, lateral magnocellular nucleus of the nidopallium; M, mesopallium; N, nidopallium; nXIIts; tracheosyringeal portion of the twelfth cranial nerve nucleus; RA, robust nucleus of the arcopallium; St, striatum; Thal., thalamus; X, area X of the striatum. Dotted boxes represent the locations of 200×200 μm cell density sampling areas within (a) HVC (shown in Figs. 5A and 6A), (b) adjacent nidopallial shelf, (c) area X and (d) surrounding striatum (shown in Figs. 7B, 8B and 9B). Solid boxes represent the areas shown in Figs. 2-3 (for HVC) and 7A, 8A and 9A(for X). Rostral orientation is to the right, dorsal to the top.
The anterior forebrain pathway (AFP; Fig. 1, grey connections) consists of sequential projections from HVC to Area X of the medial striatum (X), to the medial part of the dorsolateral nucleus of the thalamus (DLM), and to the lateral magnocellular nucleus of the nidopallium (LMAN), which projects back to X as well as to RA in the vocal-motor pathway (Fig. 1). Lesions and pharmaceutical blocking of AFP nuclei during the subsong phase prematurely reduces song variability and ultimately disrupts the vocal learning ability (Bottjer et al. 1984, Kao and Brainard 2006, Olveczky et al. 2005, Scharff and Nottebohm 1991, Sohrabji et al. 1990). While the AFP is active during both juvenile and adult song production (Hessler and Doupe 1999, Jarvis et al. 1998), it has a primary role in the production of song during juvenile song learning. We also note the shelf and cup areas adjacent to HVC and RA, respectively (not shown); these areas are part of the central auditory pathways but are not involved in vocal learning or production, and they are considered conserved across birds, regardless of the presence of the song control system (Mello et al. 1998, Vates et al. 1996, Wild 1993).
The nuclei of the song system are distinctly present by 20 dph (Bottjer et al. 1985), but their cell populations and projections undergo marked changes during the song learning period. These changes (see review in Bottjer 2004) include the increases in volume of neuronal structures (Gahr and Metzdorf 1999), the loss of some neuronal cell populations (Konishi and Akutagawa 1985, Korsia and Bottjer 1989), and the incorporation of newly-formed neurons in HVC and X (Alvarez-Buylla and Kirn 1997, Alvarez-Buylla et al. 1990, Goldman and Nottebohm 1983). Neuronal turnover is associated with experimental prolonging of the song learning period (Wilbrecht et al. 2006) and the decline in neurogenesis is associated with increased stereotypy of the song (Pytte et al. 2007). Also important are increases in neuronal somata sizes and dendritic arborizations, changes in neuropil spacing (Bottjer et al. 1985, Bottjer et al. 1986), and the formation and maturation of several specific projections (Foster and Bottjer 1998, Konishi and Akutagawa 1985).
The distinct cytoarchitectonic and connectivity characteristics of the song system are presumed to be driven by gene regulatory differences in the circuit, relative to the neighboring tissues. Studies of single or few molecular markers provided early hints that the song system nuclei are distinct in terms of gene expression (Agate et al. 2007, Chen et al. 2005, Denisenko-Nehrbass et al. 2000, Gahr and Metzdorf 1997, Kim and Arnold 2005, Kim et al. 2004, London et al. 2003, Pinaud et al. 2007, Shahbazi et al. 2011, Velho et al. 2007, Wada et al. 2004, Wade 2000, Wade et al. 2005). The use of high-throughput approaches, in particular microarray screenings (Jarvis et al. 2002, Kato M. and Okanoya 2010, Li et al. 2007, Lovell et al. 2008, Tomaszycki et al. 2009), has greatly expanded our knowledge of differential gene expression and molecular regulatory pathways within the song system. Known specializations of song nuclei in adults include genes encoding a diversity of structural and functional proteins (e.g., ion channels, membrane receptors, cytoskeletal and cell-adhesion proteins), whose expression is likely related to the maintenance of the adult song control circuitry and the production of highly stereotyped song. In contrast, despite some early efforts (summarized in Clayton 1997), our knowledge of gene expression regulation during the song learning period is still scarce. In consequence, a clear picture has not emerged on how molecular changes during development may relate to the morphological and functional maturation of the system.
Here we used in situ hybridization to examine whether the expression of known robust molecular markers of song nuclei in adult zebra finches undergoes developmental changes in juveniles. Our main focus was on nucleus HVC, as it is a key nucleus that interfaces with both the direct pathway and the AFP (Reiner et al. 2004b), and plays central roles in the encoding of motor sequences during the production of learned song (Hahnloser et al. 2002, Long et al. 2010, Yu and Margoliash 1996). Furthermore, a large set of markers of adult HVC have been identified (Lovell et al. 2008), many of which have been implicated in processes like the recruitment of newly-formed neurons, the establishment of neuronal connections, and/or the maturation of specific neuronal populations, based on data from other experimental organisms. We chose a subset of previously undescribed markers to examine in the current report, based on the relatively strong expression patterns in the adult HVC. The majority of these genes belong to functional groups whose developmental expression has not yet been examined in zebra finches. We found that during the song learning period several HVC markers undergo marked regulation within song nuclei or in adjacent areas outside of the song circuitry, culminating in the adult patterns. These observations considerably expand our knowledge of regulatory events within the song system during the song learning period, suggesting some intriguing genes and associated molecular pathways as targets for future mechanistic studies.
METHODS
Animals and brain tissue
Animal use was approved by the OHSU's IACUC, protocol B11310. Adult male zebra finches (Taeniopygia gutatta; >90 dph) were obtained from a commercial supplier and maintained in our colony, while juveniles were born and raised in our breeding colony to 20, 35 or 50 dph. Males were identified based on plumage, or in the case of 20-day-old birds, males were identified based on molecular genotyping with sex-specific PCR primers using DNA from feathers (Soderstrom et al. 2007, Vucicevic et al. 2013). Adult males were isolated in sound-proof chambers overnight and sacrificed by decapitation at 2 hr after lights on the following morning, after monitoring to ensure that singing did not occur during this period. To minimize possible adverse effects of stress, juvenile birds were not sound isolated. Instead, they were kept in their family cages throughout development, then taken from the aviary and sacrificed within a few minutes by decapitation. The brains of all birds were dissected and blocked sagitally with tissue-tek and frozen in a dry icepropanol slurry. Brains (right hemispheres only) were sectioned at 10 μm with a Leica cryostat and mounted on charged slides. Slides were stored at −80°C until processed.
To identify appropriate brain sections that included HVC and X the position of these nuclei were determined along the medial-lateral axis by examination of regularly spaced sections at 200 μm under darkfield illumination (for visualization of fiber pathways) and adjacent sections stained with Nissl (cresyl violet) followed by examination under brightfield. We also confirmed the boundaries of song nuclei by processing subsets of the serial sections for in situ hybridization for strong molecular markers of these song nuclei. This was particularly important for 20 and 35 dph brains, where the combination of darkfield and Nissl stain are often insufficient for identification of the unambigious boundaries and extent of these nuclei. We also verified that all HVCs on the sections selected for molecular studies measured a minimum of 250 μm in dorsal-ventral thickness, thus avoiding the medial-most thin region that could correspond to paraHVC, which lacks RA-projecting neurons (Olson et al. 2011).
In situ hybridization (ISH)
Digoxigenin-labeled riboprobes were generated from the ESTIMA cDNA library of brain transcripts (Replogle et al. 2008). A full list of the clones used and a summary of their functions (from Entrez Gene) is provided in Supplemental Table 1. In short, cDNAs were isolated from bacterial stock and following cleaning with a PCR purification kit, antisense riboprobes were generated from the DNA template with T7 RNA polymerase using a digoxigenin(DIG)-UTP RNA labeling kit (Roche) at 37°C for 5 hours. Riboprobes were then hybridized to brain sections following Mello et al. (1997) using an optimized non-radioactive detection protocol for DIG-labeled riboprobes, as detailed in Carleton et al. (Carleton et al. 2014). Brain sections were fixed in 3% phosphate-buffered paraformaldehyde for 5 min, followed by a brief rinse in 0.1 M PBS. Sections were then acetylated for 10 min (0.25% acetic anhydride in 1.4% triethanolamine and dH2O), rinsed in 2x SSPE, and alcohol-series dehydrated. Tissue was covered in hybridization solution (50% formamide, 2x SSPE, 2 μg/μl tRNA, 1 μg/μl BSA, 1 μg/μl Poly A, and ~25 nl/μl riboprobe per slide), coverslipped, and immersed overnight in mineral oil at 65-67°C, depending on optimized probe-specific conditions. The next day slides were rinsed in chloroform and de-coverslipped in 2x SSPE. Post-hybridization washes included 1 hr in 2x SSPE plus 50% formamide, and then two washes for 30 min in 0.1x SSPE at the same temperature as the hybridization, with agitation at 10 min intervals. Slides were removed to a Tris-HCl, NaCl buffer with 0.3% Triton X-100 (TNT) and sections were framed with a Dako Pen, blocked with 8.3 ng/μl BSA in TNT (TNB) for 30 min, followed by incubation with an anti-DIG alkaline phosphatase conjugated antibody (1:600; Roche) in TNB for 2 hrs. The slides were then rinsed in TNT, and incubated with ~200 μl per slide of 0.21 g/L 5-bromo-4-chloro-3-indolyl-phosphate with 0.42 g/L nitroblue tetrazolium in Tris buffer (BCIP/TNB; Perkin-Elmer) at room temperature until a detectable signal was visible (1-3 days). Slides were finally rinsed in TNT for 15 min, and then fixed in 3% phosphate-buffered paraformaldehyde for 5 min, followed by two brief rinses in deionized water. Slides were allowed to air-dry before coverslips were placed with Aqua-Mount mounting medium. For consistency of each experiment, each gene was run in a single batch composed of slides selected from each brain at a medial-lateral level previously confirmed to contain HVC or X. Controls included sense strand hybridizations and probe omission, both of which yielded no background labeling, and gad2 (gad65 ), which yields consistent robust expression at all ages examined. For all probes, specificity is verified by BLAT-alignment analysis of the ESTIMA collection sequences against the zebra finch genome, using UCSC's genome browser; hybridization conditions and initial adult expression assessments for a subset of probes in the present study have been previously established (Lovell et al, 2008). Serial digital images for a subset of the genes reported here have been included in the online Zebra finch Expression Brain Expression Atlas (ZEBrA; www.zebrafinchatlas.org).
Brain mapping, imaging and analysis
Brain nuclei were mapped and cells were counted with a Lucivid-illuminated Nikon E-600 microscope yoked to a microcomputer equipped with Neurolucida software (ver. 5.05.4, MicrobrightField, Inc.). Whenever possible several independent criteria were used to delineate the borders of song nuclei on the sections analyzed. For adults and 50-day-old birds, song nuclei on sections processed for ISH could be identified directly under darkfield illumination, or in adjacent or close Nissl-stained sections under brightfield. For the earlier ages, these criteria needed to be combined with mapping of song nuclei boundaries on closely adjacent brain sections based on the expression of molecular markers.
Quantitative analysis was performed under brightfield at 40x magnification, with the aid of Neurolucida software. In short, we created an outline of each brain section with the major song nuclei identified, and then placed a 200×200 μm box in the center of each nucleus, making sure that all corners fit entirely within the boundaries of the selected nucleus (Fig. 1; placement of counting windows indicated by small rectangles a-d). All cells in each box that met an inclusion criterion consisting of detectable labeling with cytoplasmic distribution (typical donut shape) and a relatively clear nucleus were counted. Here we were interested in quantifying how the expression profiles of different genes changed such that these genes became molecular markers of song nuclei in adults (i.e., differential expression relative to the adjacent tissue). Thus, we also performed measurements with the same counting window adjacent to the song nuclei, in the immediate surrounding tissues ca. 200 μm from the margin of the nucleus. For HVC, this represents the nidopallial shelf area; for X this represents the medial striatum immediately ventral to X. Labeled cell counts for each brain region are reported in units of cells per mm2 (density of labeled cells). We note that several factors may affect these counts, including changes in cell size, number and density (number of cells per area), all of which have been shown to occur in song nuclei of finches across developmental ages, as well as whether all cells or only subsets or specific cell types express a given marker. Further analyses that are beyond the scope of our present report would be required to clearly differentiate among these possibilities. We also note that we were not concerned in determining changes in the total number of cells expressing our markers, thus we did not consider it necessary to employ a reconstructive stereological approach.
To quantify possible age-related effects on the levels of gene expression of individual cells, we used a densitometric approach to measure grayscale (GS) values over labeled cells. We first created a background correction based on an image of a blank slide to correct for illumination gradients across the microscope field of view, and then photographed all the sections labeled with the same gene under the same exposure and lighting conditions at 20x magnification. This provided a series of TIFF images of whole HVCs and Xs with an ample number of cells for quantification. Image files were then converted to grey scale in Adobe Photoshop and opened in IMAGEJ (NIH, ver. 1.42q). We next located the areas of interest and outlined cells (n = 20 per area) with the free hand outline tool to measure the optical intensity in a GS range of 0(black)-255(white), and recorded the mean GS values per section. To perform this step, we zoomed in to a close up view of the image and then panned the field of view within a predefined region of tissue within the central regions of interest, following a grid pattern to avoid double counting. The first 20 cells identified in the close up view were analyzed, to avoid biases in cell selection based on expression intensity. The values were first inverted (so larger values represent greater expression) and then corrected by subtracting the inverted expression level of the slide background (without tissue). The data were then normalized to the average adult expression levels to create a cell labeling index relative to adults (1.0 means expression levels similar to that in adults). Our selection criteria required that cells showed a visible cytoplasmic labeling surrounding a non-stained nucleus. Since we directly identified and outlined labeled cells, we avoided the issue of including in our analysis non-cellular tissue artifacts that often occur when using a thresholding approach to densitometry.
To measure the effects of age on labeled cell counts and cell labeling index we used ANOVA with birds’ age as discrete independent variables: 20, 35, 50 dph and adults. Because age is a measure of regular intervals along a continuous axis, we then used a post-hoc analysis of differences between subsequent transitions of 20 to 35 dph, 35 to 50 dph and 50 dph to adults, in order to identify the age with the most pronounced change in gene expression. To account for multiple comparisons in this last analysis we used Student's t-tests with an alpha of 0.05 divided by the number of intervals to account for multiple comparisons (α =0.0167). Statistical analyses were performed with JMP 9.
RESULTS
The main focus of our study was HVC, for which a large set of highly differential markers has been identified in adults. To gain insight into molecular changes that occur in this nucleus during the song learning period, and that result in the adult expression patterns, we initially conducted a qualitative age comparison of known HVC markers between 20 dph juveniles and adults (n = 2 males per age group). Based on the outcome, we categorized the patterns into 4 groups (Table 1): Group A, where expression appeared higher in adults (representative examples in Fig. 2A-G); Group B, where expression appeared higher in juveniles (representative examples in Fig. 3A-B); Group C, where expression appeared equivalent between juveniles and adults, but large age differences in expression were apparent in the adjacent shelf area, such that the HVC border was clearly distinguishable in adults but not in juveniles (for example see Fig. 3C); and Group D, with similar expression between adults and juveniles in HVC and adjacent tissue (not shown). Although our focus was on HVC, we also examined changes in RA, X and LMAN (Table 1) in cases where HVC markers were differentially expressed in these other nuclei and when data were available from the HVC sections. Lastly, we also examined TAC1, a highly robust marker of X in adults.
Table 1.
Qualitative assessment of gene expression differences in song nuclei of juvenile (20 day old) vs. adult male zebra finches.
| GENE | HVC | shelf | X | Str. | LMAN | Nido. | RA | Arco. |
|---|---|---|---|---|---|---|---|---|
| A. HVC higher in adults | ||||||||
| ADAM23 | ↑ | = | ↑ | = | ↑ | = | ||
| COL12A1* | ↑ | = | ↑ | = | ||||
| COL21A1* | ↑ | ↓ | ↑ | ↓ | ||||
| CXCR7 | ↑ | = | ↑ | ↑ | ||||
| MPZL1 | ↑ | ↑ | ↑ | = | ↑ | ↑ | ||
| PVALB | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ||
| SLC8A1 | ↑ | = | = | = | ↓ | = | ||
| B. HVC higher in juveniles | ||||||||
| CACNG3 | ↓ | ↓ | ↑ | = | ||||
| CNTN4 | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ||
| FLRT2 | ↓ | ↓ | = | ↓ | ↓ | ↓ | ||
| KCNT2 | ↓ | ↓ | ↑ | ↓ | ||||
| LGMN* | ↓ | = | ||||||
| LRRTM2* | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ||
| NRP1* | ↓ | = | ↓ | = | ||||
| SAP30L* | ↓ | = | = | ↓ | ↑ | = | ||
| C. Shelf higher in, juveniles | ||||||||
| MAP1B* | = | ↓ | ↑ | ↓ | ↓ | ↓ | ||
| NDRG4 | = | ↓ | ↑ | = | ||||
| RCAN2 | = | ↓ | = | ↓ | = | ↓ | ||
| SEMA6A | = | ↓ | = | = | ↓ | ↓ | = | ↑ |
| SV2B | = | ↓ | ↑ | ↓ | = | ↓ | ||
| D. Marker across ages | ||||||||
| CADPS2 | = | = | = | = | ||||
| CDH6 | = | = | ||||||
| KCNA1 | = | = | = | ↓ | ↑ | ↓ | ||
| NETO1 | = | = | ||||||
| NROB1 | = | = | ||||||
| PPM1E | = | = | ↓ | = | ||||
| SCUBE1 | = | = | ||||||
| SEMA3E | = | = | = | = | = | = | ||
| SYT4 | = | = |
↑ indicates an expression increase, ↓ indicates an expression decrease, = indicates comparable expression between the ages. Empty fields indicate expression was not evaluated, in most cases because the gene is not differentially expressed in that nucleus in adults. Grey background represents the genes/nuclei for which we performed a detailed expression quantification. Serial images for most genes are available in ZEBrA, exceptions are indicated by an asterisk.
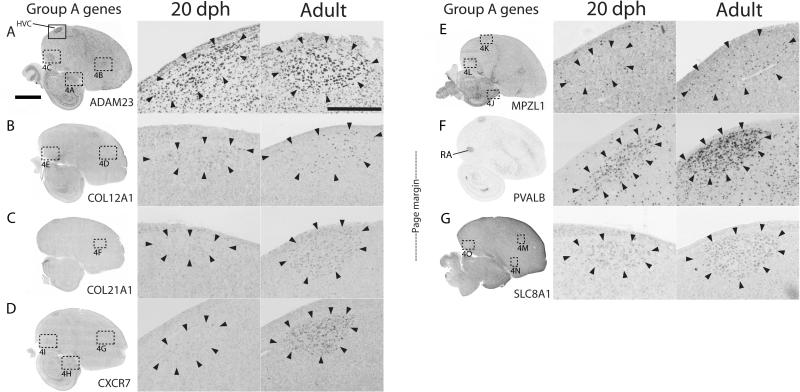
Figure 2.
HVC expression patterns of Group A genes. A-G: Shown are views of whole parasagittal brain sections of adult males at the level of major song nuclei, processed for in situ hybridization to illustrate general expression patterns (left panels), as well as detailed views of HVC at 20 dph (middle panels) and in adults (right panels). The solid box in A shows the location of the magnified images of HVC in the middle and right panels; dotted boxes show the locations of several panels in Fig. 4. Arrowheads indicate the approximate boundaries of HVC; scale bars: 5 mm for left panels and 500 μm for middle and right panels.
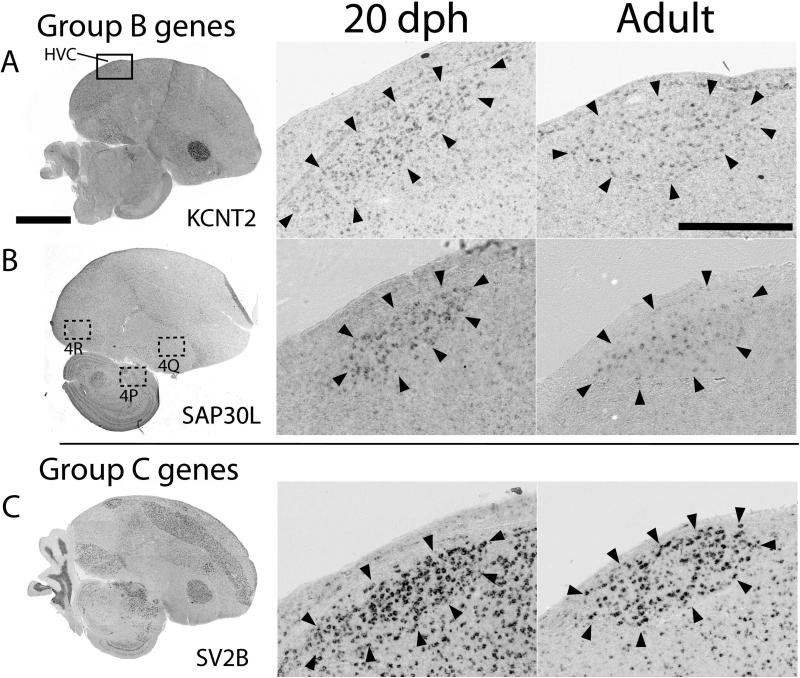
Figure 3.
HVC expression patterns of select Group B and C genes. A-C: Shown are views of whole parasagittal brain sections of adult males at the level of major song nuclei, processed for in situ hybridization to illustrate general expression patterns (left panels), as well as detailed views of HVC at 20 dph (middle panels) and in adults (right panels). The solid box in A shows the location of the magnified images of HVC in the middle and right panels; dotted boxes show the locations of several panels in Fig. 4. Arrowheads indicate the approximate boundaries of HVC; scale bars: 5 mm for left panels and 500 μm for middle and right panels.
We next conducted a quantitative analysis of expression at multiple ages spanning the song learning period in juveniles (20, 35 and 50 dph) and adults (n = 4 per age group). We focused mostly on Group A, as that set contains the most robust positive markers of adult HVC, but we also analyzed some genes from Groups B and C that had apparent marked age differences; Group D will be analyzed in future studies of the early ontogeny of the song system. Our quantitative analyses consisted of performing labeled cell counts (expressed in number of labeled cells per area) in HVC (Fig. 1, box a) and in the adjacent shelf (Fig. 1, box b), and optical density (OD) assessments to generate a cell labeling index (OD levels per labeled cell, normalized to average levels of adults, as detailed in Methods). We also performed quantitative analysis for some HVC markers we found to be robust markers of other song nuclei, with apparent developmental changes; this included three markers of X, two markers of RA, and five markers of LMAN. We detected significant changes for several genes, and the quantitative analysis was generally consistent with the qualitative assessment. Below, we first briefly describe the general brain distribution of each gene in adult males, since in most cases this is the first documentation of their expression in finches and/or avian species (examples in Fig. 4; further details on expression patterns in the Zebra finch Expression Brain Atlas, www.zebrafinchatlas.org), followed by the quantitative analyses of age-dependent changes in HVC and surrounding shelf (Figs. 5 and 6), X and adjacent striatum (Figs. 7-9), and other song nuclei (Fig. 10); statistical analyses are presented in Tables 2-4.
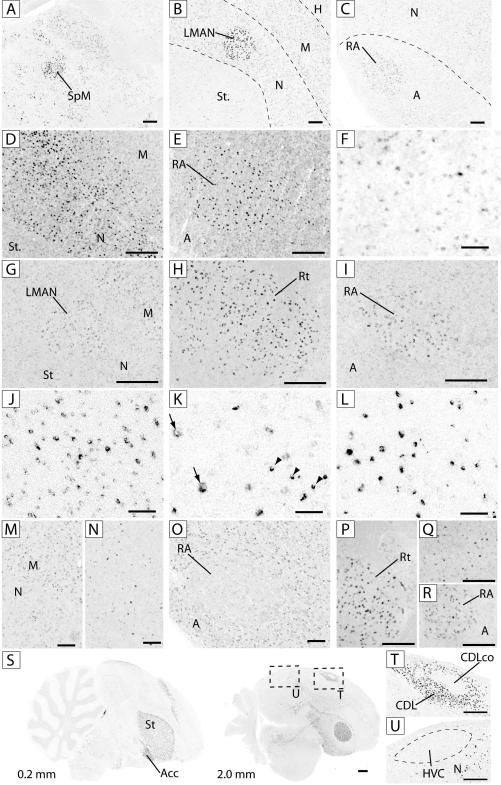
Figure 4.
Expression patterns of select genes in the brain and song system. Shown are views of detailed areas or whole sections of adult males, processed for in situ hybridization. A-C: ADAM23 in the thalamus (A) and rostral (B) and caudal (C) telencephalon; D-E: COL12A1 in the rostral (D) and caudal (E) telencephalon: F: COL21A1 in the nidopallium; G-I: CXCR7 in the rostral telencephalon (G), thalamus (I) and arcopallium (I); J-L: MPZL1 in the optic tract (J), HVC (K; arrows point to large neuronal-like cells and arrowheads indicate small oligodendrocyte-like cells) and RA; M-O: SLC8A1 in rostral telencephalon (M), striatum (N) and arcopallium (O); P-R: SAP30L in thalamus (P), striatum (Q) and arcopallium (R); S: TAC1, views of parasagittal sections at 0.2 mm (left ) and 2.0 mm (right) from the midline; dashed squares show location of panels 4T and U; T: TAC1 in dorsolateral corticoid area (CDL) and its core (CDLco); U: TAC1 in area around HVC. Scale bars: 50 μm for F, J, K and L, 250 μm for all other panels; dashed lines represent approximate boundaries of brain subdivisions and nuclei. See Figs. 2 and 3 for location of panels, and Fig. 1 legend for anatomical abbreviations.
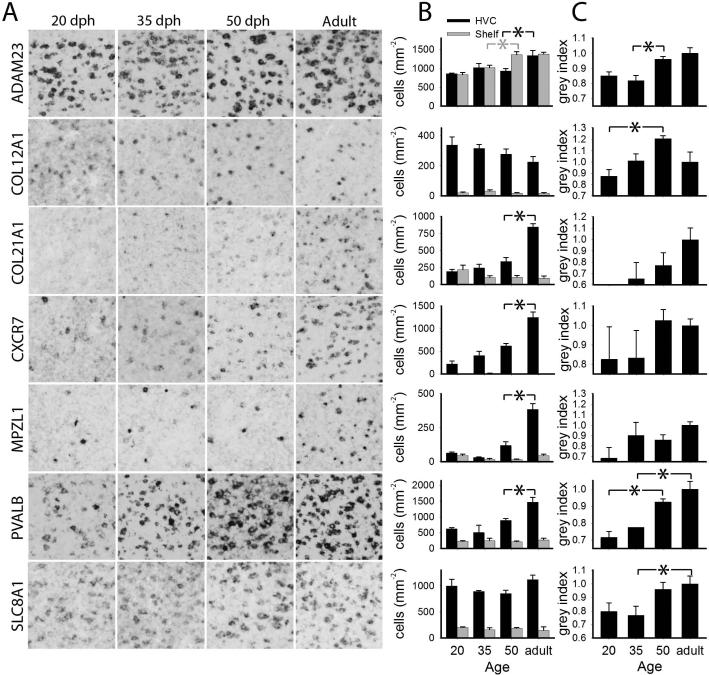
Figure 5.
Quantitative analysis of age-related changes in gene expression in HVC for Group A genes. A: detail views of 200 × 200 μm boxes within HVC for the age groups measured; B: labeled cell counts per unit area in HVC (black) and shelf (grey) with age; C: cell labeling index within HVC. Plotted in B and C are means + sem; significant age group differences are indicated with a black or grey asterisk (*) for HVC or shelf, respectively.
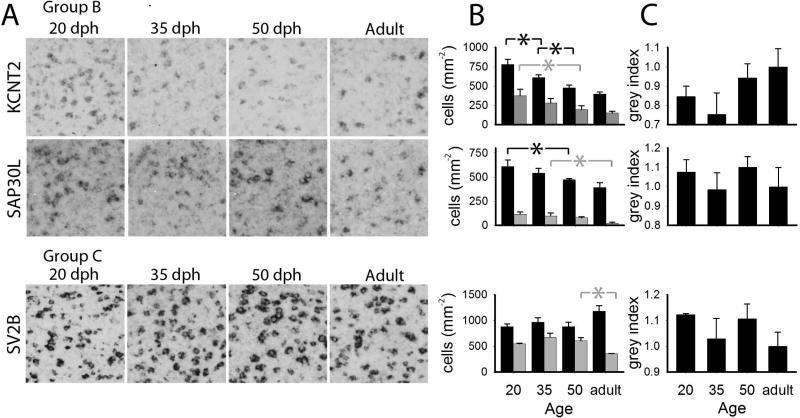
Figure 6.
Quantitative analysis of age-related changes in gene expression in HVC for select Group B and C genes. A: detailed views of 200 × 200 μm boxes within HVC for the age groups measured. B: labeled cell counts per area in HVC (black) and shelf (grey); C: cell labeling index within HVC. Plotted in B and C are means + sem; significant age group differences are indicated with a black or grey asterisk (*) for HVC or shelf, respectively.
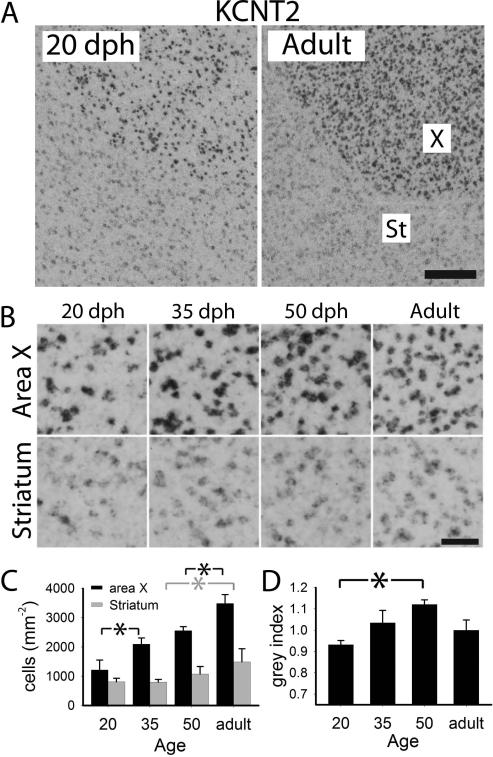
Figure 7.
Quantitative analysis of age-related changes of KCNT2 in area X A: views of X and adjacent striatum of 20 dph and adult males; B: detailed views of X and striatum across age groups; C: labeled cell counts per area in X (black) and adjacent striatum (grey); D: cell labeling index in X. Plotted in C and D are means + sem; significant age group differences are indicated with a black or grey asterisk (*) for X or striatum, respectively. Scale bars: 100 μm (A), 25 μm (B).
Figure 9.
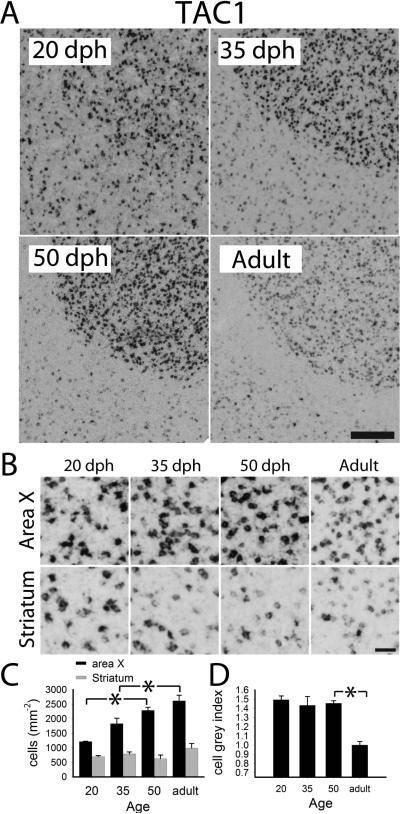
Quantitative analysis of age-related changes of TAC1 in area X. A: views of X and adjacent striatum of 20, 35, 50 dph and adult males. B: detailed views of X and striatum across age groups; C: labeled cell counts per area in X (black) and adjacent striatum (grey); D: cell labeling index in X. Plotted in C and D are means + sem; significant age group differences are indicated with a black asterisk (*) for X. Scale bars: 100 μm (A), 25 μm (B).
Figure 10.
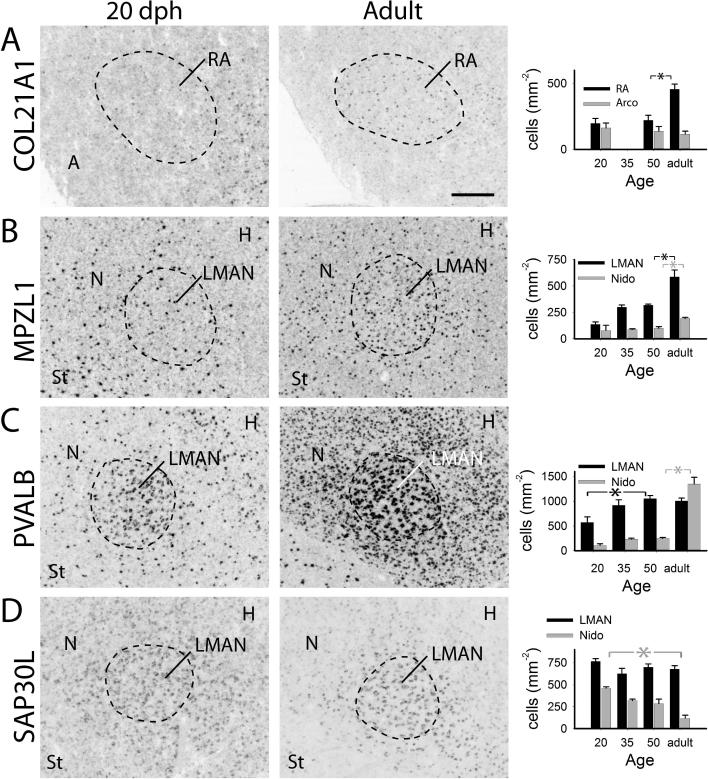
Age-related changes in expression of select genes in RA and LMAN. Each row shows gene expression in song nuclei RA (A) or LMAN (B-D) and adjacent tissue for 20 dph (left) and adult (middle) males, and plots of labeled cell counts per area (right) in RA or LMAN (black) and surrounding tissue (grey) A: COL12A1; B: MPZL1; C: PVALB; D: SAP30L. Plotted are means + sem; significant differences are indicated with a black or grey asterisk (*) for RA/LMAN cells or surrounding tissue, respectively; arco: arcopallium, nido: nidopallium; for other anatomical abbreviations, see Fig. 1 legend.
Table 2.
ANOVA of age-related changes between 20 dph and adults in zebra finch HVC and adjacent shelf. See text for a description of variables.
| Gene | Group | F, HVC cell counts | P, HVC cell counts | F, shelf cell counts | P, shelf cell counts | F, HVC labeling index | P, HVC labeling index |
|---|---|---|---|---|---|---|---|
| ADAM23 | A | F3,11 = 4.6 | 0.025 | F3,11 = 4.6 | 0.025 | F3, 11 = 8.11 | 0.0039 |
| COL12A1 | A | F3,10 = 1.6 | 0.24 | F3,10 = 1.3 | 0.32 | F3, 11 = 5.03 | 0.02 |
| COL21A1 | A | F3,11 = 32.0 | 0.0001 | F3,11 = 2.1 | 0.16 | F3, 6 = 2.3 | 0.18 |
| CXCR7 | A | F3,10 = 19.8 | 0.0001 | F3,10 = 1.8 | 0.21 | F3, 9 = 1.4 | 0.31 |
| MPZL1 | A | F3,9 = 27.5 | 0.0001 | F3,10 = 2.9 | 0.093 | F3, 12 = 2.40 | 0.12 |
| PVALB | A | F3,9 = 8.4 | 0.0055 | F3,9 = 0.36 | 0.78 | F3, 7 = 14.4 | 0.0022 |
| SLC8A1 | A | F3, 11 = 1.8 | 0.22 | F3, 11 = 0.43 | 0.74 | F3, 11 = 3.85 | 0.042 |
| KCNT2 | B | F3,10 = 15.1 | 0.0005 | F3,10 = 3.2 | 0.069 | F3, 9 = 1.5 | 0.27 |
| SAP30L | B | F3,11 = 3.6 | 0.049 | F3,11 = 4.9 | 0.022 | F3, 11 = 0.51 | 0.69 |
| SV2B | C | F3,12 = 2.6 | 0.10 | F3,12 = 7.4 | 0.0046 | F3, 12 = 1.13 | 0.38 |
Table 4.
ANOVA results of age-related changes between 20 dph and adult in labeled cell counts from zebra finch LMAN, non-cup adjacent arcopallium, RA and adjacent nidopallium for several genes.
| Gene | Structure | Age range available | F, p | Effect description |
|---|---|---|---|---|
| COL12A1 | RA | 20, 50, adult | F2, 6 = 2.51, p = 0.16 | Trend decrease 50-adult |
| COL12A1 | Arco. | 20, 50, adult | F2, 6 = 0.94, p > 0.05 | |
| COL21A1 | RA | 20, 50, adult | F2, 7 = 14.68, p = 0.0031 | Sig. Increase 50-adult |
| COL21A1 | Arco. | 20, 50, adult | F2, 7 = 0.59, p > 0.05 | |
| MPZL1 | LMAN | 20, 35, 50, adult | F3, 8 = 14.8, p = 0.0012 | Sig. Increase 50-adult |
| MPZL1 | Nido. | 20, 35, 50, adult | F3, 8 = 7.32, p = 0.0111 | Sig. Increase 50-adult |
| PVALB | LMAN | 20, 35, 50, adult | F3, 10 = 5.15, p = 0.0207 | Sig. Increase 20-50 dph |
| PVALB | Nido. | 20, 35, 50, adult | F3, 10 = 82.96, p<0.0001 | Sig. Increase 50-adult |
| SAP30L | LMAN | 20, 35, 50, adult | F3, 11 = 1.87, p = 0.19 | Trend decrease 20-adult |
| SAP30L | Nido. | 20, 35, 50, adult | F3, 11 = 17.64, p < 0.0002 | Sig. decrease 20-adult |
| SLC8A1 | LMAN | 35, 50, adult | F2, 8 = 2.59, p = 0.14 | Trend increase 20-adult |
| SLC8A1 | Nido. | 35, 50, adult | F2, 8 = 1.80, p = 0.21 | |
| SV2B | LMAN | 20, 35, 50, adult | F3, 10 = 1.90, p = 0.19 | |
| SV2B | Nido. | 20, 35, 50, adult | F3, 10 = 3.18, p = 0.07 | Trend decrease 50-adult |
ADAM23
This gene had a broad and homogenous pallial distribution, expression in several midbrain nuclei, in particular the nucleus spiriformis medialis (SpM; Fig. 4A), moderate expression in pallial layers with lower expression in the striatum (Fig. 4B), and enriched expression in LMAN (Fig. 4B) and RA (Fig. 4C). We observed a slight age-dependent increase in cellular labeling within HVC (Fig. 5A). Quantitative analysis revealed significant increase in labeled cell counts in HVC between 50 dph and adult, with parallel increases in the shelf between 35-50 dph (Fig. 5B). A significant increase in the cell labeling index within HVC (Fig. 5C) was likely the main contributing factor to this marker being a more robust HVC marker in adults than in juveniles.
COL12A1
Expression was low throughout much of the brain, but enriched in the nidopallium (Fig. 4D). Compared to the surrounding arcopallium, it was distinctly higher in RA (Fig. 4E), where it was seen in a discrete population of strongly labeled cells. We observed an apparent developmental decrease in labeled cells and increase in the intensity of labeling in HVC (Fig. 5A). The labeled cell counts in HVC, but not the shelf, showed a trend for a decline across age groups (Fig. 5B). In contrast, the cell labeling index in HVC increased significantly with age (Fig. 5C), with a distinct peak at 50 dph. We also quantified cell counts in RA and the surrounding arcopallium, where there were not significant age-related changes.
COL21A1
While expression was low throughout the brain, it was relatively high in parts of the arcopallium (not shown), moderate in the nidopallium (Fig. 4F), sparse through the striatum (not shown), and enriched in RA relative to the adjacent arcopallium (Fig. 10A). Within HVC, expression was very weak at 20 dph and increased markedly until adulthood (Fig. 5A). There was a significant increase in labeled cell counts between 50 dph and adulthood in HVC but not the shelf (Fig. 5B), and a trend for an age-related increase in the cell labeling index in HVC (Fig. 5C). There was also an apparent increase in expression in RA (Fig. 10A, left and middle); quantification showed a developmental increase in labeled cell counts in RA, particularly between 50 dph and adult (Fig. 10A, right), but not in the arcopallium ventral to RA (excluding the cup region).
CXCR7
We observed very low expression throughout the brain, with some enrichment in the mesopallium (Fig. 4G) and high expression in several thalamic nuclei, including rotundus (Fig. 4H) as well as marked enrichments in LMAN (Fig. 4G) and RA (Fig. 4I), with little to no expression in the adjacent areas. Within HVC, there was a marked developmental increase in expression (Fig 5A). The labeled cell counts in HVC reflected the qualitative assessment, with a significant increase between 50 dph and adult (Fig. 5B); labeled cells in the shelf were not detectable (Fig. 5B) and the cell labeling index in HVC did not significantly differ across ages (Fig. 5C).
MPZL1
Expression was broadly distributed, being stronger in the white matter, including fiber tracts, laminae and commissures, particularly in the brainstem and cerebellum; expression was seen in strongly labeled cells with small somata resembling oligodendrocytes (Fig. 4J) consistent with gene function (Roubelakis et al. 2007). Within HVC, expression occurred in cells resembling oligodendrocytes (Fig. 4K, arrowheads), but also in some larger cells of possible neuronal identity (Fig. 4K, arrows). Similar cellular pattern and enrichment were seen in RA (Fig. 4L) and in LMAN and surrounding nidopallium (Fig. 10B, middle). HVC expression in 20 dph brains was very low and comparable to the adjacent shelf, consisting of very few small, but well-labeled cells; a marked increase in these occurred after 50 dph (Fig. 5A), as confirmed by quantitative analysis (Fig. 5B), culminating in the high counts seen in adults. Labeled cell counts in the shelf and cell labeling index in HVC did not significantly change with age (Fig. 5B-C). We also noted age increases in expression in both LMAN and adjacent nidopallium (Fig. 10B, left and middle), confirmed by quantitative analysis (Fig. 10B, right).
PVALB
In accordance with previously described protein (Braun et al. 1991, Wild et al. 2001, Zuschratter et al. 1985) and mRNA (Hara et al. 2012) expression, we observed a wide distribution consisting of sparse, strongly labeled large cells, as well as smaller, more weakly labeled cells in both pallium and striatum (not shown). Expression was highly enriched in several brainstem nuclei, in Purkinje cells and the molecular layer of the cerebellum, in primary sensory areas (field L2a and entopallium), and in song nuclei RA (Fig. 2F, left), LMAN (Fig. 10C) and interfacialis (not shown). In HVC, we observed a subset of large cells with very strong expression, as well as smaller cells that were more numerous (Fig. 5A, Adult). Expression was lower in 20 dph juveniles than in adults (Fig. 5A), and there was a significant age-related increase in labeled cell counts between 50 dph and adult in HVC but not shelf (Fig. 5B); the cell labeling index in HVC increased more gradually between 20 dph and adulthood (Fig. 5C). Expression also increased in LMAN and adjacent nidopallium (Fig. 10C, left and middle), as confirmed by quantitative analysis (Fig. 10C, right).
SLC8A1
Expression was most robust in the mesopallium (Fig. 4M), and much sparser in other pallial layers; the striatum was populated by a sparse cell type with strong labeling (Fig. 4N). It was markedly enriched in HVC but not in other song nuclei, and markedly downregulated in RA compared to the rest of the arcopallium (Fig. 4O). It appeared more highly expressed in adult than juvenile HVC (Fig. 5A); while there were no significant age changes in labeled cell counts in HVC or shelf (Fig. 5B), there was a significant increase in the cell labeling index within HVC, most pronounced between 35 and 50 dph (Fig. 5C). In LMAN and adjacent nidopallium the labeled cell counts remained statistically constant.
KCNT2
As previously described (Lovell et al. 2013), expression was low and sparse throughout the pallium, with HVC being a positive marker (higher expression) relative to its surround, and RA a negative marker (lower expression) relative to the surround (not shown); expression was also slightly higher in the striatum, with very robust enrichment in X (Fig. 7A, right). We observed an age-related decrease in expression in HVC (Fig. 6A left); quantitative analysis showed a gradual age-related decline in labeled cell counts in both HVC and shelf (Fig. 6B), and a trend for an increase in the cell labeling index within HVC (Fig. 6C), resulting in the distinct appearance of adult HVC. The enrichment in X was much more robust in adults than in juveniles (Fig. 7A and B). Indeed, we observed marked age increases in labeled cell counts in X that paralleled a similar but smaller change in the striatum (Fig. 7C), and a significant increase in the cell labeling index in X that peaked at 50 dph (Fig. 7D).
SAP30L
It was found to be a distinct marker of rotundus (Rt; Fig. 4P) and other thalamic nuclei, low to moderate in pallial regions, and in a sparsely distributed cell population in the striatum (Fig. 4Q); it was markedly enriched in RA (Fig. 4R) and in LMAN and the surrounding nidopallial region (Fig. 10D, Middle). We observed an age-dependent decline in HVC (Fig. 6A), which was confirmed by a significant decrease in labeled cell counts in HVC between 20 and 50 dph, paralleled by a less marked decrease in the shelf (Fig. 6B); the cell labeling index did not significantly change across ages (Fig. 6C). We also noted that this gene was a more robust marker of adult than juvenile LMAN (Fig. 10D left and middle). While labeled cell counts in this song nucleus were constant across ages, we noted a strong decrease in the adjacent nidopallium excluding the shell of LMAN (Fig. 10D), consistent with the emergence of a marker pattern by a decrease in labeling of surrounding tissue.
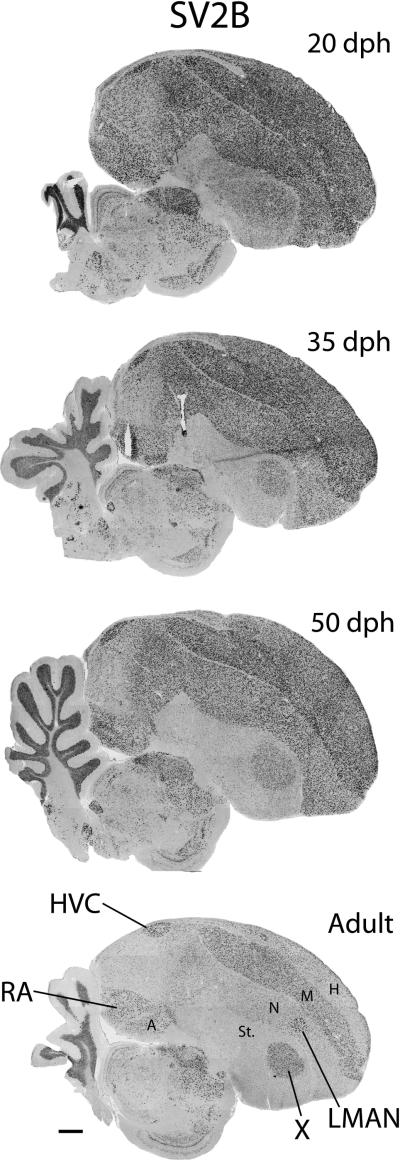
SV2B
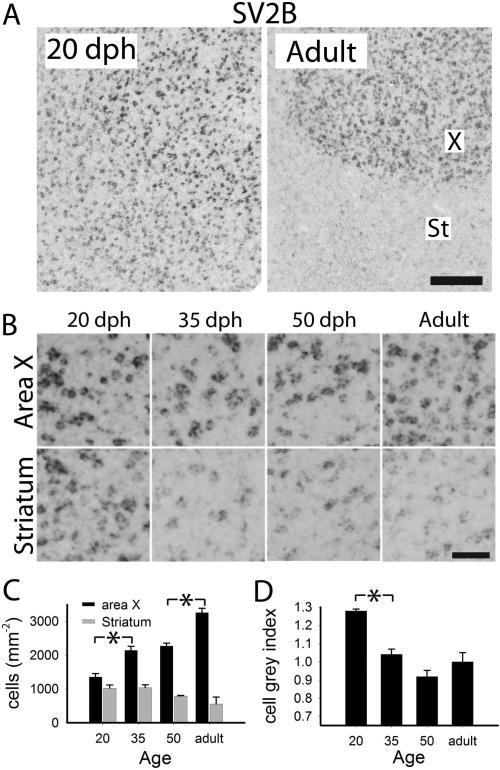
Expression was enriched in the mesopallium and arcopallium, lower in other pallial areas, and very low in most of the striatum (Fig. 11, bottom); high enrichments were seen in parts of the cerebellum and in several brainstem nuclei (not shown), as well as in song nuclei DLM (not shown), LMAN and X, but not RA (Fig. 11, bottom). We noted that this gene was a more robust marker of several song nuclei in adults than in juveniles, an effect that appeared to be largely due to general declines throughout several broad brain subdivisions, including the areas surrounding specific song nuclei (Fig. 11). Quantitative analysis showed no significant age-related changes in labeled cell counts or cell labeling index in HVC (Fig. 6B and C), but a significant decline in cell counts in the shelf between 50 dph and adults (Fig. 6B), leading to the highly differential expression in HVC compared to shelf of adults. As for the age-related change in X (Figs. 8A and B), we detected significant increases in labeled cell counts between 20 and 35 dph, and between 50 dph and adults (Fig. 8C), even though we also saw a significant early decrease in the cell labeling index in this nucleus between 20 and 35 dph (Fig. 8D). Interestingly, we observed no significant age-related decrease in labeled cell counts in the striatum outside of X (Fig. 8C); we thus interpret the apparent marked decline in expression in this area (Fig. 8A and B) as due to a decrease in cell expression levels. Similar findings were noted for LMAN, where the nucleus stood out more clearly in adults relative to 20 dph birds, but labeled cell counts did not differ across age groups (not shown).
Figure 11.
Age-related changes in expression of SV2B. Shown are views of whole parasagittal brain sections at ~2.0 mm from the midline, processed for in situ hybridization. For anatomical abbreviations, see Fig. 1 legend.
Figure 8.
Quantitative analysis of age-related changes of SV2B in area X. A: views of X and adjacent striatum of 20 dph and adult males. B: detailed views of X and striatum across age groups; C: labeled cell counts per area in X (black) and adjacent striatum (grey); D: cell labeling index in X. Plotted in C and D are means + sem; significant age group differences are indicated with a black or grey asterisk (*) for X or striatum, respectively. Scale bars: 100 μm (A), 25 μm (B).
TAC1
Expression was highly enriched in the striatum, particularly the medial portion, including the nucleus accumbens (Acc; Fig. 4S, left), as well as in the dorsolateral corticoid area (CDL; Fig. 4S, right), except for its core (CDLco; Fig. 4T). Expression was much sparser in other pallial areas, and practically absent in LMAN, RA and HVC (Fig. 4U); in the latter, this contrasts with expression in sparse cells in the rostral shelf. We observed high enrichment in X, in contrast with a surrounding striatal ring of lower expression (Fig. 9A, lower right), and apparent developmental changes in X and adjacent St (Fig. 9A and B). Quantitative analysis revealed significant increases in labeled cell counts within X across several ages, but a parallel late decrease (between 50 dph and adults) in cell labeling index in X (Fig. 9C), which may help explain the stronger marker appearance of X in 50 dph than adults (Fig. 9A lower panels). There was also a detectable age-related change in labeled cell counts in the striatum outside of X (Fig. 9C), due to a ~200 μm thick halo region of very low expression that occurred in adults (Fig. 9A, bottom right), but which was not evident in juveniles.
DISCUSSION
Here we establish that over the song learning period HVC and other major song control nuclei of songbirds undergo dynamic changes in the expression of markers associated with a broad range of molecular and cellular functions. Of the markers examined, roughly 70% showed some change between 20 dph and adult (Table 1). Given that these are a representative subset of a much larger set of adult markers (Li et al. 2007, Lovell et al. 2008, Wada et al. 2006), the number of genes that change expression with development could number in the hundreds, or possibly thousands. This would be consistent with the main conclusion from a meta-analysis of the large set of microarray studies associated with the Songbird Neurogenomics (SoNG) initiative (Drnevich et al. 2012), where most genes in the arrays used showed changes in some context (age, brain region, etc.), while only a small minority (~9% of 18,848 genes) appeared to be stable. We also found that the developmental changes fall into distinct groups, according to the timing and direction of the change. These results are largely consistent with early findings of waves of regulated gene expression during key phases of the song learning period (see Clayton 1997), but we also reveal different patterns of regulation, adding to our understanding of how the song system changes during this developmental period. Since we examined only a subset of developmentally regulated genes, it is possible that other genes not yet examined may have yet other developmental trajectories not described here.
Our study establishes several markers (e.g., SLC8A1, KCNT2 and SAP30L) that can be used for the reliable identification of HVC in 20 dph birds, when these nuclei are not clearly visible by cytoarchitectonic features or myelination. Previously, only a limited set of such markers was available, including the androgen receptor, which plays an important role in the masculinization of the circuit (Nottebohm 1980), and retinaldehyde dehydrogenase (ALDH1A2, a.k.a. zRalDH) involved in the local synthesis of retinoic acid (Denisenko-Nehrbass et al. 2000, Gahr and Metzdorf 1997, Kim and Arnold 2005, Kim et al. 2004, Olson et al. 2011). Importantly, the molecular events described here could be possibly associated with developmental changes in the morphological and functional properties of song nuclei and/or in the juvenile vocalizations, but causal links remain to be established.
Of note, the developmental changes we observed in gene expression could be associated with various and complex changes in cell composition and morphology that are known to occur in song nuclei during the song learning period. In HVC, for example, the overall cell density (cells counts per area) increases with age (Bottjer et al. 1985, Bottjer et al. 1986), while the density of HVC-X projecting neurons decreases due to expansion of the nucleus and wider spacing among cells (Gahr and Metzdorf 1999), and HVC-RA projecting cells increase due to neuronal addition (Dewulf and Bottjer 2005, Walton et al. 2012). A precise definition of how these factors affected the patterns we described would require finer analysis involved double-labeling approaches to define cell type specificity of the markers. We note, though, that in several cases we have detected changes in the cell labeling index, which is reflective of expression levels per cell and thus are not affected by the dynamics of cell incorporation, growth or death.
The distinct features of the song nuclei, such as the distinct cytoarchitectonics, heavy myelination and differential expression of several molecular markers compared to adjacent areas, suggest that some of the processes that operate within the song system circuitry differ markedly from those in other brain areas. As discussed below, significant clues about the functional role of developmentally regulated genes and how they might be related to distinct song system features can be obtained by close examination of when and where their expression patterns change.
Gene upregulation in early phases of song learning
Several markers showed higher expression in HVC prior to the beginning of active song learning, decreasing thereafter (Group B), consistent with a postulated early phase of gene regulation (Clayton 1997). Of note, several of these genes represent markers that can be of great help for identifying and delineating HVC in juveniles, when the cytoarchitectonic features of this nucleus are yet fully defined. Because of this time-course and their known functional roles in other systems, these genes are likely associated with processes that are active early during song learning and may become suppressed in adulthood. Transcriptional regulation is one example. As previously reported, the retinoid-generating enzyme, ALDH1A2 is a robust marker of RA at 20 dph but absent in this nucleus at later ages (Denisenko-Nehrbass et al. 2000). Retinoic acid receptors regulate the transcription of a range of targets involved in neuronal generation and differentiation (Chiang et al. 1998, Lane and Bailey 2005, Luo et al. 2009), thus multiple genes of relevance to the song system are likely up-regulated early in RA in association with this signaling pathway. From the present study, SAP30L plays roles in gene silencing as a component of a larger complex that represses DNA-binding histones (Viiri et al. 2009) and regulates the development of the cardiovascular system (Teittinen et al. 2012). Although its role in nervous system development has not to our knowledge been defined, its expression suggests a role as an early transcriptional regulator within HVC.
Other genes in this group play key roles in nervous system development and thus could play analogous roles in the formation and maturation of the song control circuitry. For example, FLRT2 regulates the migration of newly-formed neuronal cells into the subcortical plate during the development of the mammalian cortex (Yamagishi et al. 2011), and thus could influence the incorporation of early populations of newly-formed neurons into the song control circuitry. NRP1 is a semaphorin co-receptor (with plexin) that, when present, switches the axonal guidance properties of growing neuronal processes from repulsive to attractive (Jongbloets and Pasterkamp 2014). Thus, it could be of high significance for establishing specific connections of the song system, while minimizing connectivity in adjacent non-song system areas where its expression is low.
Some genes associated with cell excitability and cation homeostasis also undergo developmental decreases, suggesting that specific electrophysiological processes that operate within parts of the song system circuitry during early phases are downregulated or suppressed as the system matures. KCNT2, a sodium-activated potassium channel (subfamily T, member 2), was previously proposed to play important roles in song plasticity, due to its marked expression in adult X (Lovell et al. 2013). We now show that while its expression in X increases dramatically over the song learning period, both in terms of its expression index and cell numbers, expression in HVC decreases. KCNT2 expression in mammalian auditory areas has been linked to modulation of neuronal spike timing (Yang et al. 2007); by extension, its marked developmental decrease in HVC could be associated with changes in neuronal firing patterns that likely occur as the coding of learned song patterns becomes established in this nucleus (Hahnloser et al. 2002, Nottebohm et al. 1976, Yu and Margoliash 1996). The opposite directional changes in X and HVC could reflect the transition in the control of song production that occurs from the anterior forebrain pathway in juveniles to the direct motor pathway of adults (Aronov et al. 2008). CACNG3 is a subunit of voltage-gated calcium channels (De Waard et al. 1996, Singer et al. 1991), and its developmental downregulation in HVC could reflect changes in intracellular calcium responses to cell depolarization as the song system matures. Alternatively, CACNG3 is also a trans-membrane AMPA receptor modulator (Kato A. S. et al. 2010); together with several related genes that also control the trafficking of AMPA receptors, it is highly enriched and regionally expressed in the mammalian brain, showing differential changes through development (Tomita et al. 2003). The developmental down regulation of CACNG3 in HVC could thus be associated with altered plasticity of glutamatergic transmission (Sager et al. 2009). This is particularly relevant as numerous glutamate receptors have strong differential expression in song nuclei of vocal learning birds (Wada et al. 2004), a feature that likely results from modulatory events during development.
Gene upregulation in late phases of song learning and in adults
Another major type of change and a main focus of the present study consisted of genes whose expression in song nuclei increases during the song learning period, culminating with the strong marker patterns of adults. Based on the functions of this set of genes, their developmental upregulation in HVC might be associated with modulation of age-related rates of neuronal incorporation and maturation in HVC (Wilbrecht and Kirn 2004), as well as with projections and synaptic connections that are formed or mature during late stages of the song learning period. Particularly relevant would be the connections of HVC-to-RA projection neurons, which are continuously formed into adulthood (Wilbrecht and Kirn 2004). ADAM23, for example, a metalloprotease expressed during rodent brain development (Lin et al. 2008), is involved in cleaving membrane-bound proteins important for cell adhesion, cell migration, neurite extension and axonal guidance (Reiss and Saftig 2009), and suppresses neuronal differentiation in P19 cells (Wang et al. 2012). CXCR7, a chemokine receptor (to CXCL12), is involved in the establishment of cortical interneurons at multiple stages of brain development (Sanchez-Alcaniz et al. 2011, Wang et al. 2011); its downregulation leads to an abnormal accumulation of migrating interneurons in the mammalian cortical plate. We also note the intriguing marked upregulation of two collagen genes; given the role this gene family plays in other tissues, these markers may be involved in the stabilization of cell-substrate interactions, which could be of relevance to the incorporation of newly-formed neurons. Lastly, we note the upregulation of MPZL1 (myelin protein zero-like 1), possibly reflecting the myelination of specific projections as the song system matures.
Other upregulated genes may be more closely associated with the differentiation and maturation of neuronal cells in HVC. Among these, PVALB (parvalbumin) is a calcium buffering protein and SLC8A1 encodes a Na+/Ca2+ exchanger that effectively extrudes calcium from the cytoplasm; the marked upregulation of these genes may reflect the emergence of mechanisms that allow for a tight regulation of intracellular calcium levels in the rapidly firing song nuclei in adults. PVALB expressing cells become surrounded by perineuronal nets in the song system nuclei (Balmer et al. 2009), a specialization of extracellular space associated with brain tissue maturation and which is highly developed in adult HVC. TAC1 (tachykinin 1, a.k.a. Substance P) is a modulatory neuropeptide with broad roles in pain perception, behavioral modulation (aggression, anxiety), and learning and memory (Liu et al. 2011), among others. Immunohistochemical characterization of the TAC1 gene product in X suggests localization in woolly fiber terminals of medium spiny neurons, but provide little evidence of staining in the soma to confirm its source in X (Carrillo and Doupe 2004, Reiner et al. 2004a). In contrast, the in situ data that we present here show robust TAC1 positive neurons that are numerous in X, and its strong expression and increase in expression suggest a role in modulating the learning of song as well.
Interestingly, some upregulated genes peaked in their expression at 50 dph, and then declined with further maturation. These include COL12A1 and TAC1, which showed a peak expression in HVC and X respectively. This developmental pattern resembles that previously described for other genes, including NF-M, which peaks in HVC between 35-50 dph (Velho et al. 2007), ALDH1A2, which peaks in HVC between 35-50 dph (Olson et al. 2011) and CNTNAP2, a target of the song-related gene FOXP2 which undergoes increased expression in male LMAN between 50-75 days (Panaitof et al. 2010). We suggest that such genes may be associated with processes that peak around that age (for example, when vocal plasticity is high), but then declines into adulthood.
Gene regulation in areas adjacent to song nuclei
Yet another observed pattern consisted of genes that were stably expressed within song nuclei but declined markedly in adjacent areas or throughout the rest of the brain across the developmental ages examined, emerging as robust song nuclei markers in adults. To our knowledge this type of expression change has not been previously described in finches. We suggest that these cases reflect molecular processes characteristic of early postnatal development that become largely suppressed throughout the brain, but are specifically retained within song nuclei into adulthood. An example is SV2B, a representative synaptic vesicle protein that plays important roles in exocytosis and the release of neurotransmitters, and whose loss results in reduced neurotransmission and synaptic vesicle release, and altered protein composition of synaptic vesicles (Morgans et al. 2009). SV2B is a strong marker of the four major song nuclei in adult finches, a pattern mainly achieved by age-dependent reductions throughout the striatum, nidopallium and hyperpallium, while expression in song nuclei HVC, LMAN and X is largely retained. Similarly, TAC1 decreased substantially in the striatum, particularly in a halo around X, and SAP30L showed large decreases in the nidopallium but not in LMAN. This overall developmental pattern suggests the idea that, compared to the rest of the brain, the adult song nuclei maintain some features characteristic of developmental processes. An intriguing possibility to be tested in the future is that these features could be related to some aspects of plasticity that are retained in adults, e,g, the continued replacement of some specific neuronal populations (Goldman and Nottebohm 1983, Wilbrecht et al. 2006) or the need for continued vocal practice to maintain the adult song (Leonardo and Konishi 1999, Nordeen and Nordeen 1992).
Conclusions
We have expanded our knowledge of zebra finch song system markers that undergo expression changes over the juvenile song learning period, and identified additional useful markers of song nuclei at early developmental ages. We have also identified examples of a notable decline in expression in adjacent tissues in parallel with retained expression within song nuclei; this previously overlooked developmental pattern suggests that some properties of the adult song circuitry that may allow for the maintenance of adult learned song may borrow from earlier developmental processes that occur broadly across the brain. Future directions to test the dual roles of these genes during development and song maintenance (e.g. through conditional knockout experiments) may inform how vocal learning emerged from a non-vocal learning ancestor. Some of the markers with the most marked changes are from families not widely known for their expression in the brain, such as collagens, bringing novel insights into how gene expression regulation may contribute to cell phenotype diversity in the brain. An important next step to establish functional links will be to examine whether manipulating the expression of specific genes affects the maturation of the song circuitry and/or the learning and maturation of birdsong.
Supplementary Material
Supplemental Table 1. Summary of brain transcripts used in this study. Abbreviations are based on HUGO terminology, confirmed by sequence and syntactic alignment of zebra finch genes associated with the clones. Also included are the Human Entrez gene IDs, the Clone IDs, the Gene Name and the Entrez Summary Description, when available.
Table 3.
ANOVA of age-related changes between 20 dph and adult in zebra finch area X and adjacent striatum. See text for a description of variables.
| Gene | F, X labeled cell counts | p, X labeled cell counts | F, Str labeled cell counts | p, Str labeled cell counts | F, X cell labeling index | p, X cell labeling index |
|---|---|---|---|---|---|---|
| KCNT2 | F3,11 = 53.2 | 0.0001 | F3,11 = 5.2 | 0.017 | F3, 11 = 3.2 | 0.067 |
| SV2B | F3,11 = 50.3 | 0.0001 | F3,11 = 3.3 | 0.062 | F3, 11 = 20.2 | <0.0001 |
| TAC1 | F3,11 = 14.1 | 0.0004 | F3,11 = 2.0 | 0.18 | F3, 11 = 15.6 | 0.0003 |
ACKNOWLEDGMENTS
This research was supported by National Institute of Health grants R24-GM092842 (CVM) and F32-NS062609 (CRO). LH was supported by a Howard Hughes Medical Institute Undergraduate Science Education Grant through Lewis and Clark College.
Footnotes
The authors declare that they have no competing interests.
REFERENCES
- Agate RJ, Hertel M, Nottebohm F. FnTm2, a novel brain-specific transcript, is dynamically expressed in the song learning circuit of the zebra finch. J Comp Neurol. 2007;504:127–148. doi: 10.1002/cne.21441. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Kirn JR. Birth, migration, incorporation, and death of vocal control neurons in adult songbirds. J Neurobiol. 1997;33:585–601. [PubMed] [Google Scholar]
- Alvarez-Buylla A, Kirn JR, Nottebohm F. Birth of projection neurons in adult avian brain may be related to perceptual or motor learning. Science. 1990;249:1444–1446. doi: 10.1126/science.1698312. [DOI] [PubMed] [Google Scholar]
- Aronov D, Andalman AS, Fee MS. A specialized forebrain circuit for vocal babbling in the juvenile songbird. Science. 2008;320:630–634. doi: 10.1126/science.1155140. [DOI] [PubMed] [Google Scholar]
- Balmer TS, Carels VM, Frisch JL, Nick TA. Modulation of perineuronal nets and parvalbumin with developmental song learning. J Neurosci. 2009;29:12878–12885. doi: 10.1523/JNEUROSCI.2974-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottjer SW. Developmental regulation of basal ganglia circuitry during the sensitive period for vocal learning in songbirds. Ann N Y Acad Sci. 2004;1016:395–415. doi: 10.1196/annals.1298.037. [DOI] [PubMed] [Google Scholar]
- Bottjer SW, Glaessner SL, Arnold AP. Ontogeny of brain nuclei controlling song learning and behavior in zebra finches. J Neurosci. 1985;5:1556–1562. doi: 10.1523/JNEUROSCI.05-06-01556.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottjer SW, Halsema KA, Brown SA, Miesner EA. Axonal connections of a forebrain nucleus involved with vocal learning in zebra finches. J Comp Neurol. 1989;279:312–326. doi: 10.1002/cne.902790211. [DOI] [PubMed] [Google Scholar]
- Bottjer SW, Miesner EA, Arnold AP. Forebrain lesions disrupt development but not maintenance of song in passerine birds. Science. 1984;224:901–903. doi: 10.1126/science.6719123. [DOI] [PubMed] [Google Scholar]
- Bottjer SW, Miesner EA, Arnold AP. Changes in neuronal number, density and size account for increases in volume of song-control nuclei during song development in zebra finches. Neurosci Lett. 1986;67:263–268. doi: 10.1016/0304-3940(86)90319-8. [DOI] [PubMed] [Google Scholar]
- Braun K, Scheich H, Heizmann CW, Hunziker W. Parvalbumin and calbindin-D28K immunoreactivity as developmental markers of auditory and vocal motor nuclei of the zebra finch. Neuroscience. 1991;40:853–869. doi: 10.1016/0306-4522(91)90017-i. [DOI] [PubMed] [Google Scholar]
- Carleton JB, Lovell PV, McHugh A, Marzulla T, Horback KL, Mello CV. An optimized protocol for high-throughput in situ hybridization of zebra finch brain. Cold Spring Harb Protoc. 2014 doi: 10.1101/pdb.prot084582. 2014: pdb prot084582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo GD, Doupe AJ. Is the songbird Area X striatal, pallidal, or both? An anatomical study. J Comp Neurol. 2004;473:415–437. doi: 10.1002/cne.20099. [DOI] [PubMed] [Google Scholar]
- Chen X, Agate RJ, Itoh Y, Arnold AP. Sexually dimorphic expression of trkB, a Z-linked gene, in early posthatch zebra finch brain. Proc Natl Acad Sci U S A. 2005;102:7730–7735. doi: 10.1073/pnas.0408350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MY, Misner D, Kempermann G, Schikorski T, Giguere V, Sucov HM, Gage FH, Stevens CF, Evans RM. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. Neuron. 1998;21:1353–1361. doi: 10.1016/s0896-6273(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Clayton DF. Role of gene regulation in song circuit development and song learning. J Neurobiol. 1997;33:549–571. [PubMed] [Google Scholar]
- De Waard M, Gurnett CA, Campbell KP. Structural and functional diversity of voltage-activated calcium channels. Ion Channels. 1996;4:41–87. doi: 10.1007/978-1-4899-1775-1_2. [DOI] [PubMed] [Google Scholar]
- Denisenko-Nehrbass NI, Jarvis E, Scharff C, Nottebohm F, Mello CV. Sitespecific retinoic acid production in the brain of adult songbirds. Neuron. 2000;27:359–370. doi: 10.1016/s0896-6273(00)00043-x. [DOI] [PubMed] [Google Scholar]
- Dewulf V, Bottjer SW. Neurogenesis within the juvenile zebra finch telencephalic ventricular zone: a map of proliferative activity. J Comp Neurol. 2005;481:70–83. doi: 10.1002/cne.20352. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Kuhl PK. Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- Drnevich J, et al. Impact of experience-dependent and -independent factors on gene expression in songbird brain. Proc Natl Acad Sci U S A 109 Suppl. 2012;2:17245–17252. doi: 10.1073/pnas.1200655109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eales LA. Song learning in zebra finches: some effects of song model availability on what is learnt and when. Anim Behav. 1985;33:1293–1300. [Google Scholar]
- Foster EF, Bottjer SW. Axonal connections of the high vocal center and surrounding cortical regions in juvenile and adult male zebra finches. J Comp Neurol. 1998;397:118–138. [PubMed] [Google Scholar]
- Gahr M, Metzdorf R. Distribution and dynamics in the expression of androgen and estrogen receptors in vocal control systems of songbirds. Brain Res Bull. 1997;44:509–517. doi: 10.1016/s0361-9230(97)00233-5. [DOI] [PubMed] [Google Scholar]
- Gahr M, Metzdorf R. The sexually dimorphic expression of androgen receptors in the song nucleus hyperstriatalis ventrale pars caudale of the zebra finch develops independently of gonadal steroids. J Neurosci. 1999;19:2628–2636. doi: 10.1523/JNEUROSCI.19-07-02628.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SA, Nottebohm F. Neuronal production, migration, and differentiation in a vocal control nucleus of the adult female canary brain. Proc Natl Acad Sci U S A. 1983;80:2390–2394. doi: 10.1073/pnas.80.8.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnloser RH, Kozhevnikov AA, Fee MS. An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature. 2002;419:65–70. doi: 10.1038/nature00974. [DOI] [PubMed] [Google Scholar]
- Hara E, Rivas MV, Ward JM, Okanoya K, Jarvis ED. Convergent differential regulation of parvalbumin in the brains of vocal learners. PLoS One. 2012;7:e29457. doi: 10.1371/journal.pone.0029457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessler NA, Doupe AJ. Singing-related neural activity in a dorsal forebrainbasal ganglia circuit of adult zebra finches. J Neurosci. 1999;19:10461–10481. doi: 10.1523/JNEUROSCI.19-23-10461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immelman K. Song development in the zebra finch and other estrildid finches. In: Thorpe WH, Hinde RA, editors. Bird vocalizations: their relations to current problems in biology and psychology. Cambridge U.P.; London: 1969. p. xv, 394. [Google Scholar]
- Jarvis ED. Learned birdsong and the neurobiology of human language. Ann N Y Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. For whom the bird sings: context-dependent gene expression. Neuron. 1998;21:775–788. doi: 10.1016/s0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, et al. A framework for integrating the songbird brain. J Comp Physiol A. 2002;188:961–980. doi: 10.1007/s00359-002-0358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloets BC, Pasterkamp RJ. Semaphorin signalling during development. Development. 2014;141:3292–3297. doi: 10.1242/dev.105544. [DOI] [PubMed] [Google Scholar]
- Kao MH, Brainard MS. Lesions of an avian basal ganglia circuit prevent context-dependent changes to song variability. J Neurophysiol. 2006;96:1441–1455. doi: 10.1152/jn.01138.2005. [DOI] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Yu H, Nisenbaum ES, Bredt DS. TARPs differentially decorate AMPA receptors to specify neuropharmacology. Trends Neurosci. 2010;33:241–248. doi: 10.1016/j.tins.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Kato M, Okanoya K. Molecular characterization of the song control nucleus HVC in Bengalese finch brain. Brain Res. 2010;1360:56–76. doi: 10.1016/j.brainres.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Kim YH, Arnold AP. Distribution and onset of retinaldehyde dehydrogenase (zRalDH) expression in zebra finch brain: lack of sex difference in HVC and RA at early posthatch ages. J Neurobiol. 2005;65:260–268. doi: 10.1002/neu.20192. [DOI] [PubMed] [Google Scholar]
- Kim YH, Perlman WR, Arnold AP. Expression of androgen receptor mRNA in zebra finch song system: developmental regulation by estrogen. J Comp Neurol. 2004;469:535–547. doi: 10.1002/cne.11033. [DOI] [PubMed] [Google Scholar]
- Konishi M, Akutagawa E. Neuronal growth, atrophy and death in a sexually dimorphic song nucleus in the zebra finch brain. Nature. 1985;315:145–147. doi: 10.1038/315145a0. [DOI] [PubMed] [Google Scholar]
- Korsia S, Bottjer SW. Developmental changes in the cellular composition of a brain nucleus involved with song learning in zebra finches. Neuron. 1989;3:451–460. doi: 10.1016/0896-6273(89)90204-3. [DOI] [PubMed] [Google Scholar]
- Lane MA, Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol. 2005;75:275–293. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Leonardo A, Konishi M. Decrystallization of adult birdsong by perturbation of auditory feedback. Nature. 1999;399:466–470. doi: 10.1038/20933. [DOI] [PubMed] [Google Scholar]
- Li X, Wang XJ, Tannenhauser J, Podell S, Mukherjee P, Hertel M, Biane J, Masuda S, Nottebohm F, Gaasterland T. Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc Natl Acad Sci. 2007;104:6834–6839. doi: 10.1073/pnas.0701619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Luo J, Redies C. Differential expression of five members of the ADAM family in the developing chicken brain. Neuroscience. 2008;157:360–375. doi: 10.1016/j.neuroscience.2008.08.053. [DOI] [PubMed] [Google Scholar]
- Liu XM, Shu SY, Zeng CC, Cai YF, Zhang KH, Wang CX, Fang J. The role of substance P in the marginal division of the neostriatum in learning and memory is mediated through the neurokinin 1 receptor in rats. Neurochem Res. 2011;36:1896–1902. doi: 10.1007/s11064-011-0511-5. [DOI] [PubMed] [Google Scholar]
- London SE, Boulter J, Schlinger BA. Cloning of the zebra finch androgen synthetic enzyme CYP17: a study of its neural expression throughout posthatch development. J Comp Neurol. 2003;467:496–508. doi: 10.1002/cne.10936. [DOI] [PubMed] [Google Scholar]
- Long MA, Jin DZ, Fee MS. Support for a synaptic chain model of neuronal sequence generation. Nature. 2010;468:394–399. doi: 10.1038/nature09514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Carleton JB, Mello CV. Genomics analysis of potassium channel genes in songbirds reveals molecular specializations of brain circuits for the maintenance and production of learned vocalizations. BMC Genomics. 2013;14:470. doi: 10.1186/1471-2164-14-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong “transcriptomics”: neurochemical specializations of the oscine song system. PLoS One. 2008;3:e3440. doi: 10.1371/journal.pone.0003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Wagner E, Drager UC. Integrating retinoic acid signaling with brain function. Dev Psychol. 2009;45:139–150. doi: 10.1037/0012-1649.45.1.139. [DOI] [PubMed] [Google Scholar]
- Marler P. Birdsong: The acquisition of a learned motor skill. Trends Neurosci. 1981;4:88–94. [Google Scholar]
- Mello CV, Jarvis ED, Denisenko N, Rivas M. Isolation of song-regulated genes in the brain of songbirds. Methods Mol Biol. 1997;85:205–217. doi: 10.1385/0-89603-489-5:205. [DOI] [PubMed] [Google Scholar]
- Mello CV, Vates GE, Okuhata S, Nottebohm F. Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata). J Comp Neurol. 1998;395:137–160. [PubMed] [Google Scholar]
- Morgans CW, Kensel-Hammes P, Hurley JB, Burton K, Idzerda R, McKnight GS, Bajjalieh SM. Loss of the Synaptic Vesicle Protein SV2B results in reduced neurotransmission and altered synaptic vesicle protein expression in the retina. PLoS One. 2009;4:e5230. doi: 10.1371/journal.pone.0005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordeen KW, Nordeen EJ. Auditory feedback is necessary for the maintenance of stereotyped song in adult zebra finches. Behav Neural Biol. 1992;57:58–66. doi: 10.1016/0163-1047(92)90757-u. [DOI] [PubMed] [Google Scholar]
- Nottebohm F. The origins of vocal learning. American Naturalist. 1972;106:116–140. [Google Scholar]
- Nottebohm F. Testosterone triggers growth of brain vocal control nuclei in adult female canaries. Brain Res. 1980;189:429–436. doi: 10.1016/0006-8993(80)90102-x. [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Kelley DB, Paton JA. Connections of vocal control nuclei in the canary telencephalon. J Comp Neurol. 1982;207:344–357. doi: 10.1002/cne.902070406. [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- Olson CR, Rodrigues PV, Jeong JK, Prahl DJ, Mello CV. Organization and development of zebra finch HVC and paraHVC based on expression of zRalDH, an enzyme associated with retinoic acid production. J Comp Neurol. 2011;519:148–161. doi: 10.1002/cne.22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olveczky BP, Andalman AS, Fee MS. Vocal experimentation in the juvenile songbird requires a basal ganglia circuit. PLoS Biol. 2005;3:e153. doi: 10.1371/journal.pbio.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaitof SC, Abrahams BS, Dong H, Geschwind DH, White SA. Languagerelated Cntnap2 gene is differentially expressed in sexually dimorphic song nuclei essential for vocal learning in songbirds. J Comp Neurol. 2010;518:1995–2018. doi: 10.1002/cne.22318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenning AR, et al. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science. 2014;346:1256846. doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud R, Saldanha CJ, Wynne RD, Lovell PV, Mello CV. The excitatory thalamo–“cortical” projection within the song control system of zebra finches is formed by calbindin-expressing neurons. J Comp Neurol. 2007;504:601–618. doi: 10.1002/cne.21457. [DOI] [PubMed] [Google Scholar]
- Pytte CL, Gerson M, Miller J, Kirn JR. Increasing stereotypy in adult zebra finch song correlates with a declining rate of adult neurogenesis. Dev Neurobiol. 2007;67:1699–1720. doi: 10.1002/dneu.20520. [DOI] [PubMed] [Google Scholar]
- Reiner A, Laverghetta AV, Meade CA, Cuthbertson SL, Bottjer SW. An immunohistochemical and pathway tracing study of the striatopallidal organization of area X in the male zebra finch. J Comp Neurol. 2004a;469:239–261. doi: 10.1002/cne.11012. [DOI] [PubMed] [Google Scholar]
- Reiner A, Perkel DJ, Mello CV, Jarvis ED. Songbirds and the revised avian brain nomenclature. Ann N Y Acad Sci. 2004b;1016:77–108. doi: 10.1196/annals.1298.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K, Saftig P. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol. 2009;20:126–137. doi: 10.1016/j.semcdb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Replogle K, et al. The Songbird Neurogenomics (SoNG) Initiative: communitybased tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper A, Zann R. The Onset of Song Learning and Song Tutor Selection in Fledgling Zebra Finches. Ethology. 2006;112:458–470. [Google Scholar]
- Roubelakis MG, Martin-Rendon E, Tsaknakis G, Stavropoulos A, Watt SM. The murine ortholog of the SHP-2 binding molecule, PZR accelerates cell migration on fibronectin and is expressed in early embryo formation. J Cell Biochem. 2007;102:955–969. doi: 10.1002/jcb.21334. [DOI] [PubMed] [Google Scholar]
- Sager C, Tapken D, Kott S, Hollmann M. Functional modulation of AMPA receptors by transmembrane AMPA receptor regulatory proteins. Neuroscience. 2009;158:45–54. doi: 10.1016/j.neuroscience.2007.12.046. [DOI] [PubMed] [Google Scholar]
- Sanchez-Alcaniz JA, Haege S, Mueller W, Pla R, Mackay F, Schulz S, Lopez-Bendito G, Stumm R, Marin O. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Scharff C, Nottebohm F. A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: implications for vocal learning. J Neurosci. 1991;11:2896–2913. doi: 10.1523/JNEUROSCI.11-09-02896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M, Schmidt M, Carruth LL. Distribution and subcellular localization of glucocorticoid receptor-immunoreactive neurons in the developing and adult male zebra finch brain. Gen Comp Endocrinol. 2011;174:354–361. doi: 10.1016/j.ygcen.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Singer D, Biel M, Lotan I, Flockerzi V, Hofmann F, Dascal N. The roles of the subunits in the function of the calcium channel. Science. 1991;253:1553–1557. doi: 10.1126/science.1716787. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Qin W, Leggett MH. A minimally invasive procedure for sexing young zebra finches. J Neurosci Methods. 2007;164:116–119. doi: 10.1016/j.jneumeth.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabji F, Nordeen EJ, Nordeen KW. Selective impairment of song learning following lesions of a forebrain nucleus in the juvenile zebra finch. Behav Neural Biol. 1990;53:51–63. doi: 10.1016/0163-1047(90)90797-a. [DOI] [PubMed] [Google Scholar]
- Teittinen KJ, et al. SAP30L (Sin3A-associated protein 30-like) is involved in regulation of cardiac development and hematopoiesis in zebrafish embryos. J Cell Biochem. 2012;113:3843–3852. doi: 10.1002/jcb.24298. [DOI] [PubMed] [Google Scholar]
- Tomaszycki ML, Peabody C, Replogle K, Clayton DF, Tempelman RJ, Wade J. Sexual differentiation of the zebra finch song system: potential roles for sex chromosome genes. BMC Neurosci. 2009;10:24. doi: 10.1186/1471-2202-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS. Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol. 2003;161:805–816. doi: 10.1083/jcb.200212116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vates GE, Broome BM, Mello CV, Nottebohm F. Auditory pathways of caudal telencephalon and their relation to the song system of adult male zebra finches. J Comp Neurol. 1996;366:613–642. doi: 10.1002/(SICI)1096-9861(19960318)366:4<613::AID-CNE5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Vates GE, Nottebohm F. Feedback circuitry within a song-learning pathway. Proc Natl Acad Sci. 1995;92:5139–5143. doi: 10.1073/pnas.92.11.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vates GE, Vicario DS, Nottebohm F. Reafferent thalamo–“cortical” loops in the song system of oscine songbirds. J Comp Neurol. 1997;380:275–290. [PubMed] [Google Scholar]
- Velho TA, Lovell P, Mello CV. Enriched expression and developmental regulation of the middle-weight neurofilament (NF-M) gene in song control nuclei of the zebra finch. J Comp Neurol. 2007;500:477–497. doi: 10.1002/cne.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viiri KM, Heinonen TY, Maki M, Lohi O. Phylogenetic analysis of the SAP30 family of transcriptional regulators reveals functional divergence in the domain that binds the nuclear matrix. BMC Evol Biol. 2009;9:149. doi: 10.1186/1471-2148-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucicevic M, Stevanov-Pavlovic M, Stevanovic J, Bosnjak J, Gajic B, Aleksic N, Stanimirovic Z. Sex determination in 58 bird species and evaluation of CHD gene as a universal molecular marker in bird sexing. Zoo Biol. 2013;32:269–276. doi: 10.1002/zoo.21010. [DOI] [PubMed] [Google Scholar]
- Wada K, et al. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Sakaguchi H, Jarvis ED, Hagiwara M. Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J Comp Neurol. 2004;476:44–64. doi: 10.1002/cne.20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J. TrkB-like immunoreactivity in the song system of developing zebra finches. J Chem Neuroanat. 2000;19:33–39. doi: 10.1016/s0891-0618(00)00051-x. [DOI] [PubMed] [Google Scholar]
- Wade J, Tang YP, Peabody C, Tempelman RJ. Enhanced gene expression in the forebrain of hatchling and juvenile male zebra finches. J Neurobiol. 2005;64:224–238. doi: 10.1002/neu.20141. [DOI] [PubMed] [Google Scholar]
- Walton C, Pariser E, Nottebohm F. The zebra finch paradox: song is little changed, but number of neurons doubles. J Neurosci. 2012;32:761–774. doi: 10.1523/JNEUROSCI.3434-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, Pleasure SJ, Behrens T, Rubenstein JL. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun Y, Qiao S. ADAM23 knockdown promotes neuronal differentiation of P19 embryonal carcinoma cells by up-regulating P27KIP1 expression. Cell Biol Int. 2012;36:1275–1279. doi: 10.1042/CBI20120154. [DOI] [PubMed] [Google Scholar]
- Wilbrecht L, Kirn JR. Neuron addition and loss in the song system: regulation and function. Ann N Y Acad Sci. 2004;1016:659–683. doi: 10.1196/annals.1298.024. [DOI] [PubMed] [Google Scholar]
- Wilbrecht L, Williams H, Gangadhar N, Nottebohm F. High levels of new neuron addition persist when the sensitive period for song learning is experimentally prolonged. J Neurosci. 2006;26:9135–9141. doi: 10.1523/JNEUROSCI.4869-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild JM. Descending projections of the songbird nucleus robustus archistriatalis. J Comp Neurol. 1993;338:225–241. doi: 10.1002/cne.903380207. [DOI] [PubMed] [Google Scholar]
- Wild JM. Functional neuroanatomy of the sensorimotor control of singing. Ann N Y Acad Sci. 2004;1016:438–462. doi: 10.1196/annals.1298.016. [DOI] [PubMed] [Google Scholar]
- Wild JM, Williams MN, Suthers RA. Parvalbumin-positive projection neurons characterise the vocal premotor pathway in male, but not female, zebra finches. Brain Res. 2001;917:235–252. doi: 10.1016/s0006-8993(01)02938-9. [DOI] [PubMed] [Google Scholar]
- Williams H. Birdsong and singing behavior. Ann N Y Acad Sci. 2004;1016:1–30. doi: 10.1196/annals.1298.029. [DOI] [PubMed] [Google Scholar]
- Yamagishi S, et al. FLRT2 and FLRT3 act as repulsive guidance cues for Unc5-positive neurons. EMBO J. 2011;30:2920–2933. doi: 10.1038/emboj.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Desai R, Kaczmarek LK. Slack and Slick K(Na) channels regulate the accuracy of timing of auditory neurons. J Neurosci. 2007;27:2617–2627. doi: 10.1523/JNEUROSCI.5308-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AC, Margoliash D. Temporal hierarchical control of singing in birds. Science. 1996;273:1871–1875. doi: 10.1126/science.273.5283.1871. [DOI] [PubMed] [Google Scholar]
- Zeigler HP, Marler P. Behavioral Neurobiology of Birdsong.. Ann N Y Acad Sci; Proceedings of a conference; New York, New York, USA. 12-14 December 2002; 2004. pp. xiii–xvii.pp. 1–788. [PubMed] [Google Scholar]
- Zeigler HP, Marler P. Neuroscience of birdsong. Cambridge University Press; 2008. [Google Scholar]
- Zuschratter W, Scheich H, Heizmann CW. Ultrastructural localization of the calcium-binding protein parvalbumin in neurons of the song system of the zebra finch, Poephila guttata. Cell Tissue Res. 1985;241:77–83. doi: 10.1007/BF00214628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Summary of brain transcripts used in this study. Abbreviations are based on HUGO terminology, confirmed by sequence and syntactic alignment of zebra finch genes associated with the clones. Also included are the Human Entrez gene IDs, the Clone IDs, the Gene Name and the Entrez Summary Description, when available.