Abstract
Recently, Alazami and colleagues identified 33 putative candidate disease genes for neurogenetic disorders. One such gene was DPH1, in which a homozygous missense mutation was associated with a 3C syndrome-like phenotype in four patients from a single extended family. Here we report a second homozygous missense variant in DPH1, seen in four members of a founder population, and associated with a phenotype initially reminiscent of Sensenbrenner syndrome. This post-publication ‘match’ validates DPH1 as a gene underlying syndromic intellectual disability with short stature and craniofacial and ectodermal anomalies, reminiscent of, but distinct from, 3C and Sensenbrenner syndromes. This validation took several years after the independent discoveries due to the absence of effective methods for sharing both candidate phenotype and genotype data between investigators. Sharing of data via web-based anonymous data exchange servers will play an increasingly important role towards more efficient identification of the molecular basis for rare Mendelian disorders.
Keywords: DPH1, Sensenbrenner, intellectual disability, rare disorders, Matchmaker Exchange
Although individually rare (defined as affecting fewer than 200 000 people in the United States or fewer than 1 in 2 000 people in Europe), millions of people worldwide are affected by rare disorders. Of an estimated 7 000 rare disorders, the genetic basis is still unknown for approximately half of them (http://www.omim.org). Genetic diagnosis is a critical first step in providing patients with appropriate care and families with accurate and timely genetic counseling. The advent of next generation sequencing technologies has accelerated the rate of novel gene discovery in single-gene disorders [Beaulieu et al., 2014], but several challenges still exist for rare disorders. Novel gene discovery, with subsequent provision of clinical genetic testing, requires definitive evidence of causality, often derived from identifying multiple pathogenic variants in a common gene underlying the same disorder. Finding additional, unrelated patients becomes increasingly difficult as the rarity of the disorder increases. In addition, the scarcity of patients with rare disorders can also limit the ability to determine the complete phenotypic spectrum associated with variants of a particular gene, and hence the understanding of the phenotype may be biased by the earliest publications of the gene in question.
Genes underlying rare disorders have historically typically been successfully identified through collection of small to medium sized cohorts of patients; frequently requiring international collaboration. For ultra-rare phenotypes in particular this has frequently been accomplished at “unknown” syndrome sessions at international meetings or by means of personal communication between research groups sharing unpublished phenotypes or genetic information. This approach is far from systematic and can result in significant delays in new gene discovery and publication, which consequently delays the understanding of rare disorders. As a result, affected patients and families are often unable to access clinical tests, such as carrier testing (of particular importance in rare autosomal recessive disorders in founder populations) or prenatal diagnosis. An alternative, and evolving, approach is to use a matchmaking process that allows for the systematic sharing of annotated phenotypic data without the need to reveal confidential data [Gottlieb et al., 2015]. Once common phenotypic presentations are identified between patients, genetic data can be shared in the hopes of identifying pathogenic variants disrupting a common gene [Gottlieb et al., 2015]. A reverse approach to matchmaking can also be implemented using an online-based tool such as GeneMatcher [Sobreira et al., 2015] where investigators can post genes of interest. Data remains confidential because the database cannot be searched and no identifiable data is collected. Investigators are only notified once a match is made, and can then decide whether or not to share their data.
In this report, we outline a situation in which the implementation of matchmaking could have successfully connected two cohorts of patients identified in our respective North American and Saudi Arabian centers, with phenotypic similarities and pathogenic variants in the same gene: DPH1 (MIM# 603527). Instead, these phenotypic similarities and a common mutated gene were only brought to light after the publication of a report of single mutations in multiple novel candidate genes for several autosomal recessive disorders [Alazami et al., 2015], resulting in a post-publication match process. While reporting of novel phenotypes in case reports has and will continue to be a highly necessary activity in clinical genetics, this approach will not be sufficient to further identify the underlying molecular basis for the hundreds of ‘unsolved’ rare disorders. Given that it is time consuming, and not always appropriate, to report single mutations in putative novel genes in the medical literature, post-publication matchmaking is an inherently inefficient, slow and haphazard process. Matching techniques based on sharing of candidate phenotype and genotype data via web-based anonymous data exchange servers could greatly improve the efficiency of pathogenic variant discovery in rare disorders by systematically connecting clinician researchers at an earlier stage in the process, thereby facilitating more timely and conclusive publications.
Over the course of the last decade, four individuals (two siblings and two sporadic probands – patients 1–4) from a North American genetic isolate were identified with developmental delay, dysmorphic features (including scaphocephaly and sparse scalp hair), and frequent short stature (Table 1, Fig. 1 and Supporting Information). The phenotype was considered most consistent with Sensenbrenner syndrome (cranioectodermal dysplasia; MIM# 218330), however there were key differences identified even in the early stages, including the presence of developmental delay that is atypical of Sensenbrenner syndrome. Our study patients also did not have significant retinal or hepatic disease, and only one patient had mild renal disease rendering classic Sensenbrenner syndrome an unlikely diagnosis, suggesting that the clinical features seen in these patients potentially represent a novel rare disorder.
Table 1.
Clinical features of both North American and Saudi Arabian patients
| North American Patients | Saudi Arabian Patients | # of patients with features | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Features | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Developmental delay/Intellectual Disability | + | + | + | + | + | + | + | + | 8/8 |
| Unusual skull shape (including scaphocephaly or prominent forehead) with or without craniosynostosis | + | + | + | + | + | + | + | + | 8/8 |
| Dysmorphic features (hypertelorism, downslanting palperbral fissure, epicanthal folds, low-set ears, depressed nasal bridge and micrognathia) | + | + | + | + | + | + | + | + | 8/8 |
| Sparse hair on scalp/eyelashes, eyebrows | + | + | + | + | + | + | + | + | 8/8 |
| Short stature | + | + | + | - | + | + | + | + | 7/8 |
| Renal anomaly | + | - | - | - | + | + | + | - | 4/8 |
| Central nervous system malformations (Dandy-Walker malformation, cerebellar vermis hypoplasia and posterior fossa cyst) | ND | ND | - | - | + | + | + | + | 4/6 |
| Ventricular septal defect (heart) | - | - | - | - | + | + | - | + | 3/8 |
| Early Lethality (age of death) | - | - | - | - | - | 8m | 4y | 18m | 3/8 |
Figure 1.
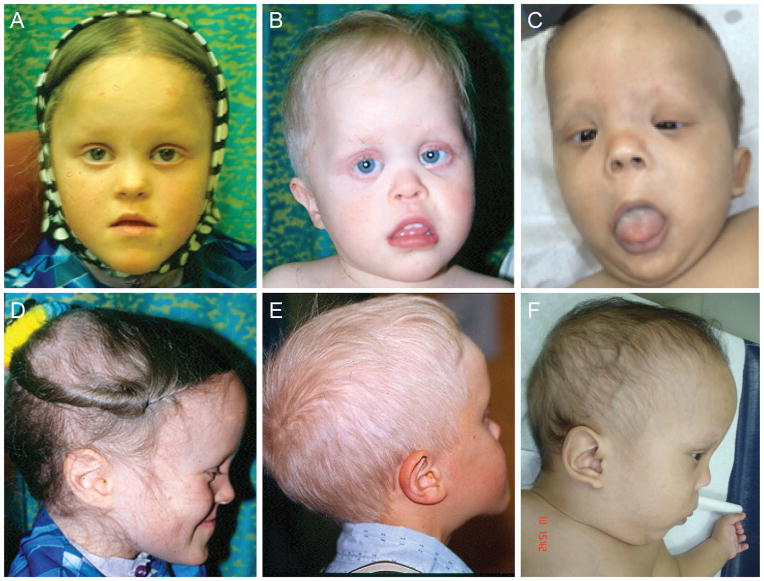
Clinical pictures of patients with homozygous missense variants in DPH1. A,D: Patient 1, age 8. B,E: Patient 2, age 3 (frontal) and age 8 (lateral). C,F: Saudi Arabian patient (Patient 5), age 14 months. Note midface hypoplasia, sparse eyebrows, epicanthal folds, upturned nose and downturned mouth corners. A prominent forehead is seen in all patients, with elongated skull shape in patients 1 and 2, and trigoncephaly in patient 5.
Patient 1 was born to second cousin parents. She was suspected to have Down syndrome in the neonatal period and has a history of mild to moderate developmental delay. She walked at 15 months and developed full sentences at age 2. On examination, she was found to have short stature, sparse hair and eyebrows, scaphocephaly, abnormal toenails and pes planus (Fig. 1). She has a history of absence and generalized seizures that have been easily controlled. Although she has had normal renal function and imaging, recurrent hematuria and proteinuria on urinalysis in the context of a possible diagnosis of Sensenbrenner syndrome, prompted a renal biopsy that revealed evidence of mild focal interstitial nephritis and other non-specific changes. Now age 23, she has short stature, anxiety and mild-moderate cognitive issues such that she is not able to live fully independently. She however has been medically stable, with no ongoing evidence of renal disease. Patient 2 is the younger brother of patient 1. He presented to genetics prior to his sister owing to his more obvious developmental delay and more striking dysmorphic features. He walked at 18 months and spoke words at age 2. He has had ongoing academic concerns, performing at a grade 2 level at age 12. He presented with short stature and multiple dysmorphic features including prominent forehead, scaphocephaly, sparse eyebrows, sparse and coarse scalp hair, epicanthal folds, small hands and hypoplastic toenails (Fig. 1). In addition, he has slight esotropia, mild hearing loss and a history of increased urinary frequency but no obvious signs of retinal or renal disease. Now at age 18, he has moderate cognitive disability and short stature, but has been medically healthy. Patient 3 is another child belonging to the same genetic isolate. She was born to second cousins and is not closely related to patients 1 and 2. She was referred to genetics in the immediate newborn period for multiple dysmorphic features (referring physician questioned Down syndrome and achondroplasia). She also had developmental delay where at age 8 she was found to have a full scale IQ (intelligence quotient) of 48. She presented with short stature, sagittal craniosynostosis and dysmorphic features including scaphocephaly, sparse hair, multiple dental anomalies (including 2 natal teeth lost at age 3), epicanthal folds and hypoplastic toenails. She has a history of enuresis, but normal renal function to date. High myopia is present without retinal degeneration. Cranial imaging via CT scan was done and was essentially normal. Patient 4 was initially assessed at 17 months of age due to a history of failure to thrive, global developmental delay and dysmorphic features. Her parents are second cousins and are also members of the same genetic isolate. She was most recently assessed at 4 years of age at which time her height, weight and head circumference were at the 90th, 50th and >97th centiles respectively. She had a history of sleep apnea, bilateral inguinal hernia repairs but was otherwise healthy with an unremarkable review of systems. By 4 years of age, she was accessing additional developmental services and was diagnosed with moderate global developmental delays as well as attention deficit hyperactivity disorder - inattentive type. Her dysmorphic features included relative macrocephaly with a tall and prominent forehead, sparse eyebrows, downslanting palpebral fissures, and a triangular face. Her clinical features were similar to the other three patients. Cranial MRI, abdominal ultrasound and array CGH were normal.
At the start of this work, only three of the four North American patients had been ascertained. The causative variant for these patients was expected to lie within a homozygous chromosomal region inherited from a common ancestor. To find this homozygous region, the genetic study of North American families was conducted with approval from the Conjoint Health Research Ethics Board at the University of Calgary. A large region spanning ~9.8 Mb was found on chromosome 17 using a 10 K SNP array (Supp. Figure S1) and confirmed using microsatellite markers (Supp. Figure S2A). Exome sequencing was then performed on patient 3 using the SureSelect Human All Exon v2 kit (Agilent Technologies) followed by paired-end sequencing on an Illumina GAII. Variants were treated as candidate causative variants if they were homozygous, present in the homozygous region identified on chromosome 17, absent from databases housing common variants (dbSNP, exome variant server and the 1000 Genomes Project) and resulted in frameshift, nonsense, nonsynonymous or splice-site changes. Using these criteria, only one candidate pathogenic variant was uncovered, NM_001383.3 (DPH1):c.17T>A (ClinVar SCV000223815, http://www.ncbi.nlm.nih.gov/clinvar/). This variant results in the substitution of lysine for a highly conserved methionine (p.Met6Lys). In addition, it was predicted to be benign by PolyPhen-2 (version 2.2.2r398 (http://genetics.bwh.harvard.edu/pph2/); HumVar score 0.142 (sensitivity: 0.90, specificity: 0.71)) [Adzhubei et al., 2010] and tolerated by SIFT (http://sift.jcvi.org/www/SIFT_aligned_seqs_submit.html; score of 1.00 (median sequence conservation score 3.50 across 6 sequences) [Kumar et al., 2009] using default parameters as set by Alamut Visual version 2.6.0 (Interactive Biosoftware, Rouen, France). Repeat variant assessment using current databases and prediction tools are consistent with the original outcomes, and in addition the variant is absent from the ExAC database. Sanger sequencing confirmed that this variant was present in all three patients. We then identified an additional sporadic patient (Patient 4) from the same genetic isolate (Table 1, Fig. 1 and Supporting Information), and Sanger sequencing revealed she also carried the variant in homozygosity.
Haplotype analysis including patient 4 revealed only a small homozygous region of ~1.9 Mb on chromosome 17 surrounding DPH1 that was shared by all four patients (Supp. Figure S2B). A custom TaqMan allelic discrimination assay (Applied Biosystems) was developed to genotype the c.17T>A DPH1 variant in healthy individuals from the genetic isolate to which the patients belonged. The A variant was never seen in a homozygous state, but it was seen in 5 of 1098 chromosomes, giving a minor allele frequency of 0.46% in the population. This is consistent with the c.17T>A variant being a founder mutation within this population. Together, these data suggest that the c.17T>A variant likely underlies this disorder for these patients belonging to this North American genetic isolate, however the possibility that this variant simply represented a rare variant in this population that is in tight linkage disequilibrium with the actual disease-causing variant could not be confidently excluded. Furthermore, we were unable to find additional patients outside of the genetic isolate with similar clinical presentations and pathogenic variants in DPH1 to support the suspected causality of the discovered gene. This variant was initially identified in 2010, and over the years attempts were made to identify similarly affected patients through informal presentation at ‘unknown’ sessions at meetings; formal presentation of the variant at scientific conferences with available internet searchable abstract (http://www.ichg2011.org/cgi-bin/showdetail.pl?absno=21415) and more recently (November, 2014) a submission to GeneMatcher [Sobreira et al., 2015] without success.
In parallel, Alazami et al. identified a single homozygous missense variant NM_001383.3(DPH1):c.701T> C, which results in the substitution of a highly conserved amino acid (p.Leu234Pro) predicted to be possibly damaging and deleterious by PolyPhen-2 and SIFT, segregating in four affected individuals (patients 5–8) belonging to a previously published extended consanguineous Saudi Arabian family with clinical features thought to most closely resemble 3C (cranio-cerebello-cardiac, Ritscher-Schinzel; MIM# 220210) syndrome but with renal involvement [Seidahmed et al, 2011]. Although the DPH1-associated disorder found in the North American genetic isolate was originally classified as a different disorder (a variant of Sensenbrenner syndrome), the presence of DPH1 pathogenic variants in both sets of patients prompted a reappraisal of the clinical features of all patients (Table 1 and Fig. 1). Differences exist both within and between the two sets of patients, however, common features seen in most patients include short stature, intellectual disability, ectodermal features (sparse hair on the scalp and eyebrows) and unusual skull shape (prominent forehead with trigonocephaly or scaphocephaly, with or without confirmed craniosynostosis) and dysmorphic features including epicanthal folds and upturned nose. Central nervous system malformations and early lethality, in particular, were more frequent in the Saudi Arabian patients, which could reflect severity of pathogenic variant, or other genetic factors (either background/modifying factors, or the presence of other highly penetrant mutations). Together, these data show that recessive pathogenic variants in DPH1 are causative of a developmental phenotype in humans. These data also indicate that the two cohorts of patients may represent different points on a phenotypic spectrum, although the identification of further families will be required to fully appreciate the spectrum of DPH1-related disease.
Consistent with DPH1 variants causing developmental phenotypes in patients investigated in this report, pathogenic variants in Dph1 result in developmental defects in mice. Dph1-deficient mice die perinatally and show restricted growth, developmental defects, cleft palate and craniofacial abnormalities [Chen and Behringer, 2004; Yu et al., 2014]. DPH1 was originally recognized as a tumour suppressor [Phillips et al., 1996] and known as OVCA1 (ovarian cancer-associated gene 1) but was later discovered to be required for the biosynthesis of diphthamide, a posttranslational modification of a conserved histidine residue of eukaryotic elongation factor 2 (EEF2). This modification is specifically targeted by diphtheria toxin to impair protein translation, resulting in cell death [Van Ness et al., 1980] and has been hypothesized to stabilize codon/anticodon interactions for efficient translation [Ortiz et al., 2006; Liu et al., 2012]. Interestingly, DPH1 is included in the Miller-Dieker syndrome (MDS; MIM# 247200) critical deletion region, prompting the investigation of the role of DPH1 in craniofacial development in a recent publication [Yu et al., 2014]. Conditional ablation of Dph1 in neural crest cells resulted in craniofacial abnormalities with hypoplastic nasal bone and lower jaw [Yu et al., 2014], supporting a role for DPH1 in craniofacial development and lending support for DPH1 pathogenic variants contributing to the craniofacial anomalies seen in patients investigated in this report.
One of the remaining major challenges for the clinical genetics community is to better understand the full spectrum of rare Mendelian disorders. In the absence of conclusive functional data, the identification of similar patients with comparable phenotypes is frequently necessary to confirm pathogenicity of novel gene variants. This is difficult for rare disorders that often occur in the context of founder effect and consanguinity and has in part prompted several publications [Najamabadi et al., 2011; Puffenberger et al., 2012; Alazami et al., 2015] that jointly identified nearly one hundred candidate autosomal recessive disorder genes based on single or few pedigrees, typically with a single homozygous pathogenic variant. One such publication identified DPH1 as a candidate gene associated with an autosomal recessive developmental disorder. An important consideration is at what point these candidate genes become definitively disease-associated, frequently resulting in the delineation of a novel genetic syndrome, thereby ending the diagnostic odyssey for affected patients and allowing for clinical testing and potentially targeted therapy. In this report, we validated DPH1 as disease-associated through a post-publication match made possible by the identification of a second novel homozygous missense variant in DPH1 that provides additional support that dysfunction of this gene causes an autosomal recessive developmental disorder. Both groups had identified their candidate variants two to five years prior to this publication and had tried several conventional means to identify further cases. This match in fact came to light when one of the authors received an alert to a new article reporting 33 novel candidate genes for several autosomal recessive disorders [Alazami et al., 2015], and in reviewing this paper while stuck in traffic at a stoplight, came across a gene of interest DPH1 buried within the paper. While this approach was successful in this case, it is not likely to be scalable or reliable, in addition to being against the distracted driving laws of many jurisdictions. In this case, the systematic sharing of both phenotype and genotypic data [Gottlieb et al., 2015; Sobreira et al 2015] prior to publication could have helped to connect the two cohorts of patients with pathogenic variants in DPH1. The phenotypes while in the end, highly overlapping, are also disparate and may have prevented a pre-publication match if only phenotypes were considered, while sharing of genetic data would have resulted in a match. This will likely continue to be common for such rare diseases and highlights the importance of sharing both phenotypic and genotypic data in order to fully benefit from pre-publication matching opportunities. This is particularly relevant in this new era of ‘reverse dysmorphology’ where the phenotypic overlap is studied in detail, only after identification of the shared genetic basis; a term first coined in relation to copy number variation [Firth et al., AJHG 2009] and will likely continue to be the case due to accelerated pathogenic variant discovery facilitated by next generation sequencing technologies.
Supplementary Material
Acknowledgments
We thank the patients and their families for participation in this study. This work was supported in part by Basil O’Connor Starter Scholar Research Award Grant No. 5-FY09-529 from the March of Dimes Foundation to K.M.B., the Alberta Children’s Hospital Foundation to J.S.P., F.P.B, and A.M.I. and National Institute of Health Grant No. R01 HL085197 to C.O. C.M.L. was supported by the CIHR Training Program in Genetics, Child Development, and Health at the University of Calgary.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–161. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Chen C-M, Behringer RR. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 2004;18:320–332. doi: 10.1101/gad.1162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb MM, Arenillas DJ, Maithripala S, Maurer ZD, Tarailo Graovac M, Armstrong L, Patel M, van Karnebeek C, Wasserman WW. GeneYenta: A Phenotype-Based Rare Disease Case Matching Tool Based on Online Dating Algorithms for the Acceleration of Exome Interpretation. Hum Mutat. 2015;36:432–438. doi: 10.1002/humu.22772. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Liu S, Bachran C, Gupta P, Miller-Randolph S, Wang H, Crown D, Zhang Y, Wein AN, Singh R, Fattah R, Leppla SH. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proceedings of the National Academy of Sciences. 2012;109:13817–13822. doi: 10.1073/pnas.1206933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, Zecha A, Mohseni M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem. 2006;281:32639–32648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- Phillips NJ, Ziegler MR, Radford DM, Fair KL, Steinbrueck T, Xynos FP, Donis-Keller H. Allelic deletion on chromosome 17p13.3 in early ovarian cancer. Cancer Res. 1996;56:606–611. [PubMed] [Google Scholar]
- Puffenberger EG, Jinks RN, Sougnez C, Cibulskis K, Willert RA, Achilly NP, Cassidy RP, Fiorentini CJ, Heiken KF, Lawrence JJ, Mahoney MH, Miller CJ, et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS ONE. 2012;7:e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidahmed MZ, Alkuraya FS, Shaheed M, Zahrani Al M, Manea Al W, Mansour F, Mustafa T, Farid G, Salih MA. Ritscher-Schinzel (cranio-cerebello-cardiac, 3C) syndrome: report of four new cases with renal involvement. Am J Med Genet A. 2011;155A:1393–1397. doi: 10.1002/ajmg.a.33966. [DOI] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New Tools for Mendelian Disease Gene Identification: PhenoDB Variant Analysis Module; and GeneMatcher, a Web-Based Tool for Linking Investigators with an Interest in the Same Gene. Hum Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem. 1980;255:10717–10720. [PubMed] [Google Scholar]
- Yu Y-R, You L-R, Yan Y-T, Chen C-M. Role of OVCA1/DPH1 in craniofacial abnormalities of Miller-Dieker syndrome. Human Molecular Genetics. 2014;23:5579–5596. doi: 10.1093/hmg/ddu273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.