Summary
Three subsets of invariant natural killer T (iNKT) cells have been identified, NKT1, NKT2 and NKT17, which produce distinct cytokines when stimulated, but little is known about their localization. Here, we have defined the anatomic localization and systemic distribution of these subsets and measured their cytokine production. Thymic NKT2 cells that produced interleukin-4 (IL-4) at steady state were located in the medulla and conditioned medullary thymocytes. NKT2 cells were abundant in the mesenteric lymph node (LN) of BALB/c mice and produced IL-4 in the T cell zone that conditioned other lymphocytes. Intravenous injection of α-galactosylceramide activated NKT1 cells with vascular access, but not LN or thymic NKT cells, resulting in systemic interferon-γ and IL-4 production, while oral α-galactosylceramide activated NKT2 cells in the mesenteric LN, resulting in local IL-4 release. These finding indicate that the localization of iNKT cells governs their cytokine response both at steady state and upon activation.
Introduction
Invariant natural killer T (iNKT) cells are a specialized subset of T cells that recognize CD1d molecules presenting lipid antigens (Bendelac et al., 2007). When stimulated with the agonistic lipid α-galactosylceramide (αGalCer), they rapidly secrete high amounts of several cytokines, and there is growing interest in exploiting αGalCer as an immunological adjuvant (Carreno et al., 2014; Singh et al., 2014; Venkataswamy et al., 2014). iNKT cells also secrete cytokines at steady state and early after infection to influence the development and activation of surrounding immune cells (Engel and Kronenberg, 2014; Lee et al., 2013). Despite being essentially monospecific, iNKT cells nonetheless display substantial functional heterogeneity, with subsets producing different cytokines having distinct tissue localization preferences (Coquet et al., 2008; Doisne et al., 2009; Doisne et al., 2011; Michel et al., 2007; Terashima et al., 2008; Watarai et al., 2012). Recently, we showed that the three major functionally distinct subsets of iNKT cells that exist in mice (NKT1, NKT2 and NKT17 cells) express distinct transcription factor profiles: T-bet, GATA-3 or RORγt (with distinct levels of promyelocytic leukemia zinc finger (PLZF)) and that this generally correlates with their cytokine response upon activation (interferon-γ (IFN-γ), interleukin-4 (IL-4), or IL-17, respectively) (Lee et al., 2013). However, little is known about where these subsets of iNKT cells are localized during steady state and upon activation with aGalCer, and identifying these cells by current methods can be challenging.
iNKT cells can be recognized by staining with CD1d tetramers and by intracellular staining for the lineage specific transcription factor promyelocytic leukaemia zinc finger (PLZF) (Kovalovsky et al., 2008; Savage et al., 2008). These two markers, however, are not readily applicable to immunofluorescence imaging, as CD1d tetramer binding requires live cells for optimal sensitivity and PLZF is also expressed in subsets of γδ T cells, myeloid cells, and stem cells. For these reasons, conventional methods using fresh frozen or paraformaldehyde-fixed tissues to stain for iNKT cells raises issues of sensitivity and specificity. Several reports have tried to visualize iNKT cells using immunofluorescence. Bendelac and colleagues used CD1d tetramers to directly stain frozen tissue sections of Vα14 transgenic (Vα14Tg) mice and showed that iNKT cells are mainly localized in the extravascular area or T cell zone of spleen and lymph node (LN) (Thomas et al., 2011). This technique, however, was not sensitive enough to detect endogenous iNKT cells in wild-type (WT) mice and may have preferentially visualized NKT2 cells expressing high numbers of surface T cell receptors (TCRs), which are abundant in Vα14Tg mice. Batista and colleagues used TCRβ and NK1.1 instead of CD1d tetramers to detect splenic iNKT cells and showed that most are in the marginal zone or red pulp of the spleen (Barral et al., 2012). However, splenic TCRβ+NK1.1+ T cells represent only NKT1 cells, and not NKT2 or NKT17 cells, and some conventional memory T cells also express NK1.1. In other studies, Vα14Tg mice were used as a source of donor cells and congenic markers were used after adoptive transfer for immunofluorescence or intravital imaging (Barral et al., 2010; Chang et al., 2012). However, adoptive transfer may not recapitulate the natural distribution of iNKT populations and in some of these experiments, iNKT cells were isolated using TCRβ and CD1d tetramer binding, which could impact the activation state of the donor cells. Most recently, Leadbetter and colleagues made thick tissue sections using a vibrating microtome and directly stained sections using CD1d tetramer, which showed that iNKT cells are dispersed throughout the splenic parenchyma (King et al., 2013). This study successfully visualized endogenous iNKT cells using CD1d tetramer, but did not separate subsets or validate the sensitivity of staining method. Previous studies have not directly visualized thymic iNKT cells, and studies that addressed the localization of iNKT cells in spleen or LN did not provide data on distribution of iNKT subsets.
Here we report an ex vivo technique to visualize endogenous iNKT cells in thymus, spleen and LN using CD1d tetramers and various transcription factors. We employed a histocytometric algorithm to analyze the data and validate sensitivity. We found that iNKT subsets are not symmetrically distributed throughout tissues: thymic NKT2 cells were mainly located in the medullary area, splenic NKT1 cells preferentially resided in red pulp, whereas splenic NKT2 cells were abundant in the T cell zone. We also found that each subset had unique peripheral distribution, with some inter-strain variation. Notably, the mesenteric LNs (mLNs) of BALB/c mice were substantially enriched for NKT2 cells.
We show that the distribution of iNKT subsets affected the cytokine milieu in lymphoid tissues. At steady state, NKT2 cells in the thymic medulla and T cell zone of the mLN conditioned lymphocytes to express pSTAT6, consistent with production of IL-4 by the NKT2 subset. Intravenous (IV) injection of αGalCer rapidly activated NKT1 cells in the splenic red pulp and liver, which induced systemic IFN-γ and IL-4 responses, but not iNKT cells in LN, thymus, or other tissues. On the other hand, oral administration of αGalCer activated NKT2 cells in mLN, which induced a local IL-4 response. As most iNKT cells do not circulate (Lynch et al., 2015; Thomas et al., 2011), these results show how the steady state localization of iNKT cells determines their systemic or local cytokine responses at steady state and upon activation, providing conceptual challenges to the design of therapeutic protocols in humans.
Results
iNKT cell subsets can be localized with CD1d tetramer immunofluorescence and histocytometry
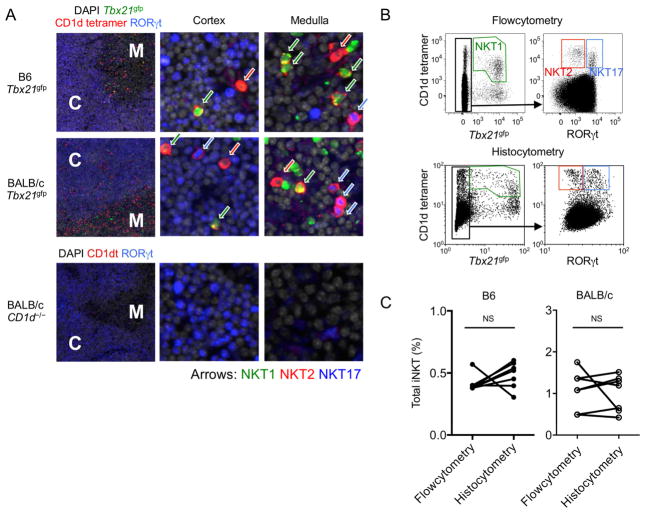
We developed a CD1d tetramer immunofluorescence technique as illustrated in Supplementary Figure 1A and described in experimental procedures. We used reporter mice for the transcription factor T-bet (Tbx21gfp) or stained for intracellular RORγt together with CD1d tetramers to identify NKT1 and NKT17 cells respectively. RORγt was also highly expressed on cortical DP thymocytes, which demarcated the cortex (He et al., 1998) (Figure 1A). In this staining, NKT2 cells were defined as positive for CD1d tetramer but negative for both Tbx21gfp and RORγt and, in a separate staining, we confirmed these cells were all positive for PLZF (data not shown). We used Cd1d−/− mice as a specificity control, in which we detected minimal background staining (Figure 1B, lower panel). To address whether this method detects all iNKT subsets efficiently, we employed a previously described histocytometric algorithm, by which quantitative data from immunofluorescence images are converted into two-dimensional dot plots (Gerner et al., 2012) (Supplementary Figure 1B). Figure 1B shows a side-by-side comparison of the thymic phenotype of iNKT cells by flow cytometry and histocytometry, which separately evaluated each thymic lobe from a single mouse. When we compared the frequency of total iNKT cells (Figure 1C) and the fraction in each subset (Supplementary Figure 1C) between flow cytometry and histocytometry, no significant differences were observed. These results demonstrate the utility of this method to visualize distinct subsets of iNKT cells, with a sensitivity and specificity comparable to flow cytometry.
Figure 1.
iNKT cells localized with CD1d tetramer immunofluorescence and histocytometry. (A) Thymi of B6 and BALB/c Tbx21gfp mice and BALB/c Cd1d−/− mouse were stained with CD1d tetramer (red), RORγt (blue) and DAPI (grey) to visualize and distinguish NKT1, NKT2 and NKT17 cells. Arrows indicate NKT1 (green), NKT2 (red) and NKT17 (blue) cells. M; medulla, C; cortex. (B) Each thymic lobe from BALB/c Tbx21gfp mouse was subjected to histocytometric analysis after immunofluorescence staining or to flow cytometry analysis. Representative plots are shown among 4 different experiments. (C) Frequencies of total iNKT cells in thymus obtained from histocytometry and flow cytometry were compared in 7 different sections obtained from 4 different B6 or BALB/c Tbx21gfp mice. NS; not significant (paired t-test). See also Figure S1.
Medullary distribution of NKT2 cells determines localized pSTAT6 expression
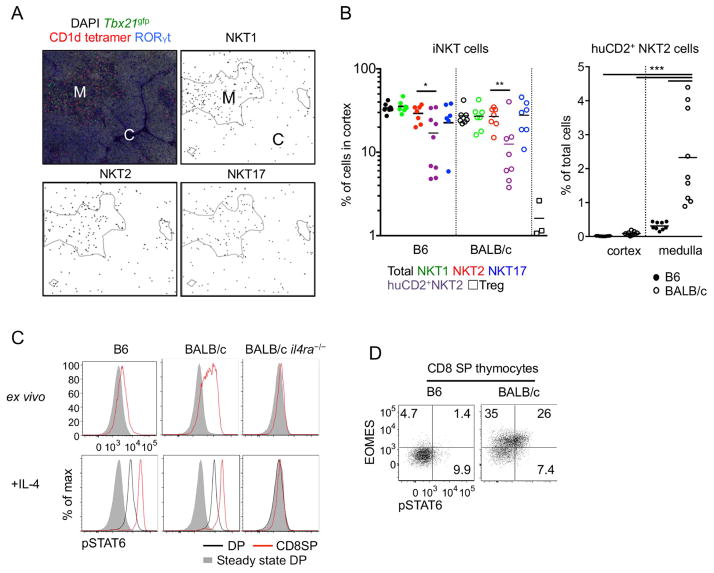
Using the histocytometric algorithm, we tracked the localization of iNKT subsets and quantified them throughout the thymic cortex and medulla (Figure 2A). We found medullary frequencies of iNKT cells averaged 2.7% in C57BL/6 (B6) mice and 5.6% in BALB/c mice, whereas in the cortex, they were 0.27% and 0.32% respectively (Supplementary Figure 2D). Although the medullary frequency of iNKT cells was about 10 times higher than that of cortex, the thymic cortex occupies, on average, approximately 80% of the cut section (Irla et al., 2013b) and its cellularity is 1.7 fold higher than that of the medulla (data not shown). Correcting for this, we estimate that on average 30% of all iNKT cells were in the thymic cortex regardless of subset or mouse strain (Figure 2B), in contrast to most mature cell populations that primarily reside in the medulla. To directly compare this localization pattern to that of another mature cell lineage in thymus we analyzed the localization of thymic regulatory T (Treg) cells using Foxp3gfp mice (Supplementary Figure 2A). We found that only 1.5% of Treg cells resided in the thymic cortex as shown previously (Fontenot et al., 2005) (Figure 2B). This difference suggests that the cortical localization of iNKT cells is not due to a passive overflow of medullary lymphocytes, but is actively regulated process.
Figure 2.
Medullary distribution of NKT2 cells determines localized pSTAT6 expression. (A) Thymi of BALB/c Tbx21gfp mice were stained with indicated markers and analyzed localization of each subset using histocytometric algorithm. Representative figures from at least 5 independent experiments are shown. M; medulla, C; cortex. (B) Cortical frequencies of thymic iNKT cells in B6 and BALB/c Tbx21gfp mice were analyzed (left) and frequencies of human CD2 (huCD2)+ NKT2 cells were calculated in thymic medulla (right). Pooled data from 7 to 9 different sections using 4 different mice are shown. *p=0.032, **p<0.016, ***p<0.0005 (unpaired t-test). Each dot represents an individual section and horizontal bars indicate mean values. (C) Histograms of pSTAT6 expression of double positive (DP) and CD8 single positive (SP) thymocytes are shown from indicated mouse strains with or without IL-4 (10ng/ml) treatment for 30 minutes ex vivo. Representative results of 5 independent experiments are shown. All histograms have the same axis scales. (D) CD8 SP thymocytes from B6 and BALB/c mouse were stained for Eomesodermin (Eomes) and pSTAT6. Numbers indicate frequency of cells in each quadrant and the two plots have the same axis scales. Representative results of five independent experiments are shown. See also Figure S2.
To track the localization of IL-4 producing cells among total NKT2 cells, we immunostained human CD2 (huCD2) in Tbx21gfp mice crossed to KN2+/− mice that express huCD2 on the surface of cells secreting IL-4 (Supplementary Figure 2B and 2C). We observed that the majority of huCD2+ NKT2 cells in both B6 and BALB/c mice were localized in the medulla. However, in BALB/c mice, IL-4 producing NKT2 cells represented 2.3% of medullary thymocytes (Figure 2B, right panel), which was 7.4-fold higher than in B6 mice. This may explain why NKT2 cells condition CD8 single positive (SP) thymocytes to express Eomesodermin (Eomes) in BALB/c but not B6 mice (Lee et al., 2013). We performed flow cytometry with antibodies to the phosphorylated form of the transcription factor STAT6 (pSTAT6) to see the range of influence that IL-4 had on thymocytes (Figure 2C and 2D). pSTAT6 was expressed in medullary SP thymocytes in BALB/c mice, and this was associated with Eomes upregulation. Cortical DP thymocytes were competent for pSTAT6 expression upon in vitro IL-4 stimulation, but negative for pSTAT6 in vivo at steady state indicating that IL-4 exerts its effects primarily on medullary thymocytes due to the selective localization of NKT2 cells to the medulla.
iNKT subsets have unique peripheral distribution, with some interstrain variation
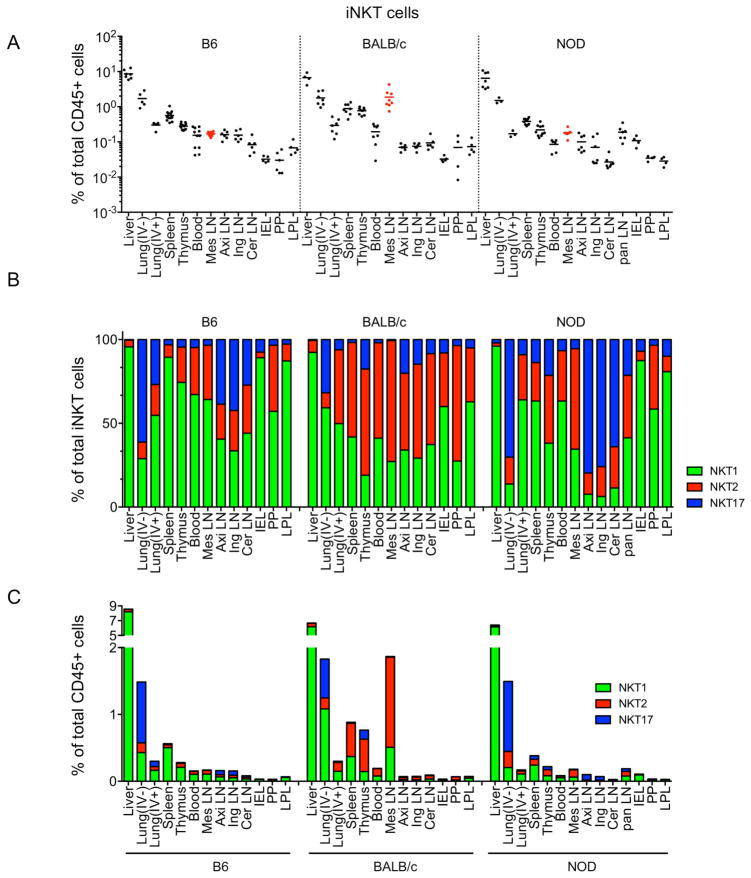
We further identified the tissue distribution of iNKT subsets in three different inbred strains of mice using flow cytometry (Figure 3A). We analyzed the frequency of iNKT cells among total CD45+ cells or frequencies of each subset among total iNKT cells. An intravenous (IV) labeling method was used to quantify iNKT cells residing within lung parenchyma (IV−) or inside the blood vasculature (IV+) (Supplementary Figure 3). The liver contained the highest frequency of iNKT cells in all three strains, followed by the lung (IV−), spleen and thymus. iNKT cells were generally less abundant in LN and intestinal tissues. Subset analysis revealed that NKT1 cells were particularly enriched in liver and NKT17 cells in lung (IV−) and LNs (Figure 3B and Supplementary Figure 3), consistent with previous publications (Michel et al., 2007; Webster et al., 2014). mLNs were notably distinct, showing a high frequency of iNKT cells in BALB/c mice and relative abundance of NKT2 cells in all strains (Figure 3A and 3B). Among total CD45+ lymphocytes, NKT1 cells were overrepresented in the liver, NKT17 cells in the lung (IV−) and NKT2 cells in BALB/c mLN (Figure 3C). Collectively these results show that iNKT subsets are uniquely distributed in peripheral lymphoid and non-lymphoid organs with some inter-strain variation, especially in the mLN.
Figure 3.
iNKT subsets have unique peripheral distribution, with some interstrain variation. Frequencies of total iNKT cells among total CD45+ cells (A), mean frequencies of iNKT subsets among total iNKT cells (B) and frequency of iNKT subsets among total CD45+ cells (C) are shown in indicated organs and strains of mice. IV−; intravenous antibody unlabeled, IV+; intravenous antibody labeled, Mes; Mesenteric, Axi; Axillary, Ing; Inguinal, Cer; Cervical, Pan; Pancreas, IEL; Intra-Epithelial Lymphocytes, PP; Peyer’s Patches, LPL; Lamina Propria Lymphocytes. Each symbol represents an individual mouse (n = 4 ~ 9) and horizontal bars indicate mean values. Pooled data from 8 independent experiments are shown. See also Figure S3.
NKT2 cells in BALB/c mLN produce IL-4 and condition lymphocytes at steady state
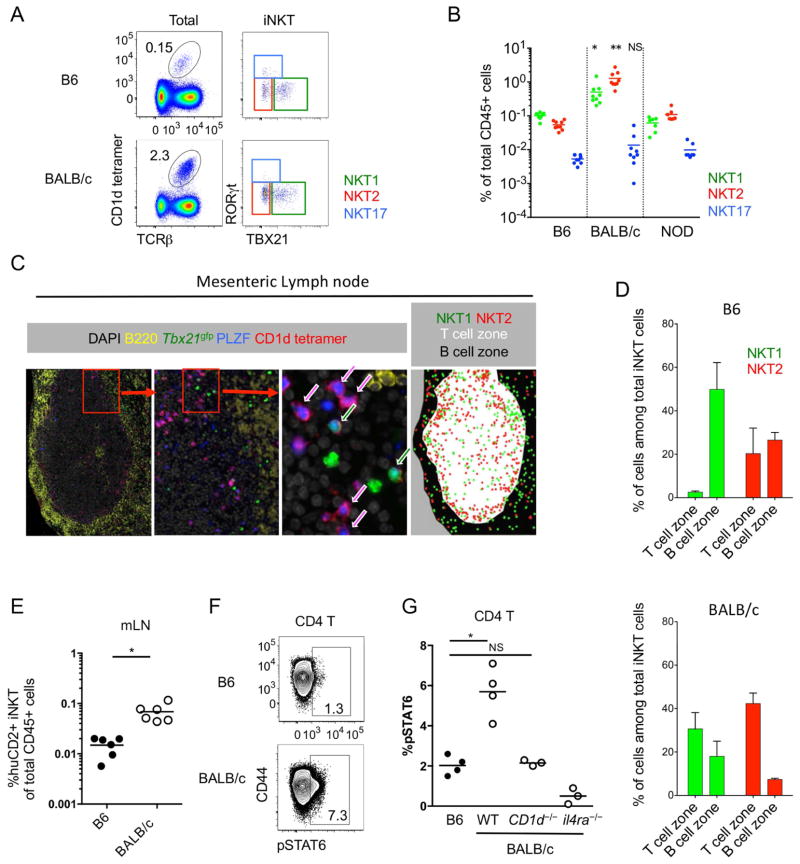
As mLNs were unique in their iNKT subset distribution, we further analyzed their properties. We previously showed that the transcription factor Nur77 is a marker of antigen receptor signaling and that in Nur77gfp reporter mice, GFP is upregulated in iNKT cells after synthetic lipid challenge (Lee et al., 2013; Moran et al., 2011). Interestingly, NKT2 cells in the mLNs showed higher GFP expression in BALB/c Nur77gfp mice (Supplementary Figure 4A), and BALB/c mice contained a 10-fold higher frequency of NKT2 cells than B6 or NOD mice (Figure 4A and 4B). To observe the distribution of iNKT cells within the LN, CD1d tetramer histocytometry was performed. We discriminated the B cell area (or follicle), T cell area (or paracortex), and medulla. In both inguinal and mesenteric LNs, BALB/c iNKT cells were more enriched in the T cell zone compared to B6 (Figure 4C and D and Supplementary Figure 4B and 4C). Consistent with a previous report (Gray et al., 2012), we found that some NKT17 cells are located in the subcapsular area (Supplementary Figure 4B). In the mLN, BALB/c NKT2 cells were particularly enriched in the T cell zone, (Figure 4C and 4D) and analysis of KN2 mice showed an 8-fold higher frequency of IL-4 producing iNKT cells in BALB/c mice compared to those of B6 mice (Figure 4E). Consistent with this, a proportion of T cells in the mLN of BALB/c mice constitutively expressed pSTAT6 (Figure 4F and 4G), unlike T cells in spleen, inguinal LN (iLN) or liver (Supplementary Figure 4D and 4E). pSTAT6 expression in B cells was not different between B6 and BALB/c mice (data not shown), consistent with our observations that BALB/c NKT2 cells were preferentially localized in the T cell zone. This result suggests that in the mLN, like in the thymus, NKT2 cells produce IL-4 to influence neighboring T cells, as evidenced by pSTAT6 expression at steady state.
Figure 4.
NKT2 cells in BALB/c mLN produce IL-4 and condition lymphocytes at steady state. (A) Dot plots show total lymphocytes (left) or iNKT cells (right) from mesenteric lymph nodes (mLNs) of B6 (top) and BALB/c (botton) mice. Numbers indicated frequency of cells in adjacent gates and all plots have the same axis scales. Representative data from at least five independent experiments are shown. (B) Statistical analysis of frequencies of each iNKT subset among total lymphocytes in mLN of B6 (n = 9), BALB/c (n = 9) and NOD (n = 7) mice are shown. *p=0.0014, **p<0.0001 (one-way analysis of variance (ANOVA)). NS, not significant. Horizontal bars indicate mean values. (C) mLN of BALB/c Tbx21gfp mice was stained with indicated antibodies and CD1d tetramer. Arrows indicate NKT1 cells (green) and NKT2 cells (red). (D) Average frequencies of each NKT subset among total iNKT cells in T and B cell zones were analyzed in B6 (n = 3) and BALB/c (n = 3) mice. Error bars indicate standard deviation. (E) Human CD2 (huCD2) positive iNKT cells among total lymphocytes in mLN were analyzed in B6 (n = 6) and BALB/c (n = 6) mice. *p=0.0009 (unpaired t-test). Each dot represents an individual mouse and horizontal bars indicate mean values. (F) Flow cytometry plots show pSTAT6 expression of CD4 T cells in mLN. Numbers indicated percentage of cells in adjacent gates and the two plots have the same axis scales. (G) Statistical analysis of pSTAT6 expressing CD4 T cells in B6 and BALB/c mice (wild type (WT), CD1d−/− and il4ra−/−) (n = 3 – 4) is shown. *p=0.001 (unpaired t-test). NS, not significant. Each dot represents an individual mouse and horizontal bars indicate mean values. See also Figure S4.
Splenic NKT1 cells are mainly localized in red pulp, while NKT2 cells are in T cell zone
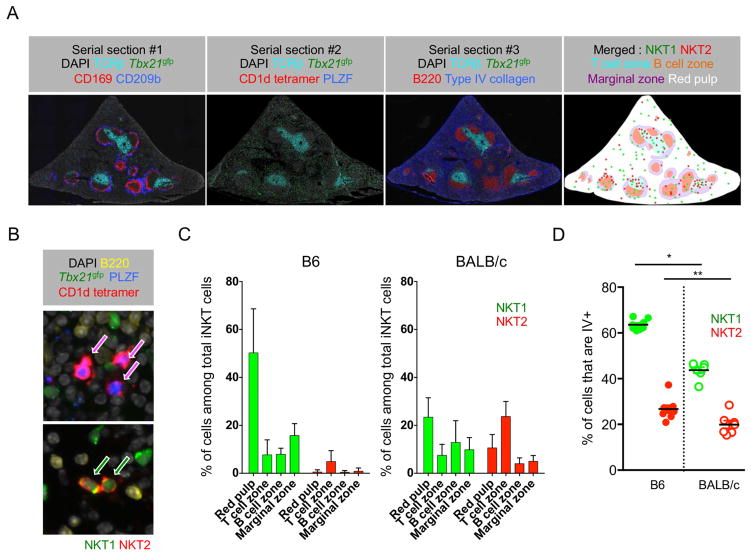
In spleen, we performed CD1d tetramer immunofluorescence with various anatomical markers in serially sectioned tissues. As shown in Figure 5A, section 2 was stained with CD1d tetramer and sections 1 and 3 were stained with TCRβ, B220, type IV collagen, CD169 and CD209b. These markers aid in defining T and B cell zones, the marginal zone (CD169 and CD209b), and splenic red pulp (type IV collagen). We differentiated NKT1 and NKT2 cells using Tbx21gfp reporter mice and co-staining for PLZF (Figure 5B). We found that NKT1 cells localized preferentially in the red pulp, whereas NKT2 cells were in the T cell zone of the white pulp (Figure 5C). We also performed IV antibody labeling of lymphocytes, which defines cells in blood, splenic red pulp, and marginal zone (Anderson et al., 2014). Consistent with the imaging analysis, we observed that NKT1 cells were more frequently IV+ than NKT2 cells (Figure 5D). There was some inter-strain variation, in that a higher frequency of B6 NKT1 cells were IV+ or defined as “vascular associated” compared to BALB/c mice, which was not seen in B cells or CD4 T cells (Supplementary Figure 5). Collectively, these results show NKT1 and NKT2 cells have distinct anatomic localization within spleen.
Figure 5.
Splenic NKT1 cells are mainly localized in red pulp, while NKT2 cells are in T cell zone. (A) Five micron serial sections of spleen of BALB/c Tbx21gfp mice were stained with indicated combinations of antibodies and CD1d tetramer. A merged image in far right shows combined boundaries of image #1 and image #3 with green and red dots representing NKT1 and NKT2 cells from image #2. Representative images of four independent experiments are shown. (B) Representative images of NKT2 cells in T cell zone (top) and NKT1 cells in red pulp (red) are shown. (C) Bar graphs show average frequencies of iNKT subsets in each anatomic region among total iNKT cells in B6 (n = 4) and BALB/c (n = 7) mice. Error bars show standard deviation. (D) Frequencies of splenic NKT1 and NKT2 cells labeled with intravenous (IV) anti-CD45 staining are shown in B6 (n = 8) and BALB/c (n = 7) mice. *p<0.0001, **p=0.015 (unpaired t-test). Each dot represents an individual mouse and horizontal bars indicate mean values. See also Figure S5.
Vascular localized NKT1 cells respond rapidly to αGalCer challenge
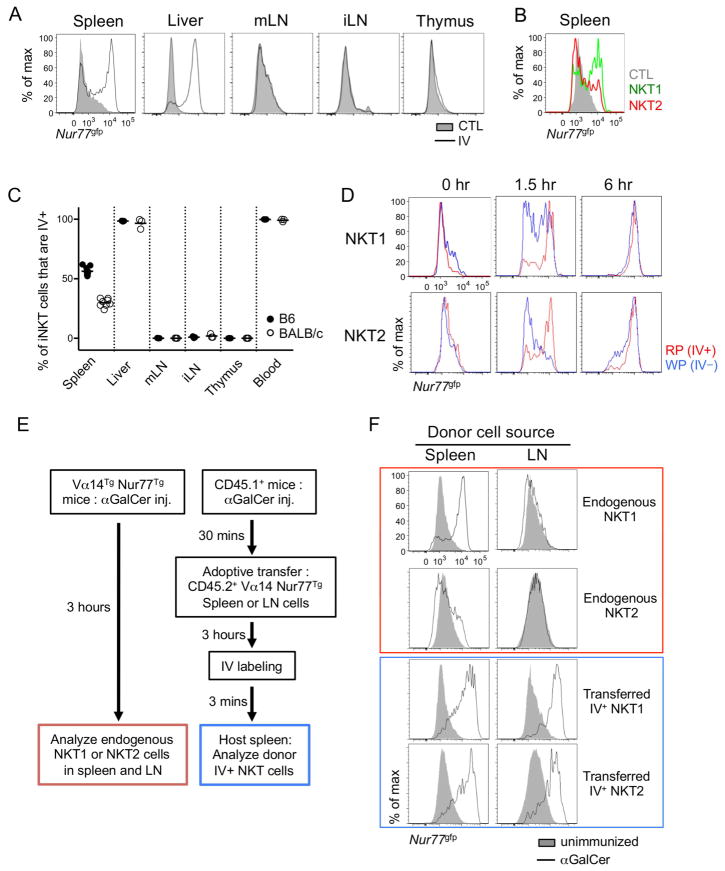
Next, we asked how these differences in tissue and anatomic localization of iNKT subsets influence activation upon αGalCer challenge. Since the subsets produce distinct cytokines (NKT17 produce IL-17, NKT2 produce IL-4, and NKT1 produce IFNγ and some IL-4 depending on stimulation conditions), and CD69 expression is non-specifically upregulated on many lymphocyte populations after αGalCer, we chose to measure the activation of iNKT cells using the Nur77gfp reporter mouse, in which GFP expression is rapidly and highly upregulated on iNKT cells when stimulated via TCR engagement (Supplementary Figure 6A) (Holzapfel et al., 2014a; Moran et al., 2011). When we compared GFP expression in various organs 3 hours after IV αGalCer injection, iNKT cells in thymus had not upregulated GFP, as described in previous report (Pellicci et al., 2003). Surprisingly, iNKT cells in the LN did not respond at all, whereas most of hepatic iNKT cells and a majority of splenic iNKT cells responded (Figure 6A). LN iNKT cells did not show any GFP upregulation at time points up to 6 hours after injection (data not shown) suggesting that they did not become activated through the TCR after lipid injection. In the spleen, a higher percentage of NKT1 cells expressed Nur77gfp after αGalCer challenge than NKT2 cells (Figure 6B). This difference did not reflect a cell intrinsic feature, as NKT2 cells efficiently upregulated Nur77gfp expression after ex vivo activation (Supplementary Figure 6B). Interestingly, the percent of iNKT cells activated in each tissue was roughly correlated to the frequency of cells labeled by IV antibody staining in that tissues (Figure 6C) and thus we hypothesized that only vascular associated iNKT cells responded immediately after IV αGalCer challenge. To test this hypothesis, we compared the endogenous response of iNKT cells in red pulp (IV+) or white pulp (IV−) and found that both NKT1 and NKT2 cells in the red pulp responded quickly after αGalCer injection, whereas cells in white pulp showed delayed activation (Figure 6D and Supplementary Figure 6C). Furthermore, we adoptively transferred enriched iNKT cells into αGalCer preinjected mice as outlined in Figure 6E and harvested the spleens of host mice after 3 hours. We performed CD1d tetramer based magnetic bead enrichment to detect rare donor cells (Supplementary Figure 6D) and found that both LN iNKT cells and splenic NKT2 cells responded well when they were localized in the splenic red pulp, confirming that localization rather than a type of cells, determines the response to IV injected lipid (Figure 6F). As serum concentrations of IL-4 after αGalCer injection peaked at 1.5 hours (Sullivan et al., 2010b), these results suggest that iNKT cells in the white pulp are not major contributors to the serum cytokine response.
Figure 6.
Vascular localized NKT1 cells respond rapidly to αGalCer challenge. (A) BALB/c Nur77gfp mouse was intravenously (IV) injected with αGalCer and indicated organs were collected 3 hours later. Overlaid histograms show Nur77gfp expression in total iNKT cells from αGalCer IV injected and uninjected control (CTL) mice. Representative histograms of at least five independent experiments are shown. All histograms have the same axis scales. mLN, mesenteric lymph node; iLN, inguinal lymph node. (B) Overlaid histogram shows Nur77gfp expression in NKT1 (green) and NKT2 (red) cells 3 hours after αGalCer IV injection in BALB/c mice. Representative histogram of at least five independent experiments is shown. (C) Percentages of iNKT cells labeled with IV anti-CD45 antibody in corresponding organs of B6 (n = 3 ~ 8) and BALB/c (n = 3 ~ 7) mice are shown. Each dot represents an individual mouse and horizontal bars indicate mean values. (D) BALB/c Nur77gfp mice were IV injected with αGalCer and labeled with IV ant-CD45 antibody after indicated time periods. Three minutes later, mice were sacrificed and analyzed for Nur77gfp expressions. All histograms have the same axis scales. Representative results of three independent experiments are shown. RP, red pulp; WP, white pulp. (E) Schematic representation shows experimental procedure for (F). Vα14Tg Nur77gfp (left) or CD45.1+ congenic B6 (right) mice were IV injected with αGalCer and the latters were transferred with LN cells of Vα14Tg Nur77gfp mice. Three hours later, mice were sacrificed after CD45.1+ congenic B6 recipient mice were labeled with IV anti-CD45 antibody for 3 minutes. (F) Histograms show Nur77 expression of NKT1 and NKT2 cells of spleen and LN of Vα14Tg Nur77gfp mice (red) and donor iNKT cells in splenic red pulp of host mice immunized (blue). Filled histogram shows Nur77 expression of NKT1 and NKT2 cells in unimmunized Vα14Tg Nur77gfp mice. All histograms have the same axis scales. Representative data of three mice are shown. See also Figure S6.
mLN NKT2 cells respond to oral administration of αGalCer
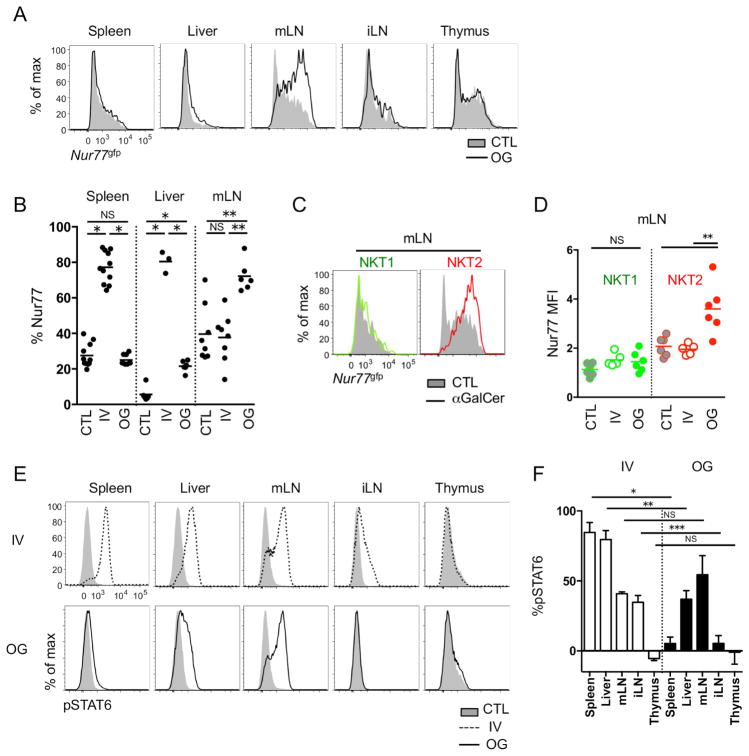
Various routes of αGalCer delivery have been shown to differentially affect immune responses (Furlan et al., 2003; Miyamoto et al., 2001). As IV injection of αGalCer did not activate LN resident iNKT cells, we tested whether they could be activated by oral gavage of αGalCer. In contrast to IV injection, oral administration of αGalCer specifically upregulated Nur77gfp in mLN NKT cells and to a much lesser extent iNKT cells in liver, but not in the spleen or thymus (Figure 7A and 7B). When we compared iNKT subsets, only NKT2 cells upregulated Nur77gfp (Figure 7C and 7D) and, consistent with this finding, IL-4 was produced from these NKT2 cells (Supplementary Figure 7A). NKT1 cells did not produce IFN-γ or IL-4 upon oral administration of αGalCer, although they produce both after IV injection (Supplementary Figure 7A). IV injection of αGalCer rapidly activated iNKT cells in spleen and liver, and serum increases of both IFN-γ and IL-4 could be detected as previously described (Supplementary Figure 7B) (Sullivan et al., 2010a). The systemic release of IL-4 had effects at distant sites, as evidenced by the observation that T cells in LNs upregulated pSTAT6 (Figure 7E and 7F) even though LN NKT cells were not activated by IV injection of αGalCer (Figure 6A). However, after oral gavage, there was no serum surge of IL-4 (Supplementary Figure 7B) and pSTAT6 upregulation was detected only in mLN and somewhat in liver, but not in spleen or iLN (Figure 7E and 7F). These results indicate that oral gavage of lipid antigens can be used to modulate the regional cytokine milieu in mLN without systemic effects.
Figure 7.
mLN NKT2 cells respond to oral administration of αGalCer. (A) BALB/c Nur77gfp mice were oral gavaged (OG) with 2μg of αGalCer and Nur77gfp expressions in total iNKT cells were analyzed after 24 hours. All histograms have the same axis scales. Representative histograms of at least five independent experiments are shown. mLN, mesenteric lymph node; iLN, inguinal lymph node. (B) Statistical analyses show frequencies of iNKT cells expressing Nur77gfp in spleen, liver and mLN of unimmunized control (CTL), intravenously (IV) αGalCer injected and oral gavaged (OG) BALB/c mice (n = 3 ~ 9). *p<0.0001, **p=0.0002 (one-way analysis of variance (ANOVA)). NS, not significant. Each dot represents an individual mouse and horizontal bars indicate mean values. (C and D) Overlay histograms show Nur77gfp expressions in NKT1 (green) and NKT2 (red) cells in BALB/c mice 24 hours after αGalCer oral gavage. Representative histograms of three independent experiments (C) and its statistical analysis (n = 6) (D) are shown. All histograms have the same axis scales. **p=0.001 (unpaired t-test). NS, not significant. Each dot represents an individual mouse and horizontal bars indicate mean values. (E) Histograms show pSTAT6 expression in CD4 T cells 3 hours after IV injection (IV, top) or 24 hours after oral gavage (OG, bottom) of αGalCer. All histograms have the same axis scales. (F) Bar graphs show average percent point increases of pSTAT6 expression in CD4 T cells after IV injection or OG of αGalCer in BALB/c mice (n = 3 ~ 6). *p<0.0001, **p=0.0014, ***p=0.0033 (unpaired t-test). NS, not significant. Error bars show standard deviation. See also Figure S7.
Discussion
In this report, we provided an in-depth analysis of the tissue distribution and anatomic localization of iNKT cells. For detailed analysis, we developed a CD1d tetramer-based histocytometry approach, which provided an unbiased tool for analysis of immunofluorescence data. As a result, we showed that IL-4 producing NKT2 cells are enriched in the thymic medulla and the T cell zone of mLNs. In both of these locations, they appear to condition neighboring thymocytes and lymphocytes, as evidenced by the expression of pSTAT6 at steady state. We also showed that different routes of antigen delivery could activate iNKT cells in different anatomic locations. IV injection of αGalCer activated NKT1 cells in spleen and liver with systemic IFN-γ and IL-4 release, whereas oral gavage of αGalCer stimulated NKT2 cells in mLN with only local IL-4 effects only. Collectively these results show that subset localization of iNKT cells is crucial to determine which cytokines are produced and whether a systemic or regional cytokine response ensues.
In the thymus, T-bet, GATA-3 and RORγt regulate lineage differentiation of iNKT cells from a common progenitor (Constantinides and Bendelac, 2013; Lee et al., 2013). Previously, we showed that NKT2 cells secrete IL-4 at steady state in BALB/c mice, which conditioned SP thymocytes to express Eomes, stimulated thymic dendritic cell to secrete CCL17 and CCL22 and caused B cells to produce immunoglobulin E (Lee et al., 2013). In this paper, we showed that NKT2 cells, and in particular IL-4-producing NKT2 cells, mainly reside in the thymic medulla and their IL-4 effect was confined to medullary thymocytes. The medulla is a crucial environment for the development of iNKT cells, as their numbers were sharply reduced in relb−/− thymi, which lack medullary epithelial cells (White et al., 2014). As NK1.1+ iNKT cells were most strongly affected, we infer that NKT1 cells in particular depend on medullary epithelial cells for their development, which is to be expected since such cells produce IL-15, a factor previously shown to be required for T-bet upregulation in iNKT cells (Castillo et al., 2010; Gordy et al., 2011; White et al., 2014). Interestingly, iNKT cells also promote the maturation of medullary epithelial cells, through production of RANKL (White et al., 2014). Since NK1.1+ cells (NKT1) do not produce RANKL, it is possible that NKT2 or NKT17 cells provide this function. Our data would suggest that both NKT2 and NKT17 cells are localized in the appropriate environment for this.
Regardless of subset or strain, an estimated 30% of NKT cells were localized in the thymic cortex, in contrast to Treg cells, of which only 1.5% were localized in the cortex. As genetic evidence has shown that iNKT cells are positively selected from cortical DP thymocytes expressing RORγt (Egawa et al., 2005), two possibilities can explain these observations: iNKT cells might differentiate in the cortex then migrate into the medulla, or they might first migrate into the medulla, differentiate into various subsets and a fraction of them may move back to the cortex. In the former scenario, younger mice would have a higher frequency and older mice would have a lower frequency of cortical iNKT cells. However, this was not the case when we analyzed cortical localization of iNKT cells from 3 or 15 weeks old mice (data not shown). The latter scenario seems more likely, considering that the medulla is required for differentiation to the T-bet+ NKT1 subset (White et al., 2014), and that CCR7 a chemokine receptor known to be involved in medullary localization is expressed on “uncommitted” PLZFhi iNKT cells, but not on differentiated NKT1, NKT2, or NKT17 cells (data not shown). Other chemokine receptors might regulate localization as well. For example, CXCR3 is expressed exclusively on NKT1 cells and its ligand CXCL10 produced by medullary thymic epithelial cells was suggested to direct iNKT cell medullary localization and long-term residency (Berzins et al., 2006; Drennan et al., 2009). However, our analysis showed that not only NKT1 cells but also NKT2 and NKT17 cells preferentially resided in the medulla, indicating that CXCR3 is not the only factor regulating medullary localization of iNKT cells. Further studies are required to identify factors regulating cortico-medullary or medullo-cortical migration of iNKT cells and to determine if cortical iNKT cells serve a particular function.
We extended our analysis of subset localization into other lymphoid organs. The mLN of BALB/c mice were particularly enriched with NKT2 cells, especially in the T cell zone. These cells also had an increased MFI of Nur77gfp in the steady state, suggesting their antigen receptors were being stimulated, and they produced IL-4 at steady state, which conditioned surrounding T cells to phosphorylate STAT6. One possible explanation of these observations is that microbial antigens from the gut are carried to the mLN where they stimulate NKT2 cells. A previous report showed that stimulation of intestinal iNKT cells during the neonatal period by a sphingolipid from Bacteroides fragilis suppressed subsequent accumulation and pathogenic activation of iNKT cells in the colon and lung (An et al., 2014; Olszak et al., 2012). In addition, iNKT cells from germ free mice showed reduced expression of CD5 and CD69 at steady state and general hypo-responsiveness upon αGalCer challenge. B6 mice from Taconic, compared to those from Jackson Laboratory, have an increased frequency of iNKT cells in spleen and gut that express TCR Vβ7 and CD127, and the difference disappears with co-housing (Wingender et al., 2012). Collectively, these results suggest that intestinal antigens provide homeostatic signals to iNKT cells. However, it remains to be determined if the intestinal microbiome regulates inter-strain differences in iNKT cell development and/or activation in the mLN.
Recently, two more subsets of iNKT cells have been described; NKTFH (follicular helper NKT) and NKT10 cells expressing transcription factor Bcl6 and E4BP4 respectively (Chang et al., 2012; Lynch et al., 2015; Sag et al., 2014). NKTFH cells are localized in germinal center and provide help to B cells. NKT10 cells are localized in white adipose tissue and secrete IL-10 to create immunoregulatory environment. We did not analyze these cells as they are not found in lymphoid organs at steady state or induced at early timepoints after immunization.
IV administration of αGalCer immediately activated iNKT cells with vascular access but not iNKT cells in LN or thymus. It is not clear why these iNKT cells are not responsive, but we showed it was not an intrinsic feature of LN iNKT cells (Figure 6F). It is possible that serum components such as lipid binding proteins (van den Elzen et al., 2005) restrict αGalCer distribution to the vascular compartment, or that antigen presenting cells in the LN are not specialized to present lipid antigens.
Oral antigenic intake induces tolerogenic T cell responses (Pabst and Mowat, 2012). However, co-administration of oral αGalCer together with protein antigens induced a protective immune response against tumor cells expressing the cognate antigens (Silk et al., 2004). The adjuvant effects were not seen by IV injection of αGalCer, suggesting that routes of antigenic delivery are critical. In this paper, we provided mechanistic insight as to how oral αGalCer exerts its effects; oral αGalCer specifically activates NKT2 cells in mLN to produce IL-4.
After oral gavage, it took about 7 hours before iNKT cells began to be activated in mLN and the response peaked at 24 hours (data not shown). We think this delay may be due to the requirement of antigen presenting cells to traffic from intestine to mLN. In mLN, NKT2 cells might share an anatomic niche with migrating dendritic cells or migrating dendritic cells might be specialized to activate only NKT2 cells, but not NKT1 cells via their expression patterns of co-stimulatory molecules or secreted factors. However, this is not consistent with previous findings that migrating dendritic cells in the mLN favor the differentiation of Th1 and/or Th17 cells, but not Th2 cells (Cerovic et al., 2014).
Inflammatory bowl diseases such as Crohn’s disease and ulcerative colitis are chronic relapsing inflammatory disorder involving the gastrointestinal tract (Kaser et al., 2010). Immunologically, pathogenic Th1 and Th2 responses characterize these two diseases respectively. Our finding that oral αGalCer administration induced an IL-4 rich environment in the mLN, has clinical implications, as oral αGalCer might be able to suppress pathogenic Th1 cell activation in Crohn’s disease, while it might worsen Th2 driven ulcerative colitis. We also showed that oral αGalCer induced an IL-4 effect confined to mLN, which could minimize potential systemic side effects. IV injection of αGalCer eventually induces NKT10 cells secreting IL-10 long after immunization in spleen (Sag et al., 2014) and if this is the same case in mLN after oral gavage of αGalCer, it might be helpful to suppress both Th1 or Th2 driven inflammation as IL-10 is critical for suppressing pathogenic T cell activation in the gut (Saraiva and O’Garra, 2010).
The systemic distribution of iNKT cells in humans is different from that of mice. In humans, iNKT cells are not enriched in liver, and the small intestine is a major organ where iNKT cells are localized early in life (Loh et al., 2014). In human clinical trials, IV αGalCer injection failed to increase serum IL-4 or IFN-γ in a majority of subjects (Giaccone et al., 2002), which has been attributed to their low numerical abundance, as humans have a lower proportion of iNKT cells in blood than mice, albeit with two-log phase variability between individuals (Lee et al., 2002). However, our results suggest that not only numerical issues, but also qualitative issues such as the localization and subset differentiation are crucial for in vivo activation and cytokine secretion from iNKT cells upon αGalCer challenge. As parabiosis experiments suggest that iNKT cells do not circulate (Lynch et al., 2015; Thomas et al., 2011), IV injection of αGalCer may not be an ideal therapeutic strategy if the majority of human iNKT cells are not readily accessible to the vasculature. In this context, the type and localization of human iNKT cells and the most efficient immunization routes to activate them must be clarified.
Experimental Procedures
Mice
B6 (C57BL/6NCr) mice were purchased from the National Cancer Institute. BALB/c (BALB/cJ or BALB/cByJ), Cd1d−/− BALB/cJ (C.129S2-Cd1tm1Gru/J), Il4ra−/− BALB/cJ (BALB/c-Il4ratm1Sz/J) and NOD (NOD/ShiLtJ) mice were from Jackson laboratory. B6 and BALB/c Tbx21gfp and Tbx21gfp KN2+/− mice were described previously (Lee et al., 2013). Vα14Tg Nur77gfp TCR transgenic B6 mice were described before (Holzapfel et al., 2014b). All the mouse experiments were performed under protocols approved by the Institutional Animal Care and Use Committee of the University of Minnesota.
Immunofluorescence
For CD1d tetramer immunofluorescence, fresh thymic lobes, whole lymph nodes or 1–2 mm thickness spleen trans-sections were overnight incubated in 50μl phosphate buffered saline (PBS) solution containing 1:10~1:25 dilution of R-Phycoerythrin (PE) labeled PBS-57 loaded CD1d tetramer in 2% fetal calf serum containing PBS at 4°C. Next day, tissues were washed three times with PBS, fixed with 4% paraformaldehyde (PFA) for one hour and snap frozen. Five micrometer tissue sections were blocked with 5% bovine serum albumin and goat sera (Jackson Laboratory) for 1 hour at 25°C and stained with anti-PE antibody (Novus Biologicals, rabbit polyclonal) followed by goat anti-rabbit AF555 (Life Technology). For human CD2 immunofluorescence, tyramide based amplification was done according to manufacturer’s instruction (PerkinElmer) after staining with anti-human CD2 (RPA2) antibody. RORγt (RORg2, Millipore or Q31-378, BD) and PLZF (D9, SantaCruz or R17-809, BD) were stained followed by goat anti-hamster IgG (Jackson Laboratory) and goat anti-mouse IgG1 (Southern Biotech).
Histocytometry
Hitocytometric analysis was performed as described previously, with modifications (Gerner et al., 2012). Briefly, fluorochrome intensities of each single region of interest were quantified using ImageJ and data were exported into Excel and Prism software for two-dimensional plotting. XY coordinates of gated populations were overlaid on original images to analyze localization. For comparison between histocytometry and flow cytometry in thymus, we obtained 30 to 50 thymic stitch images containing a total of 5×104 ~ 105 cells and adjusted the medullary fraction to 20% as previously described (Irla et al., 2013a).
Flow cytometry
Single-cell suspensions were prepared from thymi, spleens and lymph nodes and hepatic mononuclear cells were separated by percoll gradient. Biotinylated PBS-57 loaded or unloaded CD1d monomers were obtained from the tetramer facility of the US National Institutes of Health. For intracellular staining, single cell suspensions were surface stained, fixed and permeabilized with eBioscience Foxp3 staining buffer set. For pSTAT6 staining, cells were fixed with 1.6% PFA (25°C) and methanol (−20°C) for 10 mi nutes respectively and stained with surface and intracellular markers. For in vitro IL-4 culture, total thymocytes were incubated for 30 minutes in 10ng/ml IL-4 (eBioscience). For isolation of lymphocytes from lung parenchyma, we labeled circulating cells as described before (Anderson et al., 2014) and chopped tissues were digested with collagenase D (Roche). All antibodies were from eBioscience, BD or Biolegend, unless indicated. Cells were analyzed on an LSR II (Becton Dickinson) and data were processed with FlowJo software (TreeStar).
Enzyme-linked immunosorbent assay
A mouse IL-4 and IFN-γ ELISA MAX kit (Biolegend) was used for quantification of serum cytokines.
In vitro stimulation of iNKT cells
Splenocytes of Vα14Tg Nur77gfp TCR transgenic mouse were resuspended in RPMI media containing 10% FCS at 5×106/ml concentrations. PE conjugated PBS-57 loaded CD1d tetramer or PE conjugated empty CD1d tetramer was added in media and cultured for 4 hours at 37°C. GFP expression was determined by flow cytomet ry analysis after in vitro stimulation.
Lipid injection
Two micrograms of α-galactosylceramide (Avanti Polar Chemical, KRN7000) were dissolved in DMSO (1mg/ml), diluted in PBS and intravenously injected or oral gavaged. Control mice were administrated with DMSO/PBS. Mice were analyzed after 3 or 24 hours respectively if not otherwise indicated.
Statistical analysis
Prism software (Graphpad) was used for statistical analysis. ANOVA and t-test (paired or unpaired, two-tailed) were used for data analysis and the generation of P values.
Supplementary Material
Supplementary Figure 1 related to Figure 1. Workflow pipeline to quantify localization of iNKT subsets. (A) Schematic representation showing CD1d tetramer immunofluorescence (IF) technique. PFA, paraformaldehyde. (B) Fluorescence intensities were quantified in each single cell identified by ImageJ and data were transferred into Prism software to gate cells. XY coordinates of gated cells were overlaid on an original image and the distribution of cells were analyzed. ROIs, region of interests. (C) Three to four different B6 and BALB/c mice were analyzed for their thymic frequencies of each subset of iNKT cells among total thymocytes either by flowcytometry (FC) or CD1d tetramer histocytometry (HC). NS, not significant (paired t-test).
Supplementary Figure 2 related to Figure 2. Regulatory T cells and IL-4 producing NKT cells reside in thymic medulla. (A) Thymi of Foxp3gfp mice were stained with RORγt and G8.8 to analyze the localization of regulatory T cells (Tregs). An arrow indicates a Treg in the cortex. C, cortex; M, medulla. (B and C) Thymi of Tbx21gfp KN2+/− mice were stained with CD1d tetramer and huCD2 molecule (B) and analyzed by histocytometric algorithms (C). Numbers indicate frequency of cells in adjacent gates. (D) Compilation of cortical and medullary frequencies of total and each iNKT subset are shown for B6 (4 mice, 7 sections) and BLAB/c (4 mice, 8 secretions) iNKT cells and B6 regulatory T cells (Treg, 3 mice, 3 sections). Each dot represents an individual section and horizontal bars indicate mean values.
Supplementary Figure 3 related to Figure 3. Flowcytometry profiles of iNKT cells in multiple organs. Representative flowcytometry dot plots and gating strategies are shown in various organs. Numbers indicate frequency of cells in adjacent gates and all dot plots have the same axis scales. Representative data of at least five independent experiments are shown. IV, intravenous; mLN, mesenteric lymph node; iLN, inguinal lymph node; IEL, intraepithelial lymphocyte.
Supplementary Figure 4 related to Figure 4. mLN NKT2 cells condition lymphocytes to express pSTAT6. (A) Nur77gfp expression in each subset of iNKT cells is shown in indicated organs at steady state. Representative histograms of at least 3 independent experiments are shown. All histograms have the same axis scales. iLN, inguinal lymph node; mLN, mesenteric lymph node. (B) iLN of B6 Tbx21gfp mice was stained with indicated antibodies and CD1d tetramer. Arrows indicate NKT1 cells (green) and NKT17 cells (blue). (C) Average frequencies of each NKT subset among total iNKT cells in T cell zone, B cell zone and medulla of iLN were analyzed in B6 (n = 6) and BALB/c (n = 6) mice. Error bars indicate standard deviation. (D) pSTAT6 expressions in CD4 T cells from spleen and iLN of B6 and BALB/c mice are shown at steady state. Numbers indicated frequencies of cells at adjacent gates and dot plots have the same axis scales. (E) Frequencies of pSTAT6 positive CD4 T cells in indicated organs are shown in B6 (n = 3 ~ 8), BALB/c (n = 3 ~ 17) and BALB/c il4ra−/− (n = 3) mice. Each dot represents an individual mouse and horizontal bars indicate mean values. Error bars show standard deviation. *p<0.0001, **p<0.001 (one-way analysis of variance (ANOVA)). NS, not significant
Supplementary Figure 5 related to Figure 5. Intravital labeling of splenocytes in B6 and BALB/c mice. B6 and BALB/c mice were intravenously (IV) injected with anti-CD45 antibody and sacrificed 3 minutes later. Splenic B cells and CD4 T cells were analyzed antibody labeled frequencies in B6 (n = 8) and BALB/c (n = 7) mice. NS, not significant (unpaired t-test).
Supplementary Figure 6 related to Figure 6. iNKT cell localization determines cellular response to αGalCer. (A) BALB/c Nur77gfpKN2+/− mice were intravenously (IV) injected with αGalCer and analyzed for Nur77gfp, surface human CD2 (huCD2) and intracellular IFN-γ expressions after 3 hours. Representative data of at least 3 independent experiments are shown. Numbers indicate frequencies of cells in each quadrant and dot plots have the same axis scales. (B) Splenocytes from BALB/c Nur77gfp mice were in vitro culture with CD1d tetramer for 4 hours and measured Nur77gfp expression in NKT1 (green) and NKT2 (red) cells. Frequencies of GFP expression were normalized at 0.1μg/ml tetramer concentrations in three independent experiments. Error bars indicate standard deviation. (C) BALB/c Nur77gfp mice were IV injected with αGalCer and analyzed Nur77gfp expression in each subset at indicated time points and compared Nur77gfp fold increased between cells in red pulp (RP, IV antibody labeled) or white pulp (WP, IV antibody unlabeled). Error bars indicate standard deviation. P values indicate unpaired t-tests. (D) Vα14 TCR transgenic NKT cells from spleen and lymph node (LN) were transferred into αGalCer immunized host mice. Donor iNKT cells in spleen were analyzed after CD1d tetramer pull down. Numbers indicated frequency of cells in adjacent gates and dot plots have the same axis scales.
Supplementary Figure 7 related to Figure 7. Oral gavage of αGalCer stimulates NKT2 cells in mLN to produce IL-4. (A) BALB/c KN2+/− mice were administrated with αGalCer either by intravenous (IV) injection or oral gavage (OG) and analyzed NKT1 (green) and NKT2 (red) cells in spleen and mesenteric lymph node (mLN) for their expression of human CD2 (huCD2) and intracellular IFN-γ at 3 and 24 hours later respectively. Representative histograms of 3 independent experiments are shown. All histograms have the same axis scales. (B) BALB/c mice were measured their serum concentrations of IL-4 and IFN-γ by ELISA after either IV injection or oral gavage of αGalCer at indicated time points (n = 5). Error bars indicate standard deviation.
Highlights.
IL-4 secreting NKT2 cells are localized in the thymic medulla
Splenic NKT1 cells are in red pulp and NKT2 cells in T cell zone
IV injection of αGalCer activates NKT1 cells in spleen and liver
Oral administration of αGalCer stimulates NKT2 cells in mesenteric LN
Acknowledgments
We thank Michael Y. Gerner (NIH) for technical comments. The authors declare no competing financial interests. This research was supported by NIH grants R37-AI39560 (to K.A.H.), RO1-AI075168 (to S.C.J.) and K99-AI114884 (to Y.J.L).
Footnotes
Author Contributions
Y.J.L. designed and performed experiments, analyzed data and wrote the manuscript; H.W., G.J.S. and V.P. performed experiments and provided input for interpretation; S.C.J. provided input for research design and interpretation; and K.A.H. conceptualized the research, directed the study, analyzed data and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, Masopust D. Intravascular staining for discrimination of vascular and tissue leukocytes. Nature protocols. 2014;9:209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral P, Polzella P, Bruckbauer A, van Rooijen N, Besra GS, Cerundolo V, Batista FD. CD169(+) macrophages present lipid antigens to mediate early activation of iNKT cells in lymph nodes. Nature immunology. 2010;11:303–312. doi: 10.1038/ni.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral P, Sanchez-Nino MD, van Rooijen N, Cerundolo V, Batista FD. The location of splenic NKT cells favours their rapid activation by blood-borne antigen. The EMBO journal. 2012;31:2378–2390. doi: 10.1038/emboj.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annual review of immunology. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Berzins SP, McNab FW, Jones CM, Smyth MJ, Godfrey DI. Long-term retention of mature NK1.1+ NKT cells in the thymus. Journal of immunology. 2006;176:4059–4065. doi: 10.4049/jimmunol.176.7.4059. [DOI] [PubMed] [Google Scholar]
- Carreno LJ, Kharkwal SS, Porcelli SA. Optimizing NKT cell ligands as vaccine adjuvants. Immunotherapy. 2014;6:309–320. doi: 10.2217/imt.13.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo EF, Acero LF, Stonier SW, Zhou D, Schluns KS. Thymic and peripheral microenvironments differentially mediate development and maturation of iNKT cells by IL-15 transpresentation. Blood. 2010;116:2494–2503. doi: 10.1182/blood-2010-03-277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerovic V, Bain CC, Mowat AM, Milling SW. Intestinal macrophages and dendritic cells: what’s the difference? Trends in immunology. 2014;35:270–277. doi: 10.1016/j.it.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, Hogan JJ, Cerundolo V, Tangye SG, Bittman R, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nature immunology. 2012;13:35–43. doi: 10.1038/ni.2166. [DOI] [PubMed] [Google Scholar]
- Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Current opinion in immunology. 2013 doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, Berzins SP, Smyth MJ, Godfrey DI. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci U S A. 2008;105:11287–11292. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doisne JM, Becourt C, Amniai L, Duarte N, Le Luduec JB, Eberl G, Benlagha K. Skin and peripheral lymph node invariant NKT cells are mainly retinoic acid receptor-related orphan receptor (gamma)t+ and respond preferentially under inflammatory conditions. Journal of immunology. 2009;183:2142–2149. doi: 10.4049/jimmunol.0901059. [DOI] [PubMed] [Google Scholar]
- Doisne JM, Soulard V, Becourt C, Amniai L, Henrot P, Havenar-Daughton C, Blanchet C, Zitvogel L, Ryffel B, Cavaillon JM, et al. Cutting edge: crucial role of IL-1 and IL-23 in the innate IL-17 response of peripheral lymph node NK1.1-invariant NKT cells to bacteria. Journal of immunology. 2011;186:662–666. doi: 10.4049/jimmunol.1002725. [DOI] [PubMed] [Google Scholar]
- Drennan MB, Franki AS, Dewint P, Van Beneden K, Seeuws S, van de Pavert SA, Reilly EC, Verbruggen G, Lane TE, Mebius RE, et al. Cutting edge: the chemokine receptor CXCR3 retains invariant NK T cells in the thymus. Journal of immunology. 2009;183:2213–2216. doi: 10.4049/jimmunol.0901213. [DOI] [PubMed] [Google Scholar]
- Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Engel I, Kronenberg M. Transcriptional control of the development and function of Valpha14i NKT cells. Current topics in microbiology and immunology. 2014;381:51–81. doi: 10.1007/82_2014_375. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Dooley JL, Farr AG, Rudensky AY. Developmental regulation of Foxp3 expression during ontogeny. The Journal of experimental medicine. 2005;202:901–906. doi: 10.1084/jem.20050784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan R, Bergami A, Cantarella D, Brambilla E, Taniguchi M, Dellabona P, Casorati G, Martino G. Activation of invariant NKT cells by alphaGalCer administration protects mice from MOG35-55-induced EAE: critical roles for administration route and IFN-gamma. European journal of immunology. 2003;33:1830–1838. doi: 10.1002/eji.200323885. [DOI] [PubMed] [Google Scholar]
- Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histocytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–376. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccone G, Punt CJ, Ando Y, Ruijter R, Nishi N, Peters M, von Blomberg BM, Scheper RJ, van der Vliet HJ, van den Eertwegh AJ, et al. A phase I study of the natural killer T-cell ligand alpha-galactosylceramide (KRN7000) in patients with solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:3702–3709. [PubMed] [Google Scholar]
- Gordy LE, Bezbradica JS, Flyak AI, Spencer CT, Dunkle A, Sun J, Stanic AK, Boothby MR, He YW, Zhao Z, et al. IL-15 regulates homeostasis and terminal maturation of NKT cells. Journal of immunology. 2011;187:6335–6345. doi: 10.4049/jimmunol.1003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Friend S, Suzuki K, Phan TG, Cyster JG. Subcapsular sinus macrophage fragmentation and CD169+ bleb acquisition by closely associated IL-17-committed innate-like lymphocytes. PloS one. 2012;7:e38258. doi: 10.1371/journal.pone.0038258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YW, Deftos ML, Ojala EW, Bevan MJ. RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel KL, Tyznik AJ, Kronenberg M, Hogquist KA. Antigen-dependent versus -independent activation of invariant NKT cells during infection. Journal of immunology. 2014a;192:5490–5498. doi: 10.4049/jimmunol.1400722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel KL, Tyznik AJ, Kronenberg M, Hogquist KA. Antigen-dependent versus -independent activation of invariant NKT cells during infection. Journal of immunology. 2014b;192:5490–5498. doi: 10.4049/jimmunol.1400722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irla M, Guenot J, Sealy G, Reith W, Imhof BA, Serge A. Three-dimensional visualization of the mouse thymus organization in health and immunodeficiency. Journal of immunology. 2013a;190:586–596. doi: 10.4049/jimmunol.1200119. [DOI] [PubMed] [Google Scholar]
- Irla M, Guenot J, Sealy G, Reith W, Imhof BA, Serge A. Three-dimensional visualization of the mouse thymus organization in health and immunodeficiency. Journal of immunology. 2013b;190:586–596. doi: 10.4049/jimmunol.1200119. [DOI] [PubMed] [Google Scholar]
- Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annual review of immunology. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IL, Amiel E, Tighe M, Mohrs K, Veerapen N, Besra G, Mohrs M, Leadbetter EA. The mechanism of splenic invariant NKT cell activation dictates localization in vivo. Journal of immunology. 2013;191:572–582. doi: 10.4049/jimmunol.1300299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature immunology. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PT, Putnam A, Benlagha K, Teyton L, Gottlieb PA, Bendelac A. Testing the NKT cell hypothesis of human IDDM pathogenesis. The Journal of clinical investigation. 2002;110:793–800. doi: 10.1172/JCI15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nature immunology. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh L, Ivarsson MA, Michaelsson J, Sandberg JK, Nixon DF. Invariant natural killer T cells developing in the human fetus accumulate and mature in the small intestine. Mucosal immunology. 2014;7:1233–1243. doi: 10.1038/mi.2014.13. [DOI] [PubMed] [Google Scholar]
- Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, Besra G, Vomhof-Dekrey EE, Tighe M, Koay HF, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nature immunology. 2015;16:85–95. doi: 10.1038/ni.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, Wong CH, Schneider E, Dy M, Leite-de-Moraes MC. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. The Journal of experimental medicine. 2007;204:995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 2001;413:531–534. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. The Journal of experimental medicine. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal immunology. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci DG, Uldrich AP, Kyparissoudis K, Crowe NY, Brooks AG, Hammond KJ, Sidobre S, Kronenberg M, Smyth MJ, Godfrey DI. Intrathymic NKT cell development is blocked by the presence of alpha-galactosylceramide. European journal of immunology. 2003;33:1816–1823. doi: 10.1002/eji.200323894. [DOI] [PubMed] [Google Scholar]
- Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. The Journal of clinical investigation. 2014;124:3725–3740. doi: 10.1172/JCI72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nature reviews Immunology. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk JD, Hermans IF, Gileadi U, Chong TW, Shepherd D, Salio M, Mathew B, Schmidt RR, Lunt SJ, Williams KJ, et al. Utilizing the adjuvant properties of CD1d-dependent NK T cells in T cell-mediated immunotherapy. The Journal of clinical investigation. 2004;114:1800–1811. doi: 10.1172/JCI22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Quispe-Tintaya W, Chandra D, Jahangir A, Venkataswamy MM, Ng TW, Sharma-Kharkwal S, Carreno LJ, Porcelli SA, Gravekamp C. Direct incorporation of the NKT-cell activator alpha-galactosylceramide into a recombinant Listeria monocytogenes improves breast cancer vaccine efficacy. British journal of cancer. 2014;111:1945–1954. doi: 10.1038/bjc.2014.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Nagarajan NA, Wingender G, Wang J, Scott I, Tsuji M, Franck RW, Porcelli SA, Zajonc DM, Kronenberg M. Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. Journal of immunology. 2010a;184:141–153. doi: 10.4049/jimmunol.0902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Nagarajan NA, Wingender G, Wang J, Scott I, Tsuji M, Franck RW, Porcelli SA, Zajonc DM, Kronenberg M. Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. Journal of immunology. 2010b;184:141–153. doi: 10.4049/jimmunol.0902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K, Iwamura C, Nakajima H, Nakayama T, Taniguchi M. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. The Journal of experimental medicine. 2008;205:2727–2733. doi: 10.1084/jem.20080698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SY, Scanlon ST, Griewank KG, Constantinides MG, Savage AK, Barr KA, Meng F, Luster AD, Bendelac A. PLZF induces an intravascular surveillance program mediated by long-lived LFA-1-ICAM-1 interactions. The Journal of experimental medicine. 2011;208:1179–1188. doi: 10.1084/jem.20102630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elzen P, Garg S, Leon L, Brigl M, Leadbetter EA, Gumperz JE, Dascher CC, Cheng TY, Sacks FM, Illarionov PA, et al. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. [DOI] [PubMed] [Google Scholar]
- Venkataswamy MM, Ng TW, Kharkwal SS, Carreno LJ, Johnson AJ, Kunnath-Velayudhan S, Liu Z, Bittman R, Jervis PJ, Cox LR, et al. Improving Mycobacterium bovis bacillus Calmette-Guerin as a vaccine delivery vector for viral antigens by incorporation of glycolipid activators of NKT cells. PloS one. 2014;9:e108383. doi: 10.1371/journal.pone.0108383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, Yoshida H, Kubo M, Kawamoto H, Koseki H, Taniguchi M. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10:e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster KE, Kim HO, Kyparissoudis K, Corpuz TM, Pinget GV, Uldrich AP, Brink R, Belz GT, Cho JH, Godfrey DI, Sprent J. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal immunology. 2014;7:1058–1067. doi: 10.1038/mi.2013.122. [DOI] [PubMed] [Google Scholar]
- White AJ, Jenkinson WE, Cowan JE, Parnell SM, Bacon A, Jones ND, Jenkinson EJ, Anderson G. An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. Journal of immunology. 2014;192:2659–2666. doi: 10.4049/jimmunol.1303057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, Braun J, Mazmanian SK, Kronenberg M. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 related to Figure 1. Workflow pipeline to quantify localization of iNKT subsets. (A) Schematic representation showing CD1d tetramer immunofluorescence (IF) technique. PFA, paraformaldehyde. (B) Fluorescence intensities were quantified in each single cell identified by ImageJ and data were transferred into Prism software to gate cells. XY coordinates of gated cells were overlaid on an original image and the distribution of cells were analyzed. ROIs, region of interests. (C) Three to four different B6 and BALB/c mice were analyzed for their thymic frequencies of each subset of iNKT cells among total thymocytes either by flowcytometry (FC) or CD1d tetramer histocytometry (HC). NS, not significant (paired t-test).
Supplementary Figure 2 related to Figure 2. Regulatory T cells and IL-4 producing NKT cells reside in thymic medulla. (A) Thymi of Foxp3gfp mice were stained with RORγt and G8.8 to analyze the localization of regulatory T cells (Tregs). An arrow indicates a Treg in the cortex. C, cortex; M, medulla. (B and C) Thymi of Tbx21gfp KN2+/− mice were stained with CD1d tetramer and huCD2 molecule (B) and analyzed by histocytometric algorithms (C). Numbers indicate frequency of cells in adjacent gates. (D) Compilation of cortical and medullary frequencies of total and each iNKT subset are shown for B6 (4 mice, 7 sections) and BLAB/c (4 mice, 8 secretions) iNKT cells and B6 regulatory T cells (Treg, 3 mice, 3 sections). Each dot represents an individual section and horizontal bars indicate mean values.
Supplementary Figure 3 related to Figure 3. Flowcytometry profiles of iNKT cells in multiple organs. Representative flowcytometry dot plots and gating strategies are shown in various organs. Numbers indicate frequency of cells in adjacent gates and all dot plots have the same axis scales. Representative data of at least five independent experiments are shown. IV, intravenous; mLN, mesenteric lymph node; iLN, inguinal lymph node; IEL, intraepithelial lymphocyte.
Supplementary Figure 4 related to Figure 4. mLN NKT2 cells condition lymphocytes to express pSTAT6. (A) Nur77gfp expression in each subset of iNKT cells is shown in indicated organs at steady state. Representative histograms of at least 3 independent experiments are shown. All histograms have the same axis scales. iLN, inguinal lymph node; mLN, mesenteric lymph node. (B) iLN of B6 Tbx21gfp mice was stained with indicated antibodies and CD1d tetramer. Arrows indicate NKT1 cells (green) and NKT17 cells (blue). (C) Average frequencies of each NKT subset among total iNKT cells in T cell zone, B cell zone and medulla of iLN were analyzed in B6 (n = 6) and BALB/c (n = 6) mice. Error bars indicate standard deviation. (D) pSTAT6 expressions in CD4 T cells from spleen and iLN of B6 and BALB/c mice are shown at steady state. Numbers indicated frequencies of cells at adjacent gates and dot plots have the same axis scales. (E) Frequencies of pSTAT6 positive CD4 T cells in indicated organs are shown in B6 (n = 3 ~ 8), BALB/c (n = 3 ~ 17) and BALB/c il4ra−/− (n = 3) mice. Each dot represents an individual mouse and horizontal bars indicate mean values. Error bars show standard deviation. *p<0.0001, **p<0.001 (one-way analysis of variance (ANOVA)). NS, not significant
Supplementary Figure 5 related to Figure 5. Intravital labeling of splenocytes in B6 and BALB/c mice. B6 and BALB/c mice were intravenously (IV) injected with anti-CD45 antibody and sacrificed 3 minutes later. Splenic B cells and CD4 T cells were analyzed antibody labeled frequencies in B6 (n = 8) and BALB/c (n = 7) mice. NS, not significant (unpaired t-test).
Supplementary Figure 6 related to Figure 6. iNKT cell localization determines cellular response to αGalCer. (A) BALB/c Nur77gfpKN2+/− mice were intravenously (IV) injected with αGalCer and analyzed for Nur77gfp, surface human CD2 (huCD2) and intracellular IFN-γ expressions after 3 hours. Representative data of at least 3 independent experiments are shown. Numbers indicate frequencies of cells in each quadrant and dot plots have the same axis scales. (B) Splenocytes from BALB/c Nur77gfp mice were in vitro culture with CD1d tetramer for 4 hours and measured Nur77gfp expression in NKT1 (green) and NKT2 (red) cells. Frequencies of GFP expression were normalized at 0.1μg/ml tetramer concentrations in three independent experiments. Error bars indicate standard deviation. (C) BALB/c Nur77gfp mice were IV injected with αGalCer and analyzed Nur77gfp expression in each subset at indicated time points and compared Nur77gfp fold increased between cells in red pulp (RP, IV antibody labeled) or white pulp (WP, IV antibody unlabeled). Error bars indicate standard deviation. P values indicate unpaired t-tests. (D) Vα14 TCR transgenic NKT cells from spleen and lymph node (LN) were transferred into αGalCer immunized host mice. Donor iNKT cells in spleen were analyzed after CD1d tetramer pull down. Numbers indicated frequency of cells in adjacent gates and dot plots have the same axis scales.
Supplementary Figure 7 related to Figure 7. Oral gavage of αGalCer stimulates NKT2 cells in mLN to produce IL-4. (A) BALB/c KN2+/− mice were administrated with αGalCer either by intravenous (IV) injection or oral gavage (OG) and analyzed NKT1 (green) and NKT2 (red) cells in spleen and mesenteric lymph node (mLN) for their expression of human CD2 (huCD2) and intracellular IFN-γ at 3 and 24 hours later respectively. Representative histograms of 3 independent experiments are shown. All histograms have the same axis scales. (B) BALB/c mice were measured their serum concentrations of IL-4 and IFN-γ by ELISA after either IV injection or oral gavage of αGalCer at indicated time points (n = 5). Error bars indicate standard deviation.