SUMMARY
The introduction of vaccination in the 1950s significantly reduced the morbidity and mortality of pertussis. However, since the 1990s, a resurgence of pertussis has been observed in vaccinated populations, and a number of causes have been proposed for this phenomenon, including improved diagnostics, increased awareness, waning immunity, and pathogen adaptation. The resurgence of pertussis highlights the importance of standardized, sensitive, and specific laboratory diagnoses, the lack of which is responsible for the large differences in pertussis notifications between countries. Accurate laboratory diagnosis is also important for distinguishing between the several etiologic agents of pertussis-like diseases, which involve both viruses and bacteria. If pertussis is diagnosed in a timely manner, antibiotic treatment of the patient can mitigate the symptoms and prevent transmission. During an outbreak, timely diagnosis of pertussis allows prophylactic treatment of infants too young to be (fully) vaccinated, for whom pertussis is a severe, sometimes fatal disease. Finally, reliable diagnosis of pertussis is required to reveal trends in the (age-specific) disease incidence, which may point to changes in vaccine efficacy, waning immunity, and the emergence of vaccine-adapted strains. Here we review current approaches to the diagnosis of pertussis and discuss their limitations and strengths. In particular, we emphasize that the optimal diagnostic procedure depends on the stage of the disease, the age of the patient, and the vaccination status of the patient.
INTRODUCTION
Before childhood vaccination was introduced in the 1950s and 1960s, pertussis, or whooping cough, was a major cause of infant death worldwide (1). Widespread vaccination significantly reduced morbidity and mortality due to pertussis; however, in the last 20 years, the disease has resurged in many highly vaccinated populations (2). A number of causes have been proposed for the resurgence of pertussis, including improved diagnosis, increased awareness, waning immunity, and adaptation of the causative agent of pertussis, Bordetella pertussis (3). Pertussis resurgence is probably multifactorial, and the relative contributions of each factor may differ between countries.
The first pertussis vaccines were composed of whole, inactivated bacteria. In the 1980s and 1990s, these whole-cell vaccines (WCVs) were replaced by more defined acellular vaccines (ACVs) comprised of one to five purified B. pertussis antigens (4). Monocomponent pertussis vaccines contain pertussis toxin (Ptx) only, while multicomponent pertussis vaccines contain one or more additional antigens, including filamentous hemagglutinin (FHA), fimbriae (Fim), and/or pertactin (Prn). ACVs cause fewer side effects than WCVs. However, it has become clear that immunity induced by ACVs is less long-lasting than that induced by WCVs (5–8). Thus, the switch from WCVs to ACVs has increased the role of waning immunity in the resurgence of pertussis. ACVs induce higher levels of antibodies against Ptx than WCVs do, and this has complicated the serodiagnosis of pertussis, which is based mainly on levels of Ptx antibodies (9).
One of the hallmarks of the pertussis resurgence is that the largest increases are found in adolescents and adults (10). It has been estimated that 15% of adults with prolonged cough (>3 weeks) are infected by B. pertussis (11). Seroprevalence studies have revealed a very high circulation of B. pertussis among adolescents and adults in vaccinated populations, with estimated yearly infection frequencies varying between 1% and 9% (12, 13).
The resurgence of pertussis highlights the importance of standardized, sensitive, and specific laboratory diagnosis, the lack of which is responsible for the large differences observed in pertussis notifications between countries (14, 15). Among other factors, a reliable comparison of the pertussis burdens in different countries is important to assess the effects of different vaccines and vaccination schedules. Laboratory diagnosis is also important to distinguish between the several etiologic agents of pertussis-like diseases, which involve both viruses and bacteria (16). It has been shown that a proportion of cases of pertussis-like cough may be caused by adenovirus, parainfluenza viruses, respiratory syncytial virus, Mycoplasma pneumoniae, and Chlamydophila pneumoniae (17). The diverse etiology of coughing also includes noninfectious conditions. A reliable and specific pertussis diagnosis may prevent unnecessary and expensive diagnostic procedures. Furthermore, if pertussis is diagnosed in a timely manner, antibiotic treatment of the patient can be considered to mitigate the symptoms, and also to prevent transmission. During an outbreak, timely detection of B. pertussis is particularly important, as it allows prophylactic treatment of infants too young to be (fully) vaccinated, for whom pertussis is a severe, sometimes fatal disease. Finally, reliable diagnosis of pertussis is required to reveal trends in the (age-specific) disease incidence, which may point to changes in vaccine efficacy, waning immunity, and the emergence of vaccine-adapted strains (3).
Here we review current approaches to the diagnosis of pertussis and discuss their limitations and strengths. In particular, we show that the optimal diagnostic procedure depends on the stage of the disease and the issues to be addressed.
ETIOLOGIC AGENTS OF PERTUSSIS
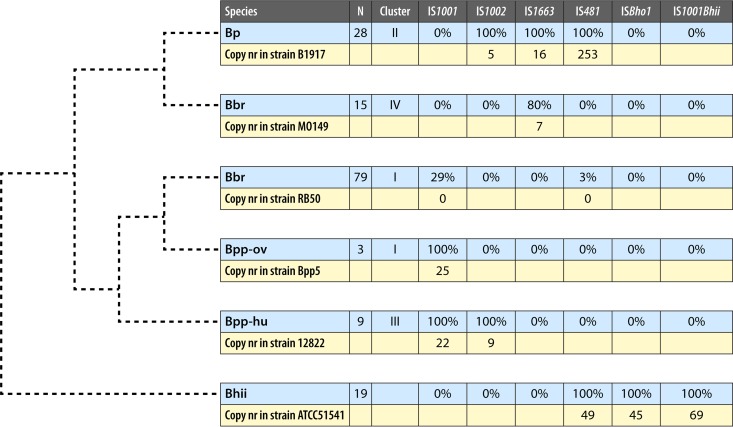
By definition, the etiologic agent responsible for pertussis infection is B. pertussis. However, pertussis-like symptoms can be caused by several other Bordetella species, including Bordetella parapertussis, Bordetella bronchiseptica, and Bordetella holmesii. The genetic relationships among the Bordetella species are shown in Fig. 1. B. pertussis, B. parapertussis, and B. bronchiseptica are closely related, whereas B. holmesii forms a distinct branch and is more closely related to the fowl pathogen Bordetella avium (18). Genetic analysis has shown that B. bronchiseptica strains form two distinct clusters, clusters I and IV, which are preferentially isolated from animals and humans, respectively (19). Interestingly, B. pertussis (cluster II) was found to be more closely related to B. bronchiseptica strains preferentially isolated from humans (cluster IV), suggesting that B. pertussis evolved from a human-adapted B. bronchiseptica lineage. As observed for B. bronchiseptica, B. parapertussis strains form two lineages, one of which clusters together with B. bronchiseptica strains preferentially isolated from animals (cluster I) and the other of which forms a distinct branch (cluster III). Cluster I and cluster III B. parapertussis strains are exclusively isolated from ovines and humans and are designated B. parapertussisOV and B. parapertussisHU, respectively (19, 20). B. holmesii has acquired DNA from B. pertussis, which, among others, contains genes for iron acquisition (18). It has been suggested that this horizontal gene transfer event may have contributed to increased circulation of B. holmesii among humans (18). Of the four Bordetella species associated with cough in humans, B. pertussis is most frequently isolated from patients suspected of having pertussis.
FIG 1.
Genetic relationships among Bordetella species associated with cough in humans, along with the distribution of IS elements. The figure is based on the work of Diavatopoulos et al. (19) and updated with genomic sequences. The phylogenetic tree (dashed lines) shows relative genetic relationships and does not indicate true genetic distances. In the blue rows, the percentage of strains containing a particular IS element is indicated for each species. N, number of strains analyzed. The percentages for ISBho1 and IS1001Bhii were determined by BLAST searches. The yellow rows show the copy numbers of IS elements, which were determined using representative strains for which complete closed genomes were available. Only copies containing at least 90% of the IS element with at least 95% identity were scored. Abbreviations: Bp, B. pertussis; Bbr, B. bronchiseptica; Bpp-ov, B. parapertussis isolated from sheep; Bpp-hu, B. parapertussis isolated from humans; Bhii, B. holmesii.
CLINICAL SYMPTOMS
Classical pertussis disease is divided into three stages: the catarrhal, paroxysmal, and convalescence stages. The catarrhal stage is characterized by nonspecific symptoms similar to those of the common cold. The paroxysmal phase, with the classical whooping cough, is the hallmark of the disease. Paroxysms may be followed by vomiting. Profuse production of mucus, which is removed by coughing, is also observed in pertussis patients. It is generally assumed that B. pertussis causes a more severe disease than that observed with B. parapertussis (21) or B. holmesii (22), while B. bronchiseptica is often found in the immunocompromised host (23, 24). Although classical pertussis can be diagnosed reliably based on clinical symptoms, infections of hosts primed by vaccination or infection often follow an atypical course, as disease is mitigated by partial immunity. In this case, laboratory diagnosis has to supplement clinical diagnosis. As discussed below, the optimal (in terms of sensitivity and specificity) diagnosis of pertussis changes with age and the degree of host immunity.
DIAGNOSIS
Case Definition
Clinical case definitions of pertussis (15, 25–27) require the presence of one or more typical clinical symptoms, such as paroxysmal cough for at least 2 weeks, inspiratory whoop, posttussive emesis, and sometimes, depending on the case definition, apnea and/or cyanosis. Clinical case definitions vary by country and/or institution (28, 29). The specificity of case definitions is negatively influenced by the time between infection and diagnosis, by previous vaccination or infection, and by increasing age of patients (30). Furthermore, the sensitivity of clinical diagnosis is low for adolescents and adults, due to the mitigated presentation of the disease.
Direct Fluorescent-Antibody Assay
Direct fluorescent-antibody assay (DFA) of nasopharyngeal samples is a simple and rapid method that relies on microscopic visualization of fluorescent antibodies directed toward B. pertussis cells. Since both the sensitivity and specificity of this assay are low (31), DFA diagnosis should always be supported by culture, PCR, or serology.
Culture
Nature of clinical specimens, sampling materials, and methods of sample collection.
Despite its low sensitivity compared to that of PCR, culture is the gold standard for pertussis diagnosis. Although in severe cases B. pertussis is also found in the lower respiratory tract (32), the preferred colonization site of B. pertussis is the upper respiratory tract. Both for culturing and for PCR, samples taken from the nasopharynx are optimal (33, 34), and these can be obtained by aspiration or by swabs (35). Sampling of aspirates is often regarded as cumbersome and requires skilled personnel, but it may give better yields (36) than sampling via nasopharyngeal swabs, which are mostly used. Swabs should have a thin flexible shaft to be able to reach the posterior nasopharyngeal area and should be composed of Dacron or nylon if both culture and PCR are to be performed. Cotton swabs contain substances that are toxic for B. pertussis, affecting its viability when plated, but they might be used for PCR purposes only. Calcium alginate swabs are appropriate only for culture, because they inhibit PCRs (37, 38). Flocked swabs can also be used; the brush-like structure has the potential to improve both sample collection and release of specimens. A study of nasopharyngeal sampling of Streptococcus pneumoniae revealed higher bacterial loads by quantitative PCR (qPCR) when nylon flocked swabs were used than when Dacron swabs were used (39). However, although flocked swabs are promising, no data have yet been presented for the recovery of B. pertussis cells by use of flocked swabs, although they were used in one study (40). Oral fluid was used to diagnose pertussis by PCR in a limited study, and it was found to be as sensitive as nasopharyngeal swabs (C. Heuvelman and F. R. Mooi, unpublished data). Sampling of oral fluid is less stressful for the patient than sampling of the nasopharynx, but it is unsuitable for culture due to the high level of contamination with resident microbiota.
Review of culture media, methods, and conditions.
Bordet-Gengou and Regan-Lowe agars are the media of choice for culture of clinical specimens to detect B. pertussis. Addition of the antibiotic cephalexin has been recommended to inhibit growth of contaminating bacteria. However, since cephalexin has been suggested to also inhibit growth of B. holmesii (41), the addition of methicillin or oxacillin or plates with and without cephalexin should be used. Most critical for optimal sensitivity of culture is rapid specimen transport (<24 h) in a suitable transport medium (42). If transport of specimens is involved, enrichment media are recommended, such as Regan-Lowe transport medium or Stainer-Scholte broth. The shelf life of Regan-Lowe medium is longer (8 weeks) than that of Bordet-Gengou fluid medium (1 week), which should be freshly made.
Growth of bordetellae is accomplished by incubation of agar plates at 35 to 37°C in a high-humidity environment with low levels (<4%) of CO2. Incubation periods of up to 12 days are recommended for optimal sensitivity, as growth of B. pertussis and B. holmesii may be retarded (43). B. bronchiseptica usually grows faster, with visible colonies after 1 to 3 days, while B. parapertussis shows an intermediate growth rate. Growth should be checked daily to prevent overgrowth by contaminating microorganisms. After growth, bordetellae can be identified by biochemical reactions, agglutination with specific sera, or, preferably, PCR.
Bordetellae can be distinguished biochemically by oxidase, urease, and citrate utilization and nitrate reduction and microbiologically by growth rate, motility, and production of a brown pigment when grown on tyrosine agar.
The routine use of culture for diagnosis of pertussis has declined since the introduction of PCR methods (16). This is unfortunate, as strains are required for more in-depth analyses, such as whole-genome sequencing, transcriptomics, and proteomics (44–47). A convenient way to obtain isolates, without compromising PCR, is to streak a nasopharyngeal swab on selective media after elution of the swab with water or physiological salt for PCR. In our hands, B. pertussis colonies can be recovered from 10% to 30% of swabs which give a positive PCR result (Heuvelman and Mooi, unpublished).
PCR Assays
Introduction.
PCR assays have become an established method for detection and identification of causative agents of pertussis (48–50). These assays have evolved from conventional or block-based PCR to real-time PCR and from monoplex (singleplex) PCR to multiplex PCR with at least an additional internal control. Conventional PCR employs 2 primers that generate fairly large DNA fragments (amplicons) to allow their visualization on agarose gels. Visualization of amplicons is accomplished by capillary electrophoresis or by agarose gel electrophoresis, which requires staining of DNA with an intercalating agent, such as ethidium bromide or the less hazardous stain SYBR green. The sensitivity and specificity of PCR may be enhanced by subsequent amplicon hybridization or by hybridization of internally situated labeled oligonucleotides. A variant is conventional (hemi)nested PCR, in which one or two extra primers are directed to an internal fragment of the first amplicon in a second round of PCR. Conventional PCRs, as opposed to real-time PCRs, have the disadvantage that they are prone to contamination due to the required post-PCR analysis. This is especially true for the nested PCRs, which have to be subjected to a second PCR.
The conventional PCR assays have generally been replaced by real-time PCR methods, which mostly use hydrolysis of labeled probes to release a reporter that produces an accumulating fluorescence signal with every amplification cycle to enable monitoring of the PCR results in real time. Real-time PCR amplicons are usually chosen to be short (<200 bp), often allowing only space for the forward and reverse primers and the internal probe. Short real-time products are more efficiently amplified and allow shorter elongation times, resulting in faster results. Instead of using fluorescent probes, specific amplification of PCR can also be detected by high-resolution melt (HRM) analysis. HRM analysis starts after the PCR process, by heating the sample to denaturate the amplicon, which is stained with an intercalating fluorescent dye. The decrease in fluorescence is measured during the increase in temperature until the PCR product is completely denatured and single stranded and the intercalating dye can no longer bind. Another method for amplifying DNA is isothermal loop-mediated amplification (LAMP). This method is carried out by using a DNA polymerase with high strand displacement activity and autocycling at a constant temperature. Two sets of target-specific forward and reverse primers are used (51, 52). To initiate LAMP, all four primers are employed to form a dumbbell structure (51). Subsequently, only the inner primers are used for amplification by strand displacement DNA synthesis, resulting in stem-loop DNA products which can be visualized with an intercalating agent, such as SYBR green. Due to the use of multiple primer sets, LAMP is highly specific (53). However, the use of multiple primers per target increases the chance of primer-primer interactions and decreases the possibilities for multiplexing. Another qualitative isothermal assay is DNA helicase-dependent amplification (HDA). A single-stranded DNA is formed by use of DNA helicase, facilitating primer hybridization and the subsequent extension by DNA polymerase (54).
Following the development of real-time multiplex PCR for detection of one or more related pathogens, the latest trend in PCR-based detection is the syndromic approach. This involves development of assays that are symptom based and can detect multiple respiratory pathogens, including B. pertussis (55–58). A disadvantage of the latter approach can be that multiple sets of primers may reduce the sensitivity of detection.
PCR samples.
For PCR, the same swab as that used for culture can be used (59). If only PCR is performed, swabs can be sent dry (60). Liquid transport medium should be avoided because of the potential for contamination of the liquid medium during transit should the liquid wash over the handle, which may have been contaminated by mishandling during specimen collection (61). Swabs can be suspended in physiological saline or molecular-grade water and boiled to release DNA (but see the paragraph on culture, above). To decrease inhibition of PCRs, purification of DNA is recommended. Extraction of DNA/RNA from clinical samples can be performed manually or with a fully automated system. The manual procedures include phenol-chloroform extraction, use of chaotropic agents, such as guanidine thiocyanate, and use of resins for purification by column extraction. For manual extraction, column purification kits are recommended (62) because of their yield and ability to remove inhibitors. Ion-exchange chromatography methods perform well for extraction of DNA, although their effective removal of inhibitors varies (63).
In the contemporary clinical laboratory, fully automated extraction systems are becoming the standard. Automated systems offer several advantages over manual handling by reduction of the risk of contamination, reduction of PCR inhibition, and standardization. In a comparison of automated and manual extractions, no significant differences were found in recovery (64), as confirmed by Caro et al. (65).
IS elements as targets for PCR.
Insertion sequence (IS) elements are mobile DNA fragments of approximately 1,300 bp that have terminal inverted repeats and contain an open reading frame encoding a transposase (tnpA). IS elements are generally present in multiple copies in genomes, presenting excellent targets for highly sensitive PCR detection. Although IS elements specific for different Bordetella species have been found, it should be noted that, by their very nature, IS elements may be transferred between different species. Indeed, there is strong evidence that B. pertussis, B. bronchiseptica, B. parapertussis, and B. holmesii exchanged IS elements (18, 19). The distribution of IS elements within the Bordetella species associated with cough in humans and their estimated copy numbers are shown in Fig. 1. Copy numbers are based on the complete, closed genome sequences of representative strains.
IS481 (66) and IS1001 (67) were assumed to be specific for B. pertussis and B. parapertussisHU, respectively, in which they occur at copy numbers of 253 and 22, respectively. However, both IS elements have also been found in other Bordetella species. IS481 is present in all B. holmesii isolates analyzed to date, with a copy number of approximately 49. IS481 was probably acquired by B. holmesii from B. pertussis, together with genes involved in iron uptake (18). IS481 is also found in up to 3% of B. bronchiseptica strains belonging to cluster I (Fig. 1). IS1001 is found not only in human B. parapertussis strains but also in ovine B. parapertussis strains, at copy numbers of approximately 25 (18, 68, 69). Furthermore, IS1001 is also present in 29% of B. bronchiseptica strains belonging to cluster I.
IS1002 (69) is found in all B. pertussis and B. parapertussisHU strains analyzed to date, with approximately 5 and 9 copies, respectively. In addition to IS481, two other IS elements have been detected in B. holmesii strains: ISBho1 (previously designated bhoA [18]) and IS1001Bhii (18). These are present at approximate copy numbers of 45 and 69, respectively. IS1663 (47) is present at approximately 16 copies in all analyzed B. pertussis strains. It is also found in 80% of cluster IV B. bronchiseptica strains. The copy number in a representative of cluster IV B. bronchiseptica strains was found to be seven.
Numerous PCR assays to detect bordetellae have been described, including conventional PCR (70–79), (semi)nested PCR (80–84), real-time PCR (85–89), and LAMP (53) assays. Many studies have shown that due to the presence of multiple copies in the Bordetella genomes, PCRs targeting IS elements are highly sensitive. The IS elements IS481 and IS1001 are the most used targets for detection of B. pertussis and B. parapertussis, respectively, by PCR (66, 67). For simultaneous detection of B. pertussis and B. parapertussis, multiplex PCR assays targeting both IS elements have been developed (90–101).
The specificities of IS481 and IS1001 PCRs have been studied widely by analysis of genetically related pathogens or pathogens that occupy the respiratory tract (102). No PCR positivity for these targets has been observed outside the Bordetella genus. However, as discussed above, both IS481 and IS1001 have been found to be present in other Bordetella species (Fig. 1) (103–106), necessitating the inclusion of additional targets to increase specificity. For example, for discrimination of B. pertussis, B. parapertussis, and B. bronchiseptica, the ptxP promoter region was utilized in many studies (107–110), but it showed a lower sensitivity than that with the high-copy-number IS sequences (111).
To overcome nonspecific cross-reactions with other members of the Bordetella genus, dual targets for B. pertussis, utilizing IS481 and one other target, such as IS1002, the pertussis toxin promoter (ptxP), or the open reading frame BP0283, have been shown to be useful (112–118). The combination of IS481 and the promoter region for pertussis toxin, ptxP, has been used most often (101, 119). However, ptxP PCR suffers from drawbacks, because due to sequence variation over the ptxP region (120, 121), false-negative results may arise (122). The purpose of dual-target PCR is to discriminate B. pertussis from other Bordetella species and thus to increase the specificity. Using two multicopy targets would ensure a higher degree of sensitivity.
For optimal sensitivity of multitarget PCRs, IS1002 may be used in addition to IS481 and IS1001. In this case, positive results for both IS1002 and IS481 are indicative of B. pertussis, while a positive result only for IS481 is indicative of B. holmesii. Furthermore, if both IS1001 and IS1002 PCRs are positive, this is indicative of B. parapertussisHU, while either a positive IS1001 or IS1002 PCR is indicative of a B. bronchiseptica infection, as both IS elements are carried only by B. parapertussisHU. As the copy number of IS481 is significantly larger than those of the other Bordetella IS elements discussed here, a high IS481 quantification cycle (Cq) value (i.e., the number of amplification cycles needed to produce a positive signal) must be interpreted with caution, because the lower limit of detection (LLOD) of IS elements with lower copy numbers may have been passed (Fig. 2). B. holmesii infections may also be identified by targeting ISBho1 or IS1001Bhii, both of which are specific for B. holmesii (99, 100).
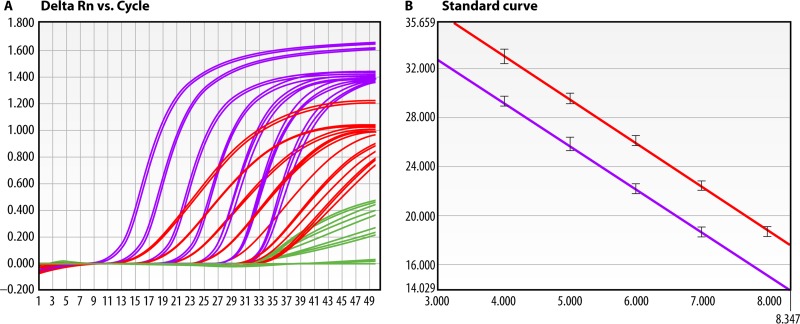
FIG 2.
Significant differences in target copy numbers in multiplex PCRs may result in incorrect identification at high Cq values. Cycle threshold (Cq) values range from 1 to 50 and are depicted on the x axis. The threshold (horizontal line) is arbitrary and just above the curve breakpoints. (A) Amplification curves for multiplex PCR targeting IS481 (purple) and IS1002 (red) in the presence of an internal control (phocine herpesvirus [green]) of a clinical sample positive for B. pertussis. At high concentrations of B. pertussis DNA, amplification of the internal control (green) is outcompeted for reaction components. (B) At low concentrations of DNA, a result interpretation error may arise because of the difference in the lower limits of detection for IS481 (high copy number) and IS1002 (low copy number). In this case, IS481 PCR shows a positive signal over the lowest concentrations, whereas IS1002 PCR is negative. A single amplification curve for IS481 can also indicate a B. holmesii infection (see the text). At high Cq values, this multiplex PCR may give a false B. holmesii result when B. pertussis is in fact the causative agent. (Courtesy of Lieuwe Roorda [reprinted with permission].)
In addition to IS elements, other, single-copy sequences specific for a particular Bordetella species have been used, including the ptxA (123, 124), cyaA (125), prn (126, 127), recA, carB, and bhur (128) genes and a region upstream of the outer membrane porin gene (129). Genes of unknown function, designated BP283, BP485 (130), and BP3385 (131), have also been targeted for PCR.
As discussed above, if IS481 PCR is combined with a single-copy target, this may lead to an increased risk of interpretation errors. Furthermore, the genomes of bordetellae are not static, and strains may differ significantly in gene content (132–134). Thus, with single-copy targets, a negative result may be due to gene loss.
In summary, the extremely high copy number of IS481 facilitates a high sensitivity of B. pertussis detection but, at low DNA concentrations, leads to a proportion of B. pertussis IS481 PCR-positive results which cannot be confirmed by an additional PCR with another target present at a lower copy number. However, in case of a controlled positive PCR, when contamination can be excluded, it may still be regarded as proof for an infection with B. pertussis.
Commercial PCR assays.
Several commercial assays are available for detection of B. pertussis and B. parapertussis (135–138). Most assays target IS481 and IS1001 in a real-time multiplex PCR format; examples are the assays available from Elitech, Eragen, and Focus Diagnostics (135), Cepheid (136), and Diagenode (137). The PCR kit available by Argene (137) targets IS481 only. All assays contain an internal control, and some assays also use an extraction control (Focus, Argene). Comparisons of these assays were performed with in-house real-time PCR assays for detection of B. pertussis (135–137). The sensitivities of the commercial assays were found to range from 96% to 98% compared to the in-house assays. With the GenoQuick PCR kit for detection of B. pertussis and B. parapertussis (139), dipstick hybridization is performed to visualize bands of specific sizes, but the sensitivity of this system is unknown. Another assay is provided by Qiagen (138), employing 4 separate real-time PCRs, for detection of IS481, IS1001, FHA (the filamentous hemagglutinin gene), and the internal control. Differentiation of B. pertussis, B. parapertussis, and B. bronchiseptica (but not B. holmesii) is performed by HRM analysis of FHA amplicons. Evaluation of 6 commercial PCR assays for detection of multiple respiratory pathogens, including B. pertussis (140), showed high specificities but low sensitivities.
Although various commercial assays approximate the sensitivity of in-house PCR assays, the lack of specificity of IS481 for B. pertussis is not always addressed. Another drawback of commercial assays is that they may not be adjusted in a timely fashion when novel sequence data show that the target is polymorphic, not specific, or not found in all strains. The latter is particularly relevant for B. pertussis, which shows gene loss over time (134, 141). However, for laboratories with less experience in molecular biology, performance of some commercial PCR assays may be acceptable, because they can detect a larger portion of B. pertussis infections than culture alone. Although some primer sets for detection of Bordetella are FDA cleared (Focus), none of the complete assays discussed here has received FDA clearance. CE-IVD-marked commercial kits for detection of Bordetella can be provided by Focus and Argene.
Contamination prevention.
With PCR diagnosis, contamination is a concern (142, 143). Carryover of clinical samples may occur (34), or contamination of samples with PCR amplicons (144) or even aerosolized vaccine (145) is possible. Most contaminations are caused by post-PCR handling of samples, which is necessary with conventional PCR methods, and most notably in nested PCRs, which require opening of vials after the first PCR to initiate a second round of PCR.
To prevent contamination of clinical samples, physical separation of pre- and post-PCR rooms, air pressure control, and the use of uracil-N9-glycosylase for removal of contaminating amplicons may be implemented.
The implementation of real-time PCR methods has greatly reduced the risk of contamination, as, in principle, the PCR system is a closed system requiring no post-PCR handling, even if DNA melting curves obtained by high-resolution melting are used to differentiate between amplicons. Several commercial assays are also closed systems, and some can be used with direct clinical samples, thus reducing or nearly eliminating contamination risk.
Internal controls.
The inclusion of internal controls in PCR assays offers a means to monitor the reliabilty of PCRs (146). The recommendations for controlled PCR assays are as follows: inclusion of at least one negative (no template) control per PCR run for monitoring carryover of amplification products or samples, inclusion of a positive control in each PCR run to ensure the validity of the PCR mixture, and addition of an internal control to each PCR vial to detect inhibition of PCR. When a large proportion of samples is expected to be positive, e.g., in an outbreak setting, alternate inclusion of a negative control with every clinical sample is recommended. Positive controls can consist of purified DNA or suspensions of B. pertussis cells. Inhibition can be measured by spiking of negative samples with low concentrations of the positive control in a second round of PCR, but inclusion in a multiplex PCR is more efficient. Various internal controls have been constructed for this purpose (147–149). If the internal control is added to the sample prior to DNA/RNA extraction, it can be used to monitor the extraction procedure as well. In some studies, a control for adequate sampling is included by coamplification of human DNA (79, 150, 151) and may also be used as an inhibition control. Since the input of human DNA cannot be controlled, it may be less suitable as an inhibition control.
PCR assay validation.
An excellent guideline for validation of laboratory-developed molecular assays for infectious diseases was presented by Burd in 2010 (152), and the following validation parameters are recommended: a study of the reportable range or linearity of PCR amplification of serial dilutions and assessments of the lower limit of detection, the analytical sensitivity, and the reproducibility or precision of intra-assay variation. These concepts are illustrated in Fig. 3. Thorough validation of (semi)quantitative real-time PCR is also supported by the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines (153), which recommend detailed validation description in publications. In the past, many publications lacked sufficient detail to allow the reader to appreciate the quality of the presented PCR assays.
FIG 3.
Analytical validation of a multiplex PCR for detection of B. pertussis, targeting IS481 and IS1002 in the presence of an internal control (phocine herpesvirus). (A) A 1:10 dilution series of B. pertussis DNA ranging from approximately 105 to 10−2 cell per PCR. The LLODs for IS481 (purple) and IS1002 (red) are 10−2 and 1 cell, respectively. Intra-assay reproducibility is demonstrated by triplicate reactions. The heights of curves (δRn) are dependent on the probe labels. (B) The efficiency and linearity of assays are given by standard curves. The efficiency of PCR is deducted from the slope of the standard curve and is 94% for IS481 PCR and 92% for IS1002 PCR. Linearity is the range over which an increase in input DNA results in an increase of amplified product. The linear range of IS481 PCR comprises 103 to 10−1 cell, and that of IS1002 PCR is 104 to 1 cell and includes the true LLOD. (Courtesy of Lieuwe Roorda [reprinted with permission].)
The accuracy or trueness of the test is investigated by comparison of different methods or by comparison between measured Cq values and actual copy number estimates.
IS481 is present as more than 200 copies in the genome of B. pertussis, and theoretically, PCR may detect as few as 0.02 B. pertussis cell. Whether this is meaningful in surveillance or diagnosis is under debate (154). Based on a comparison of multitarget PCR and single-target PCR, a cutoff Cq value (>35) for IS481 PCR has been suggested (155). Assessment of a reportable range of Cq values would enable a more precise quantification and determination of whether whole B. pertussis bacterial cells are detected (>200 copies).
It should be noted that a positive PCR result may not always be clinically relevant, as PCR does not distinguish between viable and nonviable bacteria. The persistent presence of B. pertussis DNA has been detected by PCR after treatment of patients with antibiotics for up to 21 days (80, 156).
Whether or not the patient should be retested for the presence of B. pertussis after antibiotic treatment depends on the clinical picture and factors such as possible resistance to antibiotics. This decision is best left to the clinician treating the patient.
Quality assessment.
Yearly participation in an external quality control (QC) program is highly recommended to maintain adequate sensitivity and specificity of the PCR assay in use. Proficiency panels distributed to European laboratories showed variations in sensitivity of 1,000- to 10,000-fold and showed misidentifications of B. bronchiseptica, B. holmesii, and B. parapertussis (157). A quality assessment program in France revealed sensitivities of 0.2 to 2 CFU/μl and a mean specificity of 94.3% (65). However, despite the fact that real-time PCRs were used, the rates of false positivity ranged from 0% to 18.7%. A study conducted among U.S. laboratories which use real-time PCRs also revealed some cross-contamination. Only B. pertussis was distributed and at least 1 false-positive result was reported for 5% of the contributing laboratories (158). Distribution of a panel of DNA samples of B. pertussis, B. parapertussis, and B. holmesii among national reference laboratories in 25 European countries revealed that several laboratories were unable to discriminate between DNAs from different bordetellae and that several of the assays used lacked sufficient sensitivity (159). A recent performance exercise among U.S. public health laboratories showed that 79% differentiated B. pertussis and B. holmesii and 72% identified B. parapertussis (160). In a recent study, correct Bordetella species identification was evaluated between U.S. commercial laboratories and the CDC. This study demonstrated 83.4% agreement between two U.S. commercial laboratories and the CDC and little misidentification of Bordetella species during the 2012 U.S. epidemic (161).
These findings underline the importance of continued monitoring of PCR assays by well-defined interlaboratory quality control programs. Apart from the contributions to quality assessment schemes, repeatedly performed standard curves should be made by the laboratory to maintain and monitor the quality of the real-time PCR assay. This should be done on a yearly basis or with every new batch of primers/probes. In addition, Cq values for controls should be monitored, e.g., in a Levy-Jenkins plot (152), to ensure sufficient sensitivity and reproducibility of the assay.
Serodiagnosis of Pertussis
Serodiagnosis is among the earliest techniques used to confirm the clinical diagnosis of pertussis. Many of the problems associated with serodiagnosis encountered in the early days still persist, such as the interference of previous vaccinations or previous infections with serodiagnosis, cross-reactivity with other Bordetella species or perhaps other bacteria, and the variable response to B. pertussis antigens. However, by using purified antigens, in particular Ptx, serodiagnosis has become the most sensitive way to establish infections by B. pertussis of sufficient duration to have mounted an immune response, i.e., relatively late in disease.
Clinical case definitions of pertussis have limited specificity (162–164). Therefore, in the following, for assessment of the sensitivities of serodiagnosis methods, we used studies, as much as possible, in which clinical suspicion of pertussis was confirmed by positivity of culture and/or PCR for B. pertussis.
Enzyme-linked immunosorbent assay (ELISA).
After establishment of B. pertussis as the etiologic agent of whooping cough by Bordet and Gengou in 1906 (165), for a period of circa 80 years the complement fixation (CF) assay and the bacterial agglutination (BA) assay were the tests most used for serodiagnosis of the disease (166–178). With the CF assay, it was demonstrated that the dynamic phase of the immune response to infection starts 1 to 4 weeks after the onset of symptoms, reaching peak levels 4 to 7 weeks after the onset of symptoms, with the latest response in very young immune-naive children (168). Because of the high prevalence of BA and CF titers in population sera, serodiagnosis was taken to require the demonstration of significant (≥4-fold) increases of titers in serum pairs, i.e., in acute- and convalescent-phase sera, obtained at presentation and 2 to 4 weeks later, respectively. However, for the CF assay, it was suggested that the finding of strong CF activity in a first (single) serum could be considered supportive for diagnosis: high CF titers induced by infection were shown to decrease again quite rapidly, and sera of individuals who had a history of whooping cough in the previous 2 to 30 years mostly contained low CF activity, with none containing strong CF activity (168).
Since the 1980s, the CF and BA assays have gradually been replaced by ELISAs for serodiagnosis of pertussis. For different ELISAs, various B. pertussis antigens have been used to detect antibodies. In direct comparisons, ELISAs were shown to have a better diagnostic performance than that of the CF and BA assays (179–183). Concomitantly, the Chinese hamster ovary (CHO) cell assay was developed, in which antibodies to Ptx were quantified based on their ability to neutralize the toxic effect of Ptx on CHO cells, which results in cell clustering (184, 185). However, in paired sera from patients with pertussis, significant increases in Ptx antibodies were detected more often with an ELISA than with the CHO cell assay (186–188). An additional advantage of ELISA over other immunoassays is its ability to differentiate IgM, IgA, and IgG antibodies. Also, compared with the older immunoassays, the accuracy of quantitative measurements with ELISAs is much higher, allowing the definition of more precise diagnostic cutoff values for significant increases of antibody levels in paired sera and of diagnostic cutoff values for high antibody levels in single sera.
Suitability of IgM, IgA, and IgG antibodies for diagnosis.
In young unvaccinated children with culture-confirmed pertussis, IgM responses occurred as frequently as or more frequently than IgG responses (186, 189, 190). However, IgM responses were slow and often absent in vaccinated children and adults with pertussis, i.e., in those who were primed with antigens of B. pertussis (191, 192).
In studies in which IgM, IgA, and IgG antibodies were measured in sera from patients with well-documented pertussis and with various ages and vaccination histories, the common finding was that the IgG parameters were most sensitive (31, 180, 182, 191, 193, 194). Likewise, in studies in which IgG and IgA antibodies were measured, IgG parameters were more sensitive than IgA parameters, and combinations of IgG and IgA parameters (with “and/or” interpretation) did not enhance or only slightly enhanced the sensitivity (186, 187, 195–198). In young children aged <4 years, IgA responses to infection may be very low or even absent (199), and in the first 10 to 15 years of life, IgA responses to B. pertussis infection tend to increase with age (186, 196). Also, the prevalence of B. pertussis-specific IgA antibody in the population tends to increase with age (199, 200). In several studies, IgA-Ptx was shown to be less sensitive than IgA-FHA (187, 198, 201, 202).
Despite the shortcomings of IgA levels for the diagnosis of pertussis, interest in measurement of IgA antibodies for serodiagnosis remains, because primary vaccinations with WCVs or ACVs in the first year of life induce IgM and IgG antibodies but do not induce IgA antibodies (177, 203–206). Boosting with ACV at the age of 4 or 9 years in some cases induced low levels of IgA antibodies to antigens contained in the vaccine, somewhat more in WCV-primed children than in ACV-primed children (204). Booster vaccination of adolescents and adults with ACV has been shown to induce IgG as well as IgA antibodies, although IgA responses were less frequent and less strong (207).
In conclusion, measuring IgG antibodies to B. pertussis antigens gives the best results in terms of sensitivity for all age groups. Measuring IgA may be useful to distinguish between recent vaccination and recent infection.
ELISAs based on antigens present in acellular vaccines.
Several studies have compared the abilities of ELISAs based on different B. pertussis antigens to detect increases in specific antibodies by using paired sera from patients with mostly culture-confirmed pertussis. The ability was similar or slightly superior for Ptx-based ELISAs compared to FHA-based ELISAs (177, 179, 193, 198, 206, 208, 209) and was considerably better for Ptx-based ELISAs than for ELISAs based on Prn, fimbriae (177, 198, 202, 206, 208), outer membrane protein extracts, or sonicates of B. pertussis cells (189). In young children with pertussis, IgG responses to fimbriae occurred only in those who had been primed by vaccination with vaccines containing fimbriae (210).
ELISAs based on antigens not present in acellular vaccines.
The introduction of ACVs raised the possibility of distinguishing between vaccination and infection by using antigens not present in the vaccine. Several B. pertussis antigens absent from ACVs have been tested for use in serodiagnosis, including adenylate cyclase toxin (ACT), the catalytic domain of ACT, comprising the N-terminal 400 amino acids (CatACT), Bordetella resistance to killing protein A (BrkA), a peptidoglycan-associated lipoprotein (PAL; BP3352), B. pertussis lipooligosaccharide (LOS-Bp), and B. parapertussis lipopolysaccharide (LPS-B.para).
In paired sera from unvaccinated children with pertussis or parapertussis, responses of IgG antibodies to ACT occurred. However, responses were low or even absent in children with pertussis who had been vaccinated with WCV or ACV (211), and vaccination with WCV or 5-component ACV did not induce IgG-ACT (211, 212). The authors suggested that the poor IgG-ACT response in vaccinated children can be explained by a preferential response to antigens for which they were primed, a phenomenon known as original antigenic sin (213). In adults with pertussis, IgG-ACT concentrations increased 3- to 4-fold, and high concentrations persisted during a follow-up of 28 months, as did titers to FHA and Prn, while titers to Ptx decreased. Due to similarities between the C-terminal region of ACT and other bacterial toxins (e.g., alpha hemolysin of Escherichia coli) (212), low titers of IgG-ACT in human sera may be due to cross-reactions. Given these findings, the authors concluded that intact ACT cannot be used for serodiagnosis of pertussis.
In another ELISA-based study, the usefulness of five nonvaccine antigens for the serological diagnosis of pertussis was compared to that of the conventional vaccine antigens, Ptx and FHA (191). The nonvaccine antigens included the catalytic domain of the adenylate cyclase toxin (CatACT), the C-terminal region of FHA (C-FHA), LOS, PAL, and the BrkA protein. The serological responses of individuals with culture-confirmed pertussis were compared to those of adults with no recent history of a coughing disease. Antibody responses to LOS, PAL, and BrkA were not found to be useful for the serodiagnosis of pertussis. The ELISA based on the detection of anti-Ptx IgG was the most sensitive (92.2%) test for serodiagnosis of pertussis. Compared to the results for anti-Ptx IgG, measuring antibodies against nonvaccine antigens resulted in lower sensitivities (sensitivities for anti-CatACT IgG, anti-C-FHA IgG, and anti-LOS IgA of 62.8%, 39.2%, and 29.4%, respectively).
Cross-reactivities of antibodies against B. pertussis with other pathogens.
B. pertussis, B. parapertussis, and B. bronchiseptica are highly related and share most of their surface-exposed proteins (47). It is to be expected, therefore, that infection with B. parapertussis or B. bronchiseptica will result in cross-reacting antibodies against B. pertussis. However, B. pertussis does contain unique antigens, as it is the only Bordetella species that produces Ptx. Until now, no indications have been found that other antigens can induce antibodies that cross-react with Ptx.
Cross-reacting sera have been well documented in the literature. Antibodies to WCV B. pertussis in ELISA cross-react with B. parapertussis and B. bronchiseptica (214, 215). Cross-reacting antibodies are induced by both FHA and Prn. Increases of antibodies to B. pertussis FHA occurred in 63% of children with culture-confirmed B. parapertussis infection (23). In serial sera from 23 symptomatic patients from whom B. parapertussis was cultured or who had a family member with culture-confirmed infection with B. parapertussis, antibody responses to B. pertussis-derived FHA, Prn, and Fim2 occurred in 83%, 65%, and 35% of the patients, respectively (206). Surprisingly, antibodies to Ptx were detected in three patients. Since B. parapertussis does not produce Ptx, the presence of Ptx antibodies suggests a coinfection of B. parapertussis and B. pertussis. In another study, paired sera from 8 of 11 patients with culture-proven B. parapertussis showed an antibody response to FHA, while none of the patient sera bound to Ptx (187).
The amino acid sequence of FHA is similar to those of immunogenic surface-exposed proteins of nontypeable Haemophilus influenzae, and antibodies to FHA recognize FHA-like proteins of nontypeable (i.e., nonencapsulated) H. influenzae, and vice versa (216). However, in paired sera from elderly patients with bacteriologically documented infection with nonencapsulated H. influenzae, significant increases of IgG-FHA antibodies were absent (217).
The absence of Ptx antibodies in sera which do contain antibodies to other B. pertussis antigens is often taken as evidence for infection by Bordetella species other than B. pertussis, or even by nonbordetella respiratory pathogens. For example, in single sera from 54 soldiers with a long-lasting cough, high levels of antibodies to B. pertussis FHA were found in the absence of high levels of antibodies to Prn and Ptx in 15 sera; 8 sera had high reactivity to B. pertussis FHA and Prn and not to Ptx (218). In follow-up sera from unvaccinated children, obtained at ages 2, 4, 6, 13, and 30 months, ≥2-fold increases of IgGs to B. pertussis FHA and Prn occurred between the ages of 13 and 30 months in 20/44 and 10/44 children, respectively, while none had detectable IgG-Ptx antibodies at those time points. Furthermore, in children from whom sera had been obtained at ages 13 months and 6 years, ≥2-fold increases of IgGs against B. pertussis FHA and Prn in the absence of detectable IgG-Ptx occurred in 11/14 and 8/14 children, respectively (219). In serial sera from 71 children, obtained before, during, and after vaccination with a monocomponent Ptx-containing ACV vaccine, maternally derived IgG-Prn and IgG-FHA declined in the first 3 months of life. After the age of 1 year, IgG-Bp-FHA and IgG-Bp-Prn reemerged at relatively low titers, at such a rate that at 36 months, all 71 children had detectable IgG-Bp-FHA and 58 of 71 children had detectable IgG-Bp-Prn, while in this study period, none of the children had had symptoms compatible with pertussis (220). Between the ages of 1 and 2 years, 25% and 31% of children had significant increases (≥3-fold) of IgG-Bp-FHA and IgG-Bp-Prn, respectively (endpoint titrations); between 2 and 3 years, those percentages were 21% and 13%.
In conclusion, the only antigen as yet which is specific for B. pertussis in serological assays is Ptx. Antibody responses to Ptx have sporadically been observed in patients with bacteriologically documented infection with B. parapertussis (199, 206), but perhaps in those cases, coinfection with B. pertussis was missed. Such coinfections have been documented to occur (21, 198).
Accuracy and comparability using reference sera.
At the Center for Biologics Evaluation and Research (CBER) of the Food and Drug Administration (FDA), standard reference sera with defined contents of IgG and IgA antibodies to Ptx, FHA, and Prn have been made available for several decades. For ELISAs using those reference standards, results can be expressed in CBER ELISA units (EU) per milliliter (175, 221, 222). More recently, at the request of the WHO and using the CBER reference sera, “the first international standard for pertussis antiserum” was prepared, which is defined to contain the following per ampoule: 335 IU IgG-Ptx, 65 IU IgA-Ptx, 130 IU IgG-FHA, 65 IU IgA-FHA, 65 IU IgG-Prn, and 42 IU IgA-Prn (223). This WHO standard, IS 06/140, can be obtained from The National Institute for Biological Standards and Control (NIBSC), London, United Kingdom.
Various methods to transform optical densities (ODs) measured in ELISA to units by calibration with a reference serum have been investigated; these include reference line units (the slope of the dilution curve for the patient serum is adapted to the slope of the reference curve; median OD reading), nonparallel line units (no adaptation of slopes; low OD reading at x axis crossings), parallel line units (adaptation of both curves to the same degree; median OD reading), single-point reference line units (one dilution of test serum), and endpoint titer reading at the dilution of test serum with an OD close to the background (224). The assay using reference line units had the lowest intra-assay coefficients of variation (CVs) (4 to 7% versus 6 to 31% for the others) and the lowest interassay CVs (12 to 14% versus 12 to 47% for the others), the assay using single-point reference line units was second best, and the assay using endpoint titer calculation resulted in the highest CVs.
Investigators with CBER (FDA) showed that ELISAs that measured IgGs to Ptx, FHA, Prn, and serotype 2 and 3 fimbriae and used CBER reference standards and the reference line method for calculation had CVs that were consistently <20% for sera with antibody concentrations ≥4 times higher than the minimal level of detection. Results from two laboratories correlated well, with R values of ≥0.93 for the four IgG-ELISAs (222). ELISAs of that format were used in the various field trials of ACVs and WCVs that were conducted toward the end of the last century to provide accurate and comparable measurements of antibody responses as well as for assessments of vaccine efficacy through contributing to case finding (198, 201, 206, 225–228).
In a collaborative study, 33 participating laboratories from the United States, Canada, Australia, Japan, and various countries in Europe each used their own ELISAs to measure IgGs to Ptx (32 labs), FHA (30 labs), Prn (17 labs), and fimbriae (13 labs) in 21 samples in ways that allowed calculation of intra- and interassay CVs over a range of values, from low to high (221). For less than half of the ELISAs, CVs of <20% for 75% of the samples were reached. Assays measuring fimbrial antibodies were the least precise. The best comparisons between laboratories occurred in the 10 laboratories using the CBER protocol (222), applying CBER reference sera, with various dilutions of each reference serum as well as the test serum, and using the reference line method for calculation. Correlation of IgG-Ptx ELISAs with the IgG-Ptx ELISA of the organizing lab varied between 0.766 and 0.992, with 13 IgG-Ptx ELISAs having correlation values of ≥0.950 (among which 8 of 9 laboratories used the CBER protocol).
In another study, using an Italian IgG-Ptx ELISA performed according to the CBER protocol as a reference and with a panel of 150 sera with a broad range of IgG-Ptx values as the test panel, it was shown that IgG-Ptx ELISAs from 7 other European countries correlated quite well, with correlation coefficients varying from 0.84 to 0.92 (229). Retesting of sera with moderate to high antibody values also gave good results, with two exceptions. The first involved the (only) commercial IgG-Ptx ELISA, in which one dilution of reference serum and one dilution of test serum were used. The second involved the (only) ELISA that used coating of Ptx after precoating with fetuin in combination with two dilutions of test serum, one of which, depending on the OD found, was used to calculate the concentration relative to the reference line. The laboratory using the last ELISA later changed its format by deleting the lower of the two dilutions of test serum and replacing the in-house reference serum with a reference serum that was carefully calibrated to the CBER standard reference serum (230). Because precoating with fetuin enhances the sensitivity of Ptx ELISA (i.e., less Ptx is required) (197, 231, 232), this procedure was continued.
In a study comparing four IgG-Ptx ELISAs with different sources of Ptx, excellent correlations were found. Three of four participants used the same reference serum (the CBER standard or the equivalent WHO reference serum). Among those was the only ELISA (from the CDC) in which one dilution of test samples was used (233). The authors concluded that using the same reference serum may be the dominant important parameter for comparability. In the United States, an IgG-Ptx ELISA was developed by using a single dilution of patient serum and expressing results in CBER EU per milliliter or WHO reference IU per milliliter (equivalent to the CBER value), and the prespecified criteria for precision, linearity, and accuracy were met for samples containing 50 to 200 IU/ml, i.e., the range considered most relevant for single-serum diagnosis (234). In conclusion, we concur that using the same reference serum may be the dominant important parameter for comparability.
Cutoff points for significant increases of antibody in paired sera.
Cutoff points for significant increases of antibody in paired sera used in various studies included 1.5-fold (17, 187), 2-fold (31, 198, 201, 209, 225, 228, 235–238), 3-fold (193, 194, 208), and 4-fold (9, 182). Because of the limited accuracy of ELISA (high CVs) at low values (222), a criterion for the minimal level to be reached in the second serum is included in most definitions for significant dynamics, i.e., minimally 4 times the detection level or, specifically for IgG-Ptx ELISA, minimally 20 CBER EU/ml or 20 WHO IU/ml. With paired sera from patients with culture- or PCR-confirmed pertussis, the sensitivities of such cutoff points for increases of IgG-Ptx varied from 70% to 92% (9, 179, 186, 198, 201, 208). However, for children who contracted culture-confirmed pertussis in the year following vaccination with a Ptx-containing ACV, the diagnostic sensitivity of increases of IgG-Ptx in paired sera was much lower and was related to the presence of vaccine-induced IgG-Ptx (187, 201, 209).
Interestingly, in the choice of cutoff points for increases of antibody indicating infection, the accuracy of the immunoassay appears to have been the most important consideration. Rarely, cutoff points have been established by comparing dynamics in paired sera from pertussis patients and control patients. In one Swedish study, the value for 3 standard deviations (SD) of the differences in IgG-Ptx levels in serum pairs from 55 healthy adults, measured by an ELISA using endpoint titrations for calculation, was 2.6-fold, leading to the decision to consider ≥3-fold increases as indicative of infection (194). In serial sera from blood donors sampled over a period of 3 months, variations of antibodies to Ptx, FHA, and Prn, measured by an ELISA performed according to the CBER protocol, i.e., using the reference line method for calculation, were less than 40%, which was taken to support consideration of an increase of ≥2-fold as indicative of infection (239).
However, it is not precisely known which degree of increase is characteristic of a specific immune response. Ideally, dynamics in paired sera from patients with pertussis and patients with pertussis-like illness not caused by B. pertussis should be compared. Such controls were implicitly present in a study of serological data from a large routine diagnostic laboratory, in which cluster analysis was done of IgG-Ptx dynamics in paired sera from 2,455 patients suspected of pertussis, as measured with an IgG-Ptx ELISA that was calibrated with the CBER reference standard and that used a single-point reference line for calculation (230). These patients could be taken either to have pertussis or to have another infection or disease that causes pertussis-like coughing. Two-component cluster analysis of subgroups with low or moderate levels of IgG-Ptx in the first serum yielded very sharp distinctions between a Gaussian distribution of increases and decreases of antibody around a median of 0-fold and a Gaussian distribution of increases around a median of 12-fold, resulting in a nearly perfect receiver operating characteristic (ROC) curve, with an area under the curve (AUC) of 0.999. Cutoff points at 1.5- and 2.0-fold had specificities of 83% and 95%, respectively, and increases of 1.5- to 2.0-fold fell well within the distribution of the “negative” cluster; a 3-fold cutoff point had a specificity of 99.4% and the highest cumulative sensitivity plus specificity.
Cutoff points for absolute values of IgG-Ptx in single sera.
In specific settings and in specific studies, the interpretation of absolute values of antibody levels in single sera as indicative of recent infection has been done by using various cutoff points, e.g., the mean plus 3 SD of the antibody parameter under investigation in control sera (182, 237, 240), the mean plus 2 SD (191, 238), or the 99.9th percentile (240). Cutoff points proposed for IgG-Ptx are 200 CBER EU/ml (i.e., the upper tolerance limit of the 99th percentile of IgG-Ptx levels in 239 controls) (241), 75 CBER EU/ml (i.e., the mean plus 2 SD plus 20% for the IgG-Ptx levels in 271 blood donors) (242), 94 CBER EU/ml (97.5th percentile for 5,400 population sera) (243), and 125 CBER EU/ml (i.e., the 99th percentile for 7,756 population sera) (9, 229).
In second (late) sera of serum pairs from patients with culture- or PCR-confirmed pertussis, the sensitivities of such cutoff points for absolute values of IgG-Ptx with a preset high specificity varied from 66% to 92.2% (186, 191, 242). In paired sera from 89 patients with culture- or PCR-confirmed pertussis, IgG-Ptx levels of ≥125 CBER EU/ml were found in 11 first (early) sera (12%) and 70 second (late) sera (79%) (9, 229). For single sera obtained at presentation from patients with culture-confirmed pertussis, the sensitivity of the applied cutoff for IgG-Ptx of 200 CBER EU/ml depended on the duration of disease at presentation: the sensitivity was 36% for patients with a disease duration of <2 weeks at presentation and 67% for patients with a disease duration of ≥2 weeks at presentation (244). Indeed, there are large interindividual variations of times between the onset of symptoms, onset of the IgG-Ptx response, and time to reach peak values (245). Australian investigators showed that in patients with PCR-confirmed pertussis, the median time for IgG-Ptx to rise to ≥66 CBER EU/ml was 33 days; the accuracy of serology (cutoff point, 94 EU/ml) was optimal 2 to 8 weeks after the onset of symptoms (196).
In population sera sampled in Sweden in 1997 (<1 year after the nation-wide introduction of vaccination against pertussis) and in 2007, proportions of individuals with IgG-Ptx levels of ≥100 CBER EU/ml declined strongly in children aged <10 years, increased in adolescents aged 14 to 18 years, from 3 to 6%, and remained stable (2 to 3%) in older individuals (246). High levels of IgG-Ptx antibodies induced by infection start to decrease again within a few months, in a biphasic manner, initially quickly and later, gradually, more slowly (196, 242, 245, 247, 248). The decline of IgG-Ptx is faster than that of other antibodies induced by infection with B. pertussis (182, 202, 211). Nevertheless, in some patients, high levels persist for more than 1 year; for example, 31% of adults and 23% of children with pertussis, who all had IgG-Ptx levels of ≥125 CBER EU/ml in sera obtained shortly after infection, still had values of ≥125 EU/ml in sera obtained 12 months later. A longer follow-up of the children showed that the proportion with high levels decreased to 19% after 1 to 2 years and to 0% after 3 to 4 years (245). In a Danish study of serial sera from patients with culture- or PCR-confirmed pertussis, the median half-life of postinfection IgG-Ptx was 221 days, and 1 year later, 4% of sera still had IgG-Ptx values of ≥100 CBER EU/ml (242). In an Australian study of patients with PCR-confirmed pertussis, the median time for IgG-Ptx values of ≥94 CBER EU/ml to decline below that diagnostic cutoff was 200 days, and in 22 of 45 patients from whom sera were available and were collected between 5 and 12 months after the onset of disease, IgG-Ptx levels at that time were still ≥94 IU/ml (196).
In countries in which the large majority of children are vaccinated against pertussis, the seroprevalence of IgG-Ptx levels above diagnostic cutoff points may be high in the first months after the 3rd or 4th vaccination, thereby temporarily compromising the positive predictive value of high IgG-Ptx levels for diagnosis of infection. Indeed, high levels of antibodies induced by vaccination with WCV or ACV in the first year of life decline rapidly and in several studies have been shown to be near or below detection levels within 1.5 to 4 years (249–252). However, antibodies induced by a preschool booster dose may reach higher levels or may persist longer. Investigators in the United Kingdom modeled the decay curve for IgG-Ptx found in follow-up oral fluid samples from children after vaccination with 3 doses of ACV in the first month of life and after the preschool booster at 3 to 5 years of age (253). It was shown that 1 year after the primary series, the positive predictive values for IgG-Ptx levels of >70 arbitrary units (the cutoff for recent infection) at a priori chances of 0.1, 0.3, and 0.5 were 93%, 98%, and 99%, respectively. High values induced by the preschool booster persisted longer: 3 years after the booster, positive predictive values for IgG-Ptx levels of >70 arbitrary units at a priori chances of 0.1, 0.3, and 0.5 were 67%, 89%, and 95%, respectively.
In adolescents and adults boosted with ACV, the duration of interference of vaccine-induced high IgG-Ptx levels with serodiagnosis of infection depends on the Ptx content of the vaccine used. Three and 5 years after boostering of adolescents with a 3-component ACV containing 8 μg Ptx, high IgG-Ptx levels induced by vaccination had declined at such a rate that differences in distributions of IgG-Ptx in boosted and nonboosted groups had vanished completely (254). In the APERT study, a longitudinal follow-up of adults and adolescents boosted with the same three-component ACV containing 8 μg Ptx showed that 1 year after boosting, only 18% of the vaccine-induced peak level remained (207). After boosting of adults with an ACV containing only 2.5 μg of Ptx, practically all participants showed an IgG-Ptx response, but only 8 of 102 individuals developed a peak response of ≥94 IU/ml, and after 6 months, none were above that cutoff (255). Modeling suggested that 75 days after Tdap administration, all levels would already be <94 IU/ml. After boosting of adults with a 4-component ACV containing 5 μg Ptx, the median peak level of 80 CBER EU/ml reached after 4 weeks had declined to 20 EU/ml 1 year later (256). In contrast, boosting of adults with TdaP-IPV containing 20 μg Ptx resulted in a half-life for postvaccination IgG-Ptx of 508 days, and 1 year after the booster, 30% of individuals still had values of ≥100 CBER EU/ml (242). Boosting with a two-component ACV containing 25 μg Ptx resulted in a median IgG-Ptx level of 314 CBER EU/ml after 4 weeks, which declined to a median of 76 EU/ml at 1 year, with a minimal further decline in the 3 years thereafter (257). A similarly high induction of IgG-Ptx and decline after 1 year were noted after boosting with various aP vaccines containing 25 to 40 μg Ptx (256).
Cluster analysis for defining IgG-Ptx cutoff points for absolute values in single sera.
Because population sera may contain sera from individuals with recent infections with B. pertussis, use of those sera for determination of the specificity of cutoff points for infection may be associated with underestimation. In an attempt to solve this problem, 2-, 3-, and 4-component mixture models (i.e., models searching for 2, 3, or 4 discernible distributions) were applied to IgG-Ptx distributions in population sera sampled in the United States in the years 1991 to 1994, from 5,400 individuals aged 10 to 49 years (243). For this age range, moderate to high levels of IgG-Ptx were believed to be induced by infection rather than by childhood vaccination. Both a 3-component mixture and a 4-component mixture could be fitted with practically equal statistical powers. From the lowest to highest cluster, the four components constituted 83.8% (all values below the lower level of quantitation, i.e., <20 CBER EU/ml), 8.4%, 3.6%, and 4.2% of all measurements, respectively. The results of the 4-component model rather than the 3-component model were further analyzed because the highest cluster in the 4-component mixture was taken to represent the individuals with recent infection with B. pertussis. The 99th percentile of the third cluster (94 EU/ml) was proposed as the IgG-Ptx cutoff for serodiagnosis of pertussis in single sera. However, the hypothesis that, for population sera, IgG-Ptx values induced by recent infection will cluster is problematic. Given the pattern of decline of IgG-Ptx levels after infection, such clustering would require clustering of the time elapsed between the onset of infection and sampling of the serum. It is highly unlikely that this will occur in population sera sampled over a period of 3 years. It is therefore doubtful that these findings will be reproduced in future collections of population sera. Considering all sera, the value of 94 EU/ml constituted the 97.5th percentile.
In another study, two-component cluster analysis was applied to sera from patients suspected of having pertussis which had been submitted to a large routine diagnostic laboratory over a period of 6 years (230). In these sera, IgG-Ptx had been measured with a CBER reference-calibrated ELISA. Patients vaccinated with high-level IgG-Ptx-inducing vaccines were excluded. Sera from patients who had symptoms for less than 100 days were selected (14,452 sera; one serum sample per patient). Two distributions of log2-transformed concentrations were found, with little overlap. The lowest distribution was taken to represent patients in whom cough of recent onset was not caused by infection with B. pertussis, while the highest distribution was taken to represent patients with cough of recent onset caused by infection with B. pertussis. The resulting ROC curve had an AUC of 0.993. A value of 68 CBER EU/ml as the cutoff point had the highest cumulative sensitivity and specificity (96.4% and 95.7%, respectively). The values of 62, 94, and 125 EU/ml as cutoff points had specificities of 95%, 97.7%, and 98.8%, respectively. For serodiagnosis, the authors proposed that values of ≥125 EU/ml (in combination with clinical signs and symptoms compatible with infection with B. pertussis) be considered diagnostic; values in the range of 62 to 125 EU/ml be considered “suspect,” which does not with certainty confirm the diagnosis and should be followed up by investigation of a second serum to be obtained 2 weeks later to determine if an increase in Ptx antibodies occurred; and values of <62 EU/ml be considered “within the normal range,” which does not exclude the diagnosis and also should be followed up by investigation of a second serum to be obtained 2 weeks later. In situations in which the a priori chance is estimated to be high, e.g., in an outbreak setting (195), a lower diagnostic cutoff point may be considered.
IgG-Ptx antibodies in oral fluids.
In the United Kingdom, for older children suspected of having pertussis, a high correlation was found between IgG-Ptx levels in oral fluid, as measured with an IgG-Ptx capture ELISA, and IgG-Ptx levels in serum, as measured with an ELISA performed according to the CBER protocol (258). Using cutoff points for IgG-Ptx of 100 EU/ml for serum and 70 arbitrary units (aU) for oral fluid, it was shown that 80% of seropositive individuals were also positive by the oral fluid assay and that 97% of the seronegative individuals were also negative by the oral fluid assay. The oral fluid assay was positive for 3.6% of asymptomatic controls. Subsequently, in the United Kingdom, patients that were notified of clinically diagnosed pertussis, without laboratory confirmation, were offered IgG-Ptx testing of oral fluid free of charge (259).With this strategy, the number of laboratory-confirmed cases increased from 1,852 (confirmed by culture, PCR, and/or serology) to 2,443 due to the addition of 591 cases confirmed only (and only tested) by the oral fluid assay.
Commercially available ELISAs.
While in-house IgG-Ptx ELISAs accurately distinguished WHO reference preparations and gave results comparable to the expected values, commercial kits using mixtures of antigens did not appear to be able to give results that correlated with those obtained with the WHO reference preparations (223). Comparison of the performances of five commercial ELISAs for detection of antibodies to B. pertussis (two using a mixture of Ptx and FHA and three using an unknown coating antigen) with in-house ELISAs measuring IgG and IgA antibodies to Ptx and FHA according to the CBER protocol resulted in the conclusion that these ELISAs needed further improvement and standardization (260). In a study of convalescent-phase sera from 41 patients with culture- or PCR-confirmed pertussis and from 65 control patients with other diseases, the discriminative powers of the IgG-Ptx components of three commercial ELISAs and of the in-house IgG-ELISA were practically identical (AUC of ROC curves, 0.92 to 0.93) (261). However, the cutoff point for interpretation of absolute values of IgG-Ptx given by the manufacturer yielded extremely low specificities (<50%) for two of the three commercial IgG-Ptx ELISAs, was suboptimal for the other commercial IgG-Ptx ELISA (sensitivity, 68%; specificity, 97%), and was nearly optimal for the in-house IgG-Ptx ELISA (sensitivity, 80%; specificity, 97%). In a comparison of IgG-Ptx ELISAs from 7 European countries, the only participating commercial IgG-Ptx ELISA correlated quite well (R2 = 0.89) with the reference assay, i.e., the CBER reference-standardized Italian IgG-Ptx ELISA (229).
FACTORS THAT INFLUENCE THE SENSITIVITY OF DIAGNOSTIC METHODS
The gold standard of B. pertussis laboratory diagnosis is culture, and the sensitivity of culture depends on the stage of disease in which the sample is taken, the vaccination status of the patient, the age of the patient, and culture methods. For assessment of the sensitivity of culture, various clinical case definitions are used, and sensitivities have been calculated for various patient and age groups, such as in outbreak settings (schools), vaccine efficacy trials (infants), or a population with sporadic cases (all age groups). The sensitivity of culture also depends on the bacterial load, as demonstrated by studies using semiquantitative PCRs (40, 111, 262–264). It was found that the culture sensitivity decreased with increasing Cq values (and hence a decreasing bacterial DNA presence) for real-time PCR (85, 265). It was also shown that, in general, the bacterial loads in older patients were smaller than those in infants and children (264).
Several studies have shown that culture sensitivity is high early in disease, and in infants and unvaccinated children, but decreases with the duration of illness and with an increasing age of patients (95, 199, 264). For older and vaccinated children, adolescents, and adults, culture is less useful, especially if the time after onset of infection exceeds 3 to 4 weeks (264).
PCR is much more sensitive than culture for detecting B. pertussis. In all studies, culture-positive samples were always PCR positive. Thus, when PCR is compared to culture, the sensitivity of PCR is 100%. The sensitivity of culture compared to PCR ranges from 26% to 85% (93, 150, 265–268). It has been shown that the sensitivity of culture with regard to PCR sensitivity decreases with an increasing duration of disease, and thus shows a steeper decline than that of PCR (267). With PCR, 89% of patients (<5 years of age) are found to be PCR positive 14 days after the onset of paroxysmal cough, and positive PCR results can be achieved up to 38 days after infection (269). After that, laboratory confirmation is generally possible only by serology (Fig. 4).
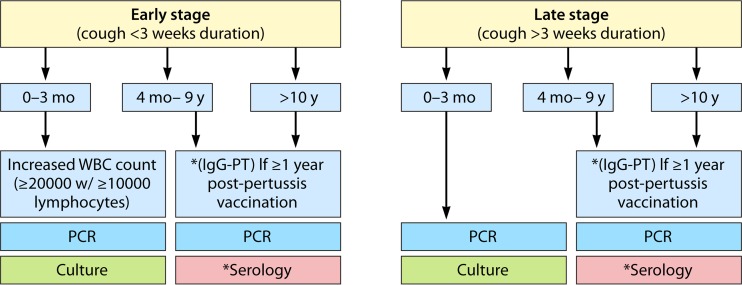
FIG 4.
Schematic flow diagram for recommended laboratory diagnosis of B. pertussis. Only for very young infants is an elevated white blood cell count (≥20,000 cells/μl) diagnostic for a B. pertussis infection. The depicted algorithm was proposed by Cherry et al. (29), except that we added the recommendation of performing PCR for the older age group and the late stage of disease, as PCR may in some cases be positive after more than 3 weeks of cough, albeit by detection of DNA of nonviable B. pertussis cells. Mo, month; y, year; WBC, white blood cell; IgG, immunoglobulin G; PT, pertussis toxin.
With serological diagnosis, it was found that positivity (IgA and IgG titers) increased with an increasing age and duration of disease, while PCR and culture positivity decreased (199). In comparisons of PCR and serology, several studies showed a limited overlap in positivity for both methods (270–273), ranging from approximately 8% to 44%, while the overlap in positivity for culture and serology was even lower (0% to <5%).
Many factors affect the accuracy and sensitivity of serodiagnosis of pertussis, such as the time elapsed since the last vaccination or infection, whether only the primary series or additional booster vaccinations were given, the antigen content of the vaccine, serology, and age. Thus, there is no one-size-fits-all method, and sensitivity and accuracy may have to be balanced, e.g., by using age-specific cutoffs. Furthermore, cutoff values may be increased or decreased depending on the question addressed. Lower cutoffs should be used when false-negative results may have serious health consequences, e.g., when unvaccinated or incompletely vaccinated infants are involved.
Regarding the choice of antigen, given the data presented in this review, serodiagnosis of infection with B. pertussis can best be performed by measuring IgG-Ptx antibodies in serum. Furthermore, a quantitative IgG-Ptx ELISA should be used, calibrated with the CBER standard serum (lot 3) or the WHO international IgG-Ptx standard serum (CBER EU are equivalent to WHO IU). Recently, a similar recommendation was given by experts from European reference laboratories (274). Cutoff points for absolute values of IgG-Ptx in single sera within the range of 62 to 200 IU/ml are associated with specificities between 95% and 99%. Sensitivity depends on the duration of disease at the time of sampling and may reach 70 to 90% late in disease. Due to high levels of vaccine-induced IgG-Ptx, single-serum diagnosis is not reliable for 1 to 3 years after vaccination with Ptx-containing vaccines, with the period of interference being shortest after primary vaccinations in the first year of life and longest after boostering of adults with vaccines used in childhood, i.e., those with a high Ptx content. If the IgG-Ptx level is below the chosen cutoff, the diagnosis of pertussis can be neither confirmed nor denied, and a second serum obtained at least 2 weeks later and 4 to 6 weeks after the onset of disease (and 6 to 8 weeks after the onset of disease for very young immune-naive children) (168) should be investigated. Increases of ≥3-fold in paired sera or any increase to a value above the cutoff for absolute values in single sera can then be considered to confirm the diagnosis of pertussis.
CONCLUSIONS AND RECOMMENDATIONS
The increase in booster vaccinations with Ptx-containing ACVs will increasingly reduce the accuracy of IgG-Ptx-based serodiagnosis. Further investigations into the serodiagnostic value of measurement of IgA antibody parameters in recently vaccinated individuals suspected of having pertussis should be performed. Also, other antigens not contained in ACVs should be searched for and tested to replace Ptx for serodiagnosis of pertussis in recently vaccinated individuals. The complete genome sequences of more than 400 B. pertussis strains will be helpful in the selection of highly immunogenic antigens.
The Global Pertussis Initiative (29) proposed the utilization of 3 different age-related case definitions for pertussis, with distinctive diagnostic criteria for the age categories of 0 to 3 months, 4 months to 9 years, and ≥10 years. The resulting diagnostic algorithm recommends culture and PCR for the first age group (0 to 3 months), irrespective of cough duration, and PCR for the age group of 4 months to 9 years with a cough duration of <3 weeks. Serology is recommended in all other cases (Fig. 4). We also recommend PCR to complement serology for older age groups and for later disease, as DNA may be present in some cases for up to 38 days after onset (269).
In summary, on the time scale of different stages of B. pertussis infection, different diagnostic methods should be employed (Fig. 5). PCR is most sensitive and should always be included, independent of the stage of disease, to complement culture in the early stage and serology in the later stage. In case of a positive PCR result in the absence of distinctive symptoms, a positive culture, or positive serology, this result can be valued as a true indication of infection when all quality criteria for the assay have been met and contamination can be excluded.
FIG 5.
Relative diagnostic sensitivities of culture (green), PCR (blue), serology (red), and clinical diagnosis (orange) during different stages of B. pertussis infection. The represented sensitivities were idealized for clarity. As PCR may detect DNA of nonviable bacteria, positive PCR results may be obtained for 1 to 2 weeks longer than positive culture results.
Biographies

Anneke van der Zee, Ph.D., is a Medical Molecular Microbiologist at Maasstad Hospital, Rotterdam, The Netherlands. She studied at Utrecht University and received her doctoral degree in 1996, studying pertussis at the National Institute for Public Health and the Environment, Bilthoven, The Netherlands. Starting in 1996, she was involved in the development of molecular microbiology laboratory diagnosis at St. Elisabeth Hospital, Tilburg, The Netherlands, and since 2009, she has been leading the Molecular Diagnostics Unit at Maasstad Hospital, which develops and implements molecular assays for diagnosis of infectious diseases, hemato-oncology, tumor therapy, and pharmacogenetics. Her main interest is the diagnosis and epidemiology of infectious diseases.

Joop F. P. Schellekens, M.D., Ph.D., is a Clinical Microbiologist at the Certe Laboratory for Infectious Diseases, Groningen, The Netherlands, which serves eight hospitals in the northern part of the Netherlands. He received his M.D. and Ph.D. at the University and Academic Hospital of Utrecht, The Netherlands. From 1991 to 2004, he was Deputy Head of the Laboratory of Infectious Diseases of the National Institute of Public Health and the Environment, Bilthoven, The Netherlands, during which his primary focus was epidemiology and laboratory diagnosis of infectious diseases. At his present position, his primary focus is the laboratory diagnosis and management of patients with infectious diseases.

Frits R. Mooi, Ph.D., is currently retired. He was Project Leader on pertussis at the National Institute for Public Health and the Environment, The Netherlands, from 1986 to 2015. He obtained his Ph.D. in molecular microbiology from the Free University Amsterdam in 1982. In 1999, he was appointed a (part-time) Professor in Molecular Microbiology at the Department of Medical Microbiology, University of Utrecht. His research focused on the molecular epidemiology and evolution of pathogens, including Escherichia coli, Bordetella pertussis, Moraxella catarrhalis, and Vibrio cholerae.
REFERENCES
- 1.Roush SW, Murphy TV, Vaccine-Preventable Disease Table Working Group. 2007. Historical comparisons of morbidity and mortality for vaccine-preventable diseases in the United States. JAMA 298:2155–2163. doi: 10.1001/jama.298.18.2155. [DOI] [PubMed] [Google Scholar]
- 2.Mills KH, Ross PJ, Allen AC, Wilk MM. 2014. Do we need a new vaccine to control the re-emergence of pertussis? Trends Microbiol 22:49–52. doi: 10.1016/j.tim.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Mooi FR, Van Der Maas NA, De Melker HE. 2014. Pertussis resurgence: waning immunity and pathogen adaptation—two sides of the same coin. Epidemiol Infect 142:685–694. doi: 10.1017/S0950268813000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berbers GA, de Greeff SC, Mooi FR. 2009. Improving pertussis vaccination. Hum Vaccin 5:497–503. doi: 10.4161/hv.8112. [DOI] [PubMed] [Google Scholar]
- 5.Witt MA, Arias L, Katz PH, Truong ET, Witt DJ. 2013. Reduced risk of pertussis among persons ever vaccinated with whole cell pertussis vaccine compared to recipients of acellular pertussis vaccines in a large US cohort. Clin Infect Dis 56:1248–1254. doi: 10.1093/cid/cit046. [DOI] [PubMed] [Google Scholar]
- 6.Liko J, Robison SG, Cieslak PR. 2013. Priming with whole-cell versus acellular pertussis vaccine. N Engl J Med 368:581–582. doi: 10.1056/NEJMc1212006. [DOI] [PubMed] [Google Scholar]
- 7.Witt MA, Katz PH, Witt DJ. 2012. Unexpectedly limited durability of immunity following acellular pertussis vaccination in pre-adolescents in a North American outbreak. Clin Infect Dis 54:1730–1735. doi: 10.1093/cid/cis287. [DOI] [PubMed] [Google Scholar]
- 8.Sheridan SL, Ware RS, Grimwood K, Lambert SB. 2012. Number and order of whole cell pertussis vaccines in infancy and disease protection. JAMA 308:454–456. doi: 10.1001/jama.2012.6364. [DOI] [PubMed] [Google Scholar]
- 9.de Melker HE, Versteegh FG, Conyn-Van Spaendonck MA, Elvers LH, Berbers GA, van der Zee A, Schellekens JF. 2000. Specificity and sensitivity of high levels of immunoglobulin G antibodies against pertussis toxin in a single serum sample for diagnosis of infection with Bordetella pertussis. J Clin Microbiol 38:800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Güriş D, Strebel PM, Bardenheier B, Brennan M, Tachdjian R, Finch E, Wharton M, Livengood JR. 1999. Changing epidemiology of pertussis in the United States: increasing reported incidence among adolescents and adults, 1990–1996. Clin Infect Dis 28:1230–1237. doi: 10.1086/514776. [DOI] [PubMed] [Google Scholar]
- 11.Cherry JD. 2014. Adult pertussis in the pre- and post-vaccine eras: lifelong vaccine-induced immunity? Expert Rev Vaccines 13:1073–1080. doi: 10.1586/14760584.2014.935765. [DOI] [PubMed] [Google Scholar]
- 12.de Greeff SC, de Melker HE, van Gageldonk PGM, Schellekens JFP, van der Klis FRM, Mollema L, Mooi FR, Berbers GAM. 2010. Seroprevalence of pertussis in the Netherlands: evidence for increased circulation of Bordetella pertussis. PLoS One 5:e14183. doi: 10.1371/journal.pone.0014183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wirsing von König CH, Halperin S, Riffelmann M, Guiso N. 2002. Pertussis of adults and infants. Lancet Infect Dis 2:744–750. doi: 10.1016/S1473-3099(02)00452-8. [DOI] [PubMed] [Google Scholar]
- 14.European Centre for Disease Prevention and Control. 2013. Annual epidemiological report. Reporting on 2011 surveillance data and 2012 epidemic intelligence data. ECDC, Stockholm, Sweden. [Google Scholar]
- 15.ECDC. 2012. Guidance and protocol for the use of real-time PCR in laboratory diagnosis of human infection with Bordetella pertussis or Bordetella parapertussis. ECDC, Stockholm, Sweden: http://ecdc.europa.eu/en/publications/Publications/Guidance-protocol-PCR-laboratory-diagnosis-bordetella-pertussis-parapertussis.pdf. [Google Scholar]
- 16.Centers for Disease Control and Prevention. 2007. Outbreaks of respiratory illness mistakenly attributed to pertussis—New Hampshire, Massachusetts, and Tennessee, 2004–2006. MMWR Morb Mortal Wkly Rep 56:837–842. [PubMed] [Google Scholar]
- 17.Wirsing von König CH, Rott H, Bogaerts H, Schmitt HJ. 1998. A serologic study of organisms possibly associated with pertussis-like coughing. Pediatr Infect Dis J 17:645–649. doi: 10.1097/00006454-199807000-00013. [DOI] [PubMed] [Google Scholar]
- 18.Diavatopoulos DA, Cummings CA, van der Heide HG, van Gent M, Liew S, Relman DA, Mooi FR. 2006. Characterization of a highly conserved island in the otherwise divergent Bordetella holmesii and Bordetella pertussis genomes. J Bacteriol 188:8385–8394. doi: 10.1128/JB.01081-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diavatopoulos DA, Cummings CA, Schouls LM, Brinig MM, Relman DA, Mooi FR. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog 1:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullinane LC, Alley MR, Marshall RB, Manktelow BW. 1987. Bordetella parapertussis from lambs. N Z Vet J 35:175. doi: 10.1080/00480169.1987.35433. [DOI] [PubMed] [Google Scholar]
- 21.He Q, Viljanen MK, Arvilommi H, Aittanen B, Mertsola J. 1998. Whooping cough caused by Bordetella pertussis and Bordetella parapertussis in an immunized population. JAMA 280:635–637. doi: 10.1001/jama.280.7.635. [DOI] [PubMed] [Google Scholar]
- 22.Yih WK, Silva EA, Ida J, Harrington N, Lett SM, George H. 1999. Bordetella holmesii-like organisms isolated from Massachusetts patients with pertussis-like symptoms. Emerg Infect Dis 5:441–443. doi: 10.3201/eid0503.990317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mastrantonio P, Stefanelli P, Giuliano M, Herrera Rojas Y, Ciofi degli Atti M, Anemona A, Tozzi AE. 1998. Bordetella parapertussis infection in children: epidemiology, clinical symptoms, and molecular characteristics of isolates. J Clin Microbiol 36:999–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woolfrey BF, Moody JA. 1991. Human infections associated with Bordetella bronchiseptica. Clin Microbiol Rev 4:243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CDC. 2014. Pertussis/whooping cough (Bordetella pertussis) 2014 case definition. CDC, Atlanta, GA: http://wwwn.cdc.gov/nndss/script/casedef.aspx?CondYrID=950&DatePub=1/1/2014%2012:00:00%20AM. [Google Scholar]
- 26.ECDC. 2012. Pertussis. ECDC, Stockholm, Sweden: http://ecdc.europa.eu/en/activities/surveillance/euvac/case_definition/Pages/pertusis.aspx. [Google Scholar]
- 27.WHO. 2008 WHO-recommended surveillance standard of pertussis. http://www.who.int/immunization/monitoring_surveillance/burden/vpd/surveillance_type/passive/pertussis_standards/en/.
- 28.Cherry JD, Grimprel E, Guiso N, Heininger U, Mertsola J. 2005. Defining pertussis epidemiology: clinical, microbiologic and serologic perspectives. Pediatr Infect Dis J 24(Suppl 5):S25–S34. doi: 10.1097/01.inf.0000160926.89577.3b. [DOI] [PubMed] [Google Scholar]
- 29.Cherry JD, Tan T, Wirsing von König CH, Forsyth KD, Thisyakorn U, Greenberg D, Johnson D, Marchant C, Plotkin S. 2012. Clinical definitions of pertussis: summary of a Global Pertussis Initiative roundtable meeting, February 2011. Clin Infect Dis 54:1756–1764. doi: 10.1093/cid/cis302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashita N, Akaike H, Teranishi H, Kawai Y, Ouchi K, Kato T, Hayashi T, Okimoto N. 2013. Diagnostic value of symptoms and laboratory data for pertussis in adolescent and adult patients. BMC Infect Dis 13:129. doi: 10.1186/1471-2334-13-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steketee RW, Wassilak SG, Adkins WN Jr, Burstyn DG, Manclark CR, Berg J, Hopfensperger D, Schell WL, Davis JP. 1988. Evidence for a high attack rate and efficacy of erythromycin prophylaxis in a pertussis outbreak in a facility for the developmentally disabled. J Infect Dis 157:441–449. doi: 10.1093/infdis/157.3.441. [DOI] [PubMed] [Google Scholar]
- 32.Paddock CD, Sanden GN, Cherry JD, Gal AA, Langston C, Tatti KM, Wu KH, Goldsmith CS, Greer PW, Montague JL, Eliason MT, Holman RC, Guarner J, Shieh WJ, Zaki SR. 2008. Pathology and pathogenesis of fatal Bordetella pertussis infection in infants. Clin Infect Dis 47:328–338. doi: 10.1086/589753. [DOI] [PubMed] [Google Scholar]
- 33.Bejuk J, Begovac J, Bace A, Kuzmanovic-Sterk N, Aleraj B. 1995. Culture of Bordetella pertussis from three upper respiratory tract specimens. Pediatr Infect Dis J 14:64–65. [DOI] [PubMed] [Google Scholar]
- 34.Marcon MJ, Hamoudi AC, Cannon HJ, Hribar MM. 1987. Comparison of throat and nasopharyngeal swab specimens for culture diagnosis of Bordetella pertussis infection. J Clin Microbiol 25:1109–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.CDC. 2011. Pertussis (whooping cough) specimen collection. CDC, Atlanta, GA: http://www.cdc.gov/pertussis/clinical/diagnostic-testing/specimen-collection.html. [Google Scholar]
- 36.Hallander HO, Reizenstein E, Renemar B, Rasmuson G, Mardin L, Olin P. 1993. Comparison of nasopharyngeal aspirates with swabs for culture of Bordetella pertussis. J Clin Microbiol 31:50–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cloud JL, Hymas WC, Carroll KC. 2002. Impact of nasopharyngeal swab types on detection of Bordetella pertussis by PCR and culture. J Clin Microbiol 40:3838–3840. doi: 10.1128/JCM.40.10.3838-3840.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadowsky RM, Laus S, Libert T, States SJ, Ehrlich GD. 1994. Inhibition of PCR-based assay for Bordetella pertussis by using calcium alginate fiber and aluminum shaft components of a nasopharyngeal swab. J Clin Microbiol 32:1054–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dube FS, Kaba M, Whittaker E, Zar HJ, Nicol MP. 2013. Detection of Streptococcus pneumoniae from different types of nasopharyngeal swabs in children. PLoS One 8:e68097. doi: 10.1371/journal.pone.0068097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vestrheim DF, Steinbakk M, Bjørnstad ML, Moghaddam A, Reinton N, Dahl ML, Grude N, Sandvena P. 2012. Recovery of Bordetella pertussis from PCR-positive nasopharyngeal samples is dependent on bacterial load. J Clin Microbiol 50:4114–4115. doi: 10.1128/JCM.01553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pittet LF, Emonet S, Schrenzel J, Siegrist CA, Posfay-Barbe KM. 2014. Bordetella holmesii: an under-recognised Bordetella species. Lancet Infect Dis 14:510–519. doi: 10.1016/S1473-3099(14)70021-0. [DOI] [PubMed] [Google Scholar]
- 42.Hoppe JE, Schwaderer J. 1989. Direct plating versus use of transport medium for detection of Bordetella species from nasopharyngeal swabs. Eur J Clin Microbiol Infect Dis 8:264–265. doi: 10.1007/BF01965276. [DOI] [PubMed] [Google Scholar]
- 43.Katzko G, Hofmeister M, Church D. 1996. Extended incubation of culture plates improves recovery of Bordetella spp. J Clin Microbiol 34:1563–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bart MJ, Harris SR, Advani A, Arakawa Y, Bottero D, Bouchez V, Cassiday PK, Chiang CS, Dalby T, Fry NK, Gaillard ME, van Gent M, Guiso N, Hallander HO, Harvill ET, He Q, van der Heide HG, Heuvelman K, Hozbor DF, Kamachi K, Karataev GI, Lan R, Lutyńska A, Maharjan RP, Mertsola J, Miyamura T, Octavia S, Preston A, Quail MA, Sintchenko V, Stefanelli P, Tondella ML, Tsang RS, Xu Y, Yao SM, Zhang S, Parkhill J, Mooi FR. 2014. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. mBio 5:e01074. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Gouw D, Hermans PW, Bootsma HJ, Zomer A, Heuvelman K, Diavatopoulos DA, Mooi FR. 2014. Differentially expressed genes in Bordetella pertussis strains belonging to a lineage which recently spread globally. PLoS One 9:e84523. doi: 10.1371/journal.pone.0084523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Gouw DSD, de Jonge MI, Hermans PWM, Wessels HJCT, Zomer A, Yantorno OM, Diavatopoulos DA, Mooi FR. 2014. The vaccine potential of Bordetella pertussis biofilm-derived membrane proteins. Emerg Microbes Infect 3:e58. doi: 10.1038/emi.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeño-Tárraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 48.Ewanowich CA, Chui LWL, Paranchych MG, Peppler MS, Marusyk RG, Albrittons WL. 1993. Major outbreak of pertussis in northern Alberta, Canada: analysis of discrepant direct fluorescent-antibody and culture results by using polymerase chain reaction methodology. J Clin Microbiol 31:1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loeffelholz M. 2012. Towards improved accuracy of Bordetella pertussis nucleic acid amplification tests. J Clin Microbiol 50:2186–2190. doi: 10.1128/JCM.00612-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riffelmann M, Wirsing von Konig CH, Caro V, Guiso N. 2005. Nucleic acid amplification tests for diagnosis of Bordetella infections. J Clin Microbiol 43:4925–4929. doi: 10.1128/JCM.43.10.4925-4929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Notomi T, Mori Y, Tomita N, Kanda H. 2015. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 53:1–5. doi: 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- 53.Kamachi K, Toyoizumi-Ajisaka H, Toda K, Soeung SC, Sarath S, Nareth Y, Horiuchi Y, Kojima K, Takahashi M, Arakawa Y. 2006. Development and evaluation of a loop-mediated isothermal amplification method for rapid diagnosis of Bordetella pertussis infection. J Clin Microbiol 44:1899–1902. doi: 10.1128/JCM.44.5.1899-1902.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vincent M, Xu Y, Kong H. 2004. Helicase-dependent isothermal DNA amplification. EMBO Rep 5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Kruijssen AM, Templeton KE, van der Plas RN, van Doorn HR, Claas EC, Sukhai RN, Kuijper EJ. 2007. Detection of respiratory pathogens by real-time PCR in children with clinical suspicion of pertussis. Eur J Pediatr 166:1189–1191. doi: 10.1007/s00431-006-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Wang L, Fan J, Kraft A, Bose ME, Tiwari S, Van Dyke M, Haigis R, Luo T, Ghosh M, Tang H, Haghnia M, Mather EL, Weisburg WG, Henrickson KJ. 2008. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J Clin Microbiol 46:3063–3072. doi: 10.1128/JCM.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDonough EA, Barrozo CP, Russell KL, Metzgar D. 2005. A multiplex PCR for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and Bordetella pertussis in clinical specimens. Mol Cell Probes 19:314–322. doi: 10.1016/j.mcp.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Kong F, Yang Y, Gilbert GL. 2008. A multiplex PCR-based reverse line blot hybridization (mPCR/RLB) assay for detection of bacterial respiratory pathogens in children with pneumonia. Pediatr Pulmonol 43:150–159. doi: 10.1002/ppul.20749. [DOI] [PubMed] [Google Scholar]
- 59.Chan EL, Antonishyn N, McDonald R, Maksymiw T, Pieroni P, Nagle E, Horsman GB. 2002. The use of TaqMan PCR assay for detection of Bordetella pertussis infection from clinical specimens. Arch Pathol Lab Med 126:173–176. [DOI] [PubMed] [Google Scholar]
- 60.He Q, Mertsola J, Soini H, Viljanen MK. 1994. Sensitive and specific polymerase chain reaction assays for detection of Bordetella pertussis in nasopharyngeal specimens. J Pediatr 124:421–426. doi: 10.1016/S0022-3476(94)70365-5. [DOI] [PubMed] [Google Scholar]
- 61.Mandal S, Tatti KM, Woods-Stout D, Cassiday PK, Faulkner AE, Griffith MM, Jackson ML, Pawloski LC, Wagner B, Barnes M, Cohn AC, Gershman KA, Messonnier NE, Clark TA, Tondella ML, Martin SW. 2012. Pertussis pseudo-outbreak linked to specimens contaminated by Bordetella pertussis DNA from clinic surfaces. Pediatrics 129:e424–e430. doi: 10.1542/peds.2011-1710. [DOI] [PubMed] [Google Scholar]
- 62.Rantakokko-Jalava K, Jalava J. 2002. Optimal DNA isolation method for detection of bacteria in clinical specimens by broad-range PCR. J Clin Microbiol 40:4211–4217. doi: 10.1128/JCM.40.11.4211-4217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Riffelmann M, Schmetz J, Bock S, Wirsing von Koenig CH. 2008. Preparation of Bordetella pertussis DNA from respiratory samples for real-time PCR by commercial kits. Eur J Clin Microbiol Infect Dis 27:145–148. doi: 10.1007/s10096-007-0403-4. [DOI] [PubMed] [Google Scholar]
- 64.Dundas N, Leos NK, Mitui M, Revell P, Rogers BB. 2008. Comparison of automated nucleic acid extraction methods with manual extraction. J Mol Diagn 10:311–316. doi: 10.2353/jmoldx.2008.070149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caro V, Guiso N, Alberti C, Liguori S, Burucoa C, Couetdic G, Doucet-Populaire F, Ferroni A, Papin-Gibaud S, Grattard F, Réglier-Poupet H, Raymond J, Soler C, Bouchet S, Charreau S, Couzon B, Leymarie I, Tavares N, Choux M, Bingen E, Bonacorsi S. 2009. Proficiency program for real-time PCR diagnosis of Bordetella pertussis infections in French hospital laboratories and at the French National Reference Center for Whooping Cough and other Bordetelloses. J Clin Microbiol 47:3197–3203. doi: 10.1128/JCM.00817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McLafferty MA, Harcus DR, Hewlett EL. 1988. Nucleotide sequence and characterization of a repetitive DNA element from the genome of Bordetella pertussis with characteristics of an insertion sequence. J Gen Microbiol 134:2297–2306. [DOI] [PubMed] [Google Scholar]
- 67.van der Zee A, Agterberg C, van Agterfeld M, Peeters M, Mooi FR. 1993. Characterization of IS1001, an insertion sequence element of Bordetella parapertussis. J Bacteriol 175:141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Zee A, Groenendijk H, Peeters M, Mooi FR. 1996. The differentiation of Bordetella parapertussis and Bordetella bronchiseptica from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. Int J Syst Bacteriol 46:640–647. doi: 10.1099/00207713-46-3-640. [DOI] [PubMed] [Google Scholar]
- 69.van der Zee A, Mooi FR, van Embden J, Musser J. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J Bacteriol 179:6609–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aoyama T, Tamura C, Takeuchi Y, Kamimura T, Imaizumi A. 1997. Rapid, sensitive and specific diagnosis of Bordetella pertussis using the polymerase chain reaction. Acta Paediatr 39:44–47. doi: 10.1111/j.1442-200X.1997.tb03554.x. [DOI] [PubMed] [Google Scholar]
- 71.Dragsted DM, Dohn B, Madsen J, Jensen JS. 2004. Comparison of culture and PCR for detection of Bordetella pertussis and Bordetella parapertussis under routine laboratory conditions. J Med Microbiol 53:749–754. doi: 10.1099/jmm.0.45585-0. [DOI] [PubMed] [Google Scholar]
- 72.Glare EM, Paton JC, Premier RR, Lawrence AJ, Nisbet IT. 1990. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol 28:1982–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grimprel E, Bégué P, Anjak I, Betsou F, Guiso N. 1993. Comparison of polymerase chain reaction, culture and Western immunoblot serology for diagnosis of Bordetella pertussis infections. J Clin Microbiol 31:2745–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He Q, Mertsola J, Soini H, Skurnik M, Ruuskanen O, Viljanen MK. 1993. Comparison of polymerase chain reaction with culture and enzyme immunoassay for diagnosis of pertussis. J Clin Microbiol 31:642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lichtinghagen R, Diedrich-Glaubitz R, von Hörsten B. 1994. Identification of Bordetella pertussis in nasopharyngeal swabs using the polymerase chain reaction: evaluation of detection methods. Eur J Clin Chem Clin Biochem 32:161–167. [DOI] [PubMed] [Google Scholar]
- 76.Loeffelholz MJ, Thompson CJ, Long KS, Gilchrist MJ. 1999. Comparison of PCR, culture, and direct fluorescent-antibody testing for detection of Bordetella pertussis. J Clin Microbiol 37:2872–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nelson S, Matlow A, McDowell C, Roscoe M, Karmali M, Penn L, Dyster L. 1997. Detection of Bordetella pertussis in clinical specimens by PCR and a microtiter plate-based DNA hybridization assay. J Clin Microbiol 35:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olcen P, Bäckman A, Johansson B, Esbjorner E, Tornqvist E, Dygraves J, McPheat WL. 1992. Amplification of DNA by the polymerase chain reaction for the efficient diagnosis of pertussis. Scand J Infect Dis 24:339–345. doi: 10.3109/00365549209061340. [DOI] [PubMed] [Google Scholar]
- 79.Wadowsky RM, Michaels RH, Libert T, Kingsley LA, Ehrlich GD. 1996. Multiplex PCR-based assay for detection of Bordetella pertussis in nasopharyngeal swab specimens. J Clin Microbiol 34:2645–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bidet P, Liguori S, De Lauzanne A, Caro V, Lorrot M, Carol A, Faye A, Guiso N, Bingen E, Bonacorsi S. 2008. Real-time PCR measurement of persistence of Bordetella pertussis DNA in nasopharyngeal secretions during antibiotic treatment of young children with pertussis. J Clin Microbiol 46:3636–3638. doi: 10.1128/JCM.01308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bäckman A, Johansson B, Olcen P. 1994. Nested PCR optimized for detection of Bordetella pertussis in clinical nasopharyngeal samples. J Clin Microbiol 32:2544–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Donno A, Quattrocchi M, Ansaldi F, Campa A, Rollo MC, Gabutti G. 2006. Direct detection of Bordetella pertussis and Bordetella parapertussis: comparison of polymerase chain reaction and culture. J Int Med Res 34:367–373. doi: 10.1177/147323000603400405. [DOI] [PubMed] [Google Scholar]
- 83.Farrell DJ, Daggard G, Mukkur TK. 1999. Nested duplex PCR to detect Bordetella pertussis and Bordetella parapertussis and its application in diagnosis of pertussis in nonmetropolitan Southeast Queensland, Australia. J Clin Microbiol 37:606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reizenstein E, Lindberg L, Möllby R, Hallander HO. 1996. Validation of nested Bordetella PCR in pertussis vaccine trial. J Clin Microbiol 34:810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Farrell DJ, McKeon M, Daggard G, Loeffelholz MJ, Thompson CJ, Mukkur TK. 2000. Rapid-cycle PCR method to detect Bordetella pertussis that fulfills all consensus recommendations for use of PCR in diagnosis of pertussis. J Clin Microbiol 38:4499–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fry NK, Tzivra O, Li YT, McNiff A, Doshi N, Maple PA, Crowcroft NS, Miller E, George RC, Harrison TG. 2004. Laboratory diagnosis of pertussis infections: the role of PCR and serology. J Med Microbiol 53:519–525. doi: 10.1099/jmm.0.45624-0. [DOI] [PubMed] [Google Scholar]
- 87.Gullsby K, Hallander HO, Bondeson K. 2007. Performance of Bordetella pertussis IS481 real-time PCR in a vaccine trial setting. APMIS 115:1370–1375. doi: 10.1111/j.1600-0463.2007.00774.x. [DOI] [PubMed] [Google Scholar]
- 88.Knorr L, Fox JD, Tilley PA, Ahmed-Bentley J. 2006. Evaluation of real-time PCR for diagnosis of Bordetella pertussis infection. BMC Infect Dis 6:62. doi: 10.1186/1471-2334-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Poddar SK. 2004. Rapid detection of Bordetella pertussis by real-time PCR using SYBR green I and a LightCycler instrument. J Clin Lab Anal 18:265–270. doi: 10.1002/jcla.20035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van der Zee A, Agterbeerg C, Peeters M, Schellekens J, Mooi FR. 1993. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J Clin Microbiol 31:2134–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cloud JL, Hymas WC, Turlak A, Croft A, Reischl U, Daly JA, Carroll KC. 2003. Description of a multiplex Bordetella pertussis and Bordetella parapertussis LightCycler PCR assay with inhibition control. Diagn Microbiol Infect Dis 46:189–195. doi: 10.1016/S0732-8893(03)00045-2. [DOI] [PubMed] [Google Scholar]
- 92.Sloan LM, Hopkins MK, Mitchell PS, Vetter EA, Rosenblatt JE, Harmsen WS, Cockerill FR, Patel R. 2002. Multiplex LightCycler PCR assay for detection and differentiation of Bordetella pertussis and Bordetella parapertussis in nasopharyngeal specimens. J Clin Microbiol 40:96–100. doi: 10.1128/JCM.40.1.96-100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grogan JA, Logan C, O'Leary J, Rush R, O'Sullivan N. 2011. Real-time PCR-based detection of Bordetella pertussis and Bordetella parapertussis in an Irish paediatric population. J Med Microbiol 60:722–729. doi: 10.1099/jmm.0.030049-0. [DOI] [PubMed] [Google Scholar]
- 94.Kösters K, Riffelmann M, Wirsing von Konig CH. 2001. Evaluation of a real-time PCR assay for detection of Bordetella pertussis and B. parapertussis in clinical samples. J Med Microbiol 50:436–440. [DOI] [PubMed] [Google Scholar]
- 95.Lind-Brandberg L, Welinder-Olsson C, Lagergard T, Taranger J, Trollfors B, Zackrisson G. 1998. Evaluation of PCR for diagnosis of Bordetella pertussis and Bordetella parapertussis infections. J Clin Microbiol 36:679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ménard A, Lehours P, Sarlangue J, Bébéar C, Mégraud F, de Barbeyrac B. 2007. Development of a real-time PCR for the identification of Bordetella pertussis and Bordetella parapertussis. Clin Microbiol Infect 13:419–423. doi: 10.1111/j.1469-0691.2006.01659.x. [DOI] [PubMed] [Google Scholar]
- 97.Müller FM, Heininger U, Schnitzler N, Kockelkorn P, Cloot O, Lorenz C, Haase G. 1998. Discrimination of Bordetella parapertussis and Bordetella pertussis organisms from clinical isolates by PCR using biotin-labelled oligonucleotide probes. Mol Cell Probes 12:213–217. doi: 10.1006/mcpr.1998.0173. [DOI] [PubMed] [Google Scholar]
- 98.Reference deleted.
- 99.Tatti KM, Sparks KN, Boney KO, Lucia Tondella M. 2011. Novel multitarget real-time PCR assay for rapid detection of Bordetella species in clinical specimens. J Clin Microbiol 49:4059–4066. doi: 10.1128/JCM.00601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Templeton KE, Scheltinga SA, van der Zee A, Diederen BMW, Kruijssen AM, Goossens H, Kuijper E, Claas ECJ. 2003. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J Clin Microbiol 41:4121–4126. doi: 10.1128/JCM.41.9.4121-4126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu Y, Xu Y, Hou Q, Yang R, Zhang S. 2010. Triplex real-time PCR assay for detection and differentiation of Bordetella pertussis and Bordetella parapertussis. APMIS 118:685–691. doi: 10.1111/j.1600-0463.2010.02644.x. [DOI] [PubMed] [Google Scholar]
- 102.Birkebaek NH, Heron I, Skjodt K. 1994. Bordetella pertussis diagnosed by polymerase chain reaction. APMIS 102:291–294. doi: 10.1111/j.1699-0463.1994.tb04878.x. [DOI] [PubMed] [Google Scholar]
- 103.Loeffelholz MJ, Thompson CJ, Long KS, Gilchrist MJR. 2000. Detection of Bordetella holmesii using Bordetella pertussis IS481 PCR assay. J Clin Microbiol 38:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Register KB, Sanden GN. 2006. Prevalence and sequence variants of IS481 in Bordetella bronchiseptica: implications for IS481-based detection of Bordetella pertussis. J Clin Microbiol 44:4577–4583. doi: 10.1128/JCM.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reischl U, Lehn N, Sanden GN, Loeffelholz MJ. 2001. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J Clin Microbiol 39:1963–1966. doi: 10.1128/JCM.39.5.1963-1966.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tizolova A, Guiso N, Guillot S. 2013. Insertion sequences shared by Bordetella species and implications for the biological diagnosis of pertussis syndrome. Eur J Clin Microbiol Infect Dis 32:89–96. doi: 10.1007/s10096-012-1718-3. [DOI] [PubMed] [Google Scholar]
- 107.Afonina I, Metcalf M, Mills A, Mahoney W. 2008. Evaluation of 5′-MGB hybridization probes for detection of Bordetella pertussis and Bordetella parapertussis using different real-time PCR instruments. Diagn Microbiol Infect Dis 60:429–432. doi: 10.1016/j.diagmicrobio.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 108.He Q, Schmidt-Schläpfer G, Just M, Matter HC, Nikkari S, Viljanen MK, Mertsola J. 1996. Impact of polymerase chain reaction on clinical pertussis: Finnish and Swiss experiences. J Infect Dis 174:1288–1295. doi: 10.1093/infdis/174.6.1288. [DOI] [PubMed] [Google Scholar]
- 109.Reizenstein E, Johansson B, Mardin L, Abens J, Möllby R, Hallander HO. 1993. Diagnostic evaluation of polymerase chain reaction discriminative for Bordetella pertussis, B. parapertussis, and B. bronchiseptica. Diagn Microbiol Infect Dis 17:185–191. doi: 10.1016/0732-8893(93)90094-N. [DOI] [PubMed] [Google Scholar]
- 110.Schmidt-Schläpfer G, Liese JG, Porter F, Stojanov S, Just M, Belohradsky BH. 1997. Polymerase chain reaction (PCR) compared with conventional identification in culture for detection of Bordetella pertussis in 7153 children. Clin Microbiol Infect 3:462–467. doi: 10.1111/j.1469-0691.1997.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 111.Fry NK, Duncan J, Wagner K, Tzivra O, Doshi N, Litt DJ, Crowcroft N, Miller E, George RC, Harrison TG. 2009. Role of PCR in the diagnosis of pertussis infection in infants: 5 years' experience of provision of a same-day real-time PCR service in England and Wales from 2002 to 2007. J Med Microbiol 58:1023–1029. doi: 10.1099/jmm.0.009878-0. [DOI] [PubMed] [Google Scholar]
- 112.André P, Caro V, Njamkepo E, Wendelboe AM, Van Rie A, Guiso N. 2008. Comparison of serological and real-time PCR assays to diagnose Bordetella pertussis infection in 2007. J Clin Microbiol 46:1672–1677. doi: 10.1128/JCM.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nikbin VS, Shahcheraghi F, Lotfi MN, Zahraei SM, Parzadeh M. 2013. Comparison of culture and real-time PCR for detection of Bordetella pertussis isolated from patients in Iran. Iran J Microbiol 5:209–214. [PMC free article] [PubMed] [Google Scholar]
- 114.Qin X, Galanakis E, Martin ET, Englund JA. 2007. Multitarget PCR for diagnosis of pertussis and its clinical implications. J Clin Microbiol 45:506–511. doi: 10.1128/JCM.02042-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rodgers L, Martin SW, Cohn A, Budd J, Marcon M, Terranella A, Mandal S, Salamon D, Leber A, Tondella ML, Tatti K, Spicer K, Emanuel A, Koch E, McGlone L, Pawloski L, Lemaile-Williams M, Tucker N, Iyer R, Clark TA, Diorio M. 2013. Epidemiologic and laboratory features of a large outbreak of pertussis-like illnesses associated with cocirculating Bordetella holmesii and Bordetella pertussis—Ohio, 2010–2011. Clin Infect Dis 56:322–331. doi: 10.1093/cid/cis888. [DOI] [PubMed] [Google Scholar]
- 116.Roorda L, Buitenwerf J, Ossewaarde JM, van der Zee A. 2011. A real-time PCR assay with improved specificity for detection and discrimination of all clinically relevant Bordetella species by the presence of distribution of three insertion sequence elements. BMC Res Notes 4:11. doi: 10.1186/1756-0500-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Spicer KB, Salamon D, Cummins C, Leber A, Rodgers LE, Marcon MJ. 2014. Occurrence of three Bordetella species during an outbreak of cough illness in Ohio: epidemiology, clinical features, laboratory findings, and antimicrobial susceptibility. Pediatr Infect Dis J 3:e162–e167. [DOI] [PubMed] [Google Scholar]
- 118.Tatti KM, Wu KH, Tondella ML, Cassiday PK, Cortese MM, Wilkins PP, Sanden GN. 2008. Development and evaluation of dual-target real-time polymerase chain reaction assays to detect Bordetella spp. Diagn Microbiol Infect Dis 61:264–272. doi: 10.1016/j.diagmicrobio.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 119.Qin X, Turgeon DK, Ingersoll BP, Monsaas PW, Lemoine CJ, Tsosie T, Stapp LO, Abe PM. 2002. Bordetella pertussis PCR: simultaneous targeting of signature sequences. Diagn Microbiol Infect Dis 43:269–275. doi: 10.1016/S0732-8893(02)00405-4. [DOI] [PubMed] [Google Scholar]
- 120.Kallonen T, Mertsola J, Mooi FR, He Q. 2012. Rapid detection of the recently emerged Bordetella pertussis strains with the ptxP3 pertussis toxin promoter allele by real-time PCR. Clin Microbiol Infect 18:E377–E379. doi: 10.1111/j.1469-0691.2012.04000.x. [DOI] [PubMed] [Google Scholar]
- 121.Mooi FR, van Loo IHM, van Gent M, He Q, Heuvelman CJ, Bart MJ, de Greeff SC, Diavatopoulos DA, Teunis PF, Nagelkerke NJ, Mertsola J. 2009. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis 15:1206–1213. doi: 10.3201/eid1508.081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nygren M, Reizenstein E, Ronaghi M, Lundeberg J. 2000. Polymorphism in the pertussis toxin promoter region affecting the DNA-based diagnosis of Bordetella infection. J Clin Microbiol 38:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Houard S, Hackel C, Herzog A, Bollen A. 1989. Specific identification of Bordetella pertussis by the polymerase chain reaction. Res Microbiol 140:477–487. doi: 10.1016/0923-2508(89)90069-7. [DOI] [PubMed] [Google Scholar]
- 124.Mastrantonio P, Stefanelli P, Giuliano M. 1996. Polymerase chain reaction for the detection of Bordetella pertussis in clinical nasopharyngeal aspirates. J Med Microbiol 44:261–266. doi: 10.1099/00222615-44-4-261. [DOI] [PubMed] [Google Scholar]
- 125.Douglas E, Coole JG, Parton R, McPheat W. 1993. Identification of Bordetella pertussis in nasopharyngeal swabs by PCR amplification of a region of the adenylate cyclase gene. J Med Microbiol 38:140–144. doi: 10.1099/00222615-38-2-140. [DOI] [PubMed] [Google Scholar]
- 126.Mäkinen J, Viljanen MK, Mertsola J, Arvilommi H, He Q. 2001. Rapid identification of Bordetella pertussis pertactin gene variants using LightCycler real-time polymerase chain reaction combined with melting curve analysis and gel electrophoresis. Emerg Infect Dis 7:952–958. doi: 10.3201/eid0706.010606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vincart B, De Mendonça R, Rottiers S, Vermeulen F, Struelens MJ, Denis O. 2007. A specific real-time PCR assay for the detection of Bordetella pertussis. J Med Microbiol 56:918–920. doi: 10.1099/jmm.0.46947-0. [DOI] [PubMed] [Google Scholar]
- 128.Walsh P, Overmeyer C, Kimmel L, Feola M, Pusavat J, Nguyen TA, Kuan S, Emery K, Rosengreen M, Mordechai E, Adelson ME. 2008. Prevalence of Bordetella pertussis and Bordetella parapertussis in samples submitted for RSV screening. West J Emerg Med 9:135–140. [PMC free article] [PubMed] [Google Scholar]
- 129.Li ZM, Jansen DL, Finn TM, Halperin SA, Kasina A, O'Connor SP, Aoyama T, Manclark CR, Brennan MJ. 1994. Identification of Bordetella pertussis infection by shared-primer PCR. J Clin Microbiol 32:783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Probert WS, Ely J, Schrader K, Atwell J, Nossoff A, Kwan S. 2008. Identification and evaluation of new target sequences for specific detection of Bordetella pertussis by real-time PCR. J Clin Microbiol 46:3228–3231. doi: 10.1128/JCM.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Register KB, Nicholson TL, Guthrie JL. 2010. Evaluation of specificity of BP3385 for Bordetella pertussis detection. J Clin Microbiol 48:3334–3337. doi: 10.1128/JCM.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brinig MM, Cummings CA, Sanden GN, Stefanelli P, Lawrence A, Relman DA. 2006. Significant gene order and expression differences in Bordetella pertussis despite limited gene content variation. J Bacteriol 188:2375–2382. doi: 10.1128/JB.188.7.2375-2382.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heikkinen E, Kallonen T, Saarinen L, Sara R, King AJ, Mooi FR, Soini JT, Mertsola J, He Q. 2007. Comparative genomics of Bordetella pertussis reveals progressive gene loss in Finnish strains. PLoS One 2:e904. doi: 10.1371/journal.pone.0000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.King AJ, van Gorkom T, Pennings JL, van der Heide HG, He Q, Diavatopoulos D, Heuvelman K, van Gent M, van Leeuwen K, Mooi FR. 2008. Comparative genomic profiling of Dutch clinical Bordetella pertussis isolates using DNA microarrays: identification of genes absent from epidemic strains. BMC Genomics 9:311. doi: 10.1186/1471-2164-9-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hassan F, Hays L, Bell J, Selvarangan R. 2014. Evaluation of 3 analyte-specific reagents for detection of Bordetella pertussis and Bordetella parapertussis in clinical specimens. Diagn Microbiol Infect Dis 80:181–184. doi: 10.1016/j.diagmicrobio.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 136.Lanotte P, Plouzeau C, Burucoa C, Grélaud C, Guillot S, Guiso N, Garnier F. 2011. Evaluation of four commercial real-time PCR assays for detection of Bordetella spp. in nasopharyngeal aspirates. J Clin Microbiol 49:3943–3946. doi: 10.1128/JCM.00335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kenicer J, Hardie A, Hamilton F, Gadsby N, Templeton K. 2014. Comparative evaluation of the Diagenode multiplex PCR assay on the BD Max system versus a routine in-house assay for detection of Bordetella pertussis. J Clin Microbiol 52:2668–2670. doi: 10.1128/JCM.00212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Koidl C, Bozic M, Burmeister A, Hess M, Marth E, Kessler HH. 2007. Detection and differentiation of Bordetella spp. by real-time PCR. J Clin Microbiol 45:347–350. doi: 10.1128/JCM.01303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cohen-Bacrie S, Bertin F, Gassiot AS, Prère MF. 2010. Rapid molecular genetic assay for direct identification of Bordetella from patients specimens. Pathol Biol 58:52–54. doi: 10.1016/j.patbio.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 140.Pillet S, Lardeux M, Dina J, Grattard F, Verhoeven P, Le Goff J, Vabret A, Pozzetto B. 2013. Comparative evaluation of six commercialized multiplex PCR kits for the diagnosis of respiratory infections. PLoS One 8:e72174. doi: 10.1371/journal.pone.0072174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kallonen T, Grondahl-Yli-Hannuksela K, Elomaa A, Lutynska A, Fry NK, Mertsola J, He Q. 2011. Differences in the genomic content of Bordetella pertussis isolates before and after introduction of pertussis vaccines in four European countries. Infect Genet Evol 11:2034–2042. doi: 10.1016/j.meegid.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 142.Lievano FA, Reynolds MA, Waring AL, Ackelsberg J, Bisgard KM, Sanden GN, Guris D, Golaz A, Bopp DJ, Limberger RJ, Smith PF. 2002. Issues associated with and recommendations for using PCR to detect outbreaks of pertussis. J Clin Microbiol 40:2801–2805. doi: 10.1128/JCM.40.8.2801-2805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Weber DJ, Miller MB, Brooks RH, Brown VM, Rutala WA. 2010. Healthcare worker with “pertussis”: consequences of a false-positive polymerase chain reaction test result. Infect Control Hosp Epidemiol 31:306–307. doi: 10.1086/650760. [DOI] [PubMed] [Google Scholar]
- 144.Taranger J, Trollfors B, Lind L, Zackrisson G, Beling-Holmquist K. 1994. Environmental contamination leading to false-positive polymerase chain reaction for pertussis. Pediatr Infect Dis J 13:936–937. [DOI] [PubMed] [Google Scholar]
- 145.Salimnia H, Lephart PR, Asmar BI, Prebelich D, Paulson E, Fairfax MR. 2012. Aerosolized vaccine as an unexpected source of false-positive Bordetella pertussis PCR results. J Clin Microbiol 50:472–474. doi: 10.1128/JCM.01250-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meade BD, Bollen A. 1994. Recommendation for use of the polymerase chain reaction in the diagnosis of Bordetella pertussis infections. J Med Microbiol 41:51–55. doi: 10.1099/00222615-41-1-51. [DOI] [PubMed] [Google Scholar]
- 147.Herwegh S, Carnoy C, Wallet F, Loïez C, Courcol RJ. 2005. Development and use of an internal positive control for detection of Bordetella pertussis by PCR. J Clin Microbiol 43:2462–2464. doi: 10.1128/JCM.43.5.2462-2464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Koidl C, Bozic M, Berg J, Stöcher M, Mühlbauer G, Marth E, Kessler HH. 2005. Addition of a homologous internal control to a real-time PCR assay for detection of Bordetella pertussis. Clin Chem 51:2404–2406. doi: 10.1373/clinchem.2005.052183. [DOI] [PubMed] [Google Scholar]
- 149.Müller FM, Schnitzler N, Cloot O, Kockelkorn P, Haase G, Li Z. 1998. The rationale and method for constructing internal control DNA used in pertussis polymerase chain reaction. Diagn Microbiol Infect Dis 31:517–523. doi: 10.1016/S0732-8893(98)00043-1. [DOI] [PubMed] [Google Scholar]
- 150.Fry NK, Tzivra O, Duncan J, Doshi N, Harrison TG. 2004. Consensus real-time PCR assay for Bordetella pertussis including an internal process control provides specific diagnosis, rapid turnaround time, high diagnostic yield and enhanced surveillance. Clin Microbiol Infect 10(Suppl 3):7. [Google Scholar]
- 151.Lingappa JR, Lawrence W, West-Keefe S, Gautom R, Cookson BT. 2002. Diagnosis of community-acquired pertussis infection: comparison of both culture and fluorescent-antibody assays with PCR detection using electrophoresis or dot blot hybridization. J Clin Microbiol 40:2908–2912. doi: 10.1128/JCM.40.8.2908-2912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Burd EM. 2010. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev 23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 154.Papenburg J, Fontela P. 2009. What is the significance of a high cycle threshold positive IS481 PCR for Bordetella pertussis? Pediatr Infect Dis J 28:1143. doi: 10.1097/INF.0b013e3181bd4e1f. [DOI] [PubMed] [Google Scholar]
- 155.Guthrie JL, Seah C, Brown S, Tang P, Jamieson F, Drews SJ. 2008. Use of Bordetella pertussis BP3385 to establish a cutoff value for an IS481-targeted real-time PCR assay. J Clin Microbiol 46:3798–3799. doi: 10.1128/JCM.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Edelman K, Nikkari S, Ruuskanen O, He Q, Viljanen M, Mertsola J. 1996. Detection of Bordetella pertussis by polymerase chain reaction and culture in the nasopharynx of erythromycin-treated infants with pertussis. Pediatr Infect Dis J 15:54–57. doi: 10.1097/00006454-199601000-00012. [DOI] [PubMed] [Google Scholar]
- 157.Muyldermans G, Soetens O, Antoine M, Bruisten S, Vincart B, Doucet K, Populaire F, Fry NK, Olcen P, Scheftel JM, Senterre JM, van der Zee A, Riffelmann M, Pierard D, Lauwers S. 2005. External quality assessment for molecular detection of Bordetella pertussis in European laboratories. J Clin Microbiol 43:30–35. doi: 10.1128/JCM.43.1.30-35.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tatti KM, Martin SW, Boney KO, Brown K, Clark TA, Tondella ML. 2013. Qualitative assessment of pertussis diagnostics in United States laboratories. Pediatr Infect Dis J 32:942–945. doi: 10.1097/INF.0b013e3182947ef8. [DOI] [PubMed] [Google Scholar]
- 159.Dalby T, Fry NK, Krogfelt KA, Jensen JS, He Q. 2013. Evaluation of PCR methods for the diagnosis of pertussis by the European surveillance network for vaccine-preventable diseases (EUVAC.NET). J Clin Microbiol Infect Dis 32:1285–1289. doi: 10.1007/s10096-013-1874-0. [DOI] [PubMed] [Google Scholar]
- 160.Williams MM, Taylor TH Jr, Warshauer DM, Martin MD, Valley AM, Tondella ML. 2015. Harmonization of Bordetella pertussis real-time PCR diagnostics in the United States in 2012. J Clin Microbiol 53:118–123. doi: 10.1128/JCM.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Burgos-Rivera B, Lee AD, Bowden KE, Faulkner AE, Seaton BL, Lembke BD, Cartwright CP, Martin SW, Tondella ML. 2015. Evaluation of level of agreement in Bordetella species identification in three U.S. laboratories during a period of increased pertussis. J Clin Microbiol 53:1842–1847. doi: 10.1128/JCM.03567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ghanaie RM, Karimi A, Sadeghi H, Esteghamti A, Falah F, Armin S, Fahimzad A, Shamshiri A, Kahbazi M, Shiva F. 2010. Sensitivity and specificity of the World Health Organization pertussis clinical case definition. Int J Infect Dis 14:e1072–e1075. doi: 10.1016/j.ijid.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 163.Patriarca PA, Biellik RJ, Sanden G, Burstyn DG, Mitchell PD, Silverman PR, Davis JP, Manclark CR. 1988. Sensitivity and specificity of clinical case definitions for pertussis. Am J Public Health 78:833–836. doi: 10.2105/AJPH.78.7.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Strebel PM, Cochi SL, Farizo KM, Payne BJ, Hanauer SD, Baughman AL. 1993. Pertussis in Missouri: evaluation of nasopharyngeal culture, direct fluorescent antibody testing, and clinical case definitions in the diagnosis of pertussis. Clin Infect Dis 16:276–285. doi: 10.1093/clind/16.2.276. [DOI] [PubMed] [Google Scholar]
- 165.Bordet J, Gengou O. 1906. Le microbe de la coqueluche. Ann Inst Pasteur 20:731–741. [Google Scholar]
- 166.Bordet J, Gengou O. 1907. Note complémentaire sur le microbe de la coqueluche. Ann Inst Pasteur 21:720–726. [Google Scholar]
- 167.Weichsel M, Douglas HS. 1937. Complement fixation tests in pertussis. J Clin Invest 16:15–22. doi: 10.1172/JCI100833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Donald AB. 1938. The diagnosis of whooping-cough. Br Med J 2:613–618. doi: 10.1136/bmj.2.4054.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Evans DG, Maitland HB. 1939. Agglutination as a diagnostic test for whooping cough. J Pathol 48:468–470. doi: 10.1002/path.1700480219. [DOI] [Google Scholar]
- 170.Kendrick PL, Gottshall RY, Anderson HD, Volk VK, Bunney WE, Top FH. 1969. Pertussis agglutinins in adults. Public Health Rep 84:9–15. doi: 10.2307/4593487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.British Medical Journal. 1970. Diagnosis of whooping cough: comparison of serological tests with isolation of Bordetella pertussis. A combined Scottish study. Br Med J 4:637–639. [PMC free article] [PubMed] [Google Scholar]
- 172.Bradstreet CMP, Tannahill AJ, Edwards JMB, Benson PF. 1972. Detection of Bordetella pertussis antibodies in human sera by complement-fixation and immunofluorescence. J Hyg (Cambridge) 70:75–83. doi: 10.1017/S0022172400022117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Macaulay ME. 1979. The serological diagnosis of whooping cough. J Hyg (Cambridge) 83:95–102. doi: 10.1017/S0022172400025857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Macaulay ME. 1981. The IgM and IgG response to Bordetella pertussis vaccination and infection. J Med Microbiol 14:1–7. doi: 10.1099/00222615-14-1-1. [DOI] [PubMed] [Google Scholar]
- 175.Manclark MC, Meade BD, Burstyn DG. 1986. Serological response to Bordetella pertussis, p 388–394. In Rose NR, Friedman H, Fayhey JL (ed), Manual of clinical laboratory immunology, 3rd ed American Society for Microbiology, Washington, DC. [Google Scholar]
- 176.Guiso N, Grimprel E, Anjak I, Bégué P. 1993. Western blot analysis of antibody responses of young infants to pertussis infection. Eur J Clin Microbiol Infect Dis 12:596–600. doi: 10.1007/BF01973637. [DOI] [PubMed] [Google Scholar]
- 177.Mink CM, O'Brien CH, Wassilak S, Deforest A, Meade BD. 1994. Isotype and antigen specificity of pertussis agglutinins following whole-cell pertussis vaccination and infection with Bordetella pertussis. Infect Immun 62:1118–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Heininger U, Stehr K, Schmitt-Grohe S, Überall M, Lorenz C, Rost R, Cherry JD. 1995. Der Mikroagglutinationstest-ein einfaches und sensitives Verfahren zur Serodiagnostik von Pertussisinfektionen. Klin Paediatr 207:277–280. doi: 10.1055/s-2008-1046551. [DOI] [PubMed] [Google Scholar]
- 179.Aoyama T, Kato T, Takeuchi Y, Kato K, Morokuma K, Hirai T. 1997. Simple, speedy, sensitive, and specific serodiagnosis of pertussis by using a particle agglutination test. J Clin Microbiol 35:1859–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Granström M, Lindberg AA, Askelöf P, Hederstedt B. 1982. Detection of antibodies in human serum against the fimbrial haemagglutinin of Bordetella pertussis by enzyme-linked immunosorbent assay. J Med Microbiol 15:85–96. doi: 10.1099/00222615-15-1-85. [DOI] [PubMed] [Google Scholar]
- 181.Lawrence AJ, Paton JC. 1987. Efficacy of enzyme-linked immunosorbent assay for rapid diagnosis of Bordetella pertussis infection. J Clin Microbiol 25:2102–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Long SS, Welkon CJ, Clark JL. 1990. Widespread silent transmission of pertussis in families: antibody correlates of infection and symptomatology. J Infect Dis 161:480–486. doi: 10.1093/infdis/161.3.480. [DOI] [PubMed] [Google Scholar]
- 183.Mertsola J, Ruuskanen O, Kuronen T, Viljanen MK. 1983. Serologic diagnosis of pertussis: comparison of enzyme-linked immunosorbent assay and bacterial agglutination. J Infect Dis 147:252–257. doi: 10.1093/infdis/147.2.252. [DOI] [PubMed] [Google Scholar]
- 184.Hewlett EL, Sauer KT, Myers GA, Cowell JL, Guerrant RL. 1983. Induction of a novel morphological response in Chinese hamster ovary cells by pertussis toxin. Infect Immun 40:1198–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gillenius P, Jäätmaa E, Askelöf P, Granström M, Tiru M. 1985. The standardization of an assay for pertussis toxin and antitoxin in microplate culture of Chinese hamster ovary cells. J Biol Stand 13:61–66. doi: 10.1016/S0092-1157(85)80034-2. [DOI] [PubMed] [Google Scholar]
- 186.Granström G, Wretlind B, Salenstedt CR, Granström M. 1988. Evaluation of serologic assays for diagnosis of whooping cough. J Clin Microbiol 26:1818–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hallander HO, Storsaeter J, Möllby R. 1991. Evaluation of serology and nasopharyngeal cultures for diagnosis of pertussis in a vaccine efficacy trial. J Infect Dis 163:1046–1054. doi: 10.1093/infdis/163.5.1046. [DOI] [PubMed] [Google Scholar]
- 188.Trollfors B, Krantz I, Sigurs N, Taranger J, Zackrisson G, Roberson R. 1988. Toxin-neutralizing antibodies in patients with pertussis, as determined by an assay using Chinese hamster ovary cells. J Infect Dis 158:991–995. doi: 10.1093/infdis/158.5.991. [DOI] [PubMed] [Google Scholar]
- 189.Mertsola J, Ruuskanen O, Kuronen T, Meurman O, Viljanen MK. 1990. Serologic diagnosis of pertussis: evaluation of pertussis toxin and other antigens in enzyme-linked immunosorbent assay. J Infect Dis 161:966–971. doi: 10.1093/infdis/161.5.966. [DOI] [PubMed] [Google Scholar]
- 190.Novotny P, Macaulay ME, Hart TC, Skvaril F. 1991. Analysis of antibody profiles in children with whooping cough. Dev Biol Stand 73:267–273. [PubMed] [Google Scholar]
- 191.Watanabe M, Connelly B, Weiss AA. 2006. Characterization of serological responses to pertussis. Clin Vaccine Immunol 13:341–348. doi: 10.1128/CVI.13.3.341-348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Granström M, Granström G, Lindfors A, Askelöf P. 1982. Serologic diagnosis of whooping cough by an enzyme-linked immunosorbent assay using fimbrial hemagglutinin as antigen. J Infect Dis 146:741–745. doi: 10.1093/infdis/146.6.741. [DOI] [PubMed] [Google Scholar]
- 193.Zackrisson G, Arminjon F, Krantz I, Lagergård T, Sigurs N, Taranger J, Trollfors B. 1988. Serum antibody response to filamentous hemagglutinin in patients with clinical pertussis measured by an enzyme-linked immunosorbent assay. Eur J Clin Microbiol Infect Dis 7:764–770. doi: 10.1007/BF01975044. [DOI] [PubMed] [Google Scholar]
- 194.Zackrisson G, Krantz I, Lagergård T, Larsson P, Sekura R, Sigurs N, Taranger J, Trollfors B. 1988. Antibody response to pertussis toxin in patients with clinical pertussis measured by enzyme-linked immunosorbent assay. Eur J Clin Microbiol Infect Dis 7:149–154. doi: 10.1007/BF01963068. [DOI] [PubMed] [Google Scholar]
- 195.Horby P, Macintyre CR, McIntyre PB, Gilbert GL, Staff M, Hanlon M, Heron LG, Cagney M, Bennett C. 2005. A boarding school outbreak of pertussis in adolescents: value of laboratory diagnostic methods. Epidemiol Infect 133:229–236. doi: 10.1017/S0950268804003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.May ML, Doi SA, King D, Evans J, Robson JM. 2012. Prospective evaluation of an Australian pertussis toxin IgG and IgA enzyme immunoassay. Clin Vaccine Immunol 19:190-197. doi: 10.1128/CVI.05430-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nagel J, de Graaf S, Schijf-Evers D. 1985. Improved serodiagnosis of whooping cough caused by Bordetella pertussis by determination of IgG anti-LPF antibody levels. Dev Biol Stand 61:325–330. [PubMed] [Google Scholar]
- 198.Wirsing von König CH, Schmitt HJ. 1996. Epidemiologic aspects and diagnostic criteria for a protective efficacy field trial of a pertussis vaccine. J Infect Dis 174:S281–S286. doi: 10.1093/infdis/174.Supplement_3.S281. [DOI] [PubMed] [Google Scholar]
- 199.van der Zee A, Agterberg C, Peeters M, Mooi F, Schellekens J. 1996. A clinical validation of Bordetella pertussis and Bordetella parapertussis polymerase chain reaction: comparison with culture and serology using samples from patients with suspected whooping cough from a highly immunized population. J Infect Dis 174:89–96. doi: 10.1093/infdis/174.1.89. [DOI] [PubMed] [Google Scholar]
- 200.Deen JL, Mink CA, Cherry JD, Christenson PD, Pineda EF, Lewis K, Blumberg DA, Ross LA. 1995. Household contact study of Bordetella pertussis infections. Clin Infect Dis 21:1211–1219. doi: 10.1093/clinids/21.5.1211. [DOI] [PubMed] [Google Scholar]
- 201.Giammanco A, Taormina S, Genovese M, Mangiaracina G, Giammanco G, Chiarini A. 1997. Serological responses to infection with B pertussis. Dev Biol Stand 89:213–220. [PubMed] [Google Scholar]
- 202.Heininger U, Cherry JD, Stehr K. 2004. Serologic response and antibody-titer decay in adults with pertussis. Clin Infect Dis 38:591–594. doi: 10.1086/381439. [DOI] [PubMed] [Google Scholar]
- 203.Burstyn DG, Baraff LJ, Peppler MS, Leake RD, St Geme J, Manclark CR. 1983. Serological response to filamentous hemagglutinin and lymphocytosis-promoting toxin of Bordetella pertussis. Infect Immun 41:1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hendrikx LH, Öztürk K, de Rond LG, de Greeff SC, Sanders EA, Berbers GA, Buisman AM. 2011. Serum IgA responses against pertussis proteins in infected and Dutch wP or aP vaccinated children: an additional role in pertussis diagnostics. PLoS One 6:e27681. doi: 10.1371/journal.pone.0027681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nagel J, Poot-Scholtens EJ. 1983. Serum IgA antibody to Bordetella pertussis as an indicator of infection. J Med Microbiol 16:417–426. doi: 10.1099/00222615-16-4-417. [DOI] [PubMed] [Google Scholar]
- 206.Stehr K, Cherry JD, Heininger U, Schmitt-Grohé S, Uberall M, Laussucq S, Eckhardt T, Meyer M, Engelhardt R, Christenson P. 1998. A comparative efficacy trial in Germany in infants who received either the Lederle/Takeda acellular pertussis component DTP (DTaP) vaccine, the Lederle whole-cell component DTP vaccine, or DT vaccine. Pediatrics 101:1–11. doi: 10.1542/peds.101.1.1. [DOI] [PubMed] [Google Scholar]
- 207.Le T, Cherry JD, Chang SJ, Knoll MD, Lee ML, Barenkamp S, Bernstein D, Edelman R, Edwards KM, Greenberg D, Keitel W, Treanor J, Ward JI, APERT Study. 2004. Immune responses and antibody decay after immunization of adolescents and adults with an acellular pertussis vaccine: the APERT Study. J Infect Dis 190:535–544. doi: 10.1086/422035. [DOI] [PubMed] [Google Scholar]
- 208.Isacson J, Trollfors B, Taranger J, Lagergård T. 1995. Acquisition of IgG serum antibodies against two Bordetella antigens (filamentous hemagglutinin and pertactin) in children with no symptoms of pertussis. Pediatr Infect Dis J 14:517–521. doi: 10.1097/00006454-199506000-00009. [DOI] [PubMed] [Google Scholar]
- 209.Trollfors B, Lagergård T, Gunnarsson E, Taranger J. 2003. Determination of pertactin IgG antibodies for the diagnosis of pertussis. Clin Microbiol Infect 9:585–589. doi: 10.1046/j.1469-0691.2003.00552.x. [DOI] [PubMed] [Google Scholar]
- 210.Hallander HO, Ljungman M, Jahnmatz M, Storsaeter J, Nilsson L, Gustafsson L. 2009. Should fimbriae be included in pertussis vaccines? Studies on ELISA IgG anti-Fim2/3 antibodies after vaccination and infection. APMIS 117:660–671. doi: 10.1111/j.1600-0463.2009.02521.x. [DOI] [PubMed] [Google Scholar]
- 211.Cherry JD, Xing DXL, Newland P, Patel K, Heininger U, Corbel MJ. 2004. Determination of serum antibody to Bordetella pertussis adenylate cyclase toxin in vaccinated and unvaccinated children and in children and adults with pertussis. Clin Infect Dis 38:502–507. doi: 10.1086/381204. [DOI] [PubMed] [Google Scholar]
- 212.Arciniega JL, Hewlett EL, Johnson FD, Deforest A, Wassilak SGF, Onorato IM, Manclark CR, Burns DL. 1991. Human serological response to envelope-associated proteins and adenylate cyclase toxin of Bordetella pertussis. J Infect Dis 163:135–142. doi: 10.1093/infdis/163.1.135. [DOI] [PubMed] [Google Scholar]
- 213.Fazekas S, Webster RG. 1966. Disquisitions on original antigenic sin. I. Evidence in man. J Exp Med 124:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Goodman YE, Wort AJ, Jackson FL. 1981. Enzyme-linked immunosorbent assay for detection of pertussis immunoglobulin A in nasopharyngeal secretions as an indicator of recent infection. J Clin Microbiol 13:286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Viljanen MK, Ruuskanen O, Granberg C, Salmi TT. 1982. Serological diagnosis of pertussis: IgM, IgA and IgG antibodies against Bordetella pertussis measured by enzyme-linked immunosorbent assay (ELISA). Scand J Infect Dis 14:117–122. doi: 10.3109/inf.1982.14.issue-2.08. [DOI] [PubMed] [Google Scholar]
- 216.Barenkamp SJ, Leininger E. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect Immun 60:1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Trollfors B, Burman L, Lagergård T, Leinonen M, Taranger J. 1996. No cross-reactivity with filamentous hemagglutinin of Bordetella pertussis in sera from patients with nontypable Haemophilus influenzae pneumonia. Pediatr Infect Dis J 15:558–559. [DOI] [PubMed] [Google Scholar]
- 218.Vincent JM, Cherry JD, Nauschuetz WF, Lipton A, Ono CM, Costello CN, Sakaguchi LK, Hsue G, Jackson LA, Tachdjian R, Cotter PA, Gornbein JA. 2000. Prolonged afebrile nonproductive cough illnesses in American soldiers in Korea: a serological search for causation. Clin Infect Dis 30:534–539. doi: 10.1086/313707. [DOI] [PubMed] [Google Scholar]
- 219.Carlsson RM, Claesson BA, Lagergärd Trollfors B. 1999. Acquisition of serum antibodies against filamentous hemagglutinin and pertactin unrelated to Bordetella pertussis infection. Clin Microbiol Infect 5:709–712. doi: 10.1111/j.1469-0691.1999.tb00520.x. [DOI] [Google Scholar]
- 220.Isacson J, Trollfors B, Hedvall G, Taranger J, Zackrisson G. 1995. Response and decline of serum IgG antibodies to pertussis toxin, filamentous hemagglutinin and pertactin in children with pertussis. Scand J Infect Dis 27:273–277. doi: 10.3109/00365549509019021. [DOI] [PubMed] [Google Scholar]
- 221.Lynn F, Reed GF, Meade BD. 1996. Collaborative study for the evaluation of enzyme-linked immunosorbent assays used to measure human antibodies to Bordetella pertussis antigens. Clin Diagn Lab Immunol 3:689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Meade BD, Deforest A, Edwards KM, Romani TA, Lynn F, O'Brien CH, Swartz CB, Reed GF, Deloria MA. 1995. Description and evaluation of serologic assays used in a multicenter trial of acellular pertussis vaccines. Pediatrics 96(Suppl):570–575. [PubMed] [Google Scholar]
- 223.Xing D, Wirsing von König CH, Newland P, Riffelmann M, Meade BD, Corbel M, Gaines-Das R. 2009. Characterization of reference materials for human antiserum to pertussis antigens by an international collaborative study. Clin Vaccine Immunol 16:303–311. doi: 10.1128/CVI.00372-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Reizenstein E, Hallander HO, Blackwelder WC, Kühn I, Ljungman M, Möllby R. 1995. Comparison of five calculation modes for antibody ELISA procedures using pertussis serology as a model. J Immunol Methods 183:279–290. doi: 10.1016/0022-1759(95)00067-K. [DOI] [PubMed] [Google Scholar]
- 225.Gustafsson L, Hallander HO, Olin P, Reizenstein E, Storsaeter J. 1996. A controlled trial of a two-component acellular, a five-component acellular, and a whole-cell pertussis vaccine. N Engl J Med 334:349–355. doi: 10.1056/NEJM199602083340602. [DOI] [PubMed] [Google Scholar]
- 226.Heininger U, Schmidt-Schläpfer G, Cherry JD, Stehr K. 2000. Clinical validation of a polymerase chain reaction assay for the diagnosis of pertussis by comparison with serology, culture, and symptoms during a large pertussis vaccine efficacy trial. Pediatrics 105:E31. doi: 10.1542/peds.105.3.e31. [DOI] [PubMed] [Google Scholar]
- 227.Schmitt HJ, von König CH, Neiss A, Bogaerts H, Bock HL, Schulte-Wissermann H, Gahr M, Schult R, Folkens JU, Rauh W, Clemens R. 1996. Efficacy of acellular pertussis vaccine in early childhood after household exposure. JAMA 275:37–41. [PubMed] [Google Scholar]
- 228.Simondon F, Iteman I, Preziosi MP, Yam A, Guiso N. 1998. Evaluation of an immunoglobulin G enzyme-linked immunosorbent assay for pertussis toxin and filamentous hemagglutinin in diagnosis of pertussis in Senegal. Clin Diagn Lab Immunol 5:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Giammanco A, Chiarini A, Maple PA, Andrews N, Pebody R, Gay N, Olander RM, Fivet-Groyne F, Baron S, Tischer A, Swidsinski S, Schellekens J, Reizenstein E. 2003. European Sero-Epidemiology Network: standardisation of the assay results for pertussis. Vaccine 22:112–120. doi: 10.1016/S0264-410X(03)00514-0. [DOI] [PubMed] [Google Scholar]
- 230.de Greeff SC, Teunis P, de Melker HE, Mooi FR, Notermans DW, Elvers B, Schellekens JF. 2012. Two-component cluster analysis of a large serodiagnostic database for specificity of increases of IgG antibodies against pertussis toxin in paired serum samples and of absolute values in single serum samples. Clin Vaccine Immunol 19:1452–1456. doi: 10.1128/CVI.00229-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sekura RD, Fish F, Manclark CR, Meade B, Zhang YL. 1983. Pertussis toxin. Affinity purification of a new ADP-ribosyltransferase. J Biol Chem 258:14647–14651. [PubMed] [Google Scholar]
- 232.Zackrisson G, Lagergård T, Lönnroth I. 1986. An enzyme-linked-immunosorbent assay method for detection of immunoglobulins to pertussis toxin. Acta Pathol Immunol Scand Sect C 94:227–231. [DOI] [PubMed] [Google Scholar]
- 233.Kapasi A, Meade BD, Plikaytis B, Pawloski L, Martin MD, Yoder S, Rock MT, Coddens S, Haezebroeck V, Fievet-Groyne F, Bixler G, Jones C, Hildreth S, Edwards KM, Messonnier NE, Tondella ML. 2012. Comparative study of different sources of pertussis toxin (PT) as coating antigens in IgG anti-PT enzyme-linked immunosorbent assays. Clin Vaccine Immunol 19:64–72. doi: 10.1128/CVI.05460-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Menzies SL, Kadwad V, Pawloski LC, Lin TL, Baughman AL, Martin M, Tondella ML, Meade BD, Pertussis Assay Working Group. 2009. Development and analytical validation of an immunoassay for quantifying serum anti-pertussis toxin antibodies resulting from Bordetella pertussis infection. Clin Vaccine Immunol 16:1781–1788. doi: 10.1128/CVI.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Greco D, Salmaso S, Mastrantonio P, Giuliano M, Tozzi AE, Anemona A, Ciofi degli Atti ML, Giammanco A, Panei P, Blackwelder WC, Klein DL, Wassilak SG. 1996. A controlled trial of two acellular vaccines and one whole-cell vaccine against pertussis. Progetto Pertosse Working Group. N Engl J Med 334:341–348. [DOI] [PubMed] [Google Scholar]
- 236.Granström G, Granström M. 1994. Effect of erythromycin treatment on antibody responses in pertussis. Scand J Infect Dis 26:453–457. doi: 10.3109/00365549409008619. [DOI] [PubMed] [Google Scholar]
- 237.Strebel P, Nordin J, Edwards K, Hunt J, Besser J, Burns S, Amundson G, Baughman A, Wattigney W. 2001. Population-based incidence of pertussis among adolescents and adults, Minnesota, 1995–1996. J Infect Dis 183:1353–1359. doi: 10.1086/319853. [DOI] [PubMed] [Google Scholar]
- 238.Wirsing von König CH, Gounis D, Laukamp S, Bogaerts H, Schmitt HJ. 1999. Evaluation of a single-sample serological technique for diagnosing pertussis in unvaccinated children. Eur J Clin Microbiol Infect Dis 18:341–345. doi: 10.1007/PL00015016. [DOI] [PubMed] [Google Scholar]
- 239.Wirsing von König CH, Postels-Multani S, Bock HL, Schmitt HJ. 1995. Pertussis in adults: frequency of transmission after household exposure. Lancet 346:1326–1329. doi: 10.1016/S0140-6736(95)92343-8. [DOI] [PubMed] [Google Scholar]
- 240.Senzilet LD, Halperin SA, Spika JS, Alagaratnam M, Morris A, Smith B, Sentinel Health Unit Surveillance System Pertussis Working Group. 2001. Pertussis is a frequent cause of prolonged cough illness in adults and adolescents. Clin Infect Dis 32:1691–1697. doi: 10.1086/320754. [DOI] [PubMed] [Google Scholar]
- 241.Marchant CD, Loughlin AM, Lett SM, Todd CW, Wetterlow LH, Bicchieri R, Higham S, Etkind P, Silva E, Siber GR. 1994. Pertussis in Massachusetts, 1981–1991: incidence, serologic diagnosis, and vaccine effectiveness. J Infect Dis 169:1297–1305. doi: 10.1093/infdis/169.6.1297. [DOI] [PubMed] [Google Scholar]
- 242.Dalby T, Petersen JW, Harboe ZB, Krogfelt KA. 2010. Antibody responses to pertussis toxin display different kinetics after clinical Bordetella pertussis infection than after vaccination with an acellular pertussis vaccine. J Med Microbiol 59:1029–1036. doi: 10.1099/jmm.0.020826-0. [DOI] [PubMed] [Google Scholar]
- 243.Baughman AL, Bisgard KM, Edwards KM, Guris D, Decker MD, Holland K, Meade BD, Lynn F. 2004. Establishment of diagnostic cutoff points for levels of serum antibodies to pertussis toxin, filamentous hemagglutinin, and fimbriae in adolescents and adults in the United States. Clin Diagn Lab Immunol 11:1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yih WK, Lett SM, des Vignes FN, Garrison KM, Sipe PL, Marchant CD. 2000. The increasing incidence of pertussis in Massachusetts adolescents and adults, 1989–1998. J Infect Dis 182:1409–1416. doi: 10.1086/315863. [DOI] [PubMed] [Google Scholar]
- 245.Versteegh FGA, Mertens PLJM, de Melker HE, Roord JJ, Schellekens JFP, Teunis PFM. 2005. Age-specific long-term course of IgG antibodies to pertussis toxin after symptomatic infection with Bordetella pertussis. Epidemiol Infect 133:737–748. doi: 10.1017/S0950268805003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hallander HO, Andersson M, Gustafsson L, Ljungman M, Netterlid E. 2009. Seroprevalence of pertussis antitoxin (anti-PT) in Sweden before and 10 years after the introduction of a universal childhood pertussis vaccination program. APMIS 117:912–922. doi: 10.1111/j.1600-0463.2009.02554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Teunis PF, van der Heijden OG, de Melker HE, Schellekens JF, Versteegh FG, Kretzschmar ME. 2002. Kinetics of the IgG antibody response to pertussis toxin after infection with B. pertussis. Epidemiol Infect 129:479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hallander HO, Ljungman M, Storsaeter J, Gustafsson L. 2009. Kinetics and sensitivity of ELISA IgG pertussis antitoxin after infection and vaccination with Bordetella pertussis in young children. APMIS 117:797–807. doi: 10.1111/j.1600-0463.2009.02530.x. [DOI] [PubMed] [Google Scholar]
- 249.Esposito S, Agliardi T, Giammanco A, Faldella G, Cascio A, Bosis S, Friscia O, Clerici M, Principi N. 2001. Long-term pertussis-specific immunity after primary vaccination with a combined diphtheria, tetanus, tricomponent acellular pertussis, and hepatitis B vaccine in comparison with that after natural infection. Infect Immun 69:4516–4520. doi: 10.1128/IAI.69.7.4516-4520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Giuliano M, Mastrantonio P, Giammanco A, Bottone M, Piscitelli A, Di Tommaso S, Robino L, Basso F, D'Orazio P. 1997. Antibody kinetics and long-term sero-prevalence in the Italian clinical trial of acellular pertussis vaccines. Dev Biol Stand 89:275–278. [PubMed] [Google Scholar]
- 251.Guiso N, Njamkepo E, Vié le, Sage F, Zepp F, Meyer CU, Abitbol V, Clyti N, Chevallier S. 2007. Long-term humoral and cell-mediated immunity after acellular pertussis vaccination compares favourably with whole-cell vaccines 6 years after booster vaccination in the second year of life. Vaccine 25:1390–1397. doi: 10.1016/j.vaccine.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 252.Mallet E, Matisse N, Mathieu N, Langue J, Boisnard F, Soubeyrand B, Pentavac Study Group . 2004. Antibody persistence against diphtheria, tetanus, pertussis, poliomyelitis and Haemophilus influenzae type b (Hib) in 5–6-year-old children after primary vaccination and first booster with a pentavalent combined acellular pertussis vaccine: immunogenicity and tolerance of a tetravalent combined acellular pertussis vaccine given as a second booster. Vaccine 22:1415–1422. doi: 10.1016/j.vaccine.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 253.Fry NK, Litt DJ, Duncan J, Vaghji L, Warrener L, Samuel D, Andrews N, Harnden A, Harrison TG. 2013. Modelling anti-pertussis toxin IgG antibody decay following primary and preschool vaccination with an acellular pertussis vaccine in UK subjects using a modified oral fluid assay. J Med Microbiol 62:1281–1289. doi: 10.1099/jmm.0.062000-0. [DOI] [PubMed] [Google Scholar]
- 254.Edelman K, He Q, Mäkinen J, Sahlberg A, Haanperä M, Schuerman L, Wolter J, Mertsola J. 2007. Immunity to pertussis 5 years after booster immunization during adolescence. Clin Infect Dis 44:1271–1277. doi: 10.1086/514338. [DOI] [PubMed] [Google Scholar]
- 255.Pawloski LC, Kirkland KB, Baughman AL, Martin MD, Talbot EA, Messonnier NE, Tondella ML. 2012. Does tetanus-diphtheria-acellular pertussis vaccination interfere with serodiagnosis of pertussis infection? Clin Vaccine Immunol 19:875–880. doi: 10.1128/CVI.05686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Keitel WA, Muenz LR, Decker MD, Englund JA, Mink CM, Blumberg DA, Edwards KM. 1999. A randomized clinical trial of acellular pertussis vaccines in healthy adults: dose-response comparisons of 5 vaccines and implications for booster immunization. J Infect Dis 180:397–403. doi: 10.1086/314869. [DOI] [PubMed] [Google Scholar]
- 257.Riffelmann M, Littmann M, Hülsse C, von König CH. 2009. Antibody decay after immunisation of health-care workers with an acellular pertussis vaccine. Eur J Clin Microbiol Infect Dis 28:275–279. doi: 10.1007/s10096-008-0625-0. [DOI] [PubMed] [Google Scholar]
- 258.Litt DJ, Samuel D, Duncan J, Harnden A, George RC, Harrison TG. 2006. Detection of anti-pertussis toxin IgG in oral fluids for use in diagnosis and surveillance of Bordetella pertussis infection in children and young adults. J Med Microbiol 55:1223–1228. doi: 10.1099/jmm.0.46543-0. [DOI] [PubMed] [Google Scholar]
- 259.Campbell H, Amirthalingam G, Fry NK, Litt D, Harrison TG, Wagner K, Crowcroft NS, Miller E. 2014. Oral fluid testing for pertussis, England and Wales, June 2007–August 2009. Emerg Infect Dis 20:968–975. doi: 10.3201/eid2006.131069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kösters K, Riffelmann M, Dohrn B, von König CH. 2000. Comparison of five commercial enzyme-linked immunosorbent assays for detection of antibodies to Bordetella pertussis. Clin Diagn Lab Immunol 7:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Schellekens JFP, Boshuizen HC, Verbakel JMM, Elvers LH, Boshuis GL, van Houte AJ, Meijer BC, Peeters MF. 2001. Serodiagnosis of pertussis with commercial ELISAs, abstr D-1403, p 163 Int Conf Antimicrob Agents Chemother, Chicago, IL, 22 to 25 September 2001. [Google Scholar]
- 262.DeVincenzo JP, Guyton C, Rea H, Elmore E, Patel S, Wynn L, Harrison L, El Saleeby CM, Bagga B. 2013. Molecular detection and quantification of pertussis and correlation with clinical outcomes in children. Diagn Microbiol Infect Dis 76:10–15. doi: 10.1016/j.diagmicrobio.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 263.Erlandsson A, Bäckman A, Nygren M, Lundeberg J, Olcén P. 1998. Quantification of Bordetella pertussis in clinical samples by colorimetric detection of competitive PCR products. APMIS 106:1041–1048. doi: 10.1111/j.1699-0463.1998.tb00256.x. [DOI] [PubMed] [Google Scholar]
- 264.Nakamura Y, Kamachi K, Toyoizumi-Ajisaka H, Otsuka N, Saito R, Tsuruoka J, Katsuta T, Nakajima N, Okada K, Kato T, Arakawa Y. 2011. Marked difference between adults and children in Bordetella pertussis DNA load in nasopharyngeal swabs. Clin Microbiol Infect 17:365–370. doi: 10.1111/j.1469-0691.2010.03255.x. [DOI] [PubMed] [Google Scholar]
- 265.Gao F, Mahoney JC, Daly ER, Lamothe W, Tullo D, Bean C. 2014. Evaluation of a multitarget real-time PCR assay for detection of Bordetella species during a pertussis outbreak in New Hampshire in 2011. J Clin Microbiol 52:302–306. doi: 10.1128/JCM.01656-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Abu Raya B, Bamberger E, Gershtein R, Peterman M, Srugo I. 2012. The laboratory diagnosis of Bordetella pertussis infection: a comparison of semi-nested PCR and real-time PCR with culture. Eur J Clin Microbiol Infect Dis 31:619–622. doi: 10.1007/s10096-011-1327-6. [DOI] [PubMed] [Google Scholar]
- 267.Holberg-Petersen M, Jenum PA, Mannsåker T, Melby KK. 2011. Comparison of PCR with culture applied on nasopharyngeal and throat swab specimens for the detection of Bordetella pertussis. Scand J Infect Dis 43:221–224. doi: 10.3109/00365548.2010.538855. [DOI] [PubMed] [Google Scholar]
- 268.Leite D, Morozetti Blanco R, Vieira de Melo LC, Fiorio CE, Martins LM, Ibelli Vaz TM, Fernandes SA, Tavares Sacchi C. 2013. Implementation and assessment of the use of real-time PCR in routine diagnosis for Bordetella pertussis detection in Brazil. Arch Pediatr Infect Dis 1:196–202. [Google Scholar]
- 269.Palmer CM, McCall B, Jarvinen K, Nissen MD. 2007. Bordetella pertussis PCR positivity, following onset of illness in children under 5 years of age. Commun Dis Intell Q Rep 31:202–205. [DOI] [PubMed] [Google Scholar]
- 270.Cengiz AB, Yildirim I, Ceyhan M, Seçmeer G, Gür D, Kara A. 2009. Comparison of nasopharyngeal culture, polymerase chain reaction (PCR) and serological test for diagnosis of pertussis. Turk J Pediatr 51:309–316. [PubMed] [Google Scholar]
- 271.Chia JH, Su LH, Lin PY, Chiu CH, Kuo AJ, Sun CF, Wu TL. 2004. Comparison of multiplex polymerase chain reaction, culture, and serology for the diagnosis of Bordetella pertussis infection. Chang Gung Med J 27:408–415. [PubMed] [Google Scholar]
- 272.Haaheim H, Vorland L, Gutteberg TJ. 2001. Laboratory diagnosis of respiratory diseases: PCR versus serology. Nucleosides Nucleotides Nucleic Acids 20:1255–1258. doi: 10.1081/NCN-100002530. [DOI] [PubMed] [Google Scholar]
- 273.Yildirim I, Ceyhan M, Kalayci O, Cengiz AB, Secmeer G, Gur D, Pelton S. 2008. Frequency of pertussis in children with prolongued cough. Scand J Infect Dis 40:314–319. doi: 10.1080/00365540701689659. [DOI] [PubMed] [Google Scholar]
- 274.Guiso N, Berbers G, Fry NK, He Q, Riffelmann M, Wirsing von König CH, EU Pertstrain Group. 2011. What to do and what not to do in serological diagnosis of pertussis: recommendations from EU reference laboratories. Eur J Clin Microbiol Infect Dis 30:307–312. doi: 10.1007/s10096-010-1104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]