Abstract
Background
Moths rely heavily on pheromone communication for mate finding. The pheromone components of most moths are modified from the products of normal fatty acid metabolism by a set of tissue-specific enzymes. The turnip moth, Agrotis segetum uses a series of homologous fatty-alcohol acetate esters ((Z)-5-decenyl, (Z)-7-dodecenyl, and (Z)-9 tetradecenyl acetate) as its sex pheromone components. The ratio of the components differs between populations, making this species an interesting subject for studies of the enzymes involved in the biosynthetic pathway and their influence on sex pheromone variation.
Results
Illumina sequencing and comparative analysis of the transcriptomes of the pheromone gland and abdominal epidermal tissue, enabled us to identify genes coding for putative key enzymes involved in the pheromone biosynthetic pathway, such as fatty acid synthase, β-oxidation enzymes, fatty-acyl desaturases (FAD), fatty-acyl reductases (FAR), and acetyltransferases. We functionally assayed the previously identified ∆11-desaturase [GenBank: ES583599, JX679209] and FAR [GenBank: JX679210] and candidate acetyltransferases (34 genes) by heterologous expression in yeast. The functional assay confirmed that the ∆11-desaturase interacts with palmitate and produces (Z)-11-hexadecenoate, which is the common unsaturated precursor of three homologous pheromone component acetates produced by subsequent chain-shortening, reduction and acetylation. Much lower, but still visible, activity on 14C and 12C saturated acids may account for minor pheromone compounds previously observed in the pheromone gland. The FAR characterized can operate on various unsaturated fatty acids that are the immediate acyl precursors of the different A. segetum pheromone components. None of the putative acetyltransferases that we expressed heterologously did acetylate any of the fatty alcohols tested as substrates.
Conclusions
The massive sequencing technology generates enormous amounts of candidate genes potentially involved in pheromone biosynthesis but testing their function by heterologous expression or gene silencing is a bottleneck. We confirmed the function of a previously identified desaturase gene and a fatty-acyl reductase gene by heterologous expression, but the acetyltransferase postulated to be involved in pheromone biosynthesis remains illusive, in spite of 34 candidates being assayed. We also generated lists of gene candidates that may be useful for characterizing the acetyl-CoA carboxylase, fatty acid synthetase and β-oxidation enzymes.
Keywords: Agrotis segetum, Pheromone biosynthesis, Transcriptome, Desaturase, Fatty-acyl reductase, Acetyltransferase, Functional assay, Yeast expression
Background
Moths rely heavily on sex pheromones for mate finding [1]. Usually the females produce and emit the pheromone from a specialized structure, the sex pheromone gland located at the intersegmental membrane between the 8th and 9th abdominal segment, associated with the ovipositor at the end of the adult female abdomen [2]. Over the last five decades, sex pheromones have been identified from more than 700 species [3, 4]. The pheromone compounds are mostly fatty acid derivatives, with carbon chain length C10-C18, with 0–4 double bonds [3], and an oxygenated functional group (alcohol, aldehyde, acetate ester) [3–5]. A substantial portion is made up by acetate esters [3]. Most moths use a combination of two or more compounds in a specific ratio, constituting a more or less species-specific blend. The pheromone biosynthesis pathways have been studied extensively and are well documented in many moth species [6, 7]. Characterization of the enzymes involved in the process of pheromone biosynthesis not only helps to understand the evolution of sexual communication and speciation, but could ultimately also aid in pest control by allowing the design of drugs that block the biosynthetic machinery or by allowing the synthetic biologist to produce species-specific pheromones for mass trapping or mating disruption in biological systems like cell factories or genetically modified plants [8–10].
During the last two decades, desaturases introducing double bonds into the acyl chain in ∆6 [11], ∆9 [12, 13], ∆10 [14], ∆11 [12, 13, 15–17] and ∆14 [18] position have been cloned from many moth species, and their functions have been characterized in various heterologous expression systems. Also an omega-desaturase that introduces a double bond in the methyl terminal carbon has been cloned and characterized [19]. In some cases, two double bonds can be produced by one desaturase [17] or by the consecutive activities of two desaturases [11]. These studies highlight desaturases as key players for the diversity of pheromone structures in moth species and their role in reproductive isolation and speciation. A variety of desaturases in combination with limited chain-shortening or chain-elongation account for the diversity of double bond isomerism observed among moth sex pheromone compounds.
After the double bonds are in place and the acyl chain length is adjusted, the carbonyl carbon is modified to form a functional group. Firstly it requires a step that converts the fatty-acyl precursors into fatty alcohols. Great progress has been made since the first fatty-acyl reductase gene was identified in Bombyx mori [20]. Several Ostrinia spp. FARs have been characterized [21, 22]. The Ostrinia FARs are very essential to determine the final pheromone compositions [22], minimal changes in sequence can cause the pheromone component ratio to shift dramatically [23]. On the other hand, there are also FARs that are very versatile in terms of substrate specificity [24, 25], involved in the biosynthesis of multicomponent pheromones.
Fatty alcohols serve as the actual pheromone components in a number of moth species [3], but mostly, fatty alcohols will be either oxidized to aldehydes [26–28] or esterified to form acetate esters [29–31]. Acetyltransferases are important enzymes since the acetate esters are very commonly occurring pheromone components among moth species. They have not been cloned from insects yet, but have been investigated in some cases by in vivo labeling studies [32–34]. Unlike FARs, these functional group modification enzymes have not been studied extensively.
The chain-shortening pathway has not been characterized at the enzymatic level in insects, but it was noted in a couple of cases that mutations in the β-oxidation pathway did affect the final pheromone compositions. The major pheromone component of cabbage looper moth Trichoplusia ni is Z7-12:OAc, whereas a mutant strain produced a greatly increased amount of Z9-14:OAc [35, 36].
The turnip moth, A. segetum uses a series of short chain acetate esters as sex pheromone components, including the homologues (Z)-5-decenyl, (Z)-7-dodecenyl, and (Z)-9 tetradecenyl acetate (Z5-10:OAc, Z7-12:OAc, and Z9-14:OAc) [37–39]. The biosynthesis of this type of pheromone involves desaturation of palmitic acid (16C), a product of the ubiquitous fatty acid synthase machinery. The unsaturated fatty acid undergoes chain shortening, reduction, and acetylation [38]. Populations of A. segetum from different geographic areas differ in the ratio of their pheromone components [40, 41]. For instance the Swedish population has a ratio of Z9-14:OAc/Z7-12:OAc/Z5-10:OAc = 29/59/12 whereas the Zimbabwean population has a ratio of 2/20/78 [37]. This shift in ratios could be due to differences in the chain-shortening, in the FAR activity or less likely in the activity of the acetyltransferase.
An EST-library was previously constructed from the A. segetum pheromone gland, revealing candidate genes involved in pheromone production [42] and a ∆11 desaturase and a FAR involved in pheromone production were already characterized [9]. The EST library, however, contains only 2, 285 objects. We now report a more extensive database of candidate genes potentially involved in the A. segetum sex pheromone biosynthetic pathway, generated by next generation sequencing technology (NGS). We constructed transcriptome libraries from two tissues of A. segetum: pheromone gland (As_PG) and abdomen (As_AB). Furthermore, to test the function of several of the candidate genes we assayed them in a yeast heterologous expression system.
Results and discussion
Illumina sequencing, unigene assembly, and analysis of transcripts
In total 53 million raw clean reads were obtained from each (As_PG and As_AB) library, with a total of 4.8G clean nucleotides in each (Table 1). The clean reads from the two libraries were assembled into 62,165 consensus contigs (Table 2), including 22,633 distinct clusters (referred to as CL) and 39,532 distinct singletons (referred to as Unigene). These consensus contigs have a mean length of 733 nt, and N50 [43] =1,150, total length 45 Mb. Size distributions of the unigenes can be seen in Fig. 1 and their differential expression in the two tissues is displayed in Fig. 2. This transcriptome shotgun assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GBCW00000000. The version described in this paper is the first version, GBCW01000000.
Table 1.
Output statistics of sequencing
| Sample | Total raw reads | Total clean reads | Total clean nucleotides | Q20 % | N % | GC % |
|---|---|---|---|---|---|---|
| As_PG | 63,086,832 | 53,235,252 | 4,791,172,680 | 96.61 % | 0.01 % | 47.54 % |
| As_AB | 62,193,108 | 54,156,050 | 4,874,044,500 | 96.62 % | 0.01 % | 47.44 % |
Q20 percentage is proportion of nucleotides with quality value larger than 20 in reads (sequencing error rate = 1 %); N percentage is proportion of unknown nucleotides in clean reads. GC percentage is proportion of guanidine and cytosine nucleotides among total nucleotides
Table 2.
Statistics of assembly quality
| Sample | Total length (nt) | Mean length (nt) | N50 | Total consensus sequences | Distinct contigs | Distinct singletons |
|---|---|---|---|---|---|---|
| As_PG | 37,824,354 | 574 | 907 | 65,840 | 17,534 | 48,306 |
| As_AB | 36,392,640 | 618 | 1007 | 58,847 | 13,055 | 45,792 |
| All | 45,545,001 | 733 | 1150 | 62,165 | 22,633 | 39,532 |
N50 value defined as the length for which the collection of all sequences of that length or longer contains at least half of the sum of the lengths of all sequences
Fig. 1.
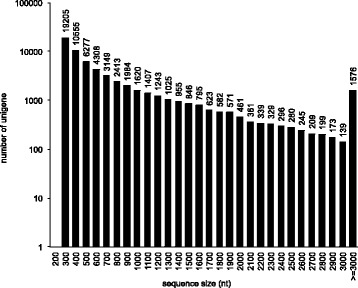
Length distribution of unigenes. The consensus sequence lengths ranging from 200 bp to more than 3,000 bp, and above each column is indicated the number of genes of each length range. The most abundant unigenes are 300 bp (19205) and the least abundant unigenes were 3000 bp (139); sequences over 3000 bp were grouped together. The number of sequences decreased as the length increased
Fig. 2.
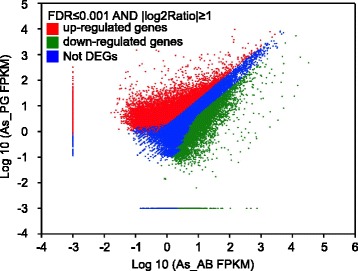
Differentially expressed unigenes displayed by FPKM in As_PG versus As_AB, in log10 scale. X-axis (As_AB) and Y-axis (As_PG) shows the logarithm value of normalized expression of each gene in FPKM (Fragments per kb per Million fragments). There are 21,965 unigenes that are up regulated (red dots), measured by As_PG(FPKM)/As_AB(FPKM) > 2. On the contrary, 14,292 unigenes are down regulated (green dots), since their As_PG(FPKM)/As_AB(FPKM) < 0.5. Most of the unigenes, 25,895, were equally expressed (blue dots) in both tissue (0.5 < As_PG(FPKM)/As_AB(FPKM) < 2)
After alignment by blastx to protein databases NCBI Nr, Swiss-Prot, KEGG and COG (e-value < 0.00001), and alignment by blastn to nucleotide database NCBI Nt, annotations were retrieved from the highest sequence similarity with the given unigenes along with their protein functional annotations. About half of the unigenes have a hit in one or more of the databases (Table 3). 14,108 unigenes hit the first record in the Nr database with Bombyx mori (Table 4). A. ipsilon got 293 hits, but this does not mean the A. segetum is closer to B. mori than A. ipsilon, it is just because the B. mori got more records deposited in GenBank than A. ipsilon. In terms of clusters of orthologous groups (COG), 19,911 unigenes were classed in one or more of the 26 COG functional categories (Fig. 3).
Table 3.
Summary of annotation results
| Nr | Nt | SwissProt | KEGG | COG | GO |
|---|---|---|---|---|---|
| 27,344 | 20,403 | 19,143 | 17,351 | 8,502 | 12,049 |
Unigenes were annotated with the databases of Nr, Nt, Swiss-Prot, KEGG, COG and GO. Then counted the number of unigenes annotated with each database. The result is summarized as the follow table
Table 4.
Nr annotations of assembled A. segetum consensus sequences
| Species | Gene numbers | Percentage |
|---|---|---|
| Bombyx mori | 14,108 | 50.56 % |
| Danaus plexippus | 7,807 | 27.98 % |
| Tribolium castaneum | 642 | 2.3 % |
| Papilio xuthus | 569 | 2.04 % |
| Helicoverpa armigera | 420 | 1.51 % |
| Agrotis ipsilon | 293 | 1.05 % |
| Other | 4063 | 14.56 % |
The first column shows the species with the highest number of similar genes in descending order, the second column indicates the number of these annotated genes, and the last column shows the percentage of genes with respect to the total annotated genes sequences
Fig. 3.
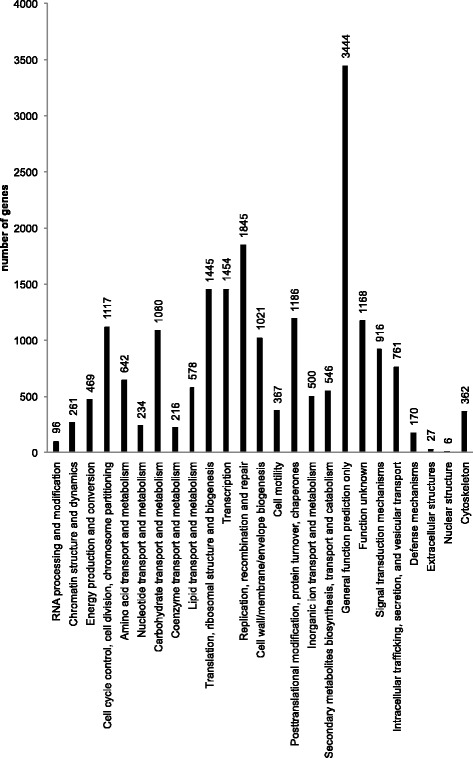
COG classification of unigenes. Histogram of COG classifications of assembled consensus sequences. Results are presented for the 25 main COG categories. The number above the column indicates number of unigenes in each category
By running the Blast2GO program [44] using Nr annotation, 11,246 (18 %) unigenes were assigned to one or more GO categories. The GO terms cellular process, binding, catalytic activity, and metabolic process were the most abundantly represented categories (over 5,000 unigenes, details see Fig. 4). These numbers and percentages are similar to the results that Gu and coauthors [45] presented from A. ipsilon.
Fig. 4.
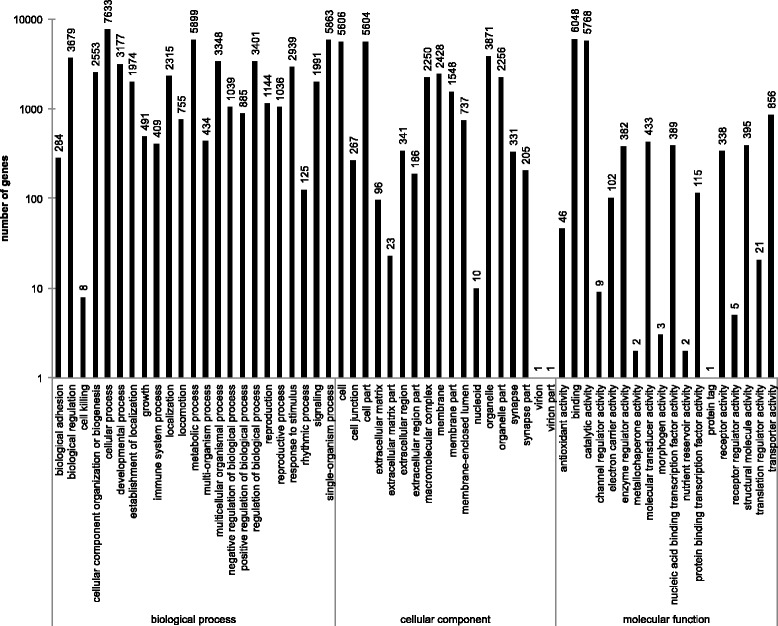
GO classification of unigenes. Histogram of GO classifications of consensus sequences. Results are summarized for the three main GO categories: biological process, cellular component and molecular function. The number on the bars represents the total number of unigenes in each category
The biosynthetic pathway (Fig. 5) leading to the pheromone components of A. segetum is similar to what has been reported for many other moth species [37, 45]. The key players among enzymes involved or postulated to be involved are desaturases, β-oxidation enzymes, fatty-acyl reductases, and acetyltransferases. Pheromone biosynthesis is reported to be under control by a pheromone biosynthesis activating neuropeptide (PBAN) [46]. In the following we present candidate genes related to each step in the biosynthetic pathway, their expression levels and their functional assay in yeast heterologous expression systems.
Fig. 5.
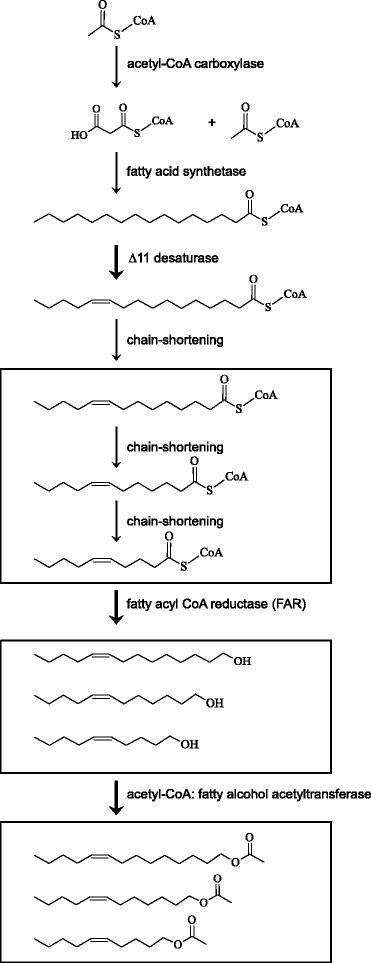
Biosynthetic pathway leading to the sex pheromone of Agrotis segetum, modified from [37]. It starts with carboxylation of acetyl-CoA to malonyl-CoA, and then they are entered to a cycle of fatty acid synthesis and end up with common fatty acids stearate and palmitate. The ∆11 desaturase inserts a double bond in the acyl chain and then the unsaturated fatty acid is subjected to three rounds of chain-shortening by β-oxidation, forming three acyl-chains different by two carbon atoms. These acyl-chains are then reduced by fatty-acyl CoA reductase (FAR) to make fatty alcohols, which are then acetylated to acetate esters, the final A. segetum pheromones. Thick arrows represent steps are functionally assayed in heterologous systems
Pheromone biosynthesis activating neuropeptide (PBAN) receptor
PBAN is released from the subesophageal ganglion (located near the brain) and is transported through hemolymph (or via the ventral nerve cord) to the pheromone gland. Upon binding to the PBAN receptor present on the pheromone gland cell membrane [47], it will induce the opening of calcium channels causing influx of extracellular calcium [7]. Then the calcium binds to calmodulin, that stimulates phosphatase (and/or kinase), which subsequently activates the FAR in the case of Bombyx mori [48] or in other cases the acetyl-CoA carboxylase [49] or the acetyltransferase [50] maybe regulated. In As_PG, we found one gene, Ase_17579 [GenBank: KJ622075], which is 97 % identical to Helicoverpa zea PBAN receptor. Its expression level is similar in As_PG and As_AB, 15.6 FPKM and 16.9 FPKM, respectively.
Acetyl-CoA carboxylase
The rate-limiting step in fatty acid biosynthesis [51] is the ATP-dependent carboxylation of acetyl-CoA to malonyl-CoA catalyzed by acetyl-CoA carboxylase (ACCase) [52], the first step in saturated long chain fatty acid biosynthesis. ACCase is a large protein with multiple catalytic activities, working coordinately and providing malonyl-CoA substrate for the biosynthesis of fatty acids [50]. In A. segetum, we found one contig Ase_7442 (KJ622074), encoding the full-length of this protein. It shares 69 % amino acid identity with the ACCase of Tribolium castaneum and 88 % aa identity with B. mori ACCase. The expression level of Ase_7442 is not significantly different in As_AB (37 FPKM) and in As_PG (26 FPKM).
Fatty acid synthase
Fatty acid synthase (FAS) is a multifunctional protein [52] that produces saturated fatty acids using malonyl-CoA and acetyl-CoA as substrate and that requires NADPH as reducing agent, in a cyclic process in which an acetyl primer undergoes a series of decarboxylative condensations with several malonyl moieties [53]. The resulting products are palmitic and stearic acid in insects, as proven by labeling studies [49, 53, 54]. We found six unigenes (KJ622068-KJ622073) that are homologous to the Aip_FAS_JX989151 [45]. In total, these unigenes are seven times more expressed (significant) in As_PG than in As_AB (FPKM 67.1 / FPKM 9).
Desaturases
Fatty acid desaturases catalyze the introduction of double bonds into acyl chains with strict regioselectivity and stereoselectivity, and can be divided into four categories [55]: 1) first desaturases, inserting a double bond into the saturated acyl chain; 2) front-end desaturases, introducing a double bond between an existing double bond and the carboxylic end; 3) omega desaturases, inserting a double bond between an existing double bond and the methyl end; 4) sphingolipid desaturases, introducing double bonds into sphingolipids which are important components of eukaryotic plasma membranes. We found 10 desaturase candidates from the two tissues, and they belong to the first desaturase (7/10), the front-end desaturase (2/10), and the sphingolipid desaturase (1/10) subfamilies. Among the first desaturase subfamily, several groups have been recognized based on phylogenetic and functional analysis and a four-letter “signature motif” has been suggested for each of them as shown in Fig. 6. These signature motifs strongly associate with the location of double bond that the desaturase is inserting in the fatty-acyl chain, with less emphasis on the chain length selectivity: ∆9_KPSE (C16 > C18) desaturases and ∆9_NPVE (C18 > C16) desaturases that are mostly involved in fatty acid metabolism [56], and ∆11,∆10 and bifunctional desaturases with the “xxxQ” motif (with a few exceptions having “xxxE” motif) exclusively involved in pheromone biosynthesis [13, 57] (Fig. 6). The Ase_1623 and Ase_4567 have the signature motif of KPSE and NPVE, respectively, that strongly suggest their roles in ordinary metabolic pathways as ∆9 desaturases. The Ase_21308 and Ase_5534 both have “xxxQ” motif, suggesting that they represent ∆11 desaturases. The Ase_5534 is the obvious candidate for pheromone biosynthesis since its expression level in As_PG is 1–2 magnitudes higher than any other candidate, and it has a very low expression level in As_AB. The functional assay confirms that it possess the ability of creating a ∆11-double bond mainly on palmitate (Fig. 7), corroborating that Ase_5534 [GenBank:KJ622051] is the desaturase responsible for pheromone production in A. segetum. This result is consistent with the recent study in which this gene was expressed in different yeast strains [9] and this desaturase was as a matter of fact found already in the EST analysis reported by Strandh et al. [42]. This desaturase introduces a double bond with Z configuration on 16:Acyl. Besides Z11-16:Acyl, some other minor products were detected: ∆11-12:Acyl, Z11-14:Acyl, E11-14:Acyl, and Z11-15:Acyl (Fig. 7). The Ase_21308 displayed no activity in our yeast expression system. Gu et al. [45] found a desaturase that is very close to our Ase_5534 (as shown in Fig. 6) and which should be the one involved in pheromone biosynthesis in A. ipsilon. Compared to [42, 45, 58], our dataset includes a larger set of desaturases isolated from the moth pheromone gland and surrounding tissues.
Fig. 6.
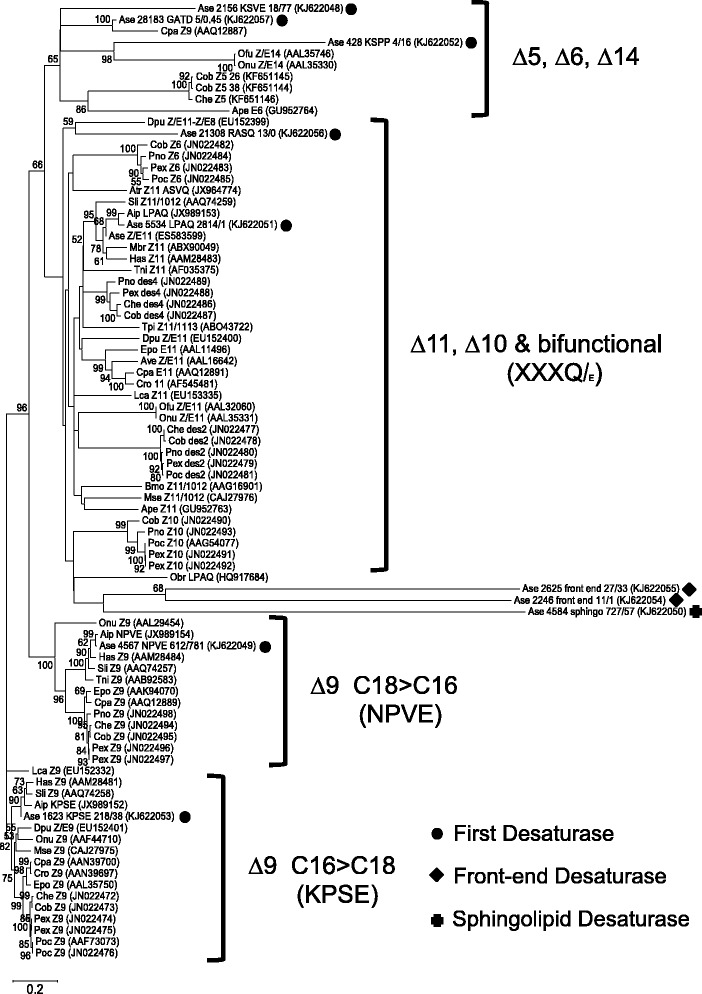
The neighbor-joining tree of selected lepidopteran desaturase genes, constructed using amino-acid sequences. Desaturases described in this study are indicated by different shapes (with signature motif displayed for the First Desaturase), followed by unigene expression levels in the gland and abdomen library, respectively (As_PG_FPKM /As_AB_FPKM). Desaturases in previous studies are named as follows: biochemical activities (if known) are indicated in connection to the species name, followed by accession number in parenthesis. Most of the desaturases used in here are First Desaturases that introduce double bond into saturated fatty acids. Among the First Desaturase, four distinctive groups formed that separate their biological functions. The ∆9 desaturases are usually used for normal fatty acid metabolism, with the “KPSE” group having preference on C16 and “NPVE” group mainly modifying C18. The ∆11,∆10 and bifunctional desaturases with the “xxxQ” motif (with a few exceptions having “xxxE” motif) exclusively involved in pheromone biosynthesis. The ∆5,∆6,∆14 group contain a mixture of different signature motifs derived from the ∆9 and ∆11 groups, and their biological function are also diverged. The tree was rooted on the ∆9-desaturase-KPSE (C16 > C18) functional class
Fig. 7.
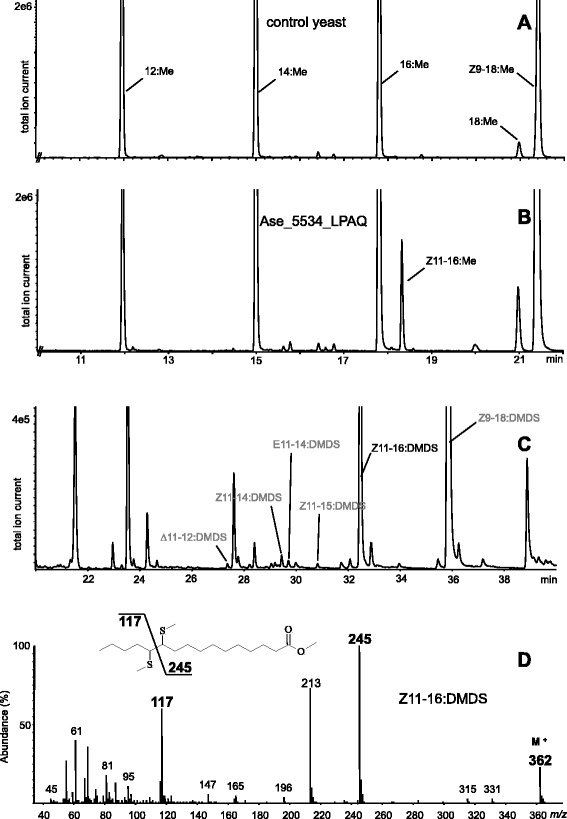
Functional characterization of the highest expressed desaturase gene, Ase_5534, in yeast expression system. GC-MS analyses of methanolyzed lipid extracts from yeast transformed with empty plasmid pYEX-CHT (a) and pYEX-CHT-Ase_5534 (b). Double bond position of the unsaturated palmitate was confirmed by DMDS derivatization (c, d)
β-oxidation enzymes
After the palmitate has undergone ∆11 desaturation in the A. segetum pheromone gland, it is subject to limited chain shortening by β-oxidation, resulting in three homologous fatty-acyl pheromone precursors with 14C, 12C, and 10C chain length (Fig. 5). Chain-shortening by β-oxidation is the action of a series of enzymes, working sequentially and forming a reaction spiral.
In the first reaction, acyl-CoA is converted to E2-enoyl-CoA, by acyl-CoA oxidases (in peroxisomes) and acyl-CoA dehydrogenases (in mitochondria). Four acyl-CoA dehydrogenases with different chain length specificities cooperate to assure that the complete degradation of all fatty acids with different chain length. The names of the four dehydrogenases, short-chain, medium-chain, long-chain, and very long-chain acyl-CoA dehydrogenases, reflect their chain-length specificities. Short-chain acyl-CoA dehydrogenase only acts on short-chain substrates like butyryl-CoA and hexanoyl-CoA. Medium-chain acyl-CoA dehydrogenase is most active with substrates from hexanoyl-CoA to dodecanoyl-CoA, whereas long-chain acyl-CoA dehydrogenase preferentially acts on octanoyl-CoA and longer-chain substrates [59]. Very-long-chain acyl-CoA dehydrogenase extends the activity spectrum to longer-chain substrates, including those having acyl chains of 22 and 24 carbon atoms [60]. We found a full spectrum of dehydrogenases and oxidases in A. segetum (Table 5). It is noteworthy that unigene6715 was expressed 72 times higher (386FPKM) in As_PG than in As_AB (5FPKM). In addition, we found two unigenes of isovaleryl-CoA dehydrogenase, which is specific for metabolism of branched-chain fatty acids [61].
Table 5.
The gene candidates found in As_PG that may be involved in p-oxidation processes
| genelD | GenBank accession | As PG FPKM | As AB FPKM | Swissprot-annotation | ko_definition |
|---|---|---|---|---|---|
| acyl-CoA dehydrogenase | |||||
| Unigene5337_All | KJ622076 | 2.49 | 0.93 | Short-chain specific acyl-CoA dehydrogenase | butyryl-CoA dehydrogenase[EC:1.3.8.1] |
| Unigenel8315_All | KJ622077 | 4.35 | 3.26 | Short-chain specific acyl-CoA dehydrogenase | butyryl-CoA dehydrogenase[EC:1.3.8.1] |
| CL3917.Contigl_All | KJ622078 | 8.02 | 14.14 | Short-chain specific acyl-CoA dehydrogenase | butyryl-CoA dehydrogenase[EC:1.3.8.1] |
| Unigenel5571_All | KJ622079 | 24.39 | 67.70 | Short/branched chain specific acyl-CoA dehydrogenase | short/branched chain acyl-CoA dehydrogenase [EC:1.3.99.12] |
| CL 1315 .Contig2_All | KJ622080 | 29.09 | 40.69 | Probable medium-chain specific acyl-CoA dehydrogenase | acyl-CoAdehydrogenase [EC: 1.3.99.3] |
| Unigene4168_All | KJ622081 | 4.04 | 3.43 | Acyl-CoA dehydrogenase family member 9 | Acyl-CoA dehydrogenase family member 9 [EC:1.3.99.-] |
| CL252.Contigl_All | KJ622082 | 10.03 | 7.93 | Very long-chain specific acyl-CoA dehydrogenase | Very long chain acyl-CoA dehydrogenase [EC:1.3.99.-] |
| CL622.Contig4_All | KJ579211 | 77.52 | 107.57 | Trifunctional enzyme subunit beta | acetyl-CoAacyltransferase [EC:2.3.1.16] |
| Unigene7769_All | KJ622083 | 3.29 | 2.51 | Isovaleryl-CoA dehydrogenase | Isovaleryl-CoA dehydrogenase [EC:1.3.8.4] |
| Unigene9593_All | KJ622084 | 8.77 | 9.31 | Isovaleryl-CoA dehydrogenase | Isovaleryl-CoA dehydrogenase [EC:1.3.8.4] |
| acyl-CoA oxidase | |||||
| CL3960.Contigl_All | KJ622085 | 16.52 | 17.33 | Probable peroxisomal acyl-CoA oxidase | acyl-CoA oxidase [EC:1.3.3.6] |
| CL4327.Contigl_All | KJ622086 | 3.85 | 4.30 | Peroxisomal acyl-CoA oxidase | acyl-CoA oxidase [EC:1.3.3.6] |
| CL6898.Contigl_All | KJ622087 | 5.75 | 11.79 | Probable peroxisomal acyl-CoA oxidase 1 | acyl-CoA oxidase [EC:1.3.3.6] |
| CL7062.Contigl_All | KJ622088 | 14.54 | 18.14 | Peroxisomal acyl-CoA oxidase 3 | acyl-CoA oxidase [EC:1.3.3.6] |
| Unigenel 9521_All | KJ622089 | 29.25 | 23.07 | Probable peroxisomal acyl-CoA oxidase 1 | acyl-CoA oxidase [EC:1.3.3.6] |
| Unigene26088_All | KJ622090 | 2.29 | 0.68 | Peroxisomal acyl-CoA oxidase 1 | acyl-CoA oxidase [EC:1.3.3.6] |
| Unigene33961_All | KJ622091 | 3.46 | 2.66 | Probable peroxisomal acyl-CoA oxidase 1 | acyl-CoA oxidase [EC:1.3.3.6] |
| Unigene4386_All | KJ622092 | 1.74 | 1.78 | Probable peroxisomal acyl-CoA oxidase 1 | acyl-CoA oxidase [EC:1.3.3.6] |
| Unigene5059_All | KJ622093 | 1.92 | 1.13 | Peroxisomal acyl-CoA oxidase 1 | acyl-CoA oxidase [EC:1.3.3.6] |
| Unigene6715_A11 | KJ622094 | 386.08 | 4.64 | Probable peroxisomal acyl-CoA oxidase 1 | acyl-CoA oxidase [EC:1.3.3.6] |
| enoyl-CoA hydratase | |||||
| CL1656.Contigl_All | KJ622095 | 16.69 | 23.69 | Methylglutaconyl-CoA hydratase | Methylglutaconyl-CoA hydratase [EC:4.2.1.18] |
| CL2503.Contigl_All | KJ622096 | 8.85 | 1.87 | Probable enoyl-CoA hydratase | enoyl-CoA hydratase [EC:4.2.1.17] |
| CL2595.Contigl_All | KJ622097 | 122.91 | 142.29 | Trifunctional enzyme subunit alpha | enoyl-CoA hydratase [EC:4.2.1.17] |
| Unigenel4514_All | KJ622098 | 4.15 | 4.35 | 3-hydroxyisobutyryl-CoA hydrolase | 3-hydroxyisobutyryl-CoA hydrolase [EC:3.1.2.4] |
| Unigenel7624_All | KJ622099 | 73.15 | 87.01 | Enoyl-CoA hydratase | enoyl-CoA hydratase [EC:4.2.1.17] |
| Unigene6308_All | KJ622100 | 5.00 | 5.89 | Enoyl-CoA hydratase domain-containing protein 3 | enoyl-CoA hydratase [EC:4.2.1.17] |
| L-3-hydroxyacyl-CoA dehydrogenase | |||||
| Unigenel2153_All | KJ622101 | 11.64 | 1.60 | 3-hydroxyacyl-CoA dehydrogenase | 3-hydroxyacyl-CoA dehydrogenase [EC1.1.1.35] |
| Unigenel5615_All | KJ622102 | 8.46 | 8.38 | 3-hydroxyacyl-CoA dehydrogenase | 3-hydroxyacyl-CoA dehydrogenase [EC1.1.1.35] |
| Unigenel7583_All | KJ622103 | 174.80 | 83.79 | Peroxisomal multifunctional enzyme | 3-hydroxyacyl-CoA dehydrogenase [EC1.1.1.35] |
| Unigenel9554_All | KJ622104 | 103.82 | 78.74 | Flydroxyacyl-CoA dehydrogenase | 3-hydroxyacyl-CoA dehydrogenase [EC1.1.1.35] |
| Unigene8215_All | KJ622105 | 8.18 | 4.22 | Probable 3-hydroxyacyl-CoA dehydrogenase | 3-hydroxyacyl-CoA dehydrogenase [EC1.1.1.35] |
| 3-ketoacyl-CoA thiolase | |||||
| CL2371.Contigl_All | KJ579207 | 10.01 | 5.43 | 3-ketoacyl-CoA thiolase | acetyl-CoA acyltransferase 2 [EC:2.3.1.16] |
| Unigene21478_A11 | KJ622106 | 4.47 | 0.05 | 3-ketoacyl-CoA thiolase | acetyl-CoA acyltransferase 2 [EC:2.3.1.16] |
| Unigene26739_All | KJ622107 | 3.32 | 0.00 | 3-ketoacyl-CoA thiolase | acetyl-CoA acyltransferase 2 [EC:2.3.1.16] |
| Unigene28250_All | KJ622108 | 122.89 | 42.24 | Trifunctional enzyme subunit beta | acetyl-CoA acyltransferase [EC:2.3.1.16] |
| Unigene30699_All | KJ622109 | 1.19 | 0.76 | 3-ketoacyl-CoA thiolase | acetyl-CoA acyltransferase 2 [EC:2.3.1.16] |
| Unigene5444_All | KJ622110 | 2.35 | 15.28 | 3-ketoacyl-CoA thiolase | acetyl-CoA acyltransferase 2 [EC:2.3.1.16] |
| A3,A2-trans-enoyl-CoA isomerase | |||||
| CL3311.Contigl_All | KJ622111 | 24.40 | 35.52 | Enoyl-CoA delta isomerase 1 | 3,2-trans-enoyl-CoAisomerase, mitochondrial [EC:5.3.3.8] |
| Unigene2565_All | KJ622112 | 3.56 | 1.88 | Enoyl-CoA delta isomerase 2 | peroxisomal 3,2-trans-enoyl-CoA isomerase [EC:5.3.3.8] |
| Δ3,5 Δ2,4-dienoyl-CoA isomerase | |||||
| Unigenel6713_All | KJ622113 | 11.43 | 18.26 | Delta(3,5)-Delta(2,4)-dienoy 1-Co A is omerase | delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase [EC:5.3.3.-] |
The second step of β-oxidation E2-enoyl-CoA is reversibly hydrated by enoyl-CoA hydratase to L-3-hydroxyacyl-CoA. Two categories of enoyl-CoA hydratases have been identified in mitochondria [62]. One is specialized for crotonyl-CoA (4C). The second one is long-chain enoyl-CoA hydratase, effectively hydrates medium-chain and long-chain substrates. Long-chain enoyl-CoA hydratase is a component enzyme of the trifunctional β-oxidation complex, which additionally exhibits long-chain activities of L-3-hydroxyacyl-CoA dehydrogenase and 3-ketoacyl-CoA thiolase [63], and resides in the inner mitochondrial membrane. We found six enoyl-CoA hydratases from A. segetum (Table 5), with similar expression level in both As_PG and As_AB.
The third reaction is the reversible dehydrogenation of L-3-hydroxyacyl-CoA to 3-ketoacyl-CoA catalyzed by L-3-hydroxyacyl-CoA dehydrogenase. Four categories of L-3-hydroxyacyl-CoA dehydrogenases have been identified in mitochondria. Long chain L-3-hydroxyacyl-CoA dehydrogenase is a component enzyme of the trifunctional β-oxidation [63], which is most active with long-chain substrates. Medium-chain and short-chain L-3-hydroxyacyl-CoA dehydrogenase, are both soluble matrix enzymes, processing medium- and short-chain substrates. These three enzymes complement each other and thus assure high rates of dehydrogenation over the whole spectrum of β-oxidation intermediates [63]. In A. segetum this group of enzymes is represented by five unigenes (Table 5), with similar expression level in both tissues.
The final step in which 3-ketoacyl-CoA is cleaved by thiolase between its α and β carbon atoms, makes the substrate two carbons shorter. Three classes of thiolase exist in motochondria: acetoacetyl-CoA thiolase or acetyl-CoA acetyltransferase (specific for acetoacetyl-CoA), 3-ketoacyl-CoA thiolase or acetyl-CoA acyltransferase (act on C4-C16), and long-chain 3-ketoacyl-CoA thiolase that is a component enzyme of the membrane-bond trifunctional β-oxidation complex, whereas the first two thiolases are soluble matrix enzymes [62]. We found six unigenes of 3-ketoacyl-CoA thiolase from A. segetum (Table 5), and all of them expressed at similar level in As_PG and As_AB.
The degradation of unsaturated fatty acids requires auxiliary enzymes like ∆3,∆2-enoyl-CoA isomerase and 2,4-dienoyl-CoA reductase to modify the structure of double bonds during the β-oxidation process to ensure a continuous flow of intermediates through the β-oxidation spiral [64]. We found two unigenes of the ∆3,∆2-enoyl-CoA isomerase, one mitochondrial type and one peroxisomal. In addition, we found a ∆3,5∆2,4-dienoyl-CoA isomerase that is specialized for processing odd-numbered double bonds [61].
Insects in general have the ability to shorten long-chain fatty acids to a specific shorter chain length [65]. Jurenka [5, 66] suggests that this kind of limited chain shortening takes place in the peroxisomes. Unlike the β-oxidation in mitochondria in which substrates are thoroughly degraded into two-carbon units, the β-oxidation in peroxisomes ceases at the formation of medium-chain fatty-acyl-CoAs, because acyl-CoA oxidase is inactive toward substrates having acyl moieties of eight or fewer carbon atoms [66]. Moth pheromone components are commonly 12C and 14C, suggesting that there is still something specific about the chain-shortening being involved in moth pheromone biosynthesis.
In the present study we found representatives of all the key players and auxiliary enzymes of β-oxidation, and some of them are very high in expression level and differentially expressed among the As_PG and As_AB (Table 5), forming promising candidates to tackle the role of β-oxidation on pheromone biosynthesis either by heterologous expression or by RNAi.
Fatty-acyl reductases
Chain-shortened fatty-acyl precursors are reduced to the corresponding alcohols by Fatty-Acyl Reductases (FARs) [20]. FARs have been cloned and characterized from several moth species [21–23] since the first one was found in B. mori [20]. We found ten full-length unigenes in both the As_PG and As_AB (Fig. 8) and two of them cluster within the pgFAR clade [25]. We expressed the two ORFs that belong to the pheromone-producing clade in our yeast expression system. The results showed that it is the one named Ase_1929 that is responsible for reducing all the three fatty-acyl precursors (Z5-10:CoA, Z7-12:CoA, Z9-14:CoA) into their corresponding fatty alcohols (Fig. 9), and it is the most abundant transcript among all these ten unigenes (Fig. 8). This result is consistent with previous findings that a single FAR takes a wide range of the fatty-acyl substrates and convert them into fatty alcohols [24, 25, 67]. Our results complement the findings of Hagström et al. [9], who presented the Ase_1929 with longer chain acyl substrates (16C) and found it to be very active on them. But what appears in Fig. 9 as if the Ase_1929 when expressed in yeast is more active towards longer chain length substrates than towards shorter ones, does not necessarily represent its actual activity in the pheromone gland. This difference could partly be due to the volatility of the substrates and products, since shorter chain methyl esters and alcohols are more volatile and thus may escape from the yeast expression system during incubation. The other unigene Ase_20982 which also clustered with the pgFAR clade, did not show any function in our heterologous expression system. The FAR [GenBank: JX989146, protein ID: AGR49323] found by Gu et al. [45] clustered very close to our Ase_1929, suggesting that it is likely involved in pheromone biosynthesis in A. ipsilon.
Fig. 8.
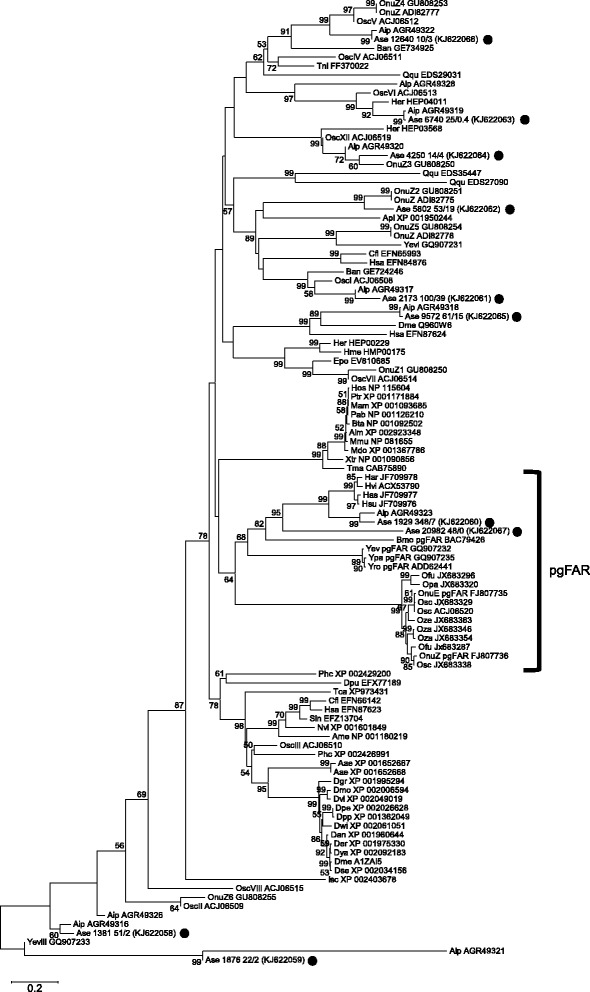
Phylogenetic relationship of FARs from arthropods, mammals and lepidoptera constructed using amino acid sequences. The pgFAR clade is marked by a black bracket, which contains previously studied functional FARs involved in moth pheromone biosynthesis. FARs identified in this study are displayed by black dots, with As_PG_FPKM and As_AB_FPKM indicated
Fig. 9.
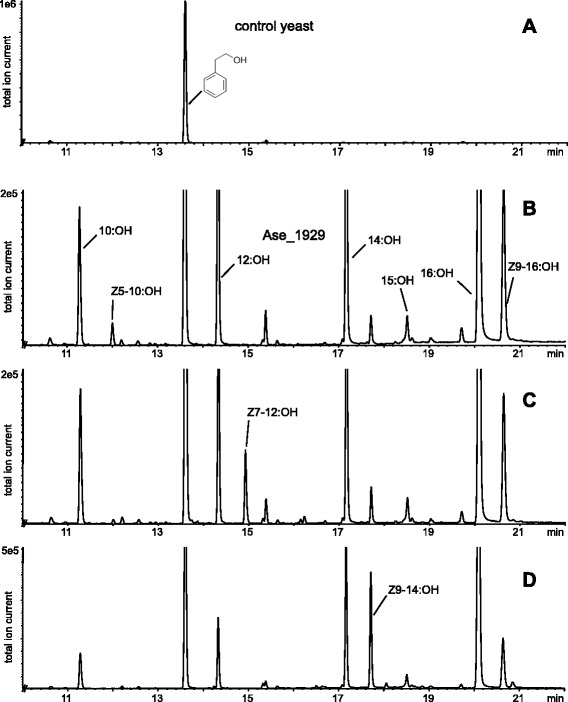
Functional assay of the highest expressed FAR in As_PG, Ase_1929 identified in this study. GC trace of hexane extract of yeast transformed by empty plasmid (a) and pYES2_CL1929 supplemented with Z5-10:Me (b), Z7-12:Me (c), Z9-14:Me (d). The control yeast produced no fatty alcohols whereas the yeast expressing Ase_1929 convert a series of fatty acids into their corresponding fatty alcohols (b-d)
Acetyltransferases
The genes involved in acetylation of fatty pheromone alcohols have not been cloned from any insect species. Acetyltransferases probably belong to a huge family of acyl CoA-utilizing enzymes whose products include a variety of chemicals, such as neurotransmitters [68], plant volatile esters [69, 70], constitutive defense compounds, waxes [71], phytoalexins, lignin, phenolics, alkaloids, and anthocyanins [72–74], which makes it very difficult to make functional predictions from primary sequence information alone. Attempts have been made but ended up with getting other member of the family [75]. This step is the last step in the A. segetum pheromone biosynthetic pathway. Most likely it does not significantly influence the ratio between the homologous acetate pheromone components [32, 33], although in several moth species the Z isomers are produced faster than the E isomers and yield more products, when the alcohol substrates were supplied to the pheromone gland homogenate [34]. Previously, a number of acetyltransferases that were cloned from plant species were studied. For example, the kiwi alcohol acetyltransferase AT9 produced butyl acetate and butyl propionate with highest catalytic activities when tested with short chain alcohols [69]. Another example is the apple alcohol acyltransferases (MpAAT1) that use coenzyme A (CoA) donors together with alcohol acceptors as substrates, which were cloned and characterized [76]. The MpAAT1 recombinant enzyme can utilize a range of alcohol substrates from short to medium straight chain (C3-C10), branched chain, aromatic and terpene alcohols. The enzyme can also utilize a range of short to medium chain CoAs [76], but alcohols longer than C10 have not been tested. Similar work was done with enzymes forming volatiles esters from banana, melon, and strawberry. The recombinant enzymes were capable of producing esters from a wide range of alcohols and acyl-CoA [70, 77, 78]. Overall, these enzymes are members of the BAHD [73, 74], generally recognized by their active site motif (HXXXD) and a conserved region (DFGWG) with likely structural significance [69]. A tBLASTn search with current published BAHDs as query against our A. segetum transcriptome got no hit, suggesting this moth may not express this gene family in its pheromone gland or they have undergone substantial evolutionary changes. But according to previously published putative acetyltransferase [45, 58] and the annotation results we found 34 candidates in our transcriptome (Table 6) and we heterologously expressed them in our yeast system. The ORF of each of these putative acetyltransferases was thus cloned into a yeast expression vector, pYES-DEST52, under the control of galactose inducible promoter. The ∆ATF1 yeast strain (knock out strain that lacking most of the acetylation activity) was transformed with an individual construct and incubated in liquid culture with the culture medium supplemented with a mixture of Z9-14:OH, Z7-12:OH and Z5-10:OH. After incubation for 2 days, the total lipids were extracted and analyzed by GC-MS for acetate esters. The results did not reveal any of insect candidate genes being capable of esterifying fatty alcohols into acetate esters (Fig. 10), whereas the strain overexpressing ATF1 was highly active (positive control).
Table 6.
List of tested acetyltransferases that are generated by annotations and by Blastx of As_PG library with previously published ([54, 55] and references therein) promising candidates as queries
| Gene object | Accession code | bp | As PG FPKM | As AB FPKM | Nr annotation | KO/COG/GO/Swissprot annotation | Species | Accession number | % Identity |
|---|---|---|---|---|---|---|---|---|---|
| CL1935 | KJ579206 | 1542 | 53.53 | 70.39 | Acetyl-CoA acetyltransferase | aeetyl-CoA C-aeetyltransferase [EC:2.3.1.9] | Danaus plexippus | EHJ68573 | 82 |
| CL2371 | KJ579207 | 1203 | 14.63 | 8.51 | Thiolase 4 | aeetyl-CoAacyltransferase 2 [EC:2.3.1.16] | Heliothis virescens | AGG55002 | 92 |
| CL2825 | KJ579208 | 771 | 6.38 | 14.16 | Hypothetical protein KGM 16,501 | Predicted acetyltransferases and hydrolases | Danaus plexippus | EHJ72951 | 87 |
| CL3492 | KJ579209 | 2094 | 16.48 | 10.62 | Crooked neck protein | peptide alpha-N-acetyltransferase [EC:2.3.1.88] | Aedes aegypti | XP001653815 | 83 |
| CL5827 | KJ579210 | 1608 | 682.84 | 198.28 | Sterol carrier protein 2/3- oxoacyl-CoA thiolase | Sterol carrier protein 2 [EC:2.3.1.176] | Spodoptera littoralis | AAT72922 | 94 |
| CL622 | KJ579211 | 1425 | 146.13 | 113.81 | Fatty acid beta-oxidation complex subunit beta | acetyl-CoA acyltransferase [EC:2.3.1.16] | Heliothis virescens | ADB57045 | 95 |
| CL7064 | KJ579212 | 543 | 76.93 | 186.14 | Acetyltransferase | Peptide alpha-N-acetyltransferase [EC:2.3.1.88] | Agrotis ipsilon | AGQ45625 | 100 |
| Unigenel520 | KJ579213 | 1191 | 136.8 | 89.85 | Thiolase 1 | aeetyl-CoA acyltransferase 2 [EC:2.3.1.16] | Heliothis virescens | AGG54999 | 93 |
| Unigene5444 | KJ579214 | 1113 | 2.35 | 15.28 | Thiolase 3 | aeetyl-CoA acyltransferase 2 [EC:2.3.1.16] | Heliothis virescens | AGG55001 | 80 |
| Unigene7635 | KJ579215 | 891 | 23.37 | 46.32 | Palmitoyl-protein thioesterase 1 like isoform XI | Predicted acetyltransferases and hydrolases | Bombyx mori | XP004931556 | 73 |
| Unigene8748 | KJ579216 | 549 | 13.51 | 9.31 | Dynactin 4 protein | Carbonic anhydrases/acetyltransferases, isoleucine patch superfamily | Bombyx mori | NP001040117 | 99 |
| CL197 | KJ579217 | 1080 | 20.88 | 20.02 | Monoacylglycerol acyl transferase | 2-acylglycerol O-acyltransferase 2 [EC:2.3.1.22] | Manduca sexta | AHH25136 | 72 |
| CL2038 | KJ579218 | 1443 | 5.54 | 30.82 | Transmembrane protein nessy | lysophospholipid acyltransferase 5 [EC:2.3.1.23 2.3.1.-] | Bombyx mori | XP004933932 | 76 |
| CL2800 | KJ579219 | 1638 | 20.71 | 27.68 | Dihydroxyacetone phosphate acyltransferase | glyceronephosphate O-acyltransferase [EC:2.3.1.42] | Bombyx mori | XP004921835 | 64 |
| Unigene21478 | KJ579220 | 540 | 4.47 | 0.05 | Thiolase 2 | aeetyl-CoA acyltransferase 2 [EC:2.3.1.16] | Heliothis virescens | AGG55000 | 73 |
| Unigene2251 | KJ579221 | 819 | 1.54 | 0.17 | Acetyltransferase | lysophosphatidate acyltransferase [EC:2.3.1.51] | Agrotis ipsilon | AGQ45623 | 97 |
| Unigene4319 | KJ579222 | 1152 | 7.58 | 11.83 | Acyltransferase AGPAT6 | lysophosphatidic acid acyltransferase/[EC:2.3.1.51] | Heliothis virescens | AGG54998 | 96 |
| CL3297 | KJ579223 | 1296 | 12.72 | 12.03 | Endophilin-A-like isoform XI | Endophilin-A | Bombyx mori | XP004929694 | 86 |
| CL3384 | KJ579224 | 1269 | 69.16 | 84.67 | Ipase 1-like | Predicted hydrolases or acyltransferases | Bombyx mori | XP004927335 | 57 |
| CL3414 | KJ579225 | 1428 | 7.08 | 16.56 | Sterol O-acyltransferase 1 | Sterol O-aeyltransferase [EC:2.3.1.26] | Danaus plexippus | EHJ66395 | 66 |
| CL3666 | KJ579226 | 1272 | 38.77 | 53.18 | Putative glycerol-3-phosphate acyltransferase | type I keratin, acidic | Danaus plexippus | EHJ77802 | 56 |
| CL3797 | KJ579227 | 1164 | 10.06 | 4.91 | Hypothetical protein KGM19212 | [EC:2.3.1.-] | Danaus plexippus | EHJ67359 | 73 |
| CL5775 | KJ579228 | 1179 | 22.36 | 14.42 | l-acylglycerol-3-phosphate O- acyltransferase ABHD5 | abhydrolase domain-containing protein 5 [EC:2.3.1.51] | Bombyx mori | XP004927229 | 85 |
| CL6732 | KJ579229 | 1071 | 7.31 | 8.66 | Epoxide hydrolase 4-like protein | Soluble epoxide hydrolase [EC:3.3.2.10] | Heliconius erato | AGC92732 | 66 |
| CL8534 | KJ579230 | 1410 | 157.51 | 175.16 | Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase, partial | 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide succinyltransferase) [EC:2.3.1.61] | Papilio polytes | BAM20532 | 93 |
| CL8651 | KJ579231 | 675 | 99.26 | 87.58 | Hypothetical protein KGM 17,353 | Pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12] | Danaus plexippus | EHJ75541 | 75 |
| CL914 | KJ579232 | 1962 | 19.4 | 17.94 | Carnitine o-acyltransferase | Carnitine O-palmitoyltransferase 2 [EC:2.3.1.21] | Bombyx mori | XP004929482 | 75 |
| CL9375 | KJ579233 | 2073 | 16.36 | 9.35 | Juvenile hormone epoxide hydrolase-like protein 3 | Predicted hydrolases or acyltransferases | Bombyx mori | NP001159619 | 60 |
| Unigenel0654 | KJ579234 | 1440 | 10.41 | 5.11 | Lipoamide acyltransferase | 2-oxoisovalerate dehydrogenase E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168] | Bombyx mori | XP004926652 | 71 |
| Unigenel5439 | KJ579235 | 924 | 0.58 | 6.57 | Serine hydrolase-like protein 2- like | [EC:3.1.-.-] | Bombyx mori | XP004926488 | 55 |
| Unigene6166 | KJ579236 | 1446 | 5.56 | 27.08 | Glycerol-3-phosphate acyltransferase 4 | Glycerol-3-phosphate O-acyltransferase 3/4 [EC:2.3.1.15] | Bombyx mori | XP004925117 | 84 |
| Unigene7548 | KJ579237 | 996 | 15.04 | 13.97 | Probable serine hydrolase-like | Predicted hydrolases or acyltransferases (alpha/beta hydrolase superfamily) | Bombyx mori | XP004924867 | 70 |
| Unigene7854 | KJ579238 | 522 | 3.92 | 1.09 | acyltransferase AGPAT5 | Lysophosphatidate acyltransferase [EC:2.3.1.51] | Heliothis virescens | AGG54997 | 83 |
| Unigene8703 | KJ579239 | 1200 | 3.71 | 3.53 | Lysophospholipid acyltransferase 1-like | Lysophospholipid acyltransferase 1/2 [EC:2.3.1.51 2.3.1.-] | Bombyx mori | XP004927037 | 74 |
Fig. 10.
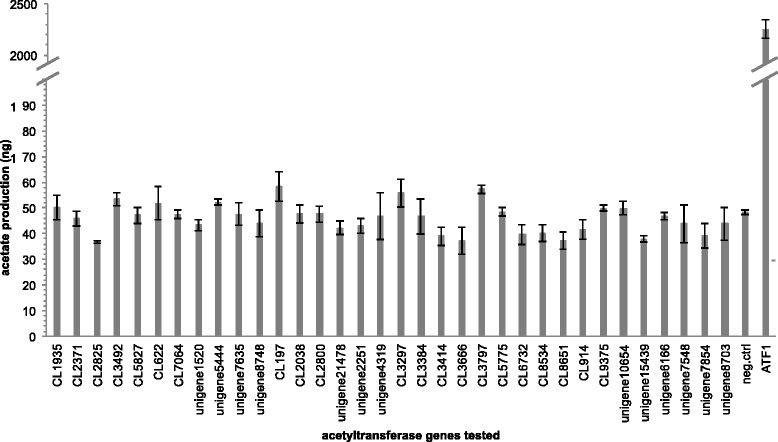
Functional assay of putative acetyltransferase genes. Y-axis represents the total amount of acetate esters (sum of Z5-10:OAc, Z7-12:OAc, and Z9-14:OAc) produced by the yeast cells transformed with candidate genes (±95 % confidence interval, n = 3). Negative control is yeast cell (∆ATF1) transformed with empty vector and the yeast strain overexpressing ATF1 gene serves as positive control. None of the 34 candidate genes produces significantly higher amount of acetate esters compared to the negative control (overlapping 95 % confidence intervals), whereas the ATF1 produces a 45-fold increase in acetate production
Conclusions
We explored the data obtained from massive sequencing of pheromone producing tissue of the turnip moth and compared the expression levels of candidate unigenes with their expression levels in the abdominal epidermal tissue. This allowed identification of key parts involved in the pheromone biosynthesis pathway such as: β-oxidation enzymes, a desaturase, a fatty-acyl reductase, putative acetyltransferases, and other components involved in the fatty-acid metabolism, like acetyl-CoA carboxylase and fatty-acid synthetase. By phylogenetic analyses of desaturases and FARs, we found the most promising candidates for each gene and confirmed their function in pheromone biosynthesis. The ∆11-desaturase is a specialist interacting preferentially with 16:CoA and producing Z11-16:CoA. The FAR we found is a generalist that can reduce a broad range of saturated and unsaturated acyl substrates from 10C to 16C. We had specifically hoped to be able to clone and characterize an acetyltransferase involved in pheromone biosynthesis that has been postulated in many studies. We tested 34 genes that were annotated to be acetyltransferases in our yeast expression system (which can successfully express plant-derived acetyltransferase [10]) but it turned out that none of them was functional in comparison to the ATF1 positive control. The nature of the acetyltransferase involved in moth pheromone biosynthesis remains illusive. In addition, we generated a list of β-oxidation enzymes, ACC and FAS that can be further tested either by heterologous expression or by RNAi.
Methods
Insects and tissue collection
Agrotis segetum were obtained from a laboratory culture, continuously maintained for more than 20 years in Lund, but repeatedly rejuvenated by addition of field-collected insects. The larvae were reared on a semisynthetic bean-based diet [79] and kept at 25 °C under a 16 h:8 h light: dark cycle. Pupae were separated by sex and placed in different jars. The third day after they emerged, 30 pheromone glands [58, 80] and abdominal epidermal tissue [6] from 3 individuals of female moths were dissected 3–4 h into scotophase [81] and stored in −80 °C freezer until RNA extraction.
RNA extraction
Total RNA from pheromone gland and abdominal tissue were extracted using the TRIzol reagent (Life Technologies, Lidingö, Sweden) according to the manufacturer’s instructions except for three additional ethanol washes before dissolving RNA in water. RNA concentration and purity were checked on NanoDrop2000 (Thermo Scientific, Saveen Werner, Malmö, Sweden).
Illumina sequencing and bioinformatic analysis
Twenty μg of total RNA from each sample were sent to BGI (Hong Kong Co., Ltd) for library construction, Illumina sequencing and subsequent bioinformatic analysis.
Reads were assembled using Trinity [82] into contigs. Then the reads are mapped back to contigs, get sequences without Ns and cannot be extended on either end. Such sequences are defined as unigenes. Then TGICL [83] is used to assemble all the unigenes from As_PG and As_AB to form a single set of non-redundant unigenes. Then gene family clustering was performed and unigenes were divided to two classes. One is clusters, which the prefix is CL and the cluster id is behind. In one cluster, there are several unigenes which similarity between them is more than 70 %. The other is singletons, which the prefix is unigene. Unigene sequences are aligned with blastdb using blastx (E-value < 0.00001). Sequence orientations are determined according to the best hit in the database. Bioinformatic data were viewed and further processed using Geneious version (6.1.6), created by Biomatters and available from http://www.geneious.com.
The calculation of unigene expression was performed using the FPKM method (Fragments Per kb per Million fragments) [84, 85], the formula is FPKM = 106C/NL/103. Set FPKM to be the expression of unigene A, and C to be number of fragments that uniquely aligned to unigene A, N to be total number of fragments that uniquely aligned to all unigenes, and L to be the base number in the CDS of unigene A. The FPKM method eliminates the influence of different gene lengths and sequencing levels on the calculation of gene expression. Therefore the calculated gene expression can be directly used for comparing the differences in gene expression between samples [85].
Functional annotations of unigenes were conducted, based on protein sequence similarity, towards the KEGG Pathway, COG [86] and Gene Ontoloty (GO) databases. Briefly, we search all unigene sequences against protein databases (NR, SwissProt, KEGG, COG) using blastx (E-value < 0.00001). Based on NR annotation, we use Blast2GO program [44] to get GO annotation of all unigenes. After getting GO annotation for every unigene, we use WEGO software [87] to do GO functional classification for all unigenes.
Phylogenetic reconstruction
Sequences used for phylogenetic reconstructions were retrieved from the GenBank (http://www.ncbi.nlm.nih.gov) database. Neighbor-Joining trees were constructed using Mega version 4.0 [88]. Briefly, multiple sequence alignments were run using the MAFFT (online version) and the output FASTA format data were plugged into Mega software. Genetic distance model was JTT, 1500 replicates, Neighbor-Joining as tree building method.
Fatty acid precursors
Z9-18:Me, 16:Me were purchased from Larodan Fine Chemicals AB (Malmö, Sweden). Z5-10:OH, Z7-12:OH, Z9-14:OH were purchased from Pherobank (Wageningen, The Netherlands, 98 % purity). Z9-14:Me, Z7-12:Me, Z5-10:Me were prepared from their corresponding alcohols by previously described methods [89]. All FAMEs and OH were dissolved in 96 % ethanol in a 0.1 M stock solution. All alcohols used as reference compounds were from our laboratory collection of pheromone compounds.
Construction of expression vector for functional assay
For the construction of yeast expression vectors containing the candidate genes, specific primers with attB1 and attB2 sites incorporated were designed for amplifying the ORF of genes of interest. The PCR products were subjected to agarose gel electrophoresis and purified using the Wizard® SV Gel and PCR Clean up system (Promega Biotech AB, Nacka, Sweden). The ORFs were subcloned into the pDONR221 vector in presence of BP clonase (Life Technologies), after confirmation by sequencing, the correct entry clones were selected and do LR reaction with pYES2-DEST52 (for FARs and acetyltransferases) or pYEX-CHT-DEST vector (for desaturases), and resulting expression clones were analyzed by sequencing.
Functional assay in yeast
The resultant recombinant expression vectors harboring the candidate genes were introduced into the INVSc (MATa HIS3 LEU2 trp1-289 ura3-52) (for FARs), the ∆ATF1 knockout strain (for acetyltransferase), or the double deficient ole1 elo1 strain (MATa elo1::HIS3 ole1::LEU2 ade2 his3 leu2 ura3) (for desaturase) of the yeast Saccharomyces cerevisiae [19] using the S.c. easy yeast transformation kit (Life Technologies). For selection of uracil (and leucine) prototrophs, the transformed yeast was allowed to grow on SC plate containing 0.7 % YNB (w/o aa, with Ammonium sulfate) and a complete drop-out medium lacking uracil (and leucine) (Formedium™ LTD, Norwich, England), 2 % glucose, 1 % tergitol (type Nonidet NP-40, Sigma-Aldrich Sweden AB, Stockholm, Sweden), 0.01 % adenine (Sigma) and containing 0.5 mM oleic acid (Sigma) as extra fatty acid source. After 2 days (7 days for ole1 elo1 strain) at 30 °C, individual colonies were picked up to inoculate 10 mL selective medium at 30 °C and grown at 300 rpm for 48 h. Yeast cultures were diluted to an OD600 of 0.4 in 10 mL fresh selective medium containing 2 % galactose (2 mM CuSO4) with supplementation of a biosynthetic precursor. Each FAME or fatty alcohol precursor was prepared at a concentration of 100 mM in 96 % ethanol and added to reach a final concentration of 0.5 mM in the culture medium [19]. In the acetyltransferase assay the yeasts were supplemented with a mixture of the three alcohols Z9-14:OH, Z7-12:OH and Z5-10:OH. Yeasts were cultured in 30 °C in a shaking incubator at 30 °C.
Fatty acid/alcohol/acetate analysis
After 48 h of incubation yeast cells were harvested by centrifugation at 3,000 rpm. For the analysis of desaturase products, total lipids were extracted using 3.75 mL of methanol/chloroform (2:1, v/v), in a glass tube. One mL of HAc (0.15 M) and 1.25 mL of water were added to the tube to wash the chloroform phase. Tubes were vortexed vigorously and centrifuged at 2000 rpm for 2 min. The bottom chloroform phase, about 1 mL, containing the total lipids, were transferred to a new glass tube. Fatty acid methylesters (FAMEs) were made from this total lipid extract. The solution of total lipids was evaporated to dryness under gentle nitrogen flow. One mL of sulfuric acid (2 % in methanol) was added to the tube, which was then vortexed vigorously and incubated at 90 °C for an hour. After incubation, 1 mL of water was added, mixed well, and then 1 mL of hexane was used to extract the FAMEs [10].
Fatty alcohols and acetates were extracted from cells using 800 μL of hexane plus sonication. After brief centrifugation, the supernatant was transferred to a new tube and subjected to GC-MS analysis.
Double bond positions were confirmed by dimethyl disulfide (DMDS) derivatization [13], followed by GC-MS analysis. FAMEs (50 μL) were transferred to a new tube and 50 μL DMDS was added and incubated at 40 °C overnight, in the presence of 5 μL of iodine (5 % in diethyl ether) as catalyst. Hexane (200 μL) was added to the sample and the reaction was neutralized by addition of 50-100 μL Na2S2O3 (5 % in water). The organic phase was recovered and concentrated under a gentle nitrogen stream to 40-50 μL.
Gas chromatography - mass spectrometry (GC-MS)
The methylesters, fatty alcohols and acetates were subjected to GC-MS analyses on a Hewlett Packard 6890 GC coupled to a mass spectrometer HP 5973. The GC was equipped with an INNOWax column (30 m × 0.25 mm i.d. × 0.25 μm film thickness, Agilent Technologies), and helium was used as carrier gas (average velocity: 33 cm/s). The MS was operated in electron impact mode (70 eV), scaning between m/z 30 and m/z 400, and the injector was configured in splitless mode at 220 °C. The oven temperature was set to 80 °C for 1 min, then increased at a rate of 10 °C/min up to 210 °C, followed by a hold at 210 °C for 15 min, and then increased at a rate of 10 °C/min up to 230 °C followed by a hold at 230 °C for 20 min.
DMDS derivatives were analyzed on an Agilent 6890 GC system equipped with HP-5MS capillary column (30 m × 0.25 mm i.d. × 0.25 μm film thickness, Agilent Technologies) coupled with an HP 5973 mass spectrometer. The oven temperature was set at 80 °C for 1 min, raised to 140 °C at a rate of 20 °C/min, then to 250 °C at a rate of 4 °C/min and held for 20 min [11].
Data were analyzed using the ChemStation software (Agilent, Technologies, USA).
Accession code
PBANr: KJ622075
ACC: KJ622074
FAS: KJ622068–KJ622073
Desaturases: KJ622048–KJ622057
β-oxidation enzymes: KJ622076–KJ622113
FARs: KJ622058–KJ622067
Acetyltransferases: KJ579206–KJ579239
Acknowledgements
The study was supported by grants from Formas and VR to Christer Löfstedt and grants from the Royal Physiographic Society in Lund to Baojian Ding. We thank Åsa Hagström for advice on and assistance with the construction of gene trees, Tomas Johansson for helpful comments on the manuscript and Erling Jirle for technical support. We thank Fredrik Andersson and Erik Hedenström at Mid Sweden University for synthesizing the Z5-10:Me, Z7-12:Me and Z9-14:Me from corresponding alcohols.
Abbreviations
- ACC
Acetyl-CoA carboxylase
- FAS
fatty acid synthase
- FAR
fatty-acyl reductase
- GC-MS
gas chromatography mass spectrometry
- PCR
polymerase chain reaction
- Z9-14:OAc
(Z)-9-tetradecenyl acetate
- Z7-12:OAc
(Z)-7-dodecenyl acetate
- Z5-10:OAc
(Z)-5-decenyl acetate
- Z9-14:OH
(Z)-9-tetradecenol
- Z7-12:OH
(Z)-7-dodecenol
- Z5-10:OH
(Z)-5-decenol
- Z9-14:Me
(Z)-9-tetradecenoic methyl ester
- Z7-12:Me
(Z)-7-dodecenoic methyl ester
- Z5-10:Me
(Z)-5-decenoic methyl ester
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BJD and CL conceived the study, BJD and CL designed the study, BJD performed the research, BJD analyzed the data, BJD and CL wrote the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Bao-Jian Ding, Email: bao-jian.ding@biol.lu.se.
Christer Löfstedt, Email: christer.lofstedt@biol.lu.se.
References
- 1.Wyatt TD. Pheromones and Animal Behaviour. Communication by Smell and Taste. Cambridge: Cambridge University Press; 2003. [Google Scholar]
- 2.Percy-Cunningham JE, MacDonald JA. Biology and ultrastructure of sex pheromone-producing glands. In: Prestwich GD, Blomquist GJ, editors. Pheromone biochemistry. Orlando Fl: Academic; 1987. pp. 27–69. [Google Scholar]
- 3.Ando T. 2015. (http://www.tuat.ac.jp/~antetsu/List_of_Sex_Pheromones_in_English(2015.06.22).pdf.
- 4.El-Sayed AM. The Pherobase: Database of Pheromones and Semiochemicals. 2011, [http://www.pherobase.net]
- 5.Jurenka R. Insect pheromone biosynthesis. Topics in Curr Chem. 2004;239:97–132. doi: 10.1007/b95450. [DOI] [PubMed] [Google Scholar]
- 6.Blomquist GJ, Jurenka R, Schal C, Tittiger C. Biochemistry and molecular biology of pheromone production. In: Gilbet LI, Latrou K, Gill SS, editors. Comprehensive molecular insect science. London: Elsevier; 2005. p. 705–51.
- 7.Tillman JA, Seybold SJ, Jurenka RA, Blomquist GJ. Insect pheromones - an overview of biosynthesis and endocrine regulation. Insect Biochem Mol Biol. 1999;29:481–541. doi: 10.1016/S0965-1748(99)00016-8. [DOI] [PubMed] [Google Scholar]
- 8.Cook SM, Khan ZR, Pickett JA. The use of push-pull strategies in integrated pest management. Annu Rev Entomol. 2007;52:375–400. doi: 10.1146/annurev.ento.52.110405.091407. [DOI] [PubMed] [Google Scholar]
- 9.Hagström ÅK, Wang HL, Liénard MA, Lassance JM, Johansson T, Löfstedt C. A moth pheromone brewery: production of (Z)-11-hexadecenol by heterologous co-expression of two biosynthetic genes from a noctuid moth in a yeast cell factory. Microb Cell Fact. 2013;12:125. doi: 10.1186/1475-2859-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding BJ, Hofvander P, Wang HL, Durrett TP, Stymne S, Löfstedt C. A plant factory for moth pheromone production. Nat Commun. 2014;5:3353. doi: 10.1038/ncomms4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HL, Liénard MA, Zhao CH, Wang CZ, Löfstedt C. Neofunctionalization in an ancestral insect desaturase lineage led to rare ∆6 pheromone signals in the Chinese tussah silkworm. Insect Biochem Mol Biol. 2010;40:742–751. doi: 10.1016/j.ibmb.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Hao GX, O’Conner M, Liu WT, Roelofs WL. Chraracterization of Z/E11- and Z9-desaturases from the obliquebanded leafroller moth, Choristoneura rosaceana. J Insect Sci. 2002;2:26. doi: 10.1673/031.002.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liénard MA, Strandh M, Hedenström E, Johansson T, Löfstedt C. Key biosynthetic gene subfamily recruited for pheromone production prior to the extensive radiation of Lepidoptera. BMC Evol Biol. 2008;8:270. doi: 10.1186/1471-2148-8-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao G, Liu W, O'Connor M, Roelofs WL. Acyl-CoA Z9 and Z10-desaturase genes from a New Zealand leafroller moth species, Planotortrix octo. Insect Biochem Mol Biol. 2002;32:961–966. doi: 10.1016/S0965-1748(01)00176-X. [DOI] [PubMed] [Google Scholar]
- 15.Liu WT, Jiao HM, O'Connor M, Roelofs WL. Moth desaturase characterized that produces both Z and E isomers of ∆11-tetradecenoic acids. Insect Biochem Mol Biol. 2002;32:1489–1495. doi: 10.1016/S0965-1748(02)00069-3. [DOI] [PubMed] [Google Scholar]
- 16.Liu WT, Rooney AP, Xue BY, Roelofs WL. Desaturases from the spotted fireworm moth (Choristoneura parallela) shed light on the evolutionary origins of novel moth sex pheromone desaturases. Gene. 2004;342:303–311. doi: 10.1016/j.gene.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Moto K, Suzuki MG, Hull JJ, Kurata R, Takahashi S, Yamamoto M, Okano K, Imai K, Ando T, Matsumoto S. Involvement of a bifunctional fatty-acyl desaturase in the biosynthesis of the silkmoth, Bombyx mori, sex pheromone. Proc Natl Acad Sci U S A. 2004;101:8631–8636. doi: 10.1073/pnas.0402056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roelofs WL, Liu W, Hao G, Jiao H, Rooney AP, Linn CE., Jr Evolution of moth sex pheromones via ancestral genes. Proc Natl Acad Sci U S A. 2002;99(21):13621–13626. doi: 10.1073/pnas.152445399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding BJ, Liénard MA, Wang HL, Zhao CH, Löfstedt C. Terminal fatty-acyl-CoA desaturase involved in sex pheromone biosynthesis in the winter moth (Operophtera brumata) Insect Bioch Mol Biol. 2011;41:715–722. doi: 10.1016/j.ibmb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Moto K, Yoshiga T, Yamamoto M, Takahashi S, Okano K, Ando T, Nakata T, Matsumoto S. Pheromone gland-specific fatty-acyl reductase of the silkmoth, Bombyx mori. Proc Natl Acad Sci U S A. 2003;100(16):9156–9161. doi: 10.1073/pnas.1531993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antony B, Fujii T, Moto K, Matsumoto S, Fukuzawa M, Nakano R, Tatsuki S, Ishikawa Y. Pheromone-gland-specific fatty-acyl reductase in the adzuki bean borer, Ostrinia scapulalis (Lepidoptera: Crambidae) Insect Biochem Mol Biol. 2009;39(2):90–95. doi: 10.1016/j.ibmb.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Lassance JM, Groot AT, Liénard MA, Antony B, Borgwardt C, Andersson F, Hedenström E, Heckel DG, Löfstedt C. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature. 2010;466:486–489. doi: 10.1038/nature09058. [DOI] [PubMed] [Google Scholar]
- 23.Lassance JM, Liénard MA, Antony B, Qian S, Fujii T, Tabata J, Ishikawa Y, Löfstedt C. Functional consequences of sequence variation in the pheromone biosynthetic gene pgFAR for Ostrinia moths. Proc Natl Acad Sci U S A. 2013;110:3967–3972. doi: 10.1073/pnas.1208706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liénard MA, Hagström ÅK, Lassance JM, Löfstedt C. Evolution of multicomponent pheromone signals in small ermine moths involves a single fatty-acyl reductase gene. Proc Natl Acad Sci U S A. 2010;107:10955–10960. doi: 10.1073/pnas.1000823107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagström ÅK, Liénard MA, Groot AT, Hedenström E, Löfstedt C. Semi-selective fatty acyl reductases from four heliothine moths influence the specific pheromone composition. PLoS ONE. 2012;7(5):e37230. doi: 10.1371/journal.pone.0037230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teal PEA, Tumlinson JH. Properties of cuticular oxidases used for sex pheromone biosynthesis by Heliothis zea. J Chem Ecol. 1998;14:2131–2145. doi: 10.1007/BF01014254. [DOI] [PubMed] [Google Scholar]
- 27.Fang N, Teal PEA, Tumlinson JH. Correlation between glycerolipids and pheromone aldehydes in the sex pheromone gland of female tobacco hornworm moths, Manduca sexta (L.) Arch Insect Biochem Physiol. 1995;4:321–336. doi: 10.1002/arch.940300403. [DOI] [Google Scholar]
- 28.Hoskovec MA, Luxová A, Svatoš A, Boland W. Biosynthesis of sex pheromones in moths: stereochemistry of fatty alcohol oxidation in Manduca sexta. Tetrahedron. 2002;58:9193–9201. doi: 10.1016/S0040-4020(02)01199-7. [DOI] [Google Scholar]
- 29.Bestmann HJ, Herrig M, Attygalle AB. Terminal acetylation in pheromone biosynthesis by Mamestra brassicae L. (Lepidoptera:Noctuidae) Experientia. 1987;43:1033–1034. [Google Scholar]
- 30.Teal PEA, Tumlinson JH. The role of alcohols in pheromone biosynthesis by two noctuid moths that use acetate pheromone components. Arch Insect Biochem Physiol. 1987;4:261–269. doi: 10.1002/arch.940040404. [DOI] [Google Scholar]
- 31.Zhu JW, Zhao CH, Bengtsson M, Löfstedt C. Reductase specificity and the ratio regulation of E/Z isomers in the pheromone biosynthesis of the European corn borer, Ostrinia nubilalis (Lepidoptera: Pyralidae) Insect Biochem Mol Biol. 1996;26:171–176. doi: 10.1016/0965-1748(95)00079-8. [DOI] [Google Scholar]
- 32.Morse D, Meighen E. Biosynthesis of the acetate ester precursor of the spruce budworm sex pheromone by an acetyl CoA: Fatty alcohol acetyltransferase. Insect Biochem. 1987;17:53–59. doi: 10.1016/0020-1790(87)90143-0. [DOI] [Google Scholar]
- 33.Zhao CH, Lu F, Bengtsson M, Löfstedt C. Substrate specificity of acetyltransferase and reductase enzyme systems used in pheromone biosynthesis by Asian corn borer, Ostrinia furnacalis. J Chem Ecology. 1995;21:1495–1510. doi: 10.1007/BF02035148. [DOI] [PubMed] [Google Scholar]
- 34.Jurenka RA, Roelofs WL. Characterization of the acetyltransferase used in pheromone biosynthesis in moths: Specificity for the Z isomer in tortricidae. Insect Biochem. 1989;19(7):639–644. doi: 10.1016/0020-1790(89)90098-X. [DOI] [Google Scholar]
- 35.Haynes KF, Hunt RE. A mutation in pheromonal communication system of cabbage looper moth, Trichoplusia ni. J Chem Ecol. 1990;16:1249–1257. doi: 10.1007/BF01021023. [DOI] [PubMed] [Google Scholar]
- 36.Jurenka RA, Haynes KF, Adlof RO, Bengtsson M, Roelofs WL. Sex pheromone component ratio in the cabbage looper moth altered by a mutation affecting the fatty acid chain-shortening reactions in the pheromone biosynthetic pathway. Insect Biochem Mol Biol. 1994;24:373–381. doi: 10.1016/0965-1748(94)90030-2. [DOI] [Google Scholar]
- 37.Wu WQ, Zhu JW, Millar J, Löfstedt C. A comparative study of sex pheromone biosynthesis in two strains of the turnip moth, Agrotis segetum, producing different ratios of sex pheromone components. Insect Biochem Mol Biol. 1998;28:895–900. doi: 10.1016/S0965-1748(98)00063-0. [DOI] [Google Scholar]
- 38.Löfstedt C, Elmfors A, Sjögren M, Wijk E. Confirmation of sex pheromone biosynthesis from (16-D3) palmitic acid in the turnip moth using capillary gas chromatography. Experientia. 1986;42:1059–1061. doi: 10.1007/BF01940732. [DOI] [Google Scholar]
- 39.Löfstedt C, Van Der Pers JNC, Löfqvist J, Lanne BS, Appelgren M, Bergstrom G, Thelin B. Sex pheromone components of the turnip moth, Agrotis segetum: chemical identification, electrophysiological evaluation and behavioral activity. J Chem Ecol. 1982;8:1305–1321. doi: 10.1007/BF00987764. [DOI] [PubMed] [Google Scholar]
- 40.Löfstedt C, Löfquist J, Lanne BS, Van Der Peers JNC, Hansson BS. Pheromone dialects in European turnip moths Agrotis segetum. Oikos. 1986;46:250–257. doi: 10.2307/3565474. [DOI] [Google Scholar]
- 41.Tòth M, Löfstedt C, Blair BW, Cabello T, Farag AI, Hansson BS, Kovalev BG, Maini S, Nesterov EA, Pajor I, Sazonov AP, Shamshev IV, Subchev M, Szöcs G. Attraction of male turnip moths Agrotis segetum (lepidoptera, noctuidae) to sex-pheromone components and their mixtures at 11 sites in Europe, Asia, and Africa. J Chem Ecol. 1992;18(8):1337–1347. doi: 10.1007/BF00994360. [DOI] [PubMed] [Google Scholar]
- 42.Strandh M, Johansson T, Ahren D, Löfstedt C. Transcriptional analysis of the pheromone gland of the turnip moth, Agrotis segetum (Noctuidae), reveals candidate genes involved in pheromone production. Insect Mol Biol. 2008;17(1):73–85. doi: 10.1111/j.1365-2583.2008.00782.x. [DOI] [PubMed] [Google Scholar]
- 43.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 45.Gu SH, Wu KM, Guo YY, Pickett JA, Field LM, Zhou JJ, Zhang YJ. Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics. 2013;14:636. doi: 10.1186/1471-2164-14-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Millar J, Löfstedt C. Hormonal regulation of sex pheromone biosynthesis in the turnip moth, Agrotis segetum. Archs Insect Biochem Physiol. 1995;30:41–59. doi: 10.1002/arch.940300104. [DOI] [Google Scholar]
- 47.Choi MY, Fuerst EJ, Rafaeli A, Jurenka R. Identification of a G protein-coupled receptor for pheromone biosynthesis activating neuropeptide from pheromone glands of the moth Helicoverpa zea. Proc Natl Acad Sci U S A. 2003;100:9721–9726. doi: 10.1073/pnas.1632485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto S, Ozawa R, Nagamine T, Kim GH, Uchiumi K, Shono T, Mitsui T. Intracellular transduction in the regulation of pheromone biosynthesis of the silkworm, Bombyx mori: suggested involvement of calmodulin and phosphoprotein phosphatase. Biosci Biotechnol Biochem. 1995;59:560–562. doi: 10.1271/bbb.59.560. [DOI] [PubMed] [Google Scholar]
- 49.Jurenka RA, Jacquin E, Roelofs WL. Stimulation of pheromone biosynthesis in the moth Helicoverpa zea: Action of a brain hormone on pheromone glands involves Ca2+ and cAMP as second messengers. Proc Natl Acad Sci U S A. 1991;88:8621–8625. doi: 10.1073/pnas.88.19.8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mas E, Llòria J, Quero C, Camps F, Fabriàs G. Control of the biosynthetic pathway of Sesamia nonagrioides sex pheromone by the pheromone biosynthesis activating neuropeptide. Insect Biochem Mol Biol. 2000;30:455–459. doi: 10.1016/S0965-1748(00)00008-4. [DOI] [PubMed] [Google Scholar]
- 51.Pape ME, Lopezcasillas F, Kim KH. Physiological regulation of acetyl-CoA carboxylase gene expression: effects of diet, diabetes, and lactation on acetyl-CoA carboxylase mRNA. Arch Biochem Biophys. 1988;267(1):104–109. doi: 10.1016/0003-9861(88)90013-6. [DOI] [PubMed] [Google Scholar]
- 52.Volpe JJ, Vagelos PR. Saturated fatty acid biosynthesis and its regulation. Annu Rev Biochem. 1973;42:21–60. doi: 10.1146/annurev.bi.42.070173.000321. [DOI] [PubMed] [Google Scholar]
- 53.Tang JD, Charlton RE, Jurenka RA, Wolf WA, Phelan PL, Sreng L, Roelofs WL. Regulation of Pheromone Biosynthesis by a Brain Hormone in 2 Moth Species. Proc Natl Acad Sci U S A. 1989;86(6):1806–1810. doi: 10.1073/pnas.86.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bjostad LB, Roelofs WL. Biosynthesis of sex pheromone components and glycerolipid precursors from sodium [1–14C] acetate in redbanded leafroller moth. J Chem Ecol. 1984;10:681–691. doi: 10.1007/BF00994228. [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, Kanehisa M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J Lipid Res. 2008;49:183–191. doi: 10.1194/jlr.M700377-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Knipple DC, Rosenfield CL, Nielsen R, You KM, Jeong SE. Evolution of the integral membrane desaturase gene family in moths and flies. Genetics. 2002;162(4):1737–1752. doi: 10.1093/genetics/162.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeong SE, Rosenfield CL, Marsella-Herrick P, Man You K, Knipple DC. Multiple acyl-CoA desaturase-encoding transcripts in pheromone glands of Helicoverpa assulta, the oriental tobacco budworm. Insect Biochem Mol Biol. 2003;33:609–622. doi: 10.1016/S0965-1748(03)00043-2. [DOI] [PubMed] [Google Scholar]
- 58.Vogel H, Heidel AJ, Heckel DG, Groot AT. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genomics. 2010;11:29. doi: 10.1186/1471-2164-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ikeda Y, Okamura-Ikeda K, Tanaka K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J Biol Chem. 1985;260:1311–1325. [PubMed] [Google Scholar]
- 60.Izai K, Uchida Y, Orii T, Yamamoto S, Hashimoto T. Novel fatty acid beta-oxidation enzymes in rat liver mitochondria. I. Purification and properties of very-long-chain acyl-coenzyme A dehydrogenase. J Biol Chem. 1992;267:1027–1033. [PubMed] [Google Scholar]
- 61.Schulz H. Oxidation of fatty acids in eukaryotes. In: Vance DE, Vance J, editors. Biochemistry of Lipids, Lipoproteins and Membranes. 5. Amsterdam: Elsevier; 2008. pp. 131–154. [Google Scholar]
- 62.Kunau WH, Dommes V, Schulz H. beta-oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: a century of continued progress. Prog Lipid Res. 1995;34:267–342. doi: 10.1016/0163-7827(95)00011-9. [DOI] [PubMed] [Google Scholar]
- 63.Uchida Y, Izai K, Orii T, Hashimoto T. Novel fatty acid beta-oxidation enzymes in rat liver mitochondria. II. Purification and properties of enoyl-coenzyme A (CoA) hydratase/3-hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase trifunctional protein. J Biol Chem. 1992;267:1034–1041. [PubMed] [Google Scholar]
- 64.Schreurs M, Kuipers F, van der Leij FR. Regulatory enzymes of mitochondrial beta-oxidation as targets for treatment of the metabolic syndrome. Obes Rev. 2010;11:380–388. doi: 10.1111/j.1467-789X.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 65.Stanley-Samuelson DW, Jurenka RA, Cripps C, Blomquist GJ, de Renobales M. Fatty acids in insects: composition, metabolism, and biological significance. Arch Insect Biochem Physiol. 1988;9:1–33. doi: 10.1002/arch.940090102. [DOI] [Google Scholar]
- 66.Hashimoto T. Peroxisomal β-oxidation: enzymology and molecular biology. Ann N Y Acad Sci. 1996;804:86–98. doi: 10.1111/j.1749-6632.1996.tb18610.x. [DOI] [PubMed] [Google Scholar]
- 67.Liénard MA, Löfstedt C. Functional flexibility as a prelude to signal diversity?: role of a fatty acyl reductase in moth pheromone evolution. Commun Integr Biol. 2010;3(6):586–588. doi: 10.4161/cib.3.6.13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Itoh N, Slemmon J, Hawke D. Cloning of Drosophila choline acetyltransferase cDNA. Proc Natl Acad Sci U S A. 1986;83:4081–4085. doi: 10.1073/pnas.83.11.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Günther CS, Chervin C, Marsh KB, Newcomb RD, Souleyre EJ. Characterisation of two alcohol acyltransferases from kiwifruit (Actinidia spp.) reveals distinct substrate preferences. Phytochem. 2011;72:700–710. doi: 10.1016/j.phytochem.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 70.Beekwilder J, Alvarez-Huerta M, Neef E, Verstappen FWA, Bouwmeester HJ, Aharoni A. Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol. 2004;135:1865–1878. doi: 10.1104/pp.104.042580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalscheuer R, Stoveken T, Luftmann H, Malkus U, Reichelt R, Steinbüchel A. Neutral lipid biosynthesis in engineered Escherichia coli: jojoba oil-like wax esters and fatty acid butyl esters. Appl Environ Microbiol. 2006;72:1373–1379. doi: 10.1128/AEM.72.2.1373-1379.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uthoff S, Stoveken T, Weber N, Vosmann K, Klein E, Kalscheuer R, Steinbüchel A. Thio wax ester biosynthesis utilizing the unspecific bifunctional wax ester synthase/acyl coenzyme A: diacylglycerol acyltransferase of Acinetobacter sp. strain ADP1. Appl Environ Microbiol. 2005;71:790–796. doi: 10.1128/AEM.71.2.790-796.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.St-Pierre B, De Luca V. Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. In: Ibrahim R, Varin L, De Luca V, Romeo JT, editors. Recent Advances in Phytochemistry Evolution of Metabolic Pathways. 2000, 34: 285–315
- 74.D’Auria J. Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 75.Fujii T, Ito K, Katsuma S, Nakano R, Shimada T, Ishikawa Y. Molecular and functional characterization of an acetyl-CoA acetyltransferase from the adzuki bean borer moth Ostrinia scapulalis (Lepidoptera: Crambidae) Insect Biochem Mol Biol. 2009;40:74–78. doi: 10.1016/j.ibmb.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 76.Souleyre EJF, Greenwood DR, Friel EN, Karunairetnam S, Newcomb RD. An alcohol acyl transferase from apple (cv. Royal Gala), MpAAT1, produces esters involved in apple fruit flavor. FEBS J. 2005;272:3132–3144. doi: 10.1111/j.1742-4658.2005.04732.x. [DOI] [PubMed] [Google Scholar]
- 77.Yahyaoui FE, Wongs-Aree C, Latche A, Hackett R, Grierson D, Pech JC. Molecular and biochemical characteristics of a gene encoding an alcohol acyl-transferase involved in the generation of aroma volatile esters during melon ripening. Eur J Biochem. 2002;269:2359–2366. doi: 10.1046/j.1432-1033.2002.02892.x. [DOI] [PubMed] [Google Scholar]
- 78.Aharoni A, Keizer LC, Bouwmeester HJ, Sun Z, Alvarez-Huerta M, Verhoeven HA, Blaas J, van Houwelingen AM, De Vos RC, van der Voet H, Jansen RC, Guis M, Mol J, Davis RW, Schena M, van Tunen AJ, O'Connell AP. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell. 2000;12:647–662. doi: 10.1105/tpc.12.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu JW, Löfstedt C, Bengtsson BO. Genetic variation in the strongly canalized sex pheromone communication system of the European corn borer Ostrinia nubilalis Hübner (Lepidoptera; Pyralidae) Genetics. 1996;144:757–766. doi: 10.1093/genetics/144.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma PWK, Ramaswamy SB. Biology and ultrastructure of sex pheromone-producing tissue. In: Blomquist GJ, Vogt RG, editors. Insect pheromone biochemistry and molecular biology. London: Elsevier; 2003. pp. 19–51. [Google Scholar]
- 81.Rosén W. Endogenous control of circadian rhythms of pheromone production in the turnip moth, Agrotis segetum. Arch Insect Biochem Physiol. 2002;50:21–30. doi: 10.1002/arch.10026. [DOI] [PubMed] [Google Scholar]
- 82.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, Tsai J, Quackenbush J. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 84.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 85.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 87.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 89.Bjostad BL, Roelofs LW. Sex pheromone biosynthetic precursors in Bombyx mori. Insect Biochem. 1984;14:275–278. doi: 10.1016/0020-1790(84)90060-X. [DOI] [Google Scholar]


