Abstract
Tumors continually shed DNA into the blood where it can be detected as circulating tumor DNA (ctDNA). While this phenomenon has been recognized for decades, techniques that are sensitive and specific enough to robustly detect ctDNA have only become available recently. Quantification of ctDNA represents a new approach for cancer detection and disease burden quantification that has the potential to revolutionize response assessment and personalized treatment in radiation oncology. Analysis of ctDNA has many potential applications, including detection of minimal residual disease following radiotherapy, non-invasive tumor genotyping, and early detection of tumor recurrence. Ultimately ctDNA-based assays could lead to personalization of therapy based on identification of somatic alterations present in tumors and changes in ctDNA concentrations before and after treatment. In this review, we will discuss methods of ctDNA detection and clinical applications of ctDNA-based biomarkers in radiation oncology, with a focus on recently developed techniques that employ next generation sequencing for ctDNA quantification.
Introduction
The field of radiation oncology is currently going through a revolution as new technologies allow treatment of patients with more precision than ever before. While much progress has been made in improving delivery of radiotherapy, response assessment after treatment continues to rely primarily on imaging for most types of cancers. While imaging-based response assessment has significant clinical value, this approach has suboptimal sensitivity for detecting minimal residual disease, suffers from a lag between treatment and when tumor responses can be detected, is expensive, and often exposes patients to additional ionizing radiation. For these reasons, there continues to be an unmet need for developing blood-based biomarkers for cancers that are treated with radiotherapy. While such biomarkers exist for a few cancer types (e.g. PSA for prostate cancer), they do not for most cancers. The ideal biomarker would be readily generalizable, such that it could be applied to all cancer types.
While most work on cancer biomarkers has focused on detection of proteins, recently circulating tumor DNA (ctDNA) has emerged as a plasma-based biomarker with the potential to redefine prediction and monitoring of response to anti-cancer therapies. The cell-free component of whole blood has long been recognized to contain fragmented DNA molecules1 that are collectively referred to as cell-free DNA (cfDNA). These molecules are derived from dying cells and in healthy individuals mainly originate from hematopoietic cells2–4, although diverse tissues contribute to the total cfDNA pool. Concentrations of cfDNA in plasma of healthy patients range from 1–10ng/ml. Patients with advanced malignancies can display elevated levels of cfDNA, which led several early studies to suggest that measurement of total cfDNA concentration may be a useful biomarker for detection and monitoring of cancers.5–9 However, further work revealed that a variety of non-malignant physiological states and stresses can lead to dramatic elevations of cfDNA concentrations, including exercise, trauma, infection, and inflammation.10–12 Furthermore, as detailed below, the fraction of cfDNA that is derived from tumors in cancer patients is often extremely low. Thus, measurement of total cfDNA concentration does not have sufficient specificity or sensitivity to serve as a reliable cancer biomarker.
Circulating Tumor DNA
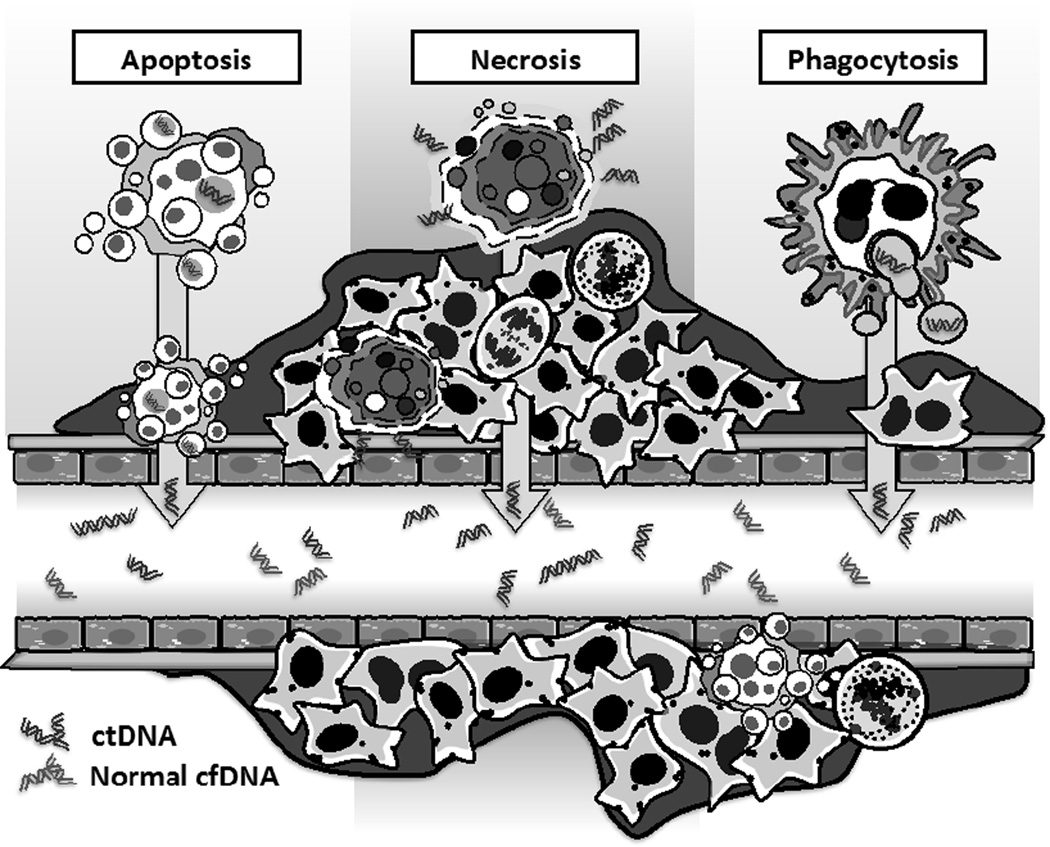
Malignant tumors continually shed DNA into the circulation.13–18 The mechanisms of release of nucleic acid into the blood are not well understood, but are thought to result from necrosis, apoptosis, and release of DNA by phagocytes that have engulfed tumor cells (Figure 1).17,19–21 Importantly, the vast majority of cfDNA found in plasma of cancer patients usually originates from non-malignant cells. This is often the case even for patients with metastatic disease. For example, in patients with metastatic NSCLC pre-treatment ctDNA percentages range from ~1%–5%, and drop if active treatment is initiated.16,18,22 Similarly, when Dawson et al. quantified ctDNA in metastatic breast cancer patients, the median mutant allele fraction was ~4%.14 This problem is magnified in patients with earlier stages of cancer and lower disease burdens, as well as in settings where no gross disease remains after therapy and microscopic residual disease must be detected. Thus, only highly sensitive detection techniques are likely to be of clinical utility.13,23 Below we describe alternative methods to detect ctDNA with higher sensitivity. These methods are compared and contrasted in Table 1.
Figure 1. Sources of circulating tumor DNA.
Tumor DNA is released into blood predominantly by tumor cell death mechanisms including necrosis and apoptosis. Phagocytes that have engulfed tumor cells may also actively release tumor DNA into the extracellular space. ctDNA, circulating tumor DNA; cfDNA, cell-free DNA.
Table 1.
Comparison of methods for detecting circulating tumor DNA.
| Allele- specific PCR |
Digital PCR |
NGS amplicon based |
WGS | WES | CAPP-Seq | |
|---|---|---|---|---|---|---|
| Method | Preferentially amplifies rare mutant DNA molecules | Counts mutant molecules via partitioning of DNA molecules | Deep sequencing of PCR amplicons | Deep sequencing of entire genome | Deep sequencing of exome | Targeted hybrid-capture |
| Detection limit (as % ctDNA) | ~0.1–1% | ~0.01% | ~0.01–2.0% | ~1% | ~5% | ~0.01% |
| Advantages |
|
|
|
|
|
|
| Limitations |
|
|
|
|
|
|
Abbreviations: ctDNA, circulating tumor DNA; SCNA, somatic copy number alteration; NGS, next generation sequencing; WGS, whole genome sequencing; WES, whole exome sequencing; CAPP-Seq, Cancer Personalized Profiling by Deep Sequencing.
Allele-specific PCR-based methods for ctDNA detection
Initial efforts to quantitate ctDNA focused on detection using PCR-based methods such as quantitative PCR or PCR approaches that can preferentially amplify low levels of mutant alleles in the presence of large amounts of wild type DNA. Examples of the latter include COLD-PCR (coamplification at lower denaturation temperature), peptide nucleic acids (PNAs), locked nucleic acids (LNAs) and amplification refractory mutation systems (ARMS). These approaches have analytic sensitivities in the range of 0.1%–1%.24 Studies using such approaches documented the presence of somatic mutations in genes such as KRAS or APC in the plasma of patients with a variety of cancer types.25–27 Similarly, it was shown that PCR-based methods could detect microsatellite alterations in cfDNA.28 Advantages of PCR-based approaches include low cost and relative easy of use. However, the key limitation of these methods is that they are usually not sufficiently sensitive to detect ctDNA in patients with early stage disease or advanced patients who are responding to therapy. This led to the conclusion in a number of early studies that ctDNA was unlikely to be a useful biomarker, which for a time period decreased interest in the field21,29,30.
Digital PCR-based ctDNA detection methods
In the last several years, there has been a renewed interest in developing ctDNA as a clinical biomarker due to the addition of two major classes of techniques for ctDNA detection. The first of these classes was digital PCR (dPCR), which allows more precise quantitation of minor mutant allele fractions than the aforementioned PCR-based techniques. Two prominent examples of dPCR-based methods are beads, emulsion, amplification, and magnetics (BEAMing27) and droplet digital PCR (ddPCR27,31). Both methods involve separation of template molecules into thousands of tiny droplets that each represent isolated reaction chambers, which contain at most one template molecule. Paired with allele specific PCR assays, this allows extremely precise enumeration of the number of total template molecules that carry a mutation of interest and can achieve detection limits as low as 0.01%. Using these methods, ctDNA has been detected in a wide range of cancer patients.22,32–34
The dPCR techniques are currently the gold standard methods for detection of specific mutations within ctDNA given their high sensitivity, specificity, and relatively low cost. However, an important limitation of these approaches is that only one or a small number of mutations can be interrogated at a time. Due to the low concentration of ctDNA found in many patients, this means that often an entire 10–20mL blood draw needs to be exhausted for evaluation of only a handful of mutations. Additionally, in order to use dPCR to monitor treatment responses, at least one mutation must first be identified via analysis of tumor tissue, thus usually requiring an initial invasive procedure.
Next Generation sequencing (NGS)-based ctDNA detection assays
Due to the practical limitation of the number of genomic positions that can be interrogated by dPCR, a number of groups recently focused efforts on developing NGS-based approaches for ctDNA detection. NGS allows for massively parallel sequencing of hundreds of millions of DNA fragments from a single sample and thus opens the possibility of genotyping many potential mutations at one time. The NGS-based approaches can be categorized based on the number of genomic regions they interrogate as either being “focused” or “broad”.
The focused NGS-based approaches have mainly employed PCR amplicon-based strategies. Specifically, PCR is used to amplify a handful of regions of the genome and the resulting amplified products are subjected to NGS. For example, Forshew and colleagues developed Tagged-Amplicon Sequencing (TAm-Seq) and used it to detect mutations in plasma samples from patients with advanced ovarian cancer, showing that this technique can achieve a detection limit of ~2%.15 In parallel, Narayan and colleagues developed an amplicon-based method that incorporates an error suppression approach. Focusing on hotspots in EGFR, BRAF, and KRAS, they showed they could detect mutations in these genes in the plasma of some lung cancer patients.35 Finally, Kinde et al. developed an amplicon-based approach called the Safe-Sequencing System (Safe-SeqS), which employs a barcoding approach to suppress errors and thus increase sensitivity while maintaining high specificity.36 This method was recently successfully applied to detect ctDNA in a large variety of tumor types.13 Key advantages of the amplicon-based approaches are that they are relatively inexpensive and sensitive, especially if error suppression strategies are employed. Limitations of these approaches include interrogation of relatively few genomic loci due to difficulties related to multiplexing of PCR assays, the inability to detect somatic copy number alterations (SCNAs), and the inability to detect breakpoints or rearrangements without patient-specific optimization of amplicons.
The “broad” NGS-based ctDNA detection methods include whole exome sequencing (WES) and whole genome sequencing (WGS). WES involves sequencing of all exons within the genome using approaches routinely employed for tumor genotyping. Murtaza et al. published the first application of this approach and conducted serial sampling of plasma from 6 patients with metastatic breast, ovarian and non-small-cell lung cancer who were undergoing systemic therapy. Multiple time points were analyzed and samples were preselected to contain high percentages of ctDNA (between 5% and 55%). Results revealed that many of the mutations detected in the plasma matched those in biopsies of metastatic deposits. Additionally, they found that new mutations arose as patients developed resistance to therapy.37 The major limitation of WES for ctDNA analysis is that it requires relatively high ctDNA burden in order to be economical and thus currently has potential utility in the research setting rather than in routine clinical use.
Finally, several groups have performed WGS directly on cfDNA from cancer patients. Leary et al. used WGS and identified copy number alterations and/or chromosomal rearrangements in 9 of 10 patients with colorectal and breast cancers, with ctDNA fractions ranging between 1.4% and 48% of total cfDNA. In a modeling exercise, maximum sensitivity of this approach was predicted to be >90% at a specificity >99% when ctDNA levels were ≥0.75%.38 Other groups have similarly performed WGS on plasma samples from patients with metastatic prostate cancer or hepatocellular carcinoma, indicating that this approach is widely applicable.39,40 Strengths of the WGS approach include that it does not restrict the sequencing space. Limitations include high cost due to the amount of sequencing required, a relatively high analytical sensitivity limit of ~1%, and dependence primarily on copy number variation analysis with inadequate sensitivity to detect specific single nucleotide variations (SNVs), insertions or deletions (indels), or rearrangments when ctDNA concentrations are not very high.38,39
Development of CAPP-Seq
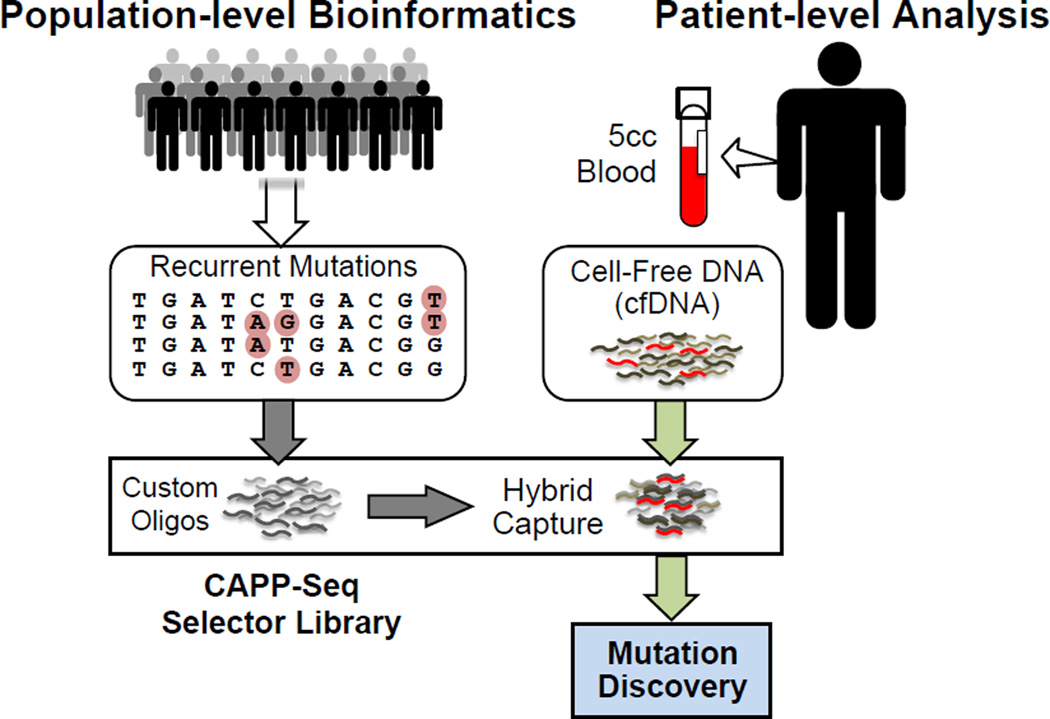
In order to take advantage of the key strengths of both the “focused” and “broad” ctDNA detection approaches described above, we recently developed a novel method called Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq). This approach allows ultra-specific and -sensitive quantification of tumor-derived ctDNA (detection limit ~0.01%), is economical, and is generalizable to nearly all tumor types (Figure 2).16 The method allows detection of SNVs, indels, SCNAs, and gene rearrangements in a single assay. CAPP-Seq employs a multi-phase bioinformatics approach using existing genomic data to design a variably sized “selector” consisting of tagged nucleic acids that allow enrichment for recurrently mutated regions in the cancer of interest via hybrid capture.16,23 The same selector is applied to all patients with a given tumor type, allowing identification of multiple mutations per tumor without the need for personalization of assays. To use CAPP-Seq for detecting or monitoring ctDNA, a sequencing library is prepared from cfDNA, the selector is applied to enrich for genomic regions of interest, and the resulting enriched library is subjected to NGS.
Figure 2. Design and Implementation of CAPP-Seq.
Bioinformatic analysis of cancer WES or WGS data is used to select recurrently mutated genomic regions for inclusion in a CAPP-Seq selector consisting of biotinylated oligonucleotides. The same CAPP-Seq selector can then be applied in nearly all patients with a given cancer type. The selector is used for hybrid capture followed by next-generation sequencing of circulating tumor DNA. WES, whole exome sequencing; WGS, whole genome sequencing; CAPP-Seq, Cancer Personalized Profiling by Deep Sequencing.
We initially developed CAPP-Seq for NSCLC and showed that in our initial cohort it detected ctDNA in 100% of patients with stage II–IV and in 50% of patients with stage I disease, with 96% specificity.16 Strengths of CAPP-Seq include that it can simultaneously cover thousands of distinct genomic regions, can detect all major classes of somatic alterations, does not require prior knowledge of a tumor’s mutations due to its breadth of coverage, does not require patient-specific optimization, is economical, and has very high analytic sensitivity. Limitations include that it is somewhat more time intensive than amplicon-based approaches and that, similar to WES or the focused NGS-based methods, it can only detect alterations in genomic regions that are covered.
Applications of ctDNA analysis in radiation oncology
The development of novel methods for ctDNA detection offers the opportunity to investigate the potential clinical utility of incorporating ctDNA analysis into treatment strategies for patients receiving radiotherapy. One major advantage of ctDNA compared to other cancer biomarkers is that it is easily generalizable. For example, protein-based biomarkers depend on the identification of specific proteins that are overexpressed by a cancer of interest and subsequent optimization of detection strategies that are usually antibody-based. For this reason, a protein-based biomarker for one cancer type will usually not readily extend to other cancer types. In contrast, since all cancers contain somatic alterations, analysis of ctDNA could easily be applied to nearly any tumor type. Thus, it is possible that successful integration of ctDNA analysis into treatment paradigms for one cancer type could allow for rapid extension of the approach to many other cancer types. We envision a number of different potential applications of ctDNA assessment in patients treated with radiotherapy and will describe these below.
Treatment response assessment
One potential application of ctDNA detection in radiation oncology is for treatment response assessment. Currently, imaging represents the mainstay of response assessment after radiation therapy. However, surveillance imaging is not without risks; the tests typically performed, CT and/or PET-CT, expose patients to additional ionizing radiation which, although significantly lower than therapeutic radiotherapy doses, often expose a larger fraction of the body and for patients with frequent surveillance and/or long follow-up can summate to a clinically meaningful risk of carcinogenesis.41–43 Indeed, for an increasing number of diseases, the risks of surveillance imaging are felt to potentially outweigh the benefits.44 Additionally, post- radiotherapy imaging studies are often difficult to interpret due to normal tissue changes such as scarring induced by treatment. This may complicate the interpretation of post-treatment scans and lead to significant subjectivity in determining treatment response or disease status.
As opposed to imaging studies, analysis of ctDNA produces unambiguous numerical results that can be tracked over time. For example, studies in patients with advanced melanoma, ovarian, breast, lung and colon cancers have shown precise tumor burden quantification with ctDNA, with increases in ctDNA concentration strongly correlating with disease progression and ctDNA decline correlating with successful treatment.14–16,32,45,46 Thus, ctDNA analysis appears to offer an attractive adjunct or potentially an alternative to standard follow-up imaging in patients with advanced disease.
Additionally, early results suggest that ctDNA analysis will likely prove to be more sensitive than conventional imaging studies for detection of disease progression. For example, Dawson et al. showed that in metastatic breast cancer patients with rising ctDNA levels during surveillance, progression was identified on average 5 months before imaging.14 Similarly, several recent studies focusing on EGFR mutant advanced NSCLC have shown that in many cases, progression can be detected via rises in ctDNA months before changes on imaging studies.18,47 Thus, ctDNA quantification offers several potential benefits over standard imaging for cancer surveillance.
Few studies to date have explored the application of ctDNA analysis for surveillance after radiation therapy. We recently provided several examples of using CAPP-Seq to distinguish between normal tissue changes and residual disease in NSCLC patients treated with fractionated radiation therapy or stereotactic ablative radiotherapy.16 These results suggest that analysis of ctDNA may have clinical utility by aiding the interpretation of post-RT imaging studies. Alternatively, if it is shown that ctDNA analysis has sufficient sensitivity, one could envision using it as the primary surveillance assay and performing imaging only when there is evidence of increasing ctDNA levels or patients develop new symptoms. The latter approach is analogous to the use of prostate specific antigen to follow patients treated with definitive radiotherapy for prostate cancer and thus has clinical precedent.
Detection of Minimal Residual Disease
The concept of minimal residual disease (MRD) is well established in hematologic malignancies and refers to microscopic deposits of malignant cells present after therapy in patients who do not have any symptoms or imaging findings confirming their presence. MRD is responsible for tumor relapse and can only be detected with modern and extremely sensitive molecular methods.48 While similar methods currently do not exist for most solid tumors, quantification of ctDNA could potentially allow for the detection of MRD after treatment with radiotherapy, because it is possible that patients with residual ctDNA after definitive treatment will be enriched for patients who will ultimately develop recurrence. One example that suggests this may be feasible comes from a recent study of ctDNA in patients with Stage II colorectal cancer treated with surgery. Tie et al. reported that in their cohort, 5 out of 6 patients with detectable post-operative ctDNA developed recurrence while only 5 out of 72 patients with no residual ctDNA recurred.49 These data suggest that residual ctDNA after curative intent treatment may provide a means to identify the subset of patients at highest risk for recurrence.
The most significant body of literature indicating that a similar approach may be fruitful in patients treated with RT comes from the analysis of circulating Epstein Barr virus (EBV) DNA in patients with nasopharyngeal carcinoma (NPC) treated with chemoradiotherapy. Since NPC is strongly associated with EBV, circulating EBV DNA levels are extremely high in many patients before RT and can be used as a biomarker to assess treatment response. Successful treatment results in loss of detectable circulating EBV DNA, while detection of EBV DNA in the circulation after therapy portends significantly worse overall and relapse-free survival.50–53 Other studies have similarly quantitated circulating human papillomavirus DNA in head and neck cancer patients.54,55 Analysis of ctDNA from other cancers is analogous to the analysis of circulating EBV and HPV DNA from head and neck cancers, except that the analyte comes from the cancer genome instead of a cancer-associated virus. For example, our initial description of CAPP-Seq included an example of a patient with Stage III NSCLC who was definitively treated with chemoradiotherapy.16 Post-treatment imaging revealed an apparent complete response, but ctDNA remained stable, suggesting progression of micrometastatic disease that compensated for the volume of tumor eliminated by radiation therapy. This patient developed metastases in multiple organs several months later, clinically confirming the ctDNA result. Thus, ctDNA analysis could potentially facilitate the identification of patients with MRD after RT. In this scenario, this would facilitate the development of clinical trials that test escalating therapy for patients (e.g. with adjuvant chemotherapy, targeted therapy, or immunotherapy) and de-escalation of therapy for patients without residual disease (e.g. by eliminating adjuvant chemotherapy in clinical situations where it is currently given to all patients).
Analysis of ctDNA kinetics for radiotherapy response prediction
Acute changes in ctDNA concentrations during radiotherapy or immediately following treatment might also have prognostic or predictive value. Since ctDNA release is a result of cell death, cell killing by radiation therapy could potentially be monitored via changes in ctDNA concentration. Given that the half-life of ctDNA in the circulation is on the order of 0.5–2 hours32, only tumor cells that died several hours prior to sample collection contribute to ctDNA levels and early changes in ctDNA levels after initiation of radiation therapy might be useful for predicting the ultimate response to treatment. Indeed, Lo et al. showed that for nasopharyngeal carcinoma treated definitively with radiation therapy, plasma EBV DNA levels rose during the first week of treatment (suggesting an increase in cell death) and subsequently declined (suggesting decreasing tumor burden).56 In some patients the initial rise occurred immediately following treatment initiation, while in others the rise occurred later in the week. It is therefore possible that ctDNA changes early during a course of radiotherapy may predict treatment outcome and that early analysis of ctDNA kinetics during treatment could allow clinicians to modify radiotherapy and/or add adjuvant systemic therapy, thus facilitating delivery of truly personalized radiation therapy regimens.
Non-invasive tumor genotyping
The application of ctDNA analysis that is likely to achieve routine clinical use first is tumor genotyping via the plasma, an approach referred to as non-invasive tumor genotyping. This application is particularly attractive in situations where tumor biopsy is medically risky or all biopsy tissue was exhausted in establishing a diagnosis, which are two scenarios frequently encountered in patients who are candidates for radiation therapy. Additionally, since blood draws are easily repeated, non-invasive tumor genotyping via ctDNA analysis allows monitoring of genomic evolution of tumors over the course of therapy.
Numerous studies have provided evidence that this application is feasible. For example, Murtaza et al. performed serial ctDNA analysis for patients with advanced cancers using WES, and showed emergence of mutant alleles at the time of treatment failure, including a PIK3CA activating mutation following paclitaxel treatment in one patient and a truncating RB1 mutation in a patient who developed acquired resistance to cisplatin.37 Analysis of ctDNA has also been used to identify specific somatic resistance mutations, primarily in EGFR and KRAS, following anti-EGFR therapy in lung and colorectal cancer patients.13,22,33,34 For example, Diaz et al. showed that KRAS mutations arise in previously wild-type colorectal cancer patients 5–6 months following treatment initiation with anti-EGFR monoclonal antibodies.33 Similarly, Misale et al. showed that in such patients, resistance mutations could be detected in the plasma as early as 10 months before radiographic disease progression.34 Thus, ctDNA analysis provides a noninvasive approach for monitoring evolution of the tumor genome and predicting resistance to systemic therapy. Detection of resistance mutations prior to clinical progression provides an opportunity for exploring if early intervention, by for example adding another agent that targets the resistance mutations or using an agent with a different resistance profile, could improve outcomes.
Non-invasive genotyping could also be useful for identifying currently unexplored resistance mechanisms. This is particularly true for radiotherapy, where resistance has been linked to mutations in pro-survival, tumor suppressor, reactive oxygen species, cell cycle checkpoint, and telomerase pathways.57–59 However, there is a paucity of literature comparing mutation profiles in tumors before radiation therapy and after recurrence, in large part due to the difficulty of obtaining tumor tissue at both time points. Thus, use of ctDNA analysis in patients whose tumors recur after definitive radiation therapy could enable the identification of novel resistance mechanisms and ultimately lead to strategies for improving radiation therapy outcomes.
Conclusions and Future Directions
Analysis of ctDNA is a noninvasive approach to detect, genotype and quantify tumor burden that has the potential to be incorporated into the practice of radiation oncology. Clinical applications include prediction of radiation treatment response, post-treatment disease surveillance, and resistance mutation detection. We envision that in the future, ctDNA assays may be used to personalize radiation therapy treatment based on ctDNA levels, kinetics and tumor genetics. In the future, ctDNA analysis may also complement or in some cases supplement traditional imaging and pathology-based staging systems for risk stratification and selection of therapy. Given the large number of potential applications, it is anticipated that there will be a significant number of studies published on ctDNA in the upcoming years exploring the utility of ctDNA in a variety of different clinical contexts. Identification of the optimal technical approach for ctDNA assessment in different clinical situations will be critical. Finally, as with any proposed cancer biomarker, well-designed and prospective clinical trials will need to be performed to fully explore the clinical utility of ctDNA in patients treated with radiation therapy.
Acknowledgements
This work was supported by the US Department of Defense (M.D.), the US National Institutes of Health Director’s New Innovator Award Program (M.D.; 1-DP2-CA186569), the Ludwig Institute for Cancer Research (M.D., A.A.A.), the CRK Faculty Scholar Fund (M.D.), and the Radiological Society of North America (E.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial & competing interests disclosure
MD and AA are inventors on patent applications related to the CAPP-Seq technology and are co-founders and consultants for CAPP Medical. No writing assistance was utilized in the production of this manuscript.
References
- 1.Mandel P, Metais P. *Les Acides Nucleiques Du Plasma Sanguin Chez Lhomme. Cr Soc Biol. 1948;142:241–243. [PubMed] [Google Scholar]
- 2.Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer research. 1975;35:2375–2382. [PubMed] [Google Scholar]
- 3.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clinica chimica acta; international journal of clinical chemistry. 2001;313:139–142. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 4.Rogers JC, Valeri CR, Boldt D, Kornfeld S. Excretion of DNA by Phytohemagglutinin-Stimulated Lymphocytes. J Clin Invest. 1972;51 doi: 10.1073/pnas.69.7.1685. A80-&. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wimberger P, Roth C, Pantel K, Kasimir-Bauer S, Kimmig R, Schwarzenbach H. Impact of platinum-based chemotherapy on circulating nucleic acid levels, protease activities in blood and disseminated tumor cells in bone marrow of ovarian cancer patients. International journal of cancer Journal international du cancer. 2011;128:2572–2580. doi: 10.1002/ijc.25602. [DOI] [PubMed] [Google Scholar]
- 6.Boddy JL, Gal S, Malone PR, Harris AL, Wainscoat JS. Prospective study of quantitation of plasma DNA levels in the diagnosis of malignant versus benign prostate disease. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:1394–1399. doi: 10.1158/1078-0432.CCR-04-1237. [DOI] [PubMed] [Google Scholar]
- 7.Jung K, Stephan C, Lewandowski M, et al. Increased cell-free DNA in plasma of patients with metastatic spread in prostate cancer. Cancer letters. 2004;205:173–180. doi: 10.1016/j.canlet.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Kamat AA, Baldwin M, Urbauer D, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer. 2010;116:1918–1925. doi: 10.1002/cncr.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarzenbach H, Stoehlmacher J, Pantel K, Goekkurt E. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Annals of the New York Academy of Sciences. 2008;1137:190–196. doi: 10.1196/annals.1448.025. [DOI] [PubMed] [Google Scholar]
- 10.Atamaniuk J, Vidotto C, Tschan H, Bachl N, Stuhlmeier KM, Muller MM. Increased concentrations of cell-free plasma DNA after exhaustive exercise. Clinical chemistry. 2004;50:1668–1670. doi: 10.1373/clinchem.2004.034553. [DOI] [PubMed] [Google Scholar]
- 11.Laktionov PP, Tamkovich SN, Rykova EY, et al. Extracellular circulating nucleic acids in human plasma in health and disease. Nucleosides, nucleotides & nucleic acids. 2004;23:879–883. doi: 10.1081/NCN-200026035. [DOI] [PubMed] [Google Scholar]
- 12.Lo YM, Rainer TH, Chan LY, Hjelm NM, Cocks RA. Plasma DNA as a prognostic marker in trauma patients. Clinical chemistry. 2000;46:319–323. [PubMed] [Google Scholar]
- 13.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Science translational medicine. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. The New England journal of medicine. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 15.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Science translational medicine. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 16.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nature medicine. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature reviews Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 18.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer research. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 20.Choi JJ, Reich CF, 3rd, Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology. 2005;115:55–62. doi: 10.1111/j.1365-2567.2005.02130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 23.Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30:3390–3393. doi: 10.1093/bioinformatics/btu549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milbury CA, Li J, Makrigiorgos GM. PCR-based methods for the enrichment of minority alleles and mutations. Clinical chemistry. 2009;55:632–640. doi: 10.1373/clinchem.2008.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1994;3:67–71. [PubMed] [Google Scholar]
- 26.Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. British journal of haematology. 1994;86:774–779. doi: 10.1111/j.1365-2141.1994.tb04828.x. [DOI] [PubMed] [Google Scholar]
- 27.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nature medicine. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- 29.Daniotti M, Vallacchi V, Rivoltini L, et al. Detection of mutated BRAFV600E variant in circulating DNA of stage III-IV melanoma patients. International journal of cancer Journal international du cancer. 2007;120:2439–2444. doi: 10.1002/ijc.22598. [DOI] [PubMed] [Google Scholar]
- 30.Morgan SR, Whiteley J, Donald E, et al. Comparison of KRAS Mutation Assessment in Tumor DNA and Circulating Free DNA in Plasma and Serum Samples. Clinical medicine insights Pathology. 2012;5:15–22. doi: 10.4137/CPath.S8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical chemistry. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nature medicine. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz LA, Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narayan A, Carriero NJ, Gettinger SN, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer research. 2012;72:3492–3498. doi: 10.1158/0008-5472.CAN-11-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 38.Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Science translational medicine. 2012;4:162ra54. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heitzer E, Ulz P, Belic J, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome medicine. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clinical chemistry. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 41.Tarin TV, Sonn G, Shinghal R. Estimating the risk of cancer associated with imaging related radiation during surveillance for stage I testicular cancer using computerized tomography. The Journal of urology. 2009;181:627–632. doi: 10.1016/j.juro.2008.10.005. discussion 32-3. [DOI] [PubMed] [Google Scholar]
- 42.Frush DP, Donnelly LF, Rosen NS. Computed tomography and radiation risks: what pediatric health care providers should know. Pediatrics. 2003;112:951–957. doi: 10.1542/peds.112.4.951. [DOI] [PubMed] [Google Scholar]
- 43.Fazel R, Krumholz HM, Wang Y, et al. Exposure to low-dose ionizing radiation from medical imaging procedures. The New England journal of medicine. 2009;361:849–857. doi: 10.1056/NEJMoa0901249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pingali SR, Jewell SW, Havlat L, et al. Limited utility of routine surveillance imaging for classical Hodgkin lymphoma patients in first complete remission. Cancer. 2014;120:2122–2129. doi: 10.1002/cncr.28698. [DOI] [PubMed] [Google Scholar]
- 45.Shinozaki M, O'Day SJ, Kitago M, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:2068–2074. doi: 10.1158/1078-0432.CCR-06-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bidard FC, Madic J, Mariani P, et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. International journal of cancer Journal international du cancer. 2014;134:1207–1213. doi: 10.1002/ijc.28436. [DOI] [PubMed] [Google Scholar]
- 47.Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer. 2014;120:3896–3901. doi: 10.1002/cncr.28964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schrappe M. Detection and management of minimal residual disease in acute lymphoblastic leukemia. Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2014;2014:244–249. doi: 10.1182/asheducation-2014.1.244. [DOI] [PubMed] [Google Scholar]
- 49.Tie JKI, Wang Y, et al. Circulating tumor DNA (ctDNA) as a marker of recurrence risk in stage II colon cancer (CC) Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32(suppl) abstr 11015. [Google Scholar]
- 50.Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. The New England journal of medicine. 2004;350:2461–2470. doi: 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
- 51.Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer research. 1999;59:1188–1191. [PubMed] [Google Scholar]
- 52.Wang WY, Twu CW, Chen HH, et al. Long-term survival analysis of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA levels. Cancer. 2013;119:963–970. doi: 10.1002/cncr.27853. [DOI] [PubMed] [Google Scholar]
- 53.Twu CW, Wang WY, Liang WM, et al. Comparison of the prognostic impact of serum anti-EBV antibody and plasma EBV DNA assays in nasopharyngeal carcinoma. International journal of radiation oncology, biology, physics. 2007;67:130–137. doi: 10.1016/j.ijrobp.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 54.Cao H, Banh A, Kwok S, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. International journal of radiation oncology, biology, physics. 2012;82:e351–e358. doi: 10.1016/j.ijrobp.2011.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahn SM, Chan JY, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA otolaryngology-- head & neck surgery. 2014;140:846–854. doi: 10.1001/jamaoto.2014.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lo YM, Leung SF, Chan LY, et al. Kinetics of plasma Epstein-Barr virus DNA during radiation therapy for nasopharyngeal carcinoma. Cancer research. 2000;60:2351–2355. [PubMed] [Google Scholar]
- 57.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 58.Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morrison R, Schleicher SM, Sun Y, et al. Targeting the mechanisms of resistance to chemotherapy and radiotherapy with the cancer stem cell hypothesis. Journal of oncology. 2011;2011:941876. doi: 10.1155/2011/941876. [DOI] [PMC free article] [PubMed] [Google Scholar]