Abstract
The extremely high prevalence of HIV/AIDS in sub-Saharan Africa and limitations of current antiretroviral medicines demand new tools to optimize therapy such as pharmacogenomics for person-to-person variations. African populations exhibit greater genetic diversity than other world populations, thus making it difficult to extrapolate findings from one population to another. Nevirapine, an antiretroviral medicine, displays large plasma concentration variability which adversely impacts therapeutic virological response. This study, therefore, aimed to identify sources of variability in nevirapine pharmacokinetics and pharmacodynamics, focusing on genetic variation in CYP2B6 and CYP1A2. Using a cross-sectional study design, 118 HIV-infected adult Zimbabwean patients on nevirapine-containing highly active antiretroviral therapy (HAART) were characterized for three key functional single nucleotide polymorphisms (SNPs), CYP2B6 c.516G>T (rs3745274), CYP2B6 c.983T>C (rs28399499), and CYP1A2 g.-163C>A (rs762551). We investigated whether genotypes at these loci were associated with nevirapine plasma concentration, a therapeutic biomarker, and CD4 cell count, a biomarker of disease progression. CYP2B6 and CYP1A2 were chosen as the candidate genes based on reports in literature, as well as their prominence in the metabolism of efavirenz, a drug in the same class with nevirapine. Nevirapine plasma concentration was determined using LC-MS/MS. The mean nevirapine concentration for CYP2B6 c.516T/T genotype differed significantly from that of 516G/G (p < 0.001) and 516G/T (p < 0.01) genotypes, respectively. There were also significant differences in mean nevirapine concentration between CYP2B6 c.983T > C genotypes (p = 0.04). Importantly, the CYP1A2 g.-163C>A SNP was significantly associated with the pharmacodynamics endpoint, the CD4 cell count (p = 0.012). Variant allele frequencies for the three SNPs observed in this Zimbabwean group were similar to other African population groups but different to observations among Caucasian and Asian populations. We conclude that CYP2B6 c.516G>T and CYP2B6 c.983T>C could be important sources of nevirapine pharmacokinetic variability that could be considered for dosage optimization, while CYP1A2 g.-163C>A seems to be associated with HIV disease progression. These inter- and intra-population pharmacokinetic and pharmacodynamics differences suggest that a single prescribed dosage may not be appropriate for the treatment of disease. Further research into a personalized nevirapine regimen is required.
Introduction
Highly Active Antiretroviral Therapy (HAART) has changed the prognosis of HIV infection from a fatal disease to a chronic illness. Effective antiretroviral (ARV) therapy largely depends on adequate drug exposure to suppress viral replication and allow the immune system to recover (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2009). However, approximately 35% of HIV-infected patients on HAART still fail to achieve durable virological suppression, while 45%–80% of those who do achieve virological suppression develop toxicities related to the ARV drugs (Cressey and Lallemant, 2007; Dandara et al., 2014a; Smyth et al., 2012; World Health Organization, 2010). These differences in treatment outcomes are attributed to inter-individual differences in the metabolism of ARV drugs (Pozniak et al., 2011; Rivero et al., 2007).
There are several ARV drugs for different stages of the HIV life cycle. Nevirapine is one of the major components of first-line HAART and has been shown to exhibit huge differences in the plasma concentration achieved in different patients on the same dose (Daar and Singer, 2005; Sánchez et al., 2011). Genetic variation in drug metabolizing enzymes and transporters has been shown to contribute to inter-individual variability in drug therapy (Ingelman-Sundberg et al., 2007; Kesselring et al., 2009), and has been of intensive focus and debate in the H3Africa initiative on knowledge translation in an African continental context (Ahmed et al. 2014; Dandara et al., 2014b).
The distribution of genetic variants differs between populations and there is a lack of consensus on the role of many genetic variants that are linked to drug metabolism (Campbell and Tishkoff, 2008). African populations have largely been omitted in drug pharmacogenomics studies, but in the few studies done, African populations have been shown to exhibit the greatest genetic diversity (Alessandrini and Pepper, 2014).
In Zimbabwe, HAART is administered as a first-line triple therapy consisting of tenofovir (TDF), nevirapine (NVP) or efavirenz (EFV), and lamivudine (3TC). According to the Ministry of Health and Child Care, NVP is the primary constituent of HAART in nearly 80% of HIV-infected Zimbabweans on treatment. NVP is the most commonly used non-nucleoside reverse transcriptase inhibitor (NNRTI) in resource-limited countries due to its relatively low cost, high potency, and relative safety in women of child-bearing age (Guay et al., 1999; Jamisse et al., 2007). NVP is administered orally as a 200 mg twice daily dose and mainly metabolized to 3- and 8-hydroxynevirapine by CYP2B6 and 2- and 12-hydroxynevirapine by CYP3A4/5 before undergoing glucuronidation and excretion (>60%) through feces (Riska et al., 1999). CYP2B6 exhibits genetic polymorphisms that affect hepatic expression and the activity of the coded enzyme, which in turn leads to inter-individual variability in the metabolism of substrate drugs (Turpeinen et al., 2006).
Two single nucleotide polymorphisms (SNPs) in the CYP2B6 gene, c.516G>T (rs3745274) and c.983T>C (rs28399499), have been shown to affect CYP2B6 enzyme activity and NVP plasma concentration (Dickinson et al., 2014; Hoffman et al., 2001). However, this has yet to be demonstrated for the Zimbabwean population. Both variants, one in exon 4, CYP2B6 c.516T (p. 172H), and the second in exon 7, CYP2B6 c.983C (p. 328Thr), have been associated with a significant loss of function of the resultant protein, and therefore higher levels of NVP compared to the alternative alleles (Code et al., 1997; Dickinson et al., 2014; Gounden et al., 2010; Haas et al., 2009; Hofmann et al., 2008; Rotger et al., 2007).
Individuals carrying the CYP2B6 c.516T and 983C alleles are slow metabolizers with respect to CYP2B6 substrate drugs, especially the widely studied efavirenz. An association has been reported between NVP-induced hypersensitivity and the CYP2B6 c.983C allele among Mozambican (Ciccacci et al., 2013) and Malawian (Carr, 2014) patients on NVP-HAART. Hepatotoxicity leads to elevated levels of the hepatic alanine aminotransferase (ALT) as a result of hepatic cell damage. The CYP2B6 c.516G>T and c.983T>C genetic polymorphisms have also been associated with increased NVP plasma concentration and a higher incidence of hypersensitivity and hepatotoxicity during initial treatment (Dickinson et al., 2014; Murphy, 2003; Murphy and Montaner, 1996).
CYP1A2 has been reported to play some role in the metabolism of both NVP and EFV. The major polymorphism in the CYP1A2 gene is a CYP1A2 g.-163C>A SNP where the CYP1A2. g-163A variant has been associated with increased CYP1A2 inducibility, which potentially leads to increased CYP1A2 enzyme levels and increased clearance of CYP1A2 substrate drugs, including EFV and NVP (Aklillu et al., 2003; Basvi et al., 2007; Corchero et al., 2001; Han et al., 2002; Swart et al., 2012). Up to a 70-fold difference in CYP1A2 activity and a 15-fold difference in mRNA expression levels has been reported among individuals (Ikeya et al., 1989; Schweikl et al., 1993). Given the variability in CYP1A2 expression and the effects of the CYP1A2g.-163A>C SNP in EFV metabolism, the role of CYP1A2g.-163C>A SNP on NVP concentration was investigated.
The high prevalence of HIV infection in sub-Saharan Africa necessitates an understanding of the distribution of pharmacogenetically relevant alleles in populations of African origin, and their impact on HAART metabolism. The current study investigated the role of genetic variation in CYP2B6 and CYP1A2 on NVP plasma concentration, CD4 cell recovery, and alanine aminotransferase (ALT) levels among HIV-infected Zimbabwean adults on NVP-containing HAART.
Methods
Study participants and sample collection
118 unrelated, HIV-infected adult Zimbabweans of Bantu origin of which 81% (n = 96) were females; self-identified as Shona for three generations, on HAART were recruited from Newlands Clinic in Harare, Zimbabwe, to participate in a cross-sectional study. Ethical and study approval was provided by Parirenyatwa Hospital and University of Zimbabwe Joint Research Ethics Committee, as well as the Medical Research Council of Zimbabwe. This study was carried out in accordance with the guidelines of the Helsinki Declaration of 2008 and written informed consent was obtained from each participant before sample collection. Patient inclusion criteria included: (1) receiving a 200 mg twice daily NVP dose and; (2) having received NVP as a component of HAART for at least 24 weeks with reported compliance to HAART. Individuals with known liver diseases were excluded from the study.
A questionnaire was used to collect participant demographics and clinical parameters were obtained from the participants' medical records. A 4 mL blood sample was collected from each participant 6–9 hours post-dose into an EDTA-containing blood-collection tube. Plasma was collected from the whole blood sample (centrifugation at 3000 g for 10 min) within 12 hours of bleeding and stored at −80°C for NVP levels measurement, while the remainder of the sample was used for DNA isolation. Genotyping of SNPs was performed using PCR-restriction fragment length polymorphism (PCR/RFLP) and SNaPshot™ mini-sequencing.
Determination of plasma NVP concentration for correlation analysis
Steady-state plasma NVP levels were determined by LC-MS/MS (Division of Clinical Pharmacology, University of Cape Town). The assay was validated according to FDA and EMA guidelines. Plasma samples were extracted and chromatographic separation was achieved on a Luna 5 μm PFP (2), 100 A, 50 × 2 mm analytical column. An AB Sciex API 4000 mass spectrometer was operated at unit resolution in the multiple reaction monitoring (MRM) mode, monitoring the transition of the protonated molecular ions at m/z 266.9 to the product ions at m/z 198.2 for NVP, and monitoring the transition of the protonated molecular ions at m/z 270.1 to the product ions m/z 229.1 for the stable isotope-labeled NVP internal standard. The calibration curve fitted a quadratic (weighted by 1/concentration2) regression over the ranges 0.0195–20.0 μg/mL. The lower limit of NVP quantification was 0.0195 μg/mL.
Genotyping for CYP2B6 c.516G>T and CYP2B6 c.983T>C single nucleotide polymorphisms
Genomic DNA was extracted from blood samples using the GenElute™ Blood Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA) according to manufacturer's instructions. Genotyping for CYP2B6 c.516G>T was performed according to the method by Lang et al. (2001) and CYP2B6 c.983T>C as reported by Swart et al. (2013).
CYP2B6 c.516G>T genotyping involved amplification of a 526 bp fragment by polymerase chain reaction (PCR) and subsequently genotyping using RFLP. The PCR contained 0.4 μM of each of the primers, CYP2B6-4F: 5′-GGTCTGCCCATCTATAAAC-3′ and CYP2B6-4R: 5′-CTGATTCTTCACATGTCTGCG-3′, 0.2 mM dNTPs (Bioline, UK), 1X GoTaq Flexi Green buffer (Promega, Madison, WI), 1.5 mM MgCl2 (Promega), 0.5 U GoTaq Flexi DNA polymerase (Promega), and 100 ng genomic DNA in a total volume of 25 μL.
A “MyCycler™ Thermal Cycler” (Bio-Rad, Hercules, CA) was used for PCR amplification and the cycling conditions were as follows; initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 40 s, and a final extension step at 72°C for 10 min. The digestion reaction contained the following reagents: 10 μL of PCR product, 1X specific restriction buffer (Fermentas Life Sciences, Burlington, Canada), 3U BSeNI restriction enzyme (Fermentas Life Sciences), and made up to a total volume of 30 μL with sdH2O. The digestion reaction was incubated for at least 2 h at 60°C and enzyme was inactivated for 5 min at 80°C. Digestion products were separated on a 2.5% (w/v) agarose gel at 100 V for 60 min and stained with GRGreen. The expected banding pattern for the CYP2B6 c.516G variant was, 17 bp, 241 bp, 268 bp, and that of the CYP2B6 c.516T variant, 17 bp, 509 bp DNA fragments.
CYP2B6 c.983T>C was genotyped using SNaPshot minisequencing and a 2108 bp fragment was amplified on a “MyCyclerTM Thermal Cycler” (Bio-Rad) using the following conditions: initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 2 min, and a final extension at 72°C for 10 min. Each PCR reaction contained the following reagents; 0.40 μM of the CYP2B6-7F: 5′-GTGATTATTCATTAATTGGGTTC-3′ and CYP2B6-8R: 5′-TGCAATGGTTGATTGATGCTC-3′ primers (Integrated DNA Technologies, Inc., Coralville, USA) 100 ng of genomic DNA, 1X Green GoTaq Flexi Reaction Buffer (Promega Corporation), 0.2 mM dNTPs (Bioline, London, UK), 2.5 mM MgCl2 (Promega), 0.5 U of GoTaq Flexi DNA Polymerase (Promega), and made up to a volume of 25 μL with sdH2O.
Individual PCR products were purified using 1.5 U of shrimp alkaline phosphatase (SAP) (Fermentas Life Sciences) and 2 U of Exonuclease I (Fermentas Life Sciences) and incubated at 37°C for 60 min, followed by 75°C for 15 min. SNaPshot™ single base extension was performed using the “GeneAmp® PCR System 9700 version 3.08” (Applied Biosystems, Carlsbad, CA) under the following conditions; 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 30 s. The reaction contained 1 μL ABI Prism® SNaPshot™ Multiplex Kit (Applied Biosystems), 2 μM of the SNP specific internal primer CYP2B6-7FS: 5′-GCGCGACACAGAGAGAGTCTACAGGGAGA-3′ (Integrated DNA Technologies, Inc., Coralville, USA), 1 μL purified PCR product and made up to a volume of 10 μL with sdH2O.
The purification reaction was repeated using 1 U SAP and an ABI 3130xl Genetic Analyzer (Applied Biosystems) was used for capillary electrophoresis with GeneMapper© Software version 4.1 (Applied Biosystems) to analyze results. 1 μL of the SNaPshot product was electrophoresed together with 0.2 μL Liz120 size standard (Applied Biosystems) and 8.8 μL HiDi.
CYP1A2 g.-163C >A genotyping
CYP1A2 g.-163C>A genotyping involved amplification of a 920bp fragment by polymerase chain reaction (PCR) and subsequently genotyping using RFLP according to the method by Pavanello et al., (2005). The PCR contained 0.4 μM of each of the primers, CYP1A2-1F: 5′- CAACCCTGCCAATCTCAAGCA-3′ and CYP1A2-1R: 5′- AGAAGCTCTGTGGCCGAGAAG-3′, 0.2 mM dNTPs (Bioline), 1X GoTaq Flexi Green buffer (Promega), 1.2 mM MgCl2 (Promega), 0.5 U GoTaq Flexi DNA polymerase (Promega), and 100 ng genomic DNA in a total volume of 25 μL.
A “MyCyclerTM Thermal Cycler” (Bio-Rad) was used for PCR amplification and the cycling conditions were as follows; initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 40 s, and a final extension step at 72°C for 10 min. The digestion reaction contained the following reagents: 10 μL of PCR product, 1X specific restriction buffer (Fermentas Life Sciences), 3 U restriction enzyme (ApaI) (Fermentas Life Sciences), and made up to a total volume of 30 μL with sdH2O. The digestion reaction was incubated for at least 2 hours at 60°C and enzyme was inactivated for 5 min at 80°C. Digestion products were separated on a 2.5% (w/v) agarose gel at 100 V for 60 min and stained with GRGreen. The expected banding pattern was 211 bp, 709 bp for the CYP1A2 g-163C variant-and a 920 bp fragment for the CYP1A2 g.-163A variant.
Statistical analysis
Of the 118 participants, 1 was missing the CYP2B6 c.516G>T genotype, 2 others had missing CYP1A2 g.-163C>A genotypes, and another participant was not genotyped for any of the three SNPs. Only one participant was homozygous for the mutant at CYP2B6 c.983T>C and this genotype was consequently treated as missing in the statistical analyses. In addition, one participant did not have sufficient plasma to determine the NVP concentration.
We investigated the effect of the CYP2B6 c.516G>T, CYP2B6 c.983T>C, and CYP1A2 g.-163C>A genotypes on plasma NVP levels using a tobit regression model controlling for gender, age, and body mass index (BMI). The tobit model is appropriate for normally distributed responses with left and right censoring, as was the case with NVP levels. Ordinary least squares regression models were used to determine whether the three SNPs influence CD4-cell count (log transformed) and ALT expression levels (log-log transformed) while controlling for the time since HAART initiation, gender, age, and BMI. We initially fitted each regression model with all of the potential explanatory variables. The models were subsequently refined by systematically dropping variables that did not explain a significant proportion of the variation in the response.
Models were compared using likelihood ratio tests and by examining the Akaike and Bayesian information criteria. In all cases, the residuals were found to be approximately normally distributed and homoscedastic. All analyses were performed in the R Language and Environment for Statistical Computing (R Core Team, 2014). The tobit models were fitted using the tobit function in the AER library. The chi-squared goodness-of-fit test was used to determine if the genotype and allele frequencies were in Hardy-Weinberg equilibrium. The frequencies of genetic variants in the Zimbabwean group were each compared to other populations using the chi-squared goodness-of-fit test.
Results
Demographic characteristics
The demographic characteristics of the study participants are presented in Table 1. While some studies report gender-related differences in terms of virological and immunological response to HAART, others do not (Anderson, 2008; Carr et al., 2014; Dong et al., 2012; Marinho et al., 2013). In this study, we report of an over-representation of females (81%). The majority of participants (93%) received the HAART regimen of tenofovir, lamuvidine, and nevirapine (TDF/3TC/NVP), while AZT replaced TDF in the remaining 7%. An increase in median CD4-cell count was observed from baseline (192.5 cells/mL) to the time of sample collection (435 cells/mL), which was on average 48 months post treatment initiation. The HIV-infected patients had been on treatment from a minimum of 24 months to a maximum of 144 months.
Table 1.
Demographic and Laboratory or Disease Characteristics of Patients on Nevirapine Treatment
| Demographic characteristics | |
|---|---|
| Total number of participants, N | 118 |
| Samples with NVP plasma levels, n/N (freq) | 112/118 (0.95) |
| Samples with NVP plasma conc. <10 μg/mL * | 64/112 (0.57) |
| Samples with NVP plasma conc. <3 μg/mL** | 5/11 (0.04) |
| Sex | |
| Female n (freq) | 96 (0.81) |
| Mean Age in years ± SD (range) | 40.8 ± 9.9 (18–68) |
| Mean BMI ± SD (range) | 24.1 ± 4.8 (14–39) |
| Mean weight in kg ± SD (range) | 64.2 ± 12.3 (41–103) |
| Mean height in m ± SD (range) | 1.62 ± 0.1 (1.41–1.85) |
| HAART combination, n (%) | |
| TDF/3TC/NVP | 110 (0.93) |
| AZT/3TC/NVP | 8 (0.07) |
| Laboratory/disease parameters | |
| Hepatotoxicity & SJS, n (freq) | 4/112 (0.04) |
| Mean ALT in ± SD (range) | 27.3 ± 18.1 (8.2–105) |
| Median duration on HAART in months (IQR) | 48 (24–75) |
| Mean plasma NVP concentration in μg/mL ± SD (range) | 9.41 ± 4 (0.082–19.9) |
| Baseline CD4, median (IQR) | 192.5 (94–292) |
| Median CD4 count at time of sample collection, (IQR) | 435 (293–608) |
| Median baseline p24, (IQR) | 7.2 (1.9–70) |
| Median p24 at time of sample collection (IQR) | 1.1 (0.7–2.0) |
Recommended minimum level confirming adherence; ** minimum level thought to ensure virologic suppression.
ALT, alanine transaminase; AZT, zidovudine; BMI, body mass index; CD4, cluster of differentiation 4; IQR, interquartile range; SD, standard deviation; TDF/3TC/NVP, tenofovir/lamivudine/NVP.
Plasma nevirapine concentration distribution
Nevirapine plasma concentration influences virological response and treatment outcome. At low concentration levels, there is likely to be poor virological suppression, which can lead to drug resistance. On the other extreme, high concentration levels can have adverse drug effects. Of the 118 patients, plasma NVP levels were measurable among 112 patients, one did not have adequate plasma sample for NVP detection and five presented with levels below the limit of quantification.
The mean plasma NVP concentration was 9.41 ± 4.00 μg/mL. Fifty-seven percent (n = 64) of the 112 participants presented with NVP plasma concentrations below the recommended adherence levels of 10 μg/mL (Cohen et al., 2008). Five patients (4%) presented with NVP plasma concentrations below 3 μg/mL (the minimum inhibitory NVP plasma concentration). Four patients reported adverse drug reactions in the form of Steven Johnson syndrome (SJS) + hepatotoxicity (n = 1) and hepatotoxicity only (n = 3) with NVP plasma concentrations of 12.5, 10.7, 12.6, and 4.98 μg/mL each. There was a 126% improvement in the median CD4-cell count from baseline to the time of sampling on average. The mean BMI (24.1 ± 4.8) of the participants at sample collection reflects a population group within the normal range. Some of the antiretroviral drugs (e.g., stavudine) have been associated with lipodystrophy (Kampira et al., 2013).
Correlation of CYP2B6 c.516G>T, CYP2B6 c.983T>C, and CYP1A2 g.-163C>A genotypes with plasma NVP levels
Plasma NVP concentrations and genotype data for CYP2B6 c.516G>T (rs3745274), CYP2B6 c.983T>C (rs28399499), and CYP1A2 g.-163C>A (rs762551) were available for 112 participants (three and two of these had NVP concentrations below and above the limits of quantification, respectively). We used multiple regression analysis to determine if the genotypes at each of the three SNPs were significantly associated with plasma NVP concentration, controlling for age, gender, and BMI (see Methods). The regression results for the most parsimonious model (i.e. omitting predictors that were not statistically significant) are reported in Table 2. The full model including all the predictors is presented in Supplementary Table S1 (supplementary material is available online at www.liebertpub.com/omi).
Table 2.
Final Tobit Regression for Nevirapine Plasma Concentration
| Estimate | Std. Error | P value | |
|---|---|---|---|
| T983C T/C | 3.719 | 1.115 | 0.000854 |
| G516T G/T | 2.016 | 0.965 | 0.036802 |
| G516T T/T | 5.173 | 1.199 | 1.60e-05 |
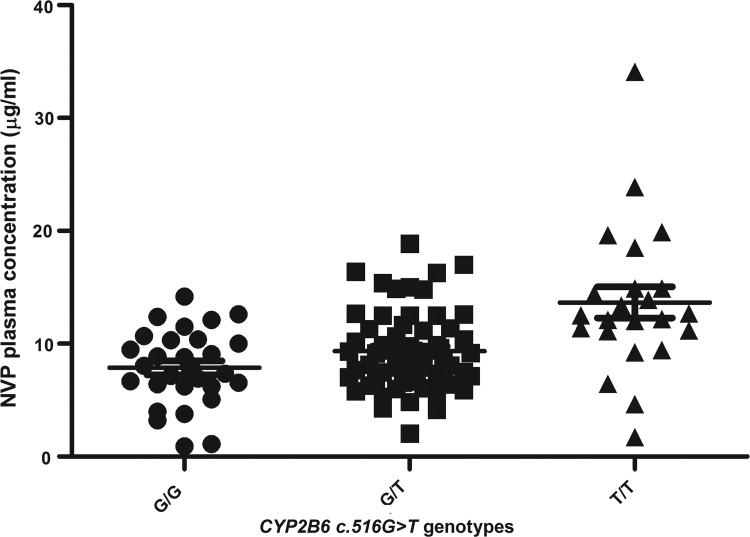
We found that the CYP2B6 516G>T SNP was significantly associated with NVP plasma concentration. The patients carrying the CYP2B6 c.516G/T heterozygous genotype presented with significantly higher mean NVP plasma concentration (p = 0.036) compared to patients carrying the CYP2B6 c.516G/G homozygous genotype. Carriers of the homozygous CYP2B6 c.516T/T genotype had an even higher mean NVP plasma concentration (p < 0.001) compared to the CYP2B6 c.516G/G homozygotes (Table 2; Fig. 1). A genotype-dose effect was observed with NVP plasma concentration increasing with copy number in carriers of the CYP2B6 c.516T allele (i.e, 516T/T >>>516G/T>>516G/G). The CYP2B6 c.983T/C heterozygotes presented with significantly higher mean NVP plasma concentration (p < 0.001) compared to the homozygous CYP2B6 c.983T/T (Table 2).
FIG. 1.
Correlation between CYP2B6 c.516G>T genotypes and nevirapine plasma concentration. CYP2B6 c.516G/G genotype presented with significantly low mean nevirapine plasma concentration compared to the CYP2B6 c.516G/T (p < 0.036) and CYP2B6 c.516G/T (p < 0.001), respectively.
Only one participant presented with the CYP2B6 c.983C/C genotype and had an NVP plasma concentration of 16.3 μg/mL. This was higher than the mean concentrations of 9.30 μg/mL, and 11.93 μg/mL, obtained for the CYP2B6 c.983T/T and CYP2B6 c.983T/C genotype carriers, respectively. The CYP1A2 g.-163C>A SNP did not show a significant association with NVP plasma concentration (Table 2). For NVP plasma concentration, the most parsimonious model only contained CYP2B6 c.516G>T and c.983T>C as significant predictors of NVP plasma concentration (Table 2). These two SNPs were significantly associated with NVP plasma concentrations irrespective of whether the other predictors were included in the model (see Table S1).
Correlation of CYP2B6 c.516G>T, CYP2B6 c.983T>C, and CYP1A2 g.163C>A genotypes with CD4-cell count and ALT
We also performed regression analyses to determine if the genotypes at any of the three SNPs were significantly associated the CD4-cell count and ALT expression levels, controlling for age, gender, BMI, and the number of months since HAART was initiated. The final model containing only significant predictors of CD4-cell count is presented in Table 3. The full model with all potential predictors is shown in Supplementary Table S2.
Table 3.
Final Regression Model for the Logarithm of CD4 Cell Count at Time of Collection
| Estimate | Std. Error | P value | |
|---|---|---|---|
| C163A A/A | −0.309 | 0.123 | 0.01398 |
| C163A C/A | −0.2539 | 0.114 | 0.02902 |
| MSHS | 0.005 | 0.001 | 0.00113 |
| Gender: female | 0.353 | 0.126 | 0.00619 |
BMI, body mass index; MSHS, months since HAART started.
We found that the CYP1A2 g.-163C>A SNP was significantly associated with CD4-cell count at the time of sample collection (the same time that NVP plasma samples were collected). Carriers of the CYP1A2 g.-163C/A (p = 0.029) and CYP1A2 g.-163A/A (p = 0.014) genotypes had significantly lower CD4-cell counts on average when compared to those with the CYP1A2 g.-163C/C genotype. The CD4-cell count was lowest among CYP1A2 g.-163A/A genotype carriers. This suggests that the CYP1A2 g.-163A allele influences CD4-cell recovery.
The simplest model included CYP1A2 g-163C>A genotypes, months since HAART started (MSHS) and gender, and accounted for 15% of the variability in the log-transformed CD4-cell counts (Table 3). Including all the other variables (CYP2B6 c.516G>T, c.983T>C, BMI, and age) in the model only accounted for an additional 3% of the variability (Supplementary Table S2). As expected, MSHS was significantly associated with CD4-cell count; the longer the MSHS, the higher the CD4 cell count (p = 0.001). The two SNPs, CYP2B6 c.516G>T and CYP2B6 c.983T>C, did not correlate with CD4-cell count (Supplementary Table S2). The association between the CYP1A2 g.-163C>A SNP and CD4-cell count was statistically significant, irrespective of whether BMI, age, and the two CYP2B6 SNPs were included in the model.
Alanine aminotransferase (ALT) expression, a measure of liver function, was not significantly associated with the CYP2B6 c.516G>T, CYP2B6 c.983T>C, and CYP1A2 g.163C>A genotypes (Supplementary Table S3). All the above NVP correlations with genotypes could be affected by putative drug interactions. However, the present study is a real-life sample, and hence, very relevant for the naturalistic study of therapeutic variation with a view to pharmacogenomics variation.
Comparison of variant allele frequencies with other world populations
The genotype distribution of the three SNPs conformed to the Hardy-Weinberg equilibrium (HWE); the genotype frequencies are given in Table 4. The three variants, CYP2B6 c.516T, CYP2B6 c.983C, and CYP1A2 g.-163A had the following frequencies, 0.47, 0.08, and 0.53 in this Zimbabwean population.
Table 4.
Comparison of CYP2B6 and CYP1A2 Allele Frequency Distribution in World Populations
| CYP2B6 c.516T (rs3745274) | CYP2B6 c.983C (rs28399499) | CYP1A2 g.-163A (rs762551) | |
|---|---|---|---|
| Zimbabweans (this study) | 0.45 | 0.09 | 0.49 |
| South Africans (Swart 2011) | 0.43 | 0.07 | 0.61a |
| Cameroonians (Swart et al., 2012) | 0.37 | NA | 0.52 |
| Malawians (Carr et al., 2014) | 0.31a | 0.08 | NA |
| Yoruba (Matimba et al., 2008) | 0.42 | 0.12 | 0.57a |
| Tanzanians (Bains et al., 2013) | 0.10a | NA | 0.49 |
| Maasai (Matimba etal., 2008) | 0.37 | 0.01a | 0.52 |
| Luhya (Ikediobi et al,. 2011) | 0.31 | 0.07 | N/A |
| Ugandan (Mukonzo et al., 2009) | 0.36 | 0.10 | NA |
| Ghanaian (Bains et al., 2013; Griese et al. 1999) | 0.48 | 0.07 | NA |
| Caucasians (Hapmap) | 0.30a | 0.00a | 0.71a |
| Asians (Hapmap) | 0.15a | 0.00a | 0.65a |
NA, not available.
Statistically significant differences when compared with Zimbabwean individuals.
Pharmacogenomically relevant genetic variants exhibit qualitative and quantitative differences in their distribution between different population groups. The frequencies of the three variant alleles investigated in the Zimbabwean population in this study, CYP2B6 c.516T, CYP2B6 c.983C, and CYP1A2 g.-163A, were, thus, compared with data from 11 other population groups from across the world, namely black South Africans, Cameroonians, Malawians, Yoruba, Tanzanians, Maasai, Luhya, Ugandans, Ghanaians, Caucasians and Asians. The Zimbabwean population group showed significant differences when compared to Caucasian and Asian population groups' at all three loci (Table 4).
Within African population groups, CYP2B6 c.516T, frequencies differed significantly (p < 0.05) between the Zimbabwean group (0.45) compared to Malawians (0.31) and Tanzanians (0.10). The CYP2B6 c.983C showed significant differences (p < 0.05) when the Zimbabwean group was compared to the Maasai (0.09 versus 0.01) while the CYP1A2 g.-163A variant showed significant differences (p < 0.05) when the Zimbabweans (0.49) were compared to South Africans (0.61) and the Yoruba from Nigeria (0.57).
Discussion
Nevirapine remains one of the cornerstones of HAART, and is extensively used in resource-limited settings because of its relatively low cost, safety in women of child-bearing age, and potency in suppression of HIV. Similar to EFV, NVP is also primarily metabolized to hydroxy-metabolites by CYP2B6 that is coded for by a polymorphic gene (Code et al., 1997). One of the problems associated with taking the 200 mg twice daily standard dose of NVP is emergence of adverse side effects, which have been reported in up to 15% of exposed patients. The adverse effects include Steven-Johnson syndrome (SJS), toxic epidermal necrolysis, and hepatotoxicity (Gatanaga et al., 2007; Steinbrook 2013; Wattanakul et al., 2013).
Anthropometry data and NVP plasma concentration
Some studies have demonstrated an association between gender and NVP plasma concentration and have suggested that it could be due to differential drug biotransformation, fat content, and exogenous and endogenous hormonal expression differences in males and females (Bersoff-Matcha et al., 2001; Dong et al., 2012; Marinho et al., 2013). Expression of liver enzymes responsible for drug metabolism has also been shown to vary with gender and BMI (Antinori et al., 2001; Bersoff-Matcha et al., 2001).
In a comparison between NVP once and twice daily dosing, Schipani et al. (2011) reported significantly increased NVP metabolism when body weight increased. Increased body weight was more associated with subtherapeutic drug exposure in once daily dosing compared to twice daily dosing (Schipani et al., 2011). However, in the present study, gender was not seen to be associated NVP plasma concentration, possibly because our cohort comprised mostly females. Contrary to Schipan et al. (2011) who recruited participants with specific weights of 50, 70, and 90 kg, in this study, selection did not consider weight, and all participants were on a twice daily dosing regimen (the recommended dose in Zimbabwe). We therefore speculate that differences in NVP metabolism due to weight and BMI only become significant within a certain and specific range. Overall, ethnic, diet, genetic, and environmental differences could be major contributors to the observed differences.
In the current study, NVP plasma concentration was not associated with age or BMI, and this is contrary to the observation by Swaminathan et al. (2011) who reported a significant association between stunting and age ≤3 years with subtherapeutic NVP levels. The difference in findings could be due to the differences in surface area to volume ratio between adults and pediatric patients. The children in the study by Swaminathan et al. (2011) were on an adult NVP formulation; hence their findings could suggest nontolerance of NVP adult formulations in pediatrics.
The mean NVP plasma concentration in the study population was 9.41 μg/mL. NVP adherence is measured among stabilized patients as plasma NVP concentration greater than 10 μg/mL which is 10 times the in vitro IC50 for HIV (Desmond et al., 2015). Nevirapine plasma concentration ranged from 0.0824 to 34.1 μg/mL. Despite all participants having reached steady state and taking the same NVP dose (200 mg twice daily), high inter-individual variation in the NVP plasma concentration was still observed; suggesting that universal dosage for NVP in treatment of infectious diseases is not advisable. Notably, five participants (4%) had plasma NVP levels below the established plasma NVP minimum inhibitory concentration (3 μg/mL) and could be at an increased risk of developing drug resistance.
CYP2B6, CYP1A2 genotypes, and NVP levels
Our data showed that CYP2B6 c.516T and 983C variants result in higher drug plasma levels for CYP2B6 substrate drugs. This argues well for the management of HIV with ART, because virological suppression is dependent on sustaining a higher drug plasma concentration. However, beyond certain drug plasma concentration, challenges of adverse drug effects start to emerge. Thus, identifying markers of drug pharmacokinetics, in this case, NVP plasma concentration could assist in promoting the use of appropriate dosages targeted to specific genotypes.
This study contributes to genetic characterization of Zimbabwean populations and determination of the effects of genetic variants on variability in NVP pharmacokinetics. If genotype-assisted dosing achieves the goal of reducing side effects, this will have a positive impact on adherence, thereby lowering the emergence of NVP resistance and treatment failure. In the present study, genetic variations in CYP2B6 gene (516T and 983C alleles) were studied in order to explore their potential role in NVP therapeutics.
The significant influence of CYP2B6 c.516G>T and CYP2B6 c.983T>C on NVP plasma concentration obtained in this study confirm reports for other populations (Dickinson et al., 2014; Rotger et al., 2005) and could assist in optimizing the efficacy of NVP use. The two CYP2B6 polymorphisms could be good candidate SNPs for a future pharmacogenomics diagnostic tool. However, seven individuals who had low NVP levels (predicted to be due to extensive CYP2B6 metabolism), were carrying either the CYP2B6 516T/T (n = 4) or the 983T/C (n = 3) genotypes. This suggests that other genes may play a role in NVP metabolism, if strict adherence to the treatment regimen can be assumed. It is worth noting that the individual with the lowest plasma NVP concentration (0.0824 μg/mL) had been on treatment for 72 months, despite carrying the CYP2B6 c.516T/T genotype. We strongly suspect that this is a result of non-adherence.
CYP1A2 genotype and CD4-cell count
We found a significant association between the CYP1A2 g.-163C>A SNP and CD4-cell count. The CYP1A2 g.-163A variant is associated with increased CYP1A2 expression, which translates into increased availability of the respective enzyme. In the presence of higher levels of CYP1A2 expression, the NVP plasma concentration is likely to be reduced, which supports virus perpetuation and therefore reduced CD4-cell counts as the virus targets CD4+ cells.
Our observation supports earlier findings by Parathyras et al. (2009), who reported two ABCB1 variants associated with improved CD4-cell recovery and functionality. The ABCB1 variants are associated with decreased ABCB1 efflux pump activity, which leads to higher drug concentration levels in cells. Given that CYP1A2 g.-163C variant is associated with lower CYP1A2 expression and therefore high NVP plasma concentration; it might be worth examining individuals with the CYP1A2 g.-163C variant for liver function and other adverse reactions associated with NVP treatment (Fauci and Folkers, 2012; Kappelhoff et al., 2005; Wattanakul et al., 2013). However, ALT expression levels and NVP plasma concentrations were not significantly associated with CYP1A2 g.-163C>A SNP.
NVP levels and adverse drug effects
Studies have found adverse drug reactions and hypersensitivity due to NVP to be present in about 6%–13% of participants (De Maat et al., 2002; Fagot et al., 2001; Kesselring et al., 2009; Wit et al., 2008). In this study, only 3% of the participants reported signs and symptoms of NVP toxicity at some point during therapy with no apparent associations of either elevated ALTs or adverse effects with NVP levels. This conflicts the observation by Dickinson et al. (2014) where NVP hypersensitivity reaction prevalence was not associated with NVP clearance. In Dickinson et al. (2014), the high prevalence of NVP-associated toxicity could have been due to their recruitment of patients who were initiating HAART. In this study, we aimed to observe the long-term effects or complications of NVP.
Among the 46 patients with high NVP concentrations (i.e., concentrations above 10 μg/mL), 12 (26%) presented with ALT levels above the average of the study cohort (27.3 U/L) possibly pointing to underlying liver disease. Elevated NVP plasma concentrations have been associated with elevated ALT levels which is a manifestation of liver injury (Dieterich et al., 2004; Torti et al., 2007). It is surprising to note that individuals with the highest NVP plasma concentrations were not necessarily the ones who experienced SJS or hepatotoxicity, possibly suggesting that NVP toxicity is a rare phenotype.
The absence of a correlation between NVP concentration and toxicity could suggest that it is not the parent drug but rather the biotransformation metabolites that could play an important role in terms of toxicity and adverse drug effects as has been reported previously (Antunes et al., 2010; Wen et al., 2009). It also suggests that, in addition to NVP plasma concentration, the duration of exposure could be an important factor that influences toxicity.
Of the four participants who developed adverse drug effects, three had ALT levels below the cohort mean levels (13.7, 21.5, and 27.1 U/L). We speculate that the participants might also have been on other unreported medications known to cause liver injury but not to elevate ALT levels. A larger sample size may be needed to demonstrate the relationship between NVP concentration and adverse effects more clearly.
Genotype and allele frequencies
Genotype and allele frequencies show significant differences between populations. For example, there is a striking difference in the frequency of CYP2B6 c.983C allele, which among African populations ranges from 1% to 12% but is absent in Asian and Caucasian populations. This finding further confirms that universal treatment of common diseases, as well as extrapolation of findings from Asian and Caucasian populations to Africans, is not always useful (Alessandrini and Pepper, 2014; Svärd et al., 2010). Generally, African populations have been treated as genetically homogenous. For a long time the Yoruba have been used as a proxy, however, there is now evidence of noticeable genetic diversity and differences across African populations (Matimba et al., 2008; Skelton et al., 2014).
Conclusions
Although all participants were on the same NVP dose, we observed large inter-individual differences in plasma NVP levels in our sample. Part of this variability could be explained by a significant association between CYP2B6 c.516T and c.983C gene variants and NVP plasma concentration. This suggests that these gene variants could be potential NVP pharmacokinetics determinants and could be used to determine a suitable dosage for each individual.
The non-uniform distribution of these variants in different populations further speaks to the limits of extrapolating genomic association studies directly to African populations from studies conducted elsewhere. We also found a significant association between CYP1A2 g-163A variant and poor CD4 T-cell recovery Thus, more investigations are needed to find the mechanisms through which CYP1A2 is involved in CD4 cell recovery.
Supplementary Material
Acknowledgments
We are thankful to the participants from Newlands OI Clinic, for none of this would have been possible without their voluntary participation. This work was supported by Medical Research Council (MRC) South Africa and National Research Foundation (NRF) grants to C. Dandara; Grant Number 2U2RTW007367 from the Fogarty International Centre, National Institutes of Health (NIH, USA) through the International Clinical, Operational and Health Services and Training Award (ICOHRTA). We are also indebted to the University of Cape Town for facilitating some of the research work.
We would also like to thank the Letten Foundation, Norway, and the University of Zimbabwe Research Support Centre for funding some of the project research costs. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1 AI068634, UM1 AI068636 and UM1 AI106701. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Ahmed AA, Mohamed AA, Guled IA, Elamin HM, and Abou-Zeid AH. (2014). Knowledge translation in Africa for 21st century integrative biology: The “know-do gap” in family planning with contraceptive use among Somali women. OMICS 18, 696–704 [DOI] [PubMed] [Google Scholar]
- Aklillu E, Carrillo JA, Makonnen E, et al. (2003). Genetic polymorphism of CYP1A2 in Ethiopians affecting induction and expression: Characterization of novel haplotypes with single-nucleotide polymorphisms in intron 1. Mol Pharmacol 64, 659–669 [DOI] [PubMed] [Google Scholar]
- Alessandrini M, and Pepper MS. (2014). Priority pharmacogenetics for the African continent: focus on CYP450. Pharmacogenomics 15, 385–400 [DOI] [PubMed] [Google Scholar]
- Anderson GD. (2008). Gender differences in pharmacological response. Intl Rev Neurobiol 83, 1–10 [DOI] [PubMed] [Google Scholar]
- Antinori A, Baldini F, Girardi E, et al. (2001). Female sex and the use of anti-allergic agents increase the risk of developing cutaneous rash associated with nevirapine therapy. AIDS 15, 1579–1581 [DOI] [PubMed] [Google Scholar]
- Antunes AM, Godinho AL, Martins ISL, Justino GC, Beland FA, and Marques MM. (2010). Amino acid adduct formation by the nevirapine metabolite, 12-hydroxynevirapine—A possible factor in nevirapine toxicity. Chem Res Toxicol 23, 888–899 [DOI] [PubMed] [Google Scholar]
- Bains RK, Kovacevic M, Plaster CA, et al. (2013). Molecular diversity and population structure at the Cytochrome P450 3A5 gene in Africa. BMC Genetics 14, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basvi PT, Dandara C, Bapiro TB, AND Hasler JA. (2007).Role of CYP 1A2*1F on CYP 1A2 activity in a black African population as determined by caffeine phenotyping. J Chin Clin Med 2, 211–214 [Google Scholar]
- Bersoff-Matcha SJ, Miller WC, Aberg JA, et al. (2001). Sex differences in nevirapine rash. Clin Infect Dis 32, 124–129 [DOI] [PubMed] [Google Scholar]
- Campbell MC, and Tishkoff SA. (2008). African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Ann Rev Genomics Human Genetics 9, 403–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr DF, Chaponda M, Castro EMC, et al. (2014). CYP2B6 c. 983T>C polymorphism is associated with nevirapine hypersensitivity in Malawian and Ugandan HIV populations. J Antimicrob Chemother 69, 3329–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccacci C, Di Fusco D, Marazzi MC, et al. (2013). Association between CYP2B6 polymorphisms and Nevirapine-induced SJS/TEN: A pharmacogenetics study. Eur J Clin Pharmacol 69,1909–1916 [DOI] [PubMed] [Google Scholar]
- Code EL, Crespi CL, Penman BW, Gonzalez FJ, Chang TK, and Waxman DJ. (1997). Human cytochrome P4502B6 interindividual hepatic expression, substrate specificity, and role in procarcinogen activation. Drug Metab Dispos 25, 985–993 [PubMed] [Google Scholar]
- Cohen K, van Cutsem G, Boulle A, et al. (2008). Effect of rifampicin-based antitubercular therapy on nevirapine plasma concentrations in South African adults with HIV-associated tuberculosis. J Antimicrob Chemother 61, 389–393 [DOI] [PubMed] [Google Scholar]
- Corchero J, Pimprale S, Kimura S, and Gonzalez FJ. (2001). Organization of the CYP1A cluster on human chromosome 15: Implications for gene regulation. Pharmacogen Genomics 11, 1–6 [DOI] [PubMed] [Google Scholar]
- Cressey TR, and Lallemant M. (2007). Pharmacogenetics of antiretroviral drugs for the treatment of HIV-infected patients: An update. Infect Genet Evolution 7, 333–342 [DOI] [PubMed] [Google Scholar]
- Daar AS, and Singer PA. (2005). Pharmacogenetics and geographical ancestry: Implications for drug development and global health. Nat Rev Genet 6, 241–246 [DOI] [PubMed] [Google Scholar]
- Dandara C, Swart M, Mpeta B, Wonkam A, and Masimirembwa C. (2014a). Cytochrome P450 pharmacogenetics in African populations: Implications for public health. Exp Opin Drug Metab Toxicol 10, 769–785 [DOI] [PubMed] [Google Scholar]
- Dandara C, Huzair F, Borda-Rodriguez A, Chirikure S, Okpechi I, Warnich L, and Masimirembwa C. (2014b). H3Africa and the African life sciences ecosystem: Building sustainable innovation. OMICS 18, 733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maat MM, Mathot RA, Veldkamp AI, et al. (2002). Hepatotoxicity following nevirapine-containing regimens in HIV-1-infected individuals. Pharmacol Res 46, 295–300 [DOI] [PubMed] [Google Scholar]
- Desmond AC, Moodley D, Conolly CA, Castel SA, and Coovadia HM. (2015). Evaluation of adherence measures of antiretroviral prophylaxis in HIV exposed infants in the first 6 weeks of life. BMC Pediatr 15, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson L, Chaponda M, Carr DF, et al. (2014). Population pharmacokinetic and pharmacogenetic analysis of nevirapine in hypersensitive and tolerant HIV-infected patients from Malawi. Antimicrob Agents Chemother 58, 706–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DT, Robinson PA, Love J, and Stern JO. (2004). Drug-induced liver injury associated with the use of nonnucleoside reverse-transcriptase inhibitors. Clin Infect Dis 38, S80–S89 [DOI] [PubMed] [Google Scholar]
- Dong BJ, Zheng Y, Hughes MD, et al. (2012). Nevirapine (NVP) pharmacokinetics (PK) and risk of rash and hepatitis among HIV-infected Sub-Saharan African Women. AIDS 26, 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagot JP, Mockenhaupt M, Bouwes-Bavinck JN, et al. (2001). Nevirapine and the risk of Stevens–Johnson syndrome or toxic epidermal necrolysis. AIDS 15, 1843–1848 [DOI] [PubMed] [Google Scholar]
- Fauci AS, and Folkers GK. (2012). Toward an AIDS-free generation. JAMA 308, 343–344 [DOI] [PubMed] [Google Scholar]
- Gatanaga H, Hayashida T, Tsuchiya K, et al. (2007). Successful efavirenz dose reduction in HIV type 1-infected individuals with cytochrome P450 2B6* 6 and* 26. Clin Infect Dis 45, 1230–1237 [DOI] [PubMed] [Google Scholar]
- Gounden V, Van Niekerk C, Snyman T, and George JA. (2010). Presence of the CYP 2 B 6 516 G>T polymorphism, increased plasma efavirenz concentrations and early neuropsychiatric side effects in South African HIV-infected patients. AIDS Res Ther 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griese EU, Asante-Poku S, Ofori-Adjel D, Mikus G, and Eichelbaum M. (1999). Analysis of the CYP2D6 gene mutations and their consequences for enzyme function in a West African population. Pharmacogen Genomics 9, 715–724 [PubMed] [Google Scholar]
- Guay LA, Musoke P, Fleming T, et at. (1999). Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet 354, 795–802 [DOI] [PubMed] [Google Scholar]
- Haas DW, Gebretsadik T, Mayo G, et al. (2009). Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J Infect Dis 199, 872–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Ouyang D, Chen X, et al. (2002). Inducibility of CYP1A2 by omeprazole in vivo related to the genetic polymorphism of CYP1A2. Br J Clin Pharmacol 54, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman SM, Nelson DR, and Keeney DS. (2001). Organization, structure and evolution of the CYP2 gene cluster on human chromosome 19. Pharmacogen Genomics 11, 687–698 [DOI] [PubMed] [Google Scholar]
- Hofmann MH, Blievernicht JK, Klein K, et al. (2008). Aberrant splicing caused by single nucleotide polymorphism c. 516G>T [Q172H], a marker of CYP2B6* 6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Therapeut 325, 284–292 [DOI] [PubMed] [Google Scholar]
- Ikediobi O, Aouizerat B, Xiao Y, Gandhi M, Gebhardt S, and Warnich L. (2011). Analysis of pharmacogenetic traits in two distinct South African populations. Human Genomics 5, 265–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya K, Jaiswal AK, Owens RA, Jones JE, Nebert DW, and Kimura S. (1989). Human CYP1A2: Sequence, gene structure, comparison with the mouse and rat orthologous gene, and differences in liver 1A2 mRNA expression. Mol Endocrinol 3, 1399–1408 [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Sim SC, Gomez A, and Rodriguez-Antona C. (2007). Influence of cytochrome P450 polymorphisms on drug therapies: Pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Therapeut 116, 496–526 [DOI] [PubMed] [Google Scholar]
- Jamisse L, Balkus J, Hitti J, et al. (2007). Antiretroviral-associated toxicity among HIV-1-seropositive pregnant women in Mozambique receiving nevirapine-based regimens. JAIDS 44, 371–376 [DOI] [PubMed] [Google Scholar]
- Kampira E, Kumwenda J, Van Oosterhout JJ, and Dandara C. (2013). Mitochondrial subhaplogroups and differential risk of stavudine-induced lipodystrophy in Malawian HIV/AIDS patients. Pharmacogenomics 14, 1999–2004 [DOI] [PubMed] [Google Scholar]
- Kappelhoff BS, van Leth F, Robinson PA, et al. (2005). Are adverse events of nevirapine and efavirenz related to plasma concentrations. Antiviral Ther 10, 489–498 [PubMed] [Google Scholar]
- Kesselring AM, Wit FW, Sabin CA, et al. (2009). Risk factors for treatment-limiting toxicities in patients starting nevirapine-containing antiretroviral therapy. AIDS 23, 1689–1699 [DOI] [PubMed] [Google Scholar]
- Marinho AT, Rodrigues PCM, Caixas U, et al. (2013). Differences in nevirapine biotransformation as a factor for its sex-dependent dimorphic profile of adverse drug reactions. J Antimicrob Chemother 69, 476–482 [DOI] [PubMed] [Google Scholar]
- Matimba A, Oluka MN, Ebeshi BU, et al. (2008). Establishment of a biobank and pharmacogenetics database of African populations. Eur J Human Genetics 16, 780–783 [DOI] [PubMed] [Google Scholar]
- Mukonzo JK, Roshammar D, Waako P, et al. (2009). A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single-dose efavirenz population pharmacokinetics in Ugandans. Br J Clin Pharmacol 68, 690–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RL. (2003). Defining the toxicity profile of nevirapine and other antiretroviral drugs. J Acquir Immune Defic Syndr 34, S15–S20 [DOI] [PubMed] [Google Scholar]
- Murphy RL, and Montaner J. (2008). Drug evaluations anti-infectives: Nevirapine: A review of its development, pharmacological profile and potential for clinical use. Expert Opin Investig Drugs 5, 1183–1199 [Google Scholar]
- Panel on Antiretroviral Guidelines for Adults and Adolescents (2009). Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; 1–161 [Google Scholar]
- Pozniak AL, Coyne KM, Miller RF, et al. (2011). British HIV Association guidelines for the treatment of TB/HIV coinfection. HIV Med 12, 517–524 [DOI] [PubMed] [Google Scholar]
- R Core Teah. (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL http://www.R-project.org/ Last access: June/15/15 [Google Scholar]
- Riska P, Lamson M, MacGregor T, et al. (1999). Disposition and biotransformation of the antiretroviral drug nevirapine in humans. Drug Metab Disposit 27, 895–901 [PubMed] [Google Scholar]
- Rivero A, Mira JA, and Pineda JA. (2007). Liver toxicity induced by non-nucleoside reverse transcriptase inhibitors. J Antimicrob Chemother 59, 342–346 [DOI] [PubMed] [Google Scholar]
- Rotger M, Tegude H, Colombo S, et al. (2007). Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Therapeut 81, 557–566 [DOI] [PubMed] [Google Scholar]
- Rotger M, Colombo S, Furrer H, et al. (2005). Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogen Genomics 15, 1–5 [DOI] [PubMed] [Google Scholar]
- Sánchez A, Cabrera S, Santos D, et al. (2011). Population pharmacokinetic/pharmacogenetic model for optimization of efavirenz therapy in Caucasian HIV-infected patients. Antimicrob Agents Chemother 55, 5314–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipani A, Wyen C, Mahungu T, et al. (2011). Integration of population pharmacokinetics and pharmacogenetics: An aid to optimal nevirapine dose selection in HIV-infected individuals. J Antimicrob Chemother 66, 1332–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweikl H, Taylor JA, Kitareewan S, Linko P, Nagorney D, and Goldstein J. (1993). Expression of CYP1A1 and CYP1A2 genes in human liver. Pharmacogenet Genomics 3, 239–249 [DOI] [PubMed] [Google Scholar]
- Skelton M, Kampira E, Wonkam A, et al. (2014). Frequency variation among sub-Saharan populations in virus restriction gene, BST-2 proximal promoter polymorphisms: Implications for HIV-1 prevalence differences among African countries. OMICS 18, 461–471 [DOI] [PubMed] [Google Scholar]
- Smyth RP, Davenport MP, and Mak J. (2012). The origin of genetic diversity in HIV-1. Virus Res 169, 415–429 [DOI] [PubMed] [Google Scholar]
- Steinbrook R. (2013). Controlling HIV/AIDS: The obstacles and opportunities ahead. JAMA 173, 11–12 [DOI] [PubMed] [Google Scholar]
- Svärd J, Spiers JP, Mulcahy F, and Hennessy M. (2010). Nuclear receptor-mediated induction of CYP450 by antiretrovirals: Functional consequences of NR1I2 (PXR) polymorphisms and differential prevalence in whites and sub-Saharan Africans. JAIDS 55, 536–549 [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Ramachandran G, Kupparam HKA, et al. (2011). Factors influencing plasma nevirapine levels: A study in HIV-infected children on generic antiretroviral treatment in India. J Antimicrob Chemother 66, 1354–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart M, Skelton M, Wonkam A, Kannemeyer L, and Dandara C. (2012). CYP1A2, CYP2A6, CYP2B6, CYP3A4 and CYP3A5 polymorphisms in two bantu-speaking populations from Cameroon and South Africa: Implications for global pharmacogenetics. Curr Pharmacogenomics Personal Med 10, 43–53 [Google Scholar]
- Torti C, Costarelli S, De Silvestri A, et al. (2007). Analysis of severe hepatic events associated with nevirapine-containing regimens. Drug Safety 30, 1161–1169 [DOI] [PubMed] [Google Scholar]
- Turpeinen M, Raunio H, and Pelkonen O. (2006). The functional role of CYP2B6 in human drug metabolism: Substrates and inhibitors in vitro, in vivo and in silico. Curr Drug Metab 7, 705–714 [DOI] [PubMed] [Google Scholar]
- Veldkamp AI, Weverling GJ, Lange JM, et al. (2001). High exposure to nevirapine in plasma is associated with an improved virological response in HIV-1-infected individuals. AIDS 15, 1089–1095 [DOI] [PubMed] [Google Scholar]
- Wattanakul T, Avihingsanon A, Manosuthi W, and Punyawudho B. (2013). Population pharmacokinetics of nevirapine in Thai HIV-infected patients. Antiviral Ther 19, 651–660 [DOI] [PubMed] [Google Scholar]
- Wen B, Chen Y, and Fitch WL. (2009). Metabolic activation of nevirapine in human liver microsomes: Dehydrogenation and inactivation of cytochrome P450 3A4. Drug Metab Disposit 37, 1557–1562 [DOI] [PubMed] [Google Scholar]
- Wit FW, Kesselring AM, Gras L, et al. (2008). Discontinuation of nevirapine because of hypersensitivity reactions in patients with prior treatment experience, compared with treatment-naive patients: The ATHENA cohort study. Clin Infect Dise 46, 933–940 [DOI] [PubMed] [Google Scholar]
- World Health Organization 2010. World health statistics (2010). World Health Organization [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.