Abstract
Our collaborative successful gene replacement therapy using AAV vectors expressing a variant of human RPGR-ORF15 in two canine models provided therapeutic proof of concept for translation into human treatment. The ORF15 sequence contained within this AAV vector, however, has ORF15 DNA sequence variations compared to the published sequence that are likely due to its unusual composition of repetitive purine nucleotides. This mutability is a concern for AAV vector production and safety when contemplating a human trial. In this study, we establish the safety profile of AAV-hIRBP-hRPGR and AAV-hGRK1-hRPGR vectors used in the initial canine proof-of-principle experiments by demonstrating hRPGR-ORF15 sequence stability during all phases of manipulation, from plasmid propagation to vector production to its stability in vivo after subretinal administration to animals. We also evaluate potential toxicity in vivo by investigating protein expression, retinal structure and function, and vector biodistribution. Expression of hRPGR is detected in the inner segments and synaptic terminals of photoreceptors and is restricted to the connecting cilium when the vector is further diluted. Treated eyes exhibit no toxicity as assessed by retinal histopathology, immunocytochemistry, optical coherence tomography, fundoscopy, electroretinogram, and vector biodistribution. Therefore, the hRPGR-ORF15 variant in our AAV vectors appears to be a more stable form than the endogenous hRPGR cDNA when propagated in vitro. Its safety profile presented here in combination with its proven efficacy supports future gene therapy clinical trials.
Introduction
Retinitis pigmentosa (RP) is a common form of genetically heterogeneous, inherited retinal diseases characterized by progressive degeneration of rod and cone photoreceptor cells and affects 1 in 4000 individuals.1,2 The X-linked form of RP (XLRP), comprising an estimated 15% of total RP cases, is among the most severe forms.3 Mutations in the gene coding for RP GTPase regulator (RPGR) cause XLRP and account for over 70% of cases.4,5 Males carrying RPGR mutations typically show signs of night blindness and restriction of visual fields in their first or second decade of life, and the disease often progresses to nearly complete vision loss in the fourth or fifth decade.6–8 Female carriers are usually not affected, but can occasionally exhibit electroretinogram (ERG) defects.6,9,10
The RPGR gene uses alternative splicing sites and polyadenylation signals producing multiple transcript variants.11 The RPGR-ORF15 variant is primarily expressed in the retina and contains exons 1–14 with a large, alternatively spliced exon ORF15 at its terminus.5,11–14 RPGR-ORF15 is localized in rod and cone photoreceptor-connecting cilia and appears to play a role in regulating protein trafficking between inner and outer segments as well as in microtubular organization. Consistent with this, RPGR-ORF15 co-immunoprecipitates with several axonemal, basal body, and microtubular transport proteins.15,16 In both RPGR-deficient12 and rd9 mutant mice,17 M-cone opsin is mislocalized and rod outer segments have a reduced level of rhodopsin.
Animal models that harbor natural RPGR mutations or transgenic RPGR mutant mice contributed to a better understanding of the disease mechanism and gave rise to potential therapeutic interventions.12,17–20 Recently, gene therapy in two RPGR-XLRP canine models that carry different ORF15 mutations21 provides proof of concept for treating RPGR mutations within the ORF15 region.22 AAV-mediated gene transfer of a full-length human RPGR-ORF15 cDNA, driven by either the human interphotoreceptor retinoid-binding protein (hIRBP) promoter or the human G-protein-coupled receptor kinase 1 (hGRK1) promoter, prevented photoreceptor degeneration and preserved retinal function in both canine models.
RPGR-ORF15 contains purine-rich repeats and encodes a repetitive glycine- and glutamic acid-rich domain of unknown function and a basic C-terminal domain. Mutations in ORF15 account for 50–60% of pathogenic RPGR mutations, and therefore this region is regarded as a mutation hotspot.4,5,23 The unusual sequence within ORF15 was suggested to promote polymerase arrest and slipped strand mispairing and therefore produces a high frequency of microdeletions, frameshifts, and premature termination of translation.5 An ORF15 sequence containing plasmid when propagated in E. coli cells is also prone to mutate.24 In fact, the ORF15 cDNA that we demonstrate successful gene therapy in the two canine models22 contain sequence variations compared with the published human RPGR reference sequence. Therefore, the stability of the ORF15 sequence within the AAV vectors containing hRPGR cDNA and the potential toxicity of the mutated sequence remain to be critical issues that have to be addressed before attempting an RPGR XLRP-based gene therapy clinical trial. In this study, we demonstrate the sequence stability of the ORF15 cDNA variant contained within AAV-hIRBP-hRPGR and AAV-hGRK1-hRPGR during plasmid and AAV vector production. We also show that ORF15 cDNA sequence remains the same in vivo. Furthermore, expression of this variant of hRPGR-ORF15 shows no toxicity in injected mouse eyes as assessed by retinal morphology and function.
Materials and Methods
Animals and ethics statement
All experiments were approved by the local Institutional Animal Care and Use Committees at the University of Florida and performed in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and National Institutes of Health regulations. All animals were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained in the University of Florida Health Science Center Animal Care Services Facilities on a 12-hr-light/12-hr-dark cycle.
Construction and packaging of AAV vectors
The assembly and cloning of the full-length human RPGR-ORF15 cDNA into pBluescript was described previously.22 The plasmid was propagated in E. coli Stbl4 (Life Technologies, Grand Island, NY). To move hRPGR-ORF15 cDNA into AAV vectors, a NotI linker-containing sequence was introduced into the pBluescript plasmid backbone. hRPGR-ORF15 cDNA was then excised utilizing these flanking NotI sites and subcloned into an AAV vector plasmid under the control of the hGRK1 promoter. To make AAV-hIRBP-hRPGR, the hGRK1 promoter was replaced with the hIRBP promoter (Fig. 1). E. coli SURE cells (Agilent Technologies, Santa Clara, CA) were used for cloning and in subsequent plasmid retransformation and propagation steps to prevent inverted terminal repeat (ITR) loss. Vector plasmids were packaged in AAV serotype 5 (AAV2/5) by transfection of HEK293 cells according to previously published methods.25
Figure 1.
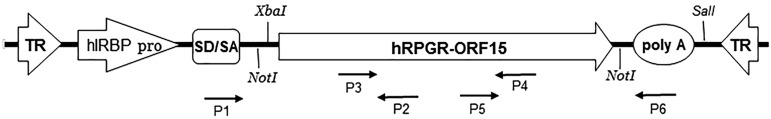
Schematic of AAV-hIRBP-hRPGR-ORF15 showing the NotI restriction sites used for moving hRPGR-ORF15 cDNA into an AAV vector, XbaI and SalI sites used for excising hRPGR-ORF15 cDNA from purified rAAV DNA, and the positions of the primers (P1–P6) used to amplify the three overlapping hRPGR-ORF15 fragments from episomal rAAV. hIRBP pro, human IRBP promoter; poly-A, polyadenylation signal; SD/SA, splice donor and acceptor sites; TR, inverted terminal repeats.
Isolation and characterization of DNA from rAAV vector genomes
Sequence characterization of viral vector genomes was carried out on nuclease-resistant DNA. Aliquots of vector preparations were treated with benzonase nuclease (Sigma-Aldrich, St. Louis, MO) to remove any residual plasmid DNA and vector genomes not packaged inside of rAAV capsids that may be carried over during vector production. Equal volumes (500 μl) of purified rAAV vector were mixed with 2× benzonase buffer and 8 μl (2600 units) of benzonase and the sample was incubated at 37°C for 60 min. Then 254 μl of 5× proteinase K buffer and 10 μl (197 μg) of proteinase K were added to remove the capsid proteins and incubated at 37°C for 60 min. Benzonase and proteinase K were subsequently removed by phenol/chloroform extraction. DNA was precipitated by adding 1/10 volume of 3 M Na acetate (pH 5.2) and 2 volumes of 100% EtOH and incubated at −25°C for 1 hr. Samples were centrifuged at 4°C for 40 min at 20,800×g, and the pellet was air-dried, then dissolved, and stored in TE buffer.
The DNA extracted from rAAV consists of an equal mixture of plus and minus single-stranded genomes. To avoid DNA transcripts arising from self-priming of the ITRs in sequencing reactions, double-stranded genomes were first produced by heating benzonase and proteinase K-treated rAAV to 90°C in a water bath and gradually reducing the temperature until room temperature was reached. The annealed rAAV DNA was then digested with XbaI and SalI to remove the ITRs (Fig. 1). The fragment containing only hRPGR-ORF15 and the poly-A signal sequence was then gel-purified and sequenced.
AAV genome recovery and characterization from injected mouse eyes
Wild-type mice injected with AAV2/5-hIRBP-hRPGR subretinally were sacrificed one month after injection. Retinas were dissected and DNA extraction was performed using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The PCR primers (Fig. 1) used for amplifying three overlapping fragments covering entire hRPGR-ORF15 cDNA are as follows: F1, 5′-TTACTTCTAGGCCTGTACGGAA (forward primer specific to AAV); R1, 5′-CAAGAGTCCCTTCTATTGGAGG; F2, 5′-GATTCTTTTTCAATGAGGAGAACA (forward primer specific to human RPGR); R2, 5′-TTTCACGTTCTCCCTCCACT; F3, 5′-GATGAGGAAGTAGAGATCCC (forward primer specific to human RPGR); R3, 5′- TCGAGTTACTTCAATTCCAAG (reverse primer specific to AAV). PCR was performed with Herculase polymerase (Agilent Technologies) with 4% DMSO (95°C for 3 min initial denature, 35 cycles of 95°C for 45 sec, 55°C for 45 sec, 72°C for 5 min, and final 72°C for 10 min). PCR fragments were gel-purified and subjected for sequencing.
Sequencing reaction
DNA sequencing was carried out at the University of Florida DNA Sequencing core. Sequencing reactions were performed using ABI Prism BigDye Terminator cycle sequencing protocols developed by Applied Biosystems (Perkin-Elmer Corp., Waltham, MA). Sequencing reactions were assembled in 20 μl reaction volume by adding 500 ng DNA, 10 pmol primer, 4 μl of ABI prism BigDye Terminator v.1.1 (BDv1.1) or 4 μl mix of BDv1.1:dGTP BDv1, 3 μl of 5× sequencing buffer, 4 μl of 1 M betaine, and 5% DMSO (Sigma-Aldrich) and amplified according to the manufacturer's recommendations. Excess dye-labeled terminators were removed using MultoScreen 96-well filtration system (Millipore, Billerica, MA). The purified extension products were dried in a SpeedVac and then suspended in Hi-di formamide. Sequencing was performed on a 50 cm POP-7 sieving matrix capillaries in an ABI Prism 3130 Genetic Analyzer or ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA) and were analyzed by ABI Sequencing Analysis software v. 5.2 and KB Basecaller.
Subretinal injection
Injections were performed on 6-week-old C57/B mice. One eye served as an uninjected control, and the contralateral eyes were injected with 1 μl AAV2/5-hIRBP-hRPGR. For toxicity and expression analysis, titer of 1.5×1011 vector genome/ml was used. Lower titer of 1010 vector genome/ml was injected to show restricted labeling in the connecting cilium and 1013 vector genome/ml was injected to show photoreceptor cell damage at higher titer. Subretinal injections were performed as previously described.26 Briefly, a 33-gauge blunt needle mounted on a 10 μl Hamilton syringe was used to inject the virus through the corneal opening introduced by a 30-gauge needle, and injections were visualized by fluorescein-positive subretinal bleb. One percent atropine eye drops and neomycin/polymyxin B/dexamethasone ophthalmic ointment were given after injection.
Electroretinography
At 4 weeks postinjection, rod- and cone-mediated ERGs were recorded separately using a UTAS Visual Diagnostic System equipped with a Big Shot Ganzfeld dome (LKC Technologies, Gaithersburg, MD). Mice were dark-adapted overnight and placed under dim red illumination (>650 nm). Dark-adapted animals were anesthetized with a mixture of 100 mg/kg ketamine, 20 mg/kg xylazine, and saline at a ratio of 1:1:3. ERGs from both eyes were recorded simultaneously. Scotopic rod recordings were performed with three increasing light intensities at −1.6, −0.6, and 0.4 log cds/m2. At each light intensity, 10 responses were recorded and averaged. Photopic cone ERGs were performed after mice were adapted to a white background light of 30 cd/m2 for 5 min. Recordings were performed with four flash intensities of 0.1, 0.7, 1.0, and 1.4 log cds/m2 in the presence of a constant 30 cd/m2 background light. Fifty responses were recorded and averaged at each intensity. Scotopic a- and b-wave amplitudes at 0.4 log cds/m2 and photopic b-wave amplitudes at 1.4 log cds/m2 from uninjected and injected eyes were averaged and used to generate average and standard deviation.
Spectral-domain optical coherence tomography
Spectral-domain optical coherence tomography (SD-OCT) was performed at one month postinjection using a high-resolution instrument (Bioptigen, Inc., Durham, NC) to measure outer nuclear (ONL) thickness and retinal morphology. Pupils of AAV2/5-hIRBP-hRPGR treated and contralateral untreated wild-type eyes were dilated with 1% atropine sulfate and 2.5% phenylephrine. The optic nerve head was centered on the OCT scan within a 1.5-mm-diameter field of view under fast fundus mode. Three hundred linear B-scans were obtained, and 30 images captured were averaged to minimize background noise and to achieve acceptable resolution. Outer nuclear thickness was measured at the same distance from the optical nerve head and four measurements were recorded from each eye and averaged. Statistical significance was determined by the paired t-test with p-values of <0.05 considered significant.
Retinal histology and immunohistochemistry
Eyes of wild-type mice injected with 1 μl of AAV2/5-hIRBP-hRPGR were enucleated at 1 month postinjection. Eyecups were fixed in 4% paraformaldehyde overnight at 4°C and then paraffin embedded and sectioned at 4 μm through the optic nerve for hematoxylin and eosin staining. Images were taken with a Carl Zeiss CD25 microscope fitted with Axio-vision release 4.6 software.
For immunohistochemistry analysis, freshly dissected eyes were put in OCT and frozen in liquid N2 directly without fixation. The eyes were cryosectioned at 10 μm thickness and used immediately for immunostaining. The sections were first fixed in 4% paraformaldehyde for 1 min, rinsed with PBS, and blocked with 2% normal goat serum 1 hr at room temperature. The RPGR antibody (Sigma-Aldrich; HPA001593) was diluted at 1:800 in PBS containing 0.05% triton X-100 and 1% BSA.
Western blot analysis
C57/BL6 eyes treated with AAV2/5-hIRBP-hRPGR and untreated controls were dissected, and the eyecups pooled and homogenized by sonication in a buffer containing 0.23 M sucrose, 2 mM EDTA, 5 mM Tris–HCl (pH 7.5), and complete protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). After centrifugation, aliquots of the extracts containing equal amounts of protein (50 μg) were separated on 4–15% SDS-PAGE, transferred onto Immobilon-FL PVDF membranes (Millipore, Temecula, CA), and probed with the same (Sigma-Aldrich; HPA001593) antibody used in immunohistochemistry. Visualization of specific bands was performed using the Odyssey Infrared Fluorescence Imaging System (Odyssey; Li-Cor, Lincoln, NE).
Vector biodistribution
At necropsy, tissues were harvested and snap-frozen in liquid nitrogen. Genomic DNA (gDNA) was extracted from retina, optic nerve, brain, parotid region, liver, and spleen using a DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions. gDNA concentrations were determined using an Eppendorf Biophotometer (Eppendorf, Hauppauge, NY). rAAV genome copies in the gDNA were quantified by real-time PCR using an ABI 7900 HT sequence detection system (Applied Biosystems) according to the manufacturer's instructions, and results were analyzed using the SDS 2.3 software. Primers and probe were designed to the SV40 poly-A of the AAV-hIRBP-hRPGR vector cassette. A standard curve was generated using plasmid DNA containing the same SV40 poly-A target sequence. PCRs with a total volume of 100 μl were performed under the following conditions: 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 sec and 60°C for 1 min.
DNA samples were assayed in triplicate. To determine whether any sample inhibited the PCR, the third replicate was spiked with target plasmid DNA at a ratio of 100 copies/μg gDNA. PCR results were considered acceptable if this replicate generated greater than 40 copies/μg gDNA. If a sample contained ≥100 copies/μg gDNA, it was considered positive for vector genomes. If a sample contained <100 copies/μg gDNA, it was considered negative for vector genomes.
Results
AAV-hIRBP-hRPGR and AAV-hGRK1-hRPGR plasmids contain sequence variations within ORF15
One potential source of sequence variation could occur during cloning and production of the vector plasmid in E. coli. The original pBluescript plasmid containing hRPGR-ORF15 cDNA was reported to contain a single in-frame triplet DNA deletion of bases 2872–2874 relative to the reference sequence (variant C; NM_001034853) (Dr. Hemant Khanna personal communication) (Fig. 2). Cloning of hRPGR-ORF15 cDNA into AAV vectors was challenging, likely because of the highly repetitive nature of the AG-rich region. We overcame this problem by screening 75 individual colonies via mini-lysate DNA isolation for the AAV-hGRK1-hRPGR construct. By direct restriction digestion of single colony DNA, we were able to identify two colonies that contained correctly sized inserts. Both colonies had the identical sequence but contained multiple differences in their ORF15 AG-rich region compared with the reference sequence (NM_001034853). All the differences are within the ORF15 domain between nucleotides 2461 and 3057 (Fig. 2). Specifically, the clone contained 7 deletions resulting in a loss of 45 basepairs (bp) but retaining the original reading frame, one 3 bp insertion, and 65 bp substitutions spread throughout the AG-rich region. These changes translate into 15 amino acid deletions, 1 amino acid insertion, and 26 amino acid substitutions. We hypothesize that these in-frame changes within the AG-rich region may not affect normal RPGR function because of its natural splicing complexity within this region, and that this ORF15 transcript retains an intact C-terminal LELK amino acid sequence.11 RPGR-ORF15 cDNA was first cloned under control of the hGRK1 promoter, and then this promoter was replaced with the hIRBP promoter to create the AAV-hIRBP-hRPGR plasmid. Both plasmids were propagated in SURE cells to prevent loss of ITRs or other rearrangements through bacterial recombination. Propagation of AAV vectors in Stbl4 cells was not feasible because the majority of ITRs were lost and the DNA yield was low. Both plasmid preparations had the identical sequence (Fig. 2).
Figure 2.
Nucleotide and amino acid sequence alignments of pBluecript-hRPGR, AAV-hIRBP-hRPGR, and AAV-hGRK1-hRPGR cDNAs with reference to the GenBank hRPGR sequence in the ORP15 domain where variations exist. Asterisks (*) indicate nucleotide or amino acid that is identical to the reference sequence, and dashes (–) indicate nucleotide or amino acid deletion.
Sequence stability of AAV-hIRBP-hRPGR and AAV-hGRK1-hRPGR plasmid DNA
To study the stability of RPGR-ORF15 sequence in both constructs, we sequenced two independent colonies each from large DNA preparations of AAV-hIRBP-hRPGR and AAV-hGRK1-hRPGR plasmids used for making AAV2/5 vectors in RPGR XLRP dog gene therapy study.22 These new plasmid preparations were identical to the original colony (Table 1). To further confirm the stability of the ORF15 cDNA, we retransformed AAV-hIRBP-hRPGR and AAV-hGRK1-hRPGR DNA back into SURE cells and sequenced five more independently isolated bacterial plasmid DNA preparations of AAV-hIRBP-hRPGR and two more similar preparations of AAV-hGRK1-hRPGR, each amplified from a different bacterial clone. All clones contained hRPGR-ORF15 cDNA sequences identical to our originally selected ORF15 clone (Table 1). Therefore, it appeared that the selected hRPGR-ORF15 cDNA from the original cloning is stable.
Table 1.
Numbers of plasmid and AAV preparations sequenced
| AAV-hIRBP-hRPGR | AAV-hGRK1-hRPGR | |
|---|---|---|
| Large-scale plasmid DNA preparations | 2 | 2 |
| Small-scale plasmid DNA preparations | 5 | 2 |
| DNA extracted from purified AAV preparations | 5 | 2 |
Sequence stability of AAV2/5-hIRBP-hRPGR and AAV2/5-hGRK1-hRPGR vector DNA
A second critical step of rAAV vector production is the replication of the input plasmid DNA in human embryonic kidney (HEK293) cells after cotransfection with helper plasmid that provides the necessary components for AAV production.27 There remains the possibility of introducing ORF15 sequence variants during this process as well because of imperfect DNA damage repair mechanisms and DNA polymerase errors during replication in the human cells caused by the repetitive AG-rich nature of the template. We therefore sequenced the hRPGR-ORF15 cDNA directly from independently produced and purified AAV vector preparations without prior PCR amplification. DNA was extracted from each AAV vector batch and the single-stranded vector DNAs annealed to create duplex DNA. The ITRs were then removed by restriction digest and fragments containing the hRPGR-ORF15 subjected to sequencing. The results from five independent AAV2/5-hIRBP-hRPGR AAV preparations and two AAV2/5-hGRK1-hRPGR AAV preparations revealed that all seven AAV vector DNAs contained the same hRPGR-ORF15 cDNA sequence as found in the plasmids used for their production (Table 1).
Sequence stability of AAV2/5-hIRBP-hRPGR vector DNA in injected eyes
Recombinant AAV vectors are packaged as either plus or minus polarity single strands with equal frequency.28 AAV-mediated gene expression relies on forming double-stranded DNA to become transcriptionally active after viral uncoating in the host cell nucleus. Complementary strand synthesis by cellular replication factors29 and reannealing of complementary DNA strands from individual infecting rAAV particles30 both are the pathways for rAAV transduction. To confirm the sequence fidelity of injected vectors in vivo after forming double-stranded DNA, we sequenced the hRPGR-ORF15 cDNA recovered from AAV-hIRBP-hRPGR vector genomes from injected mouse eyes 1 month after subretinal injection. The hRPGR-ORF15 cDNA was ∼3.5 kb and the entire cDNA was amplified using three PCRs with vector-specific primers or human-specific RPGR primers to generate overlapping fragments (Fig. 1). PCR conditions and primer combinations were optimized to amplifying the AG-rich regions. Sequencing of the three PCR fragments revealed that the hRPGR-ORF15 cDNA is identical to the original input sequence.
AAV2/5-hIRBP-hRPGR expression in vivo
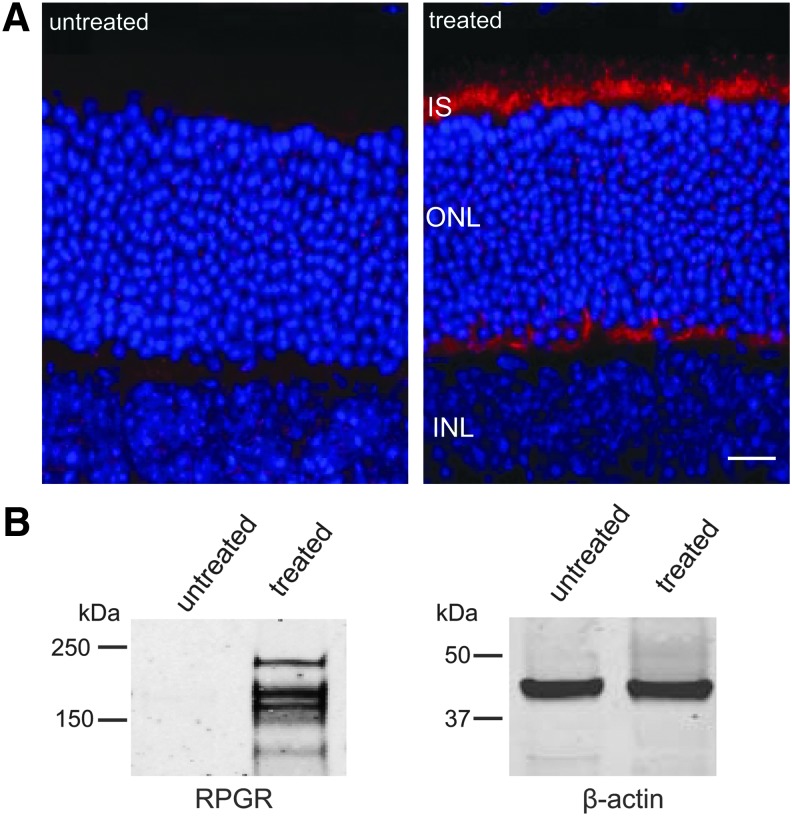
Immunolabeling with an antibody against human hRPGR-ORF15 detected robust hRPGR protein expression in normal C57/BL6 mice subretinally injected with AAV2/5-hIRBP-hRPGR (vector dose of 1.5×108 vector genomes) (Fig. 3A). Labeling was found throughout the inner segment and synaptic terminals, a pattern similar to the two canine models with ORF15 mutations treated with the same vector.22 This antibody did not cross-react with endogenous mouse hRPGR-ORF15. When the vector was further diluted 10-folds, we found that expression of hRPGR was largely restricted to the connecting cilium (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/hum). hRPGR-ORF15 has been reported to migrate in gels at 200–250 kDa.11,31 Western blot analysis using the same antibody detected three major bands between 150 and 250 kDa in injected C57/BL6 eyes, while the endogenous mouse protein was not detectable (Fig. 3B).
Figure 3.
Immunohistochemical and Western analysis of AAV-hIRBP-hRPGR expression (108 vector genomes) in the C57/BL6 mice retina. (A) Cryosections of uninjected and injected eyes were probed with a human RPGR-specific antibody. hRPGR-ORF15-specific staining is shown in red; cell nuclei are stained with DAPI (blue). INL, inner nuclear layer; IS, inner segment layers; ONL, outer nuclear layer; scale bar=20 μm. (B) Western blot of retinal extracts from uninjected and injected eyes probed with the same antibody. Three major bands between 150 and 250 kDa were detected in injected eyes.
Retinal structure and function of AAV2/5-hIRBP-hRPGR treated eyes
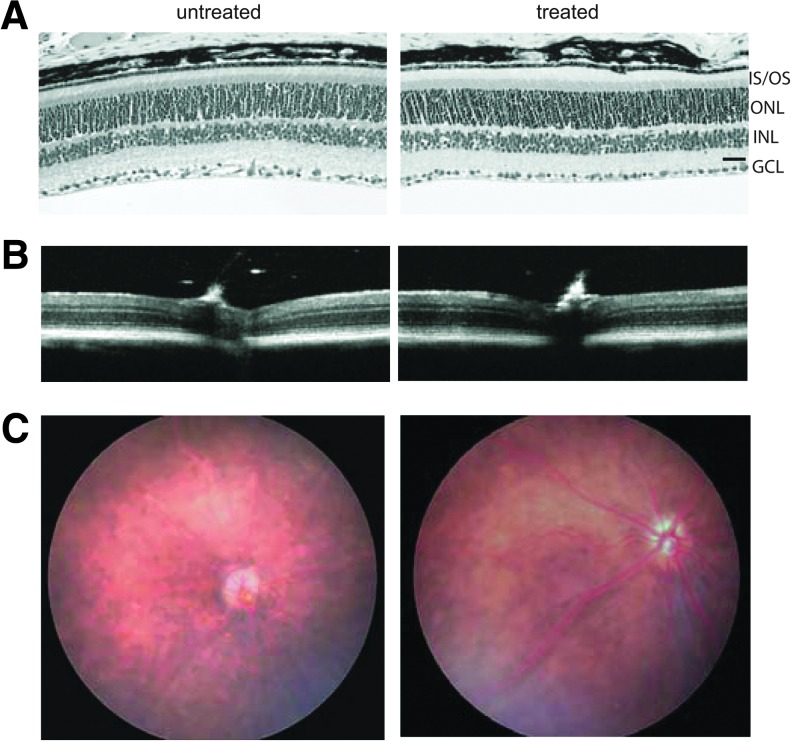
The potential toxicity of AAV2/5-hIRBP-hRPGR in vivo was assessed by retinal structure, retinal function, and vector dissemination in wild-type mice following subretinal injection. Retinal structure was analyzed by morphology, OCT, and fundoscopic examination. Paraffin-embedded sections of injected eyes (vector dose of 1.5×108 vector genomes) were stained with hematoxylin and eosin and showed normal retinal morphology with 11–12 rows of outer nuclei and normal outer and inner segment lengths (Fig. 4A), similar to their contralateral untreated controls. Comparison of retinal ONL thicknesses measured by OCT showed that untreated C57/BL6 were 0.0535±0.0011 mm, and AAV2/5-hIRBP-hRPGR-injected eyes were 0.0527±0.0029 mm (average±SEM, n=4, p>0.5) (Fig. 4B). In vivo fundoscopic examination of gross retinal anatomy visualizing retinal vessels, optic disc, and posterior retina revealed no obvious changes between vector-treated and control eyes (Fig. 4C).
Figure 4.
Analysis of retinal morphology of C57/BL6 mice injected with AAV2/5-hIRBP-hRPGR. (A) A representative image near the optic nerve head showing normal retinal morphology of the treated eye compared with the contralateral uninjected control. GCL, ganglion cell layer; INL, inner nuclear layer; IS/OS, photoreceptor inner and outer segment layers; ONL, outer nuclear layer; scale bar=20 μm. (B) Representative optical coherence tomography images showing normal outer nuclear thickness in treated eye. (C) Fundoscopic examination show normal gross retinal anatomy of a treated eye.
Retinal function was examined by electroretinography to determine any potential functional defect caused by administration of the vector. Scotopic a- and b-wave amplitudes at 0.4 log cds/m2 and photopic b-wave amplitudes at 1.4 log cds/m2 from uninjected and injected eyes were analyzed. Both scotopic (a- and b-wave amplitudes) and photopic (b-wave amplitudes only) responses showed an approximately 10% reduction in the treated eyes compared with their contralateral untreated controls (Table 2). The reduction of ERG is likely a reflection of injection-related damage as we observed previously.32,33
Table 2.
Comparison of scotopic and photopic ERG amplitudes between AAV-hIRBP-hRPGR-treated and contralateral untreated wild-type eyes
| Scotopic b wave | Scotopic a wave | Photopic b wave | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | Untreated eye | Treated eye | Treated/untreated | Untreated eye | Treated eye | Treated/untreated | Untreated eye | Treated eye | Treated/untreated |
| N=5 | 309±14 | 281±11 | 0.91 | 175±10 | 156±7 | 0.89 | 114±17 | 103±8 | 0.90 |
AAV2/5-hIRBP-hRPGR vector distribution
Quantitative real-time PCR was performed to assess the biodistribution of vector in retina, optic nerve, brain, parotid region, liver, and spleen. Vector was detected in retinas (9.1×106±1.6×106 vector genomes/μg gDNA, average±SEM, n=4) and optic nerves (2.3×106±2.1×106 vector genomes/μg gDNA, average±SEM, n=4) from injected eyes. Retinas and optic nerves from contralateral uninjected eyes and all other tissues (brain, parotid region, liver, and spleen) were all negative (<100 copies/μg gDNA).
Discussion
Although we showed in a previous study that a full-length hRPGR-ORF15 cDNA when delivered in an AAV2/5 vector prevented photoreceptor degeneration and preserved retinal structure and function in two canine RPGR-XLRP models,22 issues of the instability of the ORF15 sequence within the RPGR cDNA and its potential toxicity if mutated remained unresolved. In this study, we demonstrate that the variant hRPGR-ORF15 cDNA we employed in the canine study is stable and therefore suitable for moving forward into the IND-enabling safety studies required before a gene therapy clinical trial. The hRPGR-ORF15 cDNA sequence contained within AAV5-hIRBP-hRPGR and AAV5-hRGK1-hRPGR vectors remains unaltered during all stages of production and application since sequence analysis of multiple independently isolated and clonally distinct vector plasmid DNAs and multiple independently produced AAV vector preparations demonstrate. Moreover, we report that the hRPGR-ORF15 cDNA sequence is stable in vivo after subretinal vector injection into mice. Furthermore, the AAV-hIRBP-hRPGR vector exhibits no detectable or potential toxicity after treatment of mouse retinas as assessed by retinal structure and function and by vector biodistribution analysis. In sum, the reported studies therefore resolve several critical issues necessary before being able to move RPGR XLRP gene therapy into clinical testing.
The high mutability within ORF15 is related to its unusual composition of repetitive sequence that may promote DNA polymerase arrest and slipped-strand mispairing during replication.5 Plasmids containing RPGR-ORF15 when propagated in commonly used bacterial strains are prone to deletions and rearrangements24 (also, Drs. Hemant Khanna and Xinhua Shu, personal communication). Wu et al. found that the full-length human or mouse RPGR-ORF15 maintained their integrity in XL10 gold cells after testing various E. coli strains although the mechanism is not clear.24 Although both E. coli stbl4 and SURE cells lack components of the rec pathway that catalyzes the rearrangement and removal of DNA sequences and are recommended strains for cloning of unstable plasmids, we found that it was not feasible to grow AAV-hGRK1-hRPGR in stbl4 cells because of significant ITRs loss as well as an extremely low plasmid DNA yield. The original pBluescript plasmid containing hRPGR-ORF15 was propagated in stbl4 cells and we believe that ORF15 sequence variations were introduced during AAV-hGRK1-hRPGR cloning upon switching to SURE cells to preserve the integrity of ITRs that is necessary to package AAV genomes. The hRPGR-ORF15 sequence we cloned in another AAV vector under a mouse opsin promoter using hRPGR-ORF15 cDNA from original pBluescript plasmid contains different variations within purine-rich region from the one in AAV-hGRK1-hRPGR-ORF15 (data not shown), although the stability of this variant was not further characterized. The stability of the ORF15 variant contained within AAV-hGRK1-hRPGR was preserved when we made the AAV-hIRBP-hRPGR vector by switching promoters. Therefore, we believe that the ORF15 variant identified from the large number of colonies screened might be the result of natural selection of a stable and nontoxic sequence in SURE cells.
The hRPGR-ORF15 variant used in our study contains in-frame deletions and nucleotide changes in the AG-rich region compared with published hRPGR-ORF15 sequences. There is precedent for in-frame changes within the AG-rich region not affecting normal RPGR function: the natural splicing complexity within this region leads to a variety of transcripts that contain in-frame deletions of parts of ORF15 while retaining the C-terminal sequence terminating in the amino acid sequence LELK.11 In addition, human genetic studies have identified more than 30 sequence variants within the purine-rich region as benign polymorphisms.4,5,23,34–37 Of these, 15 are located between AA 821 to AA1019, where we see differences in our cDNA variant. The E905 deletion polymorphism4 is similar to E904–E906 deletions in our cDNA variant. All of these in-frame alterations and nucleotide polymorphism appear to have no clinical consequence. The high number of polymorphism in ORF15 suggests that there is a high level of protein diversity for RPGR in humans.36 In fact, a single, abbreviated RPGR-ORF15 variant that has an in-frame deletion of 654 bp within the purine-rich region of ORF15 reconstituted the RPGR function in vivo,38 suggesting that this repetitive region could be significantly shortened without ablating the protein function. It was also suggested that the alternating glycine and glutamic acid residues that stretch for several hundred residues are not likely to form into a compact structure but rather exist to link the N- and C-terminal domains.19 Furthermore, the length of this region varies considerably among species,5 also suggesting lack of any rigid functional constraint on the length of the repetitive region in ORF15.
Although the functional significance of the existence of multiple RPGR variants is as yet unclear, successful gene therapy in the two canine models carrying either a null or a truncation mutation by introduction of a single form of hRPGR indicates that the natural collection of RPGR variants in vivo may be functionally redundant. Previous studies11 showed that innumerable transcripts and a multitude of RPGR proteins in photoreceptors were generated by ORF15 by removing various portions of the purine-rich region as introns. They found that the purine-rich region contains multiple exonic splice enhancers known to promote splicing through interaction with serine–arginine repeat proteins. That explains why the ORF15 coding sequence without introns may still be able to be alternatively spliced. Consistent with this, we observed three major bands in AAV-hIRBP-hRPGR-injected mouse eyes by Western blot analysis, suggesting that hRPGR-ORF15 variant may be further spliced in mouse retinas. The same phenomena was also observed in retinas of Rpgr knockout mice treated with AAV expressing the full-length human RPGR-ORF15 using a human-specific RPGR antibody, or AAV expressing the full-length mouse RPGR-ORF15 by a mouse RPGR antibody.24 As it was shown that the purine-rich region spliced differently in mouse and human photoreceptor cells,11 it would be interesting to investigate whether this region is also further spliced in hRPGR-ORF15 AAV-transduced canine photoreceptors.
The findings presented in this work are in support of future gene replacement therapy studies, and address issues of vector sequence stability and in vivo safety. Careful vector titration is critical for treating the RPGR-ORF15 mutations. We noticed that too much RPGR expression causes toxicity to the retina. Virus titer usually considered safe in the mouse retina (1013 vector genome/ml) caused severe photoreceptor cell degeneration based on ERG and morphological analysis (Supplementary Fig. S2). Therefore, it might be important to adjust ectopic RPGR-ORF15 levels expressed from recombinant AAV as close as possible to the physiological levels of RPGR-ORF15 within photoreceptor cells. We show that this is feasible by employing a vector dose that restricts hRPGR expression specifically to the connecting cilium by simply diluting the vector. Future studies addressing the functional significance of the RPGR protein and the association between mutations and disease will be also essential for developing gene replacement therapies for human patients suffering from this form of RP.
Supplementary Material
Acknowledgments
We acknowledge NIH Grants P30EY021721 and R01EY-017549 and funds from the Macular Vision Research Foundation, Foundation Fighting Blindness, Overstreet Fund, and Research to Prevent Blindness, Inc., for partial support of this work.
Author Disclosure
W.W.H. and the University of Florida have a financial interest in the use of AAV therapies and own equity in a company (AGTC) that might, in the future, commercialize some aspects of this work.
References
- 1.Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol 2007;125:151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rattner A, Sun H, Nathans J. Molecular genetics of human retinal disease. Annu Rev Genet 1999;33:89–131 [DOI] [PubMed] [Google Scholar]
- 3.Bird AC. X-linked retinitis pigmentosa. Br J Ophthalmol 1975;59:177–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breuer DK, Yashar BM, Filippova E, et al. . A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet 2002;70:1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vervoort R, Lennon A, Bird AC, et al. . Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet 2000;25:462–466 [DOI] [PubMed] [Google Scholar]
- 6.Jacobson SG, Buraczynska M, Milam AH, et al. . Disease expression in X-linked retinitis pigmentosa caused by a putative null mutation in the RPGR gene. Invest Ophthalmol Vis Sci 1997;38:1983–1997 [PubMed] [Google Scholar]
- 7.Sandberg MA, Rosner B, Weigel-DiFranco C, et al. . Disease course of patients with X-linked retinitis pigmentosa due to RPGR gene mutations. Invest Ophthalmol Vis Sci 2007;48:1298–1304 [DOI] [PubMed] [Google Scholar]
- 8.Aleman TS, Cideciyan AV, Sumaroka A, et al. . Inner retinal abnormalities in X-linked retinitis pigmentosa with RPGR mutations. Invest Ophthalmol Vis Sci 2007;48:4759–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreasson S, Ponjavic V, Abrahamson M, et al. . Phenotypes in three Swedish families with X-linked retinitis pigmentosa caused by different mutations in the RPGR gene. Am J Ophthalmol 1997;124:95–102 [DOI] [PubMed] [Google Scholar]
- 10.Weleber RG, Butler NS, Murphey WH, et al. . X-linked retinitis pigmentosa associated with a 2-base pair insertion in codon 99 of the RP3 gene RPGR. Arch Ophthalmol 1997;115:1429–1435 [DOI] [PubMed] [Google Scholar]
- 11.Hong DH, Li T. Complex expression pattern of RPGR reveals a role for purine-rich exonic splicing enhancers. Invest Ophthalmol Vis Sci 2002;43:3373–3382 [PubMed] [Google Scholar]
- 12.Hong DH, Pawlyk BS, Shang J, et al. . A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3). Proc Natl Acad Sci USA 2000;97:3649–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschner R, Rosenberg T, Schultz-Heienbrok R, et al. . RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Hum Mol Genet 1999;8:1571–1578 [DOI] [PubMed] [Google Scholar]
- 14.Yan D, Swain PK, Breuer D, et al. . Biochemical characterization and subcellular localization of the mouse retinitis pigmentosa GTPase regulator (mRpgr). J Biol Chem 1998;273:19656–19663 [DOI] [PubMed] [Google Scholar]
- 15.Khanna H, Hurd TW, Lillo C, et al. . RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J Biol Chem 2005;280:33580–33587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu X, Fry AM, Tulloch B, et al. . RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum Mol Genet 2005;14:1183–1197 [DOI] [PubMed] [Google Scholar]
- 17.Thompson DA, Khan NW, Othman MI, et al. . Rd9 is a naturally occurring mouse model of a common form of retinitis pigmentosa caused by mutations in RPGR-ORF15. PLoS One 2012;7:e35865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunner S, Skosyrski S, Kirschner-Schwabe R, et al. . Cone versus rod disease in a mutant Rpgr mouse caused by different genetic backgrounds. Invest Ophthalmol Vis Sci 2010;51:1106–1115 [DOI] [PubMed] [Google Scholar]
- 19.Hong DH, Pawlyk BS, Adamian M, et al. . Dominant, gain-of-function mutant produced by truncation of RPGR. Invest Ophthalmol Vis Sci 2004;45:36–41 [DOI] [PubMed] [Google Scholar]
- 20.Huang WC, Wright AF, Roman AJ, et al. . RPGR-associated retinal degeneration in human X-linked RP and a murine model. Invest Ophthalmol Vis Sci 2012;53:5594–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Acland GM, Wu WX, et al. . Different RPGR exon ORF15 mutations in Canids provide insights into photoreceptor cell degeneration. Hum Mol Genet 2002;11:993–1003 [DOI] [PubMed] [Google Scholar]
- 22.Beltran WA, Cideciyan AV, Lewin AS, et al. . Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc Natl Acad Sci USA 2012;109:2132–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu X, Black GC, Rice JM, et al. . RPGR mutation analysis and disease: an update. Hum Mutat 2007;28:322–328 [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Hiriyanna S, Qian H, et al. . A long-term efficacy study of gene replacement therapy for RPGR-associated retinal degeneration. Hum Mol Genet 2015. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zolotukhin S, Byrne BJ, Mason E, et al. . Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 1999;6:973–985 [DOI] [PubMed] [Google Scholar]
- 26.Pang JJ, Chang B, Kumar A, et al. . Gene therapy restores vision-dependent behavior as well as retinal structure and function in a mouse model of RPE65 Leber congenital amaurosis. Mol Ther 2006;13:565–572 [DOI] [PubMed] [Google Scholar]
- 27.Li J, Samulski RJ, Xiao X. Role for highly regulated rep gene expression in adeno-associated virus vector production. J Virol 1997;71:5236–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berns KI, Adler S. Separation of two types of adeno-associated virus particles containing complementary polynucleotide chains. J Virol 1972;9:394–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrari FK, Samulski T, Shenk T, et al. . Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol 1996;70:3227–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakai H, Storm TA, Kay MA. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J Virol 2000;74:9451–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He S, Parapuram SK, Hurd TW, et al. . Retinitis pigmentosa GTPase regulator (RPGR) protein isoforms in mammalian retina: insights into X-linked retinitis pigmentosa and associated ciliopathies. Vis Res 2008;48:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang JJ, Deng WT, Dai X, et al. . AAV-mediated cone rescue in a naturally occurring mouse model of CNGA3-achromatopsia. PLoS One 2012;7:e35250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timmers AM, Zhang H, Squitieri A, et al. . Subretinal injections in rodent eyes: effects on electrophysiology and histology of rat retina. Mol Vis 2001;7:131–137 [PubMed] [Google Scholar]
- 34.Bader I, Brandau O, Achatz H, et al. . X-linked retinitis pigmentosa: RPGR mutations in most families with definite X linkage and clustering of mutations in a short sequence stretch of exon ORF15. Invest Ophthalmol Vis Sci 2003;44:1458–1463 [DOI] [PubMed] [Google Scholar]
- 35.Neidhardt J, Glaus E, Lorenz B, et al. . Identification of novel mutations in X-linked retinitis pigmentosa families and implications for diagnostic testing. Mol Vis 2008;14:1081–1093 [PMC free article] [PubMed] [Google Scholar]
- 36.Pusch CM, Broghammer M, Jurklies B, et al. . Ten novel ORF15 mutations confirm mutational hot spot in the RPGR gene in European patients with X-linked retinitis pigmentosa. Hum Mutat 2002;20:405. [DOI] [PubMed] [Google Scholar]
- 37.Rozet JM, Perrault I, Gigarel N, et al. . Dominant X linked retinitis pigmentosa is frequently accounted for by truncating mutations in exon ORF15 of the RPGR gene. J Med Genet 2002;39:284–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong DH, Pawlyk BS, Adamian M, et al. . A single, abbreviated RPGR-ORF15 variant reconstitutes RPGR function in vivo. Invest Ophthalmol Vis Sci 2005;46:435–441 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.