Abstract
Mammalian target of rapamycin (mTOR) complexes play a pivotal role in the cell. Raptor and Rictor proteins interact with mTOR to form two distinct complexes, mTORC1 and mTORC2, respectively. While the domain structure of Raptor is known, current bioinformatics tools failed to classify the domains in Rictor. Here we focus on identifying specific domains in Rictor by searching for conserved regions. We scanned the pdb structural database and constructed three protein domain datasets. Next we carried out multiple pairwise sequence alignments of the proteins in the domain dataset. By analyzing the z-scores of Rictor sequence similarity to protein sequences in the dataset, we assigned the structural and functional domains of Rictor. We found that, like Raptor, Rictor also has HEAT and WD40 domains, which could be the common motif binding to mTORC. Rictor may also have pleckstrin homology domains, which mediate cellular localization and transmit signals to downstream targets, as well as a domain that is homologous to 50S protein L17 and human 39S protein L17. This putative ribosome binding domain could mediate mTORC2–ribosome interaction.
Key words: : mTOR, protein domain, protein–protein interactions, Raptor, Rictor
1. Introduction
The mammalian target of rapamycin (mTOR) plays a pivotal role in cell metabolism, growth, proliferation, and survival (Wullschleger et al., 2006; Guertin and Sabatini, 2007). mTOR nucleates two distinct multiprotein complexes, mTOR complex 1 (mTORC1) (Kim et al., 2002) and mTOR complex 2 (mTORC2) (Sarbassov et al., 2004), each of which contains the unique adaptor protein, Raptor or Rictor, respectively (Kim et al., 2002; Sarbassov et al., 2004). mTORC1 and mTORC2 have different sensitivity to rapamycin treatment. The mTORC1 complex is sensitive and responds to a spectrum of intracellular and extracellular stimuli, such as growth factors, energy status, oxygen levels, amino acids, and inflammation (Kim et al., 2002), while mTORC2 does not.
mTORC1 and mTORC2 have distinct functions, with differential roles in mesenchymal stem cell differentiation (Martin et al., 2014). Loss of Rictor, an obligatory component of mTORC2, decreased thymic and peripheral invariant NKT cells, which was associated with defective survival. In contrast, mTORC1 regulators Raptor and Rheb are dispensable for NKT17 differentiation (Wei et al., 2014). In contrast, loss of mTOR complex 1 (deletion of Raptor) induces developmental blockage in early T-lymphopoiesis and eradicates T-cell acute lymphoblastic leukemia cells, while deficiency of Rictor, an mTORC2 component, did not have the same effect (Hoshii et al., 2014). mTORC1 and mTORC2 differentially regulate homeostasis of neoplastic and nonneoplastic human mast cells (Smrz et al., 2011). In Alzheimer's disease, mTORC1, but not mTORC2, was activated in patient's brains and the level of mTOR signaling activation was correlated with cognitive severity (Sun et al., 2013). mTORC1 mediates T-cell function by inhibiting the mTORC2 pathway and coupling immune signals and metabolic programming (Zeng et al., 2013). The cytoplasmic and nuclear distributions of mTORC1 and mTORC2 complexes are also different. The mTOR–Raptor complex is predominantly cytoplasmic, whereas the mTOR–Rictor complex is abundant in both cytoplasmic and nuclear compartments (Rosner and Hengstschlager, 2008). mTORC1 controls the spindle function during mitosis and meiosis, while mTORC2 contributes to actin-dependent asymmetric division during meiotic maturation in mice (Kogasaka et al., 2013). Rictor is an important mediator of chemotaxis and metastasis in breast cancer cells (Zhang et al., 2010), and in T lymphopoiesis (Tang et al., 2012).
How Raptor and Rictor sense the diverse upstream signals and how this information is coupled to mTOR activation and substrate access to its active site are still unclear. mTORC1 and mTORC2 may achieve their distinct functions through the different proteins that Raptor and Rictor recruit, or via phosphorylation (Frey et al., 2014). For example, mTORC1 phosphorylation sites encode the sensitivity to starvation and rapamycin (Kang et al., 2013). Rictor phosphorylation is also important. Under cellular stress, GSK-3β phosphorylates the mTORC2 component Rictor at serine-1235, a modification that interferes with the binding of Akt to mTORC2 (Chen et al., 2011). Activation of mTORC2 by growth factor signaling is linked to specific phosphorylation of its component Rictor on Thr-1135 (Gao et al., 2010). The phosphorylation of this site is induced by growth factor stimulation and expression of the oncogenic forms of Ras or PI3K (Boulbes et al., 2010).
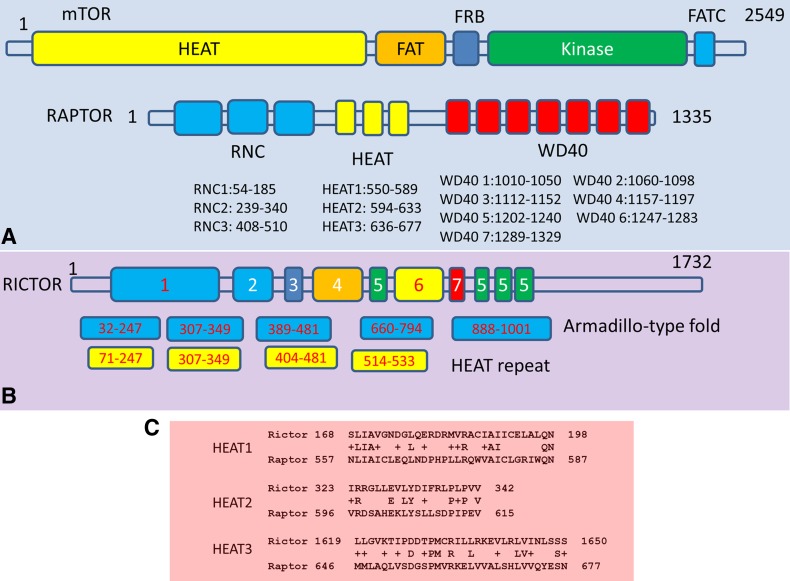
Both mTORC1 and mTORC2 are homodimers, with a small fraction of hetero-dimer formed by both Raptor and Rictor with two mTOR (Jain et al., 2014). The domain structure for mTOR includes a long HEAT, FAT, FRB, kinase, and FATC domains (Fig. 1A). The domain organization of Raptor has RNC, HEAT, and WD40 domains. The Raptor component of the mTORC1 complex binds to the N terminus of mTOR, and another Raptor monomer binds to the C-terminal of mTOR in the dimer mTORC1 complex (Yip et al., 2010). Sequence alignment of several Rictor family proteins suggested that there are seven conserved regions in the Rictor sequence (Sarbassov et al., 2004) (Fig. 1B). However, the structural and functional properties of these putative conserved regions, and thus extent of similarity (difference) to Raptor, are still unknown. In this study, we examined the sequence and structural properties of the Rictor protein using combined bioinformatics protocol. While Rictor proteins share high sequence identity within the family, their sequence homology with other proteins is limited. To overcome the requirement for high sequence homology, we searched the pdb structural database and constructed three protein domain datasets. By analyzing the z-scores of Rictor sequence similarity to protein sequences in the constructed datasets, we were able to assign its structural and functional domain. We found that Rictor also has HEAT and WD40 domains similar to Raptor, and identified a possible pleckstrin homology (PH) and ribosome binding domains.
FIG. 1.
Illustration of the sequence domains of mammalian target of rapamycin (mTOR), Raptor, and Rictor proteins. (A) mTOR has five major domains: HEAT, FACT, FRB, Kinase, and FATC. Raptor has three major domain regions: RNC, HEAT, and WD40. (B) Conserved sequence regions in Rictor and domain classification predicted using online servers. The HEAT domains were predicted by the SUPERFAMILY server (Gough et al., 2001), and the Armadillo folds were predicted by the InterPro (Jones et al., 2014). (C) Sequence alignment of the Raptor HEAT domain found matches in the Rictor sequence.
2. Methods
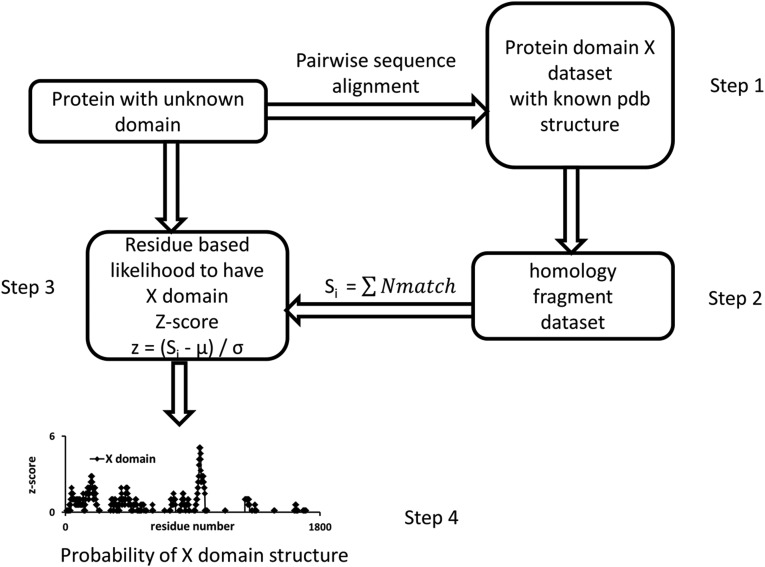
The flowchart of the domain assignment procedure is illustrated in Figure 2. In the first step, we constructed a nonredundant structural dataset of the domains. The HEAT, WD40, and PH domain containing proteins in the pdb were collected by searching the keywords “HEAT domain,” “WD40 domain,” and “PH domain,” respectively. Only protein structures containing the selected domain were chosen. We excluded structures that contain large non-HEAT, non-WD, and non-PH domains, respectively. Redundant structures with more than 95% sequence identity were discarded.
FIG. 2.
The flow chart to identify the likelihood of a protein to have a domain in the absence of a consensus sequence. In the first step, the nonredundant protein domain structure dataset X is constructed from pdb entries that have domain X. In step 2, pairwise sequence alignments between a target protein and a protein sequence in the above-selected dataset X are performed. In the third step, the z-score of a residue i to have X domain is calculated. The final probability of a protein to have domain X is checked in the last step.
In step 2, pair-wise sequence alignments between the human Rictor protein (gi|219520980|gb|AAI44510.1) and the protein sequence in the selected dataset were performed using the NCBI blast server (Madden et al., 1996). Selected proteins were then again excluded if only 10 continuous residues were detected to be similar. The final nonredundant Rictor protein homologous domain datasets are listed in Table 1.
Table 1.
Nonredundant pdb Database of Heat, WD40, and PH Domains with Sequence Homology with Human Rictor
| Heat domain | WD40 domain | PH domain | ||||
|---|---|---|---|---|---|---|
| 3ZBO | 2H4M | 4LG8 | 1U4C | 1EAZ | 2YRY | 4BBK |
| 4ATG | 2IE4 | 3FM0 | 2B4E | 1BTN | 2YS1 | 4EMO |
| 3GS3 | 2IQC | 3I2N | 3DM0 | 2Z0P | 2YS3 | 4F7H |
| 1TE4 | 2Q5D | 3ACP | 3KCI | 1AWE | 2I5F | 4H6Y |
| 1UG3 | 3IBV | 1GXR | 4DNU | 2COC | 2K2J | 1UNR |
| 4LUN | 3LWW | 3LRV | 4I0O | 4A5K | 3RCP | 1UPR |
| 4RXX | 3O2S | 3E0C | 4I79 | 1D9V | 1WJM | 1UZR |
| 4BSN | 3RRM | 3OW8 | 4V0M | 2Z4D | 1XX0 | 1V5P |
| 4KNH | 3TJ1 | 4CZV | 1NR0 | 1X1F | 2Y7B | 1V61 |
| 1PAQ | 4C0O | 4OZU | 1TRJ | 1X1G | 2Z0Q | 1V88 |
| 1YVR | 4FGV | 2H13 | 1XHM | 2ADZ | 4DIX | 1WGQ |
| 1P8Q | 4H3H | 4D0K | 2OVP | 2COA | 4KAX | 1ZYI |
| 2QK1 | 4PJU | 2CE9 | 4V0O | 2COD | 2Z0P | 2DTC |
| 3JUI | 1U6G | 2W18 | 4BH6 | 2COF | 2KIE | 2RSG |
| 3VLF | 1Z3H | 1ERJ | 2YNN | 2D9W | 2KIG | 3B77 |
| 3VYC | 2I91 | 2HES | 4PSW | 2D9Z | 3VOQ | 1NTY |
| 3W3T | 3WYF | 2A2N | 1PEV | 2DA0 | 2LUL | 2FJL |
| 3BSM | 4COP | 4X3E | 1SQ9 | 2DHI | 2×18 | 2FHX |
| 4E6H | 4GMX | 4N14 | 2DHK | 1UPQ | 2P0D | |
| 4G3A | 4KKO | 2DKP | 2LNW | 3HK0 | ||
| 4K92 | 4N5C | 2DN6 | 3PP2 | 3ODO | ||
| 1HO8 | 3U12 | |||||
In the third step, the tally scores Si of each residue were counted by their sequence matches with the proteins in the domain datasets, respectively.
 |
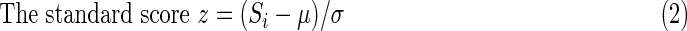 |
where z is the z-score, μ is the population mean for 1732 residues in Rictor, and σ is the standard deviation. In the Si scores, an amino acid i in Rictor is counted when it is identified in the pairwise sequence alignment. The scores Si reflect the frequency of amino acid i appearing in the homology fragment dataset (Fig. 2). Previously, a block-scoring algorithm was used to increase the homologous sequence identification (Nedelec et al., 2005). Using a similar concept, we define the likelihood of a domain distribution by examining the block distribution of the high z-score regions in the protein sequence in step 4.
3. Results and Discussion
3.1. Rictor also has HEAT and WD40 domains
The HEAT motif contains varied numbers of two antiparallel α-helix repeats that are linked by interunit loops allowing flexibility in this structure (Andrade and Bork, 1995). While there is only very limited sequence similarity between Raptor and Rictor, we suspected that there are similar domains, since both interact with the mTor protein. We first searched Rictor looking for sequence regions homologous to the HEAT and WD40 domains in Raptor. We identified three segments in Rictor that share sequence similarity with Raptor HEAT repeats. As can be seen in Figure 1b, the sequence identities are 29%, 35%, and 25% for HEAT1, HEAT2, and HEAT3 repeats, respectively. However, these matched Rictor sequence segments are not adjacent. We also tested two protein domain prediction servers. The SUPERFAMILY server (Gough et al., 2001) predicted four HEAT domain clusters, while InterPro (Jones et al., 2014) simply classified Armadillo-type fold (which include HEAT domain) in the similar regions. These methods oversimplified the domain assignment in the N-terminal and leave half of Rictor sequence in C-terminal unassigned.
To overcome the low sequence homology problem, we used a new protocol (Fig. 2). First, we searched the pdb database for sequence homology between human Rictor sequence and the sequences of known HEAT and WD40 domain structures. We found 43 nonredundant structures that have sequence matches with the human Rictor sequence. After discarding segments that have less than 10 similar amino acids, we retained a total of 168 Rictor sequence matches to HEAT-like domains. As can be seen in Table 2, the longest sequence match spans 109 residues (from 948 to 1057). The overall sequence identities matching HEAT domains range from 18% to 31%. In Figure 3A, we illustrate the homologous region in a transportin structure (PDB code 2h4m). Blast search reveals four matches (Table 2 and Fig. 3A), indicating that the Rictor protein may have HEAT repeats in these regions. After counting the frequency of the each residue in the Rictor protein sequence that appeared in the 168 Rictor sequence matches (with known HEAT domains), we calculate the standard score to obtain the probability of the residue to have a HEAT domain structure. As can be seen in Figure 3, the regions of residues 44–200, 397–430, and 937–981 are very likely to be the HEAT repeat domain, consistent with the results obtained from the SUPERFAMILY server (Gough et al., 2001). It is likely that the previously assigned domains D1 and D2 are HEAT repeats, and D3 is also likely to be a HEAT domain. It is interesting to note that the region of 937–981 has the highest HEAT domain probability. We will examine the nature of this region in next section.
Table 2.
Select Sequence Matches of Rictor with Proteins in the HEAT, WD40, and PH Domain Datasets
| Rictor sequence | PDB hit | Hit sequence | Identity % | Positive % | E-value | Domain |
|---|---|---|---|---|---|---|
| 427–502 | 3zbo | 155–227 | 21 | 40 | 1.3 | HEAT |
| 480–535 | 3zbo | 167–222 | 16 | 44 | 8.6 | |
| 138–190 | 3gs3 | 65–117 | 25 | 43 | 0.049 | |
| 720–784 | 3gs3 | 25–87 | 20 | 47 | 0.42 | |
| 5–94 | 4lun | 4–82 | 30 | 42 | 0.19 | |
| 204–250 | 4lun | 49–96 | 27 | 56 | 0.014 | |
| 887–978 | 4bsn | 35–113 | 31 | 44 | 0.013 | |
| 840–958 | 4bsn | 430–525 | 24 | 37 | 0.73 | |
| 397–457 | 3w3t | 883–945 | 23 | 44 | 0.037 | |
| 340–400 | 3w3t | 127–191 | 26 | 44 | 0.42 | |
| 948–1057 | 3w3t | 613–712 | 25 | 42 | 0.67 | |
| 1472–1564 | 3w3t | 983–1061 | 20 | 46 | 1.5 | |
| 399–507 | 4k92 | 157–262 | 27 | 38 | 1.1 | |
| 126–191 | 2h4m | 48–113 | 18 | 51 | 0.22 | |
| 764–805 | 2h4m | 351–396 | 30 | 50 | 1.4 | |
| 1148–1248 | 2h4m | 24–117 | 21 | 41 | 1.6 | |
| 430–521 | 2h4m | 515–618 | 26 | 38 | 2.9 | |
| 1595–1657 | 4ozu | 143–214 | 31 | 44 | 2.2 | WD40 |
| 1258–1353 | 4n14 | 12–110 | 23 | 39 | 0.28 | |
| 377–391 | 4n14 | 8–22 | 33 | 60 | 5.1 | |
| 1209–1231 | 4n14 | 102–124 | 35 | 56 | 5.4 | |
| 605–656 | 4n14 | 1–49 | 27 | 42 | 6.4 | |
| 1298–1373 | 2ynn | 49–128 | 27 | 43 | 0.75 | |
| 1705–1724 | 2ynn | 134–153 | 35 | 45 | 3.7 | |
| 1597–1619 | 2ynn | 244–266 | 35 | 56 | 5.9 | |
| 1135–1192 | 2ynn | 108–184 | 24 | 39 | 8.9 | |
| 1007–1081 | 1nr0 | 31–108 | 26 | 44 | 2.4 | |
| 1271–1292 | 1nr0 | 27–48 | 36 | 45 | 4.5 | |
| 1530–1576 | 1nr0 | 411–448 | 23 | 38 | 4.9 | |
| 741–834 | 1x1f | 28–137 | 27 | 41 | 0.007 | PH |
| 319–353 | 2×18 | 70–106 | 35 | 48 | 0.59 | |
| 1024–1069 | 2×18 | 52–97 | 24 | 47 | 1.2 | |
| 506–529 | 2×18 | 14–37 | 25 | 58 | 1.4 | |
| 60–74 | 2×18 | 52–66 | 40 | 60 | 1.5 | |
| 611–624 | 2×18 | 28–41 | 43 | 64 | 2.1 | |
| 452–513 | 3tca | 190–254 | 24 | 38 | 1.3 | |
| 1019–1055 | 3tca | 127–164 | 21 | 44 | 8.9 | |
| 1005–1069 | 1unr | 44–99 | 22 | 41 | 5.0 | |
| 178–207 | 1unr | 96–125 | 27 | 40 | 8.1 | |
| 248–281 | 3b77 | 57–92 | 33 | 44 | 0.6 | |
| 858–931 | 3b77 | 51–122 | 25 | 44 | 1.6 | |
| 583–1639 | 2fjl | 56–105 | 30 | 47 | 0.16 | |
| 609–630 | 2fjl | 122–143 | 36 | 54 | 2.0 |
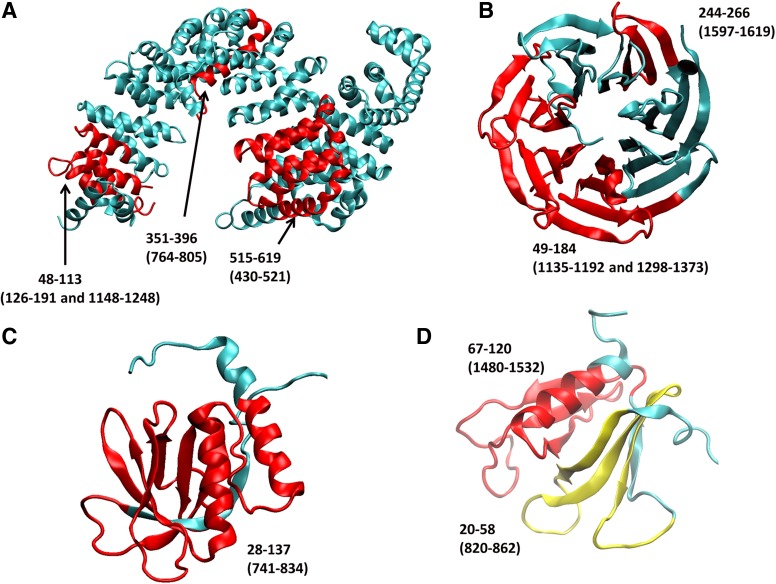
FIG. 3.
The structures of proteins that share block sequence homology with the Rictor protein. Red and yellow ribbons highlight the matched regions. Corresponding sequences of Rictor are indicated in parentheses. (A) HEAT repeats of transportin (PDB code 2h4m). (B) WD40 domain of coatomer protein COPI (PDB code 2ynn). (C) PH domain of human docking protein BRDG1 (PDB code 1x1f). (D) PH domain of human pleckstrin 2 (PDB code 1x1g).
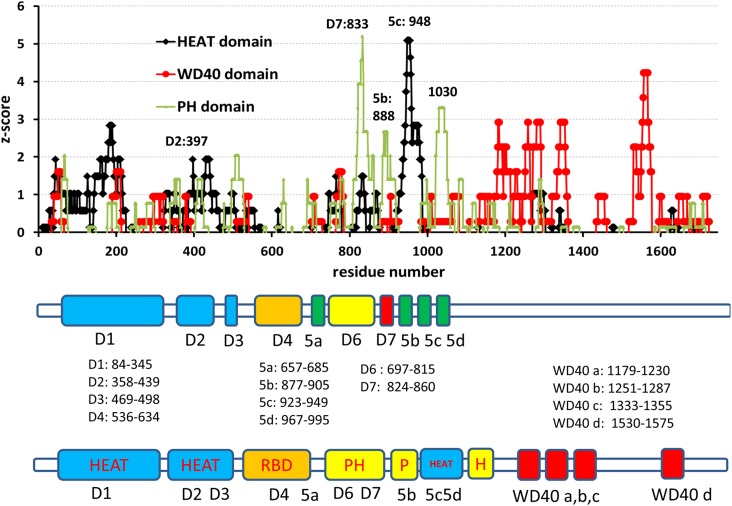
We then searched the WD40 domains using the same approach. While no matches were found using existing servers, we found more than 37 structures containing sequences homologous to the human Rictor protein (Tables 1 and 2) using pairwise sequence alignment. We collected 100 Rictor sequence matches longer than 10 residues. The longest sequence match spans from residue 1298–1373, which has 27% identity with residues 49–128 in coatomer protein COPI (Table 2 and Fig. 3B). As can be seen in Figure 3B, more than half of the WD40 repeats in the coatomer protein COPI matches the human Rictor sequence. After calculating the standard probability score of the WD40 domain structure, four clusters emerge in the c-terminal region, which are likely to be WD40 repeats. Therefore, we assign four WD domains for sequences 1179–1230, 1251–1287, 1333–1355, and 1530–1575 (Fig. 4).
FIG. 4.
Statistical analysis of the Rictor sequence matches with the domain structure dataset reveals possible HEAT, WD40, and PH domains. The top panel is the standard z-score of the three domains. The middle panel is the seven conserved regions in the Rictor sequence (Sarbassov et al., 2004). The bottom panel is the domain assignments obtained from the z-score and homology study.
3.2. Two possible PH domains
The successful identification of HEAT and WD40 domains in the human Rictor protein prompted us to examine other possible domains, especially in the D3–D7 regions, which are still undefined (Fig. 1C). We focused on the PH domain, which consists of two perpendicular anti-parallel β sheets, followed by an amphipathic helix (Lemmon, 2007). In our preliminary sequence comparisons of Rictor with its interacting proteins, we noticed some sequence similarities with the PH domain in Human Sin1, another mTorC2 subunit, and the PH domain in Akt kinase. Therefore, we used the dataset approach to look at the probability of PH domain in Rictor.
It is difficult to detect a PH domain since the loops connecting the β-strands differ greatly in length, and that there are no invariant residues within the domain. Nevertheless, we found 64 PH domains that have block sequence matches with the human Rictor protein (Table 1), with 120 matches longer than 10 residues. Unlike the short HEAT and WD40 domain, the PH domain usually spans more than 100 residues. Most of the 120 matches are not sequentially continuous residues in the Rictor protein (Table 2). However, we identified Rictor residues 741–834, which have 27% identity with human docking protein BRDG1 (PDB code 1x1f; Fig. 3C). Similarly, Rictor segment 775–831 also matches Kindlin-3 protein (PDB code 2ys3) residues 47–103 with 28% identity (Table 2). Rictor residues 741–834 cover most regions in D6 and D7 (Fig. 4). By analyzing a total of 120 sequence matches, we obtained the probabilities of the PH domain. We found that it is possible for PH domains to exist around residues 833 and 1030. Therefore, we assign the D6 regions as PH domains since the region has a high z-score and 27% sequence identity with the PH domain of human docking protein BRDG1. The nearby D7 region may also be part of the PH domain.
It is possible that Rictor contains a second PH domain. It is known that pleckstrin itself contains two PH domains, and Akt kinase also has two PH domains. An earlier study suggested that region 5a may have three other repeats beyond region 7, which were labeled as 5b, 5c, and 5d (Figs. 1 and 4). However, our domain searches do not support the putative repeat pattern. As can be seen in Figure 4, the most prominent HEAT domain signal is around residue 948, which covers regions 5c and 5d. Interestingly, the 5c/5d HEAT region inserts into two PH regions, suggesting that the second PH domain could be a split PH domain. PH domains have been found to be split, as in the case of phospholipase C-gamma (PDB code 2fjl). We found that several separated Rictor sequence regions have the potential to fold into one integrated PH domain. As indicated in Figure 4D and Table 2, Rictor residues 820–852 and 1480–1532 have 28% and 24% sequence identities with residues 20–58 and 67–120 in the c-terminal PH domain of human pleckstrin 2, respectively. While we did not find a complete match of the 5b region (residue 877–905) and region 998–1081 to one known PH domain, the standard z-scores suggest that the two separated regions could be a split PH domain, separated by the 5c5d HEAT domain.
3.3. Where is the ribosome binding domain?
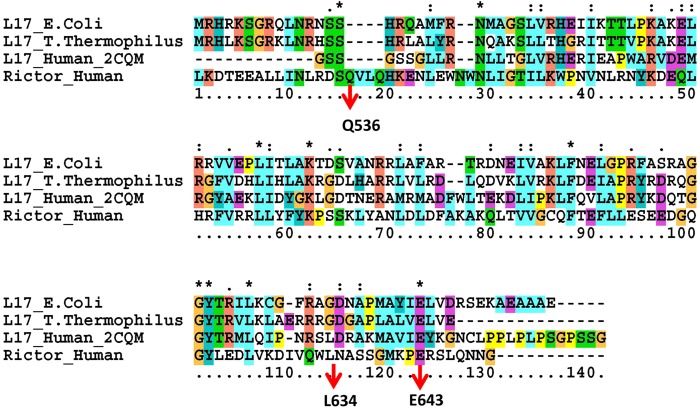
mTORC2–ribosome interaction is a likely conserved mechanism of TORC2 activation (Zinzalla et al., 2011). However, it is not clear which protein interacts with ribosome and how. The function of an unknown biological sequence can often be accurately inferred by identifying homologous sequences (Grundy, 1998). To look for Rictor–ribosome interaction, we systematically blast different Rictor domains with all pdb structures. We found that Rictor sequences 549–626 have 26% identity with residue 21–96 of 50S ribosome protein L17. This Rictor sequence covers most of the D4 domain region, indicating that this region is likely an RNA–ribosome binding domain.
To check functional relevance of the sequence homology with Rictor–ribosome binding, we compared sequence homology of bacterial 50S ribosome protein L17 and the most close mammal homolog, mitochondrial 39S ribosome protein L17. As can be seen in Figure 5, bacterial 50S ribosome protein L17 and mitochondrial 39S ribosome protein L17 are indeed homologous to Rictor sequences 536–653.
FIG. 5.
Sequence alignment of the Rictor D4 region revealed a putative ribosome binding domain. Four sequences are used in alignment: 50S ribosome protein L17 for Escherichia coli and Thermus thermophilus, human mitochondrial ribosomal protein L17 isolog (PDB code 2cqm), and the Rictor sequence in the D4 region. The red arrows indicate the residue numbers in the human Rictor protein.
3.4. Phosphorylation pattern in different Rictor domains
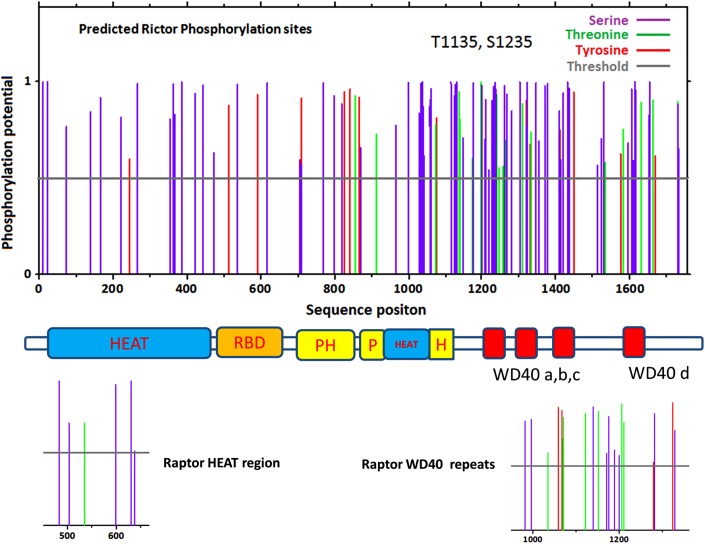
Using the NetPhos 2.0 Server (Blom et al., 1999), we scanned Raptor and Rictor for possible phosphorylation sites. In Figure 6, we plot the probabilities of Ser, Thr, and Tyr phosphorylations in Rictor and comparable domain regions in Raptor. A value larger than 0.5 indicates high probability. It is experimentally known that phosphorylation at residues T1135 and S1235 is important functionally (Gao et al., 2010; Chen et al., 2011). The webserver correctly predicted these two sites. As can be seen in Figure 6, both the N-terminal HEAT domain and the inserted HEAT domain around residues 900–1000 have several possible Ser phosphorylation sites. Raptor HEAT domains have a similar phosphorylation pattern. The putative ribosome binding domain has two Tyr and two Ser phosphorylation signals. It is interesting to note that the previously identified domain 7 region (residues 824–860) has a high probability of Tyr phosphorylation. The C-terminal WD40 regions have densely populated potential phosphorylation sites. This could be caused by the high serine residue density in the C-terminal region. Still, it is possible that phosphorylation would be an important mechanism to regulate WD40 domain interactions. It is known that, during cellular stress, GSK-3β phosphorylates Rictor's serine-1235, a modification that interferes with the binding of Akt to mTORC2 (Chen et al., 2011). Integrin-linked kinase (ILK) forms complex with Rictor in cancer but not in normal cells, and this was accompanied by ILK-dependent phosphorylation of Rictor on residue Thr1135 (Serrano et al., 2013). Our scoring indicates that Thr1135 also has high probability to be in a WD40 domain.
FIG. 6.
Predicted phosphorylation sites in Rictor and Raptor reveal that the WD40 domain could have densely populated phosphorylation sites. The top panel is the phosphorylation scores for Rictor, and the phosphorylation scores for the corresponding domains in Raptor are shown at bottom.
4. Conclusions
The difficulty of assigning a domain structure to Rictor reflects the difficulty of accurately aligning distant protein sequences, especially for those proteins with less than 25% identity. Several approaches have been proposed to address the challenge (Kececioglu et al., 2010). Homology identification could also suffer from mis-assignments because of the similarity of homologous domains in otherwise unrelated sequences (Zinzalla et al., 2011). Often the protein domain and function do not have definitive assignment because of low sequence identity, and advanced techniques from fuzzy theory have been used to classify proteins (Dombi and Kertesz-Farkas, 2009). Here, instead of designing a new algorithm to identify distant protein homology, we tested a new approach to identify specific domains in Rictor. The novelty of our protocol is in combining multiple pairwise sequence alignments with proteins in the protein domain structure dataset to obtain the likelihood of a target protein to have a specific domain. As can be seen in Table 2, the E-values from individual pairwise alignments are not in the highly significant range, which reflect (1) the lack of consensus sequence pattern for a protein domain, and (2) the low sequence homology of the target protein to a known domain. However, by combining the frequencies of these low homology hits and using z-scores to re-rank the probabilities of a protein to have a given domain, we are able to identify a domain overlooked by searches via the SUPERFAMILY (Gough et al., 2001) and InterPro (Jones et al., 2014) servers.
More extensive work is needed to check the applicability of our protocol in large-scale studies. Nevertheless, our study demonstrated one possible approach for searches for fold similarity for proteins lacking consensus sequence patterns. Exploiting this approach, we are able to define Rictor's domain arrangement for the first time. Our findings provide insights into the differences and similarities of Raptor and Rictor. Both proteins bind mTOR and drive the dimerization of mTORC complexes. We found that Rictor also has HEAT and WD40 domains, which could be the common binding motif to mTORC. We further observed that Rictor may also have PH domains. PH domains recognize specific phosphoinositides that could elicit response to cellular localization by transmitting signals to downstream targets. Rictor also contains a domain that is homologous to 50S protein L17 and human 39S protein L17, identifying an mTORC2 potential interaction site with the ribosome.
Acknowledgments
The authors are funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. P.Z. and N.Z. thank Chinese NFSC grant 30772529 and 973 program grants 2011CB933100 and 2010CB933900.
Author Disclosure Statement
No competing financial interests exist.
References
- Andrade M.A., and Bork P. 1995. HEAT repeats in the Huntington's disease protein. Nat. Genet. 11, 115–116 [DOI] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., and Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- Boulbes D., Chen C.H., Shaikenov T., et al. 2010. Rictor phosphorylation on the Thr-1135 site does not require mammalian target of rapamycin complex 2. Mol. Cancer Res. 8, 896–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H., Shaikenov T., Peterson T.R., et al. 2011. ER stress inhibits mTORC2 and Akt signaling through GSK-3beta-mediated phosphorylation of rictor. Sci. Signal 4, ra10. [DOI] [PubMed] [Google Scholar]
- Dombi J., and Kertesz-Farkas A. 2009. Applying fuzzy technologies to equivalence learning in protein classification. J. Comput. Biol. 16, 611–623 [DOI] [PubMed] [Google Scholar]
- Frey J.W., Jacobs B.L., Goodman C.A., and Hornberger T.A. 2014. A role for Raptor phosphorylation in the mechanical activation of mTOR signaling. Cell Signal 26, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Wan L., and Wei W. 2010. Phosphorylation of Rictor at Thr1135 impairs the Rictor/Cullin-1 complex to ubiquitinate SGK1. Protein Cell 1, 881–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J., Karplus K., Hughey R., and Chothia C. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313, 903–919 [DOI] [PubMed] [Google Scholar]
- Grundy W.N. 1998. Homology detection via family pairwise search. J. Comput. Biol. 5, 479–491 [DOI] [PubMed] [Google Scholar]
- Guertin D.A., and Sabatini D.M. 2007. Defining the role of mTOR in cancer. Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- Hoshii T., Kasada A., Hatakeyama T., et al. 2014. Loss of mTOR complex 1 induces developmental blockage in early T-lymphopoiesis and eradicates T-cell acute lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. USA 111, 3805–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Arauz E., Aggarwal V., et al. 2014. Stoichiometry and assembly of mTOR complexes revealed by single-molecule pulldown. Proc. Natl. Acad. Sci. USA 111, 17833–17838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.Y., et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.A., Pacold M.E., Cervantes C.L., et al. 2013. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341, 1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kececioglu J., Kim E., and Wheeler T. 2010. Aligning protein sequences with predicted secondary structure. J. Comput. Biol. 17, 561–580 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., and Ali S.M., et al. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- Kogasaka Y., Hoshino Y., Hiradate Y., et al. 2013. Distribution and association of mTOR with its cofactors, raptor and rictor, in cumulus cells and oocytes during meiotic maturation in mice. Mol. Reprod. Dev. 80, 334–348 [DOI] [PubMed] [Google Scholar]
- Lemmon M.A. 2007. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 74, 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden T.L., Tatusov R.L., and Zhang J. 1996. Applications of network BLAST server. Methods Enzymol. 266, 131–141 [DOI] [PubMed] [Google Scholar]
- Martin S.K., Fitter S., Dutta A.K., et al. 2015. Brief report: The differential roles of mTORC1 and mTORC2 in mesenchymal stem cell differentiation. Stem Cells 33, 1359–1365 [DOI] [PubMed] [Google Scholar]
- Nedelec E., Moncion T., Gassiat E., et al. 2005. A pairwise alignment algorithm which favors clusters of blocks. J. Comput. Biol. 12, 33–47 [DOI] [PubMed] [Google Scholar]
- Rosner M., and Hengstschlager M. 2008. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum. Mol. Genet. 17, 2934–2948 [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Kim D.H., et al. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- Serrano I., McDonald P.C., Lock F.E., and Dedhar S. 2013. Role of the integrin-linked kinase (ILK)/Rictor complex in TGFbeta-1-induced epithelial-mesenchymal transition (EMT). Oncogene 32, 50–60 [DOI] [PubMed] [Google Scholar]
- Smrz D., Kim M.S., Zhang S., et al. 2011. mTORC1 and mTORC2 differentially regulate homeostasis of neoplastic and non-neoplastic human mast cells. Blood 118, 6803–6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.X., Ji X., Mao X., et al. 2014. Differential activation of mTOR complex 1 signaling in human brain with mild to severe Alzheimer's disease. J. Alzheimers Dis. 8, 437–444 [DOI] [PubMed] [Google Scholar]
- Tang F., Wu Q., Ikenoue T., et al. 2012. A critical role for Rictor in T lymphopoiesis. J. Immunol. 189, 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Yang K., and Chi H. 2014. Cutting edge: discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J. Immunol. 193, 4297–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., and Hall M.N. 2006. TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- Yip C.K., Murata K., Walz T., et al. 2010. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell 38, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Yang K., Cloer C., et al. 2013. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Zhang X., Li M., et al. 2010. mTOR complex component Rictor interacts with PKCzeta and regulates cancer cell metastasis. Cancer Res. 70, 9360–9370 [DOI] [PubMed] [Google Scholar]
- Zinzalla V., Stracka D., Oppliger W., and Hall M.N. 2011. Activation of mTORC2 by association with the ribosome. Cell 144, 757–768 [DOI] [PubMed] [Google Scholar]