Abstract
The dismal clinical context of advanced-grade glioma demands the development of novel therapeutic strategies with direct patient impact. Adenovirus-mediated virotherapy represents a potentially effective approach for glioma therapy. In this research, we generated a novel glioma-specific adenovirus by instituting more advanced genetic modifications that can maximize the efficiency and safety of therapeutic adenoviral vectors. In this regard, a glioma-specific targeted fiber was developed through the incorporation of previously published glioma-specific, phage-panned peptide (VWT peptide) on a fiber fibritin-based chimeric fiber, designated as “GliomaFF.” We showed that the entry of this virus was highly restricted to glioma cells, supporting the specificity imparted by the phage-panned peptide. In addition, the stability of the targeting moiety presented by fiber fibritin structure permitted greatly enhanced infectivity. Furthermore, the replication of this virus was restricted in glioma cells by controlling expression of the E1 gene under the activity of the tumor-specific survivin promoter. Using this approach, we were able to explore the combinatorial efficacy of various adenoviral modifications that could amplify the specificity, infectivity, and exclusive replication of this therapeutic adenovirus in glioma. Finally, virotherapy with this modified virus resulted in up to 70% extended survival in an in vivo murine glioma model. These data demonstrate that this novel adenoviral vector is a safe and efficient treatment for this difficult malignancy.
Introduction
The heterogeneous tumor milieu and highly aggressive nature of glioblastoma (GBM) makes therapeutic treatments especially difficult. As a result, the prognosis for patients with this tumor entity is a mere 14.6 months even after a combination of surgery, radiotherapy, and temozolomide chemotherapy.1 This unfortunate situation does not offer much hope and is precisely why such a complicated condition requires advanced, specified treatment options heretofore unavailable.
Because of the flexibilities permitted for genetic modification and the well-characterized biology of adenovirus, the human adenovirus serotype 5 (HAd5) is applicable in promising cancer therapies as an oncolytic agent, itself, or as a cytotoxic gene(s) carrier.2–4 Despite this, glioma cells are highly variable and express little to no coxsackievirus and adenovirus receptor (CAR), the primary receptor of HAd5.3 This makes it difficult to use HAd5 in a virotherapeutic approach to glioma. For this reason, most of the current adenoviral vectors for glioma virotherapies are genetically modified to enhance their infectivity in glioma cells. These infectivity-enhanced HAd5 vectors cannot, however, distinguish between neoplastic and normal cells, which results in nonspecific viral infection and the death of peripheral, nonneoplastic cells as an unfortunate side effect.5
To overcome this current hurdle of adenovirus-mediated virotherapy against glioma, we applied new genetic modifications to the tropism-dictating fiber to engineer a virus that selectively and efficiently infects, and thereby lyses, glioma cells. Although there are several targetable glioma-specific proteins such as EGFRvIII, IL13Rα2, and CD133, the levels of their expression are highly variable in heterogeneous glioma cells.6,7 For this reason, we selected a previously published glioma-specific binding peptide, VTW peptide, originating from the phage biopanning technique.8 This technique has shown its effectiveness for selecting small peptide(s) that recognize a specific cell or a tissue type.9,10 It is a particularly suitable alternative when no clear targetable motif is known to be related to the heterogeneity of the tumor cell population.
Genetic modifications of the HAd5 fiber have typically been accomplished by adding targeting peptides to the C terminus of the fiber or by replacing CAR-binding residues on the fiber knob domain with targeting peptides, which limits the size of incorporable peptides (maximum of 10–15 amino acids) and may cause instability of the fiber because of the charge associated with incorporated peptides.11 Therefore, to increase the stability and number of targeting peptides, we replaced the entire wild-type fiber with the T4 bacteriophage-oriented fibritin trimerization domain.12 This approach is especially stable and particularly effective in the matter of incorporating a large targeting motif such as CD40L or a single-chain antibody without affecting the binding ability of the original targeting moiety, enhancing both the infectivity and specificity of a newly engineered virus.13–15
To restrict the adenoviral replication to occur only in neoplastic cells, several approaches were developed including transcriptionally regulating the expression of the replication-essential gene, early gene 1 (E1), under the control of tumor-specific promoters (TSPs; e.g., survivin, midkine, and CXCR4)16,17 and using unique cancer molecular characteristics: interaction between the E1 viral protein with Rb in neoplastic cells (the so-called E1Δ24 approach).1 We have previously shown that the survivin promoter is highly active in glioma cells yet minimally active to inactive in normal cells.16 Therefore, to further minimize any cytopathic effects mediated by virus replication in normal cells, the replication of this new glioma-targeted adenovirus was regulated by controlling the expression of the E1 gene by means of the survivin promoter. In this research, we explored the efficacy of advanced novel adenoviral genetic modifications to augment the specificity, infectivity, and exclusive replication in glioma in a way that could achieve promising therapeutic results in human clinical applications.
Materials and Methods
Cell lines
Human embryonic kidney (HEK) 293, HEK 293-F28 stably expressing Ad5 wild-type fibers, Chinese hamster ovary (CHO) cells, CHO-hCAR cells stably expressing human CAR, two glioma cell lines (U87 and U251), two nonneoplastic cell lines (NHA [normal human astrocytes] and NSC [neuronal stem cells, HB1.F3 CD]), and two neoplastic cell lines of other originated anatomical sites (A549 [human lung carcinoma cell line] and MDA-MB231-BrM2 [brain metastatic breast cancer cell line; a gift from J. Massagué]) were cultured in medium containing 10% fetal bovine serum (FBS; Hyclone, Logan, UT), penicillin (100 U/ml), and streptomycin (100 mg/ml) (Mediatech, Herndon, VA) and incubated at 37°C in 5% CO2 under humidified conditions. U87-luc was generated by transfecting pGL4 encoding firefly luciferase with Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA) and cultured in the same way as described previously. Primary human brain tumor specimen (GBM43 and GBM6) were obtained from D. James (UCSF, San Francisco, CA) in accordance with a protocol approved by the institutional review board at UCSF. The establishment and maintenance via implantation on the flank side of nude mice was previously described.18
Genetic modifications and virus production
To generate a fiber shuttle vector, pKan 566 carrying the gene encoding fiber fibritin was used.15 The nucleotide sequence for the selected glioma cell-binding peptide, VTWTPQAWFQWV, was inserted at the C terminus of fiber fibritin by a standard molecular technique, resulting in pKan-GliomaFF. With this fiber shuttle vector, two recombinant HAd5 backbones containing either an enhanced green fluorescent protein (eGFP) under the control of the cytomegalovirus (CMV) promoter in the E1-deleted region (replication-incompetent Ad5GliomaFF-CMV-GFP) or the E1 gene under the control of the survivin promoter (conditionally replicating Ad5GliomaFF-Sur E1) were constructed. The recombinant viruses were rescued and propagated in HEK293-F28 until the last propagation step, and then the last propagation in 20×175 flasks was done in HEK293 cells. Viruses were purified by two rounds of CsCl gradient ultracentrifugation and their titer was determined at 260 nm.19 The viral titer of Ad5GliomaFF was highly equivalent to that of Ad5WT (Supplementary Fig. S3; supplementary data are available online at www.liebertpub.com/hum).
Western blot
Purified adenoviral particles (1×1010) were diluted in Laemmli buffer, incubated either at room temperature (unboiled samples) or at 95°C (boiled samples) for 10 min, and loaded to a 7.5% sodium dodecyl sulfate (SDS)–polyacrylamide gel (Bio-Rad, Hercules, CA). After electrophoretic separation, samples were transferred onto a polyvinylidene difluoride (PVDF) membrane and detected with a 4D2 monoclonal anti-fiber tail antibody (diluted 1:3000) (Thermo Scientific, Rockford, IL) followed by horseradish peroxidase (HRP)-tagged anti-mouse IgG secondary antibody (diluted 1:5000) (Santa Cruz Biotechnology, Santa Cruz, CA).
Virus infectivity analysis
Cells (3×105) were plated in 24-well plates and incubated overnight. Each virus sample was diluted to a multiplicity of infection (MOI) of 300 viral particles (VP)/cell in 500 μl of infection medium containing 2% FBS in Dulbecco's modified Eagle's medium (DMEM). The cells were infected with each virus for 2 hr at 37°C. Virus-containing medium was replaced with fresh medium containing 2% FBS and cells were maintained at 37°C at atmospheric humidity and 5% CO2 for 3 days until flow cytometric analysis. For the VTW peptide inhibition assay, cells were incubated with 0–100 μg of the VTW peptide (Peptide 2.0, Chantilly, VA) for 1 hr at 37°C before viral infection.
Oncolytic efficacy
MTT assay
Viral infection was done in the same way as described previously except that an MOI of 100 VP/cell was used. The MTT assay was performed with cell proliferation kit I (MTT) according to the manufacturer's instruction (Roche, Mannheim, Germany).
Crystal violet staining
Cell were infected with serially diluted MOIs (0–100 VP/cell) and incubated for 8 days at 37°C at atmospheric humidity containing 5% CO2. After fixing with 4% paraformaldehyde, cells were stained with 500 μl of 0.1% crystal violet solution in distilled water. After several washings with water, the plates were dried completely at room temperature. Crystal violet staining was dissolved with 500 μl of 100% methanol and read with a plate reader at OD570.
Replication efficiency by quantitative polymerase chain reaction
Viral infection (MOI, 100 VP/cell) was done as described previously. The samples (cells and medium) were collected at 24-, 48-, and 72-hr intervals after viral infection and the DNAs from the samples were isolated with a DNeasy tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. qPCR was conducted with a SYBR green qPCR kit (Bio-Rad), using the primers indicated in Supplementary Table S1. Data analysis was performed by the 2–ΔΔCT method for relative quantification, and all sample values were calculated as a relative ratio to expression values from Ad5WT.
Animal study
Intracranial glioma xenograft implantation
U87MG-Luc (2.5×105 cells) or GBM43 (1×105 cells) glioma cells were implanted via cranial guide screws as described previously.18 The mice were anesthetized with a ketamine–xylazine mixture (115/17 mg/kg). A burr hole was made to facilitate stereotactic injection that was performed with a 10-μl Hamilton syringe (Hamilton, Reno, NV) with a 30-gauge needle. The small-gauge needle was mounted to a mouse-specific, stereotactic Harvard apparatus (Harvard Bioscience, Holliston, MA) to facilitate insertion and was then inserted through the burr hole to an anatomical position of 3 mm in depth. Male athymic/nude mice were obtained from Charles River Laboratory (Wilmington, MA) and were cared for in accordance with a study-specific animal protocol approved by the University of Chicago Institutional Animal Care and Use Committee. Five days after tumor implantation, mice were randomly separated into two groups and were then injected with either 10 μl of phosphate-buffered saline (PBS) or 1010 VP in 10 μl of PBS. For in vivo tumor growth analysis, mice were imaged for luciferase activity by means of intraperitoneal injection of d-luciferin (4.5 mg/animal, in 150 μl of saline), and bioluminescence analysis was performed 10 min after d-luciferin administration by using a cryogenically cooled, high efficiency, charge-coupled device camera system (Xenogen, Alameda,CA). For survival analysis, animals losing ≥30% of their body weight or having trouble ambulating, feeding, or grooming were killed by CO2 administration followed by cervical dislocation.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 4 (GraphPad Software, San Diego, CA). Sample size for each group was ≥3 and numerical data were reported as means±SEM. The Student t test was used for comparisons between two groups, and analysis of variance (ANOVA) with Tukey's post hoc test was used for comparisons between more than two groups. Kaplan–Meier survival curves were generated and the log rank test was applied to compare survival distributions. All reported p values were two-sided and were considered to be statistically significant at *p<0.05, **p<0.01, and ***p<0.001.
Results
Generation of glioma-specific adenoviral vector
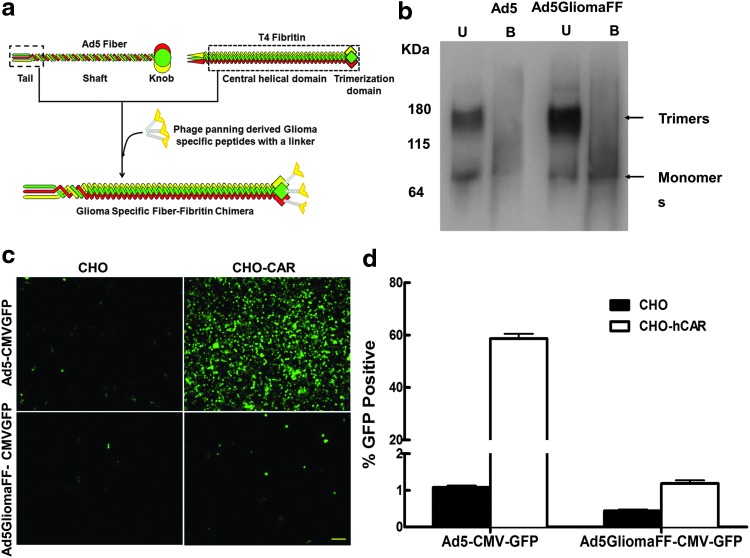
Because of the heterogeneity of glioma cells, there is no known clear surface protein to specifically target glioma cells.20,21 To achieve specifically targeted virotherapy against glioma, we applied novel genetic modifications on the viral tropism dictating fiber.2 We replaced the shaft and the knob domain of the wild-type fiber with a fiber fibritin trimerization domain to maximize the stability of the binding motif12,14,15 and incorporated a phage-panning-derived glioma-specific VTW peptide8 onto the C terminus of the fiber fibritin domain, generating a glioma-specific fiber fibritin chimera (Fig. 1a). To confirm whether the modified fiber proteins were correctly trimerized and incorporated into the viral capsids, we performed Western blot analysis with anti-HAV5-fiber tail (4D2) antibody. Two different fiber compositions from non-heat-denatured samples (trimerized fibers) and heat-denatured samples (fiber monomers) were observed at ∼180 and ∼60 kDa, respectively (Fig. 1b). The trimerization of the chimeric fiber is seen to be as efficient as that of the wild-type fiber because the ratio of the two fiber compositions is relatively equivalent for the two viruses (Fig. 1b). Thus, these data indicates that the modified fibers, GliomaFF, are correctly trimerized and incorporated into HAd5 (Fig. 1b).
Figure 1.
Generation and confirmation of the glioma-specific adenovirus. (a) Schematic diagram of a glioma-specific chimera fiber of Ad5GliomaFF. The knob and the shaft domain of HAd5 fiber were replaced with the fiber fibritin trimerization domain. The glioma-targeted chimeric fiber named GliomaFF was generated by incorporating the phage-panned glioma-specific peptides (VTW) on the C terminus of the chimeric fiber. (b) Molecular validation of a glioma-specific chimeric fiber of Ad5GliomaFF: detection of fiber trimerization and incorporation of viral particles by Western blot analysis with an HAdV-5 tail antibody (4D2). 1×1010 VP of Ad5CMV-GFP (lanes 1 and 2) or Ad5GliomFF-CMV-GFP (lanes 3 and 4) in Laemmli buffer was analyzed. Samples, identified as B (boiled, incubation at 95°C for 10 min) in lanes 2 and 4 and marked as U (unboiled, incubation at room temperature for 10 min) in lanes 1 and 3, show the denatured monomeric fiber composition and the nondenatured trimeric fiber composition, respectively. Makers indicate kilodaltons (kDa). (c) Fluorescence micrograph and (d) flow cytometric analysis of CAR-independent infection with Ad5GliomaFF-CMV-GFP. Cells were infected with Ad5GliomaFF-CMV-GFP or Ad5CMV-GFP at an MOI of 300 VP/cell for 2 hr on CAR-negative CHO cells and CAR-positive CHO-hCAR cells, and the virus-containing medium was removed and replaced with fresh medium containing 2% FBS. Fluorescence micrographs were taken 72 hr postinfection, followed by flow cytometric analysis. All images are representative of three different experiments. Scale bar, 100 μm. Each column is the average of three independent replicates. Means±SEM are plotted. Color images available online at www.liebertpub.com/hum
CAR-independent infection of Ad5GliomaFF
The deletion of the shaft and knob domains renders the virus devoid of its natural tropism but allows for novel targeting specificity by way of the fiber chimera, considering that the CAR-binding domain is located on the knob domain. First, to verify that the fiber modification results in the loss of CAR-binding ability, we analyzed the infectivity of the viruses in a human CAR (hCAR)-negative cell line, the Chinese hamster ovary (CHO) cell line, and in a CHO-derived cell line, CHO-hCAR, that stably expresses human CAR. Whereas wild-type Ad5-CMVGFP showed an approximately 60% infection rate in the CHO-hCAR cell line compared with 2% in the CHO cell line, there was no observable change in infection rate with Ad5GliomaFF-CMVGFP in either cell line (Fig. 1c and d), indicating that Ad5GliomaFF-CMVGFP infects a cell in a CAR-independent manner, confirming the loss of CAR-binding ability.
Glioma-specific and efficient infection of Ad5GliomaFF
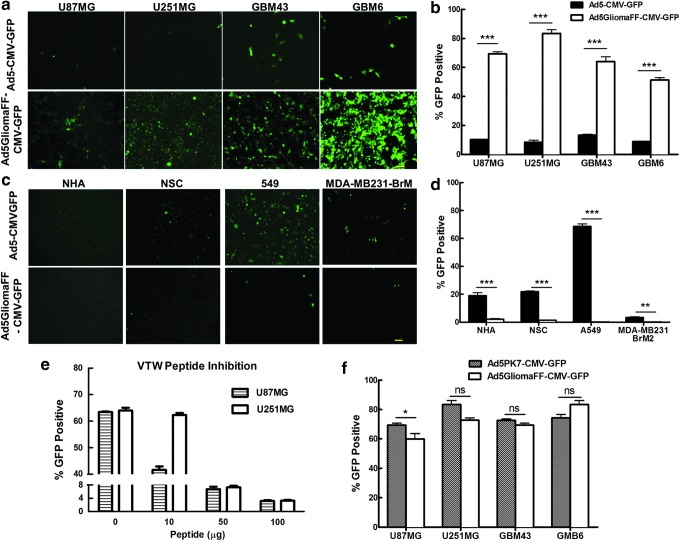
With loss of the CAR-binding ability of Ad5GliomaFF, we next investigated whether the glioma-specific fiber fibritin chimera held targeted specificity by way of fluorescence imaging techniques and flow cytometric analysis with a panel of glioma cell lines: U87MG, U251MG, GBM43, and GBM6. Whereas wild-type Ad5-CMV-GPF infected only approximately 10% of these glioma cell lines on average, Ad5GliomaFF-CMVGFP infected approximately 70% of these glioma cell lines on average (Fig. 2a and b). More importantly, this glioma-specific virus was highly efficient in the infection of cell lines obtained from patient-derived tumor xenografts: GBM43 and GBM6 (Fig. 2a and b). These two cell lines have been shown to be more clinically relevant because they have more intrinsic patient GBM molecular properties.18 To further confirm the specificity of the virus, we assessed the infectivity on a panel of nonneoplastic cells: possible by-stander target cell lines in the brain (NHA [normal human astrocyte] and NSC [neuronal stem cell]) as well as other neoplastic cell lines to establish that this virus is not cancer specific, but glioma specific: A549 (human lung adenocarcinoma epithelial cell line) and MDA-MB 231-BRM2 (brain metastatic breast cancer line). Unlike wild-type Ad5CMV-GFP, the infectivity of this glioma-specific virus showed less than 2% on this panel of cell lines (Fig. 2c and d), thereby demonstrating the specificity of Ad5GliomaFF-CMVGFP. Importantly, minimal infection of possible by-stander target cells (NHA and NSC) with the glioma-specific virus were observed (0.13 and 0.17% infection rate, respectively), affirming the safety of this virus for glioma virotherapy (Fig. 2c and d). The direct comparison demonstrated that the infection rate of Ad5GliomaFF-CMVGFP on glioma cells is statistically significant in comparison with that of the viral infection of nonneoplastic, otherwise normal cells (p<0.0001) (Supplementary Fig. S1). In addition, infection with Ad5GliomaFF-CMVGFP is dramatically decreased when the infection was performed in the present of VTW peptide, which confirmed the previously published binding specificity of VTW peptides (Fig. 2e).22
Figure 2.
Glioma-specific and efficient infection of Ad5GliomaFF. Cells were infected with Ad5GliomaFF-CMV-GFP or Ad5CMV-GFP at an MOI of 300 VP/cell for 2 hr on glioma cell lines U87 and U251, patient-derived glioma xenograft cell lines GBM43 and GBM6, nonneoplastic cells NHA (normal human astrocytes) and NSC (neuronal stem cells), and other neoplastic cell lines from various anatomical sites: A549 (human lung adenocarcinoma) and MDA-MB 231-BRM2 (brain metastatic breast cancer). The virus-containing medium was then removed and replaced with fresh medium containing 2% FBS. Fluorescence micrographs were taken 72 hr postinfection (a and c), followed by flow cytometric analysis (b and d). (e) Viral infection was done in the same way as described previously except that cells were incubated with VTW peptides for 1 hr before the infection. (f) Infection procedures were done in the same way as described previously with Ad5GliomaFF-CMV-GFP or Ad5PK7-CMV-GFP, followed by flow cytometric analysis. Scale bar, 100 μm. All images are representative of three independent experiments. Means±SEM are plotted. Color images available online at www.liebertpub.com/hum
Whenever a delivery vehicle is designed to target a specific cell population, there is often a compromise between enhanced specificity and reduced infection efficacy. Therefore, to verify the infection efficiency of this glioma-specific virus, we compared its infectivity with that of the well-established infectivity enhanced glioma virotherapeutic vector Ad5PK7-CMV-GFP, which infects a cell by binding to any anionic cell surface proteins through seven lysine residues (PK7) on the fiber. As shown in Fig. 2f, the infectivity of this new glioma-specific virus is highly comparable to that of the infectivity-enhanced virus: average infectivity of Ad5PK7-CMV-GFP was 63% and that of Ad5GliomaFF-CMV-GFP was 70% on four glioma cell lines. As such, Ad5GliomaFF-CMVGFP exclusively infects glioma cells and does so with high efficiency in a manner that can be applied safely for glioma virotherapy (Fig. 2).
The kinetics of Ad5GliomaFF infectivity
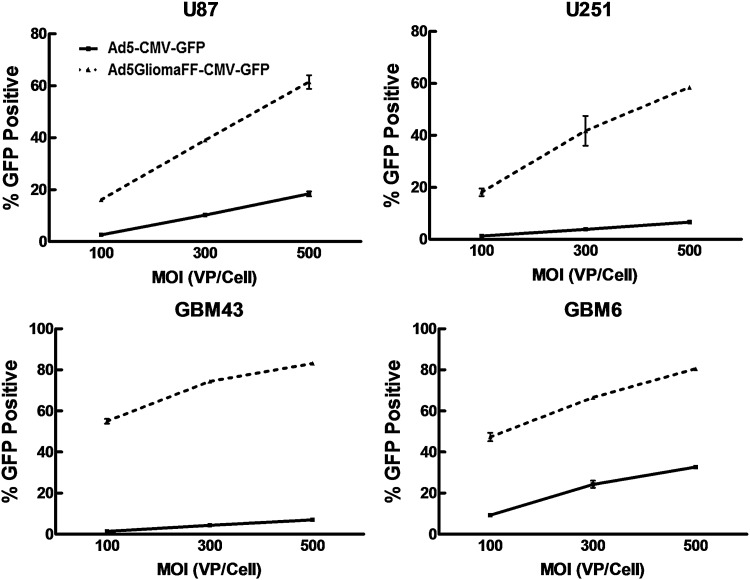
Because the modified fibers were stably trimerized and incorporated into this novel virus capsid and showed highly specific infectivity on glioma cells, we evaluated the kinetics of the viral infection to verify its infectivity efficiency in glioma cells, using various multiplicities of infection (VP/ml) on a panel of glioma cell lines. As the MOI of Ad5GliomaFF increases, the infectivity of Ad5GliomaFF shows increased infectivity on glioma cells (Fig. 3). Therefore, this novel virus has highly stable kinetics of infectivity on these glioma cell lines.
Figure 3.
Kinetics of Ad5GliomaFF infectivity. Cells were infected with Ad5GliomaFF-CMV-GFP or Ad5CMV-GFP at MOIs of 100, 200, and 300 VP/cell for 2 hr on U87MG, U251MG, GBM43, and GBM6, and the virus-containing medium was removed and replaced with fresh medium containing 2% FBS. After 72 hr, flow cytometric analysis was performed. Each data point is the average of three independent replicates. Means±SEM are plotted.
The efficient oncolytic activity of Ad5GliomaFF-SurE1 in glioma cells
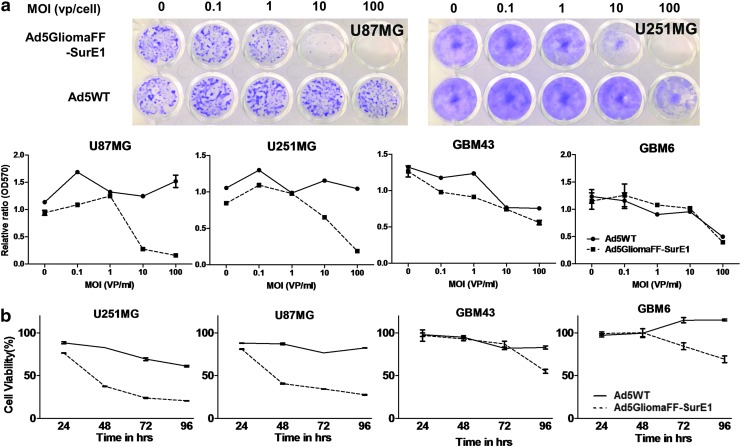
We have so far demonstrated that this novel virus is highly stable (Figs. 1b and 3) and specifically infects glioma cells with high efficiency (Fig. 2). We sought to investigate the oncolytic activity of the virus in glioma by replacing CMV-GFP (replication incompetent) with Sur-E1 (conditionally replicating under the survivin promoter). This transcriptional regulation of viral replication under the control of the survivin promoter was thought to add even more cancer-specific and safe viral characteristics for the therapeutic use of this new virus. We first performed crystal violet staining analysis on virus-infected glioma and nonglioma cell lines (Fig. 4a). Compared with Ad5WT, Ad5GliomaFF-SurE1 showed more efficient oncolytic activity on glioma cell lines, at an MOI as low as 1 VP/cell (Fig. 4a). However, there was no observable reduction of metabolic conversion rates and no cell-killing effects on the nonglioma cell line, even at an MOI as high as 100 VP/cell (Supplementary Fig. S2). To further confirm the oncolytic activity of Ad5GliomaFF-SurE1, we performed the MTT assay on two glioma cell lines (U87 and U251: highly proliferating cell lines in vitro) and two glioma cell lines obtained from patient-derived xenografts (GBM43 and GBM6: minimally proliferating cell lines in vitro) (Fig. 4b). Because the MTT assay is based on the metabolic conversion of MTT to formazan by the action of dehydrogenases, highly proliferating cell lines show more dramatic changes of metabolic conversion. Whereas Ad5WT-infected glioma cells showed a mere 10% reduction, Ad5GliomaFF-SurE1-infected glioma cells showed a highly reduced metabolic conversion rate, implying a high cell death rate. There was a 75% reduction in highly proliferating glioma cell lines and a 30–50% reduction in the patient-derived xenograft glioma cell lines in 72–96 hr (Fig. 4b). Therefore, this analysis indicates that Ad5GliomaFF-SurE1 is specifically oncolytic to glioma cells but spares nonglioma cells from the lysing effect.
Figure 4.
The efficient oncolytic activity of Ad5GliomaFF-SurE1. Cells were infected with Ad5GliomaFF-SurE1 or Ad5WT (a) at serially diluted MOI from 0 to 1000 VP/cell for 2 hr on U87MG, U251MG, GBM43, and GBM6 and the virus-containing medium was removed and replaced with fresh medium containing 2% FBS. Eight days after viral infection, cells were fixed and stained with crystal violet. Micrographs were taken with a Nikon digital camera (d5100), followed by dissolution with methanol, and read with a plate reader at OD570. Two of representative images are shown. (b) Cells were infected with Ad5GliomaFF-SurE1 or Ad5WT at an MOI of 100 VP/cell for 2 hr on U87, U251, GBM43, and GBM6 and the virus-containing medium was removed and replaced with fresh medium containing 2% FBS. At the time points of 24, 48, 72, and 96 hr, the MTT assay was performed according to the manufacturer's instruction. Each data point and column is the average of three independent replicates. Means±SEM are plotted. Color images available online at www.liebertpub.com/hum
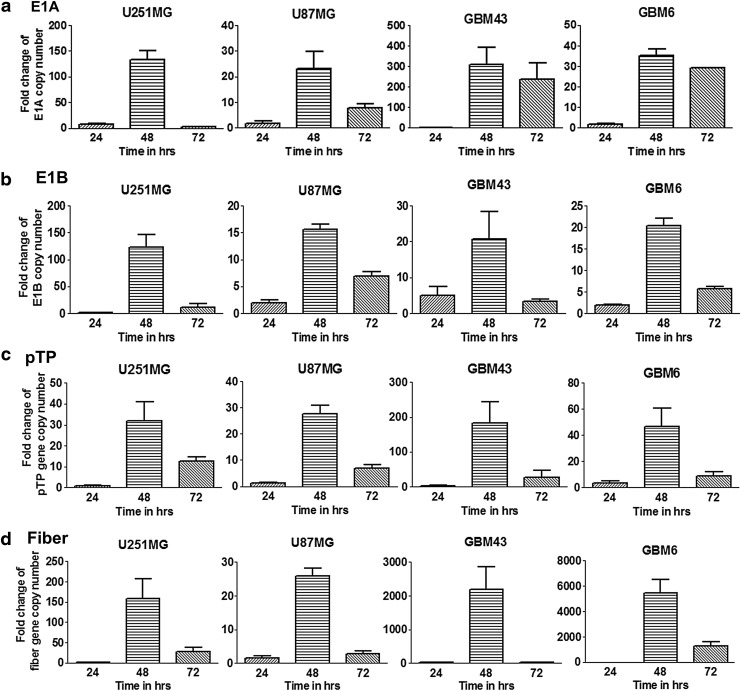
Dynamics of viral replication
Because the efficacy of virotherapy depends mainly on the viral replication-mediated oncolysis, we next investigated the dynamics of viral replication in glioma cells by quantitative PCR. To thoroughly analyze viral replication, we assessed relative gene production of the replication-dictating E1 (E1A and E1B), viral DNA replication-related precursor terminal protein (pTP), and a structural protein (fiber) from Ad5GliomaFF-SurE1-infected cells compared with Ad5WT-infected cells.
First, the transcription from the E1 gene (E1A and E1B) under the control of the survivin promoter was significantly increased in Ad5GliomaFF-SurE1-infected cells during the time period between 24 and 48 hr compared with that with Ad5WT-infected cells, implying efficient viral replication in glioma (Fig. 5a and b). Also, a viral DNA replication-related gene (precursor terminal protein, pTP) and an adenovirus structural protein gene (fiber tail region) were analyzed. Again, the transcription from both of these genes were significantly increased by Ad5GliomaFF-SurE1-infected cells during the time period between 24 and 48 hr compared with that by Ad5WT-infected cells, suggesting that efficient growth of Ad5GliomaFF-SurE1 must be present in the glioma cell lines and the patient-derived xenograft glioma cell lines (Fig. 5c and d). Therefore, Ad5GliomaFF-SurE1 can infect efficiently (Fig. 2) and transcribe viral genes effectively in these panels of glioma cell lines.
Figure 5.
Dynamics of viral replication. Cells were infected with Ad5GliomaFF-SurE1 or Ad5WT at an MOI of 100 VP/cell for 2 hr on U87, U251, GBM43, and GBM6. The virus-containing medium was removed and replaced with fresh medium containing 2% FBS. Both virus-infected cells and media were collected 24, 48, and 72 hr after viral infection and the DNA from each sample was isolated, followed by qPCR with primer sets for genes of the replication-dictating E1 (E1A (a) and E1B (b)), viral DNA replication-related precursor terminal protein (pTP (c)), and a structural protein (fiber (d)). Each column represents the average fold change in the respective gene of three independent replicates from Ad5GlimaFF-SurE1 compared with Ad5WT. Means±SEM are plotted.
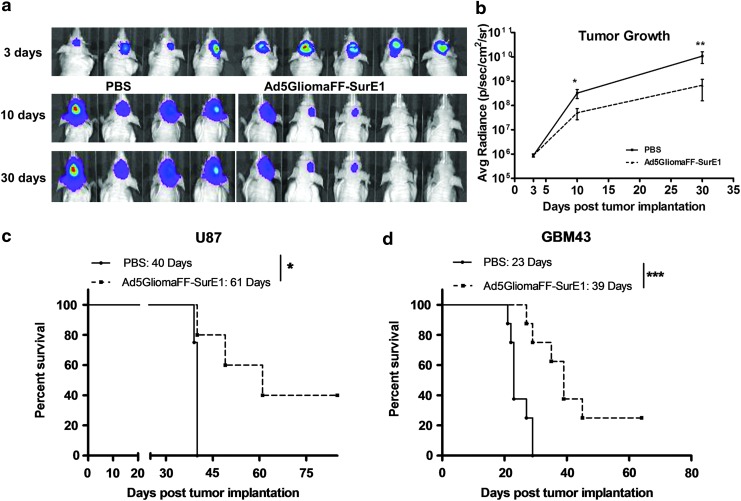
In vivo tumor growth suppression induced by Ad5GliomaFF-SurE1
Collectively, the in vitro assays showed the specificity and efficiency of Ad5GliomaFF-SurE1 in the matter of targeting and oncolysis in glioma, respectively. Hence, we next investigated tumor progression/suppression in vivo by measuring the bioluminescence of intracranially implanted U87-Luc cells, using a xenograft murine model. Four days after tumor implantation, mice were intratumorally injected with either PBS or the virus (see Materials and Methods for details). Noticeable inhibition of tumor growth was observed 5 days after viral infection via the bioluminescence images (p<0.05) and became readily apparent on day 10 after viral infection (p<0.01) (Fig. 6a and b). Therefore, the Ad5GliomaFF-SurE1-injected group showed efficient suppression of tumor growth through bioluminescence monitoring (Fig. 6).
Figure 6.
In vivo tumor growth suppression and the survival benefit conferred by Ad5GliomaFF-SurE1. Mice bearing intracranial U87-Luc tumors were treated with PBS or Ad5GliomaFF-SurE1. (a) Bioluminescence was measured 3, 10, and 30 days after tumor implantation and images were acquired at each time point. (b) Bioluminescence was plotted as average radiance versus days after tumor implantation. Coloration indicates pixel intensity of bioluminescence, which is related to the number of tumor cells. Each data point is the average of four mice (PBS-treated group) or that of five mice (Ad5GliomaFF-SurE1-treated group). Means and SE are plotted. For the survival analysis, mice bearing intracranial U87-Luc (c) or patient-derived xenograft GBM43 (d) tumors were treated with PBS or Ad5GliomaFF-SurE1 (see Materials and Methods for details). The Kaplan–Meier survival curves of treated mice were analyzed. *p<0.05, **p<0.01, and ***p<0.001. Color images available online at www.liebertpub.com/hum
The efficient oncolysis-based survival benefit conferred by Ad5GliomaFF-SurE1
As Ad5GliomaFF-SurE1 efficiently suppressed the growth of glioma, we sought to investigate further the oncolysis-mediated survival effect of Ad5GliomaFF-SurE1. In the same setting, we analyzed the survival effect of Ad5GliomaFF-SurE1 with the U87-Luc-implanted mouse model. As shown in Fig. 6a, the median survival of the Ad5GliomaFF-SurE1-treated group was extended by 50% compared with that of the PBS-treated group; the median survival of the virus-injected group was 61 days, which represented a 21-day increased median survival (p<0.05) over the PBS-injected group (40 days) (Fig. 6c).
To further investigate the oncolytic efficiency of Ad5GliomaFF-SurE1 in a more clinically relevant model, we implanted the patient-derived glioma xenograft cell line, GBM43, in the murine model. As mentioned previously, the GBM43 cell line has more intrinsic patient GBM properties and has been propagated only in the flank of nude mice.18 Even with this highly aggressive and more intrinsic patient-derived GBM model, we could see increased median survival by 16 days when mice were treated with Ad5GliomaFF-Sur-E1; 70% extended median survival as compared with the PBS-treated group (p<0.001) (Fig. 6d). Therefore, this new virus is highly specifically oncolytic to glioma and gives a more than 50% extended life span to this virus-treated group compared with the PBS-treated group.
Discussion
Although various therapies have been developed to combat GBM, there has been little progress in the development of specific and more efficient GBM-targeted therapeutic strategies without adverse effects in a clinical context.1 In our studies, the motive has been to develop a more specific, efficient, and safe virotherapeutic agent by incorporating the two imperative conditions of safety and efficient oncolysis through viral genetic modifications on both structural and functional components of the virus.
Because there is a lack of a targetable protein in the heterogeneous glioma population, we selected and incorporated phage panning-derived glioma-specific peptides (VTW) on a fiber fibritin trimerization domain that can stabilize the binding moiety of targeting peptides. Together, these two genetic modifications on the fiber domain eliminate the natural tropism of adenovirus, provide CAR-independent infection, and generate a new glioma-specific tropism for the new adenoviral vector (Ad5GliomaFF). Importantly, this new virus was able to effectively differentiate glioma cells from other cell types (both otherwise normal and neoplastic at other anatomical sites). Of note, the infection rate of cells of otherwise nonglioma differentiation was almost undetectable (less than 1%) whereas about 70% of glioma cells were infected with this virus.
In many cases, whenever a delivery vehicle is designed to specifically target a certain cell population, it is possible that it may lose its efficiency in entering the target cells, because high specificity often correlates with less efficiency. However, the infectivity of the novel virus is highly comparable to that of infectivity-enhanced Ad5PK7, which suggests that the glioma-specific virus is not only specific but also retains high efficiency for the infection of glioma cells. With highly stable kinetics of infection in addition to its highly specific and efficient infection capability, the glioma-specific virus is capable of efficient oncolysis even at a low MOI. Of note, this oncolysis was possibly due to the efficient viral replication in glioma as the production of viral genes: replication-dictating gene (E1), viral DNA replication-related gene (pTP), structural protein-producing gene (fiber) was significantly increased between 24 and 48 hr compared with Ad5WT, which further implies the high potential of the new virus for clinical therapeutic use because the clinical therapeutic efficacy depends mostly on viral replication-mediated oncolysis.3,23,24 Mechanistically, this virus specifically recognizes and infects glioma cells, replicates exponentially between 24 and 48 hr, and efficiently lyses glioma cells in 72 hr. Further, tumor progression/suppression and survival analysis in vivo showed efficient suppression of tumor growth, which gives the extended median survival of the Ad5GliomaFF-SurE1-treated group.
The efficacy of adenovirus-mediated therapeutic applications in the clinic has been shown to be minimal because most people have already been exposed to HAd5, and have generated neutralizing antibodies against the adenovirus.25,26 Also, in the matter of systemic delivery, HAd5 has been shown to have liver tropism, resulting in liver toxicity.25 However, because the majority of these neutralizing antibodies recognize either hexon (the most abundant viral protein) or fiber (the most protruded protein),26 these two domains can be easily modified to prevent antiviral responses.25,27 In our research, we replaced the wild-type fiber with the T4 phage-oriented trimer structure, to which neutralizing antibodies are highly unlikely to be present in humans, to minimize antiviral responses against the fiber antigen. Of note, we did not explore the hexon-modified approach to minimize the liver toxicity and the effect of antiviral neutralizing antibodies. This is because we injected the virus intratumorally into the brain and adenovirus cannot pass the brain–blood barrier (BBB) because of the permissive size limitation.28 Because the hexon can be targeted for antiviral responses, we can, in future research, modify the hexon to further maximize the virotherapeutic efficacy.
It was reported that virotherapeutic approaches can be augmented when virus-mediated oncolysis can induce further immune responses against tumor.29–31 In this regard, approaches that can efficiently kill tumor cells, almost to the extent of bombarding the tumor area with an oncolytic virus, were emphasized in the hope that they could induce a stronger, more potent antitumor immune response. However, these efficiency-oriented approaches, as previously said, sometimes caused the death of normal bystander cells, which also can induce unwanted immune responses against non-tumor-derived self-antigen(s). Consequentially, for more advanced and optimized virotherapy, efficacy must be achieved along with the specificity that our new, advanced approach has shown. To further enhance the antitumor immune response, synergizing with efficient oncolytic activity, we can also deliver immune modulator(s) such as PD-L1 and anti-CTLA432 on this optimized glioma-targeted virus.
In summary, we have presented here the multilevel genetic modification of a highly specific and efficient glioma-targeted virus that exponentially replicates and induces oncolysis specifically within malignant glioma cells. We showed that therapeutic treatment with this virus efficiently suppressed the growth of glioma and substantially extended median survival. We strongly believe that this approach represents a strong platform for future virotherapy, is highly translational, and could achieve promising therapeutic results in human clinical applications.
Supplementary Material
Acknowledgment
This work was supported by the National Institutes of Health (R01CA122930)
Author Disclosure
No competing financial interests exist.
References
- 1.Young JS, Kim JW, Ahmed AU, et al. . Therapeutic cell carriers: a potential road to cure glioma. Expert Rev Neurother 2014;14:651–660 [DOI] [PubMed] [Google Scholar]
- 2.Kim JW, Glasgow JN, Nakayama M, et al. . An adenovirus vector incorporating carbohydrate binding domains utilizes glycans for gene transfer. PLoS One 2013;8:e55533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nandi S, Lesniak MS. Adenoviral virotherapy for malignant brain tumors. Expert Opin Biol Ther 2009;9:737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tobias A, Ahmed A, Moon KS, et al. . The art of gene therapy for glioma: a review of the challenging road to the bedside. J Neurol Neurosurg Psychiatry 2013;84:213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng S, Ulasov IV, Han Y, et al. . Fiber-knob modifications enhance adenoviral tropism and gene transfer in malignant glioma. J Gene Med 2007;9:151–160 [DOI] [PubMed] [Google Scholar]
- 6.Pointer KB, Clark PA, Zorniak M, et al. . Glioblastoma cancer stem cells: biomarker and therapeutic advances. Neurochem Int 2014;71:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sengupta S, Thaci B, Crawford AC, et al. . Interleukin-13 receptor α2-targeted glioblastoma immunotherapy. Biomed Res Int 2014;2014:952128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C, Lo SL, Boulaire J, et al. . A peptide-based carrier for intracellular delivery of proteins into malignant glial cells in vitro. J Control Release 2008;130:140–145 [DOI] [PubMed] [Google Scholar]
- 9.Newton J, Deutscher SL. Phage peptide display. Handb Exp Pharmacol 2008;145–163 [DOI] [PubMed] [Google Scholar]
- 10.Nixon AE, Sexton DJ, Ladner RC. Drugs derived from phage display: from candidate identification to clinical practice. MAbs 2014;6:73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curiel DT. Strategies to adapt adenoviral vectors for targeted delivery. Ann N Y Acad Sci 1999;886:158–171 [DOI] [PubMed] [Google Scholar]
- 12.Krasnykh V, Belousova N, Korokhov N, et al. . Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J Virol 2001;75:4176–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alberti MO, Roth JC, Ismail M, et al. . Derivation of a myeloid cell-binding adenovirus for gene therapy of inflammation. PLoS One 2012;7:e37812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belousova N, Korokhov N, Krendelshchikova V, et al. . Genetically targeted adenovirus vector directed to CD40-expressing cells. J Virol 2003;77:11367–11377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedley SJ, Auf der Maur A, Hohn S, et al. . An adenovirus vector with a chimeric fiber incorporating stabilized single chain antibody achieves targeted gene delivery. Gene Ther 2006;13:88–94 [DOI] [PubMed] [Google Scholar]
- 16.Ulasov IV, Rivera AA, Sonabend AM, et al. . Comparative evaluation of survivin, midkine and CXCR4 promoters for transcriptional targeting of glioma gene therapy. Cancer Biol Ther 2007;6:679–685 [DOI] [PubMed] [Google Scholar]
- 17.Hardcastle J, Kurozumi K, Chiocca EA, et al. . Oncolytic viruses driven by tumor-specific promoters. Curr Cancer Drug Targets 2007;7:181–189 [DOI] [PubMed] [Google Scholar]
- 18.Ahmed AU, Thaci B, Tobias AL, et al. . A preclinical evaluation of neural stem cell-based cell carrier for targeted antiglioma oncolytic virotherapy. J Natl Cancer Inst 2013;105:968–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maizel JV, Jr, White DO, Scharff MD. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 1968;36:115–125 [DOI] [PubMed] [Google Scholar]
- 20.Jung CS, Unterberg AW, Hartmann C. Diagnostic markers for glioblastoma. Histol Histopathol 2011;26:1327–1341 [DOI] [PubMed] [Google Scholar]
- 21.Liang S, Shen G, Liu Q, et al. . Isoform-specific expression and characterization of 14-3-3 proteins in human glioma tissues discovered by stable isotope labeling with amino acids in cell culture-based proteomic analysis. Proteomics Clin Appl 2009;3:743–753 [DOI] [PubMed] [Google Scholar]
- 22.Wang C, Ning L, Wang H, et al. . A peptide-mediated targeting gene delivery system for malignant glioma cells. Int J Nanomed 2013;8:3631–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi JW, Lee JS, Kim SW, et al. . Evolution of oncolytic adenovirus for cancer treatment. Adv Drug Deliv Rev 2012;64:720–729 [DOI] [PubMed] [Google Scholar]
- 24.Pesonen S, Kangasniemi L, Hemminki A. Oncolytic adenoviruses for the treatment of human cancer: focus on translational and clinical data. Mol Pharm 2011;8:12–28 [DOI] [PubMed] [Google Scholar]
- 25.Khare R, Chen CY, Weaver EA, et al. . Advances and future challenges in adenoviral vector pharmacology and targeting. Curr Gene Ther 2011;11:241–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fausther-Bovendo H, Kobinger GP. Pre-existing immunity against Ad vectors: humoral, cellular and innate response, what's important? Hum Vaccin Immunother 2014;10:2875–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capasso C, Garofalo M, Hirvinen M, et al. . The evolution of adenoviral vectors through genetic and chemical surface modifications. Viruses 2014;6:832–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiesjema B, Hermsen HP, van Eijkeren JC, et al. . Effect of administration route on the biodistribution and shedding of replication-deficient HAdV-5: a qualitative modelling approach. Curr Gene Ther 2010;10:107–127 [DOI] [PubMed] [Google Scholar]
- 29.Wold WS, Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther 2013;13:421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi IK, Yun CO. Recent developments in oncolytic adenovirus-based immunotherapeutic agents for use against metastatic cancers. Cancer Gene Ther 2013;20:70–76 [DOI] [PubMed] [Google Scholar]
- 31.de Gruijl TD, van de Ven R. Chapter six—adenovirus-based immunotherapy of cancer: promises to keep. Adv Cancer Res 2012;115:147–220 [DOI] [PubMed] [Google Scholar]
- 32.Wainwright DA, Chang AL, Dey M, et al. . Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin Cancer Res 2014;20:5290–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.