Abstract
What happens in the early, still undetectable human malignancy is unknown because direct observations are impractical. Here we present and validate a “Big Bang” model, whereby tumors grow predominantly as a single expansion producing numerous intermixed sub-clones that are not subject to stringent selection, and where both public (clonal) and most detectable private (subclonal) alterations arise early during growth. Genomic profiling of 349 individual glands from 15 colorectal tumors revealed the absence of selective sweeps, uniformly high intra-tumor heterogeneity (ITH), and sub-clone mixing in distant regions, as postulated by our model. We also verified the prediction that most detectable ITH originates from early private alterations, and not from later clonal expansions, thus exposing the profile of the primordial tumor. Moreover, some tumors appear born-to-be-bad, with sub-clone mixing indicative of early malignant potential. This new model provides a quantitative framework to interpret tumor growth dynamics and the origins of ITH with significant clinical implications.
Introduction
The growth of human malignancies cannot be directly observed. In particular, the earliest events in the growth of a large tumor are unknown. What happens during these first cell divisions may provide clues as to how to better prevent, detect, and treat cancers. Since tumor growth is an evolutionary process and the ancestral history is recorded within tumor cell genomes1-3, detailed information on the early growth phase may be encoded in patterns of genomic intra-tumor heterogeneity (ITH) present in the final neoplasm. Specifically, in the absence of selective sweeps, it is feasible to recover the genomic profile of the primordial tumor. This task is possible because private (sub-clonal) alterations that occur early during growth should be pervasive in the final neoplasm, where pervasive refers to private alterations that are found throughout the tumor, but are not dominant. Experimentally, pervasive alterations can be detected through systematic sampling and genomic profiling of numerous regions of the same neoplasm. The initial events in neoplastic transformation are thought to occur through the step-wise accumulation of driver alterations4, whereas the growth dynamics of established neoplasms remains poorly characterized. In particular, extensive ITH and branching phylogenies revealed by cancer genomic studies5-9 suggest that the same linear paradigm does not apply to the subsequent growth of established tumors, such as colorectal carcinomas and advanced adenomas. However, the origins of ITH are unknown, and a quantitative framework to describe tumor growth dynamics is needed.
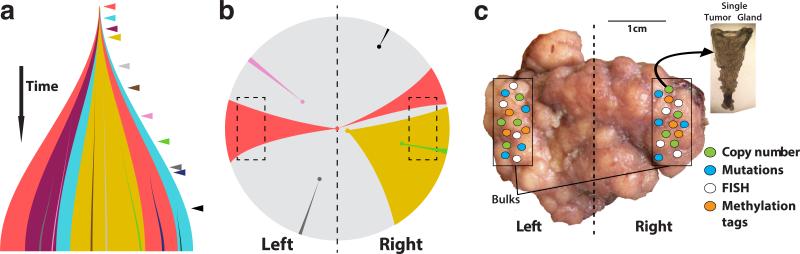
Here we propose a “Big Bang” model whereby, after the initial transformation, colorectal tumors grow predominantly as a single expansion populated by numerous intermixed sub-clones (Figure 1a). As expected, public mutations in the initiating cell will be present in all tumor cells (clonal). In contrast, while new private mutations will be continuously generated as a result of replication errors, only the earliest will be pervasive, whereas later alterations will be localized in progressively smaller tumor sub-populations. Although private mutations acquired during growth may confer survival advantages, selective sweeps that significantly alter the clonal composition of the final tumor are predicted to be extremely rare due to the rapidly expanding population and spatial constraints10-12. Hence, the timing of a mutation, rather than clonal selection for that mutation, is the primary determinant of its pervasiveness. Importantly, most observable private mutations that give rise to ITH are generated early after the transition to an advanced tumor, well before the neoplasm becomes clinically detectable. Given the absence of sequential selective sweeps, our model anticipates uniformly high levels of ITH throughout the neoplasm. Moreover, in some tumors, early sub-clone mixing followed by scattering to different distant tumor regions may occur (e.g. Figure 1a, red sub-clone). This phenomenon results in variegated tumor cell populations, where the spatial relationship between cells does not necessarily recapitulate their clonal relationship.
Figure 1. The Big Bang model of tumor growth.
(a) After initiation, a tumor grows predominantly as a single expansion populated by numerous heterogeneous sub-clones. ITH results from private mutations (colored arrowheads) continuously accumulating due to replication errors. In addition to public mutations present in the first transformed cell, private mutations acquired early persist and become pervasive in the final tumor, while remaining non-dominant (colored segments). Late-arising mutations are only present in small regions of the tumor. (b) In the Big Bang model, the pervasiveness of private mutations depends on when the mutation occurs during growth, rather than selection for that mutation. The schematic illustrates how early private alterations, despite remaining non-dominant, are pervasive within the tumor (e.g. red and yellow), and can be found in distant regions, thus appearing variegated (e.g. red). This is due to aberrant sub-clone mixing in the primordial tumor, followed by scattering during expansion. Late alterations will be restricted to small regions (e.g. black) and are essentially undetectable by conventional whole exome sequencing (WES). Distance from the dashed vertical axis indicates increasingly late mutations. Dashed boxes indicate sampled regions. (c) We sampled an average of 23 individual tumor glands (<10,000 cells) from distant regions (~0.5cm3 in size, 1cm scale shown) and bulk (left, right) samples. Samples were profiled using several genomic techniques, including copy number analysis, WES and targeted sequencing, neutral methylation tag sequencing, and FISH, providing a panoramic view of genomic alterations throughout the tumor at multiple spatial scales.
Figure 1b shows an example of the variegation predicted by the Big Bang. Progeny of the first initiating tumor cell propagate public mutations, but also acquire new private alterations (colored areas), resulting in ITH within the newly formed small primordial tumor, which can subsequently scatter to distant regions during growth. For instance, the earliest mutation in red can be scattered to opposite sides of the neoplasm during tumor expansion, despite remaining private and non-dominant. This mechanism generates patterns of genetic variegation in the tumor. Therefore, clones harboring early private mutations (red or yellow) will be more pervasive in the final tumor, whereas late arising clones will not have time to expand to a detectable size, regardless of their relative fitness advantage (pink, black, green, blue). This simple mechanism predicts that early private mutations underlie the extensive ITH commonly detected in human neoplasms. Hence, public as well as the majority of detectable private mutations occur during early tumor growth.
Here we experimentally evaluate the predictions of the Big Bang model by profiling 349 individual tumor glands sampled from opposite sides (arbitrarily defined as right and left) of 15 colorectal carcinomas and large adenomas (Supplementary Table 1) using orthogonal multi-scale genomic techniques, namely whole-genome array-based profiling of copy number aberrations (CNAs), whole exome sequencing (WES), targeted deep sequencing, fluorescent in-situ hybridization (FISH), and neutral methylation tag sequencing. By analyzing single tumor glands composed of <10,000 cells, this approach enables the detection of alterations that occur in a fraction of tumor cells with remarkable sensitivity. At this level of resolution, we find unexpected spatial structure, indicative of order amidst the apparent chaos of genomic ITH. By integrating these data in a robust statistical inference framework based on a spatial computational model of tumor growth, we also verified that most ITH detectable with current technologies arises early during tumor growth and that the genomic profile of the primordial tumor can be recovered from the present day neoplasm.
Results
Sampling individual tumor glands
Colorectal cancer (CRC) represents an optimal system in which to study tumor growth dynamics as both the normal and neoplastic colon are organized into glandular epithelial structures, where neighboring cells within a gland share a recent common ancestry13 and microenvironment, with gland fission being the primary mode of growth14,15. Glands represent natural ancestral units and are composed of nearly pure tumor populations. Here we systematically sampled an average of 23 individual tumor glands and 2 bulk fragments from the right and left side (Figure 1c) of 4 large, mitotically advanced adenomas and 11 carcinomas (Supplementary Table 1), totaling 349 tumor glands and 22 bulks. This enables the highly sensitive detection of sub-clonal alterations (i.e. < 10,000 cells per gland out of 100 billion cells in a tumor; 0.00001%).
Single gland copy number profiles reveal variegation
Copy number profiles can be used to reconstruct tumor phylogenies6,8,16, and by profiling single glands it is possible to do so with unprecedented accuracy. We exploited whole genome SNP array-based copy number data derived from individual glands (7-10 per tumor, n=127 total), the right and left bulk tumor fragments (>3cm apart) and corresponding matched normals to systematically evaluate the spatial distribution of CNAs throughout each tumor. These data revealed striking spatial patterns, which were classified as follows:
Public: found in all glands of the tumor
- Private:
- Side-specific- found in all glands from one tumor side only
- Side-variegated- found in all glands from one side and some from the opposite side
- Variegated- found in a subset of glands from both sides
- Regional- found in more than one, but not all glands from one side only
- Unique- found in a single gland
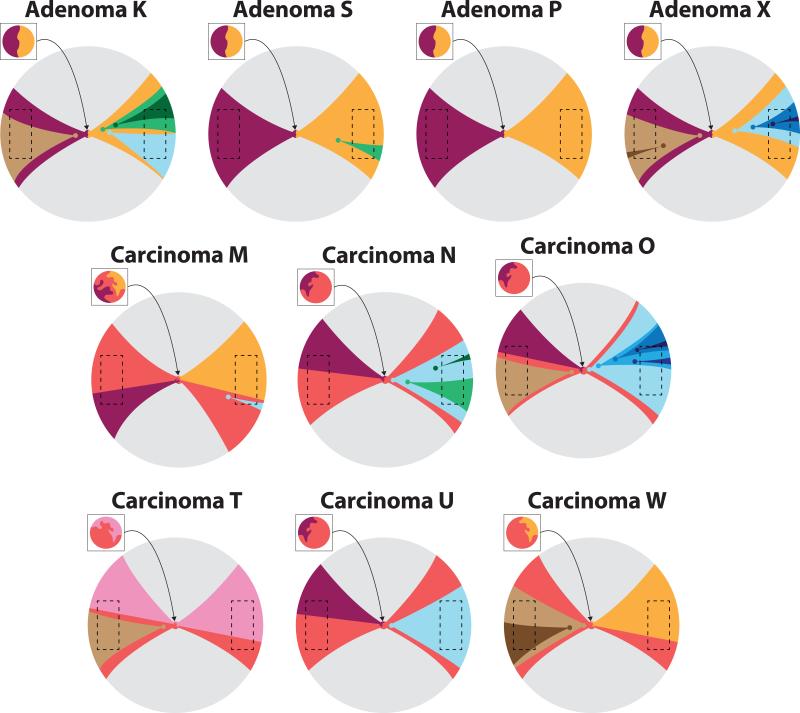
Consistent with their likely monoclonal origin from a single aberrant colon crypt17, most tumors exhibited public alterations acquired prior to initiation and hence present in all glands (Figures 2a, Supplementary Figure 1a and Supplementary Table 2). The adenomas were more chromosomally stable and less genomically complex than the carcinomas, despite their comparably large size (Supplementary Table 1). Adenomas were characterized by side-specific and unique CNAs that clearly segregated between tumor sides. In contrast, the majority of carcinomas (M, N, O, U, CA, CO and R) exhibited the same private CNA in individual glands from opposite sides of the tumor (variegated and/or side-variegated), as reflected in the underlying phylogenetic trees (Figure 2a and Supplementary Figure 2). This corresponds to the patterns of variegation presented in Figure 1b where an early private mutation originating in the primordial tumor will be scattered to distant tumor sites and will appear pervasive throughout neoplasm, despite remaining sub-clonal.
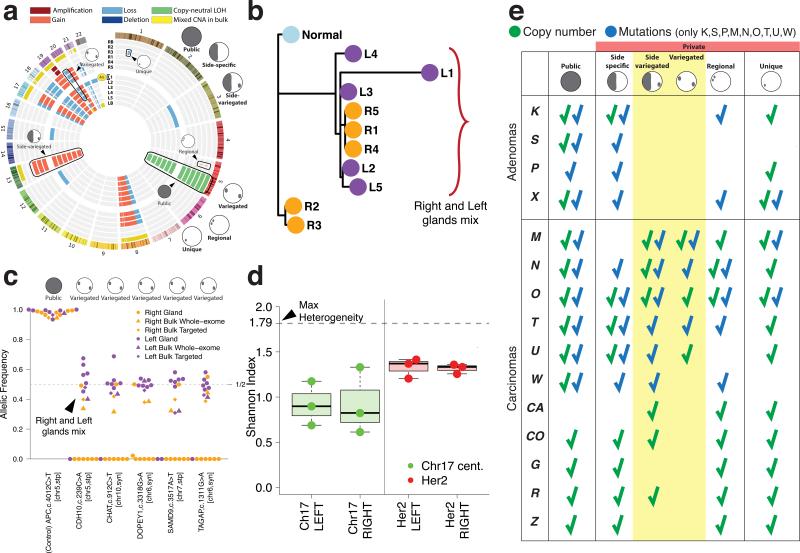
Figure 2. The spatial distribution of ITH reveals sub-clone mixing and the absence of clonal expansions.
(a) Circos plot representation of CNAs in individual glands and bulk samples for carcinoma M (shown throughout this figure). (b) Gland-level CNAs were employed to reconstruct the tumor phylogeny. Mixing of glands from opposite regions is apparent, where right and left glands are colored orange and purple, respectively. (c) Targeted sequencing of patient-specific mutations in individual glands revealed variegation in subsets of glands from opposite sides, thus confirming sub-clone mixing at the mutational level. A public APC mutation is illustrated as a clonal control (with LOH noted on chr 5). (d) FISH performed using HER2 probes (red) and corresponding chr 17 centromere (green) probes revealed high variability in copy number states between cells within a gland, as summarized by the Shannon Index. For each group, boxplots show the median, limited by the 25th (Q1) and 75th (Q3) percentiles, where whiskers represent the most extreme of the maximum or Q3+1.5(Q3–Q1) and the minimum or Q1–1.5(Q3–Q1), respectively. The maximum possible ITH value (“Max Heterogeneity”) corresponds to an index of 1.79 (99% of the FISH counts, range 0–5). (e) Summary of the characteristic spatial patterns and types of alterations in each tumor. While adenomas were characterized by low chromosomal instability and the segregation of alterations, carcinomas harbored side-variegated and variegated alterations (7/11 at the copy number level and 6/6 at the mutational level).
Such genetic variegation has been noted in leukemia18 and solid tumors19,20, but is often obscured by the prevailing approach of analyzing bulk tissue, rather than individual glands or cells. To verify that the individual glands are representative of the larger tumor mass, we profiled the right and left bulk tumor fragments (Figure 2a and Supplementary Figure 1a bulk tracks, left:LB, right:RB). We found that 99% of non-unique CNAs present in the glands were also detected in the bulk tumor, and that the majority of private gland CNAs were present as a mixture in the respective bulk (Supplementary Figure 3). All CNAs evident in a bulk sample were also detected in one or more corresponding tumor glands. Moreover, it is important to emphasize that if we had sampled only a portion of the tumor (e.g. only the right or only the left), we would have reconstructed erroneous phylogenies, as demonstrated in Supplementary Figure 4 and as noted by others21. While unlikely, we cannot exclude the possibility that the same CNA could arise independently in different glands. Hence, we also evaluated variegation at the mutational level.
Single gland sequencing confirms variegation
In order to examine mutational heterogeneity we performed WES of the bulk tumor samples (right and left) and adjacent normal tissue from each of the adenomas and for carcinomas M, N, O, T, U and W. Based on the spectrum of somatic mutations present in each bulk tumor, we selected a panel of patient-specific private mutations and known drivers of CRC for deep targeted sequencing (>600× mean target coverage) in individual glands (n=102) and the respective bulk fragments (n=20).
All sequenced tumors except for adenoma S and carcinomas O and W (MSI+), harbored public nonsense mutations in APC. Public missense mutations in KRAS were found in S, P, X, N and W, whereas public non-synonymous TP53 mutations were only found in carcinomas (M, N, and T), as previously reported4. Importantly, the mutational data corroborated the findings at the CNA level, providing further evidence for the striking segregation of sub-clones in all adenomas, whereas variegation, indicative of early sub-clone mixing, was observed in the carcinomas. A summary of the characteristic spatial patterns in each tumor is reported in Figure 2e. Here, variegation determined based on the presence of the same SNV in glands from distant tumor sides, was observed in the majority of carcinomas (Figures 2c, Supplementary Figures 1b and 5), despite being biased against detecting this phenomenon since only 7 to 10 glands were profiled per tumor.
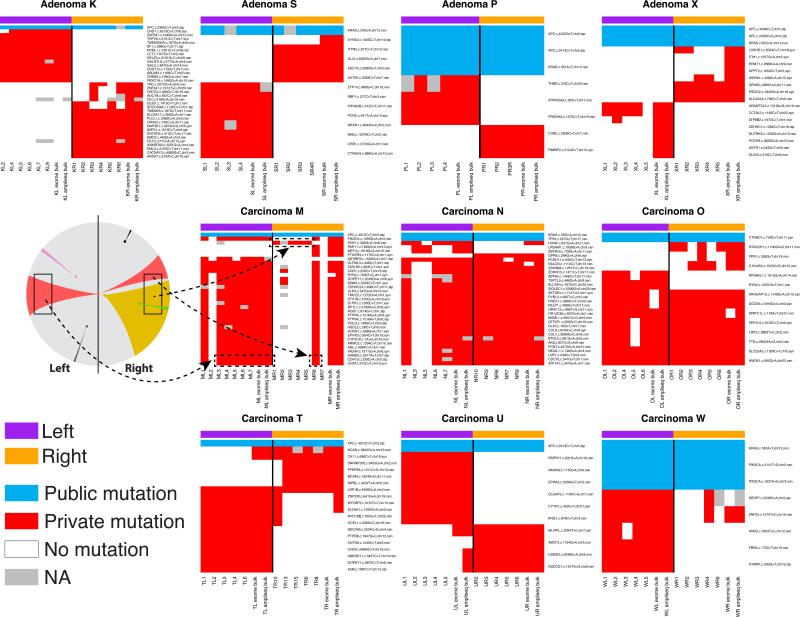
The targeted sequencing results for private mutations (red) and representative public mutations (blue) are presented in Figure 3. As shown for carcinoma M (Figure 3 and 2c), mutations in SAMD9, CDH10, and CHAT were variegated and recapitulate the predictions of the Big Bang model (Figure 1b), where a private mutation originates in the primordial tumor and subsequently scatters due to the expansion. In contrast, early public mutations in APC were found in all cells in the neoplasm, and represent a clonal control. Private mutations detectable in the bulk specimens were always present in at least one of the sampled glands, consistent with the pervasive nature of ITH. In addition, within small gland populations, any private mutation will eventually be lost or fixed. Private mutations were clonal within the gland, supporting their early acquisition, allowing sufficient time for loss or fixation via cell turnover or neutral drift22.
Figure 3. Single-gland targeted sequencing confirms the predictions of the Big Bang model and exposes variegation in carcinomas but not adenomas.
Heat maps indicate the presence of representative public and private mutations across multiple individual glands per tumor, where targeted sequencing and whole-exome sequencing of the bulk tumor is included for comparison. In all the adenomas, private mutations are confined to a single tumor side (regional and side-specific events), whereas in invasive carcinomas the same private mutation is found in distant regions of the neoplasm, despite remaining non-dominant. These patterns of genetic variegation are indicative of sub-clone mixing in the early neoplasm followed by scattering. For representative carcinoma M, the mutational data are summarized according to the schematic in Figure 1b, where variegated mutations (red) occurred early and scattered to distant tumor regions. Regional mutations (yellow) occurred later and were confined to smaller regions of the neoplasm.
Hypothetically, glands harboring the same private mutations found on opposite tumor sides (several centimeters apart) could result from alternative mechanisms such as late arising mutations and subsequent migration, or tumor cell reseeding23. However, such migration is unlikely because the private mutations were clonal within individual glands, and the migration of whole glands is improbable. Instead, sub-clone mixing is efficient in an early small malignancy characterized by loss of normal cell adhesion and disorganized growth. The ensuing expansion allows early private mutations to become fixed within glands, pervasive in the tumor, and scattered to “opposite” tumor sides, thus generating patterns of variegation. Indeed, variegation was restricted to the carcinomas (Figure 2e and 4). This observation suggests that certain malignant features, such as abnormal mobility, may be expressed very early, even before visible invasion and/or metastasis occurs, implying that some tumors are “born-to-be-bad”. An illustrative simulation demonstrates that sub-clone mixing in an early tumor followed by expansion can create complex patterns of variegation (Supplementary Figure 6). In contrast, when the same mutation arises later, sub-clones appear segregated irrespective of their relative fitness advantage.
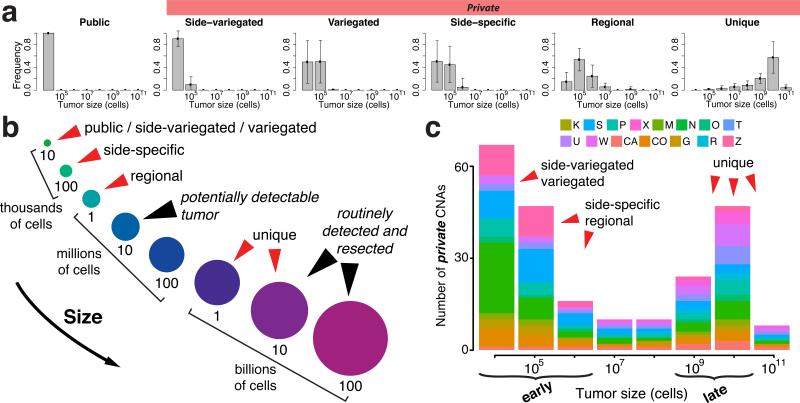
Figure 4. Inference on the genomic data verifies that most detectable ITH occurs early during tumor growth.
(a) The inferred mutational timeline is indicated for different classes of CNAs for all tumors, where time is represented in relation to tumor volume. In particular, the posterior probability distribution of tumor size (number of cells) is illustrated for each class of alteration. The results show that both public alterations, as well as the majority of private alterations (including side-specific, side-variegated, and variegated) occur very early after the transition to an advanced neoplasm, whilst the tumor is less than one million cells, whereas unique mutations occur late. Error bars represent the standard deviation. (b) A schematic representation of the mutational timeline (from panel a) illustrates that the majority of detectable non-unique alterations occur early, while the tumor is orders of magnitude smaller than can be clinically detected. As reliable estimates of cell cycle time are not available and somatic alterations depend on cell division rates rather than time, tumor size is used as surrogate of time. (c) By applying the inferred mutational timeline to the whole genome CNA profiles for each patient, it is apparent that early CNAs dominate the genomic landscape. Here, early events correspond to alterations that took place when the tumor was <106 cells and late alterations correspond to those that occurred after the tumor reached 109 cells. For simplicity, only private alterations are represented since all public alterations occur early.
Single cell profiling reveals uniformly high ITH
The fixation of private mutations within a gland could occur through stepwise selection where cells with even a slight selective advantage will sweep through the gland. In this scenario, there should be very little intra-gland heterogeneity. By contrast, a single Big Bang expansion implies that individual glands in the final tumor are relatively old populations that should exhibit similar within-gland diversity. We evaluated CN heterogeneity between physically adjacent single cells by FISH in a subset of tumor glands (n=65) and adjacent normal glands (n=22). In particular, we assayed for HER2 gene amplification, a driver event in breast and gastric cancer, which has been implicated in CRC24. These data reveal a high degree of variability in CN between physically adjacent cells within the same gland as quantified by the Shannon index19. Importantly, this diversity was uniformly high throughout the tumor (Figure 2d, Supplementary Figure 1c and Supplementary Table 3). Since mutations should fixate quickly within small populations25, this suggests the absence of recent clonal expansions within glands. Variation in CN between nearby cells is reportedly common in CRC due to chromosomal instability (CIN)26, and may be important for tumor initiation27 and progression28. Moreover, it can be used to assess genetic and phenotypic diversity in response to chemotherapy29.
Epigenetic passenger mutations were also evaluated through ultra-deep single-molecule methylation tag sequencing of individual glands (n=55), which provides an efficient means to infer cell ancestries in normal30,31 and cancerous tissues5,9. These data confirmed uniformly high ITH (Supplementary Figure 1d), reflecting similar tumor age in different glands and opposite sides of the neoplasm, in agreement with FISH. In particular, numerous mitotic sub-clones within the same gland were found in the majority (49/55) of samples (Supplementary Figure 1e), supporting the absence of recent selective sweeps, as predicted by the Big Bang model.
Statistical inference verifies the Big Bang predictions
The most striking prediction of the Big Bang tumor model is that, whereas new alterations occur continuously throughout tumor growth, the majority of private mutations that can be detected occur early after the transition to an advanced tumor, rather than as a result of the subsequent selection of de novo clones. To quantitatively test this, we extended our previously described statistical inference framework approach9 in order to take as input copy number and mutational data from multiple tumor glands and to account for sub-clone fitness differences and local microenvironmental contributions. The framework utilizes Approximate Bayesian Computation (ABC)32 and 3-dimensional mathematical modeling to infer patient-specific tumor characteristics, including the mutation rate, sub-clone fitness changes and the mutational timeline, given the observed multiple-sampling genomic data (Supplementary Figure 7). The model simulates the expansion of a tumor containing ~8 million glands, corresponding to a realistically sized neoplasm composed of ~80 billion cells with a diameter of ~5.3 centimeters, and accounts for gland proliferation in 3-dimensional space, somatic alterations (CNAs and point mutations) and sub-clone fitness changes (see Online Methods for details).
The inference results indicate that, although sub-clone fitness changes can be detected (Supplementary Figure 8a), their effects on the clonal composition of the tumor are limited, as corroborated by the presence of adjacent glands with different fitness (Supplementary Figure 8b). The magnitude of fitness changes was variable in the carcinomas, whereas the adenomas exhibited limited or no fitness differences between sub-clones. The mutation rates were also elevated in the carcinomas (10−6–10−5) as compared the adenomas (10−6) (Supplementary Figure 8a) similarly highlighting inter-tumor variability in clonal dynamics and important phenotypic differences between adenomas and carcinomas. We also employed this framework to infer the timeline during which different classes of alterations occur and quantitatively show that for each of the tumors assayed, both public and most private alterations (side-specific, side-variegated, and variegated) occurred early (Figure 4a) when the malignancy was less than 104-105 cells (Figure 4b), where size is used as a surrogate for tumor age. This is approximately 100-1000 times smaller than the size at which colorectal tumors are potentially detectable (~1 mm3 or 106 cells) and 1 million times smaller than is typical at the time of surgical resection (the source of sampled tissue). Even regional alterations tend to occur before the tumor is clinically detectable, whereas unique alterations arise later, as expected. These findings hold irrespective of tumor-specific characteristics. The same conclusions were obtained using mutational data as input to the framework (Supplementary Figure 9). By organizing the observed patient-level genomic profiles according to the inferred mutational timeline, it is evident that early sub-clonal alterations dominate the genomic landscape (Figure 4c).
Using single gland and bulk mutational profiles (shown in Figure 3) we reconstructed tumor phylogenies (see Online Methods) in order to define sub-clones, or groups of glands harboring the same private mutations. By superimposing the inferred mutational timeline for different classes of alterations (Figure 4a and Supplementary Figure 9c), we then determined the relative timing during which each sub-clone arose. This allows for the approximate reconstruction of patient-specific spatio-temporal evolutionary dynamics, as depicted schematically in Figure 5, and shows that the pervasiveness of a private mutation depends on when it arose during the expansion, rather than as a result of selection for that mutation. This schematic also illustrates that while all tumors exhibit Big Bang dynamics, early sub-clone mixing in the primordial tumor (square insets) was restricted to carcinomas.
Figure 5. Schematic representation of spatio-temporal Big Bang growth dynamics.
For each tumor profiled at the mutational level, the phylogeny was reconstructed from the single gland and bulk tumor data (see Online Methods) in order to define sub-clones, or groups of glands harboring the same private mutations. The relative timing during which each sub-clone arose was specified based on the inferred mutational timeline (Figure 4a and Supplementary Figure S9c) for the different classes of private alterations (variegated/side-specific/regional/unique). By combining information on the mutational timeline and tumor sub-clonal architecture, we can approximately reconstruct patient-specific spatio-temporal evolutionary dynamics, as shown in this schematic. Here the topographical distribution of different sub-clones is illustrated by distinct colors and distance from the tumor origin (arrowhead) indicates increasingly late alterations. Variegated and side-variegated sub-clones occurred very early within the primordial tumor (<1 million cells) and are shown within the inset square representing the zoomed-in view of the primordial neoplasm. Regional and unique sub-clones arose later and are represented outside the inset square. Dashed boxes represent regions of the tumor that were experimentally sampled. This schematic shows how in the Big Bang tumor model, the prevalence of a private mutation depends on when it arose during tumor expansion, rather than selection for that mutation. The schematic also illustrates that although all tumors exhibit Big Bang dynamics, sub-clone mixing is restricted to carcinomas, whereas adenomas are characterized by sub-clone segregation.
Clonal heterogeneity could alternatively be due to distinct local microenvironmental niches within the neoplasm that select for clones with different genomic profiles1. To investigate this scenario, we introduce microenvironmental niches in our model (Supplementary Figure 10 and Online Methods). The inferred parameters are in agreement with the results from the microenvironment-free model for both CNAs and mutations (Supplementary Figure 11), further supporting our conclusions. This follows from the fact that microenvironmental selection acts passively on existing variation. Of note, here we model the microenvironment as a static entity and do not account for the possibility that tumor cells may dynamically alter their environment, although this may play a role in later growth3. In the future, it will be of interest to examine more complex interactions between cells and their microenvironment, as well as to measure inter-clonal interactions, which have recently been described in breast cancer33,34.
Discussion
Tumor initiation is characterized by the sequential step-wise accumulation of alterations, leading to the expansion of clones with selective growth advantages, such that the fittest clone eventually dominates4. The sequential model of colorectal tumorigenesis is corroborated by epidemiological data on colorectal cancer incidence35. This model has often been postulated to describe the subsequent growth of an established tumor. In this scenario, further growth within an advanced tumor results from the acquisition of new driver mutations followed by selective sweeps and large clonal expansions. Within this model, ITH represents a transitory state between selective sweeps. As this model implies multiple sweeps, numerous drivers of tumor growth are anticipated. However, relatively few putative driver mutations have been identified in individual tumors36.
Recent studies in primary CRCs indicate that selective sweeps and large clonal expansions are infrequent after transformation13,37,38 and predict star-shaped phylogenies13,37. Studies in other cancers similarly highlight such branched phylogenies39 and punctuated clonal evolution6,40. Moreover, karyotypic chaos41, stress-induced mutational bursts42, and chromothrypsis43, a cataclysmic event involving surges of chromosomal rearrangements, have been reported. Evidently, sequential clonal evolution does not accurately describe the patterns of ITH found in human cancers.
Here we propose and test the predictions of a “Big Bang” model, whereby as a result of a single clonal expansion, most detectable ITH occurs early after the transition to an advanced tumor. In this model, due to constraints on clonal selection, private mutations are pervasive in the final neoplasm, despite remaining non-dominant. Indeed, only very strongly advantageous mutations are likely to fixate in realistic time-scales12 within rapidly expanding populations, where spatial structure delays the expansion of an advantageous mutation10-12. Such spatial constraints in solid tumors1,13,44 underline the limits with which selective forces drive the tumor expansion. Hence, both public and the majority of detectable private mutations occur early during tumor growth. Although private alterations continuously occur, only those that occur early have time to expand to a detectable size. The Big Bang model explains why ITH is pervasive in human tumors and provides a theoretical framework to describe the underlying clonal dynamics. The star-shaped phylogenies predicted by the Big Bang model are also compatible with the long-lived lineages of the cancer stem cell model45, wherein the malignancy is driven by a small number of self-renewing cells. We demonstrate that Big Bang dynamics are robust to changes in sub-clone fitness and local microenvironment, which may explain why they are observed in many tumors.
The Big Bang model explains many poorly understood features of cancer genomic data, with the following implications:
-
i.)
ITH is an inherent characteristic of colorectal tumors that arises early and continuously increases during growth, and is not significantly constrained by clonal selection. Branched phylogenies naturally follow from the Big Bang model.
-
ii.)
Significant clonal expansions or selective sweeps are extremely rare after the transition to an advanced tumor due to the dynamics and spatial constraints of the rapidly growing population, and the formation of microenvironmental niches.
-
iii.)
Both public and the majority of detectable private alterations arise early and become pervasive during tumor growth, thereby dominating the genomic structure of the neoplasm.
-
iv.)
Potentially aggressive sub-clones may remain rare or even undetectable in the primary tumor, despite their relative fitness advantage, providing a heterogeneous substrate to fuel resistance in response to treatment selective pressures.
A number of clinical implications also follow from the Big Bang model. For example, it is uncertain why certain large tumors remain localized, whereas other eventually invade and metastasize. Variegated alterations were found in the majority of invasive carcinomas, but none of the adenomas. Hence, variegation may reflect the early expression of an invasive phenotype (abnormal cell inter-mixing), such that some tumors are ‘born-to-be-bad’. This finding is compatible with a Big Bang expansion wherein malignant potential is determined early, as previously proposed46,47. Moreover, the degree of sub-clone mixing may be a readout of subsequent invasiveness, and could represent a novel biomarker for predicting which adenomas will become invasive versus those that will remain indolent. Another clinical implication that follows from the timing of mutation being the primary determinant of whether a sub-clone is pervasive in a tumor is that “dangerous” treatment-resistant clones that occur late will be undetectable, presenting obvious challenges for personalized medicine. This is in-line with recent reports that minor cell subpopulations can drive tumor growth34, and the presence of preexisting intrinsically resistant sub-clones that contribute to poor treatment response48.
Not every tumor may exhibit Big Bang dynamics and “selective bottlenecks” may be common for markedly different environments such as in the context of metastatic seeding to foreign sites or during treatment. However, for primary tumors that arose predominantly as single clonal expansions, this new model represents a theoretical framework in which to interpret cancer genomic data, and predicts that the earliest events should be pervasive in the final neoplasm. This concept shares an interesting analogy with the cosmic microwave background (CMB) of the Big Bang Universe, which is composed of scattered thermal radiation originating in the earliest phase of our universe, which subsequently streamed through the expanding cosmos. From this CMB signature it is possible to reconstruct the events that occurred right after the birth of our universe. Our findings offer a radically new way to interpret cancer genomic data, providing new insights into how primary human tumors progress, which should facilitate more effective early detection and prognostication efforts.
Online Methods
Sample collection
This study employed de-identified excess patient specimens collected in the course of routine clinical care and was approved by the local institutional review board (IRB). Individual tumor glands composed of <10,000 adjacent cells were isolated from fresh colectomy specimens following EDTA treatment, as previously described30. DNA was isolated from individual glands by incubation in a 15 μl Tris-EDTA solution with Proteinase K solution (4 hours at 56°C), followed by boiling for 5 minutes. Using this method we consistently obtain samples with >95% tumor purity. Bulk tumors and adjacent normal samples were composed of a pool of thousands of single gland were also obtained and DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen).
Analysis of copy number data
Individual glands, as well as right and left bulk tumor fragments were profiled on the OmniExpress SNP platform (Illumina) according to the manufacturer's protocol. Only samples with call rates >85% were analyzed, with an average gland call rate of 97%. Data were processed using Genome Studio software, followed by quantile normalization49 and segmentation with psCBS50, where adjacent normal tissue was employed as a baseline reference for each tumor. To define regions of aberrant copy number, we applied a threshold method based on the standard deviation, σ, calculated for the 50th central percentile of the probes sorted by the log2 relative ratio (LRR), adapted from Curtis et al. 201251. Briefly, copy number alterations were determined as follows: amplifications; LRR>6σ, gains; 2σ<LRR<6σ, heterozygous losses; 7σ<LRR<−2.5σ, and homozygous deletions; LRR<−7σ. The LRR and Beta Allele Frequency (BAF) for each array were manually inspected to verify the accuracy of the copy number calls and eventually corrected to maintain a conservative approach and to avoid overcalling ITH. Processed CN data were then used to generate inter-gland phylogenetic trees (Figure 2b, Supplementary Figure 2 and 4) using MEDICC16.
Analysis of mutational data
For all adenomas and carcinomas M, N, O, T, U and W, right and left bulk tumor fragments were subject to whole exome sequencing (WES) to a depth of coverage of 20× on the HighSeq 2000 (Illumina). For adenomas K, S and P and carcinomas M and N, the samples subsequently underwent additional sequencing to 60× coverage on the HighSeq 2500 (Illumina). For each tumor, a panel of sub-clonal mutations identified in the bulks and a set of clonal mutations, including putative drivers (for comparison) were profiled in individual tumor glands on the Ion Torrent PGM platform (Life Technologies) using custom AmpliSeq panels. Resultant data were aligned to hg19 and processed using MuTect52 for mutation calling and quantification of allelic frequencies. For the WES bulk samples, mutations were only called if the coverage exceeded 10× with 3 or more variant reads. Furthermore, to filter false positives introduced due to paralogous regions, we used BLAST to verify that the 40bp region around each mutation matched the reference genome uniquely. For the targeted sequencing data, mutations were only called if the coverage exceeded 50× with 20 or more variant reads. To avoid overcalling ITH as a result of false negatives due to low coverage, the absence of a mutation in a gland was indicated not only by a mutation not being called, but also by the presence of at least 50× coverage at the locus, of which >95% of the reads had to indicate no mutation. If a mutation was not called in a gland and there was insufficient evidence (due to low coverage) to confirm its absence, the allelic frequency was indicated as NA. Mutations for which more than half of the glands had NA values were discarded (only 4 mutations were filtered out due to this problem). The mean coverage for the targeted sequencing data was 626.58±20.2 95% CI (Supplementary Figure 12). Public canonical driver mutations (APC, KRAS, or TP53) serve as a clonal control and are reported alongside the private sub-clonal events in Figures 2c, Supplementary Figures 1b and 5. For tumor O, APC, KRAS, and TP53 mutations were not detected and a clonal CTNND1 mutation is plotted instead. Amongst the mutations reported, those for which data was available for all glands of a given tumor (i.e. no NA glands; totaling 167/194 mutations) were employed as input to the statistical inference framework for comparison with the results based on whole genome copy number profiles. We also employed the mutational profiles of individual glands to infer tumor phylogenies using MEDICC16. This allows for the identification of sub-clones or groups of glands harboring the same private mutation, where each node in the phylogeny represents a new clone (branching event). By combining the tumor phylogenies and the inferred mutational timeline for different classes of alterations based on our 3D computational model (Figure 4, Supplementary Figure 9c), we can approximately reconstruct the spatio-temporal evolutionary dynamics for each patient (Figure 5).
Analysis of neutral methylation tag data
Molecular clock analysis based on neutral methylation tag data was performed as previously described9. Briefly, DNA was extracted from individual tumor glands, subject to bisulfite conversion, followed by PCR amplification of the ZNF454 molecular clock locus and ultra-deep targeted sequencing (average >1,100× per gland) on the Roche 454/GS JR platform. Data were then processed using our custom pipeline, as previously described9.
FISH analysis
FISH analysis of the HER2 gene and chromosome 17 centromere copy number was performed using the Vysis HER-2 DNA Probe Kit (Abbott Molecular) in the M.F.P lab, which routinely performs CLIA certified HER2 assays. Fluorescence microscopy was employed to quantitatively evaluate the copy number status of 20 cells per gland for 3-6 glands from the left and 3-6 from the right side of each tumor and 20 cells from 3-4 crypts for each matched normal. Thus 120 to 240 cells were counted per tumor and 60 cells per normal. Of note, this is 6 times more cells than the 20 that are routinely counted for the diagnosis of HER2 amplification in breast cancer53. As the tissue sections employed for FISH analyses were 5 μm thick, whereas CRC cells are 8-10 μm, we verified that this did not introduce bias in estimating the number of amplified cells by analyzing multiple planes and by comparing counts from the tumor and adjacent normal glands (Supplementary Figure 13 and Supplementary Table 3).
Computational framework
We extended our previously described computational framework9 in order to i) accommodate whole genome copy number and targeted mutational data, ii) to model fitness effects, corresponding to different survival probabilities, and iii) to account for microenvironmental niches. This framework exploits Approximate Bayesian Computation (ABC), an established approach commonly used in population genetics32 in order to obtain posterior parameter distributions by fitting a computational model of tumor growth to the single gland level genomic data (Supplementary Figure 7a). The cellular automaton 3-dimensional model of tumor growth (Supplementary Figure 7b) accounts for gland growth by fission, the occurrence of CNAs and mutations, and the variable gland growth rates. The 3D position of each gland at any point in time is recorded and glands can have different survival (and growth) fitness due to copy number alterations or point mutations. In particular, we simulate the growth of a realistically sized malignancy composed of 8 million glands (~80 billion cells, 5.3 cm in diameter) and incorporate copy number alterations at a rate μ that may induce a change in fitness. As simulating changes in fitness for 80 billion cells would be computationally intractable, we assume that cells within a gland have the same fitness and that fitness changes occur at the gland level as a result of acquired somatic alterations within the gland (e.g. modal copy number changes). Beginning with a single gland with normalized fitness 1, and an associated survival probability, we simulate the possibility that deleterious, neutral, and advantageous mutations may change the fitness according to a transition distribution. The input parameters are the mutation rate (μ) and the magnitude of fitness changes (σ), where the model produces as output multi-sampling data for each simulated tumor. At the end of the simulation, glands are virtually sampled as they are physically sampled in practice from the tumor, thus maintaining information on the proximity of sub-clones. In this manner, we faithfully simulate the experimental system (which for practical reasons is restricted to sampling 7-10 glands) several thousand times.
When a CNA occurs, the fitness change is sampled from a Gaussian distribution with mean 0 and variable standard deviation, σ. This models the possibility of both advantageous and disadvantageous mutations. Higher values of σ, correspond to a greater likelihood that a new clone exhibits an increase/decrease in its fitness, whereas for σ=0 no change in fitness occurs, corresponding to the neutral model of growth in which all clones have equal fitness. Here the fitness, F, is expressed in terms of a survival increase, ranging from 1 to 5, with all simulations initiated with a single gland with F=1. At each division, a gland has probability Pα=α/F of dying, where α is set to 20%. Recent studies indicate the possibility that fitness changes in driver mutations may be as low as 1%54, but values on the order of 10% are also typically employed55. We evaluated two key parameters, namely, the magnitude of fitness changes σ ∈ {0, 0.2, 0.6}, corresponding to no change, moderate and large changes, respectively and the mutation rate μ ∈ {10−8, 10−7, 10−6, 10−5, 10−4} per gland per division. Since σ is the standard deviation of a normal distribution with mean=0, for σ=0.2 we expect a fitness increase of 10% or greater in 30.7% of the cases, whereas for σ=0.6, a fitness increase of 10% or greater is expected in 48.8% of the cases. Thus, these values correspond to a range of small to large variations in fitness. Other complex and poorly characterized processes, such as cellular migration and apoptosis within a gland are not modeled, nor is the contribution of the surrounding normal tissue or angiogenic factors.
The simulation initiates with a gland in the center of a 400×400×400 point lattice, where glands then split by fission until a volume of 8 million glands is reached. Subsequently, 5 glands from the left and 5 glands from the right side of the tumor are virtually sampled from the simulation, in accordance with the experimental sampling scheme performed on the tumor specimen. The CNA profiles of the sampled glands are saved for comparison with the actual data (Supplementary Figure 7a). We employ ABC to fit the model to the data, in order to generate posterior probability distributions of the parameters (σ, μ) for each patient, assuming uninformative uniform priors. Every CNA was associated to a binary string, indicating its presence (1) or absence (0) in each sampled gland. Public alterations were excluded from the inference as the vast majority likely occurred during pre-neoplastic stages, prior to the transition to an established neoplasm, and thus do not belong within the simulated scenario. Nevertheless, relaxing this rule yielded similar results (data not shown). Summary statistics were then computed using these binary patterns, including the number of distinct CNAs (the number of different strings), the Shannon index of the binary patterns, the total number of alterations, the number of variegated alterations and the number of side-variegated alterations. As a measure of the distance between the actual data and the simulated data, we employed the average distance of the summary statistics, normalized to mean=0 and s.d.=1. The inference framework was validated using synthetic data to demonstrate that the correct parameter value is accurately recovered in the majority of cases (Supplementary Figure 14).
To examine the influence of differences in local tumor microenvironment, we developed a version of the model in which specific CNAs were selected depending on the surrounding tumor area by incorporating static microenvironmental niches of differing size (env parameter: 5×5, 20×20, 150×150) in the simulation. Each block in the grid selects for a random CNA/mutation on a specific chromosome by inducing a high apoptotic rate (20%) for glands that do not exhibit that particular alteration, such that the overall apoptotic rate is quite high, representing positive selection. In this manner, rudimentary microenvironmental niches that select for different gland populations are represented (Supplementary Figure 10). The same approach as described above was applied to perform inference on the mutational profiles in both the niche-based and microenvironment-free models (Supplementary Figure 11) These results imply that distinct, yet static local microenvironments do not alter Big Bang dynamics. In the future, it will be of interest to examine contributions due to dynamic interactions between tumor cells and their microenvironment, as well as clonal cooperation and interference.
Supplementary Material
Acknowledgments
The project described was supported in part by an award to C.C. from the V Foundation for Cancer Research and by award numbers P30CA014089, R21CA149990 and R21CA151139 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. M.F.P. was supported by a grant from the California Institute for Regenerative Medicine (CIRM). The authors would like to acknowledge the technical assistance of Roberta Guzman.
Footnotes
Author Contributions
A.S., D.S. and C.C. designed the study, interpreted the data and constructed the model. D.S. provided clinical specimens. Z.M. and D.S processed the specimens. Z.M. generated sequencing data. P.M. and K.S. contributed data. H.K and M.F.P. performed FISH. A.S. developed and implemented the computational framework. A.S., M.P.S. and J.Z. analyzed the data with oversight from C.C. A.S., D.S. and C.C. wrote the manuscript with input from T.G. D.S. and C.C. oversaw the study. All authors read and approved the final manuscript.
The authors declare no competing final interests.
Accession Codes
The copy number data are accessible via the ArrayExpress database with accession number EMTAB-2140. The sequence data are accessible via the ArrayExpress database with accession number E-MTAB-2247. The methylation data are available via the NCBI BioProject database with accession number PRJNA230833.
References
- 1.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basanta D, Anderson ARA. Exploiting ecological principles to better understand cancer progression and treatment. Interface Focus. 2013;3:20130020. doi: 10.1098/rsfs.2013.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER, Fearon ER, Vogelstein B, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 5.Siegmund KD, et al. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4828–4833. doi: 10.1073/pnas.0810276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navin N, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlinger M, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. U.S.A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sottoriva A, Spiteri I, Shibata D, Curtis C, Tavaré S. Single-molecule genomic data delineate patient-specific tumor profiles and cancer stem cell organization. Cancer Res. 2013;73:41–49. doi: 10.1158/0008-5472.CAN-12-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korolev KS, Avlund M, Hallatschek O, Nelson DR. Genetic demixing and evolution in linear stepping stone models. Rev Mod Phys. 2010;82:1691–1718. doi: 10.1103/RevModPhys.82.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korolev KS, et al. Selective sweeps in growing microbial colonies. Phys Biol. 2012;9:026008. doi: 10.1088/1478-3975/9/2/026008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFarland CD, Korolev KS, Kryukov GV, Sunyaev SR, Mirny LA. Impact of deleterious passenger mutations on cancer progression. PNAS. 2013;110:2910–2915. doi: 10.1073/pnas.1213968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphries A, et al. Lineage tracing reveals multipotent stem cells maintain human adenomas and the pattern of clonal expansion in tumor evolution. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E2490–9. doi: 10.1073/pnas.1220353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia SB, Park HS, Novelli M, Wright NA. Field cancerization, clonality, and epithelial stem cells: the spread of mutated clones in epithelial sheets. J. Pathol. 1999;187:61–81. doi: 10.1002/(SICI)1096-9896(199901)187:1<61::AID-PATH247>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 15.Wright NA, Wright NA, Poulsom R, Poulsom R. Top down or bottom up? Competing management structures in the morphogenesis of colorectal neoplasms. Gut. 2002;51:306–308. doi: 10.1136/gut.51.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz RF, et al. Phylogenetic quantification of intra-tumour heterogeneity. PLoS Comput. Biol. 2014;10:e1003535. doi: 10.1371/journal.pcbi.1003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 18.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 19.Park SY, Gönen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin. Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thirlwell C, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–54–1454.e1–7. doi: 10.1053/j.gastro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Sprouffske K, Pepper JW, Maley CC. Accurate reconstruction of the temporal order of mutations in neoplastic progression. Cancer Prev Res (Phila) 2011;4:1135–1144. doi: 10.1158/1940-6207.CAPR-10-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 23.Comen E, Norton L, Massagué J. Clinical implications of cancer self-seeding. Nat Rev Clin Oncol. 2011;8:369–377. doi: 10.1038/nrclinonc.2011.64. [DOI] [PubMed] [Google Scholar]
- 24.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowak MA. Evolutionary Dynamics. Harvard University Press; 2006. [Google Scholar]
- 26.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 27.Nowak MA, et al. The role of chromosomal instability in tumor initiation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S Datta R, Gutteridge A, Swanton C, Maley CC, Graham TA. Modelling the evolution of genetic instability during tumour progression. Evol Appl. 2013;6:20–33. doi: 10.1111/eva.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almendro V, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep. 2014;6:514–527. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yatabe Y, Tavaré S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sottoriva A, Tavaré S. In: Proceedings of COMPSTAT 2010. Saporta G, Lechevallier Y, editors. Springer Physica-Verlag; HD: 2010. pp. 57–66. [Google Scholar]
- 32.Marjoram P, Tavaré S. Modern computational approaches for analysing molecular genetic variation data. Nat. Rev. Genet. 2006;7:759–770. doi: 10.1038/nrg1961. [DOI] [PubMed] [Google Scholar]
- 33.Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508:113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luebeck EG, Moolgavkar SH. Multistage carcinogenesis and the incidence of colorectal cancer. PNAS. 2002;99:15095–15100. doi: 10.1073/pnas.222118199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegmund KD, Marjoram P, Tavaré S, Shibata D. Many colorectal cancers are ‘flat’ clonal expansions. Cell Cycle. 2009;8:2187–2193. doi: 10.4161/cc.8.14.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kostadinov RL, et al. NSAIDs Modulate Clonal Evolution in Barrett's Esophagus. PLoS Genet. 2013;9:e1003553. doi: 10.1371/journal.pgen.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrell RA, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 40.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heng HHQ, et al. Stochastic cancer progression driven by non-clonal chromosome aberrations. J. Cell. Physiol. 2006;208:461–472. doi: 10.1002/jcp.20685. [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg SM. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- 43.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sottoriva A, et al. Cancer stem cell tumor model reveals invasive morphology and increased phenotypical heterogeneity. Cancer Res. 2010;70:46–56. doi: 10.1158/0008-5472.CAN-09-3663. [DOI] [PubMed] [Google Scholar]
- 45.Clevers H. The cancer stem cell: premises, promises and challenges. Nat. Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 46.Bernards R, Weinberg RA. Metastasis genes: A progression puzzle. Nature. 2002;418:823–823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 47.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 48.Diaz LA, Jr, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staaf J, et al. Normalization of Illumina Infinium whole-genome SNP data improves copy number estimates and allelic intensity ratios. BMC Bioinformatics. 2008;9:409. doi: 10.1186/1471-2105-9-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olshen AB, et al. Parent-specific copy number in paired tumor–normal studies using circular binary segmentation. Bioinformatics. 2011;27:2038–2046. doi: 10.1093/bioinformatics/btr329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnology. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolff AC, et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Journal of Clinical Oncology. 2006;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 54.Bozic I, et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michor F, Iwasa Y, Nowak MA. Dynamics of cancer progression. Nat. Rev. Cancer. 2004;4:197–205. doi: 10.1038/nrc1295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.