Abstract
The key moment for efficiently and accurately diagnosing dementia occurs during the early stages. This is particularly true for Alzheimer's disease (AD). In this proof-of-concept study, we applied near infrared (NIR) Raman microspectroscopy of blood serum together with advanced multivariate statistics for the selective identification of AD. We analyzed data from 20 AD patients, 18 patients with other neurodegenerative dementias (OD) and 10 healthy control (HC) subjects. NIR Raman microspectroscopy differentiated patients with more than 95% sensitivity and specificity. We demonstrated the high discriminative power of artificial neural network (ANN) classification models, thus revealing the high potential of this developed methodology for the differential diagnosis of AD. Raman spectroscopic, blood-based tests may aid clinical assessments for the effective and accurate differential diagnosis of AD, decrease the labor, time and cost of diagnosis, and be useful for screening patient populations for AD development and progression.
Keywords: Alzheimer's disease, differential diagnosis, blood serum, Raman spectroscopy, artificial neural networks
1. Introduction
Alzheimer's disease (AD) is the most widespread type of neurodegeneration-induced dementia in the elderly population worldwide [1]. It is also among the most serious health problems in industrialized nations, including the United States[1-3]. This neurodegenerative disease is progressive, incurable and lethal. More than 5.2 million Americans and 35.6 million people worldwide suffer from AD; these numbers are expected to increase dramatically by the year 2050[1-3].
AD has numerous clinical manifestations, such as a gradual loss of short-term memory, language problems, progressive difficulty performing familiar motor tasks, temporal and spatial disorientation, impairments in abstract thinking, and disturbances in behavior and personality, including sleep disturbances, depression, anxiety, psychosis and dementia[4, 5]. These symptoms are associated with significant morphological alterations of the brain tissue that are caused by processes related to the formation of amyloid beta (Aβ) plaques and neurofibrillary tangles (NFTs)[5].
The destructive pathophysiological process of AD is thought to commence many years prior to the clinical presentation with only non-specific symptoms before the clinical diagnosis of AD can be made[6]. Because the differential diagnosis for dementias relies heavily on clinical criteria, it is often a complex and difficult process[7]. The slow initiation of AD during the “preclinical” phase could provide a critical opportunity for therapeutic and disease-modifying interventions[8]. Treatments during the early stages of the disease progression would be the most effective because of the possibility of interfering with pathological process before irreversible damage occurs and keeping patients in an independent functional state for as long as possible. Therefore, the early and accurate diagnosis of AD in at-risk patients is of a great importance.
In the early stages, many types of dementia show only non-specific symptoms. For example, in Parkinson's disease (PD) patients who eventually develop dementia, the symptoms start as mild deficits in cognitive function that are similar to those observed in AD. Both disorders progress over time to the symptoms that are recognizable as mild cognitive impairment (MCI). In advanced disease, both PD and AD are associated with severe cognitive decline, and it is still unclear to what extent they can become more similar functionally over time[9]. Still, there is no single biomarker or cognitive test that can conclusively distinguish between a person having PD dementia and AD. However, both PD and AD patients show many biological abnormalities that easily distinguish them from healthy volunteers. Thus, to ensure clinical relevance and applicability, researchers seeking specific markers should focus on the differences between the relevant psychiatric disease and other neuropsychiatric disease controls rather than on the differences with healthy control cohorts. Any proposed diagnostic test for a mental disorder will require a high degree of specificity to achieve substantial clinical gains[10]. Thus, the development of an efficient and selective test to detect the presence of an AD biomarker signature in blood would have tremendous utility.
Currently, biological psychiatry does not provide any diagnostic, biomedical tests for degenerative dementias in routine clinical practice. Intensive investigation by many research teams has explored the identification of blood-based biomarkers that can be used for a clinical laboratory test, including proteomic, metabolomic and lipidomic analyses[11]. Several chemical analytes have been investigated as potential AD biomarkers, including measures of oxidative stress, metabolite profiles, lipid profiles and protein-expression profiles[12-19]. However, none of these has been accepted as a standalone diagnostic biomarker with sufficient specificity for routine diagnostic AD testing.
Such applied methods usually require substantial time, labor and financial resources and are thus difficult to transfer to the clinical laboratory; however, the results of these studies showed that potentially useful biomarkers exist. One strategy that may be able to overcome specificity and cost issues would be to simultaneously measure a complex chemical composition in terms of several classes of compounds[20, 21]. For instance, the very recent publication by Mapstone, M. et al shows that the lipid profile predicts AD development as early as the MCI stage.[18] Therefore, analyzing the panel of biomarkers allows for AD detection up to approximately two years prior to disease onset. Each single panel of biomarkers has a different sensitivity and specificity profile. A combined implementation of these panels can significantly improve the differentiation power of the diagnostic method. The emerging advances in vibrational spectroscopy and advanced statistics offer a real opportunity for probing multiple biochemical markers of disease through their overall spectroscopic signature.
In this regard, the diagnostic potential of Raman spectroscopy has recently been demonstrated for several diseases[22]. Raman spectroscopy has been applied to diagnose different types of cancer, diabetes, atherosclerosis, Alzheimer's and Parkinson's disease[23-26]. Raman spectroscopy provides specific information on the structure, conformation and composition of macromolecules, such as nucleic acids, proteins and lipids[27]. This information is unique to each molecule; therefore, Raman spectroscopy can provide “fingerprinting”-type information on the total biochemical state of blood. Raman spectroscopy could offer useful clinical tests that are simple, rapid and minimally invasive.
This preliminary study is focused on evaluating the selectivity of Raman spectroscopy by examining the spectral profiles of AD patients versus those of either subjects with other types of degenerative dementia or healthy control subjects. We propose that a selective diagnostic blood test can then be created based on a comparison of the obtained spectroscopic changes in the blood of the patient under evaluation with a developed library of Raman spectroscopic signatures for AD and other dementias.
2. Materials and methods
2.1 Clinical subjects and protocols
The cohort of subjects was recruited from neurological subspecialty clinics at the following academic medical centers at Albany Medical College (AMC: the Alzheimer's Center, the Movement Disorders Program, and the Parkinson's Disease and Movement Disorders Center. The cohort consisted of three groups of subjects. The first and second groups included unrelated patients with AD and OD, respectively. The AD group included patients diagnosed with Alzheimer's disease only. The OD group included patients diagnosed with other neurodegenerative dementias (OD): Lewy body dementia (n=5), Parkinson disease dementia (n=10), frontotemporal dementia (n=3) and progressive supranuclear palsy (n=2; a variant of frontotemporal dementia). The medical history and clinical assessments of all recruited patients were reviewed to determine their level of dementia. The clinical diagnoses were established by a trained neurologist. Dementia was defined by a Clinical Dementia Rating Scale (CDR) of .5 or more for all subjects with dementia[28]. AD was diagnosed by the NINDS-ADRDA criteria[29]. CDR was used to determine the stage of AD; a CDR of .5-1 was considered mild AD, and a CDR of 2 was considered moderate AD. The criteria for the diagnosis of Parkinson's disease included the Unified Parkinson's Disease Rating Scale[30, 31] and the criteria for Lewy body disease outlined by the DLB Consortium[32]. The criteria for the diagnosis of frontotemporal dementia (FTD) were applied as reported by Neary et al[33].
The third group consisted of age- and sex-matched control subjects who were free of any neurological and psychiatric ailments and who were in good general health with no active, major disease. The control volunteers were spouses of the patients, and therefore, their ethnic background, socioeconomic and environmental factors (e.g., age, education, race, religion, social and economic status), diet and everyday lifestyle were similar to those of the patients. All subjects were recruited at AMC during the period from 2011-2013.
The research protocol for human studies was reviewed and approved by the University at Albany Institutional Review Board (IRB) and the Albany Medical College IRB. The authorized study personal obtained written informed consent form potential study subjects prior to their participation. Table 1 summarizes the demographic information of the study subjects.
Table 1.
Summary of demographic information of the research subjects.
| Subjects | |||||
|---|---|---|---|---|---|
| Alzheimer's dementia, AD | Healthy controls, HC (n=10) | Other dementias, OD (n=18) | |||
| Moderate stage (n=10) | Mild stage (n=10) | Total AD (n=20) | |||
| Age in years ± SD* | 76±10 | 72.4±8.4 | 74±9.3 | 68±11 | 73±7 |
| Male (%) | 40 | 60 | 50 | 50 | 78 |
| Female (%) | 60 | 40 | 50 | 50 | 22 |
standard deviation
2.2 Sample preparation
A peripheral blood sample of 5 mL was collected and immediately processed into aliquots of anticoagulated whole blood, plasma and serum and then stored at −80°C until used in the analysis. EDTA was used as an anticoagulant. All blood samples were drawn and handled identically.
For Raman measurements, the blood serum sample was defrosted on ice and a 40-μL drop of the blood serum sample was placed on a microscope glass slide covered with aluminum foil and allowed to dry completely (within 5 min) under a gentle air flow. The aluminum foil was used as a substrate with a low fluorescence signal.
2.3 Raman spectroscopic measurements
The Raman spectra were collected with a Renishaw inVia confocal Raman spectrometer equipped with a research-grade Leica microscope and a 50x long-range objective (numerical aperture of 0.50) as described previously[34]. Briefly, all measurements were performed via automatic mapping using the lower plate of a Renishaw PRIOR automatic stage system . The spectra were recorded in the range of 400-1800 cm-1 under a 785 nm excitation wavelength with a diode laser using the WiRE 3.2 software. To avoid sample photodegradation, the laser power was reduced to 55 mW (50%). The excitation wavelength was chosen to reduce the fluorescence background from the sample. An automatic stage system allowed us to preprogram the mapping procedure and scan the sample within an area of 2x2 mm. The sequential spectra from 121 adjacent spots of a sample were measured with two 10 s accumulations at each point. This procedure ensures a thorough representation of the sample spot and accounts for the possible heterogeneity within the sample.
2.4 Data treatment
A total of 5808 spectra were imported to MATLAB R2012a (7.14) and preprocessed. The Raman spectra with low signal-to-noise ratios were excluded. The fluorescent background was removed using the adaptive, iteratively reweighted penalized least squares (airPLS) algorithm[35]. All spectra were normalized by the total area and were mean centered[36].
2.5 Artificial neural networks
The classification and discrimination problems between AD, HC and OD samples were solved using an artificial neural network (ANN) approach. In multivariate statistics, ANNs are simplified mathematical models that conduct information processing functions in a manner similar to that of the highly interactive human brain cortex[37]. The ANN computing structures can be described as several layers of interconnected “neurons” or nodes. Each “neuron” can perform a mathematical operation on the input values and then connect and transfer the result to the next layer. The links between the “neurons” have an associated connection weight. ANNs are trained by learning algorithms to compute the final classification results based on the connection weights. The models can recognize patterns, manage data, and learn in a manner that resembles the information organization and storage in the human brain. The internal structure of the ANN models can be modified with respect to an objective function, and the parameters of the mathematical operations can be tuned by a training algorithm[38]. The power of ANNs is in the collective behavior of many interconnected computing nodes (neurons) that enables the accurate classification and recognition of information. ANNs have been applied to solve problems related to complex data sets in many areas of science, including medicine. In neurology, ANNs have been used to aid the clinical diagnosis of neurodegenerative disorders with extrapyramidal features[39] and of Alzheimer's disease via cognitive measures analysis[40] and neuroimaging data classification[41]. ANNs have been proposed as valid computational tools for analyzing spectroscopic data when solving various clinical problems[42-44].
2.6 Genetic Algorithm
A genetic algorithm (GA) is a machine-learning technique that is designed for feature selection and extraction to identify the most useful subsets of the measured variables for discrimination and classification tasks[45]. The operation of a GA is analogous to Darwinian natural selection and genetics in biological systems[46]. GAs are based on a general adaptive optimization search methodology to select the variables with the lowest prediction error (RMSE-CE) through simulated natural selection, the action of genetic mutations and the recombination of chromosomes[47, 48]. Biology describes natural selection as the “survival of the fittest”, where an adaptation process or the evolution from one generation to another occurs via the elimination of weak elements and the retention of optimal and sub-optimal elements. In GA, a solution to a problem is a point in search space and is called a “chromosome”. Each chromosome represents a combination of meaningful features. By testing all possible solutions, the algorithm generates sets of potential solutions (populations) and ranks them according to the fitness function. Portions of the best of the identified solutions are subjected to operators such as crossover, mutation, inversion and recombination. The iterative computational process resembles natural reproduction. Only the most fit populations are allowed to breed until acceptable results are obtained. GA can handle large search spaces and is thus suitable for cases where the spectral data consist of hundreds or thousands of variables. The spectral features with the best discriminative power can be identified and can carry clinically relevant information[49].
2. Results and Discussion
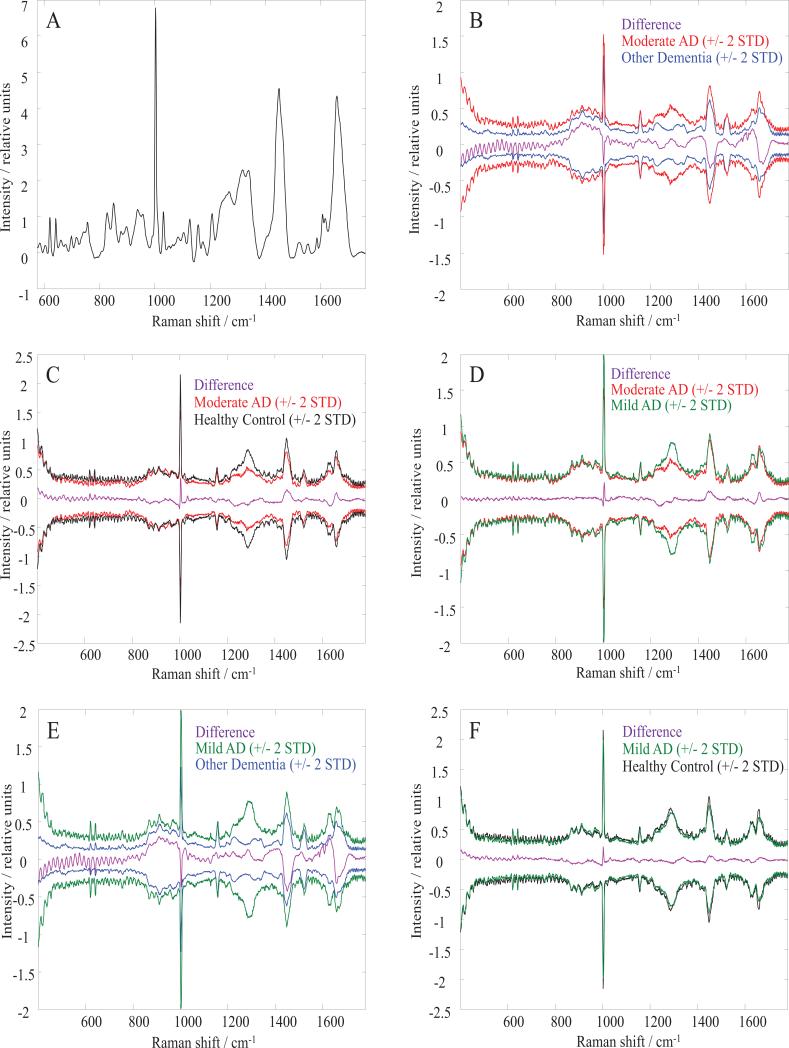
Blood samples were obtained from 10 healthy control, 20 AD and 18 OD subjects. In the AD cluster, 10 subjects were assigned to the moderate AD subgroup, while the other 10 subjects belonged to the mild AD subgroup (see Clinical subjects and protocols). Based on visual inspection, Raman spectra of blood serum originated from different groups of subjects are almost identical. Figure 1 A shows a typical blood serum Raman spectrum, the average spectrum for healthy control cohort. The difference spectra obtained by subtracting average spectra found for various groups are shown in Figures 1 B, C, D, E, and F. The difference spectra are displayed along with +/- 2 standard deviations (STD) for the group spectra that are compared. The difference spectra lay within 2 standard deviations for each compared group, suggesting the necessity for advanced statistical methods to determine spectral variability and transform hidden characteristic features into a discriminative algorithm. In the next step, we employed multivariate statistical analysis on the collected data.
Figure 1.
Average and difference Raman spectra of human blood serum.
(A) An average Raman spectrum of human blood serum obtained for healthy controls. Difference average spectra (purple lines) and spectral variations around the mean (+/-2 STD) for: (B) moderate Alzheimer's dementia and other dementia; (C) moderate Alzheimer's dementia and healthy controls; (D) moderate and mild Alzheimer's dementia cohorts; (E) mild Alzheimer's dementia and other dementia; (F) mild Alzheimer's dementia and healthy controls.
The initial data set was reduced by eliminating the Raman spectra with high variance due to additive Gaussian noise. The threshold was set to cut off 15% of the noisiest experimental Raman spectra. Further statistical analysis was performed for the remaining 85% of the Raman spectra, which were divided into four classes: healthy controls (HC), moderate AD, mild AD and other dementia (OD). The Raman spectra recorded with ~1 cm-1 spectral resolution provided more than 1000 equally spaced variables, which were highly correlated with each other. We used principle component analysis (PCA) to calculate the principle components (PCs) and to reduce the number of input variables for the subsequent advanced statistical analysis. Significant factor analysis (SFA) suggested that at least 6 PCs or significant factors were required to characterize the original spectroscopic data[50]. All classification methods used in this study were tested with between 5 and 20 PCs. Preliminary analysis demonstrated that higher classification accuracy could be obtained with a statistical method that can model the nonlinear relations between variables, such as artificial neural networks. The following machine-learning methods were applied for AD diagnostics: a multilayer perceptron (MLP) and a radial basis function (RBF) network.
The Neural Network Toolbox (MathWorks, Nattick) was used to model, test and validate the artificial neural networks. The optimal MLP architecture was determined by varying the number of hidden layers between one and two and varying the number of neurons in each layer between 10 and 600 for the first hidden layer and between 5 and 50 for the subsequent hidden layers. Additionally, we tested three transfer functions (linear, tangent sigmoidal and log sigmoidal) and five back-propagating training algorithms (Levenberg-Marquardt, Gradient descent, Powell-Beale conjugate gradient, Bayesian regularization, and Fletcher–Powell conjugate gradient). The optimal RBF architecture was determined by varying the number of neurons between 10 and 600 and varying the radius value of the function between 1 and 50. For each classification model, the HC, mild AD, moderate AD and OD data sets were randomly split into training (70%), testing (15%) and validating (15%) data subsets. The performance of the calculated classification models was confirmed using the sensitivity (true positive rate; i.e. percentage of AD spectra identified correctly) and specificity (true negative rate; i.e. percentage of control spectra, either HC or OD, identified correctly) parameters for the classification of the testing and validating data subsets. Each type of network architecture was tested ten times with a new random split of data sets each time. In total, more than 30 000 models were built. The classification models that demonstrated similar performance parameters (sensitivity and specificity) for the testing and validating data subsets were accepted for the following considerations. If the difference in sensitivity (or specificity) for the testing and validating data classifications was more than 10%, the classification model was considered to be the result of convergence to a local minima with weak generalization properties. Importantly, validation is a crucial step in developing classification models because neural networks can frequently be over trained. For example, while screening ANN architectures, we observed cases in which the accuracy of classification was close to 100% for the training data and only ~60% for the testing and validating data.
Table 2 presents the best results for the selected structures of the MLP and RBF networks for the HC vs moderate AD classification. The third column, titled “Network structure”, contains network descriptors, where the first number is the number of input neurons (the number of PCs used), the last number represents the number of output neurons, and the other numbers indicate how many neurons were used in the corresponding hidden layers.
Table 2.
Performance of the selected 9 ANN architectures in the discrimination of moderate AD from control spectroscopic data: a multilayer perceptron (MLP) and a radial basis function (RBF) networks.
| Model # | Type of network | Network structure | Sensitivity / % | Specificity / % |
|---|---|---|---|---|
| 1 | Multilayer perceptron | 5-20-20-1 | 97 | 95 |
| 2 | Multilayer perceptron | 5-50-10-1 | 96 | 96 |
| 3 | Multilayer perceptron | 5-100-1 | 96 | 95 |
| 4 | Multilayer perceptron | 7-200-20-1 | 96 | 94 |
| 6 | Radial basis function | 5-20-20-1 | 95 | 94 |
| 7 | Radial basis function | 5-50-10-1 | 96 | 95 |
| 8 | Radial basis function | 5-100-1 | 94 | 94 |
| 9 | Radial basis function | 7-200-20-1 | 94 | 94 |
MLP networks demonstrate slightly better performance than RBF networks in terms of the sensitivity and specificity of the classification models (Table 2). Note the variability of these parameters for independent implementations of the selected neural network. For example, for the 5-20-20-1 MLP network (Table 2, row 1), the observed sensitivity for the correct classification of AD spectra ranged from 89-97% for ten independent modeling events. These results demonstrate the possibility of building a classification model with parameters that indicate high performance. The observed level of instability in the classification accuracy can be attributed to two main aspects of the ANN modeling process. The first aspect involves the intrinsic properties of the experimental data set, such as the limited size of the Raman data set, the detrimental contribution of noise and the high level of similarity in Raman data. Here, we note that the ideal (100% sensitivity and specificity) discrimination between groups should not be expected because only relatively minor changes in the biochemical composition of blood serum are likely to be associated with AD progression. Additionally, because of the intrinsic inhomogeneity of serum samples that is exposed by the applied drying process, all recorded Raman spectra will not show evidence of AD. Therefore, the achieved level (more than 95% for validating data sets) for an accurately classified single Raman spectrum is a remarkable result. The second aspect is the propensity of ANN algorithms to fall into local minima, yielding a reduced generalization ability[51, 52] and leading to overtraining[53]. From a practical point of view, the ANN model with the highest validated classification accuracy can be accepted.
Table 3 shows the results of the RBF and MLP classification models for the Raman spectra. For binary models, the AD class includes a combination of the mild and moderate classes. For tertiary models, the AD class was treated as two separate classes: mild AD and moderate AD. Every single spectrum was separated from the training set and then tested as a member of the test data set using the calculated model. Usually, less than 10% of the spectra from testing data sets were misclassified. The best achieved cross-validated sensitivity and specificity parameters for healthy controls were 97 and 96% (binary classification), respectively (Table 3, lines 1-2). Slightly lower performance parameters were obtained for the three-class classification model (Table 3, lines 3-5), where the moderate and mild AD data were treated as separate classes.
Table 3.
Summary of the classification results achieved using a multilayer perceptron and a radial basis function networks for discrimination between Alzheimer's dementia and healthy controls.
| # | Class | Radial basis function | Multilayer perceptron | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CV sensitivity / % | CV specificity / % | CV sensitivity / % | CV specificity / % | ||||||
| Spectrum | Subject | Spectrum | Subject | Spectrum | Subject | Spectrum | Subject | ||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Two classes model | |||||||||
| 1 | Healthy controls | 95 | 88 | 92 | 87 | 97 | 87 | 96 | 90 |
| 2 | Alzheimer's dementia | 92 | 87 | 95 | 88 | 96 | 90 | 97 | 87 |
| Three classes model | |||||||||
| 3 | Healthy controls | 89 | 77 | 92 | 83 | 92 | 89 | 93 | 88 |
| 4 | Mild Alzheimer's dementia | 85 | 84 | 94 | 85 | 92 | 84 | 95 | 85 |
| 5 | Moderate Alzheimer's dementia | 86 | 80 | 93 | 86 | 93 | 85 | 94 | 85 |
| Healthy controls vs mild Alzheimer's dementia model | |||||||||
| 6 | Healthy controls | 96 | 90 | 89 | 87 | 95 | 92 | 91 | 91 |
| 7 | Mild Alzheimer's dementia | 89 | 87 | 96 | 90 | 91 | 91 | 95 | 92 |
| Healthy controls vs moderate Alzheimer's dementia model | |||||||||
| 8 | Healthy controls | 96 | 86 | 95 | 85 | 97 | 85 | 96 | 88 |
| 9 | Moderate Alzheimer's dementia | 95 | 85 | 96 | 86 | 96 | 88 | 97 | 85 |
| Mild vs moderate Alzheimer's dementia model | |||||||||
| 10 | Mild Alzheimer's dementia | 93 | 87 | 93 | 84 | 93 | 87 | 94 | 87 |
| 11 | Moderate Alzheimer's dementia | 93 | 84 | 93 | 87 | 94 | 87 | 93 | 87 |
Somewhat greater sensitivity and specificity levels were obtained when the above-mentioned three classes were treated in a pairwise manner. Table 3 presents the sensitivity and specificity values for the control versus mild AD (lines 6 and 7), control versus moderate AD (lines 8 and 9), and mild versus moderate AD (lines 10 and 11) classification. When considered together, these binary classifiers provided approximately 95% in the overall accuracy of AD stage determination.
After the ability of our classification methods to detect and separate AD spectra from HC spectra was established, we substituted the HC group with OD. A similar approach as above was used to model, test and validate the artificial neural networks for discrimination between AD and OD. The network with the optimal performance was able to distinguish OD with 97% specificity and 92% sensitivity, mild AD with 92% specificity and 95% sensitivity and moderate AD with 100% specificity and 93% sensitivity. The designed neural networks were optimized for pattern recognition and were capable of distinguishing between different classes with high accuracy. Additional performance parameters are presented in Table 4.
Table 4.
Summary of the classification results achieved using a multilayer perceptron method for discrimination between Alzheimer's dementia and other dementias.
| # | Class | Multilayer perceptron | |
|---|---|---|---|
| CV sensitivity / % | CV specificity / % | ||
| Two classes model | |||
| 1 | Other dementias | 95 | 98 |
| 2 | Alzheimer's dementia | 96 | 95 |
| Three classes model | |||
| 3 | Other dementias | 92 | 97 |
| 4 | Mild Alzheimer's dementia | 95 | 92 |
| 5 | Moderate Alzheimer's dementia | 93 | 100 |
| Other types of dementia vs mild Alzheimer's dementia model | |||
| 6 | Other dementias | 91 | 94 |
| 7 | Mild Alzheimer's dementia | 84 | 82 |
| Other types of dementia vs moderate Alzheimer's dementia model | |||
| 8 | Other dementias | 96 | 95 |
| 9 | Moderate Alzheimer's dementia | 92 | 99 |
Considering the results described above, we tested the possible influence of a drug effect on the accuracy of the classification. The processing procedure was the same as above. All AD subjects were divided into groups according to their medication regimen, and the optimal ANN was applied. The subjects were taking various combinations of drugs rather than one specific drug, and the cohorts were small; thus, it was not possible to precisely identify which/how a specific drug can/cannot influence the ANN. Therefore, the analysis was performed with respect to one drug (donepezil), which was the most common among our subjects. All AD spectra were split into subclasses of subjects who were taking donepezil and those who reported no medication regimen. The ANN either could not distinguish between the two classes within the same AD group or was simply “blind” and misclassified subjects on the medication regimen as medication-free subjects. This result led to the conclusion that there were no significant drug effects from donepezil on the classification models. Most probably, other medications have insignificant effects as well. However the latter conclusion would require further study involving much larger cohort of patients with a better representation of other drugs used.
Finally, all calculations were repeated using subject-wise cross-validation. Subject-wise means all individual spectra that originated from a single subject were grouped to be separately validated as one subject at a time without mixing them with all of the other individual spectra that originated from that diagnostic category. The ANN architectures that demonstrated the best results in tests where the factor of subjects was not considered (Table 2) were tested with the data sets formed by consecutively eliminating all of the Raman spectra from each subject individually. This classification was repeated 10 times for each subject. Most importantly, it was always possible to attain an ANN classification model with more than 60% correctly classified spectra. In other words, using this number as a decision threshold, we can correctly categorize all subjects. Table 3 shows the minimum and maximum (columns 3, 5, 7, 9) percentages of correctly classified spectra for the subject-wise cross-validation. The relatively low numbers and the wide range indicate that the power of the classification models depends on how the subjects were split between the training and testing data sets. A significant increase in the number of subjects will allow us to directly address the effect of population and physiological factors.
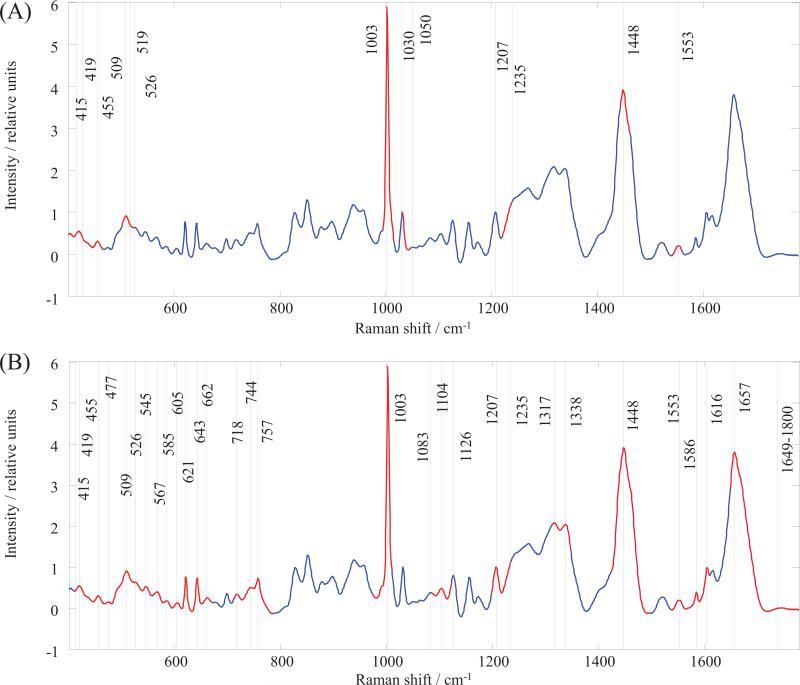
ANNs have shown that these supervised analyses can easily discriminate between all groups in question but cannot determine which wavenumbers in the Raman spectra are important; thus, they have limited use for interpreting these multidimensional spectra in biochemical terms. Therefore, a GA was used to determine the specific wavenumbers that can elucidate some characteristic biomarkers, particularly for the recently proposed blood-based biomarkers of AD[12-16, 18, 20]. We analyzed our data via GA using PLS toolbox 6.2 (Eigenvector Research, Inc., Wenatchee) within the Matlab environment[36]. A single run was selected to have 70 chromosomes for the initial population and for each population in the subsequent generations. The number of generations was set to 100, and the mutation rate was kept at a value of 0.005. The genetic algorithm allows a consideration of all possible variables within the Raman spectral dataset for the serum and of their significance for the discrimination between classes. These factors can be considered because of the reduction of the original Raman spectra to a subset(s) of wavenumbers; this reduction simplifies the spectral information for the possible identification of meaningful information. The Raman bands that yielded the best results for internal cross validation were identified from the most informative spectral regions. These regions and bands represent spectroscopic markers for the datasets in the study. The most meaningful spectral regions were identified by comparing the AD group to the HC group (Fig. 2 A) and to the OD group (Fig. 2 B). The wavenumbers of the vibrational band positions were determined within these regions. The tentative band assignments along with the borders of the corresponding spectral region and the presumptive contributions of biomolecules are given in Tables 5 and 6 for the comparison of the AD group to HC and to OD, respectively. The GA revealed more differences between AD and OD than it did for AD and HC. The GA selected 9 regions (30 bands) and 7 regions (13 bands) for the AD to OD and AD to HC comparisons, respectively. Interestingly, the 5 regions selected for the AD/HC comparison almost completely overlap with the regions from the AD/OD selections. However, one region from the AD/HC selection (# 4, Table 5) is not present, and three new regions appear in the AD/OD selections (# 2, 4, 6; Table 6). Our results are consistent with the current proteomics, metabolomics and lipidomics findings for AD biomarkers. Most of the biomolecules that we identified as contributors to the spectroscopic signature for AD have been previously reported as potential AD biomarkers. For example, a recent study by Mapstone et al[18] successfully employed lipidomic and metabolomic profiling to identify and validate a biomarker panel for pre-clinical AD detection. The featured phospholipids and metabolites, such as phosphatidylinositol, phosphatidylcholine, proline, lysine and phenylalanine, are among molecules that contribute to our spectroscopic markers. Moreover, compared with the previous studies that used elderly volunteers without dementia as a control group, we show that these biomolecules contribute to the differentiation of AD from patients with other types of dementia as well as from the healthy control group. However, a more detailed interpretation of the clinical relevance of the spectroscopic signature for AD requires further investigation.
Figure 2.
Raman spectral regions and bands identified by Genetic Algorithm (GA) as providing the highest discriminatory power between the serum samples. (A) Alzheimer's dementia versus healthy controls; (B) Alzheimer's dementia versus other dementias. The GA regions highlighted in red on the total average spectrum of the human blood serum (blue).
Table 5.
A tentative assignment of the most important regions in the Raman spectrum of blood serum for the discrimination between AD and HC, as determined by the genetic algorithm (GA).
| # | GA spectral region (cm−1) | Peak position (cm−1) +/− 5 | Tentative band assignment [22, 23, 27, 55] | Contributions [22, 23, 27, 56, 57] |
|---|---|---|---|---|
| 1 | 400-467 | 415, 419, 455 | υ (C-C), υ (S-S), δ (C-C) aliphatic chains | Proteins (amino acids: histidine, tryptophan, lysine, proline); lipids (fatty acids, triglycerides, cholesterol); carbohydrates (fructose, lactose, citric acid, galactosamine, N-acetyl-glucosamine); riboflavin; phosphatidylinositol; phosphoenolpyruvate |
| 2 | 505-523 | 509, 519, 526 | υ(S-S) | Proteins (amino acids: glycine, glutamate, phenylalanine, tryptophan, tyrosine); carbohydrates (fructose, galactosamine, N-acetyl-glucosamine); coenzyme A; phosphatidylinositol; phosphatidylserine; acetoacetate; glutathione |
| 3 | 993-1011 | 1003 | υ(C-C), υ (C-O), Aromatics ring breathing vibration | Proteins (amino acids: phenylalanine, lysine, glutamate); carbohydrates (fructose, lactose, galactosamine, N-acetyl-glucosamine); acetoacetate |
| 4 | 1030-1049 | 1031, 1050 | υ(C-C), υ(C-O) | Proteins (amino acids: valine, arginine, phenylalanine, proline, glycine, lysine, tryptophan, tyrosine); carbohydrates (fructose, lactose, mannose); fatty acids; phosphoenolpyruvate; coenzyme A |
| 5 | 1218-1237 | 1207, 1235 | υ(C-C), υ(C-H), υ(C=S), (CH2) wagging vibrations | Proteins (β sheet, Amid III, tryptophan, tyrosine, lysine, phenylalanine, glycine, proline); pyruvate |
| 6 | 1444-1462 | 1448 | δ (CH2), δ(CH2OH), δ (CH3), υ(C-C) | Proteins (amino acids: valine, phenylalanine, tryptophan, lysine, proline); fatty acids; carbohydrates (fructose, galactosamine); phosphoenolpyruvate; acetoacetate; β-carotene; glutathione |
| 7 | 1537-1556 | 1553 | υ(N=N), υ(C=C), υ(C-(NO2)) | Guanine; tryptophan; proline; palmitic acid; N-acetyl-glucosamine |
stretching mode;
bending mode;
Table 6.
A tentative assignment of the most important regions in the Raman spectrum of blood serum for the discrimination between AD and OD, as determined by the genetic algorithm (GA).
| # | GA spectral region (cm−1) | Peak position ( cm−1) +/− 5 | Tentative band assignment [22, 23, 27, 55] | Contributions [22, 23, 27, 56, 57] |
|---|---|---|---|---|
| 1 | 410-672 | 415, 419, 455, 477, 509, 519, 526 545, 567, 585, 605, 621, 643, 662 | υ(C-C), υ(C-S) aliphatic chains δ(C-C) aliphatic chains | Proteins (amino acids: histidine, tryptophan, proline, lysine, glycine); lipids (fatty acids, triglycerides); carbohydrates (fructose, lactose, citric acid, galactosamine, N-acetyl-glucosamine); riboflavin; phosphatidylinositol; phosphoenolpyruvate |
| 2 | 710-784 | 718, 744, 757 | υ(C-S) aliphatic chains υ(N+(CH3)3) υ(C-C-N+) | Proteins (amino acids: valine, glutamate, phenylalanine, tryptophan); phospholipids; phosphatidylcholine; phosphoenolpyruvate; acetoacetate; coenzyme A; acetyl coenzyme A; D-fructose-6-phosphate; riboflavin; glutathione |
| 3 | 973-1010 | 1003 | υ(C-C), υ(C-O), Aromatics ring breathing vibration | Proteins (amino acids: phenylalanine, lysine, glutamate); carbohydrates (fructose, lactose, galactosamine, N-acetyl-glucosamine); acetoacetate |
| 4. | 1086-1122 | 1083, 1104, 1126 | υ(C-C), (O-P-O), υ( C-O-C), υ(C-S) aromatic | Proteins (phenylalanine, glycine, valine, glutamate, arginine, histidine, proline); carbohydrates (β glucose, galactosamine, N-acetyl-glucosamine); acetyl coenzyme A; acetoacetate; D-fructose-6-phosphate; phosphoenolpyruvate; phospholipids |
| 5 | 1199-1235 | 1207,1235 | υ(C-C), υ(C-H), υ(C=S), (CH2) wagging vibrations | Proteins (β sheet, Amid III, tryptophan, tyrosine, phenylalanine, glycine, proline); pyruvate |
| 6 | 1311-1348 | 1317, 1338 | υ(C-(NO2)), υ(C-(SO2)-C) | Proteins (phenylalanine, tryptophan, glycine, valine, serine, glutamate, histidine, proline, lysine, adenine, α helix); carbohydrates (lactose, N-acetyl-glucosamine); phosphoenolpyruvate; glutathione; phospholipids |
| 7 | 1424-1498 | 1448 | δ(CH2), δ(CH2OH), δ(CH3), υ(C-C) | Proteins (amino acids: valine, lysine, phenylalanine, tryptophan, proline); fatty acids; carbohydrates (fructose, galactosamine); phosphoenolpyruvate; acetoacetate; β-carotene; glutathione |
| 8 | 1537-1611 | 1553, 1586 | υ(N=N), υ(C=C), υ(C-(NO2)) NH3+ | Guanine; tryptophan; proline; lysine; palmitic acid; N-acetyl-glucosamine |
| 9 | 1649-1800 | 1657 | υ(C=C), υ(C=O) | Proteins; amide I; α helix; phospholipids |
stretching mode;
bending mode;
GAs are often used as an optimization method in the design of automatic pattern classifiers[45, 46, 54]. Performance optimization can strongly affect classifier design because the choice of features has a substantial influence on several classification aspects, including the accuracy of the learned classification algorithm, the time needed to learn the classification function and the number of examples needed for learning[46]. The accuracy of the classifier outcome may be enhanced by adequate GA optimization[54]. However, such hybrid systems usually focus on either improving the classifier capabilities or feature extraction that discards the GA or ANN as an artifact of the process[45]. The ultimate goal of this work was correct classification and meaningful feature extraction. Coupling GAs and ANNs will require designing a feedback mechanism for making classification decisions while allowing the modification and/or adaptation of the feature selector and research using ANNs for evolution of the GA fitness function. For practical purposes, the classification step and the feature selection/extraction step were considered independent stages. Future research should pursue the coupling of GA and effective classification techniques and evaluate the Raman bands selected by the GA using ANN.
3. Conclusion
This proof-of-concept study demonstrated the great potential of Raman spectroscopy as a selective diagnostic tool for AD detection and differentiation. The technique is sensitive and specific relative to other types of dementia and is noninvasive because it uses conventional venipuncture to obtain the blood sample. In this study, we used the combination of Raman spectroscopy and multivariate statistics to differentiate AD patients from patients with other types of dementia and from healthy elderly volunteers with an overall accuracy 95%. We demonstrated the high probability for single subject to be correctly assigned to the particular diagnostic category. The results were verified using random ten-fold subsets of individual spectra and subject-wise cross-validation approaches. By setting the threshold to 60% of classified single spectra, all subjects except one were diagnosed correctly. The Alzheimer's disease, healthy control and other dementia groups showed similar, but not identical, Raman spectral profiles. These differences can be detected by Raman spectroscopy and distinguished using the artificial neural network approach. Genetic algorithm variable analysis combined with several cross-validation methods was performed to identify “fingerprint” regions or spectroscopic markers for selective detection of AD. Our results are consistent with current AD biomarker findings. Distinctive spectral regions that are optimal for AD differentiation were identified. The regions correspond to particular biomolecular changes. The spectroscopic markers we found display Raman signals originated from the biomolecules that have been reported to be AD biomarkers in recent studies. This proof-of-concept study was performed on a relatively small cohort of patients and successfully demonstrated the great potential of the method for Alzheimer's disease diagnostics. The developed Raman spectroscopic markers of AD open potential opportunity for the early disease diagnostics. That is one of the main objectives for our future investigation. Also study on a larger cohort of patients with a better representation of drugs used to treat AD and other dementia is required for determining possible effects of a medication regime. As well as validations in large multi-centered randomized control studies will be needed to establish effectiveness of the method for early AD diagnostics.
Multivariate data analysis of blood serum Raman spectra allows for the differentiation between patients with Alzheimer's disease, other types of dementia and healthy individuals.
Acknowledgments
The authors are grateful to Aliaksandra Sikirzhytskaya for assistance with the computer generation of the illustrations and to Vitali Sikirzhytski for assistance with the statistical analyses at the early stages of the project. The project was supported by a research grant from the New York Capital Region Research Alliance Program and the National Institute on Aging, National Institutes of Health, Grant R01AG033719 (I.K.L.).
References
- 1.Prince M, Bryce R, Albanese E, Wimo A, Ribeiro W, Ferri CP. Alzheimers Dement. 2013;9:63–75.e62. doi: 10.1016/j.jalz.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Brookmeyer R, Evans DA, Hebert L, Langa KM, Heeringa SG, Plassman BL, Kukull WA. Alzheimers Dement. 2011;7:61–73. doi: 10.1016/j.jalz.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alzheimers Dement. 2014;10:e47–e92. doi: 10.1016/j.jalz.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Monien BH, Apostolova LG, Bitan G. Expert Rev Neurother. 2006;6:1293–1306. doi: 10.1586/14737175.6.9.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farlow MR. Am J Health Syst Pharm. 1998;55(Suppl 2):S5–10. doi: 10.1093/ajhp/55.suppl_2.S5. [DOI] [PubMed] [Google Scholar]
- 6.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Jr, Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison-Bogorad M, Wagster MV, Phelps CH. Alzheimers Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helmuth L. Sci Aging Knowledge Environ. 2003;2003:oa2. doi: 10.1126/sageke.2003.43.oa2. [DOI] [PubMed] [Google Scholar]
- 8.Solomon PR, Murphy CA. Expert Rev Neurother. 2008;8:769–780. doi: 10.1586/14737175.8.5.769. [DOI] [PubMed] [Google Scholar]
- 9.Hildebrandt H, Fink F, Kastrup A, Haupts M, Eling P. Dement Geriatr Cogn Dis Extra. 2013;3:102–112. doi: 10.1159/000348350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapur S, Phillips AG, Insel TR. Mol Psychiatry. 2012;17:1174–1179. doi: 10.1038/mp.2012.105. [DOI] [PubMed] [Google Scholar]
- 11.Zellner M, Veitinger M, Umlauf E. Acta Neuropathol. 2009;118:181–195. doi: 10.1007/s00401-009-0502-7. [DOI] [PubMed] [Google Scholar]
- 12.Irizarry MC. NeuroRx. 2004;1:226–234. doi: 10.1602/neurorx.1.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns DH, Rosendahl S, Bandilla D, Maes OC, Chertkow HM, Schipper HM. J Alzheimers Dis. 2009;17:391–397. doi: 10.3233/JAD-2009-1053. [DOI] [PubMed] [Google Scholar]
- 14.Blennow K, Hampel H, Weiner M, Zetterberg H. Nat Rev Neurol. 2010;6:131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- 15.Britschgi M, Wyss-Coray T. Arch Neurol. 2009;66:161–165. doi: 10.1001/archneurol.2008.530. [DOI] [PubMed] [Google Scholar]
- 16.Kork F, Holthues J, Hellweg R, Jankowski V, Tepel M, Ohring R, Heuser I, Bierbrauer J, Peters O, Schlattmann P, Zidek W, Jankowski J. Curr Alzheimer Res. 2009;6:519–524. doi: 10.2174/156720509790147160. [DOI] [PubMed] [Google Scholar]
- 17.Doecke JD, Laws SM, Faux NG, Wilson W, Burnham SC, Lam CP, Mondal A, Bedo J, Bush AI, Brown B, De Ruyck K, Ellis KA, Fowler C, Gupta VB, Head R, Macaulay SL, Pertile K, Rowe CC, Rembach A, Rodrigues M, Rumble R, Szoeke C, Taddei K, Taddei T, Trounson B, Ames D, Masters CL, Martins RN. Arch Neurol. 2012;69:1318–1325. doi: 10.1001/archneurol.2012.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, Hall WJ, Fisher SG, Peterson DR, Haley JM, Nazar MD, Rich SA, Berlau DJ, Peltz CB, Tan MT, Kawas CH, Federoff HJ. Nat Med. 2014;20:415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Restrepo L, Stafford P, Johnston S. Neurology. 2014;82:S38.002. [Google Scholar]
- 20.Teunissen CE, Lutjohann D, von Bergmann K, Verhey F, Vreeling F, Wauters A, Bosmans E, Bosma H, van Boxtel MP, Maes M, Delanghe J, Blom HJ, Verbeek MM, Rieckmann P, De Bruijn C, Steinbusch HW, de Vente J. Neurobiol Aging. 2003;24:893–902. doi: 10.1016/s0197-4580(03)00005-8. [DOI] [PubMed] [Google Scholar]
- 21.Rosen C, Hansson O, Blennow K, Zetterberg H. Mol Neurodegener. 2013;8:20. doi: 10.1186/1750-1326-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basar G, Parlatan U, Seninak S, Gunel T, Benian A, Kalelioglu I. Int J Spectrosc. 2012;27:239–252. [Google Scholar]
- 23.Pichardo-Molina JL, Frausto-Reyes C, Barbosa-Garcia O, Huerta-Franco R, Gonzalez-Trujillo JL, Ramirez-Alvarado CA, Gutierrez-Juarez G, Medina-Gutierrez C. Lasers Med Sci. 2007;22:229–236. doi: 10.1007/s10103-006-0432-8. [DOI] [PubMed] [Google Scholar]
- 24.Koo TW, Berger AJ, Itzkan I, Horowitz G, Feld MS. Diabetes Technol Ther. 1999;1:153–157. doi: 10.1089/152091599317369. [DOI] [PubMed] [Google Scholar]
- 25.Schipper HM, Kwok CS, Rosendahl SM, Bandilla D, Maes O, Melmed C, Rabinovitch D, Burns DH. Biomark Med. 2008;2:229–238. doi: 10.2217/17520363.2.3.229. [DOI] [PubMed] [Google Scholar]
- 26.Carmona P, Molina M, Calero M, Bermejo-Pareja F, Martínez-Martín P, Toledano A. J Alzheimers Dis. 2013;34:911–920. doi: 10.3233/JAD-122041. [DOI] [PubMed] [Google Scholar]
- 27.De Gelder J, De Gussem K, Vandenabeele P, Moens L. J Raman Spectrosc. 2007;38:1133–1147. [Google Scholar]
- 28.Morris JC. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 29.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 30.Mov Disord. 2003;18:738–750. doi: 10.1002/mds.10473. [DOI] [PubMed] [Google Scholar]
- 31.Goetz CG, Tilley BC, Shaftman SR, Stebbins GT, Fahn S, Martinez-Martin P, Poewe W, Sampaio C, Stern MB, Dodel R, Dubois B, Holloway R, Jankovic J, Kulisevsky J, Lang AE, Lees A, Leurgans S, LeWitt PA, Nyenhuis D, Olanow CW, Rascol O, Schrag A, Teresi JA, van Hilten JJ, LaPelle N. Mov Disord. 2008;23:2129–2170. doi: 10.1002/mds.22340. [DOI] [PubMed] [Google Scholar]
- 32.McKeith IG, Dickson DW, Lowe J, Emre M, O'Brien JT, Feldman H, Cummings J, Duda JE, Lippa C, Perry EK, Aarsland D, Arai H, Ballard CG, Boeve B, Burn DJ, Costa D, Del Ser T, Dubois B, Galasko D, Gauthier S, Goetz CG, Gomez-Tortosa E, Halliday G, Hansen LA, Hardy J, Iwatsubo T, Kalaria RN, Kaufer D, Kenny RA, Korczyn A, Kosaka K, Lee VM, Lees A, Litvan I, Londos E, Lopez OL, Minoshima S, Mizuno Y, Molina JA, Mukaetova-Ladinska EB, Pasquier F, Perry RH, Schulz JB, Trojanowski JQ, Yamada M. Neurology. 2005;65:1863–1872. doi: 10.1212/01.wnl.0000187889.17253.b1. [DOI] [PubMed] [Google Scholar]
- 33.Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, Boone K, Miller BL, Cummings J, Benson DF. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 34.Sikirzhytskaya A, Sikirzhytski V, Lednev IK. J Biophotonics. 2014;7:59–67. doi: 10.1002/jbio.201200191. [DOI] [PubMed] [Google Scholar]
- 35.Zhang ZM, Chen S, Liang YZ. Analyst. 2010;135:1138–1146. doi: 10.1039/b922045c. [DOI] [PubMed] [Google Scholar]
- 36.Wise BM, Gallagher NB, Bro R, Shaver JM, Windig W, Koch RS, OÕSullivan D. Software. Eigenvector Research, Inc.; Wenatchee: 2011. PLS_Toolbox 6 for use with Matlab. [Google Scholar]
- 37.Basheer IA, Hajmeer M. J Microbiol Methods. 2000;43:3–31. doi: 10.1016/s0167-7012(00)00201-3. [DOI] [PubMed] [Google Scholar]
- 38.Kalderstam J, Eden P, Bendahl PO, Strand C, Ferno M, Ohlsson M. Artif Intell Med. 2013;58:125–132. doi: 10.1016/j.artmed.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Litvan I, DeLeo JM, Hauw JJ, Daniel SE, Jellinger K, McKee A, Dickson D, Horoupian DS, Lantos PL, Tabaton M. Brain. 1996;119:831–839. doi: 10.1093/brain/119.3.831. [DOI] [PubMed] [Google Scholar]
- 40.Quintana M, Guardia J, Sanchez-Benavides G, Aguilar M, Molinuevo JL, Robles A, Barquero MS, Antunez C, Martinez-Parra C, Frank-Garcia A, Fernandez M, Blesa R, Pena-Casanova J, Team NS. J Clin Exp Neuropsychol. 2012;34:195–208. doi: 10.1080/13803395.2011.630651. [DOI] [PubMed] [Google Scholar]
- 41.Aguilar C, Westman E, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, Lovestone S, Spenger C, Simmons A, Wahlund LO. Psychiatry Res. 2013;212:89–98. doi: 10.1016/j.pscychresns.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt J, Beekes M, Brauer A, Udelhoven T, Lasch P, Naumann D. Anal Chem. 2002;74:3865–3868. doi: 10.1021/ac015688s. [DOI] [PubMed] [Google Scholar]
- 43.Krafft C, Steiner G, Beleites C, Salzer R. J Biophotonics. 2009;2:13–28. doi: 10.1002/jbio.200810024. [DOI] [PubMed] [Google Scholar]
- 44.Hughes C, Iqbal-Wahid J, Brown M, Shanks JH, Eustace A, Denley H, Hoskin PJ, West C, Clarke NW, Gardner P. J Biophotonics. 2013;6:73–87. doi: 10.1002/jbio.201200126. [DOI] [PubMed] [Google Scholar]
- 45.Tong DL, Schierz AC. Artif Intell Med. 2011;53:47–56. doi: 10.1016/j.artmed.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Huang CL, Wang CJ. Expert Syst Appl. 2006;31:231–240. [Google Scholar]
- 47.Ramadan Z, Jacobs D, Grigorov M, Kochhar S. Talanta. 2006;68:1683–1691. doi: 10.1016/j.talanta.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 48.Goldberg DE. Genetic Algorithms in Search, Optimization and Machine Learning. Addison-Wesley Longman Publishing Co., Inc.; Boston: 1989. p. 372. [Google Scholar]
- 49.Baclig AC, Bakker Schut TC, O'Regan GM, Irvine AD, McLean WHI, Puppels GJ, Caspers PJ. J Raman Spectrosc. 2013;44:340–345. [Google Scholar]
- 50.Malinowski ER. Factor analysis in chemistry. 3 ed John Wiley & Sons Ltd.; New York: 2002. pp. 10–23. [Google Scholar]
- 51.Cheng B, Titterington DM. Stat Sci. 1994;9:2–30. [Google Scholar]
- 52.Everitt BS, Dunn G. Applied Multuvariate Data Analysis. 2 ed John Wiley & Sons Ltd.; Chichester: 2001. pp. 248–268. [Google Scholar]
- 53.Breiman L. Mach Learn. 1996;24:123–140. [Google Scholar]
- 54.Mantzaris D, Anastassopoulos G, Adamopoulos A. Neural Netw. 2011;24:831–835. doi: 10.1016/j.neunet.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 55.Mary MB, Umadevi M, Ramakrishnan V. Spectrochim Acta A Mol Biomol Spectrosc. 2005;61:3124–3130. doi: 10.1016/j.saa.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 56.Movasaghi Z, Rehman S, Rehman IU. Appl Spectrosc Rev. 2007;42:493–541. [Google Scholar]
- 57.Manoharan R, Wang Y, Feld MS. Spectrochim Acta A Mol Biomol Spectrosc. 1996;52:215–249. [Google Scholar]