Abstract
Purpose
The Scripps Idiopathic Diseases of huMan (IDIOM) study aims to discover novel gene-disease relationships and provide molecular genetic diagnosis and treatment guidance for individuals with novel diseases using genome sequencing integrated with clinical assessment and multidisciplinary case review.
Methods
Here we describe the IDIOM study operational protocol and initial results.
Results
121 cases underwent first tier review by the principal investigators to determine if the primary inclusion criteria were satisfied, 59 (48.8%) underwent second tier review by our clinician-scientist review panel, and 17 (14.0%) patients and their family members were enrolled. 60% of cases resulted in a plausible molecular diagnosis. 18% of cases resulted in a confirmed molecular diagnosis. 2 of 3 confirmed cases led to the identification of novel gene-disease relationships. In the third confirmed case, a previously described but unrecognized disease was revealed. In all three confirmed cases, a new clinical management strategy was initiated based on the genetic findings.
Conclusions
Genome sequencing provides tangible clinical benefit for individuals with idiopathic genetic disease, not only in the context of molecular genetic diagnosis of known rare conditions, but also in cases where prior clinical information regarding a new genetic disorder is lacking.
Keywords: clinical sequencing, rare disease, genomics, undiagnosed diseases, genome sequencing
INTRODUCTION
The traditional approach to rare, severely disabling medical conditions frequently leaves the affected individual without a diagnosis and effective treatment. Patients with such conditions can remain ill and endure a “diagnostic odyssey” for years, which is not only difficult for such individuals and their family, but can also be very cost inefficient. Many have previously suggested the utility of genomic information for diagnosing and treating such conditions, and early evidence of the successful application of whole exome sequencing (WES) and whole genome sequencing (WGS) for such purposes is emerging1–6. Of the rare likely genetic diseases that have already been described, approximately half have yet to be linked to a causal gene (termed here: “idiopathic diseases”)7. Estimates of the total number of rare likely genetic diseases based on the number of known disease causing and essential genes have resulted in predictions of between 7,000 to 15,000 disorders, suggesting many rare genetic diseases have yet to be described8. While the application of genome sequencing to the molecular genetic diagnosis of previously described rare Mendelian disorders is essentially proven, the utility of genome sequencing in novel diseases has not been systematically explored. For example, of the ~100 patients successfully diagnosed by the National Institutes of Health Intramural Undiagnosed Disease Program (NIH UDP) (20–25% of the total enrolled), 15 cases (~3.5% of total enrolled) correspond to novel gene associations for previously described diseases, and only 2 cases (<1% of total enrolled) correspond to previously unknown diseases9.
The Scripps Idiopathic Diseases of huMan (IDIOM) study was initiated in 2011. IDIOM was, in large part, modeled after the NIH UDP, with a few exceptions. The primary exception being that we focus exclusively on cases that do not fit a previously described phenotype, or cases where the disorder matches a previously described phenotype and all known genetic causes of the disorder have been ruled out. In other words, only 17 of 100 successfully diagnosed cases in the NIH UDP would have qualified for the IDIOM study (15 corresponding to a novel gene associations for previously described diseases, 2 cases corresponding to previously unknown diseases). The application of genome sequencing to molecular genetic diagnosis in this sub-population of individuals presents some unique challenges both in terms of the evaluation of cases and appropriateness for the IDIOM program, as well as for the ultimate return of genetic results. In this report we provide a description of IDIOM study procedures, the initial results from the first three years of operation, and the clinical benefit achieved by those realizing a confirmed genetic diagnosis.
METHODS
Recruitment and Screening
The Scripps Idiopathic Diseases of huMan (IDIOM) Study (IRB-11-5723) was approved by the Scripps Institutional Review Board in 2011. A separate informed consent was attained for genome sequencing vs. treatment response monitoring. Recruitment for IDIOM was done through announcements to physicians within the Scripps Health system and advocacy groups, announcements via local media10,11, and word of mouth. A number of online sources were utilized such as the CTSA Vanderbilt-hosted Researchmatch.org, “reach out” to leaders of the NIH UDP, RARE Project, and National Organization for Rare Diseases. Scripps Health hosts a landing page for the IDIOM study that includes study criteria and coordinator contact information. The trial is also listed on clinicaltrials.gov (http://clinicaltrials.gov/ct2/show/NCT01440218). Since initial financial support for our study was limited, we have been conservative with respect to our recruitment efforts so as not to be inundated with a large number of referrals that we would not have the resources to fund.
Inclusion criteria for the study are the following: (1) the patient has a grave or serious condition that is undiagnosed despite extensive medical and genetic evaluation – for patients with a likely clinical diagnosis, at minimum, a gene panel test is required to rule out known genetic causes of the disorder; (2) the patient’s condition is potentially “actionable” or amenable to treatment – this is a subjective judgment by the physician review panel that typically only excludes individuals with severe dysmorphologies; (3) the condition appears genetic in origin; (4) the patient’s anticipated life expectancy is consistent with the study timeline for sequencing; and (5) the patient has a physician champion who is willing to work with the research team and take responsibility for returning genetic results to the patient.
In order to be considered, patients or their referring physician provide a short clinical summary and all available medical records. Referrals undergo an initial review by the IDIOM study coordinator. Typically, cases with complete medical records undergo another round of internal review by core study investigators, often via email, and those that appear to meet inclusion criteria are forwarded for review by the IDIOM Clinician-Scientist Review Panel.
Clinician-Scientist Review Panel
Our clinician-scientist review panel is made up of approximately 12 practicing physicians, as well as a research team consisting of bioinformatics and genetic analysis specialists, physicians who utilize genetics extensively in clinical practice, sequencing experts, ethicists, clinical psychologists, and research nurses. The Director of the Scripps IRB is also a member of the panel. The clinical disciplines represented among the physician members include, though are not limited to, the following: neurology, rheumatology, internal medicine, allergy/immunology, cardiology, medical oncology/hematology, and gastroenterology/hepatology. For a quorum we required the presence of at least 5 physicians and 2 bioinformatics or sequencing experts. Selection of cases is made based on majority vote, however, in almost all instances to date, decisions to enroll a patient have been unanimous. The meetings typically last 1.5 hours, and between 3 and 4 cases are usually reviewed per session. We encourage and allow for, but do not require, the physician champion of patients whose cases are being reviewed to be available during the meeting (either in person or via teleconference) in order to answer questions and interface with the panel.
Consent and Enrollment
Once a case has been selected by the clinician-scientist panel, our nurse study coordinator consents the patient and family members. Usually the participants that comprise a case are those in a trio (i.e., proband, mother, and father), but occasionally other biological family members are sequenced, and in these instances, it is usually a sibling of the proband or parents.
Importantly, a primary issue raised in the literature regarding patient consent for WES and WGS studies is the identification of ‘incidental’ or ‘secondary’ findings (i.e., genomic findings of clinical relevance that are not recognized as being associated with the presenting disease/condition). We have deemed that best practices are yet to be determined by empirical data12 and thus, have elected to only return results directly relevant to the presenting indication. If we should inadvertently discover incidental findings that could have an effect on the patient’s health, the results would be reviewed by our clinician-scientist review panel, who would adjudicate how to proceed with informing the physician champion and patient13.
For selected cases, we also ask the patient’s physician champion (usually the referring physician) to sign an agreement of participation that stipulates that s/he will commit to: (a) regular interactions with the research team; (b) acceptance of responsibility for return of genomic results to the patient; (c) acceptance of any clinical decision-making on the basis of any results provided; and (d) completion of brief baseline and follow-up questionnaires and/or interviews pertaining to the study.
Sequence Data Generation, Analysis and Interpretation
Once cases have been selected and patients and family members are consented for participation, blood is drawn and brought to our lab at STSI for sequencing of the proband and biological family members. Once parentage is confirmed, WES is performed for the detection of coding variants, and low pass WGS is performed for the detection of structural and copy number variants. Target WES coverage is ~100× and target low pass WGS coverage is ~5×. Other published papers have described this data generation protocol in detail, as well as our methods for analysis and interpretation14–17. If necessary, especially for indels or variants with limited coverage in any family members, Sanger sequencing is utilized to confirm candidate causal variants. The theoretical target breakpoint resolution for low pass WGS detection of CNVs is 200bp given a bin size of 200–300bp and target coverage of 4–8×18. This resolution allows for the identification of small events such as single exon deletions. In practice, this resolution was achieved; however manual inspection of read level data of the WES and low pass WGS data was required due to false positive variant calls. Ultimately, algorithms such as Genome STRiP, which utilize population level data to account for systematic read depth biases, are necessary to improve the reliability of these results19.
Sequencing is performed in a research laboratory with results ultimately returned to the physician champion. Given the exploratory nature of these cases, results as well as the consent process include an explanation that any findings are unproven in nature and performed in an uncertified laboratory. Our case selection process, i.e. the focus on novel phenotypes and novel gene-disease relationships, eliminates from consideration individuals likely to benefit from certified laboratory tests, and we provide suggestions for the appropriate alternative commercial certified laboratories for these patients. Similarly, if a known pathogenic variant is identified, recommendations for CLIA confirmation via validated tests (rather than CLIA confirmation in a certified lab without the specific validated test) are provided.
Individualized Genomic Report and Return of Results
Once results for a specific patient have been generated, an individualized genomic report is prepared for dissemination to our Clinician-Scientist Review Panel and the patient’s physician champion. The panel has the opportunity to raise any issues related to the specific case prior to disclosing results. In all cases, one or more consultations between members of the panel and the physician champion are arranged to allow him/her to have any questions answered and have the results verbally conveyed. In cases where a plausible diagnosis is identified, the discussion with the physician champion centers on whether or not the findings should change clinical management of the patient. Any new treatment offered to the patient is ultimately the treating physician’s decision. Certified genetic counselors are available on demand if requested.
Follow-up Studies
While STSI houses the facilities required for functional studies, specialized assays are often required for appropriate functional characterization of candidate disease causative variants. Thus, to functionally characterize findings in IDIOM, appropriate follow-up studies are tailored to the disease and gene in question, and we generally do this in collaboration with other laboratories specialized in the study of a particular gene or gene family. STSI is also uniquely facile in the use of wireless sensors. Thus, when appropriate for select patients, we design and deploy N-of-1 studies for quantitative assessment of physiologic metrics. Furthermore, we have used such approaches to objectively determine whether there has been a therapeutic response to treatment once a molecular diagnosis is made and a genomically-indicated treatment initiated.
RESULTS
Referrals and Enrolled Patients
In the first three years of the IDIOM program, 121 patient referrals were received, 59 (48.8%) have undergone second tier review by our clinician-scientist review panel, and 17 (14.0%) patients and their family members have been enrolled. Referrals have come from over 16 different U.S. states, and we have received a number of international referrals.
Demographic statistics and comparisons for referred versus reviewed versus enrolled patients are shown in Table 1. Across all of the 121 patients referred to date, 31.9% were children (i.e., <18 years of age), 59.4% female, and 36.7% physician referred (versus self or family referred). As shown, however, those cases that underwent panel review and/or that were eventually enrolled were younger and more likely to be physician referred. Self/family referred subjects, if selected, were required to secure a physician champion.
Table 1. Demographic Characteristics of Referred, Reviewed and Enrolled Cases.
Demographic characteristics of referred, reviewed, and enrolled subjects.
| Variable | Referred (not Reviewed/ Enrolled) |
Panel Reviewed (not Enrolled) |
Enrolled | Referred vs. Reviewed |
Reviewed vs. Enrolled |
|---|---|---|---|---|---|
| N | 62 | 42 | 17 | NA | NA |
| Age Years (Mean, SD, Range) |
42.8 (22.7) <1 – 771 |
26.0 (18.5) <1 - 673 |
18.8 (16.0) <1 – 44.0 |
.001 | .165 |
| Gender (Female) | 63.8%2 | 47.6% | 76.5% | .124 | .043 |
| Children (<18 Years) | 15.6%1 | 35.7% | 52.9% | .054 | .222 |
| Referral from Physician | 30.6% | 31.7%4 | 70.6% | .909 | .006 |
Based on the subset of N = 32 referrals for whom we were able to obtain data;
Based on the subset of N = 47 referrals for whom we were able to obtain data;
Based on the subset of N = 39 referrals for whom we were able to obtain data;
Based on the subset of N = 41 referrals for whom we were able to obtain data
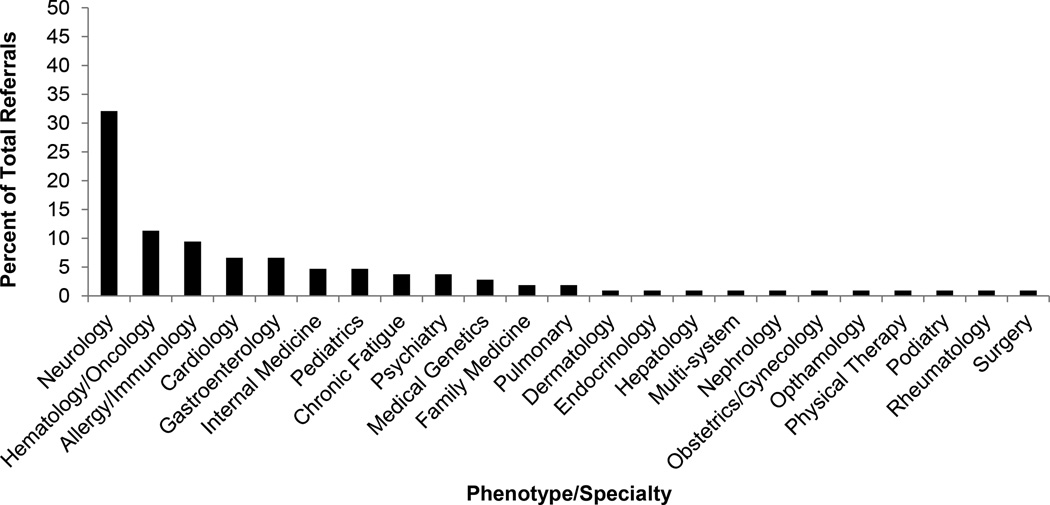
Figure 2 shows the medical specialties/phenotypes represented among the 121 patients referred to date for which clinical information was available. By far the most common category has been neurologic disorders, encompassing 32.1% of referrals, with hematology/oncology, allergy/immunology, cardiology, and gastroenterology making up the top five broad phenotypic categories.
Figure 2. Tracking of Therapy Response in IDIOM1.
Actigraphy based motion tracking demonstrates a dramatic and sustained decrease in night time myoclonic jerks due to gain of function mutation in ADCY5. Day 0 – 6 represents the last week of a three week run-in period to wash-out previous therapies. Diazepam is initiated on day 6 (arrow) with a dramatic reduction tremors sustained for over two months. Tremors are defined as movement magnitude >0 sustained for greater than 60 seconds.
The major reasons for exclusion of cases have been identification of cases that meet a previously described phenotype and exclusion of cases that do not appear to be genetic. Cases that meet a previously described phenotype, e.g. when the submitted case already has a likely clinical diagnosis, are referred to an appropriate laboratory, or appropriate test, upon exclusion. If all known genetic causes of the previously described phenotype are ruled out, the case would be reconsidered. Exclusion of cases not likely to be genetic is usually based on one of the following reasons; 1) the panel feels the condition is likely explained by an environmental insult or infectious disease, 2) unusual circumstances surrounding birth, for example pre-term birth or complications during birth – suggesting non-genetic developmental defect, 3) lack of objectively documented findings either on physical examination or testing, especially subjective symptoms that cannot be connected via dysfunction of a specific biological system.
Molecular Diagnosis
Detailed descriptions of each case and the findings are provided in the Supplemental Text. Of the cases that have been processed to date, we have arrived at a plausible molecular diagnosis in approximately 60% and a confirmed molecular diagnosis in approximately 18% of all cases (Table 2). A plausible molecular genetic diagnosis is defined as meeting the following criteria: 1) identified variants segregate in the family and reference populations in a manner consistent with the segregation of the disease in the family and incidence of the disorder in the general population, 2) variants influence the coding/splicing of a protein coding gene, and 3) the gene can be connected to the presenting phenotype through similar human disorders caused by mutation in the same gene or via genome-wide association studies, the gene can be connected to the presenting phenotype through close functional interaction with genes known to cause the presenting phenotype or similar phenotype, or the gene can be connected to the presenting phenotype via animal studies. Of the 60% of cases meeting this criteria, we have aggressively pursued functional validation in cases where the variant is of de-novo origin, further increasing confidence in the finding and maximizing yield of downstream effort with collaborators. In 2 cases so far we have completed functional and statistical confirmation of a novel gene-disease relationship via the identification of additional affected subjects with mutations in the same gene as well as functional confirmation of gene dysfunction 15,17, and in one instance a previously identified pathogenic variant was revealed. In all three cases, a new management strategy (pharmacological treatment) was initiated based on the findings, and we are currently closely monitoring this patient’s response to the therapy.
Table 2.
Genetic Diagnoses of Enrolled Cases
| Case | Phenotype | Outcome | Plausible Diagnosis |
Functional Confirmation |
|---|---|---|---|---|
| 1 | Complex Movement Disorder | ADCY5 de-novo gain of function mutation | yes | Yes |
| 2 | Lymphoproliferative Disorder | Plausible inherited candidate causative mutations in GIMAP8 | yes | not performed |
| 3 | Inflammatory Bowel Disease | Plausible inherited candidate causative mutations in MST1R | yes | not performed |
| 4 | Fibromyxoid sarcoma | No cause identified | no | N/A |
| 5 | Immunodeficiency | No cause identified | no | N/A |
| 6 | Lipomatosis and Vasculitis | Plausible inherited candidate causative mutations in NRIP1 | yes | not performed |
| 7 | Skin and Neurological Disorder | Potential, unconfirmed, diagnosis of subtype of Epidermolysis Bullosa due to inherited mutations in DST1 | yes | Ongoing |
| 8 | Developmental Delay | No cause identified | no | N/A |
| 9 | Epileptic Encephalopathy | KCNB1 de-novo missense mutation and confirmed KV2.1 dysfunction | yes | Yes |
| 10 | Developmental Delay | Plausible inherited candidate causative mutations in SYNPO | yes | ND |
| 11 | Familial Coronary Artery Disease | Plausible inherited candidate causative mutation in TDG | yes | Ongoing |
| 12 | Muscular Atrophy | No cause identified | no | N/A |
| 13 | Infantile Seizures | No cause identified | no | N/A |
| 14 | Recurrent Fever | Potential, unconfirmed, diagnosis of subtype of Chédiak–Higashi syndrome due to inherited mutation in LYST | yes | N/A |
| 15 | Congenital Hypertrichosis | Known pathogenic de-novo ABCC9 mutation identified | yes | yes |
| 16 | Suspected autosomal dominant Opitz G/BBB | ongoing | N/A | N/A |
| 17 | Plastic Bronchitis | ongoing | N/A | N/A |
Clinical Management Strategy Changes and Benefit
IDIOM1 presented with a complex movement disorder, described in detail in Chen et al. 201415, ultimately confirmed to be due to a gain of function de-novo mutation in ADCY5, and potentially modified by additional mutations in DOCK3 (unconfirmed). An N-of-1 style trial to monitor night time abnormal movements during treatment with various compounds indicated by gain of function in ADCY5 (ropinirole, carbamazepine, tetrabenazine, diazepam) with appropriate run-in and washout periods was initiated. The trial was halted upon request subsequent to initiation of the first agent, diazepam, after complete resolution of night-time myoclonic jerks. Diazepam has been previously shown to abrogate stress tolerance in ADCY5 null mice20. Figure 3 presents a dramatic and sustained resolution of abnormal movements after initiation of diazepam treatment, as captured by a movement-tracking device.
IDIOM9 presented with a complex seizure disorder with drop attacks, described in detail in Torkamani et al. 201417, ultimately confirmed to be due to a de-novo mutation in potassium channel KCNB1 which changed ion selectivity. As a result, in order to control potassium levels carefully, the subject was placed on a specialized diet and kept hydrated. Thus, a change in the clinical management strategy was initiated, with unconfirmed clinical benefit. Unfortunately, no particular anti-epileptic drug is indicated from the genetic results, and comparison to patients with a similar underlying genetic cause of epilepsy did not reveal a specific and efficacious therapeutic strategy21 (personal communication). An anecdotal reduction in drop attacks has been noted by the treating physician, though longer follow-up is required to confirm a sustained benefit.
IDIOM15 presented with hypertrichotic osteochondrodysplasia (excess hair growth on scalp, forehead, and face), a gene defect that was ultimately attributed to a known pathogenic de-novo ABCC9 mutation. Dominant missense mutations in ABCC9 cause gain-of-function channel opening and implicate a number of different channel blockers as potential therapeutic avenues22. Treatment modifications have been initiated with long-term follow-up required to confirm a sustained benefit.
DISCUSSION
The Scripps Idiopathic Diseases of huMan (IDIOM) study aims to discover novel gene-disease relationships and provide molecular genetic diagnosis and treatment options for individuals with idiopathic diseases using genome sequencing integrated with clinical assessment and multidisciplinary case review. The primary difference relative to other similar programs is our exclusive focus on novel gene-disease relationships. In fact, candidates with phenotypes similar to those previously described must have had known genes potentially mediating their phenotype ruled out before consideration as an IDIOM subject. While our protocol exclusively focuses on novel diseases or novel gene-disease relationships, it is remarkable that we demonstrate an initial rate of novel and confirmed genetic discoveries similar to that reported by programs focused on standard molecular genetic diagnosis via genome sequencing (20–25%)4,9,23,24. Although the volume, rate, and efficiency at which novel gene-disease relationships can be confirmed is lower than that achievable by standard molecular genetic diagnosis via genome sequencing, these results suggest that genome sequencing has the potential to be at least as efficacious in providing genetic diagnosis for individuals with previously undescribed disease as it is for individuals with known Mendelian disorders – with an upper limit to the diagnostic yield of ~60% (± 23% – 95% confidence interval) as informed by our plausible findings. However, we must acknowledge that a direct comparison between the diagnostic rate of these programs cannot be made since the rate depends heavily upon the ascertainment of the cohort in question.
The conversion of novel, plausible, gene-disease relationships to confirmed and validated genetic diagnoses should increase the yield of clinical genome sequencing programs above the current 20–25% diagnostic rate. In fact, individuals sequenced at clinical genome sequencing centers have initiated contact with the Scripps IDIOM investigators, subsequent to publication of our confirmed novel gene-disease relationships, bringing to light the fact that some individuals do not achieve a genetic diagnosis due to a lack of recognition of the causal variant in genome sequence data by those reporting the results, rather than any deficit in the technical identification of the causal variant. For example, we have received inquiries from two individuals with negative clinical exome results who had the same de-novo ADCY5 mutation as described in our IDIOM1 case – reported, correctly at the time of the test, as a variant of unknown significance in the clinical exome report. It remains to be seen what the relative contribution of deficits in technical variant identification vs. deficits in knowledge of gene-disease relationships is to the lack of molecular genetic diagnosis in those ~75% of individuals not receiving a genetic diagnosis after clinical genome sequencing. Ultimately, the contribution of novel gene-disease relationships to increasing the yield of clinical genome sequencing requires conversion of plausible findings to a confirmed genetic diagnosis through the identification of additional cases. This has been difficult to achieve but may be facilitated though services such as Matchmaker Exchange (matchmakerexchange.org). Comprehensive and accessible collection of phenotypic characteristics is key to achieving this goal.
In our study, all confirmed molecular genetic diagnoses achieved a tangible clinical benefit above and beyond the utility achieved simply by ending a diagnostic odyssey. However, there have been some notable challenges in the context of this project. For example, we often have families request other information, frequently referred to as secondary or “incidental findings” from their genomic analysis, and the research team has engaged in substantial discussion as to whether findings that do not pertain to the patient’s presenting condition should be assessed and reported back to the physician and/or patient. At this time, given that sequencing is performed in an uncertified laboratory for a pre-specified purpose, we do not to return information that is not directly applicable to the presenting condition. We have also observed wide variability in the extent to which our “physician champions” have knowledge of and comfort with genomic information, which has a major influence on the amount of information these clinicians then share with patients and the ultimate clinical utility of any findings. A multi-disciplinary clinician-scientist review panel has been essential to supporting physician champions as well as selection of appropriate cases most likely to benefit from genome sequencing. Finally, the transmission of information back to physician champions, clearly indicating the suggestive nature of the majority of our findings, has been essential to managing expectations of the physician champions and ultimately the enrolled subjects.
An important area of improvement, especially for this program with no specific disease focus, is to objectively link the symptoms presented by study subjects to previously described conditions. Any physician-scientist review panel is unlikely to represent specialties covering the broad range of conditions referred to the program, thus similarities to previously described conditions (as exemplified by our hypertrichotic osteochondrodysplasia case) can be missed. In a more comprehensive program, spanning molecular genetic diagnosis of undiagnosed disease and discovery of novel gene-disease relationships in idiopathic disease, automated systems for phenotype matching can provide both known genetic conditions and gene-disease relationships that should be considered for confirmatory molecular diagnosis, as well as grounding for exploration of novel gene-disease relationships through implicated biological processes and genetic networks mediating those processes. We believe this could provide a model for the unbiased application of genome sequencing across all rare genetic disorders, known and unknown.
Supplementary Material
Figure 1. Phenotypic Distribution of Case Referrals.
The distribution of phenotypes referred to the Scripps IDIOM study is plotted based on the subset of N = 106 referrals for whom we were able to obtain complete data.
Acknowledgments
We thank the patients and their families for their cooperation. This work is supported by Scripps Genomic Medicine, an NIH-NCATS Clinical and Translational Science Award (CTSA; 5 UL1 RR025774) to STSI, as well as funding from the Shaffer Family Foundation and the Anne and Henry Zarrow Foundation. Further support is from NIH-U01 HG006476 (AT).
Conflict of Interest
Ashley Scott-Van Zeeland, Eric Topol, Ali Torkamani, and Nicholas Schork declare they are co-founders and equity holders of Cypher Genomics, Inc.
REFERENCES
- 1.Jacob HJ, Abrams K, Bick DP, et al. Genomics in clinical practice: lessons from the front lines. Science translational medicine. 2013 Jul 17;5(194) doi: 10.1126/scitranslmed.3006468. 194cm195. [DOI] [PubMed] [Google Scholar]
- 2.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genetics in medicine : official journal of the American College of Medical Genetics. 2011 Mar;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, Muzny DM, Xia F, et al. Molecular Findings Among Patients Referred for Clinical Whole-Exome Sequencing. JAMA : the journal of the American Medical Association. 2014 Oct 18; doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England journal of medicine. 2013 Oct 17;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. The New England journal of medicine. 2012 Nov 15;367(20):1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava S, Cohen JS, Vernon H, et al. Clinical whole exome sequencing in child neurology practice. Annals of neurology. 2014 Oct;76(4):473–483. doi: 10.1002/ana.24251. [DOI] [PubMed] [Google Scholar]
- 7.Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nature reviews. Genetics. 2013 Oct;14(10):681–691. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 8.Cooper DN, Chen JM, Ball EV, et al. Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Human mutation. 2010 Jun;31(6):631–655. doi: 10.1002/humu.21260. [DOI] [PubMed] [Google Scholar]
- 9.Gahl WA, Markello TC, Toro C, et al. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genetics in medicine : official journal of the American College of Medical Genetics. 2012 Jan;14(1):51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbins G. The search for what's wrong with Lilly. San Diego Union Tribune. 2013 Mar 9; 2013. [Google Scholar]
- 11.Yong E. “We Gained Hope” The Story of Lilly Grossman’s Genome. National Geographic. 2013 [Google Scholar]
- 12.Tabor HK, Berkman BE, Hull SC, Bamshad MJ. Genomics really gets personal: how exome and whole genome sequencing challenge the ethical framework of human genetics research. Am J Med Genet A. 2011 Dec;155A(12):2916–2924. doi: 10.1002/ajmg.a.34357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer AN, Dimmock DP, Arca MJ, et al. A timely arrival for genomic medicine. Genetics in medicine : official journal of the American College of Medical Genetics. 2011 Mar;13(3):195–196. doi: 10.1097/GIM.0b013e3182095089. [DOI] [PubMed] [Google Scholar]
- 14.Belani R, Oliveira G, Erikson GA, et al. ASXL1 and DNMT3A mutation in a cytogenetically normal B3 thymoma. Oncogenesis. 2014;3:e111. doi: 10.1038/oncsis.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YZ, Friedman JR, Chen DH, et al. Gain-of-function ADCY5 mutations in familial dyskinesia with facial myokymia. Annals of neurology. 2014 Apr;75(4):542–549. doi: 10.1002/ana.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brownstein CA, Beggs AH, Homer N, et al. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY Challenge. Genome biology. 2014;15(3):R53. doi: 10.1186/gb-2014-15-3-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torkamani A, Bersell K, Jorge BS, et al. De Novo KCNB1 Mutations in Epileptic Encephalopathy. Annals of neurology. 2014 Aug 28; doi: 10.1002/ana.24263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome research. 2011 Jun;21(6):974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nature genetics. 2011 Mar;43(3):269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KS, Han PL. Mice lacking adenylyl cyclase-5 cope badly with repeated restraint stress. Journal of neuroscience research. 2009 Oct;87(13):2983–2993. doi: 10.1002/jnr.22119. [DOI] [PubMed] [Google Scholar]
- 21.Muona M, Berkovic SF, Dibbens LM, et al. A recurrent de novo mutation in KCNC1 causes progressive myoclonus epilepsy. Nature genetics. 2015 Jan;47(1):39–46. doi: 10.1038/ng.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harakalova M, van Harssel JJ, Terhal PA, et al. Dominant missense mutations in ABCC9 cause Cantu syndrome. Nature genetics. 2012 Jul;44(7):793–796. doi: 10.1038/ng.2324. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. Jama. 2014 Nov 12;312(18):1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaDuca H, Stuenkel AJ, Dolinsky JS, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genetics in medicine : official journal of the American College of Medical Genetics. 2014 Nov;16(11):830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.