Abstract
Nitric oxide (NO) synthesis in the retina is triggered by light stimulation. NO has been shown to modulate visual signal processing at multiple sites in the vertebrate retina, via activation of the most sensitive target of NO signaling, soluble guanylate cyclase. NO can also alter protein structure and function and exert biological effects directly by binding to free thiol groups of cysteine residues in a chemical reaction called S-nitrosylation. However, in the central nervous system, including the retina, this reaction has not been considered to be significant under physiological conditions. Here we provide immunohistochemical evidence for extensive S-nitrosylation that takes place in the goldfish and mouse retinas under physiologically relevant light intensities, in an intensity-dependent manner, with a strikingly similar pattern in both species. Pre-treatment with NEM, which occludes S-nitrosylation, or with TRIM, an inhibitor of neuronal NO synthase, eliminated the light-evoked increase in S-nitrosylated protein immunofluorescence (SNI) in the retinas of both species. Similarly, light did not increase SNI, above basal levels, in retinas of transgenic mice lacking neuronal NO synthase. Qualitative analysis of the light-adapted mouse retina with mass spectrometry revealed more than 300 proteins that were S-nitrosylated upon illumination, many of which are known to participate directly in retinal signal processing. Our data strongly suggest that in the retina, light-evoked NO production leads to extensive S-nitrosylation and that this process is a significant post-translational modification affecting a wide range of proteins under physiological conditions.
Keywords: mouse, goldfish, retina, nitric oxide, S-nitrosylation, RRID: IMSR_JAX:000664, RRID: IMSR_JAX:008519, RRID: AB_881716, RRID: AB_2168539, RRID: AB_92588, RRID: AB_218559, RRID: nif-0000-30467, RRID: rid_000081, RRID: OMICS_03354, RRID:nlx_53981
Introduction
As visual signals from photoreceptors are transmitted across the synaptic layers of the retina towards the output elements, the ganglion cells (GCs), they are subjected to modulation by a great variety of neuro active substances. One of these neuromodulators is nitric oxide (NO). Under physiological conditions, NO is synthesized from L-arginine in a light-dependent manner primarily by the neuronal-type nitric oxide synthase (nNOS), as made evident by the coincidence of light-evoked NO production with the retinal expression pattern of nNOS in many species (Blute et al., 1997; Blom et al., 2009; Giove et al., 2009). In vertebrates, the most sensitive target of NO is soluble guanylate cyclase (sGC) which is activated by nanomolar NO concentrations (Roy et al., 2008). It is also the best known path of NO action in the retina: extensive work has described NO’s modulatory effects at many stages of retinal processing via the NO→sGC→cGMP pathway (DeVries and Schwartz, 1989; Mills and Massey, 1995; McMahon and Ponomareva, 1996; Savchenko et al., 1997; Bing et al., 1997; Xin and Bloomfield, 1999; Hirooka et al., 2000; Kawai and Sterling, 2002; Yu and Eldred, 2005; Hoffpauir et al., 2006; Daniels and Baldridge, 2011). However, NO can exert biological effects via another signal transduction pathway: NO can react directly with free thiol groups of cysteine residues of proteins in a chemical reaction called S-nitrosylation (Stamler, 1994; Ahern et al., 2002). This reversible, covalent post-translational modification has been shown to alter protein function (reviewed by Ahern et al., 2002) much like phosphorylation. Thousands of proteins with potential S-nitrosylation sites have been identified, many associated with neural signaling (Seth and Stamler, 2011). For example, NR2 subunits of NMDA receptors, expressed in heterologous systems, have been shown to undergo S-nitrosylation that inhibits NMDA currents (Choi et al., 2000). However, based on NO-donor studies, S-nitrosylation is thought to require at least low micromolar NO concentrations (Ahern et al., 2002; Palmer et al., 2008), raising the question of whether it can occur under physiological conditions in the CNS (Hardingham et al., 2013). To date, detectable S-nitrosylation in neural tissue has been associated primarily with pathophysiological conditions that are characterized by aberrant, high levels of endogenous NO production (Nakamura et al., 2013): for example, elevated NO-triggered S-nitrosylation can take part in the pathogenesis of Parkinson’s and Alzheimer’s disease (Chung et al., 2004; Qu et al., 2011).
NO-selective electrodes detected ~0.2 μM NO near a single GC in the turtle retina upon NMDA application (Eldred and Blute, 2005), but little evidence supports the notion that in the healthy retina endogenous retinal NO concentrations under physiological conditions might be high enough to cause direct S-nitrosylation of proteins. To that end, in the salamander retina, Kurenny et al. (1994) described an NO-dependent process that modulated rod photoreceptor ion channels independent of sGC/cGMP. Likewise, exogenous application of NO donors at low, non-pathological concentrations, via intravitreal injection, amplified the rat electroretinogram (ERG) independent of cGMP (Vielma et al., 2010), leading to the conclusion that NO may have amplified ERG responses via protein nitrosylation. Recently, we reported evidence of a modulatory process in which endogenous retinal NO mediated activity-dependent plasticity of Mb-type bipolar cell (Mb) output in the goldfish retina through S-nitrosylation (Tooker et al., 2013).
Here we present immunohistochemical evidence describing S-nitrosylation in the adult retina under normal physiological conditions, triggered by endogenous NO. Importantly, we show that light induces S-nitrosylation with a similar pattern in goldfish and mouse retinas and the extent of immunolabeling depends on the intensity of illumination in both species. Our results are consistent with the notion that retinal NO production/release depends on the light intensity and we confirm that light-induced NO production that causes S-nitrosylation is mediated by the nNOS isoform in both goldfish and mouse. Therefore, we propose that light-evoked, NO-mediated S-nitrosylation in the vertebrate retina is a ubiquitous, dynamic modulatory process that exists across species and may function to modify the transfer of light information at multiple sites of actions within specific synaptic locations. Finally, using mass spectrometry, we provide the first descriptive account of the identities of retinal proteins that are subject to light-dependent S-nitrosylation via endogenous NO. Our findings provide a framework for future investigations of NO-mediated modulation of retinal function.
Materials and Methods
Animals
Adult male and female wild type C57BL/6J (WT; Jackson laboratories, Bar Harbor, ME, RRID: IMSR_JAX:000664) mice were used for a majority of the experiments requiring mammalian retinas. We also used retinas from adult male and female mice lacking the α isoform of neuronal nitric oxide synthase (nNOSα). These animals were generated on a C57BL/6 background by deletion of exon 6, the heme binding domain, in the nNOS gene (Gyurko et al. 2002; originally generated by P. Huang, Harvard Mass General Hospital and generously provided by S. Tobet, Colorado State University, RRID: IMSR_JAX:008519). Mice containing the genetic deletion were backcrossed to C57BL/6 mice for more than 10 generations. All mice were housed under a constant 12 h light/dark cycle with lights on at 6:00 am and were fed standard rodent chow and water ad libitum.
Adult 4- to 5-inch long Goldfish (Carassius auratus) of either sex were kept on a 12 h light/dark cycle with lights on at 10:00 am. All animals were handled in compliance with the Colorado State University Institutional Animal Care and Use Committee and all experiments and procedures met United States Public Health and Service Guidelines.
Eyecup preparation
For all experiments, animals were killed between 10:00 and 14:00 hours. Mice were anesthetized with isoflurane and decapitated before both eyes were enucleated. Goldfish were anesthetized with MS 222 before decapitation and enucleation. MS 222 stock solution (1000×) was made fresh in saturated bicarbonate buffer (pH: 7.5) which was mixed into the holding water to achieve 100 mg/l final concentration. Eyecups from both mouse and goldfish eyes were made by removing the cornea and lens. For all experiments, eyecups were maintained in bicarbonate buffered Ames’ medium (US Biological, Salem, MA) supplemented with 1.15 mM CaCl2. The pH was set to 7.45 with NaOH. The osmolarity was 290 ± 5 mOsmol for mouse eyecups and 260 ± 2 mOsmol for goldfish eyecups and in both cases the solution was gassed continuously with 95% O2/5% CO2.
Light exposure of eyecup preparations
To obtain fully light-adapted retinal tissue, the animal was subjected to ambient light for at least 1 h before the eyecups were made under a standard dissection microscope. For all other experiments, animals were dark-adapted for at least 3 h before the preparation took place under infrared illumination as previously described (Vigh et al., 2011). Quantified light exposure of the fully dark-adapted eyecups was accomplished using full-field illumination by green (λ=505 nm) LEDs (American Bright Optoelectronics, Chino, CA) with various intensities and durations. The light intensity was calibrated with an optical meter (model 1918-C, sensor 918D-SL-OD3; Newport, Irvine, CA). Dark-adapted eyecups were illuminated with 1010 photons/cm2/s for 10 s to mimic mesopic light stimulation and for 500 ms with 2.4×108 photons/cm2/s as scotopic light stimulation (Busskamp et al., 2010; Joselevitch and Kamermans; 2009). For experiments requiring incubation in specific pharmacological agents, the drug was added to the Ames’ media. One eyecup was incubated in the drug solution while the other eyecup was kept in un-altered control Ames’ medium for the same length of time as the drug-incubated eyecup in complete darkness, prior to light stimulation. Mouse and goldfish eyecups were incubated in Ames’ medium containing 1 mM N-Ethylmaleimide (NEM; Tocris Bioscience Bristol, UK) for 20 min or 50 μM 1-(2-Trifluromethylphenyl)imidazole (TRIM; Tocris Bioscience Bristol, UK) for 30 min prior to 10 s of mesopic light stimulation.
Immunohistochemistry
Light-adapted, dark-adapted and light stimulated eyecups were fixed at room temperature in freshly prepared 4% paraformaldehyde in 0.1 M phosphate-buffered saline (PBS; pH 7.45) for 20 min while protected from further light exposure. Cryostat-sectioned retinas were used for standard immunohistochemistry as previously described (Gallagher et al., 2010) with one minor adjustment: extreme care was taken to protect the retinal tissue from direct light exposure during every experimental step, beginning with fixation, as direct light can reverse and eliminate the S-nitrosylation moiety (Forrester et al., 2007). Retinal sections were permeabilized for 20 min in 0.5% Triton X-100 in 0.1 M PBS and then immediately blocked for 1 h in 5% bovine serum albumin (BSA) containing 0.5% Triton X-100 in 0.1 M PBS. The sections were incubated in blocking solution containing rabbit anti-S-nitrosocysteine (Abcam, Cat# ab50185, RRID: AB_881716) and mouse anti-PKCα primary antibodies (Enzo Life Sciences, Cat# KAM-PK020D, RRID: AB_2168539) (Table 1) overnight at room temperature. Sections were washed 3 × 15 min in 0.1 M PBS and then incubated for 2 h at room temperature in secondary antibodies donkey anti-rabbit Cy3 (Millipore, Cat# ap182C, RRID: AB_92588) and chicken anti-mouse fluorescein (Rockland, Cat# 610-9202, RRID: AB_218559), diluted 1:400 in blocking solution. Slides were washed 3 × 15 min with 0.1 M PBS and mounted in Vectashield (Vector labs, Burlingame, CA). For all immunohistochemical preparations, treated and control tissue were run in parallel, including a treated and a control retinal section on a single slide, to account and control for any inter- and intra-experimental differences.
Table 1.
Primary antibodies used.
| Antigen | Immunogen | Manufacturer, Cat. #, host species, mono- vs. polyclonal, RRID | Dilution used |
|---|---|---|---|
| S-nitrosocysteine | S-nitrosylated cysteine-KLH | Abcam, cat. #ab50185, rabbit polyclonal, RRID: AB_881716 | 1:100 |
| Protein Kinase Cα (PKCα) | Bovine Brain PKCα | Enzo Life Sciences, cat. #KAM-PK020D, mouse monoclonal, RRID: AB_2168539 | 1:200 |
Antibody Characterization
The primary antibodies used in this study are listed in Table 1.
S-nitrosocysteine
The polyclonal S-nitrosocysteine antiserum was raised in rabbit against S-nitrosylated cysteine conjugated to keyhole limpet hemocyanin (KLH) followed by purification via ion-exchange chromatography. The antibody specifically recognizes S-nitrosocysteine-BSA in immunoblotting and ELISA but does not recognize unconjugated BSA (Abcam; Cat#: ab50185, RRID: AB_881716; Table 1). The specificity of the antibody was demonstrated by Chakrabarti et al. (2010) who reported a ~70–80% loss in specific staining when S-nitrosylated human umbilical vein endothelial cells were incubated for 10 min with 100 μM DTT, a strong reducing agent capable of eliminating the nitrosothiol moiety from the cysteine. Further characterization was reported by Rossi-George and Gow (2013) who asserted that the detection of S-nitrosocysteine immunofluorescence from LPS treated BV2 cells remained present after antigen competition by pre-incubation of the primary antibody with S-nitrosoglutathione (GSNO), indicating that the antibody was specifically recognizing S-nitrosocysteine and not other nitroso compounds. Additionally, Rossi-George and Gow (2013) showed a reduction in S-nitrosocysteine immunofluorescence in BV2 cells that were treated with LPS and then subjected to strong reducing conditions using organic mercury as the reducing agent. In our hands, we saw a dramatic reduction in S-nitrosocysteine immunolabeling when we incubated the retinal tissue in 1 mM NEM for 20 min prior to light stimulation, when compared to control retinas which received the exact same light stimulation (Compare Figs. 3 and 4 with Fig. 5). This is consistent with the notion that NEM covalently binds to sulfhydryl groups with high affinity (Smyth et al., 1960) thereby preventing consecutive S-nitrosylation by NO (Hu et al., 2012).
Figure 3.
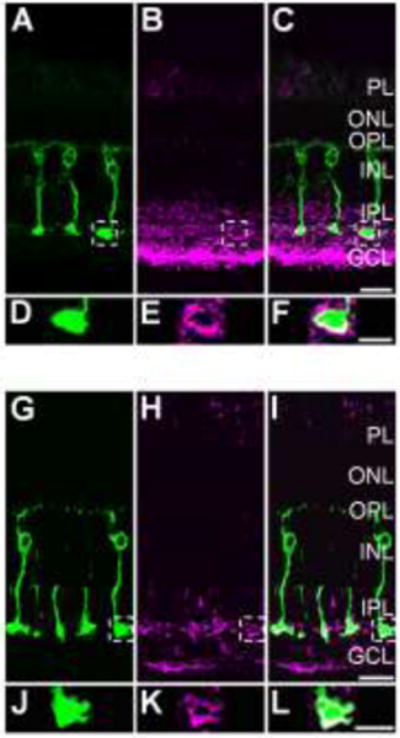
S-nitrosocysteine immunofluorescence labeling pattern in the goldfish retina is shaped by light intensity. For panels A–F, a dark-adapted goldfish retina was stimulated with rod saturating mesopic light (1×1010 photons/cm2/s, 505 nm, 10 sec). A: 40× single-plane confocal image showing PKCα positive Mbs (green) in a vertical cryostat section. B: S-nitrosocysteine immunolabel (magenta) in the same region as in A. At this light intensity, S-nitrosocysteine immunolabeling pattern is restricted to the inner retina, primarily the IPL and GCL. C: Merged images of A and B showing colocalization of S-nitrosocysteine + and PKCα + cellular structures within the IPL. D: Enlargement of the Mb terminal region located within the dashed box in A. E: Enlargement of the outlined selection in B that corresponds to the same region as the terminal outlined in A and enlarged in D. F: Enlargement of the merged image of the exact same regions as presented in D and E. Note the colocalization of the PKCα + Mb terminal with the S-nitrosocysteine immunolabel as a result of rod saturating mesopic light stimulation. For panels G–I, a dark-adapted goldfish retina was stimulated with bright scotopic light (2.4×108 photons/cm2/s, 505 nm, 500 msec). G: 40× single-plane confocal image showing PKCα positive Mbs (green) in a vertical cryostat section. H: The same region as presented in G, immunolabeled for S-nitrosocysteine (magenta). Although limited and nearly absent in most of the retinal layers, the SNI appears to be restricted to the innermost region of the IPL and some within the GCL. I: Merged image of G and H showing colocalization of PKCα + Mb terminal structures with S-nitrosocysteine immunolabel. J: Enlarged image of the area outlined by the dashed line box in G focused on the PKCα+ Mb terminal. K: Enlargement of the S-nitrosocysteine + structures within the same terminal region presented in J. L: Merged image of J and K showing precise colocalization of a PKCα + Mb terminal with an S-nitrosocysteine + cellular structure in the same optical plane. Scale bars: C, I=20 μm; F, L=10 μm.
Figure 4.
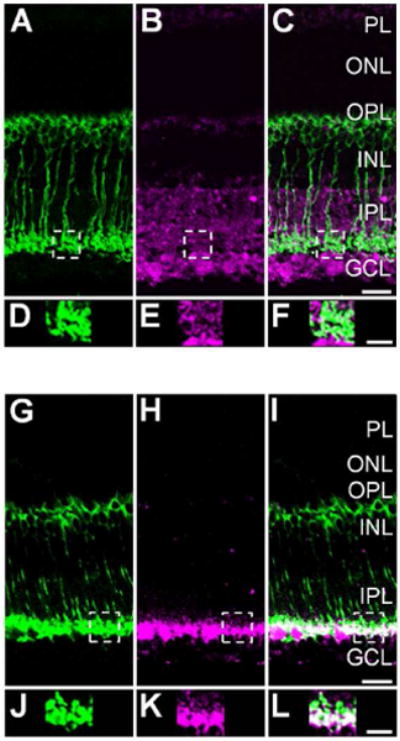
Light intensity determines the S-nitrosocysteine immunofluorescence labeling pattern in the wild-type mouse retina. For panels A–F, a dark-adapted wild-type mouse retina was stimulated with mesopic light (1×1010 photons/cm2/s, 505 nm, 10 sec). A: 40× single-plane confocal image showing PKCα positive RBCs (green) in a vertical cryostat retinal section. B: Confocal image of the S-nitrosocysteine immunolabel (magenta) in the same retinal region as illustrated in A. Stimulation with mesopic light restricted the S-nitrosocysteine immunolabeling pattern to the inner retina. C: Merged images of A and B showing colocalization of S-nitrosocysteine + and PKCα + cellular structures within the IPL. D: Enlargement of the RBC terminal region located within the dashed box in A. E: Enlargement of the outlined selection in B that corresponds to the SNI in the same region as the terminal outlined in A and enlarged in D. F: Enlargement of the merged image of the exact same regions as presented in D and E. Note the colocalization of the PKCα + RBC terminal structures with the S-nitrosocysteine immunolabel as a result of mesopic light stimulation. For panels G–I, a dark-adapted wild-type mouse retina was stimulated with bright scotopic light (2.4×108 photons/cm2/s, 505 nm, 500 msec). G: 40× single-plane confocal image showing PKCα positive RBCs (green) in a vertical cryostat section. H: The same region as presented in G, immunolabeled for S-nitrosocysteine. Scotopic light stimulation of the wild-type mouse retina appears to restrict the SNI to the innermost region of the IPL and some within the GCL. I: Merged image of G and H showing colocalization of PKCα + RBC terminal structures with S-nitrosocysteine immunolabel. J: Enlarged image of the area outlined by the dashed line box in G focused on the PKCα+ RBC terminals. K: Enlargement of the S-nitrosocysteine + structures within the same terminal region presented in J. L: Merged image of J and K showing colocalization of PKCα + RBC terminal cluster with a S-nitrosocysteine + immunolabel in the same optical plane. Scale bars: C, I=20 μm; F, L=10 μm.
Figure 5.
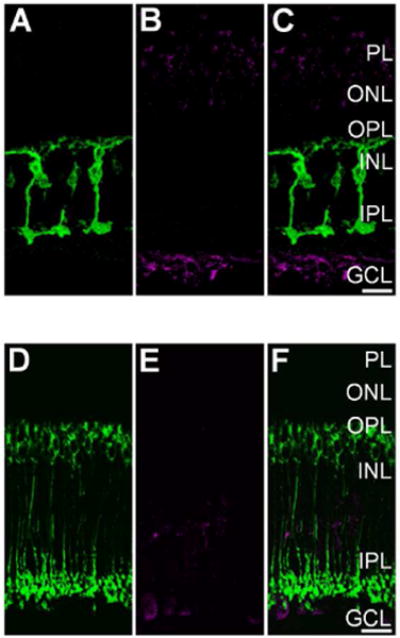
Light-evoked S-nitrosylation in the goldfish and wild-type mouse retina is prevented by pre-incubation with N-Ethylmaleimide (NEM). A: Single plane 40× confocal image showing PKCα + (green) Mbs in a vertical cryosection from a goldfish retina incubated in 1 mM NEM for 20 min prior to mesopic light stimulation (1×1010 photons/cm2/s, 505 nm, 10 sec). B: Same region as in A showing S-nitrosocysteine immunolabeling (magenta) in response to mesopic light stimulation. Note absence of immunolabel signal when compared to the untreated goldfish retina seen in Fig. 3A, B, and C. C: Merged image of confocal images from A and B showing very little colocalization between PKCα + Mbs and S-nitrosocysteine immunofluoresence. D: Single plane 40× confocal image showing PKCα + (green) RBCs in a vertical cryosection from a wild-type mouse retina incubated in 1 mM NEM for 20 min prior to mesopic light stimulation (1×1010 photons/cm2/s, 505 nm, 10 sec). E: Confocal image of the same region as presented in A illustrating that the mesopic light-evoked SNI (magenta) is dramatically reduced as compared to the untreated wild-type mouse retina (Fig. 4A, B, and C). F: Merged 40× confocal imaged of a wild-type mouse retina co-immunolabeled for PKCα and S-nitrosocysteine. Scale bars=20 μm.
Protein Kinase Cα
Bovine brain protein kinase Cα (PKCα; UniProt ID: P04409) was used as the immunogen for production of the monoclonal antiserum in mouse. The affinity purified antibody specifically recognizes a band of ~80 kDA by Western blot in mouse and fish (Enzo Life Sciences, Cat# KAM-PK020D, RRID: AB_2168539; Table 1). This primary antibody labels the appropriate pattern in the mouse that corresponds to mouse rod bipolar cells (RBCs; Greferath et al., 1990; Haverkamp and Wässle, 2000) as well as the appropriate pattern in the goldfish retina which corresponds to Mbs (Negishi et al., 1988; Suzuki and Kaneko, 1990; Yazulla and Studholme, 1992).
Confocal laser microscopy and analysis
Fluorescent images were taken using a Zeiss LSM 510 confocal microscope (Carl Zeiss, Oberkochen, Germany). Digital images were acquired separately from each laser channel and then merged to avoid crosstalk between channels. Images were acquired at 40×; for Z-stacks, 2 μm increments were used. Laser power and acquisition settings were kept identical across all retinal sections imaged. Images were compiled using Zeiss LSM Images Examiner software (Carl Zeiss, Oberkochen, Germany). For representative images, brightness and contrast were adjusted in Photoshop CS4 (version 11.0; Adobe, San Jose, CA) and all adjustments were made uniformly to the entire digital image. Quantification of immunofluorescence was performed on raw, unadjusted images of retinal sections using the Plot Profile tool in Image J (version 1.49c; NIH, Bethesda, MD, RRID: nif-0000-30467). For this analysis, we made selections of uniform size (1290 × 575 pixels) from the raw images (i.e. one selection each from the channels corresponding to S-nitrosocysteine and PKCα immunolabeling), ensuring all retinal layers were present in the vertical orientation of the selection. The intensity profile of the fluorescent signal associated with S-nitrosocysteine or PKCα immunolabel was obtained by applying the Plot Profile tool which horizontally integrated the intensity across the entire digital image selection (gray line in Fig. 1B and E and in Fig. 2B and E). The average intensity of fluorescence was calculated for each individual retinal layer from each unique plot profile after background fluorescence was subtracted, similar to the method reported by Vielma et al. (2010). In this manner, we were able to systematically quantify the intensity and pattern of S-nitrosocysteine immunofluorescence across the retinal layers (Figs. 1G and 2G). For retinal sections from eyecups stimulated with mesopic or scotopic light, we compared the extent to which the S-nitrosylation immunolabeling was distributed within the IPL, specifically, what percentage was located within the Mb/RBC terminal region. During this analysis, we divided the integral of the intensity profile curve for the region defined only by the terminals of Mbs/RBCs by the integral of the curve associated with the entire IPL. To determine the boundaries of the terminal region in the profile of the S-nitrosocysteine plot, we used the intensity profile from the PKCα fluorescence and correlated the exact pixels corresponding to the beginning and end of the terminal region. Integrals from individual images were averaged to obtain a final overall average for the integral of the intensity within the entire IPL and the intensity specific to the terminal region. Colocalization of the S-nitrosocysteine immunolabeling and the terminal region of the Mbs/RBCs was determined for individual goldfish Mb terminals or terminal clusters for mouse RBCs using images taken from a single optical plane to avoid exaggeration of the coincidence of colocalization. The JACoP plugin in the Image J software (version 1.49c; NIH, Bethesda, MD, RRID: nif-0000-30467) was used to calculate the Manders’ coefficient for 100 × 100 pixel square selections of Mb terminals or RBC terminal clusters were cut out from single-plane confocal images. Note that the Manders’ coefficient, M1, returns a value between 0 and 1, corresponding to the ratio of “summed intensities of pixels from the green image for which the intensity in the red channel is above zero to the total intensity in the green channel” and vice versa for M2 (Bolte and Cordelières, 2006). As the Manders’ coefficient is sensitive to noise and might provide a false positive correlation (Bolte and Cordelières, 2006), we used the Costes’ approach to ensure accurate measures of coincidence: the Costes’ approach, sets an automatic value for intensity threshold, thereby eliminating noise and setting the background value to zero. Statistical analysis was completed using Prism (version 6.01; GraphPad, San Diego, CA, RRID: rid_000081). Unpaired Student’s t tests were used for comparison between individual groups and one-way ANOVA with Tukey’s multiple comparison tests were performed across multiple groups. Comparisons across experiments were justified by performing the immunohistochemistry and imaging of the retinal tissue from the experimental group (i.e. drug treated) in parallel with at least one tissue slide from each group involved in the comparison (i.e. dark-adapted or control incubation without drug). All data are presented as mean +/− SEM, with p<0.05 considered significant.
Figure 1.
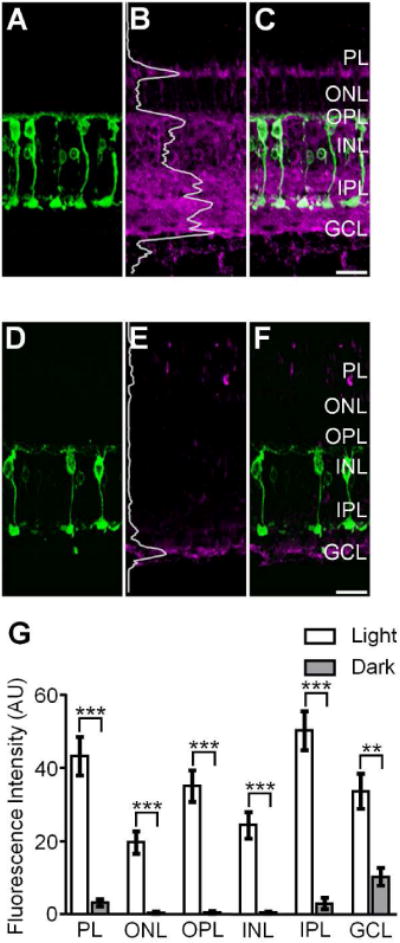
S-nitrosocysteine immunofluorescence in confocal images of vertical cryostat sections from light- and dark-adapted goldfish retinas. A, D: PKCα positive Mbs (green) in the light- and dark-adapted goldfish retina. B: Confocal image of the same region presented in A showing that in the light-adapted goldfish retina, SNI (magenta) was present in all retinal layers. Fluorescence intensity was integrated horizontally across the image (the gray superimposed trace) and indicated the relative amount and location of S-nitrosylated proteins in the light-adapted retina. C: Merged image of A and B showing colocalization of SNI and PKCα + cells as a component of the robust S-nitrosocysteine labeling pattern across all retinal layers. E: Confocal image of the same region represented in D showing SNI in the dark-adapted goldfish retina. The superimposed gray trace illustrates the fluorescence intensity plot of the SNI and indicates a significant lack of S-nitrosocysteine + label in the dark-adapted goldfish retina. Of note: confocal images in B and E were obtained using the exact same laser power settings and the intensity plot profiles in B and E are plotted on the same scale. F: Merged image of D and E illustrating the weak S-nitrosocysteine labeling pattern in select retinal layers. G: Summary graph illustrating S-nitrosylation, as determined by the fluorescence intensity of SNI, is significantly greater in every retinal layer of the light-adapted goldfish retina as compared to the dark-adapted goldfish retina. Unpaired Student’s t test, ***: p≤0.0005, **: p≤0.005, light-adapted: n=4 retinas, dark-adapted: n=2 retinas. Data is presented as mean ± SEM. PL: Photoreceptor Layer; ONL: Outer Nuclear Layer; OPL: Outer Plexiform Layer; INL: Inner Nuclear Layer; IPL: Inner Plexiform Layer; GCL: Ganglion Cell Layer. SNI: S-nitrosocysteine immunofluorescence. Scale bars=20 μm.
Figure 2.
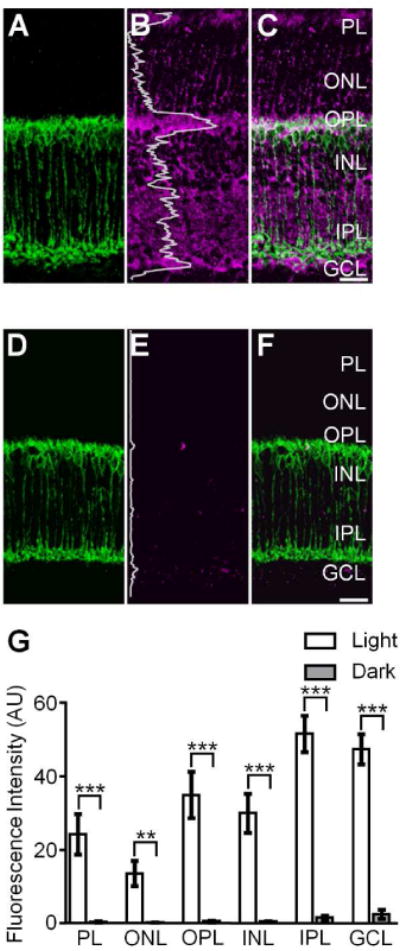
S-nitrosocysteine immunofluorescence in confocal images of vertical cryostat sections from light- and dark-adapted wild-type mouse retinas. A, D: PKCα positive rod bipolar cells (RBCs, green) in the light- and dark-adapted wild-type mouse retina. B: Confocal image of the same region presented in A showing SNI (magenta) was present in all retinal layers of the light-adapted wild-type mouse retina. The relative amount and location of S-nitrosylated proteins in the light-adapted retina was illustrated by the fluorescence intensity (the gray superimposed trace) which was integrated horizontally across the image. C: Merged image of A and B showing the robust S-nitrosocysteine labeling pattern across all retinal layers. Note the colocalization of SNI and PKCα + RBCs. E: Confocal image of the same region represented in D showing SNI in the dark-adapted wild-type mouse retina. The superimposed gray trace illustrates the fluorescence intensity plot of the SNI and indicates a substantial reduction of S-nitrosocysteine + label in the dark-adapted wild-type mouse retina as compared to the light-adapted retina. Of note: confocal images in B and E were obtained using the exact same laser power settings and the intensity plot profiles in B and E are plotted on the same scale. F: Merged image of D and E illustrating the faint and nearly absent S-nitrosocysteine labeling pattern in in the dark-adapted wild-type mouse retina. G: Summary graph illustrating S-nitrosylation, as determined by the fluorescence intensity of SNI, is significantly greater in every retinal layer of the light-adapted wild-type mouse retina as compared to the dark-adapted wild-type mouse retina. Unpaired Student’s t test, ***: p≤0.0005, **: p≤0.005, light-adapted: n=8 retinas, dark-adapted: n=6 retinas. Data is presented as mean ± SEM. Scale bars=20 μm.
Identification of S-nitrosylated Proteins
Biotin-switch assay
Retinal lysates were made by homogenizing the 2 retinas, obtained from individual light-or dark-adapted mice, in ice-cold lysis buffer (S-nitrosylation Buffer A, Cayman Chemical, Ann Arbor, MI, Item #10006520), supplemented with 1 mM Phenylmethanesulfonyl fluoride (PMSF, Sigma, St. Louis, MO). Retinal lysates were obtained in triplicate (i.e. from 3 animals) in either a dark- or light-adapted state, providing 6 total lysates to be used. The homogenized lysates were centrifuged at 1,000×g for 10 min at 4°C to remove debris. The supernatant was collected and used as the starting material for the biotin-switch assay using the S-nitrosylated Protein Detection kit (Cayman Chemical, Ann Arbor, MI, Item #10006518). The S-nitrosylated Protein Detection Kit is a modified version of the biotin-switch assay described by Jaffrey et al. (2001) in which only proteins containing the S-nitrosylation moiety are covalently labeled with a biotin in place of the nitrosothiol group. The steps in the S-nitrosylated protein detection kit were followed exactly and extreme care was taken to prevent exposure of the sample to light (fluorescent and/or sunlight) as this can cause inappropriate biotinylation and ultimately false positive protein identification (Forrester et al., 2007). Following completion of the assay per the kit, the protein samples were suspended in 200 μL of wash buffer (S-nitrosylation wash buffer, Cayman Chemical, Ann Arbor, MI, Item # 10006519) with 1 mM PMSF and stored at −20°C overnight.
NeutrAvidin resin-assisted capture of biotinylated proteins
Prior to incubation with protein sample, 100 μL of settled NeutrAvidin agarose resin (Pierce Biotechnology, Rockford, IL) was blocked by incubation with 2.5% biotin-free BSA (Sigma, St. Louis, MO) in 0.1 M PBS for 1 h at room temperature. The blocking solution was removed and the protein samples (200 μL) were incubated with the NeutrAvidin agarose resin for 2 h at room temperature. The resin was washed 5 × 20 min in 0.1 M PBS. After washing all of the unbound protein, the biotinylated proteins were removed from the resin by 4 × 20 min incubations in 150 μL of the elution buffer: 8 M guanidine-HCL (ThermoFisher Scientific), pH 1.5. All of the elution buffer fractions were collected and combined to ensure maximum protein recovery from the resin. The entire 500 μL volume of elution buffer containing the biotinylated proteins was immediately dialyzed against 0.1 M PBS for 24 h at 4°C. The eluates were precipitated from the PBS via trichloroacetic acid (TCA) protein precipitation using deoxycholate. The precipitated protein pellets were resuspended in 50 μL 0.1 M PBS and stored at −20°C. Protein concentration was determined by BCA analysis (Thermo Scientific, Rockford, IL).
Sample Preparation for LC-MS/MS Analysis
Protein samples from light-adapted (n=3) and dark-adapted (n=3) were submitted to the Proteomics and Metabolomics Facility at Colorado State University. Samples were processed for in-solution trypsin digestion as previously described (Schauer et al., 2013). Briefly, protein was precipitated out of solution in the presence of 4 volumes of 100% −20° C acetone and then resolubilized in 8 M urea, 0.2% ProteaseMAX surfactant trypsin enhancer (Promega, Madison, WI). Samples were reduced and alkylated with 5 mM dithiothreitol and 5 mM iodoacetamide. Trypsin (MS Grade, Thermo Pierce, San Jose, CA) was added at an enzyme to substrate ratio of 1:50 and incubated at 37° C for 3 h. Trypsin was deactivated with the addition of 5% trifluoroacetic acid and desalted using C18 OMIX tips (Agilent Technologies, Santa Clara, CA) using manufacturer’s instructions. Peptide eluate was dried in a vacuum evaporator and resuspended in 3% acetonitrile/0.1% formic acid at a concentration of approximately 1 μg/μL.
LC-MS/MS Analysis
Approximately 2 μg of tryptic digest for each sample was injected using an EASY nanoLC-II system (Thermo Scientific, San Jose, CA). Peptides were purified and concentrated using an online enrichment column (EASY-Column, 100 μm ID × 2 cm ReproSil-Pur C18). Subsequent chromatographic separation was performed on a reverse phase nanospray column (EASY-Column, 3μm, 75 μm ID × 100 mm ReproSil-Pur C18) using a 90 min linear gradient from 10%–35% buffer B (100% ACN, 0.1% formic acid) at a flow rate of 400 nL/min. Peptides were eluted directly into the mass spectrometer (Thermo Scientific Orbitrap Velos). The instrument was operated in Orbitrap-LTQ mode where precursor measurements were acquired in the Orbitrap (60,000 resolution) and MS/MS spectra (top 20) were acquired in the LTQ ion trap with a normalized collision energy of 35%. Mass spectra were collected over a m/z range of 400–2000 Da using a dynamic exclusion limit of 2 MS/MS spectra of a given peptide mass for 30 s (exclusion duration of 90 s). Compound lists of the resulting spectra were generated using Xcalibur 2.2 software (Thermo Scientific) with a S/N threshold of 1.5 and 1 scan/group.
Database Search and Protein Identification Criteria
Tandem mass spectra were extracted, charge state deconvoluted and deisotoped by ProteoWizard (version 3.0; MSConvert, RRID:OMICS_03354). All MS/MS samples were analyzed using Mascot (version 2.3.02; Matrix Science, London, UK). Mascot was set up to search the UniProt-KB Mus Musculus reverse-concatenated database (102,390 entries, downloaded January, 2013, RRID:nlx_53981) assuming the digestion enzyme was trypsin with a missed cleavage tolerance set to 2. Mascot was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 20 PPM. Oxidation of methionine and carbamidomethyl of cysteine were specified in Mascot as variable modifications. Search results were compiled and validated using Scaffold (version 4.4.0; Proteome Software, Portland, OR). Peptide identifications were accepted if they could be established at greater than 90.0% probability by the Scaffold Local False Discovery Rate (FDR) algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability to achieve an FDR less than 1.0% and contained at least 2 identified peptides. The number of accepted peptide identifications for each identified protein was reported as the peptide score in Table 2. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Table 2.
List S-nitrosylated proteins identified only under light-adapted conditions.
| Protein Name | Gene | Accession Number | Peptide Score |
|---|---|---|---|
| Apoptosis/Cell Death | |||
| Bcl-2-like protein 13 | Bcl2l13 | B2L13_MOUSE | 2 |
| Isoform 2 of Band 4.1-like protein 3 | Epb41l3 | E41L3_MOUSE | 10 |
| Isoform 2 of Peptidyl-prolyl cis-trans isomerase FKBP8 | Fkbp8 | FKBP8_MOUSE | 2 |
| Isoform 8 of Band 4.1-like protein 3 | Epb41l3 | E41L3_MOUSE | 2 |
| MAP kinase-activating death domain protein | Madd | A2AGQ6_MOUSE | 3 |
| Mitochondrial fission 1 protein | Fis1 | FIS1_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 | Ndufa13 | NDUAD_MOUSE | 3 |
|
Calcium Ion Regulation Calbindin |
Calb1 | CALB1_MOUSE | 4 |
| Isoform 3 of Disks large homolog 4 | Dlg4 | DLG4_MOUSE | 6 |
| Secretagogin Cardiovascular |
Scgn | SEGN_MOUSE | 7 |
| Alpha actinin 1a | Actn1 | A1BN54 MOUSE | 2 |
| Carbonic anhydrase 14 Cell Adhesion |
Ca14 | CAH14_MOUSE | 2 |
| Cadherin-2 | Cdh2 | CADH2_MOUSE | 2 |
| Cell adhesion molecule 1 | Cadm1 | CADM1_MOUSE | 8 |
| Cell adhesion molecule 2 (Fragment) | Cadm2 | CADM2_MOUSE | 10 |
| Cell adhesion molecule 3 | Cadm3 | CADM3_MOUSE | 2 |
| Contactin-1 | Cntn1 | CNTN1_MOUSE | 7 |
| Isoform 2 of Neuronal cell adhesion molecule | Nrcam | NRCAM_MOUSE | 4 |
| Limbic system-associated membrane protein | Lsamp | LSAMP MOUSE | 4 |
| Neural cell adhesion molecule 1 | Ncam1 | E9QB01_MOUSE | 2 |
| Neuroplastin | Nptn | NPTN_MOUSE | 3 |
| Peripherin-2 | Prph2 | PRPH2_MOUSE | 9 |
| Rod outer segment membrane protein 1 | Rom1 | ROM1_MOUSE | 6 |
| Signal-regulatory protein alpha | Sirpa | Q6P6I8_MOUSE | 3 |
| Transforming protein RhoA | Rhoa | RHOA MOUSE | 3 |
|
Cell Cycle/Cell Division Casein kinase II subunit beta |
Csnk2b | CSK2B_MOUSE | 2 |
| Cell cycle exit and neuronal differentiation protein 1 | Cend1 | CEND_MOUSE | 3 |
| Cell division control protein 42 homolog | Cdc42 | CDC42_MOUSE | 2 |
| Cyclin-dependent kinase 5 OS=Mus musculus | Cdk5 | CDK5_MOUSE | 2 |
| Histone H2AX | H2afx | H2AX_MOUSE | 2 |
| Isoform 2 of Septin-11 | Sep11 | SEP11_MOUSE | 2 |
| Isoform Gamma-2 of Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | Ppp1cc | PP1G_MOUSE | 3 |
| Nuclear migration protein nudC | Nudc | NUDC_MOUSE | 2 |
| Platelet-activating factor acetylhydrolase IB subunit alpha | Pafah1b1 | LIS1_MOUSE | 4 |
| Putative adenosylhomocysteinase 2 | Ahcyl1 | SAHH2_MOUSE | 2 |
| Serine/threonine-protein phosphatase | Ppp3cb | E0CZ78_MOUSE | 2 |
| Serine-protein kinase ATM | Atm | ATM_MOUSE | 2 |
|
Cell Growth/Proliferation Catenin beta-1 |
Ctnnb1 | CTNB1_MOUSE | 4 |
| Isoform C1 of Heterogeneous nuclear ribonucleoproteins C1/C2 | Hnrnpc | HNRPC MOUSE | 2 |
| Membrane-associated progesterone receptor component 1 | Pgrmc1 | PGRC1_MOUSE | 2 |
| Membrane-associated progesterone receptor component 2 | Pgrmc2 | PGRC2_MOUSE | 2 |
| Myotrophin | Mtpn | MTPN_MOUSE | 2 |
| Neuromodulin | Gap43 | NEUM_MOUSE | 2 |
| Neuronal growth regulator 1 | Negr1 | NEGR1_MOUSE | 3 |
| Neuronal membrane glycoprotein M6-a | Gpm6a | GPM6A_MOUSE | 5 |
| Prominin 1, isoform CRA_g | Prom1 | PROM1_MOUSE | 2 |
| Chaperone | |||
| 10 kDa heat shock protein, mitochondrial | Hspe1 | CH10_MOUSE | 3 |
| 26S proteasome non-ATPase regulatory subunit 5 | Psmd5 | PSMD5_MOUSE | 2 |
| Coactosin-like protein | Cotl1 | COTL1_MOUSE | 2 |
| DnaJ homolog subfamily A member 2 | Dnaja2 | DNJA2_MOUSE | 2 |
| DnaJ homolog subfamily C member 5 | Dnajc5 | DNJC5_MOUSE | 2 |
| Isoform 3 of Putative tyrosine-protein phosphatase auxilin | Dnajc6 | AUXI_MOUSE | 4 |
| Large proline-rich protein BAG6 | Bag6 | BAG6_MOUSE | 3 |
| Peptidyl-prolyl cis-trans isomerase B | Ppib | PPIB_MOUSE | 3 |
| Phosducin-like protein | Pdcl | PHLP_MOUSE | 2 |
|
Cytoskeleton Actin-related protein 2/3 complex subunit 3 |
Arpc3 | ARPC3_MOUSE | 3 |
| Alpha-adducin | Add1 | ADDA_MOUSE | 8 |
| Alpha-centractin | Actr1a | ACTZ_MOUSE | 2 |
| Destrin | Dstn | DEST_MOUSE | 4 |
| Gephyrin | Gphn | GEPH_MOUSE | 2 |
| Glycoprotein m6b, isoform CRA_g | Gpm6b | GPM6B_MOUSE | 3 |
| IQ motif and SEC7 domain-containing protein 3 | Iqsec3 | IQEC3_MOUSE | 2 |
| Isoform 2 of Rootletin | Crocc | CROCC_MOUSE | 2 |
| Isoform 4 of Nesprin-1 | Syne1 | SYNE1_MOUSE | 2 |
| Myristoylated alanine-rich C-kinase substrate | Marcks | MARCS_MOUSE | 3 |
| Protein 4.1 | Epb4.1 | A2A841_MOUSE | 12 |
| Protein kinase C and casein kinase substrate in neurons protein 2 | Pacsin2 | PACN2 MOUSE | 2 |
|
DNA Metabolism/Regulation Prohibitin |
Phb | PHB_MOUSE | 8 |
| Prohibitin-2 | Phb2 | PHB2_MOUSE | 7 |
|
Endocytosis Adaptin ear-binding coat-associated protein 1 |
Necap1 | NECP1_MOUSE | 2 |
| Dynamin-3 | Dnm3 | DYN3_MOUSE | 5 |
| Dynamin-like 120 kDa protein, mitochondrial | Opa1 | OPA1_MOUSE | 2 |
| Isoform 3 of SH3-containing GRB2-like protein 3-interacting protein 1 | Sgip1 | SGIP1_MOUSE | 3 |
| Secretory carrier-associated membrane protein 1 | Scamp1 | SCAM1_MOUSE | 2 |
| Synaptophysin | Syp | SYPH_MOUSE | 2 |
|
Energy Metabolism 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 2 variant 4 |
Pfkfb2 | B2Z892_MOUSE | 2 |
| 6-phosphofructokinase | Pfkp | Q8C605_MOUSE | 8 |
| 6-phosphofructokinase, muscle type | Pfkm | K6PF_MOUSE | 2 |
| Adenine phosphoribosyltransferase | Aprt | APT_MOUSE | 2 |
| ADP/ATP translocase 2 | Slc25a5 | ADT2_MOUSE | 3 |
| ATP synthase F(0) complex subunit B1, mitochondrial | Atp5f1 | AT5F1_MOUSE | 6 |
| ATP synthase subunit epsilon, mitochondrial | Atp5e | ATP5E_MOUSE | 2 |
| ATP synthase subunit f, mitochondrial | Atp5j2 | ATPK_MOUSE | 2 |
| ATP synthase subunit g, mitochondrial | Atp5l | ATP5L_MOUSE | 2 |
| CDGSH iron-sulfur domain-containing protein 1 | Cisd1 | CISD1_MOUSE | 3 |
| Citrate synthase, mitochondrial | Cs | CISY_MOUSE | 8 |
| Creatine kinase M-type | Ckm | KCRM_MOUSE | 2 |
| Cytochrome b-c1 complex subunit 7 | Uqcrb | Q9CQB4_MOUSE | 4 |
| Cytochrome b-c1 complex subunit 9 | Uqcr10 | QCR9_MOUSE | 2 |
| Cytochrome b-c1 complex subunit Rieske, mitochondrial | Uqcrfs1 | UCRI_MOUSE | 7 |
| Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | Cox4i1 | COX41_MOUSE | 4 |
| Cytochrome c oxidase subunit 5A, mitochondrial | Cox5a | COX5A_MOUSE | 7 |
| Cytochrome c oxidase subunit 7A2, mitochondrial | Cox7a2 | CX7A2_MOUSE | 2 |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | Dlat | ODP2_MOUSE | 6 |
| Electron transfer flavoprotein subunit alpha, mitochondrial | Etfa | ETFA_MOUSE | 4 |
| Electron transfer flavoprotein subunit beta | Etfb | ETFB_MOUSE | 3 |
| GTP:AMP phosphotransferase AK3, mitochondrial | Ak3 | KAD3_MOUSE | 2 |
| Hexokinase-2 | Hk2 | HXK2_MOUSE | 11 |
| Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | Idh3a | IDH3A_MOUSE | 8 |
| Isocitrate dehydrogenase [NAD] subunit gamma 1, mitochondrial | Idh3g | IDHG1_MOUSE | 3 |
| Isocitrate dehydrogenase [NADP], mitochondrial | Idh2 | IDHP_MOUSE | 4 |
| Isocitrate dehydrogenase 3 (NAD+) beta | Idh3b | Q91VA7_MOUSE | 6 |
| Isoform 2 of Cytochrome c1, heme protein, mitochondrial | Cyc1 | CY1_MOUSE | 6 |
| Isoform Short of Adenosine kinase | Adk | ADK_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | Ndufa10 | NDUAA_MOUSE | 5 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 | Ndufa12 | NDUAC_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4 | Ndufa4 | NDUA4_MOUSE | 4 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 | Ndufa6 | NDUA6_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | Ndufa8 | NDUA8_MOUSE | 4 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | Ndufa9 | NDUA9_MOUSE | 3 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial | Ndufb11 | NDUBB_MOUSE | 3 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 | Ndufb3 | NDUB3_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 | Ndufb4 | NDUB4_MOUSE | 3 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial | Ndufb8 | NDUB8_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 | Ndufb9 | NDUB9_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial | Ndufv1 | NDUV1_MOUSE | 5 |
| NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | Ndufv2 | NDUV2_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial | Ndufs2 | NDUS2_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | Ndufs3 | NDUS3_MOUSE | 6 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial | Ndufs7 | NDUS7_MOUSE | 2 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial | Ndufs8 | NDUS8_MOUSE | 5 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Ndufs1 | NDUS1_MOUSE | 14 |
| Phosphoglucomutase-2 | Pgm2 | PGM2_MOUSE | 2 |
| Protein Ogdhl | Ogdhl | E9Q7L0_MOUSE | 4 |
| Pyruvate carboxylase | Pcx | PYC_MOUSE | 2 |
| Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial | Pdha1 | ODPA_MOUSE | 5 |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | Pdhb | ODPB_MOUSE | 7 |
| Stomatin-like protein 2, mitochondrial | Stoml2 | STML2_MOUSE | 5 |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | Sdha | DHSA_MOUSE | 8 |
| Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | Sdhb | DHSB_MOUSE | 4 |
| Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | Suclg1 | SUCA_MOUSE | 2 |
| Succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial Glycosylation |
Sucla2 | SUCB1_MOUSE | 4 |
| Alpha-1,3/1,6-mannosyltransferase ALG2 | Alg2 | ALG2_MOUSE | 3 |
| UDP-glucose:glycoprotein glucosyltransferase 1 Immune Response |
Uggt1 | UGGG1_MOUSE | 5 |
| Neurotrimin | Ntm | NTRI_MOUSE | 3 |
| Parathymosin | Ptms | PTMS_MOUSE | 2 |
| Toll-interacting protein | Tollip | TOLIP_MOUSE | 2 |
|
Ion Channel/Transporter Calcium-binding mitochondrial carrier protein Aralar1 |
Slc25a12 | CMC1_MOUSE | 15 |
| cGMP-gated cation channel alpha-1 | Cnga1 | CNGA1_MOUSE | 7 |
| Excitatory amino acid transporter 1 | Slc1a3 | EAA1_MOUSE | 5 |
| Excitatory amino acid transporter 2 | Slc1a2 | EAA2_MOUSE | 5 |
| Isoform 2 of Protein tweety homolog 1 | Ttyh1 | TTYH1_MOUSE | 2 |
| Isoform 2 of Vesicular inhibitory amino acid transporter | Slc32a1 | VIAAT_MOUSE | 7 |
| Mitochondrial 2-oxoglutarate/malate carrier protein | Slc25a11 | M2OM_MOUSE | 2 |
| Mitochondrial carrier homolog 2 | Mtch2 | MTCH2_MOUSE | 2 |
| Mitochondrial glutamate carrier 1 | Slc25a22 | GHC1_MOUSE | 4 |
| Monocarboxylate transporter 1 | Slc16a1 | MOT1_MOUSE | 3 |
| Plasma membrane calcium-transporting ATPase 2 | Atp2b2 | AT2B2_MOUSE | 3 |
| Potassium voltage-gated channel subfamily B member 1 | Kcnb1 | KCNB1_MOUSE | 2 |
| Protein Cngb1 | Cngb1 | Q91WA8_MOUSE | 3 |
| Protein Cngb1 | Cngb1 | E9PXX0_MOUSE | 2 |
| Protein Slc24a1 | Slc24a1 | Q91WD8_MOUSE | 2 |
| Retinal-specific ATP-binding cassette transporter | Abca4 | ABCA4_MOUSE | 19 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | Atp2a2 | AT2A2_MOUSE | 6 |
| Sideroflexin-1 | Sfxn1 | SFXN1_MOUSE | 3 |
| Sideroflexin-3 | Sfxn3 | SFXN3_MOUSE | 4 |
| Sideroflexin-5 | Sfxn5 | SFXN5_MOUSE | 3 |
| Sodium- and chloride-dependent GABA transporter 1 | Slc6a1 | SC6A1_MOUSE | 3 |
| Sodium- and chloride-dependent GABA transporter 3 | Slc6a11 | S6A11_MOUSE | 4 |
| Sodium/potassium-transporting ATPase subunit beta-1 | Atp1b1 | AT1 B1_MOUSE | 8 |
| Sodium/potassium-transporting ATPase subunit beta-2 | Atp1b2 | AT1B2_MOUSE | 9 |
| Sodium/potassium-transporting ATPase subunit beta-3 | Atp1b3 | AT1B3_MOUSE | 4 |
| Sodium-coupled neutral amino acid transporter 3 | Slc38a3 | S38A3_MOUSE | 2 |
| Vesicular glutamate transporter 1 | Slc17a7 | VGLU1_MOUSE | 2 |
| Voltage-dependent anion-selective channel protein 3 | Vdac3 | VDAC3_MOUSE | 7 |
|
Lipid Transport/Synthesis/Metabolism 1-acyl-sn-glycerol-3-phosphate acyltransferase gamma |
Agpat3 | PLCC_MOUSE | 2 |
| 3-hydroxyacyl-CoA dehydrogenase type-2 | Hsd17b10 | HCD2_MOUSE | 2 |
| 3-ketoacyl-CoA thiolase A, peroxisomal | Acaa1a | THIKA_MOUSE | 4 |
| Acyl-CoA dehydrogenase family member 9, mitochondrial | Acad9 | ACAD9 MOUSE | 3 |
| Enoyl-CoA delta isomerase 1, mitochondrial | Eci1 | ECI1_MOUSE | 4 |
| Estradiol 17-beta-dehydrogenase 12 | Hsd17b12 | DHB12_MOUSE | 2 |
| Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | Hadh | HCDH_MOUSE | 3 |
| Inorganic pyrophosphatase 2, mitochondrial | Ppa2 | D3Z636_MOUSE | 2 |
| Isoform 2 of Long-chain-fatty-acid–CoA ligase 6 | Acsl6 | ACSL6_MOUSE | 6 |
| Long-chain fatty acid transport protein 1 | Slc27a1 | S27A1_MOUSE | 2 |
| Long-chain specific acyl-CoA dehydrogenase, mitochondrial | Acadl | ACADL_MOUSE | 2 |
| Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | Acadm | ACADM_MOUSE | 4 |
| Neutral cholesterol ester hydrolase 1 | Nceh1 | NCEH1_MOUSE | 2 |
| Trifunctional enzyme subunit alpha, mitochondrial | Hadha | ECHA_MOUSE | 5 |
| Trifunctional enzyme subunit beta, mitochondrial | Hadhb | ECHB_MOUSE | 3 |
| Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | Acadvl | ACADV_MOUSE | 2 |
| Very-long-chain (3R)-3-hydroxyacyl-[acyl-carrier protein] dehydratase 3 | ptplad1 | HACD3_MOUSE | 2 |
|
Neurotransmitter Regulation and Synthesis 4-trimethylaminobutyraldehyde dehydrogenase |
Aldh9a1 | AL9A1_MOUSE | 2 |
| Glutamate decarboxylase 1 | Gad1 | DCE1_MOUSE | 2 |
| Glutaminase kidney isoform, mitochondrial | Gls | GLSK_MOUSE | 10 |
| Isoform 2 of 4-aminobutyrate aminotransferase, mitochondrial | Abat | GABT_MOUSE | 7 |
| Succinate-semialdehyde dehydrogenase, mitochondrial | Aldh5a1 | SSDH_MOUSE | 5 |
|
Oxidation/Reduction Alcohol dehydrogenase class-3 |
Adh5 | ADHX_MOUSE | 2 |
| Aldehyde dehydrogenase, mitochondrial | Aldh2 | ALDH2_MOUSE | 4 |
| Biliverdin reductase A | Blvra | BIEA_MOUSE | 2 |
| Carbonyl reductase [NADPH] 3 | Cbr3 | CBR3_MOUSE | 2 |
| D-beta-hydroxybutyrate dehydrogenase, mitochondrial | Bdh1 | BDH_MOUSE | 2 |
| Glutathione peroxidase | Gpx4 | GPX41_MOUSE | 2 |
| Isoform 2 of Alpha-aminoadipic semialdehyde dehydrogenase | Aldh7a1 | AL7A1_MOUSE | 2 |
| Isoform 4 of Oxidation resistance protein 1 | Oxr1 | OXR1_MOUSE | 5 |
| NAD-dependent malic enzyme, mitochondrial | Me2 | MAOM_MOUSE | 4 |
| NADPH–cytochrome P450 reductase | Por | NCPR_MOUSE | 3 |
| Retinol dehydrogenase 12 | Rdh12 | RDH12_MOUSE | 9 |
| Saccharopine dehydrogenase-like oxidoreductase | Sccpdh | SCPDL_MOUSE | 2 |
| Serum paraoxonase/arylesterase 1 | Pon1 | PON1_MOUSE | 2 |
| Superoxide dismutase [Mn], mitochondrial | Sod2 | SODM_MOUSE | 7 |
| Thioredoxin-dependent peroxide reductase, mitochondrial | Prdx3 | PRDX3_MOUSE | 2 |
| Thioredoxin-related transmembrane protein 1 | Tmx1 | TMX1_MOUSE | 2 |
| Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase Phospholipid Transport |
Dhdh | DHDH_MOUSE | 2 |
| Phosphatidylinositol transfer protein alpha isoform | Pitpna | PIPNA_MOUSE | 2 |
|
Protein Synthesis 40S ribosomal protein S10 |
Rps10 | RS10_MOUSE | 3 |
| 40S ribosomal protein S20 | Rps20 | RS20_MOUSE | 2 |
| 40S ribosomal protein S26 | Rps26 | RS26_MOUSE | 2 |
| 60S acidic ribosomal protein P2 | Rplp2 | RLA2_MOUSE | 4 |
| 60S ribosomal protein L31 | Rpl31 | RL31_MOUSE | 2 |
| ATP-dependent RNA helicase A | Dhx9 | E9QNN1_MOUSE | 6 |
| ATP-dependent RNA helicase DDX19A | Ddx19a | DD19A_MOUSE | 2 |
| Elongation factor Tu, mitochondrial | Tufm | EFTU_MOUSE | 5 |
| Eukaryotic initiation factor 4A-III | Eif4a3 | IF4A3_MOUSE | 2 |
| Eukaryotic translation initiation factor 2 subunit 1 | Eif2s1 | IF2A_MOUSE | 3 |
|
ProteinTransport AP-1 complex subunit beta-1 |
Ap1b1 | AP1B1_MOUSE | 9 |
| AP-3 complex subunit beta-2 | Ap3b2 | AP3B2_MOUSE | 3 |
| Coatomer subunit beta’ | Copb2 | COPB2_MOUSE | 3 |
| Conserved oligomeric Golgi complex subunit 3 | Cog3 | COG3_MOUSE | 2 |
| Cytoplasmic dynein 1 intermediate chain 2 | Dync1i2 | A2BFF5_MOUSE | 4 |
| Disks large homolog 1 | Dlg1 | E9Q9H0_MOUSE | 3 |
| Endoplasmic reticulum-Golgi intermediate compartment protein 1 | Ergic1 | ERGI1_MOUSE | 2 |
| Erlin-2 | Erlin2 | ERLN2_MOUSE | 2 |
| Exportin-1 | Xpo1 | XPO1_MOUSE | 4 |
| Importin-5 | Ipo5 | IPO5_MOUSE | 3 |
| Isoform 2 of MAGUK p55 subfamily member 2 | Mpp2 | MPP2_MOUSE | 7 |
| Isoform 2 of TOM1-like protein 2 | Tom1l2 | TM1L2_MOUSE | 2 |
| Isoform B of AP-2 complex subunit alpha-1 | Ap2a1 | AP2A1_MOUSE | 2 |
| Kinectin | Ktn1 | F8VQC7_MOUSE | 2 |
| Mitochondrial import inner membrane translocase subunit TIM50 | Timm50 | TIM50_MOUSE | 2 |
| Mitochondrial import receptor subunit TOM70 | Tomm70a | TOM70_MOUSE | 3 |
| Nuclear transport factor 2 | Nutf2 | NTF2_MOUSE | 2 |
| PRA1 family protein 2 | Praf2 | PRAF2 MOUSE | 2 |
| PRA1 family protein 3 | Arl6ip5 | PRAF3_MOUSE | 2 |
| Protein ERGIC-53 | Lman1 | LMAN1_MOUSE | 5 |
| Protein MGARP (Fragment) | Mgarp | D3Z134_MOUSE | 2 |
| Protein transport protein Sec23A | Sec23a | E9Q1S3_MOUSE | 2 |
| Transmembrane emp24 domain-containing protein 10 | Tmed10 | TMEDA_MOUSE | 2 |
| Transportin-1 (Fragment) | Tnpo1 | Q3TKD0_MOUSE | 4 |
|
Proteosome Degradation/Protease 26S proteasome non-ATPase regulatory subunit 11 |
Psmd11 | PSD11_MOUSE | 2 |
| 26S proteasome non-ATPase regulatory subunit 2 | Psmd2 | PSMD2 MOUSE | 5 |
| Cathepsin B | Ctsb | CATB_MOUSE | 2 |
| Cathepsin D (Fragment) | Ctsd | F6Y6L6_MOUSE | 4 |
| Dipeptidyl peptidase 3 | Dpp3 | DPP3_MOUSE | 4 |
| E3 ubiquitin-protein ligase NEDD4 | Nedd4 | NEDD4_MOUSE | 4 |
| Isoform 2 of Protein phosphatase 1H | Ppm1h | PPM1H_MOUSE | 2 |
| Proteasome subunit alpha type-4 | Psma4 | PSA4_MOUSE | 2 |
| Proteasome subunit beta type-2 | Psmb2 | PSB2_MOUSE | 3 |
| Proteasome subunit beta type-3 | Psmb3 | PSB3 MOUSE | 2 |
| Protein DDI1 homolog 2 | Ddi2 | DDI2_MOUSE | 2 |
| Ubiquilin-2 | Ubqln2 | UBQL2_MOUSE | 3 |
| Ubiquitin carboxyl-terminal hydrolase | Usp7 | UBP7_MOUSE | 4 |
| Ubiquitin thioesterase OTUB1 | Otub1 | OTUB1_MOUSE | 6 |
|
RNA Metabolism 60 kDa SS-A/Ro ribonucleoprotein |
Trove2 | RO60_MOUSE | 2 |
| Aspartate–tRNA ligase, cytoplasmic | Dars | SYDC_MOUSE | 3 |
| CCA tRNA nucleotidyltransferase 1, mitochondrial | Trnt1 | TRNT1_MOUSE | 3 |
| Glycine–tRNA ligase | Gars | SYG_MOUSE | 3 |
| Heterogeneous nuclear ribonucleoprotein H2 | Hnrnph2 | HNRH2_MOUSE | 2 |
| Heterogeneous nuclear ribonucleoprotein U-like protein 2 | Hnrnpul2 | HNRL2_MOUSE | 2 |
| Isoleucine–tRNA ligase, mitochondrial | Iars2 | SYIM_MOUSE | 2 |
| Poly(rC)-binding protein 4 | Pcbp4 | PCBP4_MOUSE | 2 |
| Pre-mRNA-processing-splicing factor 8 | Prpf8 | PRP8_MOUSE | 2 |
| Probable ATP-dependent RNA helicase DDX6 | Ddx6 | DDX6_MOUSE | 3 |
| Probable C->U-editing enzyme APOBEC-2 | Apobec2 | ABEC2_MOUSE | 2 |
| Ribonuclease inhibitor | Rnh1 | RINI_MOUSE | 3 |
| RNA-binding protein 14 | Rbm14 | RBM14_MOUSE | 2 |
| Small nuclear ribonucleoprotein-associated protein B | Snrpb | RSMB_MOUSE | 2 |
| Splicing factor U2AF 35 kDa subunit Signal Transduction |
U2af1 | U2AF1_MOUSE | 2 |
| Diacylglycerol kinase epsilon | Dgke | DGKE_MOUSE | 2 |
| G protein-coupled receptor kinase 1 | Grk1 | RK_MOUSE | 14 |
| GTP-binding protein Di-Ras2 | Diras2 | DIRA2_MOUSE | 2 |
| Guanine nucleotide-binding protein G(i) subunit alpha-2 | Gnai2 | GNAI2_MOUSE | 4 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-3 | Gnb3 | GBB3_MOUSE | 2 |
| Guanine nucleotide-binding protein G(q) subunit alpha | Gnaq | GNAQ_MOUSE | 7 |
| Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Gnas | GNAS2_MOUSE | 3 |
| Guanine nucleotide-binding protein G(t) subunit alpha-2 | Gnat2 | GNAT2_MOUSE | 4 |
| Guanine nucleotide-binding protein G(z) subunit alpha | Gnaz | GNAZ_MOUSE | 4 |
| Guanine nucleotide-binding protein subunit alpha-14 | Gna14 | GNA14_MOUSE | 3 |
| Guanine nucleotide-binding protein subunit beta-2-like 1 | Gnb2l1 | GBLP_MOUSE | 3 |
| Guanylyl cyclase GC-E | Gucy2e | GUC2E_MOUSE | 15 |
| Isoform 1 of Regulator of G-protein signaling 9 | Rgs9 | RGS9_MOUSE | 11 |
| Isoform 2 of Guanine nucleotide-binding protein subunit beta-5 | Gnb5 | GBB5_MOUSE | 3 |
| Isoform 2 of Neurochondrin | Ncdn | NCDN_MOUSE | 2 |
| Phosphodiesterase 6A, cGMP-specific, rod, alpha | Pde6a | Q8K0A8_MOUSE | 26 |
| Protein Rap1gap | Rap1gap | A2ALS5_MOUSE | 2 |
| Ras-related protein Rab-10 | Rab10 | RAB10_MOUSE | 2 |
| Ras-related protein Rab-11B | Rab11b | RB11B_MOUSE | 4 |
| Ras-related protein Rab-18 | Rab18 | RAB18_MOUSE | 2 |
| Ras-related protein Rab-1B | Rab1b | RAB1B_MOUSE | 3 |
| Ras-related protein Rab-3C | Rab3c | RAB3C_MOUSE | 3 |
| Ras-related protein Rab-5A | Rab5a | RAB5A_MOUSE | 3 |
| Ras-related protein Rab-5B | Rab5b | RAB5B_MOUSE | 3 |
| Ras-related protein Rab-5C | Rab5c | RAB5C_MOUSE | 2 |
| Ras-related protein Rab-6B | Rab6b | RAB6B_MOUSE | 3 |
| Ras-related protein Rap-1A | Rap1a | RAP1A_MOUSE | 3 |
| Regulator of G-protein signaling 9-binding protein | Rgs9bp | R9BP_MOUSE | 3 |
| Rho GTPase-activating protein 1 | Arhgap1 | A2AH25_MOUSE | 6 |
| Rod cGMP-specific 3′,5′-cyclic phosphodiesterase subunit beta | Pde6b | PDE6B_MOUSE | 10 |
| Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform | Ppp2r2a | 2ABA_MOUSE | 3 |
| Short-wave-sensitive opsin 1 Stress Response |
Opn1sw | OPSB_MOUSE | 2 |
| Constitutive coactivator of PPAR-gamma-like protein 1 | FAM120A | F120A_MOUSE | 2 |
| Heat shock 70 kDa protein 12A | Hspa12a | HS12A_MOUSE | 6 |
| Hypoxia up-regulated protein 1 | Hyou1 | HYOU1_MOUSE | 6 |
|
Synaptic Transmission Isoform 2 of Solute carrier family 12 member 5 |
Slc12a5 | S12A5_MOUSE | 16 |
| Protein lin-7 homolog A | Lin7a | LIN7A_MOUSE | 4 |
| Synaptic vesicle glycoprotein 2B | Sv2b | SV2B_MOUSE | 4 |
| Synaptic vesicle membrane protein VAT-1 homolog-like | Vat1l | VAT1L_MOUSE | 2 |
| Synaptoporin | Synpr | SYNPR_MOUSE | 4 |
| Syntaxin-1B | Stx1b | STX1 B_MOUSE | 10 |
| Vesicle-associated membrane protein-associated protein A Transcription Regulation |
Vapa | VAPA_MOUSE | 5 |
| Alpha/beta hydrolase domain-containing protein 14B | Abhd14b | E9QN99_MOUSE | 2 |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 3, mitochondrial | Chchd3 | CHCH3_MOUSE | 3 |
| Isoform 2 of Mitochondrial inner membrane protein | Immt | IMMT_MOUSE | 19 |
| Isoform Epsilon of Lamina-associated polypeptide 2, isoforms beta/delta/epsilon/gamma | Tmpo | LAP2B_MOUSE | 7 |
| Protein arginine N-methyltransferase 1 | Prmt1 | ANM1_MOUSE | 2 |
| Transcription elongation factor B polypeptide 2 Transferase |
Tceb2 | ELOB_MOUSE | 2 |
| Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | Dlst | ODO2_MOUSE | 4 |
| Farnesyl pyrophosphate synthase | Fdps | FPPS_MOUSE | 4 |
| Isoform 2 of NAD kinase 2, mitochondrial | Nadk2 | NAKD2 MOUSE | 2 |
| Ornithine aminotransferase, mitochondrial | Oat | OAT_MOUSE | 5 |
| Phosphoserine aminotransferase | Psat1 | SERC_MOUSE | 4 |
| Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial Vesicle Transport |
Oxct1 | SCOT1_MOUSE | 12 |
| Protein NipSnap homolog 2 | Gbas | NIPS2_MOUSE | 3 |
| Protein Sptbn2 Others |
Sptbn2 | Q68FG2_MOUSE | 3 |
| Adipocyte plasma membrane-associated protein | Apmap | APMAP_MOUSE | 2 |
| C2 domain-containing protein 2-like | C2cd2l | C2C2L_MOUSE | 4 |
| Ceruloplasmin | Cp | CERU_MOUSE | 2 |
| COP9 signalosome complex subunit 1 | Gps1 | G3UXW9_MOUSE | 4 |
| Delta-aminolevulinic acid dehydratase | Alad | HEM2_MOUSE | 2 |
| DmX-like protein 2 | Dmxl2 | DMXL2_MOUSE | 4 |
| Endophilin-B2 | Sh3glb2 | SHLB2_MOUSE | 2 |
| EPM2A-interacting protein 1 | Epm2aip1 | EPMIP_MOUSE | 3 |
| ES1 protein homolog, mitochondrial | D10Jhu81e | ES1_MOUSE | 3 |
| Galectin-related protein | Lgalsl | LEGL_MOUSE | 4 |
| Gamma-crystallin A | Cryga | CRGA_MOUSE | 2 |
| Golgi apparatus protein 1 (Fragment) | Glg1 | GSLG1_MOUSE | 2 |
| Golgi reassembly stacking protein 2, isoform CRA_d | Gorasp2 | GORS2_MOUSE | 3 |
| Isochorismatase domain-containing protein 2A, mitochondrial | Isoc2a | ISC2A_MOUSE | 2 |
| LanC-like protein 2 | Lancl2 | LANC2_MOUSE | 2 |
| Leucine-rich repeat-containing protein 59 | Lrrc59 | LRC59_MOUSE | 3 |
| Low molecular weight phosphotyrosine protein phosphatase | Acp1 | PPAC_MOUSE | 2 |
| MCG140784 | Try10 | Q792Z1_MOUSE | 2 |
| NAD(P)H-hydrate epimerase | Apoa1bp | NNRE_MOUSE | 2 |
| PEX5-related protein | Pex5l | D3YYH0_MOUSE | 2 |
| Phosphoribosyl pyrophosphate synthase-associated protein 2 | Prpsap2 | KPRB_MOUSE | 2 |
| Protein Ahnak2 (Fragment) | Ahnak2 | F7DBB3_MOUSE | 3 |
| Protein NDRG3 | Ndrg3 | NDRG3_MOUSE | 2 |
| Protein SET (Fragment) | Set | SET_MOUSE | 2 |
| Tumor protein D52 (Fragment) | Tpd52 | D3Z125_MOUSE | 2 |
| Uncharacterized protein | Tubb4b-ps1 | J3QNR5_MOUSE | 2 |
| Up-regulated during skeletal muscle growth protein 5 | Usmg5 | USMG5_MOUSE | 2 |
Results
Light adaptation induces S-nitrosylation in all retinal layers
In the dark-adapted retina, the concentration of NO is present at a minimal, tonic level, however as the retina becomes light-adapted, NO synthesis is initiated and NO levels are substantially elevated (Neal et al., 1998; Walter et al., 2014). As NO levels tend to increase during light adaptation, it stands to reason that levels of S-nitrosylated proteins become elevated as well. To evaluate this possibility, we compared the levels of S-nitrosylation, by assessing S-nitrosocysteine immunofluorescence (SNI), over entire retinal sections from light-adapted and dark-adapted goldfish. For these, and all other images obtained from goldfish retinas, Mbs were identified by PKCα immunoreactivity and served to provide orientation and identification of the synaptic layers within the retina (Fig. 1A and D). In the light-adapted goldfish retina, we observed a robust SNI; the pattern of intense immunolabeling extended to all layers of the retina (Fig. 1B and C). In contrast, the overall intensity of SNI in the dark-adapted goldfish retina was drastically less than that of the light-adapted one and the pattern of labeling was also different (Fig. 1E and F). Specifically, in the dark-adapted goldfish retina, SNI was limited to the ganglion cell layer (GCL) and photoreceptor layer (PL) with greatest intensity within the GCL. Quantitative analysis of the fluorescence intensity of the S-nitrosylation signal (see Materials and Methods) revealed that, on average, the light-adapted goldfish retina (n=8 sections from 4 retinas) displayed significantly more S-nitrosylated proteins than the dark-adapted goldfish retina (Fig. 1G; n=7 sections from 2 retinas). This was consistent with previous reports describing levels of NO as being lowest when the retina is dark-adapted (Neal et al., 1998; Sekaran et al., 2005; Walter et al., 2014). These results also suggest that basal NO release in the dark-adapted retina reaches sufficient levels to cause S-nitrosylation that is detectable by immunohistochemical methods.
Considering that light-evoked NO synthesis and release has been described in a number of species, including retinas of cold blooded vertebrates and mammalian retinas (Vielma et al., 2012), we then shifted our attention to a mammalian preparation to determine whether light induces S-nitrosylation, detectable by immunohistochemical methods, in the mouse retina. For all mouse retinal sections, PKCα immunoreactivity was used to identify rod bipolar cells (RBCs) and orient the observer within the various retinal layers (Fig. 2A and D).The light-adapted mouse retina showed a robust labeling pattern of SNI, spanning all layers of the retinal section (Fig. 2B and C). This robust labeling was in stark contrast to the near absence of SNI labeling in the dark-adapted mouse retina (Fig. 2E and F). By comparison, the average intensity of SNI in individual retinal layers was significantly increased across all layers in the light-adapted retina (n=8 sections from 8 retinas) compared to dark-adapted (n=7 sections from 6 retinas) (Fig. 2G). When taken all together, these data indicate strongly that retinal S-nitrosylation is light-dependent and becomes increasingly more prevalent at higher, stronger light intensities in both goldfish and mouse retinas.
The pattern of light driven SNI across retinal layers is intensity dependent
In the natural world, a significant range of behaviorally relevant light intensities can cause simultaneous activation of both rods and cones (Wu, 1994). The overlap between the intensities at cone activation threshold and rod saturation forms the mesopic range (Krizaj, 2000). Indeed, in the goldfish retina, a rod saturating green light (1×1010 photons/cm2/s, 505 nm) will also activate cones (Joselevitch and Kamermans, 2007). Compared to baseline levels of SNI in the dark-adapted goldfish retina (Fig. 1E), SNI evoked by a 10 s exposure to mesopic light stimulus was significantly higher in the PL (Fig. 3B and C; n=9 sections from 4 retinas) (p=0.03, unpaired Student’s t test), in the OPL (p=0.01, unpaired Student’s t test), in the INL (p=0.003, unpaired Student’s t test), in the IPL (p<0.0001, unpaired Student’s t test) and in the GCL (p=0.003, unpaired Student’s t test). Although every layer, except the ONL, showed an increase in SNI intensity above basal levels, the most dramatic increase occurred within the IPL (8 fold increase from dark-adapted levels). Furthermore, mesopic green light stimulation induced an SNI labeling pattern in the IPL that clearly labeled cellular structures, particularly the large, bulbous terminals of Mbs (Fig. 3D, E and F), consistent with our previous results indicating physiologically relevant S-nitrosylation within the Mb terminal (Tooker et al., 2013). Note that SNI appears to localize to the plasma membrane of the Mb terminals. Intensity profiles of both the PKCα and the SNI revealed that 67.8 ± 3.5 % of the SNI within the IPL was located within the region associated with the Mb terminals (n=9 sections from 4 retinas). Furthermore, colocalization analysis of PKCα and S-nitrosocysteine immunolabeling within individual Mb terminals revealed that 67.2 ± 11.9 % of green pixels (PKCα) overlapped with magenta pixels (SNI) and that 75.0 ± 9.9 % of magenta pixels overlapped with green pixels (data not shown, n=10 terminals).
Recently, we reported a functional aspect of retinal S-nitrosylation responsible for mediating the modulation of bipolar cell light responses in the goldfish retina; Mb light responses to dim, rod mediated input were enhanced via S-nitrosylation upon exposure to illumination with intensity of at least 2.4×108 photons/cm2/s for 500 ms (Tooker et al., 2013). Accordingly, we reduced the intensity of the stimulus to a bright scotopic light step (2.4×108 photons/cm2/s, 505 nm, 500 ms). In the goldfish this bright scotopic light nearly fully activates rods, and the subsequent elements of the rod pathways, without reaching the threshold of cone activation (Joselevitch and Kamermans, 2009). After a 500 ms exposure to 2.4×108 photons/cm2/s, SNI was nearly fully confined to the inner retina (Fig. 3H and I) and the IPL was the only layer to show a significant increase in SNI compared to basal levels in the dark-adapted retina (p=0.01, unpaired Student’s t test, n=10 sections from 4 retinas). Importantly, 80.0 ± 2.7% of the SNI found within the IPL was restricted to the portion containing the Mb terminals. SNI was found to colocalize with structures defined by PKCα immunofluorescence (Fig. 3G, H and I, boxed area). Analysis of individual Mb terminals revealed that 61.5 ± 17.4% of green pixels overlapped magenta pixels and 82.1 ± 14.4% of magenta pixels overlapped with green pixels (n=10 terminals). Within the IPL, analysis by one-way ANOVA revealed a significant difference between fully light-adapted retinas, retinas stimulated with rod saturating mesopic light and bright scotopic light (p<0.0001, one-way ANOVA). Post hoc comparisons using Tukey’s multiple comparison test indicated that the mean intensity of SNI in the IPL was significantly higher in light-adapted retinas (50.2 ± 5.3) than in retinas stimulated with 1×1010 photons/cm2/s (23.5 ± 1.8) and both were significantly elevated over the SNI in retinas stimulated with 2.4×108 photons/cm2/s (9 ± 1.3). Furthermore, the average percentage of SNI that was associated with Mb terminals was significantly greater in retinas stimulated with 2.4×108 photons/cm2/s than in those stimulated with 1×1010 photons/cm2/s (p=0.01, unpaired Student’s t test). In the GCL, analysis by one-way ANOVA indicated a significant difference between light stimulation paradigms. Tukey’s multiple comparison test revealed a significant reduction in the mean SNI when the retina was stimulated with 2.4×108 photons/cm2/s (9.9 ± 1.4) as compared to light-adapted (33.7 ± 4.8) and 1×1010 photons/cm2/s stimulated retinas (30.8 ± 3.8). No significant difference was detected between the mean SNI in the GCL of light-adapted or 1×1010 photons/cm2/s stimulated retinas. Importantly, when we increased the stimulus duration of the bright scotopic flash to 10 s, matching the duration of the mesopic flash, we did not observe any significant difference in the SNI labeling pattern when compared to the 500 ms flash (data not shown).
Next, we addressed the question of whether the extent of S-nitrosylation is regulated in a light intensity dependent manner in the mammalian retina. To this end, we subjected wild-type mouse retinas to the exact same light stimulation paradigm as was applied to the goldfish retinas and then assessed overall retinal S-nitrosylation through S-nitrosocysteine immunofluorescence. Wild-type mouse retinas stimulated with 1×1010 photons/cm2/s (505 nm, 10 s), a mesopic light stimuli which surpasses the activation threshold for cones (~1010 photons/cm2/s) and approaches rod saturation (~1011 photons/cm2/s, Busskamp et al., 2010), displayed an SNI labeling pattern that was primarily restricted to the inner retina (Fig. 4B and C). This labeling pattern in the mammalian retina was strikingly similar to the pattern of SNI in goldfish that was evoked by the exact same flash. Compared to baseline levels of SNI in the dark-adapted mouse retina (Fig. 2E), the 1×1010 photons/cm2/s flash evoked significantly more SNI in the inner retina: INL p=0.003, IPL p=0.001 and GCL p<0.0001(unpaired Student’s t test, n=11 sections from 6 retinas). Additionally, SNI appeared to colocalize with the RBC terminal clusters (marked by PKCα immunofluorescence) in the IPL (Fig. 4D, E and F). Intensity analysis within the IPL revealed that 68.0 ± 3.7% of the SNI was restricted to the region of the IPL that contained the terminals of RBCs. Systematic analysis of RBC terminal clusters indicated that 48.3 ± 12.6% of green pixels overlapped with magenta and 78.8 ± 6.3% of magenta pixels overlapped with green (n=10 terminal regions), indicating a majority of colocalization between the two individual patterns of immunofluorescence.
When the intensity of light stimulation was reduced to a bright scotopic flash (2.4×108 photons/cm2/s, 505 nm, 500ms), the pattern of S-nitrosocysteine immunofluorescence was localized primarily in the inner retina (Fig. 4H and I). Interestingly, the bright scotopic flash significantly elevated SNI intensity, above baseline dark-adapted levels, in the IPL and in the GCL (p<0.0001, p=0.003 respectively, unpaired Student’s t test, n=7 sections from 3 retinas). Within the IPL, 81.8 ± 2.7% of the SNI label was associated with the RBC terminal region of the IPL which was significantly more than the average percentage of SNI label within the terminal region of retinas stimulated with mesopic light stimulation (p=0.03, unpaired Student’s t test). Both light flashes evoked SNI label that was restricted to the inner retina. However, by comparison, the brighter intensity light stimulation (1×1010 photons/cm2/s) induced a greater distribution of SNI in the inner retina as there was significantly more SNI label in the INL as compared to the dim light intensity (2.4×108 photons/cm2/s; p=0.009 unpaired Student’s t test). In line with these results, analysis of individual terminal regions (n=10) from retinas stimulated with bright scotopic light revealed a colocalization pattern similar to retinas stimulated with mesopic light: 38.2 ± 13.6% of green pixels overlapped with red, while 80.5 ± 16.3% of magenta pixels overlapped with green (Fig. 4J, K and L).
We compared the fluorescent intensity within the IPL and GCL across the three various light stimulated states (i.e. photopic, mesopic, bright scotopic). One-way ANOVA revealed a significant difference in the SNI intensity within the IPL between light-adapted retinas, mesopic light stimulated retinas, and scotopic light stimulated retinas (p<0.0001). Interestingly, the difference was a result of a significant increase in the mean SNI of the light-adapted retina (51.5 ± 4.8) as compared to both the mesopic (21.7 ± 3.1) and the bright scotopic light stimulated retinas (16.0 ± 2.1, Tukey’s multiple comparison test). Although the mean SNI intensity within the IPL evoked by 1×1010 photons/cm2/s was greater than the mean intensity evoked by 2.4×108 photons/cm2/s, the difference was not significant (Tukey’s multiple comparison test). Within the GCL, a significant difference was detected in SNI intensity across light-adapted retinas, retinas stimulated with a single mesopic step and retinas stimulated with a single scotopic step of light (p<0.0001, one-way ANOVA). Similar to our observation in the IPL of the mouse retinas, Tukey’s multiple comparison test indicated that the difference was due to the significant increase of SNI intensity within the GCL of the light-adapted retina (47.3 ± 4.1) as compared to retinas stimulated with either 1×1010 photons/cm2/s (24.3 ± 2.4) or 2.4×108 photons/cm2/s (17.0 ± 4.1). Again, although there was a greater level of SNI intensity measured in the GCL of retinas stimulated with mesopic light flash, it was not significantly larger than the SNI intensity measured in retinas stimulated with scotopic light stimuli.
Together, these data strongly support the notion that the overall intensity of SNI is positively correlated with the illumination levels in both the goldfish and mouse retina. Furthermore, the reported pattern of SNI is consistent with observations that NO levels in the retina increase in a manner dependent upon intensity of a light stimulus (Sekaran et al., 2005). This increase of NO is primarily mediated by nNOS, which, in general, is localized primarily to the inner retina in a variety of vertebrates, including goldfish and mouse (Eldred and Blute, 2005; Giove et al., 2009, Pang et al., 2010; Walter et al., 2014; Zhu et al., 2014).
Light-evoked S-nitrosocysteine immunofluorescence is occluded by pre-treatment with NEM
To further test our finding that light stimulation evoked increased S-nitrosylation within the fish and mouse retina, we inhibited the formation of nitrosocysteine prior to the evoked release of NO. Dark-adapted eyecups were incubated in NEM (1 mM) which irreversibly binds to free sulfhydryl groups (Smyth et al., 1960), blocking them from interacting with NO and thereby preventing the formation of S-nitrosothiols (Hu et al., 2012). In this regard, the pre-incubation with NEM prevents the formation of new S-nitrosocysteines which would be created by the light-evoked release of NO. After the incubation in NEM, goldfish and mouse retinas were stimulated for 10 s with mesopic green light (1×1010 photons/cm2/s) as this light intensity evoked a significant increase in SNI (compared to dark-adapted retinas) across multiple retinal layers in both animal models.
In the goldfish retina, the amount of SNI induced by a mesopic light flash was drastically reduced as compared to control conditions (Fig. 5B and C). By comparison, every retinal layer showed significantly less intense light-evoked SNI labeling in the NEM treated retina compared to the control: PL: p=0.01, ONL: p=0.03, OPL: p=0.008, INL: p=0.002, IPL: p<0.0001, GCL: p=0.0002 (unpaired Student’s t test, control: n=9 sections from 4 retinas, NEM: n=7 sections from 2 retinas). Furthermore, compared to the baseline level of SNI in the dark-adapted goldfish retina, NEM prevented a significant light-evoked increase in all retinal layers and, in fact, induced a significant decrease in the amount of SNI within the GCL (p=0.04, unpaired Student’s t test). The SNI labeling that is observed in the NEM-treated goldfish retina is most likely due to S-nitrosylation that was present prior to the application and incubation in NEM.
In the mouse retina, under control conditions, a 10 s exposure to mesopic green light at 1×1010 photons/cm2/s induced significantly increased levels of SNI in the INL, IPL and GCL. However, after incubation in NEM, the same illumination failed to induce elevated SNI (Fig. 5E and F). In other words, when compared to the levels of SNI evoked in the INL, IPL and GCL by mesopic stimulation under control conditions, incubation in NEM resulted in a significant reduction (INL: p=0.005 IPL: p=0.0004, GCL: p<0.0001, unpaired Student’s t test, control: n=11 sections from 6 retinas, NEM: n=7 sections from 3 retinas). Furthermore, mesopic light stimulation evoked SNI in the presence of NEM was not measurably different from that in the dark-adapted mouse retina.
Activation of nNOS is required for light dependent S-nitrosylation in the retina
In the vertebrate retina, the pattern of light-evoked NO release is intimately connected to the expression of nNOS (Eldred and Blute, 2005; Giove et al., 2009; Walter et al., 2014), and although all three NOS isoforms have been discovered in the retina, nNOS is thought to be the major source for light-evoked NO during visual responses (Vielma et al., 2012). Therefore, the light driven S-nitrosylation we observed is most likely a result of light-evoked synthesis and release of NO via nNOS positive neurons that are mostly localized to the inner retina (Eldred and Blute, 2005; Giove et al., 2009, Pang et al., 2010; Walter et al., 2014). To assess the relative contribution of nNOS to the light-evoked increase in SNI that we observed after a mesopic light flash, we incubated goldfish and mouse retinas in TRIM (50 μM), a potent inhibitor of nNOS (Handy et al., 1996).
After pre-incubation with TRIM, a 10 s light flash at intensity of 1×1010 photons/cm2/s failed to induce SNI at levels that were greater than baseline dark-adapted conditions in the goldfish retina (Fig. 6B and C; n=6 sections from 2 retinas). In fact, we obtained a similar result after incubating the goldfish retina in TRIM as when the retina was incubated in NEM prior to light stimulation: there was no significant difference in SNI intensity across all layers except for the GCL which showed a 4 fold decrease in SNI intensity, which proved to be significant compared to SNI intensity levels in the dark-adapted control (p=0.02, unpaired Student’s t test). Compared to the goldfish retina which was stimulated with the same rod saturating light flash under control conditions (Fig. 3, top panel), the TRIM treatment significantly reduced SNI intensity in all retinal layers: PL: p=0.01, ONL: p=0.03, OPL: p=0.009, INL: p=0.002, IPL: p<0.0001, GCL: p=0.0002 (unpaired Student’s t test, control: n=9 sections from 4 retinas, TRIM: n=6 sections from 2 retinas). These results were consistent with the finding that, in the goldfish retina, nNOS immunoreactivity is found in nearly all retinal layers (Liepe et al., 1994). Note that TRIM also inhibits inducible NOS (iNOS) (Handy et al., 1996). Unlike its constitutively active counterparts, iNOS expression is only elevated in the retina under pathological conditions (Knowles and Moncada, 1994) or after intense, prolonged illumination (i.e. 8 hrs, 1,200–1,400 Lux, 490–580 nm, Palamalai et al., 2006). It is highly unlikely that our 10 s light flash at intensity 1×1010 photons/cm2/s would stimulate iNOS expression and subsequent NO release. However, note the weak SNI present in both the photoreceptors as well as in GCL in the presence of TRIM that might be resulted by the endothelial NOS (eNOS) activity that has been shown to localize to the outer and inner retina (Haverkamp et al., 1999).
Figure 6.
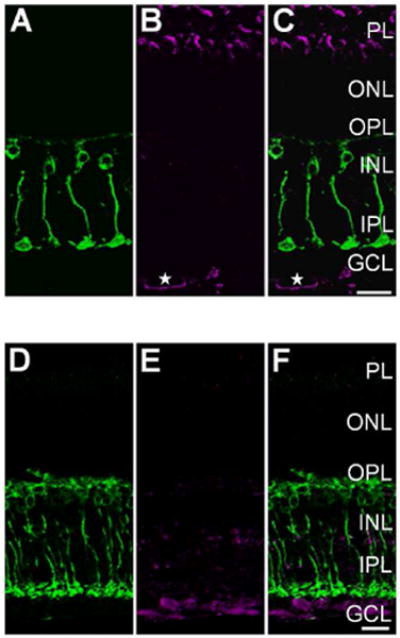
Light-evoked S-nitrosylation in the adult retina requires nNOS activation. A: Single plane 40× confocal image showing PKCα + (green) Mbs in a vertical cryosection from a goldfish retina incubated in 50 μM TRIM for 30 min prior to mesopic light stimulation (1×1010 photons/cm2/s, 505 nm, 10 sec). B: Immunolabeling, of the same region as in A, for S-nitrosocysteine (magenta). Compared to the S-nitrosocysteine immunolabeling pattern in the untreated goldfish retina (Fig. 3A, B, and C), pre-incubation with TRIM resulted in dramatically less light-driven SNI. The SNI below the GCL appears to label a structure with morphology that is consistent with a blood vessel (star). C: Merged confocal image depicting co-immunolabeling of PKCα and S-nitrosocysteine with minimal colocalization. D: Single plane 40× confocal image showing PKCα + (green) RBCs in a vertical cryosection from a wild-type mouse retina incubated in 50 μM TRIM for 30 min prior to mesopic light stimulation (1×1010 photons/cm2/s, 505 nm, 10 sec). E: S-nitrosocysteine immunolabeling (magenta) in the same region as presented in D. Notice the drastic reduction in SNI compared to that in the untreated mouse retina in Fig. 4A, B, and C. F: Merged image of the PKCα immunolabel from D and the S-nitrosocysteine immunolabel from E. Scale bars=20 μm.
In the mouse retina, TRIM significantly disrupted the pattern of SNI observed under control conditions after the mesopic light exposure (1×1010 photons/cm2/s; Fig. 6E and F; n=9 sections from 3 retinas). In the presence of TRIM the light stimulation did not significantly elevate the intensity of SNI above dark-adapted baseline levels in any retinal layer except for the GCL (p=0.03; unpaired Student’s t test, dark-adapted: n=7 sections from 6 retinas, TRIM: n=9 sections from 3 retinas). When compared side-by-side with retinas that were incubated in Ames’ media only, the TRIM incubation significantly reduced the amount of SNI that was evoked by mesopic light flash. Under control conditions, a robust SNI labeling pattern was observed in the INL, IPL and GCL (Compare Fig. 6E and F with Fig. 4B and C). After incubation in TRIM, SNI was significantly reduced in the INL, IPL and GCL (p=0.005, p<0.0001, p<0.0001, respectively; unpaired Student’s t test, control: n=11 sections from 6 retinas, TRIM: n=9 sections from 3 retinas). These data are consistent with the notion that the light stimulation is evoking nNOS dependent-NO release which drives SNI in the INL, IPL and GCL as was observed under the control conditions and further prevented with the incubation in TRIM.
While it was not significantly greater as compared to the dark control, there is still obvious SNI labeling after incubation with TRIM, particularly in the PL and the GCL in both the goldfish and mouse retinas. These results are consistent with the previous reports that some S-nitrosylation is observed in the presence of NOS inhibitors, indicating a basal, stable level of nitrosylation (Hess et al., 2005). Additionally, it is highly probable that this S-nitrosylation labeling pattern is due to NO release from endothelial NOS (eNOS), independent of the light stimulation. Note the S-nitrosocysteine labeling pattern beneath the GCL resembling the morphology of a blood vessel (Fig. 6B and C, star), an observation consistent with the general consensus that eNOS is considered to be expressed in high levels within the vascular endothelium (Knowles and Moncada, 1994).
In order to dissect the role of nNOS in contributing to light dependent S-nitrosylation in the mammalian retina, we examined the S-nitrosocysteine immunoreactivity in light- and dark-adapted retinas from mice lacking the nNOS isoform (nNOS KO). In the light-adapted nNOS KO animal, we observed a dramatic reduction in SNI across all layers of the retina (Fig. 7B and C), when compared to light-adapted WT littermates (Fig. 7E and F), with the most striking difference being the near lack of labeling in the outer retina. The SNI labeling pattern in dark-adapted retinas from nNOS KO animals (Fig. 7H and I) was remarkably similar to the dark-adapted retinas from WT littermates (Fig. 7K and L). In the absence of nNOS, it is readily apparent that light-evoked S-nitrosylation is reduced, but is it, however, entirely absent? In order to address this question we performed a one-way ANOVA comparing the intensity of the fluorescence associated with S-nitrosocysteine in the retinas of light-adapted WT animals, light-adapted nNOS KO animals and dark-adapted nNOS KO animals. The comparison between light-and dark-adapted WT animals was omitted as it had been described and analyzed above (Fig. 2G). Analysis via one-way ANOVA revealed a significant difference in SNI in all layers of the retina across the three experimental groups (PL: p<0.0001, ONL: p=0.0002, OPL, INL, IPL and GCL: p<0.0001, Fig. 7M). Tukey’s multiple comparison test revealed that in every retinal layer, the intensity of the SNI was significantly reduced for both light- and dark-adapted nNOS KO animals as compared to the light-adapted WT retina. Interestingly, in the GCL, the mean intensity of SNI was significantly higher in the light-adapted nNOS KO (10.4 ± 2.8, n=9 sections from 6 retinas) than in the dark-adapted nNOS KO retina (2.02 ± 0.6, n=6 sections from 2 retinas).
Figure 7.
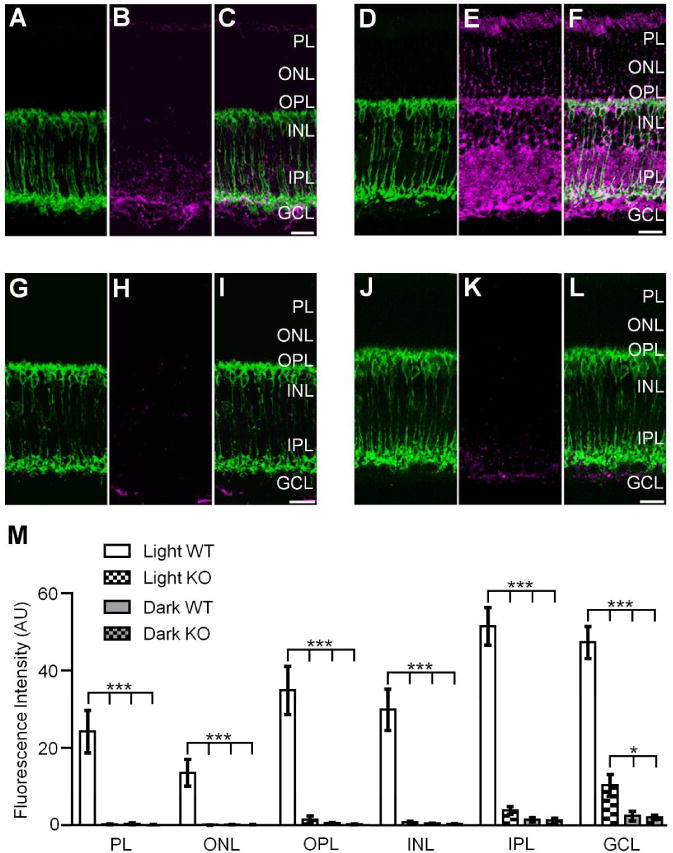
nNOS is necessary for robust S-nitrosylation in the mouse retina. 40× vertical cryostat sections of retinas from a light-adapted nNOS KO animal (top left), a light-adapted WT littermate (top right), a dark-adapted nNOS KO animal (bottom left) and a dark-adapted WT littermate (bottom right). A, D: RBCs indicated by PKCα + immunolabeling (green) in a light-adapted nNOS KO animal (A) and WT littermate (D). B, E: light-evoked S-nitrosocysteine immunofluorescence (magenta) is reduced and confined to the inner layers of the nNOS KO retina (B) as compared to the light-adapted wild-type mouse retina (E). C: Merged confocal image showing the co-immunolabeling of PKCα and S-nitrosocysteine from A and B. F: Merged image from panels D and E illustrating robust SNI in the light-adapted WT control retina. G, J: PKCα + immunofluorescence of RBCs from dark-adapted nNOS KO (G) and WT-littermate (J) retinas. SNI is nearly absent across all retinal layers in both the dark-adapted nNOS KO (H) and the dark-adapted WT (K) animals. I: Merged confocal image of panels G and H showing very little co-immunolabeling of RBCs and SNI in the dark-adapted KO animal. L: Merged confocal image of the dark-adapted WT control retina showing very little SNI. M: Summary histogram of SNI intensity across all retinal layers in light- and dark-adapted WT and nNOS KO animals. The SNI intensity is significantly reduced in all layers of the light- and dark-adapted nNOS KO retinas as compared to light-adapted WT. The intensity of SNI in the GCL of light-adapted KO animals was significantly higher than the SNI intensity quantified in both the dark-adapted KO and WT. One-way ANOVA with Tukey’s multiple comparison test, ***: p≤0.0005, *: p=0.01, light-adapted KO: n=6 retinas, dark-adapted KO: n=2 retinas, light-adapted WT: n=8 retinas, dark-adapted WT: n=6 retinas. Data is presented as mean ± SEM. Scale bars=20 μm.
Maintaining the assumption that the SNI label that is present in the dark-adapted wild-type retina is a representation of the basal levels of S-nitrosylation, we examined whether the SNI intensity observed in the light-adapted and dark-adapted retinas of nNOS KO animals was different than the SNI intensity in the dark-adapted WT retina. A one-way ANOVA comparison of the individual intensity profiles from light-adapted nNOS KO retinas, dark-adapted nNOS KO retinas, and dark-adapted WT retinas revealed there was a significant difference in the SNI labeling pattern only in the GCL (p=0.02, Fig. 7M). Tukey’s multiple comparison test revealed that in the GCL, the mean SNI intensity was significantly higher in the light-adapted nNOS KO retina (10.4 ± 2.8) compared to both the dark-adapted WT (2.4 ± 1.2) and nNOS KO (2.02 ± 0.6) retinas. In summary, it appears that the lack of nNOS significantly reduces light-evoked S-nitrosylation in the mammalian retina. However, light-adaptation evoked a small amount of S-nitrosylation (above dark-adapted baseline levels) within the GCL (Fig. 7M), which may be a result of light-evoked NO release from other sources. Furthermore, while two alternative forms of nNOS, α and γ, have been shown to be present in the mammalian retina (Giove et al., 2009), the KO animals used in these studies only eliminated expression of nNOSα (Gyurko et al. 2002). Thus even though the nNOSα isoform is considered to be the predominant source of NO release in the retina (Giove et al., 2009), the small, albeit significant, increase in SNI above baseline levels in the GCL of light-adapted nNOS KO animals may be a result of light-evoked NO release from neurons expressing the nNOSγ isoform.
Qualitative assay of S-nitrosylated retinal proteins
Although our immunohistochemical methods provide a qualitative account as to the functional nature of light-dependent S-nitrosylation in the adult retina, they fall short in providing critical information as to the identity of individual proteins being nitrosylated. One of the inherent challenges associated with attempting to identify S-nitrosylated proteins is the fragile, labile nature of the nitrosothiols moiety which can be lost when subjected to reducing conditions (Paige et al., 2008), reduced metal ions such as Cu+ (Dicks and Williams, 1996), and even to prolonged light exposure (Forrester et al., 2007). For example, using mass spectrometry (MS) an S-nitrosylated protein could be detected by a mass increase of 29 DA on an otherwise unmodified peptide (Lee et al., 2007). Nonetheless, due to the labile nature of the nitrosothiol moiety, S-nitrosylated proteins are rarely detected in MS spectra (Wang et al., 2008). Therefore, with these limitations in mind, we chose to employ a modified version (see Materials and Methods) of the biotin-switch method, originally developed by Jaffrey et al. (2001), as its reliability and specificity in identification of S-nitrosylated proteins have been well established (Jaffrey et al., 2001; Forrester et al., 2007; Seth and Stamler, 2011). The biotin-switch assay overcomes the issue of S-nitrosocysteine instability by covalently labeling only proteins containing the S-nitrosylation moiety with a biotin in place of the nitrosothiol group. The biotinylated proteins, within the retinal lysates, were enriched by resin-assisted capture before trypsin digestion and MS. Protein sequences are identified by comparison to existing protein databases (see Materials and Methods for details). Due to the limited proteomic databases that are specific to the goldfish proteome, these experiments were performed using only mouse retinas.
Our MS approach revealed 154 proteins that were S-nitrosylated only under dark-adapted conditions and 351 S-nitrosylated proteins unique to the light-adapted retina. In general, these results corroborate our immunohistochemical data suggesting that S-nitrosylation is increased in the light-adapted mammalian retina. In order to remain consistent and systematic with our approach used for interpreting our immunohistochemical results, we treated the dark-adapted MS data as providing insight into baseline S-nitrosylated proteins. In other words, we assumed that the constitutive, baseline SNI observed in the dark-adapted mouse retina (Fig. 2E and F) was (at least partly) originating from proteins which were determined by MS to be S-nitrosylated only in the dark-adapted retina. Consequently, we focused our attention on the 351 proteins that were identified as unique to the light-adapted retina. A comprehensive list of all the identified proteins can be found in Table 2.
Importantly, we detected several nitrosylated proteins in the mammalian retina that have also been detected and confirmed to be subject of S-nitrosylation, in other systems. For example, we detected channels and transporters such as: Na+/K+-ATPase, Ca2+-ATPase, and cGMP-gated cation channel (α1); enzymes such as: creatine kinase, protein kinase C, low mw phosphatase, and aldehyde dehydrogenase; redox enzymes such as: thioredoxin and glutathione peroxidase (Stamler et al., 2001; Hess et al., 2005). Due to the importance of information transfer across layers of the retina, proteins that are involved in the mechanism and regulation of exocytosis are particularly noteworthy. Based on our list of putative S-nitrosylated proteins from the light-adapted retina, we identified several proteins which play unique roles in modulating exocytosis. For example, synaptic vesicle glycoprotein, syntaxin-1B, Ras-related protein Rab-3C, excitatory amino acid transporter 1, and protein lin-7 homolog A are all involved in specific aspects of synaptic transmission and are subjected to light evoked S-nitrosylation in the mammalian retina. Syntaxin-1A and B share a conserved primary nitrosylation site at Cys (145) which, upon nitrosylation, alters the quantal size of neurotransmitter release (Palmer et al., 2008). In our experimental design and sample preparation, we did not attempt to isolate proteins with specific subcellular localization as we were not concerned with identifying any particular S-nitrosylated protein. Therefore, this approach may have hindered the identification of particular membrane bound proteins that would require a specific sample preparation for complete extraction. Nonetheless, we were able to identify several proteins known to localize to the membrane (e.g., Na+- and Cl−-dependent GABA transporter 1&3, excitatory amino acid transporter 1, E3 ubiquitin ligase, and CaM kinase-like vesicle-associated protein). Additionally, our proteomics approach identified the voltage-gated potassium channel subunit Kv 2.1, which is a significant component of the delayed rectifier K+ current in neurons (Murakoshi and Trimmer, 1999). In the mouse retina, Kv2.1 has been localized to rod-photoreceptors and horizontal cells (Klumpp et al., 1995; Pinto and Klumpp, 1998), cholinergic and dopaminergic ACs (Tian et al., 2005) in the INL and GCs in the GCL (Pinto and Klumpp, 1998). In the goldfish retina, Kv2.1 was observed from the ONL to the GCL with confirmed localization to cone inner segments, horizontal cells, Mbs, ACs and GCs (Yazulla and Studholme, 1998). Functionally, reduction of cysteine sulfhydryl groups on the N-terminal domain of Kv2.1, in vitro, resulted in sluggish channel gating and prolonged latency to channel opening (Pascual et al., 1997). However, it remains to be determined whether or not S-nitrosylation of retinal Kv2.1 results in a functionally relevant modulation of retinal information processing.
Discussion
Despite of the large number of proteins that have been identified as potential targets of S-nitrosylation by NO, there is little evidence of S-nitrosylation taking place in neural tissue under physiological conditions. The present immunohistochemical investigation was undertaken to determine if S-nitrosylation, mediated by endogenous NO, takes place under normal, physiologically relevant illumination conditions in the adult goldfish and mouse retina. The use of various light stimulation intensities, pharmacological tools and a transgenic mouse line allowed us to describe the light-dependent nature of S-nitrosylation in vertebrate retinas for the first time. Additionally, our global proteomics approach provided qualitative identification of retinal proteins that are subject to light-dependent S-nitrosylation.
Focal S-nitrosylation occurs under dim, scotopic light conditions in the retina
Over the past several decades, NO has garnered more and more attention as an important neurochemical signaling molecule responsible for modulating light responses at multiple levels of the retinal circuit. In the vertebrate retina, the synthesis and release of NO is highly coordinated with light stimulation: light stimulation has been shown to significantly increase NO in the retinas of a number of species including carp (Sekaran et al., 2005), salamander and turtle (Eldred and Blute, 2005), rabbit (Neal et al., 1998) and miniature pig (Donati et al., 1995). Synthesis and release of NO occurs at basal levels in the dark-adapted retina and is elevated in light-adapted retinas (Sekaran et al., 2005; Walter et al., 2014). Of the three NOS isoforms in the retina, the role of the endothelial type (eNOS), in mediating visual responses is unknown (Haverkamp et al., 1999; Tekmen-Clark and Gleason, 2013). The inducible (iNOS) isoform contributes to increased NO synthesis under extremely bright illumination (Palamalai et al., 2006; Piehl et al., 2007) and mediates pathological conditions, possibly through light-dependent protein nitration (Miyagi et al., 2002). Strong evidence suggests that light-evoked NO release under physiological intensities is primarily mediated by the constitutive, calcium-dependent isoform, nNOS (Blute et al., 1997; Blom et al., 2009; Giove et al., 2009). There are some differences in the expression pattern of nNOS across vertebrate species, however, nNOS expressing neurons are mostly localized to the inner retina, sending processes across the IPL in both mouse (Haverkamp and Wassle, 2000; Pang et al., 2010; Zhu et al., 2014) and goldfish (Villani and Guarnieri, 1996). This is consistent with the fact that in this study, the dimmest illumination (2.4×108 photons/cm2/s, 505 nm, for 500 ms) to trigger a detectable increase of SNI, only did so in the IPL and in the GCL in both species (Figs. 3 and 4). In our hands, lower intensities of illumination produced SNI that was indistinguishable from that seen in fully dark-adapted retinas (data not shown) suggesting that the intensity threshold for producing S-nitrosylation, detectable via immunohistochemistry, is ~2.4×108 photons/cm2/s. This intensity is the same that reliably caused S-nitrosylation dependent modulation of voltage-gated calcium currents (ICas) in goldfish Mb terminals (Tooker et al., 2013). Although the molecular target of S-nitrosylation that is responsible for ICa modulation in Mbs has not been identified, it is noteworthy that in the current study, we found light-evoked SNI clearly outlining the membranes of Mb terminals, among other structures.
As a gaseous, freely diffusible neuromodulator, the instantaneous concentration of NO in retinal tissue is inversely proportional to the distance from the source: when measured ~10 μm away from an NO producing GC with an intracellular NO concentration of ~0.2 μM, the NO concentration dropped to 10 nM (Eldred and Blute, 2005). S-nitrosylation of protein cysteine groups is thought to require more NO than is necessary for the activation of sGC (Ahern et al., 2002), thus, SNI is also expected to be restricted to regions that are in close proximity to NO production at threshold light intensities. Furthermore, some cysteine thiol side-chains have a very high affinity for reacting with NO, based on their surface availability and flanking amino acid sequence (Stamler et al., 1997), adding a level of specificity to S-nitrosylation mediated signaling.
In the mouse retina, the light sensitivity of nNOS expressing (amacrine) cells (ACs) was determined to be ~105 photons/cm2/s (Pang et al., 2010). These data indicate that nNOS expressing ACs receive rod input. In our hands, light simulation below the intensity of ~108 photons/cm2/s did not generate SNI in either mouse or goldfish retina. Furthermore, ~108 photons/cm2/s was found to produce NO-mediated functional changes via S-nitrosylation in Mb signaling based on electrophysiological evidence obtained in retinal slice preparation (Tooker et al., 2013). However, on one hand, it is important to point out that both slice electrophysiology and immunohistochemistry may overestimate the light threshold for endogenous S-nitrosocysteine formation: (1) in slice, some NO might quickly diffuse to the bath solution without exerting a measurable effect on the recorded cell; (2) our ability to detect subtle light-evoked increases in S-nitrosocysteine might be limited by the sensitivity of our antibody. On the other hand, it is also possible that NO production by nNOS+ amacrine cells in the mouse at threshold intensities (~105 photons/cm2/s) is sufficient to activate the most sensitive NO target, sGC, (activated by nanomoles of NO; Roy et al., 2008), but still be less than the levels required for S-nitrosylation (Ahern et al., 2002). Therefore, the discrepancy between the activation threshold of nNOS+ amacrine cells (~105 photons/cm2/s) and our observed threshold for SNI (~108 photons/cm2/s) may be a combinatorial effect of the light intensity-dependent nature of NO synthesis and release (Sekaran et al., 2005) and the relatively high levels of NO needed to form nitrosothiols (as compared to activating sGC, Ahern et al., 2002). To date, neither NO release, nor NO-mediated effects via the sGC pathway have been reported to be triggered in the retina by ~105 photons/cm2/s.
Due to the asymmetric excitatory input resulting in larger OFF excitation of nNOS expressing ACs, maximal NO production is expected to be most profoundly activated by bright, flickering light with frequency of 1–2 Hz (Pang et al., 2010). We have not tested the effect flickering light stimulation on retinal S-nitrosocysteine formation. Nonetheless, we could not increase SNI further than what was detected in light-adapted retinas (Figs. 1 and 2) via illumination with 1013 photons/cm2/s (λ=505 nm or 660 nm, for various durations up to 10 min) or by application of the NO donor DETA/NO (1 mM for 1 hour) in either light-adapted or dark-adapted retinas (data not shown). Taking it together, it seems difficult to determine how accurate the threshold value of 108 photons/cm2/s might be for endogenous, light-evoked S-nitrosocysteine formation.
NO signaling in processing visual information under physiological and pathological conditions
In general, much of the fundamental work describing the effects of NO in the retina focused on the traditional sGC→cGMP-dependent signaling cascade. NO has been shown to influence transmission from cones (Savchenko et al., 1997), as well as the signaling of bipolar cells (BCs; Shiells and Falk, 2002; Snellman and Nawy, 2004). The most notable effect of NO in the retina is the uncoupling of gap junctions between horizontal cells (DeVries and Schwartz, 1989; Lu and McMahon, 1997; Daniels and Baldridge, 2011) and between AII ACs and BCs (Mills and Massey, 1995).
However, evidence has shown that the classical cGMP-dependent pathway is not the exclusive route of action for NO and that modulation of cellular function can occur alternatively through S-nitrosylation reactions in the retina. For instance, exogenous NO amplified the a- and b-wave amplitude in rat electroretinograms through a cGMP-independent mechanism (Vielma et al., 2010), and recently, we reported a mechanism in which endogenous NO enhanced the sensitivity of BCs via S-nitrosylation (Tooker et al., 2013). In line with these results is the observation that GCs in nNOS KO mice show a significant deficit in their sensitivity to light stimulation compared to wild-type animals (Wang et al., 2007); however, a mechanism underlying the NO-mediated GC sensitivity was not determined.
Based on the results presented in this study, it appears that S-nitrosylation is a large scale, post-translational modification underlying retinal processes under physiologically relevant light conditions, affecting hundreds of proteins (Table 2). However, a global approach such as this most likely underestimates the actual number of retinal proteins undergoing light-dependent S-nitrosylation as it focused on the most abundant S-nitrosylated proteins in the retinal homogenates and used the most conservative proteomics approach to identify them. This might explain why certain proteins that have been implicated to undergo S-nitrosylation, such as the NR2 subunits of NMDA receptors (Choi et al., 2000) or BK calcium-dependent potassium channels (Bolotina et al., 1994) did not show up in our list of nitrosylated proteins, although they are expressed in the mouse retina (Tanimoto et al., 2012).
It is important to point out, however, that while our immunohisthochemical method and global proteomic analysis appears appropriate to detect the presence of S-nitrosylated cysteine groups on certain proteins, the resulted SNI pattern and protein identification has to be treated with caution: it cannot be interpreted as a clear indicator of S-nitrosylation evoked modulation in protein function as S-nitrosylation of any protein might not be associated with structural/functional changes. In fact, only a few examples exist where the S-nitrosylation triggered molecular mechanisms were fully elucidated (Palmer et al., 2008; Choi et al., 2000). Even if S-nitrosylation of a cysteine group does not result in a change in synaptic processing of visual function in every instance, it could serve to buffer the retinal NO and restrict its free diffusion (Wood and Garthwaite, 1994; Eldred and Blute, 2005).
Although we did not specifically direct our proteomics approach to identify a selected set of S-nitrosylated proteins, we observed an interesting set of data that corroborates a previous supposition by Kurenny et al. (1994) regarding the modulation of rod photoreceptors via NO through a cGMP-independent manner. In their report, Kurenny et al. (1994) described an increase in a non-voltage-dependent current in rod photoreceptor cells after application of NO donors, and it was proposed that NO may be modulating cGMP channels through S-nitrosylation (Kurenny et al., 1994). Along these lines, we identified the rod specific cGMP-gated cation channel α1 subunit and β1 subunit in our proteomic analysis of proteins that are S-nitrosylated in a light dependent manner (Table 2).
S-nitrosylation remains a hallmark characteristic of neurodegenerative diseases in the nervous system (Nakamura et al., 2013) as well as in the retina. When rats were given an intravitreal injection of NMDA and glycine, mimicking excitotoxic conditions often accompanying neurodegenerative diseases, retinal S-nitrosylation dramatically increased and was correlated with GC death (Manabe et al., 2005). In neurodegenerative disease, iNOS can be induced and contribute to the production of toxic levels of NO (Nakamura et al., 2013). Retinal neuronal cell death has been correlated directly to the elevated levels of NO after iNOS activation in Müller cells, under pathological conditions, and apoptosis was significantly reduced in iNOS KO animals (Goureau et al., 1999; Sennlaub et al., 2002). Interestingly, NO induced apoptosis of retinal neurons in a cGMP-independent manner, and was instead, attributed to protein nitration (Sennlaub et al., 2002), which is strong evidence implicating aberrant S-nitrosylation as the underlying mechanism.
The results presented here indicate that although aberrant S-nitrosylation may be contributing to neuronal death in certain retinopathies, in the healthy eye, S-nitrosylation can be regulated by physiologically relevant stimuli and may be important in normal visual function, for example, in regulating the sensitivity of GCs via BC output. Importantly, we provided insight into the identification of protein targets susceptible to light-dependent S-nitrosylation. These results provide the framework for further exploration into the functional implications of endogenous S-nitrosylation in modulating visual information processing in the healthy and diseased retina.
Through immunohistochemistry and qualitative proteomics, the authors demonstrate that lightdependent NO production leads to extensive S-nitrosylation in the vertebrate retina. These findings expand the role of S-nitrosylation beyond neurodegenerative diseases and strongly suggest that this process is a significant post-translational modification affecting an array of proteins under physiological conditions.
Through immunohistochemistry and qualitative proteomics, the authors demonstrate that lightdependent NO production leads to extensive S-nitrosylation in the vertebrate retina. These findings expand the role of S-nitrosylation beyond neurodegenerative diseases and strongly suggest that this process is a significant post-translational modification affecting an array of proteins under physiological conditions.
Acknowledgments
We wish to thank Lisa Wolfe and the Colorado State University Proteomics and Metabolomics Facility for technical expertise and mass spectrometry analysis of samples. We thank Paul Witkovsky for critical reading of the manuscripts and we also thank Dorothy Kociuba and Shannon Gallagher for technical assistance with immunohistochemistry.
Support: This work was supported in part by a grant from NIH R01 EY019051 (JV) and by the John H. Venable memorial scholarship (RET).
Footnotes
Associate Editor: Dr. Ian Meinertzhagen, Dalhousie University, Retina
Conflict of Interest Statement
The authors declare no conflict of interest.
Role of Authors
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: R.E.T. and J.V. Acquisition of data: R.E.T. Statistical analysis: R.E.T. Analysis and interpretation of data: R.E.T. and J.V. Drafting of the manuscript: R.E.T. and J.V. Obtained funding: R.E.T. and J.V. Study supervision: J.V.
Literature Cited
- Ahern GP, Klyachko VA, Jackson MB. cGMP and S-nitrosylation: two routes for modulation of neuronal excitability by NO. Trends Neurosci. 2002;25:510–517. doi: 10.1016/s0166-2236(02)02254-3. [DOI] [PubMed] [Google Scholar]
- Bing YE, Du JL, Yang XL. Differential effects of nitric oxide on rod and cone pathways in carp retina. Sci China C Life Sci. 1997;40:71–78. doi: 10.1007/BF02879109. [DOI] [PubMed] [Google Scholar]
- Blom JJ, Blute TA, Eldred WD. Functional localization of the nitric oxide/cGMP pathway in the salamander retina. Vis Neurosci. 2009;26:275–286. doi: 10.1017/S0952523809990125. [DOI] [PubMed] [Google Scholar]
- Blute TA, Mayer B, Eldred WD. Immunocytochemical and histochemical localization of nitric oxide synthase in the turtle retina. Vis Neurosci. 1997;14:717–729. doi: 10.1017/s0952523800012670. [DOI] [PubMed] [Google Scholar]
- Bolotina VM, Najibi S, Palacino JJ, Pagano PJ, Cohen RA. Nitric-oxide directly activates calcium-dependent potassium channels in vascular smooth-muscle. Nature. 1994;368:850–853. doi: 10.1038/368850a0. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc-Oxford. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Busskamp V, Duebel J, Balya D, Fradot M, Viney TJ, Siegert S, Groner AC, Cabuy E, Forster V, Seeliger M, Biel M, Humphries P, Paques M, Mohand-Said S, Trono D, Deisseroth K, Sahel JA, Picaud S, Roska B. Genetic Reactivation of Cone Photoreceptors Restores Visual Responses in Retinitis Pigmentosa. Science. 2010;329:413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Lekontseva O, Peters A, Davidge ST. 17 beta-Estradiol induces protein S-nitrosylation in the endothelium. Cardiovasc Res. 2010;85:796–805. doi: 10.1093/cvr/cvp368. [DOI] [PubMed] [Google Scholar]
- Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HSV, Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li XJ, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of Parkin regulates ubiquitination and compromises Parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Daniels BA, Baldridge WH. The light-induced reduction of horizontal cell receptive field size in the goldfish retina involves nitric oxide. Vis Neurosci. 2011;28:137–144. doi: 10.1017/S0952523810000490. [DOI] [PubMed] [Google Scholar]
- Devries SH, Schwartz EA. Modulation of an electrical synapse between solitary pairs of catfish horizontal cells by dopamine and 2nd messengers. J Physiol. 1989;414:351–375. doi: 10.1113/jphysiol.1989.sp017692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicks AP, Williams DLH. Generation of nitric oxide from S-nitrosothiols using protein-bound Cu2+ sources. Chem Biol. 1996;3:655–659. doi: 10.1016/s1074-5521(96)90133-7. [DOI] [PubMed] [Google Scholar]
- Donati G, Pournaras CJ, Munoz JL, Poitry S, Poitryyamate CL, Tsacopoulos M. Nitric-oxide controls arteriolar tone in the retina of the miniature pig. Invest Ophth Vis Sci. 1995;36:2228–2237. [PubMed] [Google Scholar]
- Eldred WD, Blute TA. Imaging of nitric oxide in the retina. Vision Res. 2005;45:3469–3486. doi: 10.1016/j.visres.2005.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- Gallagher SK, Witkovsky P, Roux MJ, Low MJ, Otero-Corchon V, Hentges ST, Vigh J. beta-Endorphin Expression in the Mouse Retina. J Comp Neurol. 2010;518:3130–3148. doi: 10.1002/cne.22387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giove TJ, Deshpande MM, Eldred WD. Identification of Alternate Transcripts of Neuronal Nitric Oxide Synthase in the Mouse Retina. J Neurosci Res. 2009;87:3134–3142. doi: 10.1002/jnr.22133. [DOI] [PubMed] [Google Scholar]
- Goureau O, Regnier-Ricard F, Courtois Y. Requirement for nitric oxide in retinal neuronal cell death induced by activated Muller glial cells. J Neurochem. 1999;72:2506–2515. doi: 10.1046/j.1471-4159.1999.0722506.x. [DOI] [PubMed] [Google Scholar]
- Greferath U, Grunert U, Wassle H. Rod bipolar cells in the mammalian retina show protein kinase-c-like immunoreactivity. J Comp Neurol. 1990;301:433–442. doi: 10.1002/cne.903010308. [DOI] [PubMed] [Google Scholar]
- Gyurko R, Leupen S, Huang PL. Deletion of exon 6 of the neuronal nitric oxide synthase gene in mice results in hypogonadism and infertility. Endocrinology. 2002;143:2767–2774. doi: 10.1210/endo.143.7.8921. [DOI] [PubMed] [Google Scholar]
- Handy RL, Harb HL, Wallace P, Gaffen Z, Whitehead KJ, Moore PK. Inhibition of nitric oxide synthase by 1-(2-trifluoromethylphenyl) imidazole (TRIM) in vitro: Antinociceptive and cardiovascular effects. Brit J Pharmacol. 1996;119:423–431. doi: 10.1111/j.1476-5381.1996.tb16003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham N, Dachtler J, Fox K. The role of nitric oxide in pre-synaptic plasticity and homeostasis. Front Cell Neurosci. 2013;7 doi: 10.3389/fncel.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Kolb H, Cuenca N. Endothelial nitric oxide synthase (eNOS) is localized to Muller cells in all vertebrate retinas. Vis Res. 1999;39:2299–2303. doi: 10.1016/s0042-6989(99)00007-3. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: Purview and parameters. Nat Rev Mol Cell Bio. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Hirooka K, Kourennyi DE, Barnes S. Calcium channel activation facilitated by nitric oxide in retinal ganglion cells. J Neurophys. 2000;83:198–206. doi: 10.1152/jn.2000.83.1.198. [DOI] [PubMed] [Google Scholar]
- Hoffpauir B, McMains E, Gleason E. Nitric oxide transiently converts synaptic inhibition to excitation in retinal amacrine cells. J Neurophys. 2006;95:2866–2877. doi: 10.1152/jn.01317.2005. [DOI] [PubMed] [Google Scholar]
- Hu S-Q, Ye J-S, Zong Y-Y, Sun C-C, Liu D-H, Wu Y-P, Song T, Zhang G-Y. S-Nitrosylation of Mixed Lineage Kinase 3 Contributes to Its Activation after Cerebral Ischemia. J Bio Chem. 2012;287:2364–2377. doi: 10.1074/jbc.M111.227124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Bio. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Joselevitch C, Kamermans M. Interaction between rod and cone inputs in mixed-input bipolar cells in goldfish retina. J Neurosci Res. 2007;85:1579–1591. doi: 10.1002/jnr.21249. [DOI] [PubMed] [Google Scholar]
- Joselevitch C, Kamermans M. Retinal parallel pathways: Seeing with our inner fish. Vision Res. 2009;49:943–959. doi: 10.1016/j.visres.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Kawai F, Sterling P. cGMP modulates spike responses of retinal ganglion cells via a cGMP-gated current. Vis Neurosci. 2002;19:373–380. [PubMed] [Google Scholar]
- Klumpp DJ, Song EJ, Ito S, Sheng MH, Jan LY, Pinto LH. The shaker-like potassium channels of the mouse rod bipolar cell and their contributions to the membrane current. J Neurosci. 1995;15:5004–5013. doi: 10.1523/JNEUROSCI.15-07-05004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles RG, Moncada S. Nitric-oxide synthases in mammals. Biochem J. 1994;298:249–258. doi: 10.1042/bj2980249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizaj D. Mesopic state: Cellular mechanisms involved in pre- and post-synaptic mixing of rod and cone signals. Microsc Res and Tech. 2000;50:347–359. doi: 10.1002/1097-0029(20000901)50:5<347::AID-JEMT4>3.0.CO;2-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurenny DE, Moroz LL, Turner RW, Sharkey KA, Barnes S. Modulation of ion channels in rod photoreceptors by nitric-oxide. Neuron. 1994;13:315–324. doi: 10.1016/0896-6273(94)90349-2. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lee JR, Kim YH, Park YS, Park SI, Park HS, Kim KP. Investigation of tyrosine nitration and nitrosylation of angiotensin II and bovine serum albumin with electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:2797–2804. doi: 10.1002/rcm.3145. [DOI] [PubMed] [Google Scholar]
- Liepe BA, Stone C, Koistinaho J, Copenhagen DR. Nitric-oxide synthase in müller cells and neurons of salamander and fish retina. J Neurosci. 1994;14:7641–7654. doi: 10.1523/JNEUROSCI.14-12-07641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CB, McMahon DG. Modulation of hybrid bass retinal gap junctional channel gating by nitric oxide. J Physiol. 1997;499:689–699. doi: 10.1113/jphysiol.1997.sp021961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe S, Gu ZZ, Lipton SA. Activation of matrix metalloproteinase-9 via neuronal nitric oxide synthase contributes to NMDA-induced retinal ganglion cell death. Invest Ophth Vis Sci. 2005;46:4747–4753. doi: 10.1167/iovs.05-0128. [DOI] [PubMed] [Google Scholar]
- McMahon DG, Ponomareva LV. Nitric oxide and cGMP modulate retinal glutamate receptors. J Neurophys. 1996;76:2307–2315. doi: 10.1152/jn.1996.76.4.2307. [DOI] [PubMed] [Google Scholar]
- Mills SL, Massey SC. Differential properties of 2 gap junctional pathways made by AII amacrine cells. Nature. 1995;377:734–737. doi: 10.1038/377734a0. [DOI] [PubMed] [Google Scholar]
- Miyagi M, Sakaguchi H, Darrow RM, Yan L, West KA, Aulak KS, Stuehr DJ, Hollyfield JG, Organisciak DT, Crabb JW. Evidence that light modulates protein nitration in rat retina. Mol Cell Proteomics. 2002;1:293–303. doi: 10.1074/mcp.m100034-mcp200. [DOI] [PubMed] [Google Scholar]
- Murakoshi H, Trimmer JS. Identification of the Kv2.1 K+ channel as a major component of the delayed rectifier K+ current in rat hippocampal neurons. J Neurosci. 1999;19:1728–1735. doi: 10.1523/JNEUROSCI.19-05-01728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Tu SC, Akhtar MW, Sunico CR, Okamoto SI, Lipton SA. Aberrant Protein S-Nitrosylation in Neurodegenerative Diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal M, Cunningham J, Matthews K. Selective release of nitric oxide from retinal amacrine and bipolar cells. Invest Ophth Vis Sci. 1998;39:850–853. [PubMed] [Google Scholar]
- Negishi K, Kato S, Teranishi T. Dopamine cells and rod bipolar cells contain protein-kinase c-like immunoreactivity in some vertebrate retinas. Neurosci Lett. 1988;94:247–252. doi: 10.1016/0304-3940(88)90025-0. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol Reactivity Profiling Identifies S-Nitrosylated Proteins with Unexpected Stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamalai V, Darrow R, Organisciak DT, Miyagi M. Light-induced changes in protein nitration in photoreceptor rod outer segments. Mol Vis. 2006;12:1543–1551. [PubMed] [Google Scholar]
- Palmer ZJ, Duncan RR, Johnson JR, Lian LY, Mello LV, Booth D, Barclay JW, Graham ME, Burgoyne RD, Prior IA, Morgan A. S-nitrosylation of syntaxin 1 at Cys(145) is a regulatory switch controlling Munc18-1 binding. Biochem J. 2008;413:479–491. doi: 10.1042/BJ20080069. [DOI] [PubMed] [Google Scholar]
- Pang JJ, Gao F, Wu SM. Light Responses and Morphology of bNOS-Immunoreactive Neurons in the Mouse Retina. J Comp Neurol. 2010;518:2456–2474. doi: 10.1002/cne.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual JM, Shieh CC, Kirsch GE, Brown AM. Contribution of the NH2 terminus of Kv2.1 to channel activation. Am J Phys-Cell Phys. 1997;273:C1849–C1858. doi: 10.1152/ajpcell.1997.273.6.C1849. [DOI] [PubMed] [Google Scholar]
- Piehl L, Capani F, Facorro G, Lopez EM, de Celis ER, Pustovrh C, Hager A, Coirini H, Lopez-Costa JJ. Nitric oxide increases in the rat retina after continuous illumination. Brain Res. 2007;1156:112–119. doi: 10.1016/j.brainres.2007.04.030. [DOI] [PubMed] [Google Scholar]
- Pinto LH, Klumpp DJ. Localization of potassium channels in the retina. Prog Retinal Eye Res. 1998;17:207–230. doi: 10.1016/s1350-9462(97)00011-6. [DOI] [PubMed] [Google Scholar]
- Qu J, Nakamura T, Cao G, Holland EA, McKercher SR, Lipton SA. S-Nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by beta-amyloid peptide. P Natl Acad Sci USA. 2011;108:14330–14335. doi: 10.1073/pnas.1105172108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi-George A, Gow AJ. Immunofluorescent detection of S-nitrosoproteins in cell culture. Methods. 2013;62:161–164. doi: 10.1016/j.ymeth.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Halvey EJ, Garthwaite J. An enzyme-linked receptor mechanism for nitric oxide-activated guanylyl cyclase. J Biol Chem. 2008;283:18841–18851. doi: 10.1074/jbc.M801712200. [DOI] [PubMed] [Google Scholar]
- Savchenko A, Barnes S, Kramer RH. Cyclic-nucleotide-gated channels mediate synaptic feedback by nitric oxide. Nature. 1997;390:694–698. doi: 10.1038/37803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer KL, Freund DM, Prenni JE, Curthoys NP. Proteomic profiling and pathway analysis of the response of rat renal proximal convoluted tubules to metabolic acidosis. Am J Phys-Renal Phys. 2013;305:F628–F640. doi: 10.1152/ajprenal.00210.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekaran S, Cunningham J, Neal MJ, Hartell NA, Djamgoz MBA. Nitric oxide release is induced by dopamine during illumination of the carp retina: serial neurochemical control of light adaptation. Eur J Neurosci. 2005;21:2199–2208. doi: 10.1111/j.1460-9568.2005.04051.x. [DOI] [PubMed] [Google Scholar]
- Sennlaub F, Courtois Y, Goureau O. Inducible nitric oxide synthase mediates retinal apoptosis in ischemic proliferative retinopathy. J Neurosci. 2002;22:3987–3993. doi: 10.1523/JNEUROSCI.22-10-03987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiells RA, Falk G. Potentiation of ‘on’ bipolar cell flash responses by dim background light and cGMP in dogfish retinal slices. J Physiol. 2002;542:211–220. doi: 10.1113/jphysiol.2002.019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DG, Nagamatsu A, Fruton JS. Some reactions of N-Ethylmaleimide. J Am Chem Soc. 1960;82:4600–4604. [Google Scholar]
- Snellman J, Nawy S. cGMP-dependent kinase regulates response sensitivity of the mouse On bipolar cell. J Neurosci. 2004;24:6621–6628. doi: 10.1523/JNEUROSCI.1474-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS. Redox signaling – nitrosylation and related target interactions of nitric-oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Lamas S, Fang FC. Nitrosylation: The prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: Translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Kaneko A. Identification of bipolar cell subtypes by protein-kinase c-like immunoreactivity in the goldfish retina. Vis Neurosci. 1990;5:223–230. doi: 10.1017/s0952523800000298. [DOI] [PubMed] [Google Scholar]
- Tanimoto N, Sothilingam V, Euler T, Ruth P, Seeliger MW, Schubert T. BK Channels Mediate Pathway-Specific Modulation of Visual Signals in the In Vivo Mouse Retina. J Neurosci. 2012;32:4861–4866. doi: 10.1523/JNEUROSCI.4654-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekmen-Clark M, Gleason E. Nitric oxide production and the expression of two nitric oxide synthases in the avian retina. Vis Neurosci. 2013;30:91–103. doi: 10.1017/S0952523813000126. [DOI] [PubMed] [Google Scholar]
- Tian M, Zhao JW, Yang XL, Xie JX. Voltage-gated K+ channel subunits on cholinergic and dopaminergic amacrine cells. Neuroreport. 2003;14:1763–1766. doi: 10.1097/00001756-200310060-00001. [DOI] [PubMed] [Google Scholar]
- Tooker RE, Lipin MY, Leuranguer V, Rozsa E, Bramley JR, Harding JL, Reynolds MM, Vigh J. Nitric Oxide Mediates Activity-Dependent Plasticity of Retinal Bipolar Cell Output via S-Nitrosylation. J Neurosci. 2013;33:19176–19193. doi: 10.1523/JNEUROSCI.2792-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielma AH, Delgado L, Elgueta C, Osorio R, Palacios AG, Schmachtenberg O. Nitric oxide amplifies the rat electroretinogram. Exp Eye Res. 2010;91:700–709. doi: 10.1016/j.exer.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Vielma AH, Retamal MA, Schmachtenberg O. Nitric oxide signaling in the retina: What have we learned in two decades? Brain Res. 2012;1430:112–125. doi: 10.1016/j.brainres.2011.10.045. [DOI] [PubMed] [Google Scholar]
- Vigh J, Vickers E, von Gersdorff H. Light-Evoked Lateral GABAergic Inhibition at Single Bipolar Cell Synaptic Terminals Is Driven by Distinct Retinal Microcircuits. J Neurosci. 2011;31:15884–15893. doi: 10.1523/JNEUROSCI.2959-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani L, Guarnieri T. Localization of nitric oxide synthase in the goldfish retina. Brain Res. 1996;743:353–356. doi: 10.1016/s0006-8993(96)01103-1. [DOI] [PubMed] [Google Scholar]
- Walter LT, Higa GS, Schmeltzer C, Sousa E, Kinjo ER, Rudiger S, Hamassaki DE, Cerchiaro G, Kihara AH. Functional regulation of neuronal nitric oxide synthase expression and activity in the rat retina. Exp Neurol. 2014;261:510–517. doi: 10.1016/j.expneurol.2014.07.019. [DOI] [PubMed] [Google Scholar]
- Wang GY, Van der List DA, Nemargut JP, Coombs JL, Chalupa LM. The sensitivity of light-evoked responses of retinal ganglion cells is decreased in nitric oxide synthase gene knockout mice. J Vision. 2007;7:1–13. doi: 10.1167/7.14.7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu T, Wu C, Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1353–1360. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J, Garthwaite J. Models of the diffusional spread of nitric-oxide – implications for neural nitric-oxide signaling and its pharmacological properties. Neuropharmacology. 1994;33:1235–1244. doi: 10.1016/0028-3908(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Wu SM. Synaptic transmission in the outer retina. Annu Rev Physiol. 1994;56:141–168. doi: 10.1146/annurev.ph.56.030194.001041. [DOI] [PubMed] [Google Scholar]
- Xin DY, Bloomfield SA. Comparison of the responses of AII amacrine cells in the dark-and light-adapted rabbit retina. Vis Neurosci. 1999;16:653–665. doi: 10.1017/s0952523899164058. [DOI] [PubMed] [Google Scholar]
- Yazulla S, Studholme KM. Light-dependent plasticity of the synaptic terminals of Mb bipolar cells in goldfish retina. J Comp Neurol. 1992;320:521–530. doi: 10.1002/cne.903200409. [DOI] [PubMed] [Google Scholar]
- Yazulla S, Studholme KM. Differential distribution of Shaker-like and Shab-like K+-channel subunits in goldfish retina and retinal bipolar cells. J Comp Neurol. 1998;396:131–140. doi: 10.1002/(sici)1096-9861(19980622)396:1<131::aid-cne10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Yu D, Eldred WD. Nitric oxide stimulates gamma-aminobutyric acid release and inhibits glycine release in retina. J Comp Neurol. 2005;483:278–291. doi: 10.1002/cne.20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Xu J, Hauswirth WW, DeVries SH. Genetically Targeted Binary Labeling of Retinal Neurons. J Neurosci. 2014;34:7845–7861. doi: 10.1523/JNEUROSCI.2960-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]


