Abstract
Background
Circulating Epstein-Barr virus DNA is a predictor of recurrence in patients with nasopharyngeal carcinoma. Circulating human papillomavirus (HPV) DNA has been detected in the sera of some patients with HPV-positive squamous cell carcinoma of the oropharynx (OPC). The goal of this study was to determine whether pretreatment serum HPV DNA is a useful biomarker for recurrence in patients with HPV-positive OPC.
Methods
The study included patients with newly diagnosed, previously untreated OPC. Tumor HPV status was determined by polymerase chain reaction (PCR); serum HPV DNA was detected using real-time PCR. Differences in clinical characteristics between patients positive and negative for pretreatment serum HPV DNA were described using standard descriptive statistical methods. Kaplan-Meier curves were generated and log-rank tests were used to detect significant differences in progression-free survival.
Results
A total of 262 patients were included. Patients with high N category and those with stage IV disease had higher rates of detectable pretreatment serum HPV DNA. Patients with HPV-positive tumors had better progression-free survival than patients with HPV-negative tumors. Among patients with HPV-positive tumors, those who were negative for pretreatment serum HPV DNA had better progression-free survival than those who were positive for pretreatment serum HPV DNA, but this result was not statistically significant.
Conclusions
Pretreatment serum HPV DNA was associated with higher N category and overall disease stage. However, pretreatment serum HPV DNA does not appear to have clinical utility as a marker for recurrence among patients with OPC.
Keywords: Human papillomavirus, oropharyngeal cancer, head and neck neoplasms, serum HPV DNA, survival
Introduction
Human papillomavirus (HPV) is an important cause of several cancers, including cervical cancer and squamous cell carcinoma of the oropharynx (OPC); most OPC tumors diagnosed today test positive for HPV DNA.1–5 Patients with HPV-positive tumors tend to be younger and have better prognoses than patients with HPV-negative tumors.6–8 However, this survival advantage does not appear to extend to patients who develop local-regional recurrence or distant metastatic disease. Biomarkers are needed that can identify patients with HPV-positive tumors at increased risk for these adverse outcomes.
Viral DNA has been detected in the serum of patients with virally induced tumors; for example, HPV DNA has been detected in the serum of patients with OPC and cervical cancer, hepatitis B virus DNA in the serum of patients with hepatocellular carcinoma, and Epstein-Barr virus (EBV) DNA in the serum of patients with Hodgkin disease, Burkitt lymphoma, and nasopharyngeal carcinoma (NPC).9 Higher levels of circulating EBV DNA are associated with the clinical stage of NPC and are an independent prognostic factor for recurrence and metastasis in patients with this disease.10–16 This biomarker for NPC outcomes is now being tested in a prospective multi-institutional phase III clinical trial sponsored by NRG Oncology in which serum EBV DNA levels are incorporated into the treatment decision paradigm.
To date, few studies have investigated the usefulness of circulating HPV DNA as a biomarker in cervical cancer and HPV-related OPC. However, the promising results from studies of circulating EBV DNA in patients with NPC suggest that serum HPV DNA may be a strong marker for disease extent as well as recurrence and distant metastasis in patients with HPV-related OPC. The goal of the study reported here was to determine whether pretreatment level of serum HPV DNA is a useful biomarker for recurrence in patients with HPV-positive OPC.
Materials and Methods
Patient population
This was a prospective cohort study nested within a large ongoing case-control study of head and neck cancer conducted at The University of Texas MD Anderson Cancer Center. Newly diagnosed, previously untreated patients with histologically confirmed OPC (squamous cell carcinoma of the tonsils, base of the tongue, soft palate, or oropharyngeal wall) who presented to the institution during the period from January 2006 through September 2009 were eligible for inclusion in the study. Patients with known distant metastases or immune suppression were not eligible for inclusion. At recruitment (presentation to the institution), all patients completed an Institutional Review Board-approved consent form, completed a standardized questionnaire including demographic characteristics and exposure information (including exposure to tobacco and alcohol); and donated a blood sample for molecular epidemiological testing. Patients had regularly scheduled clinical and radiologic examinations throughout their treatment and after treatment with surgeons, radiation oncologists, and medical oncologists specializing in head and neck cancer.
Determination of tumor HPV DNA status
Tumor HPV16 DNA status was determined using polymerase chain reaction (PCR). DNA was extracted using a commercially available DNA extraction kit (Qiagen Inc., Valencia, CA) from paraffin-embedded tissue. A PCR-based type-specific assay (for E6 and E7 regions) was used to determine HPV16 status. Positivity for E6 and/or E7 was considered a positive result. E6 and E7 showed excellent agreement with only 6% (8/141) of samples discordant (Κ=.83). Positive and negative controls were included and β-actin was used to ensure DNA integrity.
Extraction of HPV DNA from serum
A 30-mL blood sample was collected from each patient upon enrollment in the study prior to the start of treatment. Follow-up blood samples were also collected at 6 weeks after the end of treatment and every 6 months thereafter for up to 3 years, so each patient could have up to 7 follow-up samples available for analysis. Serum and plasma were isolated from the whole blood sample and stored at −80°C prior to analysis. HPV16 DNA was extracted from a 500-μL volume using the QIAamp Blood Kit (Qiagen Inc., Valencia, CA) for a final elution volume of 50 μL. Both serum and plasma samples were available for follow-up analysis; however, only serum samples were available for pre-treatment analysis.
Measurement of HPV16 DNA in serum
HPV16 E6 and E7 concentration were each measured in duplicate using real-time PCR amplification of the E6 and E7 regions. The assay was carried out using TaqMan Universal PCR Master Mix Kit (Applied Biosystems, Foster City, CA) with a 25-μL volume with 10 μL of template DNA. A 96-well plate format was used for the PE Applied Biosystems 7700 Sequence Detector. The β-actin gene was used to ensure DNA integrity for each sample, and multiple water blanks were included on each plate to serve as negative controls. Standard curves were generated using SiHa cells, which contain 1 to 2 copies of HPV DNA per cell. PCR conditions consisted of an initial denaturation step of 95°C for 5 minutes followed by 40 cycles of 95°C for 30 seconds, 65°C for 30 seconds, and 72°C for 40 seconds and a final elongation step of 72°C for 10 minutes.
Statistical methods and definitions of variables
Stata 12.0 (StataCorp, College Station, TX) was used for all statistical analyses. A P value of <0.05 was used to define statistical significance, and all tests were 2-sided. Categorical variables were created for smoking and alcohol drinking status (never/former/current). A patient was considered an ever-smoker if the patient had smoked at least 100 cigarettes in his or her lifetime and an ever-drinker if the patient had drunk alcoholic beverages at least once a week for a year or more during his or her lifetime. Patients who previously smoked or drank alcoholic beverages but had not done so in the year prior to OPC diagnosis were considered former-smokers and former-drinkers, respectively. Tumor site, grade, T category, N category, and overall stage were based on review of patient records, and all diagnoses were histologically confirmed. The sixth edition of the American Joint Committee on Cancer TNM staging system was used to determine disease stage at the time of presentation and work-up for all patients.
Positivity for pretreatment serum E6 and E7 HPV DNA showed excellent agreement (Κ=0.77); therefore, we combined the 2 into a single measure. We dichotomized serum HPV DNA into positive and negative; patients who were negative for both E6 and E7 DNA (0 copies of DNA per mL) were considered negative, and patients who had detectable E6 and/or E7 DNA were considered positive. For categories of DNA copy number per mL, the higher of the values for E6 and E7 DNA was counted. When a patient had HPV DNA copy number per milliliter measured in both serum and plasma post-treatment samples, the higher of the 2 values (either serum or plasma) was counted.
To determine whether differences between groups were statistically significant, Student’s t-test was used for continuous variables, and chi-square or Fisher’s exact test (when cell totals were less than 5) was used for categorical variables. Bar graphs were generated to evaluate the association between the presence of pretreatment serum HPV DNA and T and N category as well as overall TNM stage. A nonparametric test for trend across ordered groups was used to detect increasing pretreatment HPV DNA copy number per mL across T and N categories and stage.
For the progression-free survival analysis, only patients who were subsequently treated at our institution were included. Kaplan-Meier curves were generated and log-rank tests were used to detect statistically significant differences with respect to progression-free survival between patients with and without detectable pretreatment serum HPV DNA. Time to event was calculated from date of diagnosis to date of recurrence or death from any cause. Patients without recurrence or who were alive at last contact and those who were lost to follow-up were considered censored.
Results
A total of 269 patients were recruited for the study. Of these, 7 patients were excluded. Two patients were excluded because distant metastases were detected during initial evaluation (1 had HPV DNA detected in serum). Four patients were excluded because they were found not to have oropharyngeal primary tumors; of these, 1 had an oral cavity tumor with no HPV DNA detected in the pretreatment serum sample, and 3 had cervical metastases from an unknown primary tumor (1 had an HPV-positive metastasis and HPV DNA in the pretreatment serum sample; 1 had an HPV-negative metastasis without HPV DNA in the pretreatment serum sample; 1 did not have HPV DNA metastases status available but was without HPV DNA in the pretreatment serum sample). One patient was excluded because laboratory data were not available. A total of 262 patients were included in the overall analysis. For the survival analysis, only the 218 patients treated at MD Anderson were included. The median follow-up time was 67 months among all patients without progression (69 months in the subgroup with detectable pretreatment serum HPV DNA and 62 months in the subgroup without detectable pretreatment serum HPV DNA).
Table 1 shows the demographic and clinical characteristics and smoking and alcohol exposures of the patients in the study overall and by pretreatment serum HPV DNA status. Patients negative and positive for pretreatment serum HPV DNA were similar with respect to age, sex, race, and smoking and drinking status. The majority of patients (81%) had HPV-positive tumors, and patients with HPV-positive tumors were significantly more likely to have HPV DNA present in their pretreatment serum than were patients with HPV-negative tumors (P=0.011). Compared with patients negative for pretreatment serum HPV DNA, patients positive for pretreatment serum HPV DNA were more likely to present with higher N category, but not tumor size, and were more likely to present with stage IV disease. Figure 1 shows the distribution of pretreatment serum HPV DNA copies/mL by T category, N category, and overall stage. Pretreatment serum HPV E6 and E7 DNA copy number/mL increased with increasing N category and overall stage; however, no trend was observed for T category.
Table 1.
Demographic and clinical characteristics and smoking and alcohol exposures by pretreatment serum HPV DNA status
| Pretreatment serum HPV DNA status | ||||
|---|---|---|---|---|
|
|
||||
| All patients (N=262) |
E6 and E7 negative (N=114) |
E6 and/or E7 positive (N=148) |
P | |
| Age, mean (SD), years | 56.5 (9.6) | 57.4 (10.4) | 55.8 (8.8) | 0.191 |
| Age, median, years | 55 | 56.5 | 54 | 0.335 |
|
|
||||
| Sex | 0.604 | |||
| Male | 224 (85.5) | 96 (84.2) | 128 (86.5) | |
| Female | 38 (14.5) | 18 (15.8) | 20 (13.5) | |
| Race | 0.380 | |||
| White | 239 (91.2) | 102 (89.5) | 137 (92.6) | |
| Other | 23 (8.8) | 12 (10.5) | 11 (7.4) | |
| Subsite | <0.001a | |||
| Tongue | 127 (48.5) | 61 (53.5) | 66 (44.6) | |
| Base of tongue | 125 (47.7) | 43 (37.7) | 82 (55.4) | |
| Other oropharynx | 10 (3.8) | 10 (8.8) | 0 | |
| Stage | 0.004 | |||
| I–II | 20 (7.6) | 10 (8.8) | 10 (6.8) | |
| III | 26 (9.9) | 19 (16.7) | 7 (4.7) | |
| IV | 216 (82.4) | 85 (74.6) | 131 (88.5) | |
| T category | 0.495 | |||
| 1 | 76 (29.0) | 37 (32.5) | 39 (26.4) | |
| 2 | 116 (44.3) | 45 (39.5) | 71 (48.0) | |
| 3 | 41 (15.7) | 20 (17.5) | 21 (14.2) | |
| 4 | 29 (11.1) | 12 (10.5) | 17 (11.5) | |
| N category | 0.069 | |||
| 0 | 30 (11.5) | 18 (15.8) | 12 (8.1) | |
| 1–2a | 43 (16.4) | 23 (20.2) | 20 (13.5) | |
| 2b | 131 (50.0) | 52 (45.6) | 79 (53.4) | |
| 2c–3 | 58 (22.1) | 21 (18.4) | 37 (25.0) | |
| Smoking status | 0.687 | |||
| Never smoker | 119 (45.4) | 54 (47.4) | 65 (43.9) | |
| Former smoker | 95 (36.3) | 38 (33.3) | 57 (38.5) | |
| Current smoker | 48 (18.3) | 22 (19.3) | 26 (17.6) | |
| Pack-years of smoking | 0.671 | |||
| ≤10 | 154 (59.0) | 65 (57.5) | 89 (60.1) | |
| >10 | 107 (41.0) | 48 (42.5) | 59 (39.9) | |
| Missing | 1 | 1 | 0 | |
| Alcohol use status | 0.159 | |||
| Never-drinker | 69 (26.3) | 35 (30.7) | 34 (23.0) | |
| Ever-drinker | 193 (73.7) | 79 (69.3) | 114 (77.0) | |
| Grade | 0.261 | |||
| Well or moderately differentiated/keratinizing | 77 (34.4) | 38 (38.4) | 39 (31.2) | |
| Moderately poorly or poorly differentiated/non-keratinizing | 147 (65.6) | 61 (61.6) | 86 (68.8) | |
| Missing | 38 | 15 | 23 | |
| Tumor HPV DNA status | 0.011 | |||
| Negative | 27 (19.2) | 18 (28.6) | 9 (11.5) | |
| Positive | 114 (80.9) | 45 (71.4) | 69 (88.5) | |
| Missing | 121 | 51 | 70 | |
Fisher’s exact test.
Figure 1.
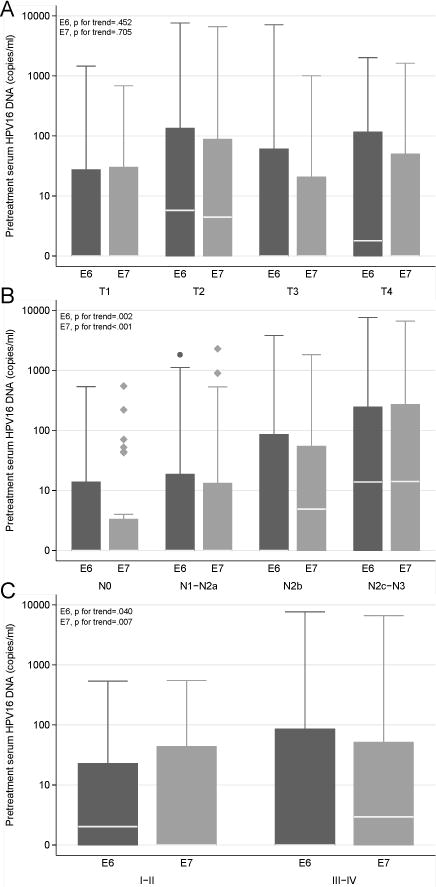
Distribution of pretreatment serum HPV E6 and E7 DNA levels among all patients by TNM categories and overall disease stage. The white lines represent median values. A) Distribution by T category (E6 P for trend = 0.452 and E7 P for trend = 0.705). B) Distribution by N category (E6 P for trend = 0.002 and E7 P for trend <0.001). C) Distribution by overall disease stage (E6 P for trend = 0.040 and E7 P for trend = 0.007).
Table 2 shows relationships between pretreatment serum HPV DNA levels and tumor HPV status and recurrence status for HPV E6 and E7 separately and combined. Patients with tumors positive for HPV E6 or E7 were more likely to have detectable pretreatment serum HPV E6 or E7 DNA than were patients with tumors negative for HPV E6 or E7, although the difference between E7-positive and E7-negative tumors was of borderline significance. Among the 218 patients treated at MD Anderson, 12 patients with HPV-positive tumors developed recurrence, and 7 of these 12 patients (58%) had HPV DNA detectable in pretreatment serum prior to treatment. Four patients with HPV-negative tumors developed recurrence, and none of these 4 patients had HPV DNA detectable in pretreatment serum prior to treatment. However, there were no significant differences in pretreatment serum HPV DNA levels between patients with and without recurrence, regardless of tumor HPV status (Table 2).
Table 2.
Tumor HPV DNA status and recurrence status by pretreatment serum HPV DNA level (copies/mL)
| Serum HPV DNA level (copies/mL)
|
||||||
|---|---|---|---|---|---|---|
| 0 | 0.1 – <10 | 10 – <100 | 100 – <1000 | ≥1000 | Pe | |
| All patients (n=262) | 114 | 23 | 59 | 47 | 19 | |
| Tumor HPV DNA status | ||||||
| E6a | 0.036 | |||||
| Negative | 20 (27.0) | 2 (16.7) | 3 (13.0) | 0 | 2 (25.0) | |
| Positive | 54 (73.0) | 10 (83.3) | 20 (87.0) | 21 (100) | 6 (75.0) | |
| E7b | 0.079 | |||||
| Negative | 20 (29.9) | 2 (13.3) | 2 (7.1) | 2 (10.5) | 1 (14.3) | |
| Positive | 47 (70.2) | 13 (86.7) | 26 (92.9) | 17 (89.5) | 6 (85.7) | |
| E6/E7c | 0.077 | |||||
| Negative | 18 (28.6) | 2 (13.3) | 4 (13.3) | 1 (4.2) | 2 (22.2) | |
| Positive | 45 (71.4) | 13 (86.7) | 26 (86.7) | 23 (95.8) | 7 (77.8) | |
| Recurrence statusd | ||||||
| All patients (N=218) | 0.677 | |||||
| No recurrence | 78 (83.9) | 18 (85.7) | 45 (90.0) | 37 (92.5) | 12 (85.7) | |
| Any recurrence | 15 (16.1) | 3 (14.3) | 5 (10.0) | 3 (7.5) | 2 (14.3) | |
| E6/E7 tumor positivec | 0.173 | |||||
| No recurrence | 33 (86.8) | 9 (75.0) | 21 (87.5) | 20 (100) | 4 (80.0) | |
| Any recurrence | 5 (13.2) | 3 (25.0) | 3 (12.5) | 0 | 1 (20.0) | |
| E6/E7 tumor negativec | 1.0 | |||||
| No recurrence | 11 (73.3) | 2 (100) | 3 (100) | 0 | 2 (100) | |
| Any recurrence | 4 (26.7) | 0 | 0 | 0 | 0 | |
Copies of serum E6 DNA in patients by tumor E6 status (E6-positive tumors may be positive or negative for E7; E6-negative tumors also E7-negative).
Copies of serum E7 DNA in patients by tumor E7 status (E7-positive tumors may be positive or negative for E6; E7-negative tumors also E6-negative).
Copies of serum DNA refers to the higher of the copy number values for E6 and E7 DNA.
Includes only patients who were treated at MD Anderson.
Fisher’s exact test.
Table 3 shows 3-year progression-free survival rates according to tumor and pretreatment serum HPV DNA status and smoking status. Although patients with HPV-positive tumors and ≤10 pack-years had better 3-year progression-free survival (91%) than patients with HPV-positive tumors and >10 pack-years (79%) and patients with HPV-negative tumors (72%), these differences were not statistically significant (P=0.146 and P=0.887, respectively, and P for trend=0.430). When patients were stratified by pretreatment serum HPV DNA status and smoking status, there were no differences in progression-free survival between subgroups. Among patients with HPV-positive tumors, those with detectable pretreatment serum HPV DNA had a worse 3-year progression-free survival rate than those without detectable pretreatment serum HPV DNA (82% vs 91%), although this difference was not statistically significant (P=0.800). The Kaplan-Meier curves for progression-free survival based on tumor and pretreatment serum HPV DNA status and smoking status are shown in Figure 2. Figure 2A shows progression-free survival of patients with HPV-positive tumors by pack-years of smoking and of patients with HPV-negative tumors. Figure 2B shows progression-free survival according to pretreatment serum HPV DNA status and pack-years of smoking, and Figure 2C shows progression-free survival according to pretreatment serum HPV DNA status among patients with HPV-positive tumors.
Table 3.
Three-year progression-free survival rates among patients with OPC by tumor HPV DNA status or pretreatment serum HPV DNA status and smoking status
| 3-year progression-free survival, % (95% CI) |
P | |
|---|---|---|
| All patients | ||
| Tumor HPV DNA status/smoking status | ||
| HPV+/≤10 pack-years | 90.5 (78.6–95.9) | Reference |
| HPV+/>10 pack-years | 78.6 (61.6–88.7) | 0.887 |
| HPV− | 71.9 (47.7–86.3) | 0.146 0.430a |
| Serum HPV DNA status/smoking status | ||
| HPV+/≤10 pack-years | 86.1 (75.6–92.3) | Reference |
| HPV+/>10 pack-years | 81.3 (66.0–90.2) | 0.905 |
| HPV− | 84.9 (75.4–91.0) | 0.879 0.977a |
| Only patients with HPV+ tumors | ||
| Serum HPV DNA status | ||
| HPV− | 91.3 (75.4–97.1) | Reference |
| HPV+ | 82.1 (69.2–90.0) | 0.800 |
Test for trend.
Figure 2.
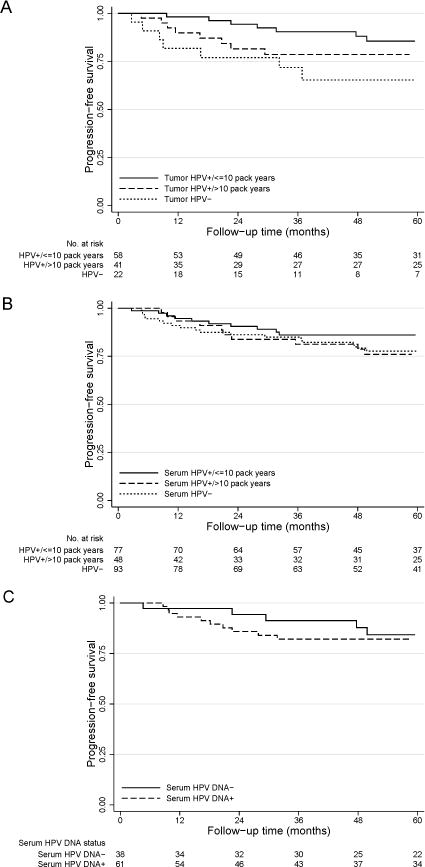
Progression-free survival for A) all patients segregated by tumor HPV status and smoking history (P for trend = 0.062), B) all patients segregated by pretreatment serum HPV DNA status and smoking history (P for trend = 0.275), and C) only patients with HPV-positive tumors segregated by pretreatment serum HPV DNA status (P=0.627). Patients with HPV-positive tumors and ≤10 pack-years of smoking had better progression-free survival than did patients with HPV-negative tumors (P=0.020), and there was a borderline significant trend across the 3 groups (P=0.062).
In an analysis of HPV DNA in follow-up blood samples, we found that of 11 patients who developed distant metastases, 4 (36%) had HPV DNA detected in serum and/or plasma following treatment (Figure 3A–D). Of the 7 patients who developed distant metastases but did not have HPV DNA detected in follow-up blood samples, 4 had HPV-positive tumors. Also of note, as previously mentioned, 2 patients were excluded from the study because distant metastases were detected during initial work-up. Both of these patients were tumor HPV DNA positive and one had a very high pretreatment serum HPV DNA level (>1000 copies/mL) while the other had none detected. Seventeen patients had a local-regional recurrence of their disease, and only 1 of these (6%) had HPV DNA detected in serum/plasma following treatment (Figure 3E). Of the 16 patients who developed local-regional recurrence but did not have HPV DNA detected in serum/plasma following treatment, 5 (31%) had HPV-positive tumors.
Figure 3.
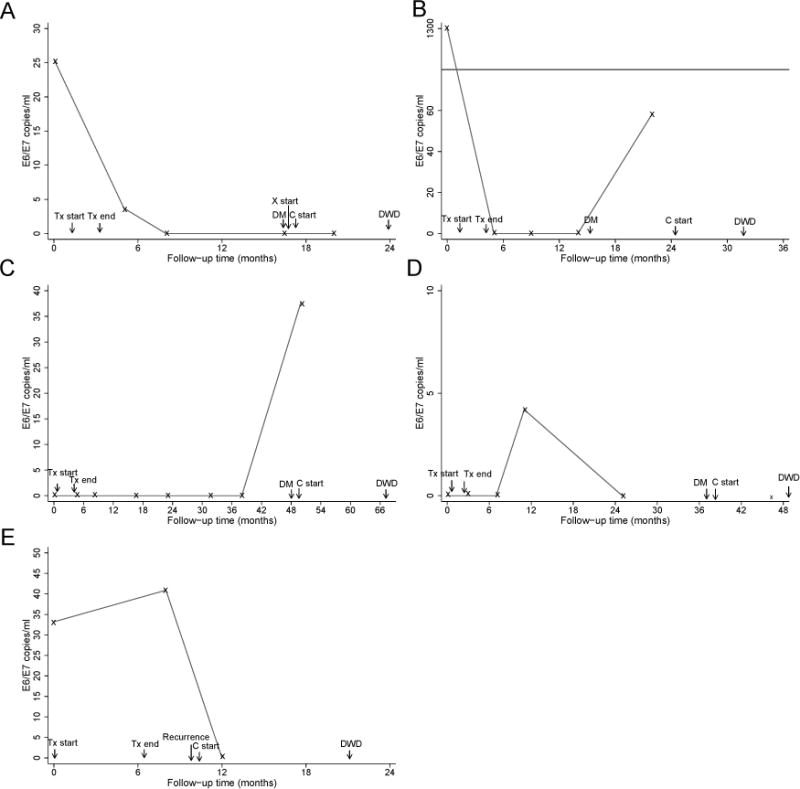
Longitudinal follow-up of 5 patients with detectable levels of serum or plasma HPV DNA after treatment. A) Patient with HPV-positive tumor and >10 pack-years of smoking. Distant metastases were detected at 12 months after treatment. HPV DNA was detectable at diagnosis and 6 weeks after treatment. B) Patient with ≤10 pack-years of smoking and tumor HPV status not available. Distant metastases were detected at 11 months after treatment. HPV DNA was detectable at diagnosis and 18 months after treatment. C) Patient with HPV-positive tumor and ≤10 pack-years of smoking. Distant metastases were detected at 46 months after treatment. HPV DNA was detectable at 46 months after treatment. D) Patient with HPV-negative tumor and ≤10 pack-years of smoking. Distant metastases were detected at 35 months after treatment. HPV DNA was detectable at 9 months after treatment. E) Patient with HPV-positive tumor and >10 pack-years of smoking. Regional metastases were detected at 4 months after treatment. HPV DNA was detectable at diagnosis and 6 weeks after treatment. “x” indicates follow-up appointments when blood was drawn. Tx, treatment; DM, distant metastases detected; C, chemotherapy; X, radiation therapy; DWD, died with disease.
Discussion
In this study, we evaluated pretreatment circulating HPV DNA as a biomarker for recurrence in a population of patients with OPC. We detected circulating HPV DNA in pretreatment serum and found that the presence of circulating HPV DNA was associated with higher N category and overall stage but not higher T category. Although patients with HPV-positive tumors with detectable pretreatment levels of circulating HPV DNA had worse progression-free survival than patients with HPV-positive tumors and no detectable circulating HPV DNA, this difference did not reach statistical significance.
To date, few studies have evaluated the association between circulating HPV DNA and outcome among patients with OPC. In a study of 13 patients with OPC, 4 of the 6 patients who had HPV DNA detected in serum developed distant metastases, whereas none of the 7 patients who did not have HPV DNA detected in serum developed distant metastases.17 Another study showed that 65% of patients with HPV-positive OPC had HPV DNA detected in plasma, whereas no patients with HPV-negative OPC and no healthy controls had HPV DNA detected in plasma.18 Moreover, in that study, pretreatment plasma HPV DNA copy number correlated with nodal metastatic tumor volume and declined rapidly after the start of treatment. However, in 3 patients who developed distant metastases, plasma HPV DNA was still detectable at the time of relapse.18 Finally, in a study that included 93 patients with OPC and squamous cell carcinoma with unknown primary tumor site, positivity for HPV DNA in saliva, plasma, or both after treatment was associated with worse recurrence-free survival, and positivity for HPV DNA in saliva or both saliva and plasma (but not plasma alone) after treatment was associated with worse overall survival.19
These results and the results of our study reported herein are consistent with prior results in patients with cervical cancer.20–24 For instance, high levels of circulating HPV DNA were detected in patients with invasive cervical cancer, while lower levels were detected in patients with carcinoma in situ.24 Sathish et al. confirmed through sequencing that the HPV type detected in cervical tumor tissue was the type found in serum from the same patients.20 Detection of HPV DNA in plasma has consistently been found to be associated with worse outcome among patients with cervical cancer. In one study, circulating HPV DNA was associated with a 15 times higher risk of recurrence and distant metastases among women with invasive cervical cancer; furthermore, the women who developed distant metastases had levels of HPV DNA in serum 3 times the levels in women who did not develop distant metastases.22 In another study, of 94 women with cervical cancer, 45% had detectable levels of serum HPV DNA prior to treatment. There were no recurrences among women who had no detectable HPV DNA in serum before treatment or who became negative for serum HPV DNA after treatment, whereas the 13 women with recurrence 10 had detectable serum HPV DNA both before and after treatment.23
Serum viral DNA was previously established as a good predictor for recurrence in both NPC and cervical cancer, and we found in the study reported herein that serum viral DNA might be a promising biomarker for OPC as well. However, in our study, the progression-free survival benefit in favor of patients who were negative for pretreatment serum HPV DNA was not statistically significant. There are several reasons why levels of serum HPV DNA may not be as strong a biomarker as levels of serum EBV DNA. The levels of serum HPV DNA that we detected were less than 8000 copies per milliliter, and the median values were 0 copies/mL for E6 and 2 copies/mL for E7. In contrast, in studies of serum EBV DNA level in patients with NPC, median copy number has been shown to be in the thousands.25–28 While HPV is maintained in an episomal state, malignant transformation is usually preceded by integration into the host genome, although it has been shown that this may not be necessary in all cervical cancers.29, 30 Integrated DNA may not be as detectable in serum as episomal DNA appears to be.31 Therefore, further studies are needed to optimize detection methods for HPV DNA to make this a viable prognostic biomarker.
This study has several limitations. First, PCR may not be the most reliable measure of tumor HPV DNA status, and thus we may have had some misclassification of patients. While this method has been used routinely in the past, a recent study found that HPV measured by PCR is not the most informative predictor of survival among patients with head and neck cancer.32 Others have reviewed the methods for detecting tumor HPV DNA currently used in clinical trials assessing treatment outcomes among patients with OPC and have suggested algorithms that include immunohistochemistry for p16 expression with in situ hybridization for HPV DNA and/or PCR.33–36 We had such data available for only 16 patients (6%) in our study; however, we are currently routinely assessing p16 overexpression and will include this measure in future studies. Second, 80% of the patients in our study had HPV-positive tumors. Having only 20% of patients with HPV-negative tumors may have reduced the power to detect meaningful associations. Third, the recurrence rate was low resulting in reduced power to observe significant associations. Future studies will benefit from increased sample size and the inclusion of p16 expression for determining tumor HPV status.
We evaluated the usefulness of serum HPV DNA level as a marker for recurrence and found that the presence of pretreatment serum HPV DNA was associated with nodal category and overall stage but not with primary tumor size. Among patients with OPC with HPV-positive tumors, patients with detectable pretreatment serum HPV DNA had better progression-free survival than patients who were negative for pretreatment serum HPV DNA, although this difference was not statistically significant. Although serum EBV DNA is a useful prognostic indicator for recurrence and metastases for NPC, in this study pretreatment serum HPV DNA did not appear to have clinical utility as a marker for recurrence for patients with OPC; however, larger studies with increased power to detect significant associations are warranted.
Acknowledgments
We thank Stephanie Deming for manuscript editing.
Financial support: This research was supported in part by the National Institutes of Health through MD Anderson’s Cancer Center Support Grant, CA016672, National Institutes of Health grant R03 CA128110-01A1 (to E.M.S.), and an institutional research grant from The University of Texas MD Anderson Cancer Center (to E.M.S.). This research was accomplished within the Oropharynx Program at The University of Texas MD Anderson Cancer Center and funded in part through the Stiefel Oropharyngeal Research Fund.
Footnotes
Conflicts of interest: None declared
References
- 1.D’Souza G, Kreimer AR, Viscidi R, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 2.Nasman A, Attner P, Hammarstedt L, et al. Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden: an epidemic of viral-induced carcinoma? Int J Cancer. 2009;125:362–6. doi: 10.1002/ijc.24339. [DOI] [PubMed] [Google Scholar]
- 3.Attner P, Du J, Nasman A, et al. The role of human papillomavirus in the increased incidence of base of tongue cancer. Int J Cancer. 2010;126:2879–84. doi: 10.1002/ijc.24994. [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi AK. Beyond cervical cancer: burden of other HPV-related cancers among men and women. J Adolesc Health. 2010;46:S20–6. doi: 10.1016/j.jadohealth.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–9. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 7.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Posner MR, Lorch JH, Goloubeva O, et al. Survival and human papillomavirus in oropharynx cancer in TAX 324: a subset analysis from an international phase III trial. Ann Oncol. 2011;22:1071–7. doi: 10.1093/annonc/mdr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 10.Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–91. [PubMed] [Google Scholar]
- 11.Leung SF, Zee B, Ma BB, et al. Plasma Epstein-Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J Clin Oncol. 2006;24:5414–8. doi: 10.1200/JCO.2006.07.7982. [DOI] [PubMed] [Google Scholar]
- 12.Lo YM, Leung SF, Chan LY, et al. Plasma cell-free Epstein-Barr virus DNA quantitation in patients with nasopharyngeal carcinoma. Correlation with clinical staging Ann N Y Acad Sci. 2000;906:99–101. doi: 10.1111/j.1749-6632.2000.tb06597.x. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari D, Codeca C, Bertuzzi C, et al. Role of plasma EBV DNA levels in predicting recurrence of nasopharyngeal carcinoma in a Western population. BMC Cancer. 2012;12:208. doi: 10.1186/1471-2407-12-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chai SJ, Pua KC, Saleh A, et al. Clinical significance of plasma Epstein-Barr Virus DNA loads in a large cohort of Malaysian patients with nasopharyngeal carcinoma. J Clin Virol. 2012;55:34–9. doi: 10.1016/j.jcv.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Hsu CL, Chan SC, Chang KP, et al. Clinical scenario of EBV DNA follow-up in patients of treated localized nasopharyngeal carcinoma. Oral Oncol. 2013;49:620–5. doi: 10.1016/j.oraloncology.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Wang WY, Twu CW, Chen HH, et al. Long-term survival analysis of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA levels. Cancer. 2013;119:963–70. doi: 10.1002/cncr.27853. [DOI] [PubMed] [Google Scholar]
- 17.Capone RB, Pai SI, Koch WM, et al. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin Cancer Res. 2000;6:4171–5. [PubMed] [Google Scholar]
- 18.Cao H, Banh A, Kwok S, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int J Radiat Oncol Biol Phys. 2012;82:e351–8. doi: 10.1016/j.ijrobp.2011.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn SM, Chan JY, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg. 2014;140:846–54. doi: 10.1001/jamaoto.2014.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sathish N, Abraham P, Peedicayil A, et al. HPV DNA in plasma of patients with cervical carcinoma. J Clin Virol. 2004;31:204–9. doi: 10.1016/j.jcv.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Kay P, Allan B, Denny L, Hoffman M, Williamson AL. Detection of HPV 16 and HPV 18 DNA in the blood of patients with cervical cancer. J Med Virol. 2005;75:435–9. doi: 10.1002/jmv.20294. [DOI] [PubMed] [Google Scholar]
- 22.Pornthanakasem W, Shotelersuk K, Termrungruanglert W, Voravud N, Niruthisard S, Mutirangura A. Human papillomavirus DNA in plasma of patients with cervical cancer. BMC Cancer. 2001;1:2. doi: 10.1186/1471-2407-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widschwendter A, Blassnig A, Wiedemair A, Muller-Holzner E, Muller HM, Marth C. Human papillomavirus DNA in sera of cervical cancer patients as tumor marker. Cancer Lett. 2003;202:231–9. doi: 10.1016/j.canlet.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Dong SM, Pai SI, Rha SH, et al. Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol Biomarkers Prev. 2002;11:3–6. [PubMed] [Google Scholar]
- 25.Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350:2461–70. doi: 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
- 26.Shao JY, Li YH, Gao HY, et al. Comparison of plasma Epstein-Barr virus (EBV) DNA levels and serum EBV immunoglobulin A/virus capsid antigen antibody titers in patients with nasopharyngeal carcinoma. Cancer. 2004;100:1162–70. doi: 10.1002/cncr.20099. [DOI] [PubMed] [Google Scholar]
- 27.Twu CW, Wang WY, Liang WM, et al. Comparison of the prognostic impact of serum anti-EBV antibody and plasma EBV DNA assays in nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2007;67:130–7. doi: 10.1016/j.ijrobp.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Lo YM, Chan LY, Chan AT, et al. Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res. 1999;59:5452–5. [PubMed] [Google Scholar]
- 29.Stanley MA, Pett MR, Coleman N. HPV: from infection to cancer. Biochem Soc Trans. 2007;35:1456–60. doi: 10.1042/BST0351456. [DOI] [PubMed] [Google Scholar]
- 30.Sathish N, Abraham P, Peedicayil A, Sridharan G, John S, Chandy G. Human papillomavirus 16 E6/E7 transcript and E2 gene status in patients with cervical neoplasia. Mol Diagn. 2004;8:57–64. doi: 10.1007/BF03260048. [DOI] [PubMed] [Google Scholar]
- 31.Mutirangura A. Serum/plasma viral DNA: mechanisms and diagnostic applications to nasopharyngeal and cervical carcinoma. Ann N Y Acad Sci. 2001;945:59–67. [PubMed] [Google Scholar]
- 32.Liang C, Marsit CJ, McClean MD, et al. Biomarkers of HPV in Head and Neck Squamous Cell Carcinoma. Cancer Res. 2012;72:5004–13. doi: 10.1158/0008-5472.CAN-11-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snow AN, Laudadio J. Human papillomavirus detection in head and neck squamous cell carcinomas. Adv Anat Pathol. 2010;17:394–403. doi: 10.1097/PAP.0b013e3181f895c1. [DOI] [PubMed] [Google Scholar]
- 34.Jordan RC, Lingen MW, Perez-Ordonez B, et al. Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol. 2012;36:945–54. doi: 10.1097/PAS.0b013e318253a2d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smeets SJ, Hesselink AT, Speel EM, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121:2465–72. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 36.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116:2166–73. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]


