Abstract
Sugar aminotransferases (SATs) are an important class of tailoring enzymes that catalyze the 5′-pyridoxal phosphate (PLP)-dependent stereo- and regiospecific installation of an amino group from an amino acid donor (typically l-Glu or l-Gln) to a corresponding ketosugar nucleotide acceptor. Herein we report the strategic structural study of two homologous C4 SATs (Micromonospora echinospora CalS13 and Escherichia coli WecE) that utilize identical substrates but differ in their stereochemistry of aminotransfer. This study reveals for the first time a new mode of SAT sugar nucleotide binding and, in conjunction with previously reported SAT structural studies, p.rovides the basis from which to propose a universal model for SAT stereo- and regiochemical control of amine installation. Specifically, the universal model put forth highlights catalytic divergence to derive solely from distinctions within nucleotide sugar orientation upon binding within a relatively fixed SAT active site where the available ligand bound structures of the three out of four representative C3 and C4 SAT examples provide a basis for the overall model. Importantly, this study presents a new predictive model to support SAT functional annotation, biochemical study and rational engineering.
Graphical Abstract
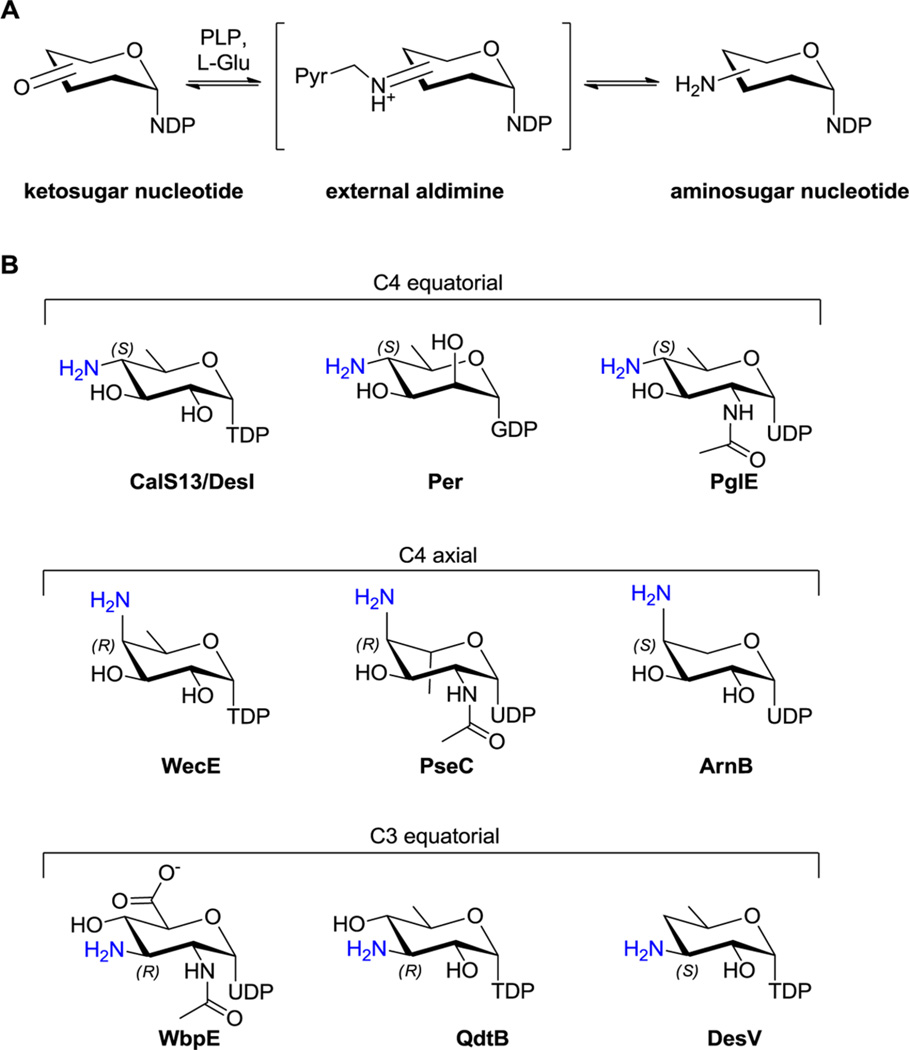
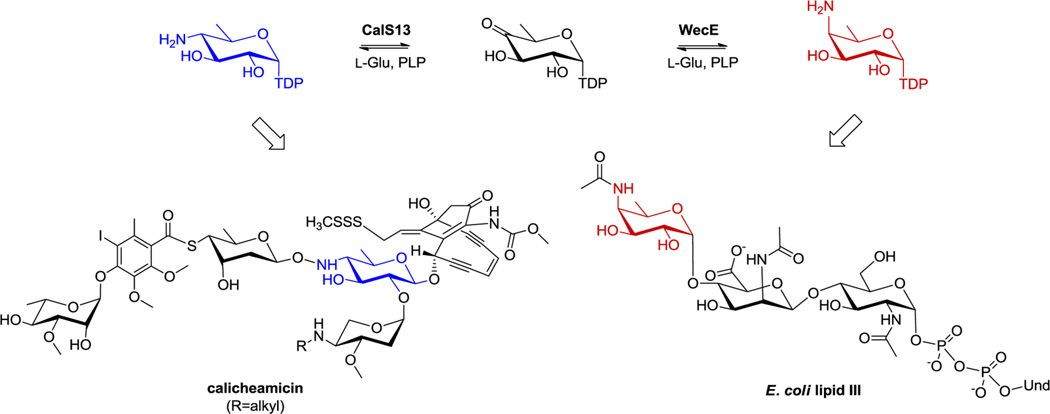
Aminosugars are prevalent in nature, where they function as precursors in primary metabolism, building blocks within biological macromolecular structures, and key contributors to biological molecular recognition events that dictate remarkably divergent phenomena such as small molecule targeting, signaling, bacterial pathogenicity, and immunological definition.1–10Biosynthetically, sugar amine installation is catalyzed by 5′-pyridoxal phosphate (PLP)-dependent sugar aminotransferases (SATs) belonging to the aspartate aminotransferase fold type I superfamily (AAT-I) (E.C. 2.6.1.X) that use an amino acid (typically l-Glu or l-Gln) as the amino donor and a free ketosugar or ketosugar nucleotide as the amino acceptor (Figure 1A).11–17 In recent years, the crystal structures of several sugar nucleotide-dependent SATs from bacteria have been described.18–26 Yet, given the diversity of substrates employed by the SATs structurally interrogated to date (Figure 1B), the general features that control the regio- and/or stereospecificity of an SAT-catalyzed amine installation remain poorly understood. To address this gap in knowledge, herein we describe a structural study of two highly homologous sugar nucleotide-dependent SATs (CalS13 and WecE) that share common substrates (TDP-4-keto-4,6-dideoxy-α-d-glucose as the acceptor and l-Glu as the amino donor) but differ in the stereochemistry of the C-4 amine installation (Figure 2A). Of these, CalS13 was demonstrated to function as a key SAT en route to the calicheamicin aryltetrasaccharide in M. echinospora via comparative genomics, genetic complementation, and in vitro biochemical characterization to provide the corresponding C-4 (S) configuration (Figure 2B).27–30 In contrast, WecE provides the corresponding C-4 (R) configuration and serves as an integral SAT in lipopolysaccharide biosynthesis.30,31 The strategic structural comparison highlighted herein provides, for the first time, a uniform structural model for the molecular basis of SAT amine installation stereospecificity wherein C-4 (R) installation is accomplished via rotation of the sugar through two distinct strategies. Specifically, in WecE, the nucleotide portion of the substrate is bound in a nearly 180° opposed orientation to that in CalS13 wherein this “nucleotide flip” mechanism in WecE affords top face amine installation to achieve the C-4 (R) product. This is a novel mechanism compared to that previously put forth based upon the ligand-bound structures of ArnB and PseC,16,19,20,26 SATs which bind the nucleotide portion of the substrate in the same manner as CalS13, wherein top face amine installation is afforded via simple rotation of the sugar (i.e., a “sugar flip” mechanism). Thus, the combined “nucleotide flip” and “sugar flip” models provide a universal blueprint for nature’s stereochemical control of SAT amine installation and a potential starting point for rational SAT engineering.
Figure 1.
(A) Scheme representing a standard SAT-catalyzed sugar transamination reaction. (B) Aminosugar nucleotide products of SATs structurally characterized to date where the amino group is colored blue (equatorial or axial designation refers to the corresponding 4C1 conformer in all cases).
Figure 2.
(A) Reactions catalyzed by CalS13 and WecE en route to calicheamicin and LPS biosynthesis, respectively.
RESULTS AND DISCUSSION
Overall Structures
The crystal structure of the CalS13/PLP/TDP-4-keto-4,6-dideoxy-α-d-glucose ternary complex was determined at 2.47 Å resolution (Table 1, PDB 4ZAS). This ternary complex was found to belong to the P2 space group with six subunits in the asymmetric unit. All six subunits contained PLP as internal aldimines of Lys202. One subunit had sufficient electron density to model the sugar nucleotide substrate (Figure S1A), three had electron density for just TDP, while the remaining two lacked interpretable electron density in the sugar nucleotide site. The root-mean-square deviation (r.m.s.d.) among different asymmetric unit members bound to PLP, PLP, and TDP; PLP and TDP-4-keto-4,6-dideoxy-α-d-glucose ranged between 0.21 and 0.25 Å, where only minor changes in active site side chain residues upon ligand binding were observed.
Table 1.
Summary of Crystal Parameters, Data Collection, and Refinement Statisticsa
| Se-Met WecE | Native WecE | CalS13 | |
|---|---|---|---|
| Crystal parameters | |||
| Space group | P2 | P1 | P2 |
| Unit-cell parameters (Å, deg) | 125.3, 87.5, 161.6 90.0, 91.1, 90.0 |
88.1, 104.0, 109.4 72.1, 73.6, 74.1 |
71.6, 88.7, 189.8, 90.0, 96.2, 90.0 |
| Data collection statistics | |||
| Wavelength (Å) | 0.98 | 0.98 | 0.98 |
| Resolution range (Å) | 50–2.70 | 50–2.24 | 50–2.47 |
| No. of reflections (measured/unique) | 275,071/90,907 | 1,781,851/233,383 | 744,102/84,933 |
| Completeness (%) | 89.4 (89.5) | 99.7 (99.2) | 100.0 (100.0) |
| Rmergeb | 0.112 (0.545) | 0.253 (1.460) | 0.117 (1.571) |
| Redundancy | 3.0 (2.9) | 7.6 (7.5) | 8.8 (8.1) |
| Mean I/sigma (I) | 10.31 (1.97) | 5.58 (1.35) | 11.61 (1.89) |
| CC1/2 | 0.99 (0.70) | 0.99 (0.74) | 0.99 (0.65) |
| Refinement and model statistics | |||
| Rcrystc/Rfreed | 0.240/0.273 | 0.204/0.246 | 0.174/0.229 |
| Ligands RSCCe | 0.91 | 0.95 | 0.92 |
| RMSD bonds (Å) | 0.002 | 0.010 | 0.008 |
| RMSD angles (deg) | 0.50 | 1.29 | 1.23 |
| B factor - protein/ligand/solvent (Å2) | 41.3/37.3/35.4 | 62.1/64.7/47.7 | 58.3/58.6/55.2 |
| No. of protein atoms | 22,501 | 23,155 | 17,112 |
| No. of waters | 288 | 686 | 502 |
| No. of auxiliary molecules in the asymmetric unitf | 8 LLP | 8 T5K, 8 PMP | 6 LLP, 3 TYD, 1 T46 |
| Ramachandran plot (%) | |||
| Favorable region | 95.4 | 98.0 | 98.0 |
| Additional allowed region | 3.7 | 2.0 | 2.0 |
| Disallowed region | 0.9 | 0.0 | 0.0 |
| PDB | 4PIW | 4ZAH | 4ZAS |
Values in parentheses are for the highest resolution shell.
Rmerge = ∑h∑i |Ii(h) − 〈I(h)〉|/∑h∑i Ii(h), where Ii(h) is the intensity of an individual measurement of the reflection and 〈I(h)〉 is the mean intensity of the reflection.
Rcryst = ∑h ‖Fobs| − |Fcalc‖/∑h |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
Rfree was calculated as Rcryst using 5.0% of randomly selected unique reflections that were omitted from the structure refinement.
Ligand RSCC: ligand real-space correlation coefficient.
Ligand abbreviations: LLP (n′-pyridoxyllysine-5′-monophosphate), TK5 (TDP-4-amino-4,6-dideoxy-α-d-galactose external aldimine), PMP (pyridoxamine-5′-phosphate), TYD (thymidine-5′-diphosphate), and T46 (TDP-4-keto-4,6-dideoxy-α-d-glucose).
WecE cocrystallization led to WecE/PLP/TDP-4-amino-4,6-dideoxygalactose ternary complexes that diffracted at 2.70 Å (Lys181 internal aldimine-containing) and 2.24 Å resolution (sugar nucleotide external aldimine-containing), belonging to the P2 and P1 space groups, respectively (Table 1, PDB: 4PIW and 4ZAH, respectively), with each containing eight subunits in the asymmetric unit. The presence of the external aldimine (Figure S1B) is consistent with the well-established reversibility of SATs16,39 and has been observed in other SAT structures.18–26 Consistent with the comparisons among other apo and ligand-bound SATs structures,18–26 there is very little conformational change between the WecE internal aldimine and external aldimine complexes (r.m.s.d. 0.31 Å). These are reserved to a slight upward shift of the pyridinium ring into the active site and very small changes in the orientation of active site residue ligand-interacting side chains.
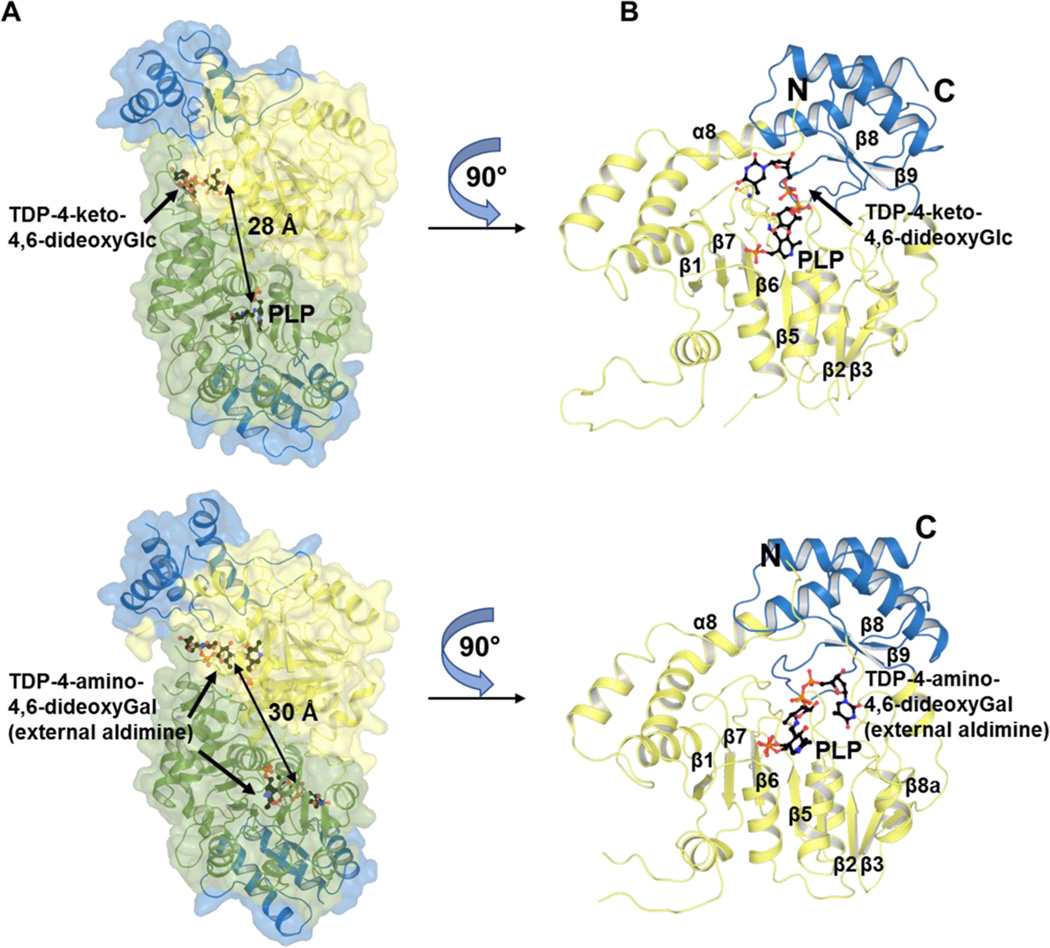
Both CalS13 and WecE belong to the AAT-I family16,17 and share many structural characteristics. Consistent with AAT-I enzymes, multimeric state calculation by PDBePISA38 indicated both CalS13 and WecE to be homodimers with an extensive dimer interface comprising an area of ~4564 Å2 (CalS13 dimer interface) and ~4405 Å2 (WecE dimer interface), respectively (Figure 3A). Each dimer assembly contains two active sites set apart by ~28–30 Å (based upon the distance between the pyridinium rings of bound PLP), with both monomers contributing critical residues to each of the active sites (Figure 3A). Analogous to other AAT-I enzymes, CalS13 and WecE have a conserved active site lysine (Lys202 and Lys181 in CalS13 and WecE, respectively) that forms the requisite internal aldimine with PLP, a conserved general acid (Asp173 and Asp152 in CalS13 and WecE, respectively) that is key to cofactor activation in the context of AAT-catalyzed transamination and a putative base (Gln176 and Gln155 in CalS13 and WecE, respectively). The overall architecture of each subunit is composed of a N-terminal “large domain” (colored yellow in Figure 3B) and a C-terminal “small domain” (colored blue in Figure 3B). The large domain contains a mixed β-sheet formed by seven β-strands (strand order β1, β7, β6, β5, β4, β2, β3) and eight β-strands (strand order β1, β7, β6, β5, β4, β2, β3, β8a) in CalS13 and WecE, respectively, where β7 is antiparallel to the rest of the β-strands in the β-sheet. This large β-sheet is flanked by α-helices on both sides. The C-terminal small domain is formed by a two-stranded antiparallel sheet (β-strands β8 and β9) surrounded by helices. The N-terminal large and C-terminal small domains are linked by a three stranded antiparallel β-hairpin. The size of the 13 α-helices in each subunit ranges from 3 to 31 residues with the largest α-helix (α8, containing a kink near residues 13–14) contributing to both the large and small domains (Figure 3B).
Figure 3.
Structure of CalS13 in complex with PLP (internal aldimine form) and keto-substrate TDP-4-keto-4,6-dideoxy-α-d-glucose (top panel) and WecE in complex with PLP (external aldimine form) and amino-product TDP-4-amino-4,6-dideoxy-α-d-galactose (bottom panel). (A) Homodimer structure where the C-terminal “small” domain is colored blue in two monomers (green and yellow) and the arrow indicates the distance between two active sites of the homodimer assembly. (B) Monomeric structure where N-terminal “large” and C-terminal “small” domains are colored yellow and blue, respectively.
Cofactor-Binding Site
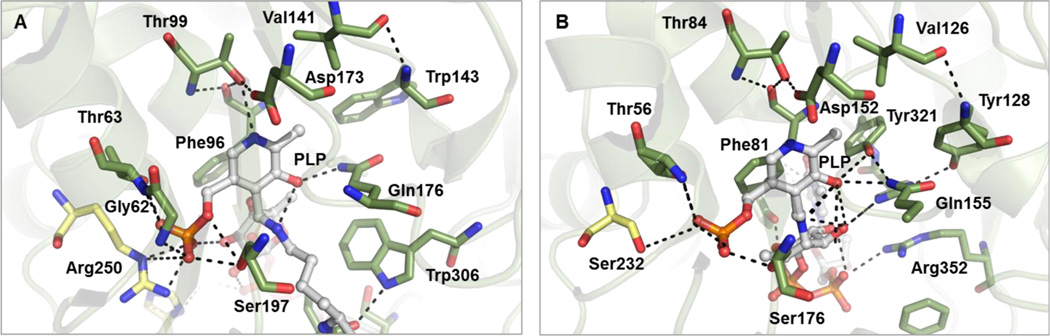
The PLP-binding site is situated within a deep cleft of the active site of each monomer. The cofactor is oriented such that the pyridinium ring is positioned above the central β-sheet of each monomer unit (Figure 3B), while the PLP phosphate moiety points toward the dimer interface. The pyridinium ring and C6 methyl of PLP are positioned for π–π stacking and hydrophobic interactions with the side chains of Phe96 and Val141 in CalS13 (Figure 4A) and Phe81 and Val126 in WecE, respectively (Figure 4B). The pyridoxal C5 hydroxyl also forms a hydrogen bond with the side chain amine of Gln176 in CalS13 and Gln155 and Tyr321 in WecE, respectively, while the cofactor N1 ring nitrogen is anchored by hydrogen bonds with the side chain of the previously discussed general acid active residue (Asp173, CalS13; Asp152, WecE) and backbone amide of Thr99 and Thr84 in CalS13 and WecE, respectively (Figure 4).16,18–26 The PLP phosphate is held in place by several direct and indirect water-mediated intrasubunit (Gly62, Thr63, and Ser197 in CalS13; Cys55, Thr56, and Ser176 in WecE) and intersubunit (Arg250 in CalS13; Ser232 in WecE) hydrogen bonding interactions (Figure 4).
Figure 4.
Cofactor binding site of (A) CalS13 and (B) WecE. The PLP internal aldimines within the CalS13 and WecE active sites are represented as ball and stick models and colored white. The active site residues from the same and adjacent subunits are colored green and yellow, respectively.
The Nucleotide Sugar-Binding Site
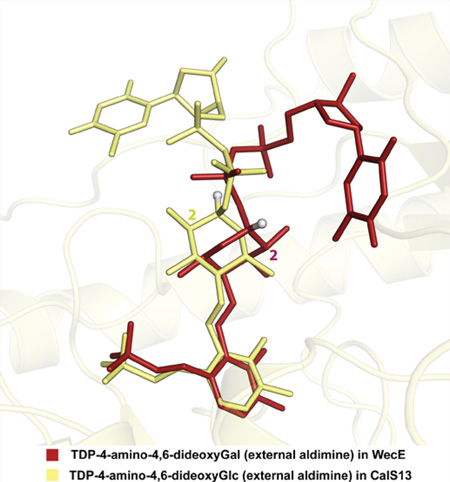
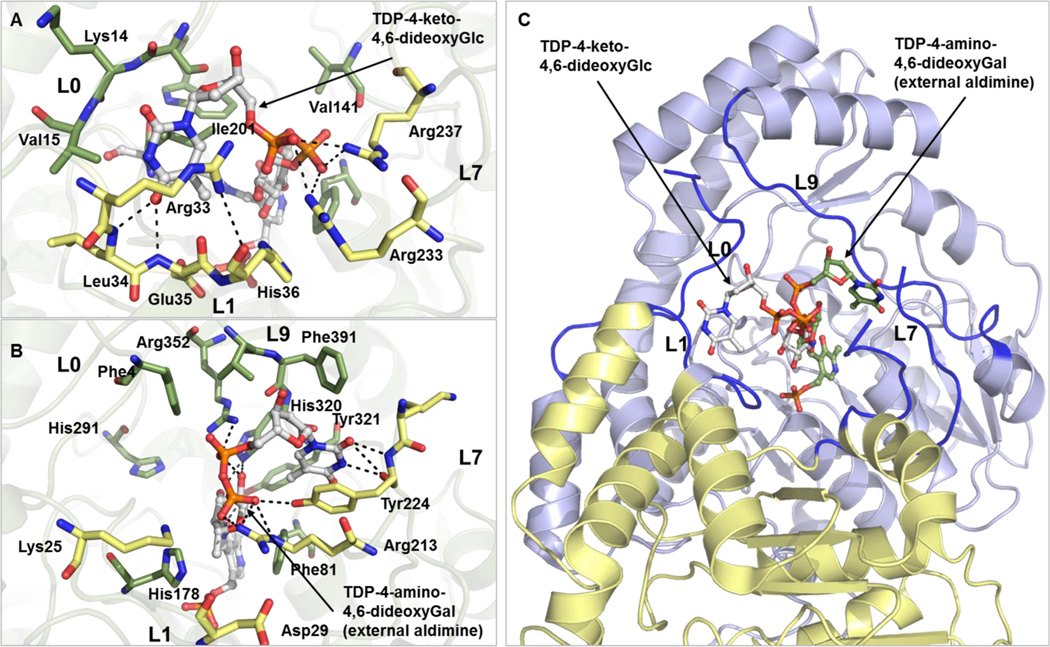
The thymidine binding pockets in CalS13 and WecE are remarkably opposed to one another by nearly 180° and this distinguishing feature defines the stereochemical outcome of aminotransfer by virtue of the sugar orientation (bottom face installation in CalS13 versus top face installation in WecE). Specifically, these distinct nucleotide interaction loops are L0 (residues Ser13-Ala16) and L1 (residues Gly32-Gly37) in CalS13 and L7 (residues Arg213–Thr225) and L9 (residues Val318–Ile322) in WecE, respectively (Figure 5). Key thymidine hydrogen bonding interactions include the thymidine exocyclic O4 with the L1 backbone amide of Leu34 and Glu35 in CalS13 and the L7 backbone amide of Tyr224 in WecE, respectively, and the thymidine endocyclic N3 with the L1 backbone carbonyls of Gly32 and L7 Tyr224 in CalS13 and WecE, respectively. Additional stability is provided by thymine aromatic stacking contributions from the L1 side chain of Arg33 and the L7 side chain of Tyr224 of CalS13 and WecE, respectively. The nucleotide ribose moiety in both CalS13 and WecE is solvent exposed and does not make any direct contacts with the enzyme. The pyrophosphate of the nucleotide sugar extends away from the ribose moiety through a deep protein cavity that leads to the cofactor binding site of the protein and is anchored to the enzyme by direct hydrogen bonding interactions with the guanidinium side chains of L7 of Arg233 and Arg237 in CalS13 (Figure 5A) and the β9 Arg352 L7, Arg213, and L7 side chain hydroxyl of Tyr224 of WecE (Figure 5B), respectively. Interestingly, CalS13 also contains a structurally equivalent L7 tyrosine (Tyr242), but this residue does not interact with the substrate pyrophosphate. Direct sugar contacts are apparent in WecE (Figure 5B) and include hydrogen bonds of the pyranose O2 and O3 with the imidazole ring of His320 (loop L9) (Figure 5B). In contrast, only water-mediated contacts are observed in CalS13 between the pyranose O2 and the L1 side chain carboxylate of Glu35 (Figure 5A). Cumulatively, the “base flip” observed in WecE as compared to CalS13 dictates the divergence in the stereochemical outcome of aminotransfer among these two highly homologous enzymes (Figure 5C), where the key pyrophosphate interactions may direct the divergence in sugar nucleotide orientation.
Figure 5.
Nucleotide sugar binding site of (A) CalS13 and (B) WecE. The substrates within the CalS13 and WecE active sites are represented as ball and stick models and colored white. The active site residues from the same and adjacent subunits are colored green and yellow, respectively. The spatial locations of loops L0, L1, L7, and L9 are labeled. (C) Overlay of the orientation of binding of TDP-4-keto-4,6-dideoxy-α-d-glucose from CalS13 and TDP-4-amino-4,6-dideoxy-α-d-galactose as external aldimine from WecE in the active site of CalS13. The TDP-4-keto-6-deoxy-α-d-glucose and the external aldimine form of TDP-4-amino-4,6-dideoxy-α-d-galactose are represented as ball and stick models colored white and green, respectively. The loops L0, L1, L7, and L9 are colored blue, and the two SAT subunits are colored blue and yellow.
A Structure-Based Model for the Stereochemical Outcome
To date, crystal structures of nine other sugar nucleotide-dependent SATs have been determined. These include the following: DesI and DesV from Streptomyces venezuelae involved in TDP-desosamine biosynthesis;20,21 Per from Caulobacter crescentus CB15 involved in the GDP-perosamine biosynthesis;22 QdtB from Thermoanaerobacterium thermosaccharolyticum involved in the biosynthesis of TDP-mycaminose;23 ArnB from Salmonella typhimurium involved in the biosynthesis of UDP-4-amino-l-arabinose;18,26 PseC from Helicobacter pylori involved in the biosynthesis of UDP-pseudaminic acid;19 AtS13 from Actinomadura melliaura, a putative C4-aminotransferase involved in the biosynthesis of TDP-4-alkylamino-2,4-dideoxypentose of AT2433 (PDB 4RXK and 4XAU;40 WbpE from Pseudomonas aeruginosa PAO1 involved in the biosynthesis of UDP-ManNAc-(3NAc)A;24 and PglE from Campylobacter jejuni (PDB: 1O61, 1O62, 1O69) involved in the biosynthesis of UDP-4-amino-4,6-dideoxy-α-d-GlcNAc. Of these, CalS13, DesI, Per, and PglE catalyze C4 equatorial amine installation (in the context of a standard 4C1 sugar conformation, Figure 1) to provide the corresponding C4-S stereocenter, while WecE, PseC, and ArnB catalyze C4 axial amine installation to provide the corresponding C4-R (WecE and PseC) or C4-S (ArnB) stereocenter. The structure elucidation of two highly homologous C4 SATs that act upon an identical substrate but differ solely in their stereochemistry of amine installation adds to this growing set of SAT structures and presents critical new information from which a universal SAR model can be proposed as highlighted below.
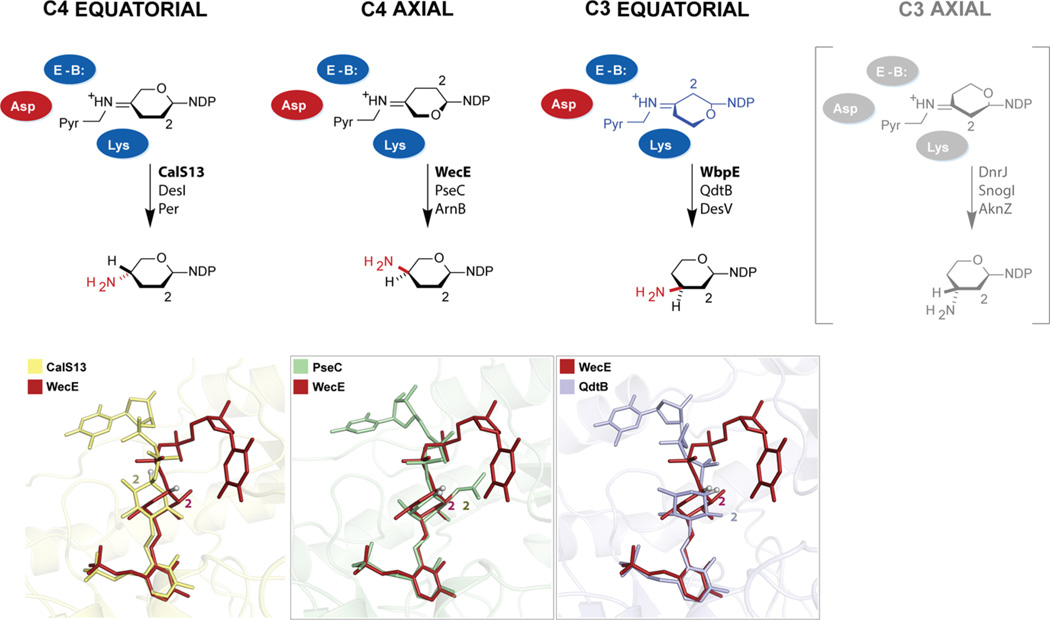
Importantly, an analysis of all nine SATs reveals structural conservation of the key residues involved in catalysis (Figure S2); specifically, the active site lysine (Lys202 and Lys181 in CalS13 and WecE, respectively) that forms the requisite PLP internal aldimine, the conserved general acid (Asp173 and Asp152 in CalS13 and WecE, respectively), and a conserved putative active site base (Gln176 and Gln155 in CalS13 and WecE, respectively). This apparent conservation of a fixed active site scaffold across SATs that catalyze diverse stereo- and regiochemically amine installation reactions suggests subtle alterations in the sugar substrate orientation to drive stereo-chemical (and possibly regiochemical) divergence. Within this context, a comparison of ligand-bound SAT structures reveals two distinct strategies to control the sugar orientation. Specifically, compared to CalS13, DesI, Per, or PglE, the inverted C4 amine installation catalyzed by PseC and ArnB is accomplished via substrate pyrophosphate rotation to ultimately achieve inverted active site entry of the sugar (i.e., a “sugar flip” mechanism, Figure 6). This strategy was first noted by Holden and co-workers via a comparison of PseC and DesI.20 Surprisingly, the current study reveals WecE to accomplish the requisite sugar reorientation via an alternative unprecedented mode of nucleotide binding (i.e., a “base flip” mechanism, Figure 6) where WecE stands as the only structurally characterized SAT to date to adopt this alternative nucleotide-binding orientation.
Figure 6.
Universal model for the stereo- and regiochemical control of an SAT-catalyzed amine installation. For each of the four possible amine installation reactions (C4 equatorial, C4 axial, C3 equatorial, and C3 axial; referring to 4C1 conformation), the fixed active-site orientation (represented by conserved colored residues) is highlighted in relation to distinct sugar orientations (upper schematic). The lower panels highlight distinctions in sugar orientation within CalS13 and WecE (emphasizing the “base flip” in WecE, left panel), the distinctions in sugar orientation within WecE and PseC (emphazing two distinct strategies to achieve sugar inversion—the PseC “sugar flip” versus the WecE “base flip”, middle panel), and distinctions in sugar orientation within C3 and C4 SATs (emphazing the C3 QdtB “sugar flip” and ~30° twist in relation to the WecE C4 “base flip” model). Highlighted on the far right (gray) is the anticipated orientation for SATs that catalyze C3 axial amine installation for which structures are currently lacking.
This model can be further extended to C3 SATs. Specifically, analysis of WdpE, QdtB, and DesV ligand bound structures reveals each to utilize the “sugar flip” mechanism reminiscent of PseC and ArnB to accomplish inverted sugar orientation to that observed for CalS13-like catalysts (Figure 6). This is consistent with the corresponding observed C3 equatorial amine stereo-chemical outcome (in the context of a standard 4C1 sugar conformation, Figure 1). Furthermore, C3 SATs adopt a ~30° twist of the sugar as compared to C4 SAT comparators to enable amine installation chemistry at C3 rather than C4. Based upon this trend, we anticipate the C3 SATs that catalyze C3 axial amine installation to bind their sugar nucleotides in an orientation reminiscent of CalS13-like catalysts but with the required a ~30° twist of the sugar to enable C3 modification. While many such putative C3 axial SATs exist (DnrJ,41 SnogI,42 and AknZ43), their three-dimensional structures have yet to be reported.
In summary, the strategic selection and structural study of two homologous C4 SATs (CalS13 and WecE) that utilize identical substrates but differ in their stereochemistry of aminotransfer reveals for the first time a new mode of SAT sugar nucleotide binding. Global analysis of the structures deriving from the current study and those previously reported provide the basis from which to propose a universal model for SAT stereo- and regiochemical control of amine installation. Specifically, this universal model highlights catalytic divergence to derive solely from distinctions within nucleotide sugar orientation upon binding within a relatively fixed SAT active site where the available ligand bound structures of the three out of four representative C3 and C4 SAT examples provide a basis for the overall model. Cumulatively, this study and the corresponding universal model provide a blueprint for future SAT engineering and biochemical study. This work also highlights the importance of structure elucidation for one or more representative members of the C3 axial set to test the SAR hypothesis put forth.
MATERIALS AND METHODS
Materials
E. coli B834 (DE3) and BL21 (DE3) competent cells were purchased from Invitrogen (Carlsbad, CA). The pET-28a E. coli expression vector and thrombin were purchased from Novagen (Madison, WI). Primers were purchased from Integrated DNA Technology (Coralville, IA). Pfu DNA polymerase was purchased from Stratagene (La Jolla, CA). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Ni-NTA superflow column and gel filtration column HiLoad 16/600 were purchased from GE Healthcare (Piscataway, NJ). Amicon Ultra centrifugal filter units were purchased from EMD Millipore (Merck, KGaA, Darmstadt, Germany). Crystal screen kits were purchased from Hampton Research (Aliso Viejo, CA), Molecular Dimensions (Altamonte Springs, FL), Rigaku (Seattle, WA), and Microlytic (Burlington, MA). All other chemicals were reagent grade or better and purchased from Sigma (St. Louis, MO). X-ray data were collected at beamline 21-ID-F (LS-CAT) in the Advanced Photon Source at Argonne National Laboratory (Chicago, IL).
Two HPLC methods (A and B) were employed for sugar nucleotide resolution, and for each, protein was removed using an Amicon Ultra centrifugal filter device prior to HPLC analysis. Method A: Analytical reverse-phase high pressure liquid chromatography (RP-HPLC) was conducted with a Gemini NX C-18 (5 µm, 250 × 4.6 mm) column (from Phenomenex, Torrance, California, USA) with a gradient of 1% B to 50% B over 30 min, 50% B for 5 min, 50% B to 1% over 1 min, 1% B for 7 min (A = 50 mM phosphate buffer pH 6 with 5 mM tetrabutylammonium bisulfate; B = acetonitrile; flow rate =1 mL min−1) and detection monitored at 254 nm. Method B: Analytical RP-HPLC was conducted with a Gemini NX C-18 (5 µm, 250 × 4.6 mm) column (from Phenomenex, Torrance, California, USA) with 2% B over 10 min, 2% B to 10% B over 10 min, 10% B to 50% B in 1 min, 50% B for 5 min, 50% B to 2% over 1 min, 2% B for 5 min (A = 50 mM triethylammonium acetate buffer pH 7; B = acetonitrile; flow rate =1 mL min−1) and detection monitored at 254 nm.
Protein Expression and Purification
The wecE gene was amplified from E. coli BL21 (DE3) strain (Invitrogen) by colony PCR using primers (forward: 5′- ATACTGGCTAGCATGATTCCATTTAACGCA reverse: 5′-TTGTTGAACTACTTTTCCTGAGAATTC) containing engineered 5′-NdeI and 3′-EcoRI restriction sites. The PCR products were subsequently digested with NdeI and EcoRI and the corresponding digested fragments purified and ligated into pET28a vector to provide expression of N-His6-WecE. For selenomethionyl and native WecE production, the pET28a-WecE construct was transformed into the E. coli methionine auxotroph strain B834 (DE3) and BL21 (DE3), respectively. Se-Met N-His6-WecE overproduction was accomplished using autoinduction medium at 25 °C following standard protocols32 while unlabeled N-His6-WecE and N-His6-CalS13 were overproduced and purified as previously described.30 The His6-tag of unlabeled and Se-Met N-His6-WecE was removed by thrombin cleavage at 4 °C overnight in buffer B (25 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM TCEP buffer containing 2 mM PLP). After proteolysis, Se-Met WecE and unlabeled WecE were purified by Superdex 200 gel filtration in buffer B and concentrated to 25 mg mL−1 and 45 mg mL−1, respectively, flash frozen in liquid nitrogen, and stored at −80 °C. N-His6-CalS13 (referred to simply as CalS13 herein) was dialyzed against 25 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM TCEP buffer containing 2 mM PLP and then purified by superdex 200 pg gel filtration in buffer B and concentrated to 14 mg mL−1, flash frozen in liquid nitrogen, and stored at −80 °C.
Protein Crystallization
Initial crystallization screens were performed using an in house screen comprising IndexHT, SaltHT, Crystal Screen HT, PegRx (Hampton Research), MIDAS, Morpheus, JCSG+ (Molecule Dimensions), Wizard screens 1–4 (Rigaku), and MCSG screens 1–4 (Microlytic) by the sitting drop method using a Mosquito dispenser (TTP labTech Hertfordshire, United Kingdom). The Se-Met WecE crystals were grown by mixing 1 µL of protein sample solution (10–25 mg mL−1, 20 mM HEPES pH 7.5, 150 mM NaCl) with 1 µL of reservoir solution (100 mM sodium acetate, 100 mM MES pH 6.5, 30% w/v PEG 2000 MME) at 20 °C using the hanging drop method. The native WecE crystals were grown by mixing 0.3 µL of protein sample solution (22.5 mg mL−1 WecE, 2 mM PLP, 2 mM TDP-4-keto-4,6-dideoxy-α-d-glucose/TDP-4-amino-4,6-dideoxy-α-d-galactose, 20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM TCEP) with 0.3 µL of reservoir solution (10% 2-propanol, 0.1 M sodium citrate pH 5.0, 26% v/v PEG 400) at 20 °C using the sitting drop method. The CalS13 crystals were grown by mixing 0.3 µL of protein sample solution (14 mg mL−1 CalS13, 2 mM PLP, 2 mM TDP-4-keto-4,6-dideoxy-α-d-glucose/TDP-4-amino-4,6-dideoxy-α-d-glucose, 20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM TCEP) with 0.3 µL of reservoir solution (200 mM ammonium sulfate, 100 mM Bis-Tris pH 6.5, 25% w/v PEG 3350) at 20 °C using the sitting drop method. The best crystals for the protein–ligand complexes appeared in the presence of TDP-4-keto-4,6-dideoxy-α-d-glucose and TDP-4-amino-4,6-dideoxy-α-d-galactose for CalS13 and WecE respectively. The Se-Met WecE crystals were cryoprotected with 27% PEG 2000 MME and 10% glycerol. Native WecE crystals were cryoprotected with 20% glycerol and 20% PEG 400. CalS13 crystals were directly frozen. All crystals mentioned above were then flash frozen in liquid nitrogen for data collection.
Data Collection and Structure Refinement
X-ray diffraction data were collected at the Life Sciences Collaborative Access Team (LS-CAT) with an X-ray wavelength of 0.98 Å at the Advanced Photon Source at Argonne National Laboratory. Data sets were indexed and scaled using HKL200033 or XDS.34 Molecular replacement was utilized to solve the phase problem of Se-Met WecE (PDB: 4PIW), using PDB 1MDO as the starting model, and then phenix.autobuild was used for automatic model building.34 For the native WecE structure (PDB: 4ZAH), molecular replacement starting from Se-Met WecE was accomplished using phenix.phaser and phenix.autobuild.35 The CalS13 structure (PDB: 4ZAS) was solved by molecular replacement as well, using an unpublished structure AtS13 (PDB: 4XAU) as the starting model.40 All the structures were completed with alternating rounds of manual model building with COOT36 and refinement with phenix.refine.35 Structure quality was then validated by Molprobity.37
Enzymatic Synthesis of TDP-4-keto-4,6-dideoxy-α-d-glucose, TDP-4-amino-4,6-dideoxyglucose, and TDP-4-amino-4,6-dideoxy-α-d-galactose
To a 1 mL solution of 10 mM TDP-α-d-glucose in 50 mM Tris buffer, pH 8, was added 10 µM RfbB,30 and the solution was incubated at 37 °C overnight. When the reaction progress was confirmed by reverse-phase HPLC (Method A) to be ≥95%, protein was removed with an Amicon Ultra centrifugal filter device (10 000 Da molecular weight cutoff, EMD Millipore) and concentrated, and crude product was used for cocrystallization. For aminosugar nucleotide production, after completion of the same 1 mL reaction, 50 mM l-Glu, 500 µM PLP, and 10 µM WecE or CalS13 were added and reactions were incubated at 37 °C overnight. Protein was removed with a Amicon Ultra centrifugal filter device (10 000 Da molecular weight cutoff, EMD Millipore), and the desired products were purified by RP-HPLC (Method B), evacuated to dryness, and resuspended in ddH2O to a concentration of roughly 20 mM. Products were confirmed by 1H NMR as previously reported.30
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health Grants U01GM098248 (GNP) and CA84374 (JST) and the National Center for Advancing Translational Sciences (UL1TR000117) and the Kresge Foundation. The Life Sciences Collaborative Access Team (LS-CAT) has been supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supplemental Figures S1 and S2. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.5b00244.
The authors declare the following competing financial interest(s): The authors report competing interests. J.S.T. is a cofounder of Centrose (Madison, WI).
REFERENCES
- 1.Kren V, Rezanka T. Sweet antibiotics - the role of glycosidic residues in antibiotic and antitumor activity and their randomization. FEMS Microbiol. Rev. 2008;32:858–889. doi: 10.1111/j.1574-6976.2008.00124.x. [DOI] [PubMed] [Google Scholar]
- 2.Slawson C, Copeland RJ, Hart GW. O-GlcNAc signaling: A metabolic link between diabetes and cancer? Trends Biochem. Sci. 2010;35:547–555. doi: 10.1016/j.tibs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gantt RW, Peltier-Pain P, Thorson JS. Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules. Nat. Prod. Rep. 2011;28:1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]
- 4.Cipolla L, Peri F. Carbohydrate-based bioactive compounds for medicinal chemistry applications. Mini Rev. Med. Chem. 2011;11:39–54. doi: 10.2174/138955711793564060. [DOI] [PubMed] [Google Scholar]
- 5.Kolarich D, Lepenies B, Seeberger PH. Glycomics, glycoproteomics and the immune system. Curr. Opin. Chem. Biol. 2012;16:214–220. doi: 10.1016/j.cbpa.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Nagaoka I, Igarashi M, Sakamoto K. Biological activities of glucosamine and its related substances. Adv. Food Nutr. Res. 2012;65:337–352. doi: 10.1016/B978-0-12-416003-3.00022-6. [DOI] [PubMed] [Google Scholar]
- 7.Gabius HJ, Kayser K. Introduction to glycopathology: The concept, the tools and the perspectives. Diagn. Pathol. 2014;9:4. doi: 10.1186/1746-1596-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison MJ, Imperiali B. The renaissance of bacillosamine and its derivatives: Pathway characterization and implications in pathogenicity. Biochemistry. 2014;53:624–638. doi: 10.1021/bi401546r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stencel-Baerenwald JE, Reiss K, Reiter DM, Stehle T, Dermody TS. The sweet spot: defining virus-sialic acid interactions. Nat. Rev. Microbiol. 2014;12:739–749. doi: 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elshahawi SI, Shaaban KA, Kharel MK, Thorson JS. A comprehensive review of glycosylated bacterial natural products. Chem. Soc. Rev. 2015 doi: 10.1039/c4cs00426d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nedal A, Zotchev SB. Biosynthesis of deoxyaminosugars in antibiotic-producing bacteria. Appl. Microbiol. Biotechnol. 2004;64:7–15. doi: 10.1007/s00253-003-1535-9. [DOI] [PubMed] [Google Scholar]
- 12.Milewski S, Gabriel I, Olchowy J. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast. 2006;23:1–14. doi: 10.1002/yea.1337. [DOI] [PubMed] [Google Scholar]
- 13.Flatt PM, Mahmud T. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Nat. Prod. Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 14.Timmons SC, Thorson JS. Increasing carbohydrate diversity via amine oxidation: Aminosugar, hydroxyaminosugar, nitrososugar, and nitrosugar biosynthesis in bacteria. Curr. Opin. Chem. Biol. 2008;12:297–305. doi: 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thibodeaux CJ, Melancon CE, 3rd, Liu HW. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romo AJ, Liu HW. Mechanisms and structures of vitamin B(6)-dependent enzymes involved in deoxy sugar biosynthesis. Biochim. Biophys. Acta. 2011;1814:1534–1547. doi: 10.1016/j.bbapap.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh S, Phillips GN, Jr, Thorson JS. The structural biology of enzymes involved in natural product glycosylation. Nat. Prod. Rep. 2012;29:1201–1237. doi: 10.1039/c2np20039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noland BW, Newman JM, Hendle J, Badger J, Christopher JA, Tresser J, Buchanan MD, Wright TA, Rutter ME, Sanderson WE, Muller-Dieckmann HJ, Gajiwala KS, Buchanan SG. Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide-modifying enzyme. Structure. 2002;10:1569–1580. doi: 10.1016/s0969-2126(02)00879-1. [DOI] [PubMed] [Google Scholar]
- 19.Schoenhofen IC, Lunin VV, Julien JP, Li Y, Ajamian E, Matte A, Cygler M, Brisson JR, Aubry A, Logan SM, Bhatia S, Wakarchuk WW, Young NM. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J. Biol. Chem. 2006;281:8907–8916. doi: 10.1074/jbc.M512987200. [DOI] [PubMed] [Google Scholar]
- 20.Burgie ES, Holden HM. Molecular architecture of DesI: A key enzyme in the biosynthesis of desosamine. Biochemistry. 2007;46:8999–9006. doi: 10.1021/bi700751d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgie ES, Thoden JB, Holden HM. Molecular architecture of DesV from Streptomyces venezuelae: A PLP-dependent transaminase involved in the biosynthesis of the unusual sugar desosamine. Protein Sci. 2007;16:887–896. doi: 10.1110/ps.062711007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook PD, Holden HM. GDP-perosamine synthase: Structural analysis and production of a novel trideoxysugar. Biochemistry. 2008;47:2833–2840. doi: 10.1021/bi702430d. [DOI] [PubMed] [Google Scholar]
- 23.Thoden JB, Schaffer C, Messner P, Holden HM. Structural analysis of QdtB, an aminotransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose. Biochemistry. 2009;48:1553–1561. doi: 10.1021/bi8022015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin A, Olivier NB, Imperiali B. Structural analysis of WbpE from Pseudomonas aeruginosa PAO1: A nucleotide sugar aminotransferase involved in O-antigen assembly. Biochemistry. 2010;49:7227–7237. doi: 10.1021/bi100805b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Straaten KE, Ko JB, Jagdhane R, Anjum S, Palmer DR, Sanders DA. The structure of NtdA, a sugar aminotransferase involved in the kanosamine biosynthetic pathway in Bacillus subtilis, reveals a new subclass of aminotransferases. J. Biol. Chem. 2013;288:34121–34130. doi: 10.1074/jbc.M113.500637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M, Sousa MC. Structural basis for substrate specificity in ArnB. A key enzyme in the polymyxin resistance pathway of Gram-negative bacteria. Biochemistry. 2014;53:796–805. doi: 10.1021/bi4015677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao LS, Ahlert J, Xue YQ, Thorson JS, Sherman DH, Liu HW. Engineering a methymycin/pikromycin-calicheamicin hybrid: Construction of two new macrolides carrying a designed sugar moiety. J. Am. Chem. Soc. 1999;121:9881–9882. [Google Scholar]
- 28.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 29.Gao Q, Zhang C, Blanchard S, Thorson JS. Deciphering indolocarbazole and enediyne aminodideoxypentose biosynthesis through comparative genomics: Insights from the AT2433 biosynthetic locus. Chem. Biol. 2006;13:733–743. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Singh S, Peltier-Pain P, Tonelli M, Thorson JS. A general NMR-based strategy for the in situ characterization of sugar-nucleotide-dependent biosynthetic pathways. Org. Lett. 2014;16:3220–3223. doi: 10.1021/ol501241a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang BY, Lee HJ, Yang YH, Joo HS, Kim BG. Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from E. coli K12. Chem. Biol. 2004;11:915–925. doi: 10.1016/j.chembiol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr, Fox BG. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr. Purif. 2005;40:256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. Xds. Acta Crystallogr., D: Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr., D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z, Hong L, Liu HW. Characterization of protein encoded by spnR from the spinosyn gene cluster of Saccharopolyspora spinosa: Mechanistic implications for forosamine biosynthesis. J. Am. Chem. Soc. 2005;27:7692–7693. doi: 10.1021/ja042702k. [DOI] [PubMed] [Google Scholar]
- 40.Singh S, Kim Y, Wang F, Bigelow L, Endres M, Kharel MK, Babnigg G, Bingman CA, Joachimiak A, Thorson JS, Phillips GN., Jr Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis. Proteins. 2015 doi: 10.1002/prot.24844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madduri K, Hutchinson CR. Functional characterization and transcriptional analysis of the dnrR1 locus, which controls daunorubicin biosynthesis in Streptomyces peucetius. J. Bact eriol. 1995;177:1208–1215. doi: 10.1128/jb.177.5.1208-1215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torkkell S, Kunnari T, Palmu K, Mäntsälä P, Hakala J, Ylihonko K. The entire nogalamycin biosynthetic gene cluster of Streptomyces nogalater: characterization of a 20-kb DNA region and generation of hybrid structures. Mol. Genet Genomics. 2001;266:276–288. doi: 10.1007/s004380100554. [DOI] [PubMed] [Google Scholar]
- 43.Räty K, Hautala A, Torkkell S, Kantola J, Mäntsälä P, Hakala J, Ylihonko K. Characterization of mutations in aclacinomycin A-non-producing Streptomyces galilaeus strains with altered glycosylation patterns. Microbiology. 2002;2002(148):3375–3384. doi: 10.1099/00221287-148-11-3375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.