Abstract
The neutral lipids diacylglycerols (DAGs) are involved in a plethora of metabolic pathways. They function as components of cellular membranes, as building blocks for glycero(phospho)lipids, and as lipid second messengers. Considering their central role in multiple metabolic processes and signaling pathways, cellular DAG levels require a tight regulation to ensure a constant and controlled availability. Interestingly, DAG species are versatile in their chemical structure. Besides the different fatty acid species esterified to the glycerol backbone, DAGs can occur in three different stereo/regioisoforms, each with unique biological properties. Recent scientific advances have revealed that DAG metabolizing enzymes generate and distinguish different DAG isoforms, and that only one DAG isoform holds signaling properties. Herein, we review the current knowledge of DAG stereochemistry and their impact on cellular metabolism and signaling. Further, we describe intracellular DAG turnover and its stereochemistry in a 3-pool model to illustrate the spatial and stereochemical separation and hereby the diversity of cellular DAG metabolism.
Keywords: Lipase, Hydrolase, Acyltransferase, Kinase, Insulin
Introduction
For a long time, diacylglycerol (DAG) has been recognized as lipid molecule which exhibits signaling function. More recently, research has unraveled that certain lipid-modifying enzymes discriminate between different stereo/regioisomers of DAG, pinpointing that different DAG isomers may have distinct cellular functions and fates. Thus, the stereochemical nature of DAG isomers by itself is a determinant for its physiological role in distinct cellular compartments and metabolic pathways.
Stereochemistry of DAG
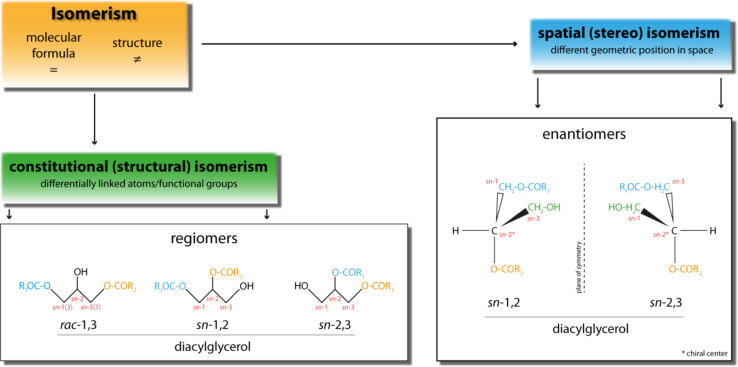
Generally, isomers (from Greek: isos—equal, mèros—part) are molecules sharing identical molecular formulas but differing in their structures. Isomers can be divided into two main groups. On the one hand, there are structural isomers which exhibit differentially linked atoms and functional groups. On the other hand, there are spatial isomers (stereoisomers) which display same linkage of atoms and functional groups but differ in their geometrical position in space. A special group of stereoisomers, named enantiomers, is related by reflection which implies that two enantiomers are not superimposable. Furthermore, enantiomers are characterized by an asymmetric or chiral carbon atom, featured by four different ligands (Fig. 1).
Fig. 1.
Schematic depiction of the different forms of isomerism of diacylglycerol. Diacylglycerols feature different forms of isomerism and can differ either in constitutional (structural) or spatial (stereo) conformation. For detailed explanation see text
DAGs are generated during a variety of metabolic reactions and have attracted much attention as important signaling molecules. Importantly, DAG represents a lipid class that exhibits different isomeric properties. Triacylglycerol (TAG), one possible metabolic precursor of DAG, contains three fatty acids (FAs) esterified to the trihydric alcohol glycerol. This implicates that TAG provides three possible sites for lipase-dependent hydrolysis, which can result in three different DAG isoforms. The stereospecific numbering (sn) designates the conformation of glycerol derivatives, hence the position of the fatty ester at the glycerol backbone (sn-1, sn-2, sn-3; according to IUPAC nomenclature). Accordingly, DAGs generated from the hydrolysis of the phospholipid (PL) headgroup are per definition sn-1,2 isomers since sn-glycerol-3-phosphate/l-α-glycerophosphate (G3P) is the basic building block of all PLs.
TAG exhibits another important property as lipase substrate, namely prochirality. Prochirality describes the condition that an achiral molecule can be converted into a chiral molecule by a one-step reaction. In case of TAG, the potential achiral carbon atom at sn-2 position becomes a chiral center by removal of the attached FA either at sn-1 or at sn-3 position.
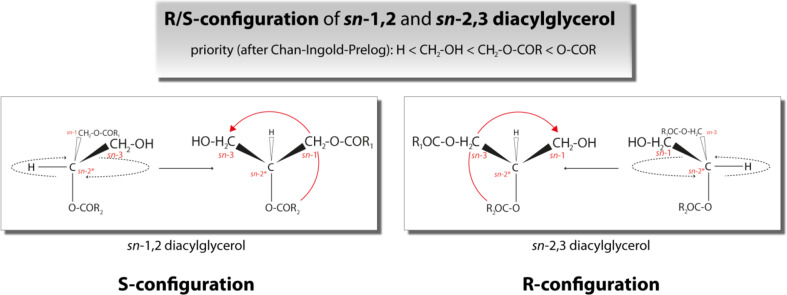
Prochiral TAG species exhibit chemically identical but enantiotopic reactive groups (e.g., oleic acid (C18:1) at sn-1 and sn-3 position). These groups (FAs) can be stereochemically discriminated during lipase-dependent hydrolysis reaction, which results in a chiral DAG product. Lipase-dependent cleavage of the FA esterified at either sn-1 or sn-3 position of a TAG molecule leads to the generation of a chiral center at the sn-2 position and to one of the two DAG enantiomers, sn-2,3 or sn-1,2 DAG, respectively. These DAG enantiomers face themselves as reflection and are not superimposable (Fig. 1). Importantly, 1,3 DAGs can be achiral or chiral, depending on the fatty acid species esterified to the sn-1 and sn-3 position. If two identical fatty acid species are esterified to both position then 1,3 DAG is achiral while if fatty esters at the sn-1 and sn-3 position are dissimilar then 1,3 DAG is chiral. Since in many studies neither fatty ester species nor their respective position on the 1,3 DAG molecule have been determined we refer to 1,3 DAG as racemic/rac-1,3 DAG throughout this review to account for unknown stereochemistry. Furthermore, enantiomeric DAGs can be classified in respect to the R/S-configuration nomenclature based on the Cahn–Ingold–Prelog (CIP) system. The CIP system is used to uniquely specify enantiomers. Therefore, priorities are assigned to all groups attached to the chiral center (CIP rules) [1, 2]. Subsequently, the lowest ranked group is set below the image plane and the other groups are counted starting at highest priority substituents. The counted sequence can be either clockwise or counterclockwise and specifies the present molecule as either R-configured (from Latin: rectus—right) or S-configured (from Latin: sinister—left). In case of DAG, sn-1,2 DAG reflects the S-configuration whereas sn-2,3 DAG is R-configured (Fig. 2).
Fig. 2.
R/S nomenclature of diacylglycerol (DAG) enantiomers according to Cahn–Ingold–Prelog convention. sn-1,2 DAG represents the S-configuration whereas sn-2,3 DAG represents the R-configuration
Upon hydrolysis of the sn-2-bound FA of a TAG molecule, the generated DAG exhibits rac-1,3 conformation. In regard to the different region of hydrolysis (sn-1/sn-3: esters of a primary alcohol; sn-2: ester of a secondary alcohol), rac-1,3 DAG is a so-called regiomer (Fig. 1). Thus, lipases can regioselectively differentiate between sn-2 and sn-1/sn-3 position or enantioselectively differentiate between sn-1 and sn-3 position. Similarly, other DAG metabolizing enzymes, such as acyltransferases may also discriminate between different isoforms of DAGs.
These differences in DAG isomerisms as well as in the selectively of DAG-generating/consuming enzymes are an important issue when DAGs which represent an intersection point between lipid and signaling metabolism are investigated. The selectivity of cellular lipases/acyltransferases as well as the impact of DAG isomerism on cellular metabolism is important to fully understand cellular functions of different DAG molecules.
Intracellular formation of DAG
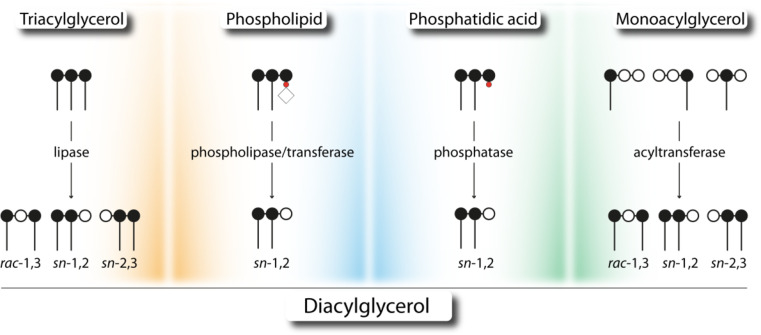
Intracellularly, several reactions contribute to the generation of DAG and are located at different subcellular compartments, including the endoplasmic reticulum (ER), the Golgi network, lipid droplets (LDs), and the plasma membrane. Therefore, either TAG, stored in cytoplasmic or ER-associated LDs or PLs, which are building blocks of cellular membranes, can act as source for lipase-, or acyltransferase-dependent generation of DAG. Additionally, DAGs are also an intermediate during de novo synthesis of TAGs either generated by acyltransferases or by phosphohydrolases. The stereo/regioselectivity of involved enzymes and the isomerism of formed DAGs are largely unknown but might be crucial for subsequent cellular reactions. The following sections describe known biochemical characteristics and stereo/regiochemical properties of enzymes involved in the formation of DAGs (Fig. 3).
Fig. 3.
Catabolic and anabolic reactions leading to the formation of diacylglycerol. Different stereo/regioisomers of diacylglycerols are generated by the hydrolysis of either triacylglycerol (lipase) or phospholipids (phospholipase), and are product of sphingomyelin synthesis (transferase reaction). Furthermore, diacylglycerol is the product of the dephosphorylation of phosphatidic acid (phosphatase) and of the esterification of monoacylglycerol by acyltransferases. Carbon atoms of the glycerol backbone are depicted as filled (esterified) or open (unesterified) circles. Phosphate group, phosphate head group, and fatty acids are depicted as red circle, open rhomb, and dash, respectively
Formation of DAG by neutral triglyceride lipases
Within most cell types, TAG turnover is crucial to balance energy storage and utilization. In whole body energy metabolism of higher organisms, this function is mainly achieved by specialized cells, named adipocytes and hepatocytes, constituents of the white adipose tissue (WAT) and the liver, respectively. In adipocytes, excessive energy is stored in form of TAG in cytoplasmic LDs. Interestingly, adipocytes usually harbor one giant LD with a size range of about 100 µm, while non-adipocytes exhibit multiple LDs with a diameter of around 1 µm. Irrespectively thereof, all LDs share basically the same architecture. The core is strictly assembled by hydrophobic lipid esters, like TAG and cholesteryl esters (CEs), and the surface is formed by a PL monolayer [3]. This monolayer harbors a variety of anchored or embedded proteins and serves as an amphipathic shield against the aqueous milieu present within the cell [4]. The most important function of adipocyte LDs is the storage and the lipase-dependent release of energy metabolites, primarily in form of FAs. This tightly regulated process of TAG degradation, known as lipolysis, is initiated by TAG lipases that generate DAGs and FAs.
Hepatocytes are a crucial intersection of the energy metabolism by receiving, remodeling, and distributing lipids from and to the systemic circulation. Hepatocytes incorporate circulating FAs into TAGs which are stored either in cytoplasmic LDs or used for very low density lipoprotein synthesis within the lumen of the endoplasmic reticulum (ER) [5]. Both, the cytoplasmic and the luminal TAG stores are accessible for different lipases.
Adipose triglyceride lipase generates rac-1,3 DAG at the LD
In 2004, adipose triglyceride lipase (ATGL) was independently identified by three laboratories. The former denoted transport secretion protein 2.2 was renamed as ATGL/patatin-like phospholipase domain containing 2 (PNPLA2) [6], desnutrin [7], and calcium-independent phospholipase A2ζ (iPLA2ζ) [8].
ATGL is one of nine PNPLA family members found in humans (PNPLA1–9) [9]. The PNPLA protein family is named after the patatin-domain which was first identified in hydrolases of the potato plant and denominated after the most abundant protein of the potato tuber, patatin. Lipid hydrolases, containing this domain, catalyze the non-selective hydrolysis of a variety of lipids, including PLs, glycolipids, DAGs, and monoacylglycerols (MAGs) [10, 11]. Orthologs of ATGL exist in almost every eukaryotic species including invertebrates, fungi, and plants.
ATGL localizes to cytosolic LDs [12] and exhibits highest hydrolytic activity for TAGs, much less for retinyl esters (REs) and no detectable activity for other neutral lipids such as DAGs, MAGs, and CEs [6, 13]. Additionally, phospholipase A2 (PLA2) activity as well as DAG transacylase activity have been also reported but the physiological relevance of these activities has not been established [8, 14].
In mice, ATGL mRNA is expressed in all examined tissues. Highest expression is observed in WAT and brown adipose tissue (BAT) and is manifold increased upon fasting. Lower expression levels are detectable in skeletal muscle, cardiac muscle, liver, and testis [6, 7, 15, 16]. Interestingly, besides nutritional regulation at the mRNA level [6, 7, 17, 18] ATGL’s activity is highly regulated at the post-transcriptional level by the interaction with an activator and inhibitor protein [19, 20]. Binding of molar amounts of comparative gene identification-58 (CGI-58; or α/β hydrolase fold domain containing protein 5, ABHD5) stimulates ATGL’s activity up to 20-fold [19, 21]. In contrast, binding of molar amounts of G0/G1 switch gene 2 (G0S2) to ATGL complete abolishes ATGL’s hydrolase activity [20, 21]. Interestingly, CGI-58 and G0S2 regulate ATGL in a non-competitive manner, although the molecular mechanism behind CGI-58- and G0S2-dependent regulation of ATGL’s activity is unknown.
The key role of ATGL in the degradation of TAGs is evident by the phenotype of ATGL-deficient mice. ATGL knockout mice exhibit enlarged adipose tissues. TAG accumulation in LDs is observable in all tissues, reaching up to tenfold increase in TAG content [22]. These observations consolidate ATGL as the rate limiting lipase in cytoplasmic TAG mobilization of adipose and non-adipose tissues.
Studies on substrate and stereospecificity revealed that ATGL hydrolyzes long-chain fatty acids of various length and saturation. Importantly, ATGL is unique among mammalian lipases by hydrolyzing TAGs selectively at sn-2 position. In the absence of its co-activator protein CGI-58, ATGL generates rac-1,3 DAGs. Interestingly, upon co-activation by CGI-58, ATGL expands its regioselectivity to the sn-1 position, generating additionally sn-2,3 DAGs. Notably, ATGL does not hydrolyze ester bonds at the sn-3 position of TAGs, hence does not generate detectable amounts of sn-1,2 DAGs [23].
Hormone-sensitive lipase hydrolyzes lipolysis-derived DAGs at the sn-3 position
Hormone-sensitive lipase (HSL) was initially identified following up the observation that WAT lipolysis is strongly inducible by hormones [24, 25]. HSL is expressed in many tissues; protein as well as mRNA expression of HSL are highest in WAT and BAT. Expression levels are barely increased upon fasting suggesting that its activity is largely regulated by post-translational events [26, 27]. The COOH terminus of HSL harbors an α/β hydrolase fold domain, containing an active site serine (Ser423) as part of a catalytic triad (Asp703, His733), responsible for hydrolytic activity [28–31]. The lipid-binding site as well as the site responsible for protein dimerization is located at the NH2-terminal region of HSL [32].
HSL-dependent activity is strongly regulated by hormones. A variety of phosphorylation events controls HSL activity by affecting both intracellular localization and protein–protein interactions. The major positive stimulus is caused by catecholamines which bind to β-adrenergic receptors during periods of nutritional deprivation (fasting). The contrary nutritional condition (feeding) inhibits HSL activity via insulin [33–39]. Even though phosphorylation events of HSL are crucial, they cause only moderate changes in enzymatic activity (~twofold). The more important factor in HSL activation is the translocation of the enzyme from the cytosol to the LD. In adipocytes, LDs are shielded by perilipin-1 which surrounds LDs, forming a barrier between lipases and respective substrates and prevents effective lipolysis [40, 41]. Upon β-adrenergic stimulation perilipin-1 is phosphorylated [42–45] which causes HSL to bind to perilipin-1. Thereby, HSL translocates to the LD surface and deploys full activity [46, 47].
The long-standing dogma that HSL acts as pacemaker of lipolysis, thereby hydrolyzing TAGs and DAGs, was disproven when mice, carrying a global deletion of HSL, showed no signs of obesity on a high-fat diet but exhibited normal bodyweight and even reduced fat mass [48, 49]. Decreased adipose mass was partially explained by reduced FA esterification, counteracting decreased lipolytic activity [50]. Noteworthy, HSL-deficient adipocytes remain catecholamine-inducible and exhibit increased FA release indicating that an additional TAG lipase must exist [48, 51, 52]. However, the most intriguing finding in HSL knockout mice was the drastic accumulation of DAG in several tissues suggesting that HSL is responsible for DAG hydrolysis [51].
Remarkably, HSL exhibits a uniquely broad substrate spectrum which includes TAGs, DAGs, MAGs, CEs, REs, and the artificial water-soluble esters of para-nitrophenol [25, 53–55]. However, in vitro studies demonstrated that the specific activity of HSL is highest against DAG which exceeds those against TAG and MAG around tenfold [53, 56]. Earlier studies reported specificity of HSL for sn-1/(3) DAGs [56], whereas most recently HSL was identified to be sn-3 specific [57]. Concerning substrate specificity, HSL exhibits preference for polyunsaturated FAs (n-3/n-6) [58].
Both ATGL and HSL together are responsible for more than 90 % of lipolytic (TAG hydrolase) activity as determined in cultured adipocytes and murine WAT [59]. This suggests that other lipases may contribute to a minor extent to TAG breakdown in adipose tissue. Several other enzymes which are known to catabolize TAG breakdown are discussed below.
Triacylglycerol hydrolase/carboxylesterase-3 hydrolyzes short-chain TAGs at the ER
Triacylglycerol hydrolase/carboxylesterase-3 (TGH/Ces3) was initially purified from porcine liver and contains an α/β hydrolase fold common for lipases. Additionally, the NH2-terminal region shows high similarity to that of proline-β-naphthylamidase which is a member of the carboxylesterase family [60, 61]. Additional structural characteristics are an NH2-terminal-located ER signal peptide, a COOH-terminal ER-retention signal (HXEL), a predicted catalytic triad formed by Ser221, Glu353 (354) and His466 (468), and a hydrophobic stretch (AA 414–429) possibly involved in lipid binding [62–68]. In contrast to ATGL and HSL, TGH/Ces3 is mainly expressed in liver and to a low extent in WAT, kidney, cardiac muscle, and small intestine [66]. TGH/Ces3 mRNA level, protein expression, and concomitant hepatic microsomal esterase activity are decreased upon treatment with glucocorticoid analogs, mainly through destabilization of mRNA [69].
Intracellularly, TGH/Ces3 localizes to the ER, especially to areas surrounding cytosolic LDs [70]. TGH/Ces3 catalyzes the hydrolysis of long-, medium-, and preferentially short-chained TAGs [60] with so far unknown stereo/regioselectivity. Besides an unclear role in adipocytes, TGH/Ces3 is affirmed to be involved in the mobilization of hepatic TAG stores, required for the assembly of apoB100-lipoproteins, which has been deduced from the phenotype of mice carrying global disruption of TGH/Ces3 gene [71].
DDHD domain containing 2 hydrolyzes TAGs in the brain
DDHD domain containing 2 (DDHD2), formerly known as KIAA0725p, is one of three sequence-related serine hydrolases which have been annotated as sn-1 specific phospholipases [72]. Mutations in DDHD2 are causative for the genetic disorder known as complex hereditary spastic paraplegia and cause intellectual disability, lower limb spasticity, and weakness.
The ~80 kDa large protein DDHD2 is ubiquitously expressed and exhibits highest expression levels in brain and adipose tissues. Within the cell, DDHD2 localizes to the Golgi network and exhibits phospholipase A1 (PLA1) activity against various PL substrates, including phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylcholine (PC), and phosphatidylethanolamine (PE) [72]. However, a recent study identifies DDHD2 as intracellular TAG lipase involved in neutral lipid metabolism of the central nervous system [73]. DDHD2-deficient mice show significantly elevated TAG levels in brain, especially neurons, but not in other peripheral tissues. Similar, TAG accumulation pattern was observed using a selective DDHD2 inhibitor. Unexpectedly, no changes in the amount or composition of brain PLs were detected in DDHD2-deficient mice [73]. The stereo/region-selectivity of DDHD2 has not been investigated. However, since DDHD2 exhibits PLA1 activity, in addition to its TAG hydrolase activity, DDHD2 may hydrolyze TAG at the sn-1 position, generating sn-2,3 DAG.
Other intracellular triglyceride lipases
Several members of the PNPLA protein family, like adiponutrin (PNPLA3), GS2 (genes sequence 2, also annotated as PNPLA4), GS2-like (PNPLA5) as well as triacylglycerol hydrolase 2/carboxylesterase ML1 (TGH2), have been reported to possess TAG hydrolase activity in vitro and thus may be involved in cellular TAG catabolism.
TGH2 mRNA is highly expressed in WAT, BAT, and liver, and mRNA expression in adipocytes is induced upon differentiation [74]. A substantial amount of TGH2 protein localizes to LDs but TGH2 is also found in cytoplasmic and microsomal fractions. TGH2 shares around 70 % identity to murine TGH/Ces3 and contains a catalytic triad (Ser221, Glu353, His466 numbered for the murine protein), a hydrophobic stretch (414–429 AA), possibly involved in lipid binding, as well as a potential COOH-terminal ER retrieval sequence (HXEL) [74]. TGH2 exhibits hydrolytic activity for para-nitrophenyl butyrate, TAGs, and MAGs but no activity for DAGs, CEs, or PLs [74]. Analyses of FA- and glycerol release of isoproterenol-stimulated 3T3–L1 cells, infected with an adenovirus expressing TGH2, suggested a minor contribution (~20 %) of TGH2 to cellular lipolysis [74]. Besides the discovery that TGH2 prefers short-chained TAGs as substrates, nothing is known about the stereo-/regioselectivity of this enzyme.
Human GS2 gene which has homologues in several mammalian genomes, except the mouse, is expressed in several tissues including WAT, skeletal muscle, cardiac muscle, liver, and skin (keratinocytes) [9, 14, 15, 75]. GS2 exhibits high homology to ATGL and contains an NH2-terminal-located catalytic dyad, including the canonical GXSXG lipase motif [14]. A variety of substrates is described for human GS2, including TAGs [14, 15], REs [14, 76], and PLs. Additionally, also a neutral lipid transacylase activity has been reported [8, 14]. Interestingly, the GS2 orthologue in rat displays lower RE hydrolase activity as well as differences in TAG hydrolase activity [14]. Whereas nothing is known about the FA preference, human GS2 is reported to hydrolyze TAGs at sn-1/(3) and sn-2 position, generating rac-1,3 and sn-1,2/(2,3) DAGs [14]. These products are not observable for rat GS2 which generates only sn-1,2/(2,3) DAGs. However, the physiological role of GS2 is unclear since no knockout model has been described and knockout mice cannot be generated (the mouse genome lacks the GS2 gene).
In contrast to GS2, GS2-like (also annotated as PNPLA5) mRNA is expressed to a low level in both mouse and human WAT, BAT, brain, and lung [9, 15]. The murine mRNA is upregulated during adipocyte differentiation and highly induced in liver samples of leptin-deficient mice [15]. In contrast, mRNA expression is markedly decreased during fasting [15]. Hydrolytic activity of GS2-like emerges to be enigmatic since non-tagged GS2-like protein exhibits no RE or TAG hydrolase activity in vitro [14] but lowers TAG accumulation when expressed in cells [15]. The stereo-/regioselectivity and the physiological relevance of GS2-like are unknown.
Adiponutrin (also annotated as PNPLA3) was first described as adipose tissue-specific transcript strongly regulated by the nutritional state [77]. Within the PNPLA family, adiponutrin shares highest sequence homology with ATGL. Adiponutrin, like all PNPLA proteins, contains a canonical serine lipase motif (GXSXG) within a catalytic dyad [10]. Adiponutrin is highly expressed in WAT, skin, and liver, and localizes to intracellular membranes and LDs [78, 79]. mRNA levels are highly increased by insulin or during adipocyte differentiation and are strongly decreased upon fasting (which is the opposite to ATGL mRNA expression) [7–9, 15, 77, 80]. Human adiponutrin exhibits TAG, DAG, MAG, and RE hydrolase as well as transacylase and PLA2 activity in vitro [8, 10, 15, 81]. Adiponutrin preferentially hydrolyzes TAG species containing long-chain unsaturated FAs (C18:1) thereby generating sn-1,2/2,3 DAGs (DAG species were not analyzed) [82]. However, overexpression or knock down of adiponutrin in mice did not affect hepatic TAG levels [83–85]. Notably, a human adiponutrin mutant (I148M) has been found to be strongly associated with nonalcoholic fatty liver disease and overexpression of the I148M mutant or a I148M knock-in in mice causes steatosis [84, 86]. In contrast to the reported hydrolytic activities, a recent study found that human and murine adiponutrin act as nutritionally regulated acyl-CoA-dependent lysophosphatidic acid acyltransferase and that this activity is elevated in the I148M mutant proteins [87]. Since decreased TAG hydrolytic activity as well as increased lysophosphatidic acid acyltransferase activity of the I148M mutant could explain the development of liver steatosis in humans, further studies are needed to clarify the physiological role of this protein.
Phospholipase C generates sn-1,2 DAGs at the plasma membrane
Phospholipases specifically hydrolyze PLs at different chemical positions. The four major classes of phospholipases are distinguished by the type of catalyzed reaction. PLA1 and PLA2 catalyze the hydrolytic cleavage of the acyl chains at respective sn-1 or sn-2 position. In contrast, phospholipase C (PLC) and D (PLD) cleave phospho-glycerol and phospho-headgroup esters, respectively. Hence, only PLCs contribute directly to the intracellular formation of DAG from glycerophospholipids.
PLC activities have been grouped according to their substrate preference into phosphatidylcholine- and phosphatidylinositol-specific PLCs. Phosphatidylcholine-specific PLC (PC-PLC) are described in many organisms including mammals, in which the number of PC-PLC isoforms depends on cell type and species [88]. Localization and activity studies in murine fibroblasts as well as human lymphocytes showed PC-PLC expression and activity at perinuclear areas and upon mitogen-mediated cell receptor stimulation a translocation to the outer leaflet of the plasma membrane [89]. The fact that the gene sequence of mammalian PC-PLC has not been identified so far displays a critical hindrance since overexpression or deletion studies of mammalian PC-PLC cannot be performed. Thus, relatively little is known about the physiological role of mammalian PC-PLCs.
So far, thirteen phosphatidylinositol-specific PLC (PI-PLC) isozymes have been identified and assigned to six subclasses, namely β1–4, γ1–2, δ1, δ3–4, ε, ζ, and η1–2 [90–93]. Virtually all PI-PLC isozymes are highly expressed in different brain regions and only a few (β3, δ1, δ3, δ4, ε) are distributed among other peripheral tissues like liver, skeletal muscle or cardiac muscle [94]. Intracellularly, the soluble PI-PLC proteins are localized mainly in the cytoplasm. Upon cell activation PI-PLCs translocate to the plasma membrane and develop catalytic activity [94]. The PI-PLC-dependent hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2), a plasma membrane-associated PL, describes a key event during regulation of a variety of cellular functions. By producing two intracellular messengers, namely sn-1,2 DAG and inositol 1,4,5-triphosphate (IP3), this reaction mediates activation of protein kinase C (PKC) as well as intracellular Ca2+ release, respectively [94]. Due to the sn-3 position of the phosphate residue of PLs, PI-PLC generates exclusively sn-1,2 DAGs [23].
Sphingomyelin synthase generates sn-1,2 DAGs at the ER and the plasma membrane
Sphingomyelin synthases (SMSs) catalyze the transfer of the phosphorylcholine residue from PC to a ceramide backbone thereby generating sphingomyelin (SM) and sn-1,2 DAG [95, 96]. So far, two mammalian enzymes, SMS1 and SMS2 have been described [97, 98]. SMS1 localizes exclusively to the luminal side of the trans-Golgi whereas SMS2 additionally localizes to the extracellular leaflet of the plasma membrane [97, 99]. DAGs, produced by SMS1/2 at the trans-Golgi, may trigger the translocation of protein kinase D (PKD) which catalyzes the formation of secretory vesicles [100, 101]. In contrast, DAGs generated at the plasma membrane are most likely utilized by SMS2 to restock SM levels which are locally reduced by sphingomyelinases [102]. SMS-related protein (SMSr) was recently identified as third member of the SMS clan. SMSr localizes to the ER and converts ceramides, generated at the ER, to ceramide-phosphoethanolamine (CPE) using PE as donor [97, 103]. However, the CPE synthesis rate is very low as compared to those of SM synthesis. Thus, SMSr is supposed to regulate ceramide concentrations at the ER [103]. Substrate specificity studies including all three enzymes showed that SMS1 and SMSr are monofunctional SM and CPE synthases, respectively, whereas SMS2 is a bifunctional enzyme, able to catalyze both, SM and CPE synthesis [104]. All three enzymes are ubiquitously expressed in mammals and regulators of SM and ceramide homeostasis. Although the SMS family members exhibit a diverse substrate specificity, the fact that these enzymes use PLs as donors, suggests that the resulting DAG intermediates are sn-1,2 isomers.
Synthesis of DAG de novo or by MGAT reaction
In addition to the catabolic generation of DAGs, two pathways contribute to the anabolic generation of DAGs. In those pathways DAG arises as intermediates of the de novo biosynthesis of TAGs and PLs. The G3P pathway is the major pathway of TAG synthesis in most tissues, predominantly in liver and WAT. The G3P-pathway involves the consecutive acylation of G3P, catalyzed by acyl-CoA-dependent glycerol-3-phosphate acyltransferase (GPAT) and acyl-CoA acylglycerol-3-phosphate acyltransferase (AGPAT). The product of these reactions is PA which is dephosphorylated to DAG by PA phosphatase (PAPase, lipins) [105–107]. Since G3P is utilized in this biosynthesis, these DAG isomers have exclusively the sn-1,2 conformation [108, 109]. In the other so-called MAG-pathway, sn-2 MAGs and long-chain fatty acyl-CoAs are esterified to sn-1,2 DAG by monoacylglycerol-acyltransferase (MGAT) [110]. This pathway plays a predominant role in enterocytes upon feeding and is also involved in the storage of TAGs in adipocytes [111, 112].
sn-1,2 DAG is generated in the G3P pathway at the ER
More than 50 years ago, it was discovered that the liver exhibits enzymatic activity that generates PA from glycerol and that DAG is a precursor for both PL and TAG biosynthesis. In fact, in the late 1950s, Kennedy and coworkers described an enzymatic activity that dephosphorylates PA to form DAG in vitro [113]. This finding completed the enzymatic sequence of TAG and PL synthesis from glycerol and is since then known as the Kennedy pathway [114, 115].
In mammals, PA dephosphorylation (3-sn-phosphatidate phosphatase/phosphohydrolase, PAP/lipin) is catalyzed by three recently identified members of the lipin family, namely lipin1, 2 and 3. Among these, lipin1 is the most extensively studied. It is highly expressed in tissues with high rates of lipid flux, like cardiac muscle, WAT, and skeletal muscle [116, 117]. In WAT and skeletal muscle, lipin1 accounts for the entire PAP activity, whereas the other two members contribute essentially to total PA dephosphorylation of liver, brain, and placenta [117, 118]. Early studies revealed that lipins locate within the cytoplasm and translocate rapidly to ER membranes upon elevated levels of intracellular FAs [119]. Loss-of-function mutations within lipin1 cause dramatic metabolic impairments, like hypertriglyceridemia and severe hepatic steatosis, as observed in fatty liver dystrophic mice [116]. The opposite effect is observed in transgenic mice overexpressing lipin1 in adipocytes. These mice exhibit increased amounts of TAG which fits to lipin1-dependent generation of DAGs as precursor for TAGs [120]. Since G3P and thus also PA are phosphorylated at sn-3 position of the glycerol backbone, their dephosphorylation generates exclusively sn-1,2 DAGs.
The MGAT reaction generates sn-1,2/2,3 DAGs at the ER in the digestive tract
MGAT enzymes catalyze the esterification of MAGs, which constitutes the first step in TAG synthesis following dietary absorption by enterocytes. So far, three enzymes are known to possess MGAT activity, MGAT1, -2, and -3. All three isoforms are located at the ER [121–125]. Besides similar intracellular localization, MGAT isoforms differ in their tissue-specific expression pattern and in their specific catalytic activities. In contrast to MGAT1 which is mainly expressed in stomach, adipose tissues, and kidney, MGAT2 and MGAT3 exhibit highest expression in small intestine [121–123, 125, 126]. MGAT3 is found exclusively in higher mammals and exhibits significantly higher specific DAG-acyltransferase activity as compared to MGAT1 and MGAT2. This indicates that MGAT3 functions as TAG synthase [127]. Furthermore, MGAT3 prefers sn-2 MAG as acyl acceptor and activated palmitic acid (C16:0) and C18:1 as acyl donors [123]. Thus, generated DAG species exhibit either sn-1,2 or sn-2,3 isomerism. The prior-mentioned lipids, sn-2 MAG, C16:0, and C18:1, are the major hydrolytic products of pancreatic triglyceride lipase/colipase (PTL)-dependent TAG hydrolysis during intestinal digestion [128, 129]. Hence, MGAT3 is thought to be main enzyme involved in the re-esterification of dietary absorbed fat within the small intestine.
MGAT2 was found to exhibit little or no selectivity concerning chain-length or level of saturation of activated FAs (FA-CoAs) [121]. Furthermore, incubation of MGAT2 with rac-1/3 MAGs results in the generation of rac-1,3 and rac-1,2/2,3 DAGs, implicating that all positions of the glycerol backbone can be esterified by MGAT2 [121]. In contrast, MGAT1 displays a stricter selectivity for glycerol position as well as for utilized FA-CoA species. MGAT1 shows high activity when incubated with long-chain unsaturated FA-CoAs, the highest with activated arachidonic acid (C20:4) [122]. Incubation with either sn-1 or sn-3 MAGs leads to the generation of rac-1,3 DAGs indicating that MGAT1 preferentially esterifies sn-1 or sn-3 position [122]. In line with these findings, incubation of sn-2 MAGs results exclusively in the generation of the rac-1,2/2,3 DAGs [122].
Extracellular formation of DAGs
Diacylglycerols are also intermediates of the extracellular lipid metabolism. DAGs are generated in the process of food digestion as a result of TAG hydrolysis by specific lipases. Additionally, DAGs are generated in the circulation by the hydrolysis of lipoprotein-associated TAGs, by the action of lipoprotein lipase (LPL) and hepatic lipase (HL). Interestingly, all stereochemically characterized TAG lipases involved in lipid digestion, such as lingual lipase (LL), PTL, and gastric lipase (GL) exhibit preference for sn-1 or sn-3 position of TAGs. The specificity of pancreatic lipase-related proteins 1 and 2 (PLRP1/2) has so far not been determined. Furthermore, only PLRP2 but not PLRP1 is catalytically active against TAG [130–133]. PTL, GL, and LL hydrolyze TAG specifically at the sn-3 position but also hydrolyze DAGs. Thus, they break down TAGs into FAs and sn-2 MAGs [129, 134–137]. An example for a positional unspecific lipase is bile salt-stimulated lipase (BSSL, =carboxyl ester lipase, CEL) which hydrolyzes all positions of TAGs, generating free glycerol and FAs [138]. In contrast to the intestinal lipases, LPL and HL which deplete circulating lipoproteins from TAGs, exhibit sn-1 specificity for TAG and also hydrolyze DAGs at the sn-2 and sn-3 position, generating either sn-3 or sn-2 MAGs, respectively [134, 135, 139, 140]. Since the majority of these lipases not only generates but also accepts DAG as further substrate, the short half-life of extracellular DAGs makes them unlikely to be involved in intracellular signaling events. However, at least in the intestine the absorbed sn-2 MAG has signaling potential, since sn-2 arachidonoyl glycerol (2-AG) is an endogenous ligand of the cannabinoid receptors 1 and 2 (CB1/2) [141, 142]. 2-AG is known to retrogradely bind and activate CB1/2. Furthermore, recent evidence has shown that in the intestinum, 2-AG inhibits gut motility and propulsion [143–145].
Intracellular metabolization of DAGs
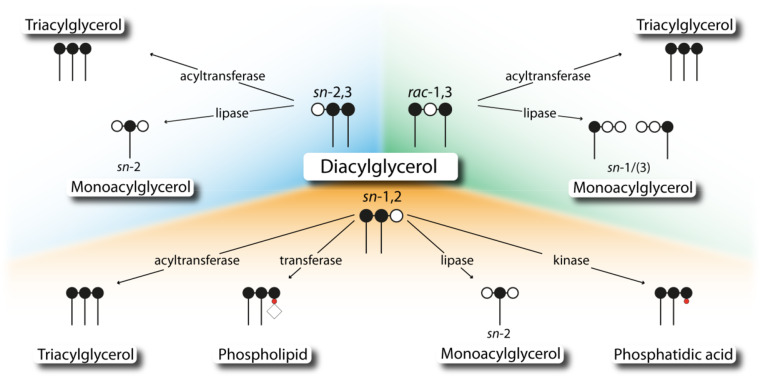
The intracellular metabolization of DAGs involves different enzymes, which in part exhibit selectivity for specific DAG isoforms. Thus, isomerism of DAG may influence (1) its degradation by lipases which leads to different regiomers of MAG species, (2) its re-esterification to TAG by DAG-specific acyltransferases, (3) its conversion to PC by CDP-choline:1,2-diacylglycerol choline phosphotransferases (CPTs), and (4) its phosphorylation by diacylglycerol kinases (DGKs) (Fig. 4).
Fig. 4.
Intracellular enzyme classes involved in diacylglycerol utilization. Different isoforms of diacylglycerol display substrates for several enzyme classes, including transferases, kinases, and lipases. Carbon atoms of the glycerol backbone are depicted as filled (esterified) or open (unesterified) circles. Phosphate group, phosphate head group, and fatty acids are depicted as red circle, open rhomb, and dash, respectively
Diacylglycerol acyltransferases are distinct in intracellular localization and enzymatic preference
The acylation of DAG is the final step of TAG synthesis. This esterification reaction uses FA-CoA as acyl donor and DAG as acyl acceptor and is catalyzed by enzymes called diacylglycerol-O-acyltransferases (DGATs). So far, two mammalian DGAT enzymes have been identified, named DGAT1 and DGAT2.
DGAT1 was initially identified due to its high sequence similarity with acyl-CoA:cholesterol acyltransferase (ACAT) enzymes [146]. DGAT1 belongs to the large family of membrane-bound O-acyltransferases whose members catalyze the transfer of FAs onto thiol or hydroxyl groups of either lipid or protein acceptors [147]. DGAT1 mRNA is highly expressed in the small intestine, adipose tissues, skeletal muscle, cardiac muscle, skin, spleen, and testis, where it localizes strictly to ER membranes [127, 148, 149]. DGAT1 contains three transmembrane-spanning domains and an active site within the COOH-terminal region, facing the ER lumen [150]. Mutagenesis of a highly conserved histidine residue (His-426) located within the COOH-terminal region and supposed to be part of the active site abolishes the ability of DGAT1 to synthesize TAG in vitro [150]. The NH2-terminal region is located in the cytoplasm, facilitates the formation of tetramers, and binds long-chain FA-CoAs [150, 151]. Noteworthy, a C-terminal deletion of DGAT1 is inactive and does not act as a dominant negative protein [152]. The fact that microsomal preparations exhibit an “overt” (cytoplasmic) and a “latent” (luminal) DGAT activity [153], both influenced by pharmacological DGAT1 inhibition in vitro or genetically deletion of DGAT1 [154], questions whether the active site of DGAT1 is entirely oriented to the lumenal side of the ER membrane. In addition to the synthesis of TAG, DGAT1 catalyzes a diversity of different acyltransferase reactions, including MGAT, wax monoester synthase, and retinol acyltransferase [155].
DGAT2 shares little similarity with DGAT1. It belongs to a seven-member family including above-mentioned MGAT1, -2, and -3 [124, 156]. All members of this family contain the highly conserved amino acid sequence HPHG, which in case of DGAT2 is thought to be part of the active site [124, 147, 157]. Additionally, DGAT2 contains a FLXLXXXn consensus sequence which functions as a neutral lipid-binding domain, also found in other neutral lipid metabolizing proteins, such as plasma cholesteryl ester transfer protein, HSL, or TGH/Ces3 [62, 157, 158]. Mutation studies on DGAT2 demonstrated that this neutral lipid-binding domain of DGAT2 is responsible for DAG binding, since DGAT2 variants with mutations in this region exhibit reduced activities [157]. Expression of DGAT2 mRNA is highest in liver, adipose tissues, mammary glands, testis, peripheral leukocytes, and cardiac muscle [124]. In cultured cells DGAT2 localizes to the ER. Upon supplementation of cells with FAs, known to induce TAG synthesis, DGAT2 also localizes to mitochondria-associated membranes (ER domains tightly interacting with mitochondria) and to LDs [149, 159]. Unlike DGAT1, DGAT2 contains two ER-spanning domains and both, the COOH- as well as NH2-terminal domain, face the cytoplasm [157]. Hence, DGAT2 is in part responsible for the “overt” DGAT activity detectable in cells. Additionally, the C-terminus of DGAT2 is supposed to be involved in directing the enzyme to LDs, since DGAT2 with a truncated or mutated C-terminus fails to co-localize with LDs [160]. These different orientations of the active site of DGAT enzymes suggest a spatial separation of DGAT1 and DGAT2-dependent TAG synthesis. Furthermore, and in contrast to DGAT1, DGAT2 does not exhibit activities towards substrates other than DAGs.
Both DGAT enzymes are involved in the intracellular TAG synthesis. Overexpression of either DGAT1 or DGAT2 in mammalian cells leads to increased DGAT activity of corresponding cell lysates [161]. Interestingly, specific enzymatic activity of DGAT1 is higher than that of DGAT2. On the contrary, cells expressing DGAT2 accumulate more TAG, contain bigger LDs, and incorporate more glycerol into the TAG moiety as compared to that of DGAT1 expressing cells [161], questioning the significance of in vitro activity data for the prediction of physiological relevance.
DGAT1 and DGAT2 differ in their acyl donor and acyl acceptor preference. Studies on murine DGAT1 have revealed preference for monounsaturated acyl donors, like C18:1-CoA, over saturated FA-CoAs, like C16:0-CoA [124, 157]; yet, another study on human DGAT1 selectivity found equal preferences for both, C16:0-CoA and C18:1-CoA [162], questioning acyl donor preference of DGAT1. Intestinal human DGAT1 prefers sn-2 MAGs over sn-3 MAGs as substrates for the synthesis of sn-1,2 DAGs [162]. For DGAT2 no preferences for degree of saturation of FA-CoAs but a preference for medium-chained FA-CoAs (C12:0) as acyl donor and short- and medium-chained DAG species (e.g., C6:0, C8:0, C12:0) as acyl acceptor has been reported [124, 156]. Furthermore, DGAT enzymes differ in regard to the isomerism of DAGs as acyl acceptor molecules. DGAT1 utilizes sn-1,2/2,3 DAGs more efficiently than rac-1,3 DAGs. In contrast, DGAT2 exhibits the opposite acyl acceptor preference and prefers rac-1,3 DAGs [23].
An additional difference between DGAT enzymes is the sensitivity against magnesium which was demonstrated by in vitro experiments. High concentrations (>50 mM) are described to entirely suppress DGAT2 activity whereas DGAT1 activity is much less affected [124].
The differences in subcellular localization as well as enzymatic properties of DGATs suggest different intracellular functions. The observation that only DGAT2 localizes to LDs together with the preference for rac-1,3 DAGs suggests a coordinated action of DGAT2 and ATGL on LDs to facilitate a TAG hydrolysis/re-esterification cycle (futile cycle), potentially remodeling sn-2 FA ester species thereby replacing long-chain unsaturated with medium-chain fatty acids. Furthermore, the different intracellular localization of DGAT1/2 suggests that different DAG pools exist for DGATs. The one at LDs is only accessible for DGAT2. The other, at ER membranes, which is mostly supplied by DAG species (sn-1,2 DAGs) derived from the de novo glycero(phospho)lipid synthesis, is accessible for both DGAT1 and DGAT2. This spatial and stereochemical separation of different DAG pools as well as of executing enzymes could be one explanation for the different phenotype of mice carrying global deletions of DGAT1 or DGAT2. The phenotypes of DGAT-deficient mice clearly demonstrate that these enzymes cannot functionally compensate for each other [161, 163]. DGAT1 knockout mice exhibit a moderate phenotype characterized by reduced TAG levels in several tissues (e.g., adipose tissues, skeletal muscle, liver) and resistance to diet-induced obesity [163]. In contrast, DGAT2 knockout mice die within the first day of life most likely as consequence of a severe skin defect and severe lipopenia [161]. The exact reason for these drastic differences of DGAT knockout mice is unknown. Recently, distinct functions of DGAT1 and DGAT2 in hepatic TAG synthesis have been reported. DGAT2 is described to mediate esterification of newly synthesized FAs, whereas DGAT1 catalyzes the synthesis of TAGs via utilization of exogenously supplied FAs [164, 165]. Whether this finding holds true for other tissues needs to be tested.
Diacylglycerol lipases hydrolyze sn-1,2 DAGs at the plasma membrane
HSL is well known to be rate limiting for DAG hydrolysis [48, 51]. Yet, there are additional intracellular lipases which possess specific hydrolytic activities for DAGs, generating MAGs and FAs. In humans, two sn-1-specific DAG lipases have been identified, named DAG lipase α and β (DAGLα and β) [166]. Both enzymes share ~30 % homology and comprise four predicted transmembrane-spanning domains as well as a serine lipase motif [166].
DAGLα is highly expressed in brain and pancreas and to a lower extent in macrophages and adipose tissues, while DAGLβ expression is highest in bone marrow, lymph nodes, spleen, and liver [166, 167]. Enzymatic characterization revealed that both enzymes hydrolyze the sn-1,2 DAG isomer (sn-2,3/rac-1,3 were not tested), exhibiting three- to eightfold higher activity for the sn-1 over the sn-2 position of DAGs [166]. Furthermore, DAGLβ prefers DAG species, containing linoleic acid (C18:2) > C18:1 > C20:4 > stearic acid (C18:0) at sn-2 position, whereas DAGLα shows equal activity against these DAG species [166].
Intracellularly, DAGLα localizes to the plasma membrane and hydrolyzes sn-1,2 DAGs generated by PLC-dependent hydrolysis of phosphatidylinositides [168]. In contrast, DAGLβ also localizes to the LDs [168]. DAGL-mediated breakdown of sn-1,2 DAGs at the plasma membrane results in the formation of sn-2 MAGs. Since PLs, the precursors of PLC-derived DAGs, exhibit a high abundance of C20:4 esterified at the sn-2 position of the glycerol backbone, the released MAG species is frequently 2-AG [169, 170]. As already mentioned above, 2-AG is the most abundant endocannabinoid in tissues and acts as ligand for cannabinoid receptors CB1 and CB2 [171]. In fact, the role of DAGLα and DAGLβ in the biosynthesis of 2-AG is evident from the phenotype of DAGLα or DAGLβ-deficient mice [167]. In line with the tissue expression patterns, DAGLα knockout mice displayed 80 % reduction in 2-AG levels in the brain, whereas DAGLβ knockout mice showed 90 % reductions in 2-AG levels in the liver [167].
The role of DAGL enzymes in the hydrolysis of plasma membrane-derived sn-1,2 DAG species is well established. To date, it is questionable whether sn-1-specific DAGL enzymes are also involved in the degradation of DAG species of other sources such as rac-1,3 DAGs which originate from lipolysis-driven TAG breakdown at the LDs or sn-1,2 DAGs which also derive from de novo synthesis at the ER membrane.
Diacylglycerol kinases are ubiquitously expressed and phosphorylate sn-1,2 DAG
Diacylglycerol kinases (DGKs) catalyze the formation of PA by phosphorylation of the free hydroxyl (–OH) group of DAG. Together with PAPases/lipins, DGKs are crucially involved in the maintenance of intracellular DAG and PA levels. So far, ten DGK isozymes have been identified in mammals [172, 173]. All mammalian DGK isozymes share two common structural features, a cysteine-rich C1 domain which is responsible for DAG binding and potentially involved in protein–protein interaction and a catalytic domain, responsible for enzymatic activity [174]. Virtually every tissue expresses at least one member of the DGK family. However, numerous tissues express several different DGK isozymes, e.g., all ten DGK isozymes can be found in brain extracts [175]. DGKs localize to multiple cellular compartments, including the nucleus, the plasma membrane, the cytoskeleton, the Golgi network, and the ER [176, 177]. Little is known about the specific functions of different DGK enzymes with regard to their organelle distribution. Whereas some isoforms translocate to the plasma membrane upon activating stimuli (most likely to funnel PLC-derived DAGs to PA), others locate inside the nucleus, presumably regulating nuclear DAG levels [174].
DGKs are selective for sn-1,2 DAGs. Most likely, this is due to their C1 domain which shares high homology to the sn-1,2 DAG-specific binding motifs C1A and C1B of PKC [178–180]. However, to date it is unclear whether DGK isoforms are also capable to phosphorylate rac-1,3 or sn-2,3 DAG isoforms. Further enzymatic properties are known for DGKε. DGKε is the shortest DGK isoform which exhibits the lowest molecular weight and localizes either to the plasma membrane or to the ER. DGKε exhibits specificity for sn-1,2 DAG species [181] containing C18:0 at the sn-1 position and C20:4 at the sn-2 position [182, 183] which is the major product of PLC-mediated phosphoinositide hydrolysis [169]. DGKε is thought to terminate 20:4-DAG signaling and to counterbalance PIP2 hydrolysis by specifically phosphorylating C20:4-containing DAGs to PA. In turn, generated PA would then be available for re-synthesis of PI and replenishment of PIP2 by consecutive phosphorylations to phosphatidylinositol 4-phosphate (PIP) and PIP2. This hypothesis is supported by the phenotype of mice carrying a global gene deletion DGKε which show downregulation of the 20:4-DAG to 20:4-PI lipid cycle [184].
Diacylglycerol choline/ethanolamine phosphotransferases specifically utilize sn-1,2 DAGs at intracellular membranes
All tissues and cell types can synthesize PC via the CDP-choline or better known as “Kennedy” pathway [114, 115]. In an analog manner, PE can be formed via the CDP-ethanolamine pathway. The final step of both pathways, namely the direct conversion of DAGs to either PC or PE is catalyzed by CPT or CDP-ethanolamine:1,2-diacylglycerol ethanolaminephosphotransferase (EPT), respectively.
In humans, two proteins exhibit CPT activity. CPT1 acts as CDP-choline-specific enzyme while CEPT1 transfers both CDP-choline as well as CDP-ethanolamine [185, 186]. Both proteins are integral membrane proteins and localize to the Golgi network and the ER membrane, respectively [187]. Human CPT1 mRNA expression is highest in testis, colon, small intestine, cardiac muscle, and spleen, whereas CEPT1 is expressed to a similar degree in all tissues examined [185, 186]. Due to the high CPT activity in cells, none of the two enzymes is thought to be rate-limiting in PC synthesis [188]. Since no purified mammalian CPT enzyme is available, information on substrate specificity is only available from structure/function analyses of CPT1 of S. cerevisiae. The yeast orthologue prefers sn-1,2 DAGs (sn-2,3/rac-1,3 were not tested), with a FA preference in the order of dipalmitolein (diC16:1) > diC16:0 = C18:1/C16:0 > C16:0/C18:1 DAG as substrates [189]. Selectivity studies using microsomal fractions of S. cerevisiae constitutively expressing human CEPT1 suggest a diC16:1 > C16:0/docosahexaenoic acid (C22:6) > C16:0/C18:1 > diC18:1 DAG species preference for CEPT1 [187]. The third enzyme, ethanolamine phosphotransferase 1 (EPT1) selectively catalyzes the transfer of CDP-ethanolamine on sn-1,2 DAGs (sn-2,3/rac-1,3 were not tested), forming PE [190], and contributes only to 5 % to cellular PC synthesis [191, 192]. Furthermore, EPT1 of S. cerevisiae prefers sn-1,2 DAGs (sn-2,3/rac-1,3 were not tested) species, with diC18:1 > diC16:1 > C16:0/C18:1 as FA chains in vitro [189]. Apart from the enzymatic activity, little is known about EPT1 expression pattern or biochemical properties. Interestingly, incubation of rat hepatocytes with radiolabeled ethanolamine results in a high specific accumulation of radioactivity in C16:0/C22:6-PE, suggesting that in mammals EPT activity may exhibit preference for C16:0/C22:6-DAGs as substrates [193].
Taken together, CPT1, CEPT1, and EPT1 are involved in the synthesis of PLs, thereby consuming sn-1,2 DAGs.
How DAG kicks into insulin signaling
Besides the capability of DAGs to negatively regulate transient receptor potential canonical (TRPC) cation channels [194, 195], dysregulations of DAG metabolism and concomitant DAG accumulation is supposed to adversely affect cellular signaling involved in the development of different disease states, like insulin resistance (IR). Intracellular accumulation of DAG is thought to be connected to altered insulin responsiveness since ectopic DAG accumulation positively correlates with disturbed insulin signaling. This adverse effect of DAGs is thought to derive from the action of the PKC family members which are known to play a crucial role in many signaling pathways which control, e.g., cellular differentiation, and cell growth. The PKC family comprises three different PKC subgroups, namely conventional (α, β1, β2 and γ; cPKC), novel (δ, ε, η and θ; nPKC) and atypical (ζ and λ/ι; aPKC) PKCs. cPKCs and nPKCs display lipid-sensitive isoforms and are usually activated by PLC-derived sn-1,2 DAGs, either Ca2+ dependent (cPKC) or independent (nPKC). The C1 domain of conventional and novel PKCs binds DAGs and the tumor promoter phorbol ester with high affinity [196–198]. Hence, the activity of cPKCs and nPKCs is highly influenced by intracellular levels of DAGs. Noteworthy, earlier studies showed that only the sn-1,2 DAG isoform has the ability to activate PKCs, the other isoforms, rac-1,3 and sn-2,3 are inactive [178–180]. Binding of cellular DAGs to members of the PKC family leads to their activation and translocation to the plasma membrane and subsequent phosphorylation of interacting proteins (such as insulin receptor substrate, IRS). It is assumed that a rise in cellular DAG levels may activate PKCs in an uncontrolled fashion.
So far, cellular effects driven by DAG accumulation have been mainly attributed to cPKCs and nPKCs. However, other “non-PKC” proteins also contain a high-affinity DAG/phorbol ester-binding C1 domain. To date a variety of DAG receptor proteins are described which belong to the families of PKD kinases, chimaerin Rac GTPase-activating proteins, Ras guanyl nucleotide-releasing proteins (RasGRPs), Munc13 scaffolding proteins, and former-mentioned DAG kinases [199–205]. Importantly, members of these families have been described to affect cellular processes like cell adhesion (chimaerins) [206], cell proliferation/transformation (RasGRPs) [207, 208], secretory vesicle priming (Munc13s) [209–212], and protein transport (PKDs) [213, 214]. Thus, biological effects driven by DAG binding to proteins of these families must be considered when cellular DAG signaling is investigated. Several reviews on the physiological role of non-PKC C1 domain-containing proteins have elaborated the current knowledge [199–205]. The emerging number of metabolic diseases as result of the first world life style brings DAG-dependent PKC signaling and related attenuation of insulin action in the focus of a multitude of present-day studies. Therefore, we focus in the following section on the PKC signaling axis in more detail.
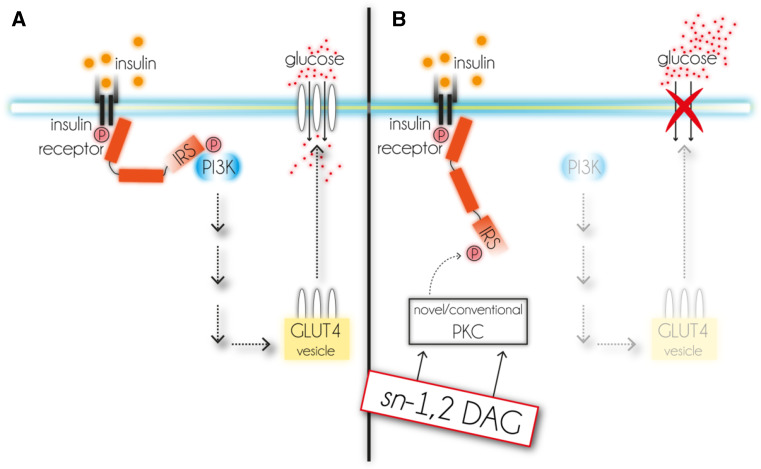
Insulin signaling in myocytes and adipocytes is initiated by the binding of insulin to its receptor at the plasma membrane. Intracellularly, this activates a tyrosine kinase which consecutively phosphorylates one of the intracellular IRS 1-4 family members on several tyrosine residues [215, 216]. Activated IRSs serve as docking station for proteins like phosphatidylinositide-3-kinase (PI3K) and induce downstream effects which finally result in the translocation of glucose-transporter type 4 (GLUT4) to the plasma membrane [217]. Only plasma membrane-associated GLUT4 facilitates glucose uptake (Fig. 5a). Defects within this signaling cascade result in a loss of insulin sensitivity and consequently to IR of affected cells/tissues.
Fig. 5.
Intracellular pathway of insulin signaling and proposed impairment by DAG. a Insulin binds to insulin receptor that activates IRS. This leads to the activation of PI3K and further downstream signaling which finally triggers the translocation of GLUT4 to the plasma membrane and enables glucose uptake. b sn-1,2 DAG activates novel and conventional PKC isoforms which phosphorylate IRS. This event inhibits downstream effector signaling and GLUT4-dependent glucose uptake. DAG diacylglycerol, GLUT4 glucose transporter 4, IRS insulin receptor substrate, P phosphorylation, PI3K phosphoinositide-3-kinase, PKC protein kinase C
Of all PKC subgroups mainly nPKCs, more precisely PKCε and PKCθ, adversely affect insulin signaling [218, 219]. Up to now several mechanisms have been identified by which nPKCs impair insulin action. Recent studies showed that PKCθ can phosphorylate IRS1 at Ser1101 which blocks insulin-stimulated tyrosine phosphorylation [220] and activation of PI3K [218]. As a consequence, GLUT4 does not translocate to the plasma membrane and glucose is not taken up (Fig. 5b).
Mouse models for deregulated DAG metabolism
Deregulation of DAG metabolism and concomitant DAG accumulation is thought to adversely affect cellular signaling and to be causally related to the development of various disease states, including IR. The phenotype of a number of genetic mouse models supports the hypothesis that increased DAG levels in insulin responsive tissues are causative for impaired insulin signaling. For example, overexpression of mitochondrial GPAT in mice leads to elevated levels of lysophosphatidic acid (LPA), DAG, and TAG in the liver and these mice develop hepatic IR in absence of a lipogenic diet or obesity [221]. Similarly, in obese Zucker rats (rats with non-functional leptin receptor) IR is associated with increased amounts of hepatic and muscle ceramide and DAG levels [222]. Another genetic mouse model arguing for DAG as mediator of IR are mice fed a high-ketogenic diet. These mice develop severe hepatic steatosis and severe hepatic IR which is associated with a 350 % increase in hepatic DAG content [223]. These mice exhibit elevated levels of activated PKCε and decreased insulin-mediated tyrosine phosphorylation of IRS2. In mice overexpressing DGAT2 specifically in the liver, hepatic TAG as well as DAG and ceramide levels are markedly increased [224, 225]. These mice were first reported to have normal hepatic insulin sensitivity [224] but were recently identified to exhibit enhanced PKCε activation, accompanied by severe hepatic IR [225]. Furthermore, an interrelation between cellular DAG levels, PKCε activation, and hepatic IR has been observed in numerous models and has been extensively reviewed elsewhere [226]. Interestingly, a recent study discovered that hepatic DAG content of cytoplasmic LDs is the best predictor of IR in obese, non-diabetic individuals [227]. In the same study, they observed distinct localization of PKCε at cytoplasmic LDs as well as enhanced activation of PKCε [227]. So far, this study is unique in connecting DAG turnover at the LD with PKC signaling.
The phenotype of a number of other genetic mouse models argues against a causative role of non-plasma membrane-derived DAGs in the development of IR. For example, HSL knockout mice accumulate large amounts of DAG in adipose and non-adipose tissues [48, 51] but generally they do not develop IR. Unfortunately, the issue of whether or not HSL-deficient mice develop IR is conflicting since depending on the genetic background, some HSL knockout strains show signs of impaired insulin signaling [228, 229] while others do not or show even increased insulin sensitivity [230, 231]. In contrast to mice, however, humans with mutated HSL develop severe diabetes which could be a result of ectopic lipid deposition in liver in combination with partial lipodystrophy [232]. Accumulation of rac-1,3 DAG, as determined in HSL-ko mice, is unlikely to be causative for the development of IR since rac-1,3 DAG isomers are known not to activate PKCs [23, 178–180].
Another mouse model with cellular DAG accumulation was generated by antisense oligonucleotides (ASO)-mediated silencing of CGI-58 expression. ASO injection into mice mainly targets the liver, adipose tissues, and kidney and does not lead to a global silencing of expression. ASO-CGI-58 mice on a high-fat diet regimen exhibit increased hepatic TAG, DAG, and ceramide levels. However, animals have been shown to exhibit improved insulin sensitivity and glucose tolerance [233]. The lack of adverse insulin signaling despite DAG accumulation in ASO-CGI-58-treated mice was explained by the sequestration of DAG molecules in LDs so that they were unable to interact with PKCs [234].
A recent study reported that overexpression of ATGL in cultured myotubes leads to increased DAG and ceramides levels which was found to be associated with impaired insulin signaling [235]. This suggests that deregulation of TAG metabolism might be linked to insulin signaling. Yet, mice globally lacking ATGL or mice overexpressing ATGL specifically in adipose tissue display also deregulated TAG metabolism, but show improved glucose tolerance [22] or are protected from IR [236], respectively, questioning a causal link between LD-associated TAG deregulation and insulin signaling.
In summary, several of the above-mentioned studies suggest a causal link between DAG, PKC signaling, and the development of IR. In this context, DAG isomerism as well as intracellular DAG compartmentation might be of crucial importance. As outlined above, a number of metabolic reactions degrade or consume DAGs. Yet, they are located at different cellular compartments and DAG isoforms involved in these reactions are stereochemically different. Earlier studies clearly demonstrated that the DAG-binding C1 domain of PKCs is highly specific for sn-1,2 DAGs. Other isoforms, like sn-2,3 or rac-1,3 DAGs, are unable to activate PKCs [178–180] and hence display no signaling properties. Due to the inability of DAG isoforms, other than sn-1,2 DAG, to activate PKCs it appears crucial to precisely determine DAG isomerism under normo-physiological and patho-physiological conditions.
3-Pool model of DAG metabolism, conclusions, and perspectives
DAG metabolism involves a battery of enzymes at spatially different cellular locations with distinct enzymatic properties and selectivities. For the sake of simplicity many studies in the past have ignored spatial and stereochemical issues and have correlated total cellular DAG levels with PKC activation as proof for causality between cellular DAG levels and DAG signaling. Future studies on DAG-producing or -consuming enzymes need to address these issues of stereoselectivity of DAG species and their cellular localization. This will help to further elaborate intracellular DAG metabolism and functions of different DAG isomers which is required to comprehend the metabolic implications of normo-physiological and pathological DAG metabolism and function. To date it is appreciated that DAGs exist in different stereo/regioisoforms and it is assumed that certain DAG isoforms exhibit specified intracellular functions at different cellular locations and processes. Furthermore, the amount of DAGs at different sites (e.g., intracellular membranes vs plasma membranes) depends on various stimuli or metabolic processes and underlies a remarkable spatiotemporal dynamic [237–239].
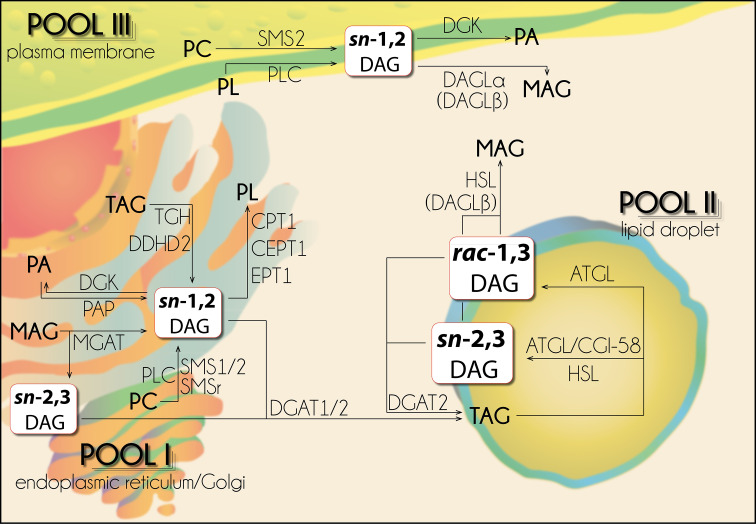
The stereochemistry of DAG isomers as well as the distinct localizations and activities of DAG-generating/consuming enzymes suggest a spatial separation of DAG pools which can be outlined in a “3-pool model” of intracellular DAG compartmentation [23]. This model describes that 3 distinct intracellular DAG pools exist at the ER/Golgi network, the LD, and the plasma membrane which differ to some extent in their DAG isomerism and are accessible for different sets of enzymes.
“Pool I” is located at the ER/Golgi network and consists of sn-1,2 DAGs. This DAG pool is generated by de novo glycero(phospho)lipid synthesis either by PAPases/lipins or MGAT enzymes, as by-product of SM synthesis catalyzed by SMSs, or as product of TAG hydrolysis by lipases like TGH/Ces3 or DDHD2. Furthermore, sn-1,2 DAGs of this pool can be utilized by DGKs forming PA, DGAT enzymes generating TAG, and PL-generating enzymes like CEPT1, CPT1, and EPT1 (Fig. 6).
Fig. 6.
“3-Pool” compartmentation model of intracellular diacylglycerols. Intracellular diacylglycerols differ in their stereo/regio conformation, compartmentation, and generating/consuming enzymes. For detailed description see text. ATGL adipose triglyceride lipase, CGI-58 comparative gene identification-58, CEPT CDP-ethanolamine/choline:1,2-diacylglycerol ethanolaminephosphotransferase CPT, CDP-choline:1,2-diacylglycerol ethanolaminephosphotransferase, DAG diacylglycerol, DAGL DAG lipase, DGAT DAG-O-acyltransferase, DGK DAG kinase, EPT CDP-ethanolamine:1,2-diacylglycerol ethanolaminephosphotransferase, HSL hormone-sensitive lipase, MAG monoacylglycerol, MGAT monoacylglycerol-O-acyltransferase, PA phosphatidic acid, PAP PA phosphohydrolase/lipin, PC phosphatidylcholine, PL phospholipid, PLC phospholipase C, SMS(r) sphingomyelin synthase (related), TAG triacylglycerol
Lipolysis-derived DAGs represent “Pool II” which is located at cytoplasmic LDs. This pool comprises rac-1,3 and sn-2,3 DAGs and is fueled by the TAG hydrolytic activities of ATGL and HSL. The rac-1,3 DAG isomer is generated by TAG hydrolysis of ATGL. The sn-2,3 DAG isomer is generated by TAG hydrolysis of HSL and of ATGL, in addition to the rac-1,3 DAG isomer, when co-activated by CGI-58. DAGs of this pool can be re-esterified to TAGs exclusively by DGAT2 or further be catabolized to sn-2 MAGs by HSL(or DAGLβ)-mediated hydrolysis (Fig. 6).
“Pool III” is located at the plasma membrane and consists of sn-1,2 DAGs. These sn-1,2 DAGs are generated either by PLC-dependent breakdown of PLs or during SM production catalyzed by SMS2. DAGs of “Pool III” can serve as precursors for the synthesis of PA through DGK-catalyzed phosphorylation or can be hydrolyzed to sn-2 MAGs by membrane-associated DAGLα (or DAGLβ). So far, only sn-1,2 DAGs of “Pool III” are established to be involved in DAG signaling events, executed by nPKC and cPKC isoforms (Fig. 6).
The differences in stereo/regiochemistry of involved DAGs as well as the topological separation of these three DAG pools may provide explanations to previously puzzling data in regard to DAG-induced signaling, lipid metabolism, and metabolic disorders. Furthermore, the involvement of so far unidentified DAG-specific binding proteins and/or DAG isomerases could interconnect different intracellular sources of DAGs and could explain seemingly contradicting findings on DAG function and signaling potential. Following a multiplicity of studies focusing on the role of DAGs and in-depth reviews of DAG metabolism and functions [240–242], the future attention will further shift to the question which role is attributed to different DAG isomers as well as to different DAG species, in respect to their FA composition, in intracellular metabolism. Unraveling these issues will finally paint a more complete picture of the interrelation between lipid metabolism, intracellular signaling, and certain disease pathologies.
Acknowledgments
We apologize to authors whose studies have not been cited due to space limitations. This work was supported by the grant P25193 (A.L.) which is funded by the Austrian Science Fund (FWF). We thank Mag. Dr. Ulrike Taschler for carefully reading the manuscript.
Abbreviations
- 2-AG
2-Arachidonoyl glycerol
- AGPAT
Acyl-CoA acylglycerol-3-phosphate acyltransferase
- ATGL
Adipose triglyceride lipase
- BAT
Brown adipose tissue
- BSSL/CEL
Bile salt-stimulated lipase/carboxyl ester lipase
- CGI-58
Comparative gene identification-58
- CE
Cholesteryl ester
- CEPT
CDP-ethanolamine/choline:1,2-diacylglycerol ethanolaminephosphotransferase
- CPT
CDP-choline:1,2-diacylglycerol ethanolaminephosphotransferase
- DAG
Diacylglycerol
- DAGL
DAG lipase
- DGAT
DAG-O-acyltransferase
- DGK
DAG kinase
- EPT
CDP-ethanolamine:1,2-diacylglycerol ethanolaminephosphotransferase
- ER
Endoplasmic reticulum
- FA
Fatty acid
- G3P
Glycerol-3-phosphate
- GL
Gastric lipase
- GLUT4
Glucose transporter 4
- GPAT
Glycerol-3-phosphate acyltransferase
- HSL
Hormone-sensitive lipase
- IR
Insulin resistance
- IRS
Insulin receptor substrate
- LD
Lipid droplet
- LL
Lingual lipase
- LPA
Lysophosphatidic acid
- LPL
Lipoprotein lipase
- MAG
Monoacylglycerol
- MGAT
Monoacylglycerol-O-acyltransferase
- PA
Phosphatidic acid
- PAP
PA phosphohydrolase/lipin
- PC
Phosphatidylcholine
- PDK
3-Phosphoinositide-dependent protein kinase
- PE
Phosphatidylethanolamine
- PI
Phosphatidylinositol
- PI3K
Phosphatidylinositide-3-kinase
- PIP
Phosphatidylinositolphosphate
- PIP2
Phosphatidylinositol 4,5-bisphosphate
- PKB/Akt
Protein kinase B
- PKC
Protein kinase C
- PKD
Protein kinase D
- PL
Phospholipid
- PLA1(2)
Phospholipase A1 (2)
- PLC
Phospholipase C
- PLD
Phospholipase D
- PLRP
Pancreatic lipase-related protein
- PNPLA
Patatin-like phospholipase domain containing
- PS
Phosphatidylserine
- PTL
Pancreatic triglyceride lipase/colipase
- RE
Retinyl ester
- SMS
Sphingomyelin synthase
- SMSr
SMS-related protein
- TGH/Ces3
Triacylglycerol hydrolase/carboxylesterase 3
- TAG
Triacylglycerol
- TRPC
Transient receptor potential canonical
- WAT
White adipose tissue
Contributor Information
Thomas Oliver Eichmann, Phone: +43/(0)316 380 1900, Email: thomas.eichmann@uni-graz.at.
Achim Lass, Email: achim.lass@uni-graz.at.
References
- 1.Prelog V, Helmchen G. Basic principles of the CIP system and proposals for a revision. Angew Chemie, Int Ed english. 1982;21:567–583. doi: 10.1002/anie.198205671. [DOI] [Google Scholar]
- 2.Cahn RS, Ingold C, Prelog V. Specifications of molecular chirality. Angew Chemie Int Ed english. 1966;5:385–415. doi: 10.1002/anie.196603851. [DOI] [Google Scholar]
- 3.Tauchi-Sato K, Ozeki S, Houjou T, et al. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 4.Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24:109–115. doi: 10.1016/S0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- 5.Gibbons GF, Wiggins D, Brown A-M, Hebbachi A-M. Synthesis and function of hepatic very-low-density lipoprotein. Biochem Soc Trans. 2004;32:59–64. doi: 10.1042/bst0320059. [DOI] [PubMed] [Google Scholar]
- 6.Zimmermann R, Strauss JG, Haemmerle G, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 7.Villena JA, Roy S, Sarkadi-Nagy E, et al. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem. 2004;279:47066–47075. doi: 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins CM, Mancuso DJ, Yan W, et al. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 9.Wilson PA, Gardner SD, Lambie NM, et al. Characterization of the human patatin-like phospholipase family. J Lipid Res. 2006;47:1940–1949. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Rydel TJ, Williams JM, Krieger E, et al. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 11.Senda K, Yoshioka H, Doke N, Kawakita K. A cytosolic phospholipase A2 from potato tissues appears to be patatin. Plant Cell Physiol. 1996;37:347–353. doi: 10.1093/oxfordjournals.pcp.a028952. [DOI] [PubMed] [Google Scholar]
- 12.Granneman JG, Moore HP, Granneman RL, et al. Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem. 2006;282:5726–5735. doi: 10.1074/jbc.M610580200. [DOI] [PubMed] [Google Scholar]
- 13.Taschler U, Schreiber R, Chitraju C, et al. Adipose triglyceride lipase is involved in the mobilization of triglyceride and retinoid stores of hepatic stellate cells. Biochim Biophys Acta Mol Cell Biol Lipids. 2015;1851:937–945. doi: 10.1016/j.bbalip.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao JG, Simon M. A comparative study of human GS2, its paralogues, and its rat orthologue. Biochem Biophys Res Commun. 2007;360:501–506. doi: 10.1016/j.bbrc.2007.06.089. [DOI] [PubMed] [Google Scholar]
- 15.Lake AC, Sun Y, Li JL, et al. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J Lipid Res. 2005;46:2477–2487. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Kershaw EE, Schupp M, Guan HP, et al. PPARgamma regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am J Physiol Endocrinol Metab. 2007;293:E1736–E1745. doi: 10.1152/ajpendo.00122.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kralisch S, Klein J, Lossner U, et al. Isoproterenol, TNFalpha, and insulin downregulate adipose triglyceride lipase in 3T3–L1 adipocytes. Mol Cell Endocrinol. 2005;240:43–49. doi: 10.1016/j.mce.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Jocken JWE, Langin D, Smit E, et al. Adipose triglyceride lipase and hormone-sensitive lipase protein expression is decreased in the obese insulin-resistant state. J Clin Endocrinol Metab. 2007;92:2292–2299. doi: 10.1210/jc.2006-1318. [DOI] [PubMed] [Google Scholar]
- 19.Lass A, Zimmermann R, Haemmerle G, et al. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Lu X, Lombes M, et al. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schweiger M, Paar M, Eder C, et al. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J Lipid Res. 2012;53:2307–2317. doi: 10.1194/jlr.M027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haemmerle G, Lass A, Zimmermann R, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 23.Eichmann TO, Kumari M, Haas J, et al. Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferase enzymes. J Biol Chem. 2012;287(49):41446–41457. doi: 10.1074/jbc.M112.400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollenberg CH, Raben MS, Astwood EB. The lipolytic response to corticotropin. Endocrinology. 1961;68:589–598. doi: 10.1210/endo-68-4-589. [DOI] [PubMed] [Google Scholar]
- 25.Vaughan M, Berger JE, Steinberg D. Hormone-sensitive lipase and monoglyceride lipase activities in adipose tissue. J Biol Chem. 1964;239:401–409. [PubMed] [Google Scholar]
- 26.Holm C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans. 2003;31:1120–1124. doi: 10.1042/bst0311120. [DOI] [PubMed] [Google Scholar]
- 27.Sztalryd C, Kraemer FB. Regulation of hormone-sensitive lipase during fasting. Am J Physiol. 1994;266:E179–E185. doi: 10.1152/ajpendo.1994.266.2.E179. [DOI] [PubMed] [Google Scholar]
- 28.Osterlund T, Danielsson B, Degerman E, et al. Domain-structure analysis of recombinant rat hormone-sensitive lipase. Biochem J. 1996;319(Pt 2):411–420. doi: 10.1042/bj3190411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osterlund T, Beussman DJ, Julenius K, et al. Domain identification of hormone-sensitive lipase by circular dichroism and fluorescence spectroscopy, limited proteolysis, and mass spectrometry. J Biol Chem. 1999;274:15382–15388. doi: 10.1074/jbc.274.22.15382. [DOI] [PubMed] [Google Scholar]
- 30.Osterlund T, Contreras JA, Holm C. Identification of essential aspartic acid and histidine residues of hormone-sensitive lipase: apparent residues of the catalytic triad. FEBS Lett. 1997;403:259–262. doi: 10.1016/S0014-5793(97)00063-X. [DOI] [PubMed] [Google Scholar]
- 31.Langin D, Laurell H, Holst LS, et al. Gene organization and primary structure of human hormone-sensitive lipase: possible significance of a sequence homology with a lipase of Moraxella TA144, an antarctic bacterium. Proc Natl Acad Sci USA. 1993;90:4897–4901. doi: 10.1073/pnas.90.11.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen WJ, Patel S, Hong R, Kraemer FB. Hormone-sensitive lipase functions as an oligomer. Biochemistry. 2000;39:2392–2398. doi: 10.1021/bi992283h. [DOI] [PubMed] [Google Scholar]
- 33.Stralfors P, Belfrage P. Phosphorylation of hormone-sensitive lipase by cyclic AMP-dependent protein kinase. J Biol Chem. 1983;258:15146–15152. [PubMed] [Google Scholar]
- 34.Shen WJ, Patel S, Natu V, Kraemer FB. Mutational analysis of structural features of rat hormone-sensitive lipase. Biochemistry. 1998;37:8973–8979. doi: 10.1021/bi980545u. [DOI] [PubMed] [Google Scholar]
- 35.Anthonsen MW, Ronnstrand L, Wernstedt C, et al. Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J Biol Chem. 1998;273:215–221. doi: 10.1074/jbc.273.1.215. [DOI] [PubMed] [Google Scholar]
- 36.Belfrage P, Fredrikson G, Olsson H, Stralfors P. Regulation of adipose tissue lipolysis through reversible phosphorylation of hormone-sensitive lipase. Adv Cyclic Nucleotide Protein Phosphorylation Res. 1984;17:351–359. [PubMed] [Google Scholar]
- 37.Belfrage P, Fredrikson G, Nilsson NO, Stralfors P. Regulation of adipose-tissue lipolysis by phosphorylation of hormone-sensitive lipase. Int J Obes. 1981;5:635–641. [PubMed] [Google Scholar]
- 38.Stralfors P, Bjorgell P, Belfrage P. Hormonal regulation of hormone-sensitive lipase in intact adipocytes: identification of phosphorylated sites and effects on the phosphorylation by lipolytic hormones and insulin. Proc Natl Acad Sci USA. 1984;81:3317–3321. doi: 10.1073/pnas.81.11.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garton AJ, Yeaman SJ. Identification and role of the basal phosphorylation site on hormone-sensitive lipase. Eur J Biochem. 1990;191:245–250. doi: 10.1111/j.1432-1033.1990.tb19116.x. [DOI] [PubMed] [Google Scholar]
- 40.Blanchette-Mackie EJ, Dwyer NK, Barber T, et al. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J Lipid Res. 1995;36:1211–1226. [PubMed] [Google Scholar]
- 41.Brasaemle DL, Rubin B, Harten IA, et al. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J Biol Chem. 2000;275:38486–38493. doi: 10.1074/jbc.M007322200. [DOI] [PubMed] [Google Scholar]
- 42.Miyoshi H, Perfield JW, 2nd, Souza SC, et al. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J Biol Chem. 2007;282:996–1002. doi: 10.1074/jbc.M605770200. [DOI] [PubMed] [Google Scholar]
- 43.Miyoshi H, Souza SC, Zhang HH, et al. Perilipin promotes HSL-mediated adipocyte lipolysis via phosphorylation-dependent and independent mechanisms. J Biol Chem. 2006;281(23):15837–15844. doi: 10.1074/jbc.M601097200. [DOI] [PubMed] [Google Scholar]
- 44.Tansey JT, Huml AM, Vogt R, et al. Functional studies on native and mutated forms of perilipins. A role in protein kinase A-mediated lipolysis of triacylglycerols. J Biol Chem. 2003;278:8401–8406. doi: 10.1074/jbc.M211005200. [DOI] [PubMed] [Google Scholar]
- 45.Sztalryd C, Xu G, Dorward H, et al. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol. 2003;161:1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egan JJ, Greenberg AS, Chang MK, et al. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc Natl Acad Sci USA. 1992;89:8537–8541. doi: 10.1073/pnas.89.18.8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su CL, Sztalryd C, Contreras JA, et al. Mutational analysis of the hormone-sensitive lipase translocation reaction in adipocytes. J Biol Chem. 2003;278:43615–43619. doi: 10.1074/jbc.M301809200. [DOI] [PubMed] [Google Scholar]
- 48.Osuga J, Ishibashi S, Oka T, et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci USA. 2000;97:787–792. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harada K, Shen WJ, Patel S, et al. Resistance to high-fat diet-induced obesity and altered expression of adipose-specific genes in HSL-deficient mice. Am J Physiol Endocrinol Metab. 2003;285:E1182–E1195. doi: 10.1152/ajpendo.00259.2003. [DOI] [PubMed] [Google Scholar]
- 50.Zimmermann R, Haemmerle G, Wagner EM, et al. Decreased fatty acid esterification compensates for the reduced lipolytic activity in hormone-sensitive lipase-deficient white adipose tissue. J Lipid Res. 2003;44:2089–2099. doi: 10.1194/jlr.M300190-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Haemmerle G, Zimmermann R, Hayn M, et al. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem. 2002;277:4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 52.Wang SP, Laurin N, Himms-Hagen J, et al. The adipose tissue phenotype of hormone-sensitive lipase deficiency in mice. Obes Res. 2001;9:119–128. doi: 10.1038/oby.2001.15. [DOI] [PubMed] [Google Scholar]
- 53.Fredrikson G, Stralfors P, Nilsson NO, Belfrage P. Hormone-sensitive lipase of rat adipose tissue. Purification and some properties. J Biol Chem. 1981;256:6311–6320. [PubMed] [Google Scholar]
- 54.Wei S, Lai K, Patel S, et al. Retinyl ester hydrolysis and retinol efflux from BFC-1beta adipocytes. J Biol Chem. 1997;272:14159–14165. doi: 10.1074/jbc.272.22.14159. [DOI] [PubMed] [Google Scholar]
- 55.Tsujita T, Ninomiya H, Okuda H. P-nitrophenyl butyrate hydrolyzing activity of hormone-Sensitive lipase from bovine adipose tissue. J Lipid Res. 1989;30:997–1004. [PubMed] [Google Scholar]
- 56.Fredrikson G, Belfrage P. Positional specificity of hormone-sensitive lipase from rat adipose tissue. J Biol Chem. 1983;258:14253–14256. [PubMed] [Google Scholar]
- 57.Rodriguez JA, Ben Ali Y, Abdelkafi S, et al. In vitro stereoselective hydrolysis of diacylglycerols by hormone-sensitive lipase. Biochim Biophys Acta. 2010;1801:77–83. doi: 10.1016/j.bbalip.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 58.Gavino VC, Gavino GR. Adipose hormone-sensitive lipase preferentially releases polyunsaturated fatty acids from triglycerides. Lipids. 1992;27:950–954. doi: 10.1007/BF02535570. [DOI] [PubMed] [Google Scholar]
- 59.Schweiger M, Schreiber R, Haemmerle G, et al. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem. 2006;281:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- 60.Lehner R, Verger R. Purification and characterization of a porcine liver microsomal triacylglycerol hydrolase. Biochemistry. 1997;36:1861–1868. doi: 10.1021/bi962186d. [DOI] [PubMed] [Google Scholar]
- 61.Matsushima M, Inoue H, Ichinose M, et al. The nucleotide and deduced amino acid sequences of porcine liver proline- beta-naphthylamidase. Evidence for the identity with carboxylesterase. FEBS Lett. 1991;293:37–41. doi: 10.1016/0014-5793(91)81147-Z. [DOI] [PubMed] [Google Scholar]
- 62.Au-Young J, Fielding CJ. Synthesis and secretion of wild-type and mutant human plasma cholesteryl ester transfer protein in baculovirus-transfected insect cells: the carboxyl-terminal region is required for both lipoprotein binding and catalysis of transfer. Proc Natl Acad Sci USA. 1992;89:4094–4098. doi: 10.1073/pnas.89.9.4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drayna D, Jarnagin AS, McLean J, et al. Cloning and sequencing of human cholesteryl ester transfer protein cDNA. Nature. 1987;327:632–634. doi: 10.1038/327632a0. [DOI] [PubMed] [Google Scholar]
- 64.Robbi M, Beaufay H. The COOH terminus of several liver carboxylesterases targets these enzymes to the lumen of the endoplasmic reticulum. J Biol Chem. 1991;266:20498–20503. [PubMed] [Google Scholar]
- 65.Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu Rev Pharmacol Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 66.Dolinsky VW, Sipione S, Lehner R, Vance DE. The cloning and expression of a murine triacylglycerol hydrolase cDNA and the structure of its corresponding gene. Biochim Biophys Acta. 2001;1532:162–172. doi: 10.1016/S1388-1981(01)00133-0. [DOI] [PubMed] [Google Scholar]
- 67.Alam M, Vance DE, Lehner R. Structure-function analysis of human triacylglycerol hydrolase by site-directed mutagenesis: identification of the catalytic triad and a glycosylation site. Biochemistry. 2002;41:6679–6687. doi: 10.1021/bi0255625. [DOI] [PubMed] [Google Scholar]
- 68.Dolinsky VW, Gilham D, Alam M, et al. Triacylglycerol hydrolase: role in intracellular lipid metabolism. Cell Mol Life Sci. 2004;61:1633–1651. doi: 10.1007/s00018-004-3426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dolinsky VW, Douglas DN, Lehner R, Vance DE. Regulation of the enzymes of hepatic microsomal triacylglycerol lipolysis and re-esterification by the glucocorticoid dexamethasone. Biochem J. 2004;378:967–974. doi: 10.1042/bj20031320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H, Wei E, Quiroga AD, et al. Altered lipid droplet dynamics in hepatocytes lacking triacylglycerol hydrolase expression. Mol Biol Cell. 2010;21:1991–2000. doi: 10.1091/mbc.E09-05-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei E, Ben Ali Y, Lyon J, et al. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metab. 2010;11:183–193. doi: 10.1016/j.cmet.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 72.Nakajima KI, Sonoda H, Mizoguchi T, et al. A novel phospholipase A1 with sequence homology to a mammalian Sec23p-interacting protein, p125. J Biol Chem. 2002;277:11329–11335. doi: 10.1074/jbc.M111092200. [DOI] [PubMed] [Google Scholar]
- 73.Inloes JM, Hsu K-L, Dix MM, et al. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc Natl Acad Sci. 2014;111:14924–14929. doi: 10.1073/pnas.1413706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okazaki H, Igarashi M, Nishi M, et al. Identification of a novel member of the carboxylesterase family that hydrolyzes triacylglycerol: a potential role in adipocyte lipolysis. Diabetes. 2006;55:2091–2097. doi: 10.2337/db05-0585. [DOI] [PubMed] [Google Scholar]
- 75.Lee WC, Salido E, Yen PH. Isolation of a new gene GS2 (DXS1283E) from a CpG island between STS and KAL1 on Xp22.3. Genomics. 1994;22:372–376. doi: 10.1006/geno.1994.1397. [DOI] [PubMed] [Google Scholar]
- 76.Gao J, Simon M. Identification of a novel keratinocyte retinyl ester hydrolase as a transacylase and lipase. J Invest Dermatol. 2005;124:1259–1266. doi: 10.1111/j.0022-202X.2005.23761.x. [DOI] [PubMed] [Google Scholar]
- 77.Baulande S, Lasnier F, Lucas M, Pairault J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem. 2001;276:33336–33344. doi: 10.1074/jbc.M105193200. [DOI] [PubMed] [Google Scholar]
- 78.Huang Y, He S, Li JZ, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci USA. 2010;107:7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He S, McPhaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kershaw EE, Hamm JK, Verhagen LA, et al. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55:148–157. doi: 10.2337/diabetes.55.01.06.db05-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pirazzi C, Valenti L, Motta BM, et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet. 2014;23:4077–4085. doi: 10.1093/hmg/ddu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286:37085–37093. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Basantani MK, Sitnick MT, Cai L, et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52:318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li JZ, Huang Y, Karaman R, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest. 2012;122:4130–4144. doi: 10.1172/JCI65179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qiao A, Liang J, Ke Y, et al. Mouse patatin-like phospholipase domain-containing 3 influences systemic lipid and glucose homeostasis. Hepatology. 2011;54:509–521. doi: 10.1002/hep.24402. [DOI] [PubMed] [Google Scholar]
- 86.Smagris E, BasuRay S, Li J, et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2014;61:108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumari M, Schoiswohl G, Chitraju C, et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab. 2012;15:691–702. doi: 10.1016/j.cmet.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H, Zhang L, Yin D, et al. Targeting phosphatidylcholine-specific phospholipase C for atherogenesis therapy. Trends Cardiovasc Med. 2010;20:172–176. doi: 10.1016/j.tcm.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 89.Ramoni C, Spadaro F, Barletta B, et al. Phosphatidylcholine-specific phospholipase C in mitogen-stimulated fibroblasts. Exp Cell Res. 2004;299:370–382. doi: 10.1016/j.yexcr.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 90.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cockcroft S. The latest phospholipase C, PLCeta, is implicated in neuronal function. Trends Biochem Sci. 2006;31:4–7. doi: 10.1016/j.tibs.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 92.Bunney TD, Katan M. Phospholipase C epsilon: linking second messengers and small GTPases. Trends Cell Biol. 2006;16:640–648. doi: 10.1016/j.tcb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 93.Suh PG, Park JI, Manzoli L, et al. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 2008;41:415–434. doi: 10.5483/BMBRep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- 94.Fukami K, Inanobe S, Kanemaru K, Nakamura Y. Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog Lipid Res. 2010;49:429–437. doi: 10.1016/j.plipres.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 95.Ullman MD, Radin NS. The enzymatic formation of sphingomyelin from ceramide and lecithin in mouse liver. J Biol Chem. 1974;249:1506–1512. [PubMed] [Google Scholar]
- 96.Voelker DR, Kennedy EP. Cellular and enzymic synthesis of sphingomyelin. Biochemistry. 1982;21:2753–2759. doi: 10.1021/bi00540a027. [DOI] [PubMed] [Google Scholar]
- 97.Huitema K, van den Dikkenberg J, Brouwers JFHM, Holthuis JCM. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tafesse FG, Ternes P, Holthuis JCM. The multigenic sphingomyelin synthase family. J Biol Chem. 2006;281:29421–29425. doi: 10.1074/jbc.R600021200. [DOI] [PubMed] [Google Scholar]
- 99.Lee NPY, Mruk DD, Xia W, Cheng CY. Cellular localization of sphingomyelin synthase 2 in the seminiferous epithelium of adult rat testes. J Endocrinol. 2007;192:17–32. doi: 10.1677/JOE-06-0002. [DOI] [PubMed] [Google Scholar]
- 100.Baron CL, Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- 101.Yeaman C, Ayala MI, Wright JR, et al. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat Cell Biol. 2004;6:106–112. doi: 10.1038/ncb1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tafesse FG, Huitema K, Hermansson M, et al. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J Biol Chem. 2007;282:17537–17547. doi: 10.1074/jbc.M702423200. [DOI] [PubMed] [Google Scholar]
- 103.Vacaru AM, Tafesse FG, Ternes P, et al. Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J Cell Biol. 2009;185:1013–1027. doi: 10.1083/jcb.200903152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ternes P, Brouwers JFHM, van den Dikkenberg J, Holthuis JCM. Sphingomyelin synthase SMS2 displays dual activity as ceramide phosphoethanolamine synthase. J Lipid Res. 2009;50:2270–2277. doi: 10.1194/jlr.M900230-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bell RM, Coleman RA. Enzymes of glycerolipid synthesis in eukaryotes. Annu Rev Biochem. 1980;49:459–487. doi: 10.1146/annurev.bi.49.070180.002331. [DOI] [PubMed] [Google Scholar]
- 106.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/S0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 107.Coleman RA, Mashek DG. Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling. Chem Rev. 2011;111:6359–6386. doi: 10.1021/cr100404w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kornberg A, Pricer WE. Enzymatic esterification of alpha-glycerophosphate by long chain fatty acids. J Biol Chem. 1953;204:345. [PubMed] [Google Scholar]
- 109.Burblitz C, Kennedy EP. Synthesis of phosphatidates in isolated mitochondria. J Biol Chem. 1954;211:951. [PubMed] [Google Scholar]
- 110.Johnston M, Schultz D, Schiller M. The utilization of the alpha-glycerophosphate and monoglyceride pathways for phosphatidyl choline biosynthesis in the intestine. Biochim Biophys Acta. 1970;218:124–133. doi: 10.1016/0005-2760(70)90099-8. [DOI] [PubMed] [Google Scholar]
- 111.Phan CT, Tso P. Intestinal lipid absorption and transport. Front Biosci. 2001;6:D299–D319. doi: 10.2741/Phan. [DOI] [PubMed] [Google Scholar]
- 112.Polheim D, David JS, Schultz FM, et al. Regulation of triglyceride biosynthesis in adipose and intestinal tissue. J Lipid Res. 1973;14:415–421. [PubMed] [Google Scholar]
- 113.Kennedy EP, Smith SW, Weiss SB. New synthesis of lecithin in an isolated enzyme system. Nature. 1956;178:594–595. doi: 10.1038/178594a0. [DOI] [PubMed] [Google Scholar]
- 114.Kennedy EP. Biosynthesis of phospholipids. Fed Proc. 1957;16:847–853. [PubMed] [Google Scholar]
- 115.Gibellini F, Smith TK. The Kennedy pathway–De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62:414–428. doi: 10.1002/iub.354. [DOI] [PubMed] [Google Scholar]
- 116.Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 117.Donkor J, Sariahmetoglu M, Dewald J, et al. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J Biol Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- 118.Harris TE, Huffman TA, Chi A, et al. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J Biol Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- 119.Cascales C, Mangiapane EH, Brindley DN. Oleic acid promotes the activation and translocation of phosphatidate phosphohydrolase from the cytosol to particulate fractions of isolated rat hepatocytes. Biochem J. 1984;219:911–916. doi: 10.1042/bj2190911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 121.Yen CL, Jr, Farese RV. MGAT2, a monoacylglycerol acyltransferase expressed in the small intestine. J Biol Chem. 2003;278:18532–18537. doi: 10.1074/jbc.M301633200. [DOI] [PubMed] [Google Scholar]
- 122.Yen CL, Stone SJ, Cases S, et al. Identification of a gene encoding MGAT1, a monoacylglycerol acyltransferase. Proc Natl Acad Sci USA. 2002;99:8512–8517. doi: 10.1073/pnas.132274899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cheng D, Nelson TC, Chen J, et al. Identification of acyl coenzyme A:monoacylglycerol acyltransferase 3, an intestinal specific enzyme implicated in dietary fat absorption. J Biol Chem. 2003;278:13611–13614. doi: 10.1074/jbc.C300042200. [DOI] [PubMed] [Google Scholar]
- 124.Cases S, Stone SJ, Zhou P, et al. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 125.Cao J, Lockwood J, Burn P, Shi Y. Cloning and functional characterization of a mouse intestinal acyl-CoA:monoacylglycerol acyltransferase, MGAT2. J Biol Chem. 2003;278:13860–13866. doi: 10.1074/jbc.M300139200. [DOI] [PubMed] [Google Scholar]
- 126.Cao J, Burn P, Shi Y. Properties of the mouse intestinal acyl-CoA:monoacylglycerol acyltransferase, MGAT2. J Biol Chem. 2003;278:25657–25663. doi: 10.1074/jbc.M302835200. [DOI] [PubMed] [Google Scholar]
- 127.Cao J, Cheng L, Shi Y. Catalytic properties of MGAT3, a putative triacylgycerol synthase. J Lipid Res. 2007;48:583–591. doi: 10.1194/jlr.M600331-JLR200. [DOI] [PubMed] [Google Scholar]
- 128.Brindley DN, Hubscher G. The effect of chain length on the activation and subsequent incorporation of fatty acids into glycerides by the small intestinal mucosa. Biochim Biophys Acta. 1966;125:92–105. doi: 10.1016/0005-2760(66)90147-0. [DOI] [PubMed] [Google Scholar]
- 129.Carriere F, Rogalska E, Cudrey C, et al. In vivo and in vitro studies on the stereoselective hydrolysis of tri- and diglycerides by gastric and pancreatic lipases. Bioorg Med Chem. 1997;5:429–435. doi: 10.1016/S0968-0896(96)00251-9. [DOI] [PubMed] [Google Scholar]
- 130.Jennens ML, Lowe ME. Rat GP-3 is a pancreatic lipase with kinetic properties that differ from colipase-dependent pancreatic lipase. J Lipid Res. 1995;36:2374–2382. [PubMed] [Google Scholar]
- 131.Thirstrup K, Verger R, Carrière F. Evidence for a pancreatic lipase subfamily with new kinetic properties. Biochemistry. 1994;33:2748–2756. doi: 10.1021/bi00176a002. [DOI] [PubMed] [Google Scholar]
- 132.Roussel A, Yang Y, Ferrato F, et al. Structure and activity of rat pancreatic lipase-related protein 2. J Biol Chem. 1998;273:32121–32128. doi: 10.1074/jbc.273.48.32121. [DOI] [PubMed] [Google Scholar]
- 133.Giller T, Buchwald P, Blum-Kaelin D, et al. Two novel human pancreatic lipase related proteins, hPLRP1 and hPLRP2. J Biol Chem. 1992;267:16509–16516. [PubMed] [Google Scholar]
- 134.Rogalska E, Ransac S, Verger R. Controlling lipase stereoselectivity via the surface pressure. J Biol Chem. 1993;268:792–794. [PubMed] [Google Scholar]
- 135.Rogalska E, Cudrey C, Ferrato F, Verger R. Stereoselective hydrolysis of triglycerides by animal and microbial lipases. Chirality. 1993;5:24–30. doi: 10.1002/chir.530050106. [DOI] [PubMed] [Google Scholar]
- 136.Paltauf F, Esfandi F, Holasek A. Stereospecificity of lipases. Enzymic hydrolysis of enantiomeric alkyl diacylglycerols by lipoprotein lipase, lingual lipase and pancreatic lipase. FEBS Lett. 1974;40:119–123. doi: 10.1016/0014-5793(74)80907-5. [DOI] [PubMed] [Google Scholar]
- 137.Jensen RG, DeJong FA, Clark RM, et al. Stereospecificity of premature human infant lingual lipase. Lipids. 1982;17:570–572. doi: 10.1007/BF02535386. [DOI] [PubMed] [Google Scholar]
- 138.Wang CS, Kuksis A, Manganaro F, et al. Studies on the substrate specificity of purified human milk bile salt-activated lipase. J Biol Chem. 1983;258:9197–9202. [PubMed] [Google Scholar]
- 139.Morley N, Kuksis A. Positional specificity of lipoprotein lipase. J Biol Chem. 1972;247:6389–6393. [PubMed] [Google Scholar]
- 140.Akesson B, Gronowitz S, Herslof B. Stereospecificity of hepatic lipases. FEBS Lett. 1976;71:241–244. doi: 10.1016/0014-5793(76)80941-6. [DOI] [PubMed] [Google Scholar]
- 141.Mechoulam R, Ben-Shabat S, Hanus L, et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem Pharmacol. 1995;50:83–90. doi: 10.1016/0006-2952(95)00109-D. [DOI] [PubMed] [Google Scholar]
- 142.Sugiura T, Kondo S, Sukagawa A, et al. 2-Arachidonoylglycerol: a possible endogenous cannabinoid receptor ligand in brain. Biochem Biophys Res Commun. 1995;215:89–97. doi: 10.1006/bbrc.1995.2437. [DOI] [PubMed] [Google Scholar]
- 143.Schicho R, Storr M. Cannabis finds its way into treatment of Crohn’s disease. Pharmacology. 2014;93:1–3. doi: 10.1159/000356512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bashashati M, Nasser Y, Keenan C, et al. Inhibiting endocannabinoid biosynthesis: a novel approach to the treatment of constipation. Br J Pharmacol. 2015;172(12):3099–3111. doi: 10.1111/bph.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Taschler U, Eichmann TO, Radner FPW et al (2015) Monoglyceride lipase-deficiency causes desensitization of intestinal cannabinoid receptor type 1 and increased colonic μ-opioid receptor sensitivity. Br J Pharmacol [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 146.Cases S, Smith SJ, Zheng YW, et al. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci USA. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci. 2000;25:111–112. doi: 10.1016/S0968-0004(99)01539-X. [DOI] [PubMed] [Google Scholar]
- 148.Yen CL, Stone SJ, Koliwad S, et al. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stone SJ, Levin MC, Zhou P, et al. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J Biol Chem. 2009;284:5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McFie PJ, Stone SL, Banman SL, Stone SJ. Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the n terminus in dimer/tetramer formation. J Biol Chem. 2010;285:37377–37387. doi: 10.1074/jbc.M110.163691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Siloto RMP, Madhavji M, Wiehler WB, et al. An N-terminal fragment of mouse DGAT1 binds different acyl-CoAs with varying affinity. Biochem Biophys Res Commun. 2008;373:350–354. doi: 10.1016/j.bbrc.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 152.Cheng D, Meegalla RL, He B, et al. Human acyl-CoA:diacylglycerol acyltransferase is a tetrameric protein. Biochem J. 2001;359:707–714. doi: 10.1042/bj3590707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Owen MR, Corstorphine CC, Zammit VA. Overt and latent activities of diacylglycerol acyltransferase in rat liver microsomes: possible roles in very-low-density lipoprotein triacylglycerol secretion. Biochem J. 1997;323(1):17–21. doi: 10.1042/bj3230017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wurie HR, Bucketts L, Zammit VA. Evidence that diacylglycerol acyltransferase 1 (DGAT1) has dual membrane topology in the endoplasmic reticulum of HepG2 cells. J Biol Chem. 2011;286:36238–36247. doi: 10.1074/jbc.M111.251900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yen CL, Monetti M, Burri BJ, Jr, Farese RV. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J Lipid Res. 2005;46:1502–1511. doi: 10.1194/jlr.M500036-JLR200. [DOI] [PubMed] [Google Scholar]
- 156.Lardizabal KD, Mai JT, Wagner NW, et al. DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J Biol Chem. 2001;276:38862–38869. doi: 10.1074/jbc.M106168200. [DOI] [PubMed] [Google Scholar]
- 157.Stone SJ, Levin MC, Jr, Farese RV. Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2. J Biol Chem. 2006;281:40273–40282. doi: 10.1074/jbc.M607986200. [DOI] [PubMed] [Google Scholar]
- 158.Alam M, Gilham D, Vance DE, Lehner R. Mutation of F417 but not of L418 or L420 in the lipid binding domain decreases the activity of triacylglycerol hydrolase. J Lipid Res. 2006;47:375–383. doi: 10.1194/jlr.M500344-JLR200. [DOI] [PubMed] [Google Scholar]
- 159.Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- 160.McFie PJ, Banman SL, Kary S, Stone SJ. Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. J Biol Chem. 2011;286:28235–28246. doi: 10.1074/jbc.M111.256008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stone SJ, Myers HM, Watkins SM, et al. Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J Biol Chem. 2004;279:11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- 162.Hiramine Y, Tanabe T. Characterization of acyl-coenzyme A: diacylglycerol acyltransferase (DGAT) enzyme of human small intestine. J Physiol Biochem. 2011;67:259–264. doi: 10.1007/s13105-010-0071-1. [DOI] [PubMed] [Google Scholar]
- 163.Smith SJ, Cases S, Jensen DR, et al. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat Genet. 2000;25:87–90. doi: 10.1038/75651. [DOI] [PubMed] [Google Scholar]
- 164.Wurie HR, Buckett L, Zammit VA. Diacylglycerol acyltransferase 2 (DGAT2) acts upstream of DGAT1, and utilises nascent diglycerides and de novo synthesised fatty acids in HepG2 cells. FEBS J. 2012;279(17):3033–3047. doi: 10.1111/j.1742-4658.2012.08684.x. [DOI] [PubMed] [Google Scholar]
- 165.Qi J, Lang W, Geisler JG, et al. The use of stable isotope-labeled glycerol and oleic acid to differentiate the hepatic functions of DGAT1 and -2. J Lipid Res. 2012;53:1106–1116. doi: 10.1194/jlr.M020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bisogno T, Howell F, Williams G, et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol. 2003;163:463–468. doi: 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gao Y, Vasilyev DV, Goncalves MB, et al. Loss of retrograde endocannabinoid signaling and reduced adult neurogenesis in diacylglycerol lipase knock-out mice. J Neurosci. 2010;30:2017–2024. doi: 10.1523/JNEUROSCI.5693-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jung K, Astarita G, Thongkham D, Piomelli D. Diacylglycerol lipase-alpha and -beta control neurite outgrowth in neuro-2a cells through distinct molecular mechanisms. Mol Pharmacol. 2011;80:60–67. doi: 10.1124/mol.110.070458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wood R, Harlow RD. Structural analyses of rat liver phosphoglycerides. Arch Biochem Biophys. 1969;135:272–281. doi: 10.1016/0003-9861(69)90540-2. [DOI] [PubMed] [Google Scholar]
- 170.Marai L, Kuksis A. Molecular species of lecithins from erythrocytes and plasma of man. J Lipid Res. 1969;10:141–152. [PubMed] [Google Scholar]
- 171.Ligresti A, Petrosino S, Di Marzo V. From endocannabinoid profiling to “endocannabinoid therapeutics”. Curr Opin Chem Biol. 2009;13:321–331. doi: 10.1016/j.cbpa.2009.04.615. [DOI] [PubMed] [Google Scholar]
- 172.Merida I, Avila-Flores A, Merino E. Diacylglycerol kinases: at the hub of cell signalling. Biochem J. 2008;409:1–18. doi: 10.1042/BJ20071040. [DOI] [PubMed] [Google Scholar]
- 173.Sakane F, Imai S, Kai M, et al. Diacylglycerol kinases: why so many of them? Biochim Biophys Acta. 2007;1771:793–806. doi: 10.1016/j.bbalip.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 174.Topham MK, Epand RM. Mammalian diacylglycerol kinases: molecular interactions and biological functions of selected isoforms. Biochim Biophys Acta. 2009;1790:416–424. doi: 10.1016/j.bbagen.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Topham MK, Prescott SM. Mammalian diacylglycerol kinases, a family of lipid kinases with signaling functions. J Biol Chem. 1999;274:11447–11450. doi: 10.1074/jbc.274.17.11447. [DOI] [PubMed] [Google Scholar]
- 176.Goto K, Hozumi Y, Nakano T, et al. Lipid messenger, diacylglycerol, and its regulator, diacylglycerol kinase, in cells, organs, and animals: history and perspective. Tohoku J Exp Med. 2008;214:199–212. doi: 10.1620/tjem.214.199. [DOI] [PubMed] [Google Scholar]
- 177.Shulga YV, Topham MK, Epand RM. Regulation and functions of diacylglycerol kinases. Chem Rev. 2011;111:6186–6208. doi: 10.1021/cr1004106. [DOI] [PubMed] [Google Scholar]
- 178.Boni LT, Rando RR. The nature of protein kinase C activation by physically defined phospholipid vesicles and diacylglycerols. J Biol Chem. 1985;260:10819–10825. [PubMed] [Google Scholar]
- 179.Rando RR, Young N. The stereospecific activation of protein kinase C. Biochem Biophys Res. 1984;122:818–823. doi: 10.1016/S0006-291X(84)80107-2. [DOI] [PubMed] [Google Scholar]
- 180.Nomura H, Ase K, Sekiguchi K, et al. Stereospecificity of diacylglycerol for stimulus-response coupling in platelets. Biochem Biophys Res Commun. 1986;140:1143–1151. doi: 10.1016/0006-291X(86)90754-0. [DOI] [PubMed] [Google Scholar]
- 181.Epand RM, Shulga YV, Timmons HC, et al. Substrate chirality and specificity of diacylglycerol kinases and the multisubstrate lipid kinase. Biochemistry. 2007;46:14225–14231. doi: 10.1021/bi701584v. [DOI] [PubMed] [Google Scholar]
- 182.Tang W, Bunting M, Zimmerman GA, et al. Molecular cloning of a novel human diacylglycerol kinase highly selective for arachidonate-containing substrates. J Biol Chem. 1996;271:10237–10241. doi: 10.1074/jbc.271.17.10237. [DOI] [PubMed] [Google Scholar]
- 183.Lung M, Shulga YV, Ivanova PT, et al. Diacylglycerol kinase ε is selective for both acyl chains of phosphatidic acid or diacylglycerol. J Biol Chem. 2009;284:31062–31073. doi: 10.1074/jbc.M109.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.De Turco EBR, Tang W, Topham MK, et al. Diacylglycerol kinase epsilon regulates seizure susceptibility and long-term potentiation through arachidonoyl- inositol lipid signaling. Proc Natl Acad Sci USA. 2001;98:4740–4745. doi: 10.1073/pnas.081536298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Henneberry AL, McMaster CR. Cloning and expression of a human choline/ethanolaminephosphotransferase: synthesis of phosphatidylcholine and phosphatidylethanolamine. Biochem J. 1999;339(Pt 2):291–298. doi: 10.1042/bj3390291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Henneberry AL, Wistow G, McMaster CR. Cloning, genomic organization, and characterization of a human cholinephosphotransferase. J Biol Chem. 2000;275:29808–29815. doi: 10.1074/jbc.M005786200. [DOI] [PubMed] [Google Scholar]
- 187.Henneberry AL, Wright MM, McMaster CR. The major sites of cellular phospholipid synthesis and molecular determinants of Fatty Acid and lipid head group specificity. Mol Biol Cell. 2002;13:3148–3161. doi: 10.1091/mbc.01-11-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.McMaster CR, Bell RM. CDP-choline:1,2-diacylglycerol cholinephosphotransferase. Biochim Biophys Acta. 1997;1348:100–110. doi: 10.1016/S0005-2760(97)00097-0. [DOI] [PubMed] [Google Scholar]
- 189.Hjelmstad RH, Morash SC, McMaster CR, Bell RM. Chimeric enzymes. Structure-function analysis of segments of sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases. J Biol Chem. 1994;269:20995–21002. [PubMed] [Google Scholar]
- 190.McMaster CR, Bell RM. CDP-ethanolamine:1,2-diacylglycerol ethanolaminephosphotransferase. Biochim Biophys Acta. 1997;1348:117–123. doi: 10.1016/S0005-2760(97)00098-2. [DOI] [PubMed] [Google Scholar]
- 191.McMaster CR, Bell RM. Phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. Regulatory insights from studies employing null and chimeric sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases. J Biol Chem. 1994;269:28010–28016. [PubMed] [Google Scholar]
- 192.McGee TP, Skinner HB, Bankaitis VA. Functional redundancy of CDP-ethanolamine and CDP-choline pathway enzymes in phospholipid biosynthesis: ethanolamine-dependent effects on steady-state membrane phospholipid composition in Saccharomyces cerevisiae. J Bacteriol. 1994;176:6861–6868. doi: 10.1128/jb.176.22.6861-6868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Samborski RW, Ridgway ND, Vance DE. Evidence that only newly made phosphatidylethanolamine is methylated to phosphatidylcholine and that phosphatidylethanolamine is not significantly deacylated–reacylated in rat hepatocytes. J Biol Chem. 1990;265:18322–18329. [PubMed] [Google Scholar]
- 194.Dietrich A, Kalwa H, Rost BR, Gudermann T. The diacylglycerol-sensitive TRPC3/6/7 subfamily of cation channels: functional characterization and physiological relevance. Pflugers Arch Eur J Physiol. 2005;451:72–80. doi: 10.1007/s00424-005-1460-0. [DOI] [PubMed] [Google Scholar]
- 195.Vazquez G, Tano JY, Smedlund K. On the potential role of source and species of diacylglycerol in phospholipase-dependent regulation of TRPC3 channels. Channels (Austin) 2010;4:232–240. doi: 10.4161/chan.4.3.12058. [DOI] [PubMed] [Google Scholar]
- 196.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Turban S, Hajduch E. Protein kinase C isoforms: mediators of reactive lipid metabolites in the development of insulin resistance. FEBS Lett. 2011;585:269–274. doi: 10.1016/j.febslet.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 198.Newton AC. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- 199.Silinsky EM, Searl TJ. Phorbol esters and neurotransmitter release: more than just protein kinase C? Br J Pharmacol. 2003;138:1191–1201. doi: 10.1038/sj.bjp.0705213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Brose N, Rosenmund C. Move over protein kinase C, you’ve got company: alternative cellular effectors of diacylglycerol and phorbol esters. J Cell Sci. 2002;115:4399–4411. doi: 10.1242/jcs.00122. [DOI] [PubMed] [Google Scholar]
- 201.Kazanietz MG. Novel “nonkinase” phorbol ester receptors: the C1 domain connection. Mol Pharmacol. 2002;61:759–767. doi: 10.1124/mol.61.4.759. [DOI] [PubMed] [Google Scholar]
- 202.Kazanietz MG. Eyes, wide shut: protein kinase C isozymes are not the only receptors for the phorbol ester tumor promoters. Mol Carcinog. 2000;28:5–11. doi: 10.1002/(SICI)1098-2744(200005)28:1<5::AID-MC2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 203.Ron D, Kazanietz MG. New insights into the regulation of protein kinase C and novel phorbol ester receptors. FASEB J. 1999;13:1658–1676. [PubMed] [Google Scholar]
- 204.Colón-González F, Kazanietz MG. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim Biophys Acta Mol Cell Biol Lipids. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 205.Yang C, Kazanietz MG. Divergence and complexities in DAG signaling: looking beyond PKC. Trends Pharmacol Sci. 2003;24:602–608. doi: 10.1016/j.tips.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 206.Herrera R, Shivers BD. Expression of alpha 1-chimaerin (rac-1 GAP) alters the cytoskeletal and adhesive properties of fibroblasts. J Cell Biochem. 1994;56:582–591. doi: 10.1002/jcb.240560419. [DOI] [PubMed] [Google Scholar]
- 207.Clyde-Smith J, Silins G, Gartside M, et al. Characterization of RasGRP2, a plasma membrane-targeted, dual specificity Ras/Rap exchange factor. J Biol Chem. 2000;275:32260–32267. doi: 10.1074/jbc.M006087200. [DOI] [PubMed] [Google Scholar]
- 208.Dupuy AJ, Morgan K, Von Lintig FC, et al. Activation of the Rap1 guanine nucleotide exchange gene, CalDAG-GEF I, in BXH-2 murine myeloid leukemia. J Biol Chem. 2001;276:11804–11811. doi: 10.1074/jbc.M008970200. [DOI] [PubMed] [Google Scholar]
- 209.Augustin I, Betz A, Herrmann C, et al. Differential expression of two novel Munc13 proteins in rat brain. Biochem J. 1999;337(Pt 3):363–371. doi: 10.1042/bj3370363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Augustin I, Rosenmund C, Südhof TC, Brose N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- 211.Aravamudan B, Fergestad T, Davis WS, et al. Drosophila UNC-13 is essential for synaptic transmission. Nat Neurosci. 1999;2:965–971. doi: 10.1038/14764. [DOI] [PubMed] [Google Scholar]
- 212.Ashery U, Varoqueaux F, Voets T, et al. Munc13-1 acts as a priming factor for large dense-core vesicles in bovine chromaffin cells. EMBO J. 2000;19:3586–3596. doi: 10.1093/emboj/19.14.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jamora C, Yamanouye N, Van Lint J, et al. Gbetagamma-mediated regulation of Golgi organization is through the direct activation of protein kinase D. Cell. 1999;98:59–68. doi: 10.1016/S0092-8674(00)80606-6. [DOI] [PubMed] [Google Scholar]
- 214.Liljedahl M, Maeda Y, Colanzi A, et al. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell. 2001;104:409–420. doi: 10.1016/S0092-8674(01)00228-8. [DOI] [PubMed] [Google Scholar]
- 215.Litherland GJ, Hajduch E, Hundal HS. Intracellular signalling mechanisms regulating glucose transport in insulin-sensitive tissues (review) Mol Membr Biol. 2001;18:195–204. doi: 10.1080/09687680110076407. [DOI] [PubMed] [Google Scholar]
- 216.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 217.Sano H, Kane S, Sano E, et al. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 218.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet. 2010;375:2267–2277. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Samuel VT, Liu ZX, Wang A, et al. Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J Clin Invest. 2007;117:739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li Y, Soos TJ, Li X, et al. Protein kinase C Theta inhibits insulin signaling by phosphorylating IRS1 at Ser(1101) J Biol Chem. 2004;279:45304–45307. doi: 10.1074/jbc.C400186200. [DOI] [PubMed] [Google Scholar]
- 221.Nagle CA, An J, Shiota M, et al. Hepatic overexpression of glycerol-sn-3-phosphate acyltransferase 1 in rats causes insulin resistance. J Biol Chem. 2007;282:14807–14815. doi: 10.1074/jbc.M611550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Turinsky J, O’Sullivan DM, Bayly BP. 1,2-Diacylglycerol and ceramide levels in insulin-resistant tissues of the rat in vivo. J Biol Chem. 1990;265:16880–16885. [PubMed] [Google Scholar]
- 223.Jornayvaz FR, Jurczak MJ, Lee HY, et al. A high-fat, ketogenic diet causes hepatic insulin resistance in mice, despite increasing energy expenditure and preventing weight gain. Am J Physiol Metab. 2010;299:E808–E815. doi: 10.1152/ajpendo.00361.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Monetti M, Levin MC, Watt MJ, et al. Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 2007;6:69–78. doi: 10.1016/j.cmet.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 225.Jornayvaz FR, Birkenfeld AL, Jurczak MJ, et al. Hepatic insulin resistance in mice with hepatic overexpression of diacylglycerol acyltransferase 2. Proc Natl Acad Sci USA. 2011;108:5748–5752. doi: 10.1073/pnas.1103451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jornayvaza F, Shulman GI, Jornayvaz FR, Shulman GI. Diacylglycerol activation of protein kinase Cepsilon and hepatic insulin resistance. Cell Metab. 2012;15:574–584. doi: 10.1016/j.cmet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kumashiro N, Erion DM, Zhang D, et al. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA. 2011;108:16381–16385. doi: 10.1073/pnas.1113359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mulder H, Sorhede-Winzell M, Contreras JA, et al. Hormone-sensitive lipase null mice exhibit signs of impaired insulin sensitivity whereas insulin secretion is intact. J Biol Chem. 2003;278:36380–36388. doi: 10.1074/jbc.M213032200. [DOI] [PubMed] [Google Scholar]
- 229.Roduit R, Masiello P, Wang SP, et al. A role for hormone-sensitive lipase in glucose-stimulated insulin secretion: a study in hormone-sensitive lipase-deficient mice. Diabetes. 2001;50:1970–1975. doi: 10.2337/diabetes.50.9.1970. [DOI] [PubMed] [Google Scholar]
- 230.Voshol PJ, Haemmerle G, Ouwens DM, et al. Increased hepatic insulin sensitivity together with decreased hepatic triglyceride stores in hormone-sensitive lipase-deficient mice. Endocrinology. 2003;144:3456–3462. doi: 10.1210/en.2002-0036. [DOI] [PubMed] [Google Scholar]
- 231.Park SY, Kim HJ, Wang S, et al. Hormone-sensitive lipase knockout mice have increased hepatic insulin sensitivity and are protected from short-term diet-induced insulin resistance in skeletal muscle and heart. Am J Physiol Metab. 2005;289:E30–E39. doi: 10.1152/ajpendo.00251.2004. [DOI] [PubMed] [Google Scholar]
- 232.Albert JS, Yerges-Armstrong LM, Horenstein RB, et al. Null mutation in hormone-sensitive lipase gene and risk of type 2 diabetes. N Engl J Med. 2014;370:2307–2315. doi: 10.1056/NEJMoa1315496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Brown JM, Betters JL, Lord C, et al. CGI-58 knockdown in mice causes hepatic steatosis, but prevents diet-induced obesity and glucose intolerance. J Lipid Res. 2010;51:3306–3315. doi: 10.1194/jlr.M010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cantley JL, Yoshimura T, Camporez JPG, et al. CGI-58 knockdown sequesters diacylglycerols in lipid droplets/ER-preventing diacylglycerol-mediated hepatic insulin resistance. Proc Natl Acad Sci USA. 2013;110:1869–1874. doi: 10.1073/pnas.1219456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Badin PM, Louche K, Mairal A, et al. Altered skeletal muscle lipase expression and activity contribute to insulin resistance in humans. Diabetes. 2011;60:1734–1742. doi: 10.2337/db10-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ahmadian M, Duncan RE, Varady KA, et al. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes. 2009;58:855–866. doi: 10.2337/db08-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Sato M, Ueda Y, Umezawa Y. Imaging diacylglycerol dynamics at organelle membranes. Nat Methods. 2006;3:797–799. doi: 10.1038/nmeth930. [DOI] [PubMed] [Google Scholar]
- 238.Kunkel MT, Newton AC. Calcium transduces plasma membrane receptor signals to produce diacylglycerol at golgi membranes. J Biol Chem. 2010;285:22748–22752. doi: 10.1074/jbc.C110.123133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ueda Y, Makino A, Murase-Tamada K, et al. Sphingomyelin regulates the transbilayer movement of diacylglycerol in the plasma membrane of Madin-Darby canine kidney cells. FASEB J. 2013;27:3284–3297. doi: 10.1096/fj.12-226548. [DOI] [PubMed] [Google Scholar]
- 240.Wakelam MJO. Diacylglycerol: when is it an intracellular messenger? Biochim Biophys Acta Mol Cell Biol Lipids. 1998;1436:117–126. doi: 10.1016/S0005-2760(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 241.Becker KB, Hannun YA (2012) Bioactive Lipids. In: Nicolaou A, Kokotos G (eds) Bioact Lipids. p 37–61
- 242.Carrasco S, Mérida I. Diacylglycerol, when simplicity becomes complex. Trends Biochem Sci. 2007;32:27–36. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]