Abstract
Pseudomonas aeruginosa glutamyl-tRNA synthetase (GluRS) was overexpressed in Escherichia coli. Sequence analysis indicated that P. aeruginosa GluRS is a discriminating GluRS and, similar to other GluRS proteins, requires the presence of tRNAGlu to produce a glutamyl-AMP intermediate. Kinetic parameters for interaction with tRNA were determined and the kcat and KM were 0.8 s−1 and 0.68 µM, respectively, resulting in a kcat/KM of 1.18 s−1 µM−1. A robust aminoacylation-based scintillation proximity assay (SPA) assay was developed and 800 natural products and 890 synthetic compounds were screened for inhibitory activity against P. aeruginosa GluRS. Fourteen compounds with inhibitory activity were identified. IC50s were in the low micromolar range. The minimum inhibitory concentration (MIC) was determined for each of the compounds against a panel of pathogenic bacteria. Two compounds, BT_03F04 and BT_04B09, inhibited GluRS with IC50s of 21.9 and 24.9 µM, respectively, and both exhibited promising MICs against Gram-positive bacteria. Time-kill studies indicated that one compound was bactericidal and one was bacteriostatic against Gram-positive bacteria. BT_03F04 was found to be noncompetitive with both ATP and glutamic acid, and BT_04B09 was competitive with glutamic acid but noncompetitive with ATP. The compounds were not observed to be toxic to mammalian cells in MTT assays.
Keywords: glutamyl-tRNA synthetase, high-throughput screening, tRNA aminoacylation, drug discovery, antibiotics
Introduction
Pseudomonas aeruginosa, a Gram-negative bacteria, is an opportunistic pathogen and a common cause of nosocomial infections (responsible for 10–15% worldwide) and the leading cause of mortality in cystic fibrosis patients.1 P. aeruginosa has the ability to acquire resistance against multiple groups of antimicrobial agents, including β-lactams, aminoglycosides, and fluoroquinolones, and multidrug resistance (MDR) is common and increasing in this organism, making treatment of infections both difficult and expensive.2 This growth in resistance has resulted in an unmet need for the development of antipseudomonal agents that have no cross-resistance with drugs currently in use.
Aminoacyl-tRNA synthetases (aaRSs) play a central role in protein biosynthesis by catalyzing the covalent attachment of an amino acid to its cognate tRNA and are crucial for cell growth and viability. The aaRS proteins from prokaryotic origins are conserved in their primary structure, yet they are sufficiently divergent from that found in eukaryotic cells as to make them suitable targets for development of selective antibacterial agents.3 Finally, the crystal structures of many of the aminoacyl-tRNA synthetases have been solved and provide useful information for use in rational drug design.
Glutamyl-tRNA synthetase (GluRS) is encoded by the gltX gene and belongs to the class I aaRSs, which contain two consensus sequences (HIGH and KMSKS) in their active site and bind ATP through a conserved structural domain (the Rossman fold).4 The class I aaRSs approach tRNA molecules from the minor groove of the acceptor stem and aminoacylate, the 3′-terminal adenosine, at the 2′-OH position, whereas class II aaRSs primarily approach the tRNA from the major groove and aminoacylate, the 3′-terminal adenosine, at the 3′-OH position.5 Different than most other aaRSs, the glutamyl-, glutaminyl-, and the arginyl-tRNA synthetases from various prokaryotic and eukaryotic organisms require the presence of the cognate tRNA to form a stable aminoacyl-adenylate that can be monitored using the ATP:PPi exchange reaction.6
In archea, Gram-positive, and in mitochondria and chloroplasts, GluRS aminoacylates both tRNAGlu and tRNAGln with glutamate.7 Glu-tRNAGln is then transformidated to form Gln-tRNAGln by a specific tRNA-dependent amidotranferase.8 The GluRS of Gram-negative bacteria from which it has been studied solely aminoacylates tRNAGlu. However, duplicate GluRS enzymes have been found in some organisms, including Acidithiobacillus ferrooxidans9 and Helicobacter pylori.10 In these organisms there are two different forms of GluRS proteins, and each possesses a distinct function. GluRS1 only aminoacylates tRNAGlu, while GluRS2 aminoacylates tRNAGln. In higher eukaryotic organisms, the cytoplasmic forms of many of the aminoacyl-tRNA synthetases are involved in the formation of multienzyme complexes. In these cells, glutamyl- and prolyl-tRNA synthetase activities are linked in a single polypeptide called Glu-ProRS.
Previously, scintillation proximity assays (SPAs) have been used to measure aminoacyl-tRNA synthetase aminoacylation activity and have been validated in high-throughput assays.11 Using a modified form of this assay, based on P. aeruginosa, GluRS we developed a high-throughput screen for inhibitors of activity. Out of 1690 natural and synthetic compounds, two of the compounds were identified as inhibitors of P. aeruginosa GluRS. These two compounds were characterized for activity against bacterial growth, mode of action, and toxicity in human cell cultures.
Materials and Methods
Materials
Oligonucleotides were from Integrated DNA Technologies (Coralville, IA). All other chemicals were obtained from either Sigma Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA). DNA sequencing was performed by Functional Bioscience (Madison, WI). Radioactive isotopes, SPA beads, and 96-well screening plates were from PerkinElmer (Waltham, MA). Escherichia coli tolC mutant, P. aeruginosa PAO200 (efflux pump mutant), and P. aeruginosa hypersensitive strain (ATCC 35151) were a kind gift from Urs Ochsner (Crestone Pharma, Boulder, CO). All other bacteria were from the American Type Culture Collection (ATCC) (Manassas, VA). Penicillin-Streptomycin Solution was from Mediatech, Inc. (Centerville, IA). The synthetic compound library was from TimTec LLC (Newark, DE), and the natural compound library was from MicroSource Discovery Systems (Gaylordsville, CT).
Gel Electrophoresis and Protein Analysis
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 4–12% polyacrylamide precast gels (Novex NuPAGE; Invitrogen, Grand Island, NY) with 3-(N-morpholino)propanesulfonic acid (MOPS) running buffer (Invitrogen). EZ-Run Rec Protein Ladder was from Fisher Scientific. Gels were stained with Simply Blue Safe Stain (Invitrogen). Protein concentrations were determined using Coomassie Protein Assay Reagent (Thermo Scientific, Waltham, MA) with bovine serum albumin as the standard.
Cloning and Purification of P. aeruginosa GluRS
The genes encoding P. aeruginosa GluRS were amplified by PCR (MJ Mini Thermo Cycler, Bio-Rad, Hercules, CA) from P. aeruginosa PAO1 (ATCC 47085) genomic DNA using the forward primer (5′-CACCATGACCACTG TTCGTACTCGCATCG-3′), which contained a 5′-CACC sequence for insertion into pET101/D-TOPO directional plasmid, and the reverse primer (5′-TCAATGGTGATG GTGATGGTGAGAACCGCCGGGAATGGCGTCGC-3′), which was designed to add six histidine amino acid residues to the C-terminus of GluRS. The PCR product was inserted into pET101/D-TOPO and transformed into E. coli Rosetta 2(DE3) Singles Competent Cells (EMD Millipore, Danvers, MA).
Bacterial cultures were grown in Terrific Broth containing 50 µg/mL of ampicillin and 50 µg/mL of chloramphenicol. Cultures overexpressing P. aeruginosa GluRS were grown at 37 °C, and expression of the target protein was induced at an optical density (A600) of 0.6–0.8 by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to 0.25 mM. Growth of bacterial cultures were continued for 2 h postinduction, and the bacteria were harvested by centrifugation (10,000 g, 30 min, 4 °C). Fraction I lysates were prepared as previously described.12 P. aeruginosa GluRS was purified to greater than 98% homogeneity using nickel–nitrilotriacetic acid (NTA) affinity chromatography (Perfect Pro, 5 Prime) followed by dialysis (two times) against a buffer containing 20 mM Hepes-KOH (pH 7.0), 40 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, and 10% glycerol. Purified proteins were fast frozen in liquid nitrogen and stored at −80 °C.
ATP:PPi Exchange Reactions
ATP:PPi exchange reactions (100 µL) were carried out at 37 °C for 20 min in 50 mM Tris-HCl (pH 7.5), 10 mM KF, 8 mM MgOAc, 1 mM dithiothreitol (DTT), 2 mM ATP, 2 mM glutamic acid (Glu), and 2 mM [32P]PPi (50 cpm/pmol) and contained 0.1 µM of P. aeruginosa GluRS without or with tRNAGlu (0.3, 0.6, or 1.2 µM). Reactions were stopped by diluting 5 µL aliquots of the reaction mix into 45 µL 50 mM Tris-HCl (pH 7.5), 10 mM KF, and 8 mM MgOAc and spotting 5 µL of the diluted solution on PEI cellulose TLC plates (Selecto Scientific, Suwanee, GA). ATP and PPi were separated using 4 M urea and 0.75 M KPi (pH 3.5) as a mobile phase.13 The plates were analyzed using a Typhoon FLA7000 laser scanning phosphorimager (GE Healthcare, Little Chalfont, UK).
Timed tRNA Aminoacylation Assays
Aminoacylation reactions were carried out at 37 °C and at time intervals between 1 and 5 min. Reactions (50 µL) contained a protein/buffer mix (35 µL) designed to yield final concentrations of 50 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 1.25 mM ATP, 1 mM spermine, 1 mM DTT, 100 µM [3H]] Glu (75 cpm/pmol), and 0.1 µM P. aeruginosa GluRS. Reactions were started by the addition of tRNA (15 µL). The tRNAGlu concentrations were varied as indicated. Reactions were stopped by the addition of 2 mL of 5% (v/v) ice-cold trichloroacetic acid (TCA), placed on ice for 10 min, then filtered through glass fiber filters (Millipore, type HA 0.45 mm). Filters were washed with 10 mL ice-cold 5% TCA, dried and counted in an LS6500 multipurpose scintillation counter (Beckman Coulter, Brea, CA). Initial velocities for aminoacylation were calculated for all tRNA concentrations, and the kinetic parameters (KM and Vmax) were determined by plotting the velocities against substrate concentration and fit to the Michaelis–Menten steady-state model using XLfit (IDBS, London, UK).
Chemical Compound Screening
tRNA aminoacylation was monitored using a scintillation proximity assay (SPA). The screening reactions were in 96-well microtiter plates (Costar). Test compounds were equilibrated by addition of 33 µL of the protein/buffer mix to 2 µL of compound (3.3 mM) dissolved in 100% DMSO as described above in the timed aminoacylation assay. The concentration of P. aeruginosa GluRS was 0.1 µM. This mixture was allowed to incubate at ambient temperature for 15 min, and reactions were then initiated by addition of 15 µL of E. coli tRNA (0.75 µM tRNAGlu), followed by incubation for 1 h at 37 °C. Reactions were stopped by the addition of 5 µL of 0.5 M EDTA. Four hundred micrograms of yttrium silicate (Ysi) poly-l-lysine–coated SPA beads (PerkinElmer) in 150 µL of 300 mM citrate buffer (pH 2.0) were added and allowed to incubate at room temperature for 1 h. The plates were analyzed using a 1450 Microbeta (Jet) liquid scintillation/luminescent counter (Wallac). Assays to determine values of the concentration that inhibits binding or activity by 50% (IC50) were as described with the test compounds serially diluted from 200 to 0.4 µM.
Microbiological Assays
Broth microdilution minimum inhibitory concentration (MIC) testing was performed in 96-well microtiter plates according to Clinical and Laboratory Standards Institute (CLSI, formerly NCCLS) guideline M7-A7.14 MIC values were determined for E. coli (ATCC 25922), E. coli tolC mutant, Enterococcus faecalis (ATCC 29212), Haemophilus influenzae (ATCC 49766), Moraxella catarrhalis (ATCC 25238), P. aeruginosa (ATCC 47085), P. aeruginosa PAO200 (efflux pump mutant), P. aeruginosa hypersensitive strain (ATCC 35151), Staphylococcus aureus (ATCC 29213), and Streptococcus pneumonia (ATCC 49619).
Time-kill studies were performed using E. faecalis and S. pneumoniae for compound BT_03F04 and S. aureus and S. pneumoniae for compound BT_04B09 based on the MIC assay results, according to CLSI document M26-A.15 Growth media was Brain Heart Infusion and Trypticase Soy Broth from Remel (Lenexa, KS) with or without 3% lysed horse blood (Hemostat Laboratories, Dixon, CA).
In Vitro Cytotoxicity Test
Eighteen hours before the assay, 25,000 NIH/3T3 cells were plated per well in 96-well plates using Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and Penicillin-Streptomycin Solution. Cells were grown under standard tissue culture conditions (5% CO2 and 37 °C). Compounds were diluted in DMSO, yielding a final DMSO concentration of 6.25% in the cell cultures. Cells were treated with the compounds or DMSO alone for 24 h. Next, the Trevigen TACS MTT Cell Proliferation Assay Kit (Gaithersburg, MD) was utilized to assess impacts on human cell proliferation and/or viability. Ten microliters of MTT reagent was added to each well and incubated under 5% CO2 at 37 °C for another 4 h. Finally, 100 µL of detergent reagent was added and incubation was continued for an additional 4 h. The optical density (A595) was determined using a Bio-Rad iMark microplate absorbance reader. Samples were carried out in triplicate. The Student two-tiered t test was utilized to assess statistical significance.
Binding Mode Assay
To determine if the compounds were competitive with ATP, IC50s were determined as described above, but containing the indicated concentrations of ATP (25, 50, 100, 250, 500, and 1000 µM) and 0.05 µM P. aeruginosa GluRS. The protein/ buffer mix was added to the compound (2 µL) and allowed to incubate at room temperature for 15 min. Final compound concentrations in the reactions ranged from 200 to 0.4 µM. The reaction was started by addition of 15 µL E. coli tRNA (0.75 µM tRNAGlu). Positive controls contained only DMSO without compound. The reactions were for 1 h at 37 °C and stopped by the addition of 5 µL of 0.5 M EDTA.
To determine if the compounds were competitive with Glu, the same assay was used. However, ATP was held at a constant concentration of 2.0 mM in assays containing indicated concentrations of Glu (25, 50, 100, 200, and 300 µM). Background amounts of free [3H]Glu in the absence of GluRS were insignificant.
Results
Sequence Analysis
The structure of GluRS with or without its substrates, Glu, ATP, or tRNAGlu, has been well studied, primarily from structural studies of GluRS from Thermus thermophilus (Tth)16 and Thermosynechococcus elongatus.17 More recently, the structure of an engineered form of GluRS from E. coli that contains two mutations (K236E/E328A) to allow crystallization has been solved.18 There is moderate amino acid sequence conservation of GluRS from P. aeruginosa when compared with the corresponding enzymes from Bacillus subtilis, E. coli, T. elongatus, or Tth, with the number of conserved residues ranging between 35 and 43%. However, when compared with human mitochondrial GluRS (hmGluRS), there is only 30% conservation of the amino acid residues, and this drops to >12% when compared to the N-terminal 676 amino acids (which contains the GluRS activity) of human cytosolic Glu-ProRS (Table S1). The active site residues that have been shown to interact with the substrates Glu and ATP are contained within the Rossmann fold in the catalytic domain. The Glu binding sites are strictly conserved between Tth and P. aeruginosa GluRS, and only moderate changes are observed when P. aeruginosa GluRS is compared with the E. coli and T. elongatus homologs (Fig. 1). In Tth GluRS, Arg5 and Arg205 (Tth numbering) interact with the glutamate γ-carboxyl group, the α-NH3+ group of the glutamate interacts with Ala7 and Glu41, and Asn191 fills the space between the crucial residues Arg5 and Tyr187 in Tth.16 In E. coli and T. elongatus GluRS, Asn191 is replaced with the smaller valine. Trp209, together with Ala7 and Tyr187, forms a hydrophobic pocket that wraps around the Glu substrate side chain methylene group in Tth.16 This Trp residue is replaced by a His in E. coli and T. elongatus GluRS. The overall ATP binding sites are also highly conserved, with only minimum alterations observed (Fig. 1).
Figure 1.
Amino acid sequence alignment of P. aeruginosa GluRS with homologs. The protein sequences were downloaded from the National Center for Biotechnology Information (NCBI). Ec, E. coli; Pa, P. aeruginosa; Tel, T. elongatus; Tth, T. thermophilus. Accession numbers for GluRS protein sequences of E. coli, P. aeruginosa, T. elongatus, and T. thermophilus are P04805, Q9XCL6, 2CFO_B, and Q72LI9, respectively. Sequence alignments were performed using Vector NTI Advance (TM) 11.0 (Invitrogen). Identical amino acid residues are white on black, and similar residues are white on gray. The positions of the HIGH and KMSKS motifs are indicated. Amino acid residues that interact with Glu (*), ATP (▲ for productive mode, △ for nonproductive mode), and tRNA (●) are indicated. The amino acid residues Arg358 and Thr444 (Tth numbering) are indicated (★).
In the absence of tRNAGlu, ATP binding by GluRS is nonproductive because the α-phosphate of ATP and the α-carboxyl groups of Glu are positioned too far apart for a reaction to occur. The presence of tRNAGlu causes conformational changes surrounding the ATP binding site, allowing ATP to bind in the productive state, which moves the α-phosphate of the ATP closer to the α-carboxyl groups of Glu, allowing the reaction for the formation of the glutamyl-AMP intermediate to occur.19 The tRNA binding site is less conserved than either the Glu or the ATP binding site. However, certain amino acids, including Arg147, which interacts with the tRNAGlu C74 phosphate; Asp44 and Arg47, which interact with the 2′-hydroxyl group of C75; and Tyr187 and Thr43, which interact with the adenosine base and the 5′-hydroxyl group of A76, are strictly conserved.
Protein Expression and Characterization
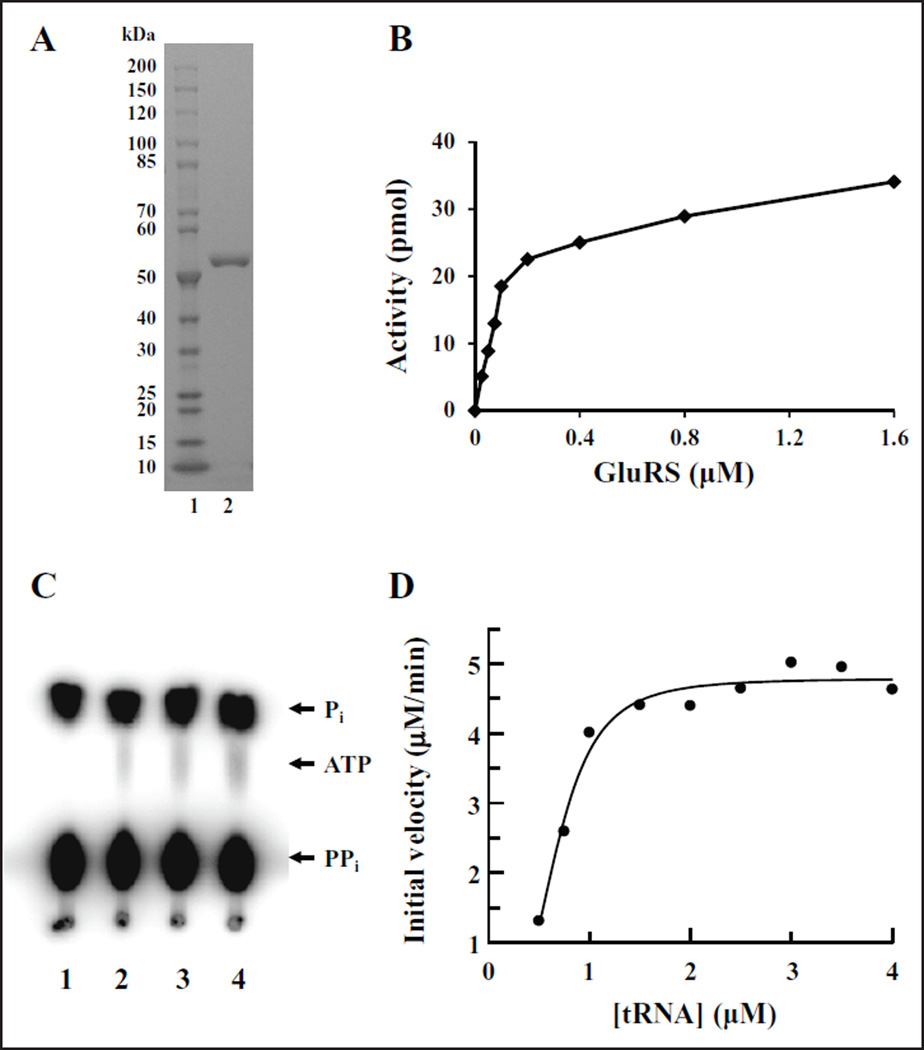
The glutamyl-tRNA synthetase gene (gltX) from P. aeruginosa was cloned, expressed, and purified to greater than 98% homogeneity (Fig. 2A). To determine the concentration of P. aeruginosa GluRS to be used in the following assays, the purified enzyme was titrated into aminoacylation assays at various concentrations (Fig. 2B). The aminoacylation of a particular tRNA with the corresponding amino acid for the majority of the aminoacyl-tRNA synthetases is a two-step reaction. In the first step, the enzyme condenses the cognate amino acid and ATP, forming an aminoacyl-adenylate intermediate with a pyrophosphate as the leaving group. This reaction is reversible in the absence of the tRNA and can be monitored using the ATP:PPi exchange reaction.20 However, GluRS, along with ArgRS and GlnRS, cannot catalyze this reaction in the absence of its cognate tRNAs for the reasons described above. We have tested P. aeruginosa GluRS, in the presence and absence of tRNA, to confirm the requirement for the tRNA using ATP:PPi exchange assays. The ATP:PPi exchange assay is indicative of the ability to form an aminoacyl-adenylate, followed by reversal of the reaction and subsequent radioactive labeling of ATP in the presence of saturating amounts of [32P]PPi (Fig. 2C). In the absence of tRNAGlu (Fig. 2C, lane 1) there was no [32P]ATP formed by the exchange of pyrophosphate observed. This indicated that no aminoacyl-adenylate had formed, or if it had formed, the GluRS/aminoacyl-adenylate complex was very stable and the reaction was not reversible. However, in the presence of increasing concentrations of tRNAGlu, increased levels of [32P]ATP were detected (Fig. 2C, lanes 2–4). This indicated that in the presence of tRNA, an aminoacyl-adenylate was formed; however, the low level of radioactive ATP formed suggests that the reversal of the reaction is not a dynamic process likely because the reaction goes to fulfillment and the tRNA is aminoacylated. Additional studies would be required to ascertain the stability of the GluRS/tRNA/aminoacyl-adenylate complex.
Figure 2.
Purification and characterization of P. aeruginosa GluRS. (A) One microgram of purified P. aeruginosa GluRS was analyzed on a 4–20% SDS-PAGE gel and the protein bands were visualized by staining with Coomassie blue. Lane 1 contains 10–200 kD protein standard; lane 2 contains P. aeruginosa GluRS. (B) P. aeruginosa GluRS was titrated into the aminoacylation assay as described in the Materials and Methods section at concentrations between 0.025 and 1.6 µM. Background activity was minimal and was subtracted from values at all concentrations of GluRS. (C) ATP:PPi exchange reaction to determine the requirement for the cognate tRNA for formation of an aminoacyl-adenylate by P. aeruginosa GluRS. The reactions contained 0, 0.3, 0.6, and 1.2 µM tRNAGlu in lanes 1, 2, 3, and 4, respectively. (D) Determination of the kinetic parameters with respect to tRNAGlu for P. aeruginosa GluRS in the aminoacylation reaction. Initial velocities were determined and the data were fit to a Michaelis–Menten steady-state model using XLfit (IDBS) to determine KM and Vmax.
Next, we determined the kinetic interactions of P. aeruginosa GluRS with its tRNA substrate. The initial rate for aminoacylation of tRNAGlu by GluRS was determined at several different concentrations of tRNAGlu (0.5, 0.75, 1, 1.5, 2, 2.5, 3, 3.5, and 4 µM) while holding the concentrations of ATP and Glu constant at 2.5 mM and 100 µM, respectively (Fig. 2D). The initial velocities were modeled by fitting them to the Michaelis–Menten steady-state model using XLfit (IDBS). The KM and kcat values were determined to be 0.68 µM and 0.8 s−1 for P. aeruginosa GluRS, which gave a kcat/KM value of 1.18 s−1 µM−1.
Screening Chemical Compounds against the Activity of P. aeruginosa GluRS
Using SPA technology, the activity of GluRS in the aminoacylation assay was screened against two chemical compound libraries. One of the compound libraries contained 800 natural products, and the other compound library contained 890 synthetic compounds. The assay detects the ability of GluRS to aminoacylate tRNAGlu, and in the presence of a chemical compound, to measure the effect of the compound on the activity of GluRS. Chemical compounds were dissolved in 100% DMSO, resulting in final DMSO concentrations in screening assays of 4%; therefore, the ability of P. aeruginosa GluRS to function in the presence of increasing amounts of DMSO was determined. There was no decrease of activity observed in the aminoacylation assays containing up to 10% DMSO (data not shown). Initial screening assays contained chemical compounds at a concentration of 132 µM and were carried out as single-point assays. Compounds observed to inhibit at least 50% of enzymatic activity were reassayed in triplicate using filter binding assays as described. These assays resulted in 14 confirmed hit compounds from the synthetic compound library; however, no hit compounds were confirmed from the natural compound library. The concentration of the compound at which 50% of the enzymatic activity was inhibited (IC50s) for all 14 compounds was determined in SPA-based aminoacylation assays. The IC50 exhibited by the hit compounds ranged from 5 to 50 µM (Table S2).
Microbiological Assays
The 14 hit compounds were tested in broth microdilution assays to determine minimum inhibitory concentrations (MICs) against a panel of 10 pathogenic bacteria, including efflux pump mutants of E. coli and P. aeruginosa and a hypersensitive strain of P. aeruginosa (Table S3). Despite the similarity in biochemical activity of the compounds, the ability to inhibit bacterial growth varied widely. Both BT_03F04 and BT_04B09 inhibit Gram-positive pathogens at moderately low concentrations. BT_03F04 exhibited MICs against E. faecalis and S. pneumoniae at 2 and 8 µg/mL, respectively, and the MIC of BT_04B09 against both S. pneumoniae and S. aureus was observed to be 32 µg/mL. BT_06A02 and BT_06H07 inhibited the E. coli and the P. aeruginosa efflux pump mutants with MICs of 8 and 32 µg/mL and 64 µg/mL, respectively. These two compounds were not considered further because of the complexity of the structures (Fig. S1). BT_06D03 inhibited the P. aeruginosa efflux pump mutant and BT_08A06 inhibited the E. coli efflux pump mutant with MICs of 32 µg/mL. This indicated that these compounds were likely gaining access to the wild-type strains of these bacteria, but at least in part because of the presence of the efflux pumps, the compounds were quickly removed from the interior of the bacteria. BT_11H07 exhibited a MIC against S. aureus of 32 µg/mL. None of the remaining compounds tested inhibit any of the pathogens tested at a concentration lower than 64 µg/mL (Table S3).
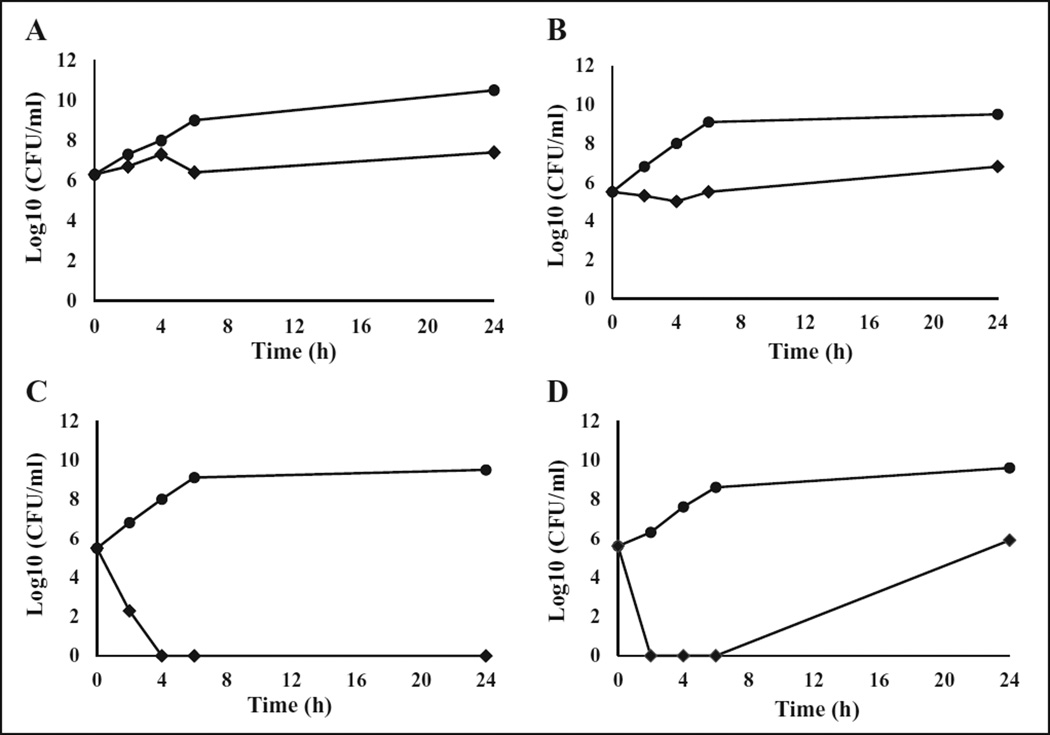
Next, time-kill studies were performed using the two compounds (BT_03F04 and BT_04EB09) to determine whether they were bacteriostatic or bactericidal. Based on the MIC results, BT_03F04 was most effective against E. faecalis and S. pneumoniae; therefore, this compound was tested against these bacteria in time-kill studies at four times the MIC between 0 and 24 h. BT_03F04 was shown to be bacteriostatic against both pathogens; it had constant growth but a decrease in colony-forming units (CFUs) of 2–3 log10 compared to the control during the times tested (Fig. 3A,B). Similarly, BT_04B09 was tested against S. pneumoniae and S. aureus based on MIC results at four times the MIC between 0 and 24 h, and it was shown to be bactericidal against both pathogens. BT_04B09 inhibited the growth of S. pneumoniae completely during all times tested (Fig. 3C), but at 24 h there was some regrowth in S. aureus due to the incapacity to completely kill all the bacteria present at this concentration, or possibly the compound lost potency over extended times (Fig. 3D).
Figure 3.
Time-kill kinetics of BT_03F04 and BT_04B09 against selected bacteria. The activity of BT_03F04 against growths of (A) E. faecalis and (B) S. pneumoniae. The activity of BT_04B09 against (C) S. pneumoniae and (D) S. aureus. Compounds were added to bacterial cultures at 4× MIC. Samples were analyzed by plating and determination of colony-forming units (CFUs) at 0, 2, 4, 6, and 24 h. Diamonds (♦) represent cultures containing the test compounds, and filled circles (●) represent control cultures grown in the absence of compounds.
Compounds BT_03F04 and BT_04B09 Were Not Toxic to Mammalian Cells
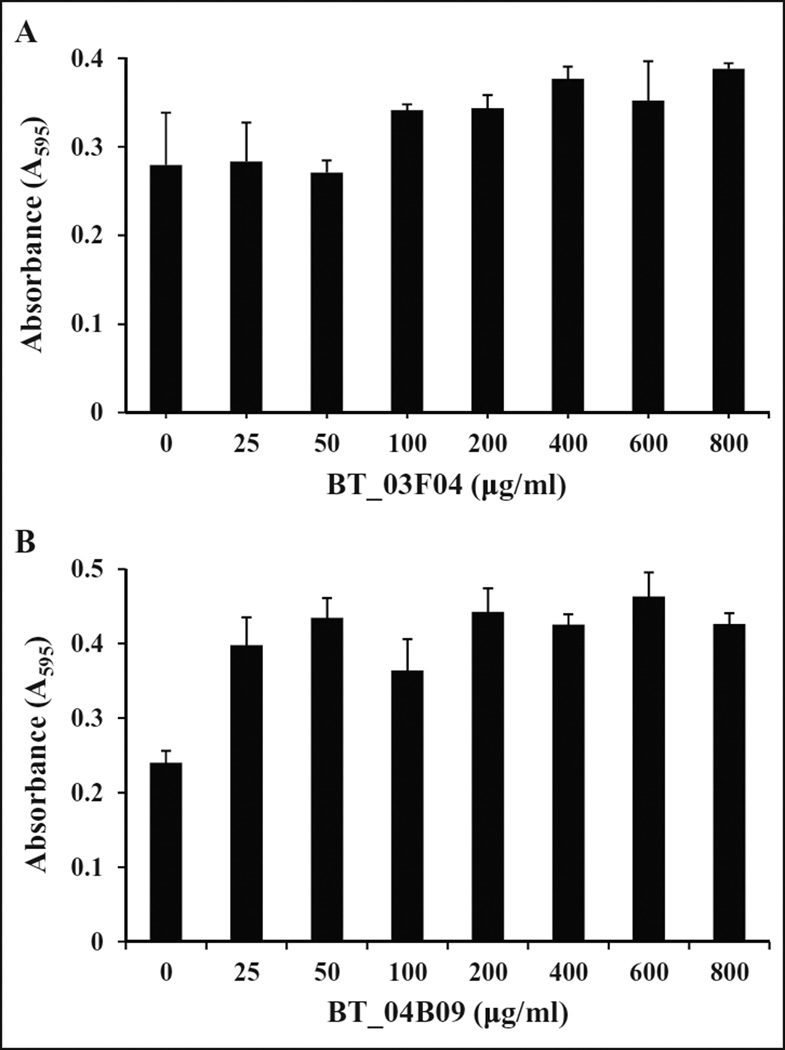
Antibiotic therapies that specifically target bacterial translation could potentially be toxic to mammalian cells given that both cytoplasmic and mitochondria GluRS are contained within the eukaryotic cell, and each share limited biochemical similarity with bacteria GluRS. MTT assays were performed to examine whether the hit compounds BT_03F04 and BT_04B09 were cytotoxic to mammalian cells. NIH/3T3 cells were treated with 25–800 µg/mL of BT_03F04 or BT_04B09 for 24 h under standard tissue culture conditions in triplicate. The compounds were not observed to be toxic to cells at any concentration tested (Fig. 4). Therefore, even at 25–400 times greater than the MIC, there was no observed cytotoxic effect, suggesting that these compounds would be amenable as therapeutics.
Figure 4.
Determination of toxicity of BT_03F04 (A) and BT_04B09 (B) in mammalian cell cultures. MTT assays were performed with the indicated dose of drug for 24 h under standard tissue culture conditions as described in the Materials and Methods section. There was no toxicity observed for either of the compounds.
Mechanism of Action
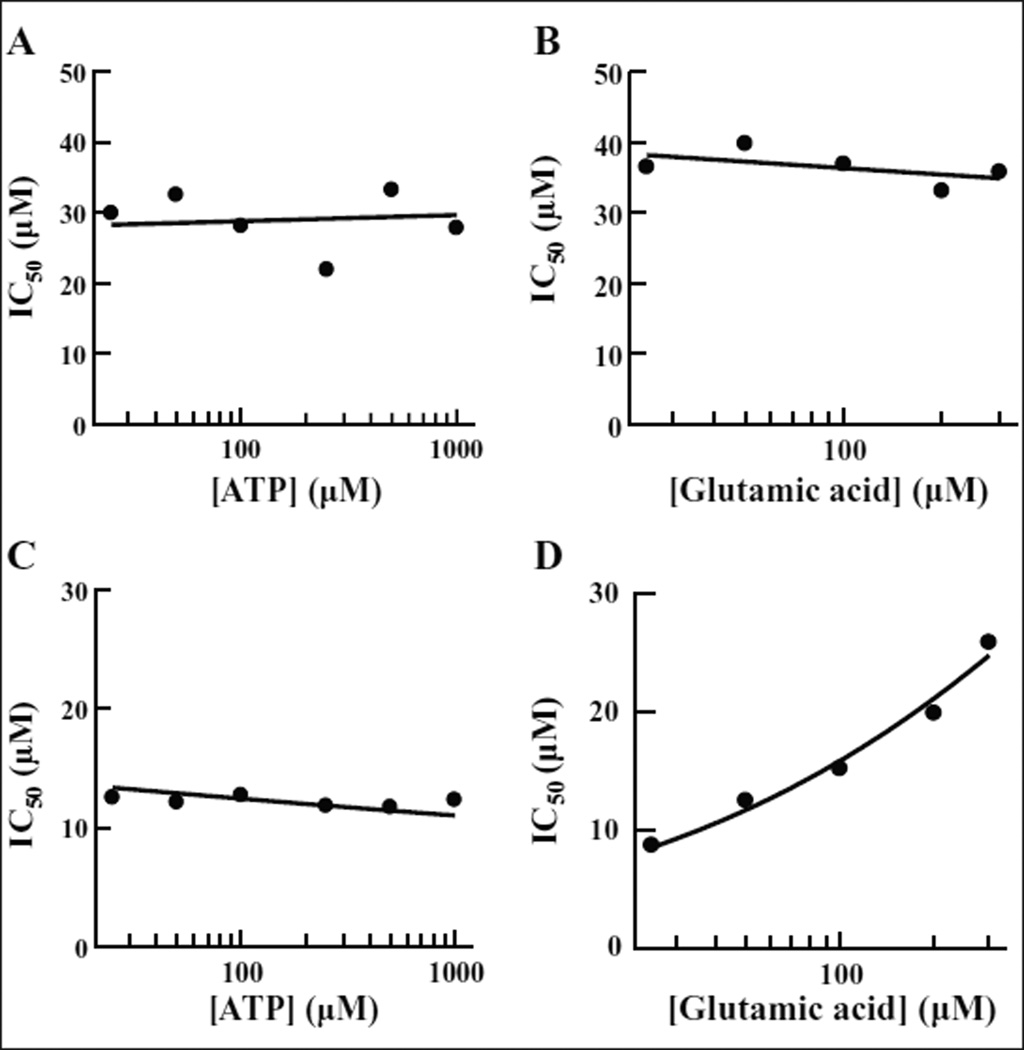
Aminoacyl-tRNA synthetases in general have three substrates: tRNA, ATP, and the amino acid. ATP and amino acids are small molecules similar in size to the compounds tested, and both have binding sites in the active site region of the synthetase. Blocking the binding site of one of these substrates would be a possible mechanism of action of an inhibitor. To determine if this was the case for either of the two hit compounds, we analyzed this as a possible mode of action. The mechanism of inhibition of both inhibitors with respect to ATP was determined using the tRNA aminoacylation assay to determine IC50 values at various ATP concentrations (25, 50, 100, 250, 500, and 1000 µM) while holding the concentration of the amino acid at 100 µM. To determine the mechanism of inhibition with respect to Glu, the same assay was used, except that ATP was held constant at saturating concentration (2.5 mM) and the IC50 was determined at different concentrations of Glu (25, 50, 100, 200, and 300 µM). The IC50s for BT_03F04 remain constant at all concentrations of both ATP and Glu (Fig. 5A,B), which is characteristic of a noncompetitive inhibitor.21 The IC50s for BT_04B09 remain constant at various concentrations of ATP; however, an increase was observed at increasing concentrations of the amino acid (Fig. 5C,D). An increase in IC50 as the concentration of the substrate is increased is characteristic of a competitive inhibitor.21 This leads us to conclude that BT_04B09 may inhibit GluRS by direct competition with the amino acid, while BT_03F04 inhibits the function of the enzyme by a different mechanism.
Figure 5.
Determination of the mode of action of BT_03F04 and BT_04B09 relative to ATP and Glu. IC50s were determined using the aminoacylation assay. Final compound concentrations in the IC50 reactions ranged from 200 to 0.4 µM. The glutamic acid concentration was fixed at 100 µM, and IC50s were determined at six different ATP concentrations ranging from 25 to 1000 µM in assays to determine competition with ATP. The ATP concentration was fixed at 2 mM, and IC50s were determined at five different amino acid (Glu) concentrations ranging from 25 to 300 µM to determine competition with the amino acid. The data were fit to the sigmoidal dose–response model using XLfit (IDBS). A lack of change of IC50 values when the substrate is increased is indicative of noncompetitive inhibition. An increase in IC50 values as substrate is increased is indicative of competitive inhibition. Reactions containing BT_03F04 were carried out in increasing concentrations of (A) ATP and (B) glutamic acid. Reactions containing BT_04B09 were carried out in increasing concentrations of (C) ATP and (D) glutamic acid.
Discussion
There are two different types of GluRS enzymes from different organisms: discriminating GluRS (D-GluRS) and nondiscriminating GluRS (ND-GluRS). Primarily structural analysis of the two types of GluRS revealed that the key feature that distinguishes D-GluRS and ND-GluRS is located in Arg358 and Thr444 (Tth numbering). In ND-GluRS, these two amino acids are replaced by the smaller glycine, Gly, which results in the relaxation of the anticodon specificity and allows recognition of both the tRNAGlu anticodon UUC and the more bulky tRNAGln anticodon UUG.22 P. aeruginosa GluRS has an Arg residue and a Ser residue corresponding to Tth Arg358 and Thr444, respectively, which are labeled with stars in Figure 1. Taken together with the fact that the P. aeruginosa genome contains a gene that encodes the glutaminyl-tRNA synthetase, we can safely propose that P. aeruginosa GluRS is a discriminating GluRS.
The catalytic mechanism for the glutamyl-, glutaminyl-, and arginyl-tRNA synthetases has drawn attention because of the unique characteristic that requires the cognate tRNA to allow the formation of a stable aminoacyl-adenylate as measured using the ATP:PPi exchange reaction. Our work has indicated that tRNA is also necessary for this reaction when catalyzed by P. aeruginosa GluRS. The crystal structures of T. thermophilus GluRS were solved with and without its substrates, Glu and ATP, and the results revealed that binding of tRNAGlu causes a conformational change in GluRS.16 The results of the conformational changes are twofold. First, the conformational change causes a shift in the ATP binding sites, which brings the ATP α-phosphate in close proximity to the α-carboxyl group of the amino acid and allows formation of the aminoacyl-adenylate. In the absence of the tRNA, the proximity of the ATP and the amino acid are not conducive to the formation of the aminoacyl-adenylate and no condensation reaction occurs.19 Second, the conformational change in GluRS resulting from tRNA binding causes the amino acid binding site to be highly selective in size, shape, and charge distribution for l-glutamic acid, which allows the enzyme to discriminate l-Glu from l-Gln, l-Asp, and d-Glu.16 This conformational change that favors Glu binding apparently acts as a proofreading function and may explain how GluRS maintains fidelity in translation without containing an editing domain, which is present in many other aminoacyl-tRNA synthetases.
In the present work, scintillation proximity assay technology was used to screen for inhibitors of the ability of P. aeruginosa GluRS to aminoacylate its cognate tRNA. The screening assays were very robust in a high-throughput format and resulted in Z′ and Z factors of approximately 0.80 and 0.74, respectively, across all plates. From almost 1700 compounds, 14 compounds (exclusively from the synthetic compound library) were identified that inhibited the activity of P. aeruginosa GluRS (Figs. 6 and S1). Two of the compounds, BT_03F04 and BT_04B09, which inhibited GluRS activity with IC50s of 21.9 and 24.9 µM, respectively, also exhibited moderate antibacterial activity against a series of Gram-positive pathogens. However, the compounds only showed low levels of activity against Gram-negative pathogens. All 14 of the hit compounds were identified using in vitro P. aeruginosa protein synthesis assays, yet none of the compounds inhibited the growth of Gram-negative organisms with any success. GluRS from the Gram-positive organisms are as similar to P. aeruginosa GluRS as the other homologs (Table S1); however, they are more similar to each other than to the Gram-negative forms of GluRS. The percent of conserved amino acids in GluRS from E. faecalis, S. pneumoniae, and S. aureus ranges from 58 to 64%, which is significantly higher than observed between P. aeruginosa GluRS and the other homologs analyzed. Therefore, even though these compounds were identified using P. aeruginosa GluRS, they may bind and affect the activity of the group of Gram-positive GluRS molecules to a greater extent. Also, GluRS from Gram-positive bacteria aminoacylate both tRNAGlu and tRNAGln, whereas in Gram-negative bacteria, GluRS only aminoacylates tRNAGlu. The inability to aminoacylate both tRNAGlu and tRNAGln in Gram-positive bacteria would certainly be additive and increase the effect of the compound on the growth of these bacteria. Even the compromised strains of E. coli and P. aeruginosa were only affected at low levels by any of the compounds, indicating that other mechanism besides efflux systems may be partially responsible for lack of activity of the compounds against the Gram-negative bacteria. This is reinforced by the fact that H. influenzae and M. catarrhalis, two Gram-negative bacteria that are more susceptible to many inhibitors because they have less robust efflux systems, were also not affected in culture by the compounds.
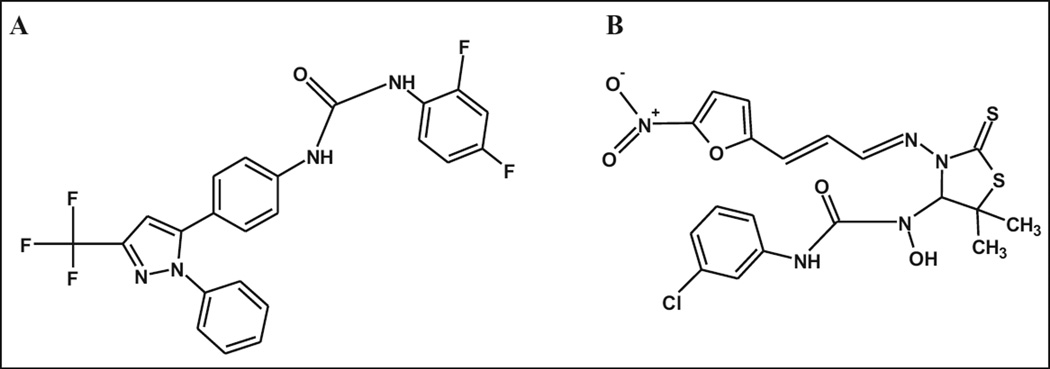
Figure 6.
The chemical structures of BT_03F04 (A) and BT_04B09 (B).
Compound BT_03F04 (1-(2,4-difluorophenyl)-3-{4-[1-phenyl-3-(trifluoromethyl)-1H-pyrazol-5-yl]phenyl}urea) contains a carboxamide group (Fig. 6A), which is present in some well-known antibiotics. Tetracycline contains a carboxamide as a side chain, whereas in BT_03F04, this motif serves as the core of the compound. We searched the PubChem BioAssay Database for the biological activity and found no biological information associated with this compound. Recent works on a number of compounds that have a carboxamide-based structure have been published. First, Ni(II) and Cu(II) complexed with hydrazine carboxamide-based compounds have been synthesized, which exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria.23 In another study, the carboxamide group of eremomycin (a glycopeptide antibiotic) was found to be a pharmacophore responsible for inhibition of the growth of certain Gram-positive bacteria and, when compared with the parent antibiotic, retained all antibacterial activity.24 Also, the carboxamide derivatives of tetramic acids based on antibacterial natural products have been shown to have bioactivity against various Gram-positive bacteria and also the Gram-negative organism H. influenzae.25 Finally, using molecular hybridization, a series of carboxamide derivatives were synthesized and shown to be active against Mycobacterium tuberculosis (MTB).26 These results, along with the activity of BT_03F04 observed against Gram-positive pathogens, indicate that further evaluation of a carboxamide-based lead series based on this compound may be advantageous.
Compound BT_04B09 (3-(3-chlorophenyl)-1-(5,5-dimethyl-3-{(E)-[(2E)-3-(5-nitrofuran-2-yl)prop-2-en-1-ylidene] amino}-2-thioxo-1,3-thiazolidin-4-yl)-1-hydroxyurea) contains both a carboxamide and a nitrofuran group (Fig. 6B). We also searched the PubChem BioAssay Database for the biological activity and again found no biological information associated with this compound. Nitrofurans are a class of synthetic broad antimicrobial spectrum antibiotics including furazolidone, furaltadone, nitrofurantoin, and nitrofurazone. The nitrofurans are active against bacteria in the enterobacteria family; however, they do not have antibacterial activity against P. aeruginosa. They also are active against Grampositive staphylococci and streptococci. Nitrofurantoin in particular has been well studied and prescribed extensively.27 A variety of side effects were noted with long-term use of this antibiotic; however, good efficacy and tolerability have been observed with short-term therapy. With the recent increase in the emergence of antibiotic resistance, the use of nitrofurantoin has increased, and it is primarily used for the treatment of lower urinary tract infections caused by multidrug-resistant pathogens.
In time-kill experiments, BT_03F04 was shown to inhibit bacterial growth in a bacteriostatic manner, while BT_04B09 inhibited bacterial growth by killing the bacteria. This is interesting in that inhibition of the aminoacylation activity of an aaRS likely leads to amino acid starvation in protein biosynthesis, thereby eliciting the stringent response resulting in static bacterial growth, but not necessarily killing bacteria. Why then was BT_04B09 observed to be bactericidal? Recent studies indicate that many aaRS proteins have secondary roles and functions; this is particularly true with eukaryotic aaRS, but has also been observed in bacteria.28 GluRS has been documented functioning in roles other than just aminoacylation of cognate and nearcognate tRNAs. There are also complex transcriptional and translational regulatory mechanisms unique to the expression of GluRS that are not well understood but appear to be affected by the enzyme itself.29 Self-regulation of transcription influences the level of gltX (GluRS) gene-derived mRNA, which has been shown to have an effect on the expression of other enzymes critical for pathways other than protein biosynthesis. From the mechanism of action studies, the two compounds were observed to inhibit GluRS by different methods. The bactericidal activity of BT_04B09 may be the result of the inhibition of a secondary function of GluRS that is not affected by BT_03F04.
Mupirocin (pseudomonic acid), the bacterial IleRS inhibitor, is currently the only clinically available aminoacyl-tRNA synthetase inhibitor presently on the market. Several glutamyl-adenylate analogs have been synthesized and were shown to exhibit varying levels of inhibition of enzymatic activity in vitro.30 At this time, however, no whole-cell antibacterial activity has been reported and none of the compounds have proceeded into the clinical stage. Both BT_03F04 and BT_04B09 have moderately good IC50 values as a starting point, and both are efficacious in inhibition of the growth of Gram-positive organisms. Increasing potency and improving efficacy against bacteria in culture via structure–activity relationship (SAR) studies will be required to guide development of a lead series based on either of these compounds.
Supplementary Material
Acknowledgments
The authors are grateful for the financial support provided by the National Institutes of Health (grant number 1SC3GM098173-01A1). The contents of this article/publication/etc. are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. Undergraduate student support was in part from the Howard Hughes Medical Institute (HHMI) Precollege and Undergraduate Science Education Program grant 52007568.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors are grateful for the financial support provided by the National Institutes of Health (grant number 1SC3GM098173-01A1). The contents of this article/publication/etc. are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. Undergraduate student support was in part from the Howard Hughes Medical Institute (HHMI) Precollege and Undergraduate Science Education Program grant 52007568.
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Driscoll JA, Brody SL, Kollef MH. The Epidemiology, Pathogenesis and Treatment of Pseudomonas aeruginosa Infections. Drugs. 2007;67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 2.Strateva T, Yordanov D. Pseudomonas aeruginosa: A Phenomenon of Bacterial Resistance. J. Med. Microbiol. 2009;58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 3.Raczniak G, Ibba M, Soll D. Genomics-Based Identification of Targets in Pathogenic Bacteria for Potential Therapeutic and Diagnostic Use. Toxicology. 2001;160:181–189. doi: 10.1016/s0300-483x(00)00454-6. [DOI] [PubMed] [Google Scholar]
- 4.Eriani G, Delarue M, Poch O, et al. Partition of tRNA Synthetases into Two Classes Based on Mutually Exclusive Sets of Sequence Motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 5.Sprinzl M, Cramer F. Site of Aminoacylation of tRNAs from Escherichia coli with Respect to the 2′- or 3′-Hydroxyl Group of the Terminal Adenosine. Proc. Natl. Acad. Sci. U. S. A. 1975;72:3049–3053. doi: 10.1073/pnas.72.8.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kern D, Lapointe J. Catalytic Mechanism of Glutamyl-tRNA Synthetase from Escherichia coli: Reaction Pathway in the Aminoacylation of tRNAGlu. Biochemistry. 1980;19:3060–3068. doi: 10.1021/bi00554a035. [DOI] [PubMed] [Google Scholar]
- 7.Schon A, Kannangara CG, Gough S, et al. Protein Biosynthesis in Organelles Requires Misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 8.Sheppard K, Akochy PM, Salazar JC, et al. The Helicobacter pylori Amidotransferase GatCAB Is Equally Efficient in Glutamine-Dependent Transamidation of Asp-tRNAAsn and Glu-tRNAGln. J. Biol. Chem. 2007;282:11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 9.Skouloubris S, Ribas de PL, De RH, et al. A Noncognate Aminoacyl-tRNA Synthetase That May Resolve a Missing Link in Protein Evolution. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salazar JC, Ahel I, Orellana O, et al. Coevolution of an Aminoacyl-tRNA Synthetase with Its tRNA Substrates. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macarron R, Mensah L, Cid C, et al. A Homogeneous Method to Measure Aminoacyl-tRNA Synthetase Aminoacylation Activity Using Scintillation Proximity Assay Technology. Anal. Biochem. 2000;284:183–190. doi: 10.1006/abio.2000.4665. [DOI] [PubMed] [Google Scholar]
- 12.Cull MG, McHenry CS. Purification of Escherichia coli DNA Polymerase III Holoenzyme. Methods Enzymol. 1995;262:22–35. doi: 10.1016/0076-6879(95)62005-2. [DOI] [PubMed] [Google Scholar]
- 13.Martinis SA, Fox GE. Non-Standard Amino Acid Recognition by Escherichia coli Leucyl-tRNA Synthetase. Nucleic Acids Symp. Ser. 1997;36:125–128. [PubMed] [Google Scholar]
- 14.Clinical Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Test for Bacteria That Grow Aerobically: Approved Guideline M7-A7. Wayne, PA: 2006. [Google Scholar]
- 15.Clinical Laboratory Standards Institute. Methods for Determining Bactericidal Activity of Antimicrobial Agents: Approved Guideline M26-A. Wayne, PA: 2002. [Google Scholar]
- 16.Sekine S, Shichiri M, Bernier S, et al. Structural Bases of Transfer RNA-Dependent Amino Acid Recognition and Activation by Glutamyl-tRNA Synthetase. Structure. 2006;14:1791–1799. doi: 10.1016/j.str.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Schulze JO, Masoumi A, Nickel D, et al. Crystal Structure of a Non-Discriminating Glutamyl-tRNA Synthetase. J. Mol. Biol. 2006;361:888–897. doi: 10.1016/j.jmb.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 18.Chongdar N, Dasgupta S, Datta AB, et al. Preliminary X-Ray Crystallographic Analysis of an Engineered Glutamyl-tRNA Synthetase from Escherichia coli. Acta Crystallogr. F Struct. Biol. Commun. 2014;70:922–927. doi: 10.1107/S2053230X14010723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekine S, Nureki O, Dubois DY, et al. ATP Binding by Glutamyl-tRNA Synthetase Is Switched to the Productive Mode by tRNA Binding. EMBO J. 2003;22:676–688. doi: 10.1093/emboj/cdg053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bullard JM, Cai YC, Demeler B, et al. Expression and Characterization of a Human Mitochondrial Phenylalanyl-tRNA Synthetase. J. Mol. Biol. 1999;288:567–577. doi: 10.1006/jmbi.1999.2708. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Prusoff WH. Relationship between the Inhibition Constant (KI) and the Concentration of Inhibitor Which Causes 50 Per Cent Inhibition (I50) of an Enzymatic Reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 22.Sekine S, Nureki O, Shimada A, et al. Structural Basis for Anticodon Recognition by Discriminating Glutamyl-tRNA Synthetase. Nat. Struct. Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 23.Chandra S, Vandana, Kumar S. Synthesis, Spectroscopic, Anticancer, Antibacterial and Antifungal Studies of Ni(II) and Cu(II) Complexes with Hydrazine Carboxamide, 2-[3-Methyl-2-Thienyl Methylene] Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015;135:356–363. doi: 10.1016/j.saa.2014.06.143. [DOI] [PubMed] [Google Scholar]
- 24.Pavlov AY, Berdnikova TF, Olsufyeva EN, et al. Carboxamides and Hydrazide of Glycopeptide Antibiotic Eremomycin: Synthesis and Antibacterial Activity. J. Antibiot. (Tokyo) 1996;49:194–198. doi: 10.7164/antibiotics.49.194. [DOI] [PubMed] [Google Scholar]
- 25.Jeong YC, Moloney MG. Synthesis and Antibacterial Activity of Monocyclic 3-Carboxamide Tetramic Acids. Beilstein. J. Org. Chem. 2013;9:1899–1906. doi: 10.3762/bjoc.9.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samala G, Devi PB, Nallangi R, et al. Development of Novel Tetrahydrothieno[2,3-c]Pyridine-3-Carboxamide Based Mycobacterium tuberculosis Pantothenate Synthetase Inhibitors: Molecular Hybridization from Known Antimycobacterial Leads. Bioorg. Med. Chem. 2014;22:1938–1947. doi: 10.1016/j.bmc.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 27.Stock I. Nitrofurantoin: Clinical Relevance in Uncomplicated Urinary Tract Infections. Med. Monatsschr. Pharm. 2014;37:242–248. [PubMed] [Google Scholar]
- 28.Martinis SA, Plateau P, Cavarelli J, et al. Aminoacyl-tRNA Synthetases: A Family of Expanding Functions. Mittelwihr, France, October 10–15, 1999. EMBO J. 1999;18:4591–4596. doi: 10.1093/emboj/18.17.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brun YV, Sanfacon H, Breton R, et al. Closely Spaced and Divergent Promoters for an Aminoacyl-tRNA Synthetase Gene and a tRNA Operon in Escherichia coli. Transcriptional and Post-Transcriptional Regulation of gltX, valU and alaW. J. Mol. Biol. 1990;214:845–864. doi: 10.1016/0022-2836(90)90340-R. [DOI] [PubMed] [Google Scholar]
- 30.Bernier S, Dubois DY, Habegger-Polomat C, et al. Glutamylsulfamoyladenosine and Pyroglutamyl-sulfamoyladenosine Are Competitive Inhibitors of E. coli Glutamyl-tRNA Synthetase. J. Enzyme Inhib. Med. Chem. 2005;20:61–67. doi: 10.1080/14756360400002007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.