Abstract
The natural history of the amoeba Dictyostelium discoideum has inspired scientific inquiry for seventy-five years. A genetically tractable haploid eukaryote, D. discoideum appeals as a laboratory model as well. However, certain rote molecular genetic tasks, such as PCR and cloning, are difficult due to the AT-richness and low complexity of its genome. Here we report on the construction of a ~20 fold coverage D. discoideum genomic library in E. coli, cloning 4 – 10 kilobase partial restriction fragments into a linear vector. End-sequencing indicates that most clones map to the six chromosomes in an unbiased distribution. Over 70% of these clones contain at least one complete open reading frame. We demonstrate that individual clones and library composition are stable over multiple replication cycles. Our library will enable numerous molecular biological applications and the completion of additional species’ genome sequences, and suggests a path towards the long-elusive goal of genetic complementation.
Keywords: Dictyostelium genomic library, linear bacteriophage plasmid
1. INTRODUCTION
The social amoeba Dictyostelium discoideum is a haploid eukaryote long established as a genetic model for numerous biological processes. These include studies of chemotaxis, cell differentiation and development, allorecognition, signal processing and pattern formation[1]. Methods for homologous recombination and random insertion mutagenesis are widely used to genetically manipulate D. discoideum[2,3]. While these approaches have proven extremely valuable in discovering and validating the genetic bases of many phenomena, much of the rote molecular biology involved can be arduous. Dictyostelium genomic DNA is notoriously AT-rich (nearly 78%), recalcitrant to in vitro amplification and in vivo cloning, and prone to recombination and secondary structure formation[4]. A major challenge in Dictyostelium research has been to faithfully clone its genomic DNA into a microbial host such as Escherichia coli.
Dynes and Firtel (1989) constructed a high depth of coverage (20×) D. discoideum genomic library in E. coli consisting of 3 – 12 kilobase (kb) inserts. In this case, the genomic DNA was cloned into a shuttle vector capable of selection and replication in Dictyostelium as well. The authors demonstrated the library’s utility for genetic complementation by restoring thymidylate synthase activity to a mutant strain, and identified a gene that encodes a functional enzyme, ThyA[5]. However, attempts to complement other phenotypes were unsuccessful, suggesting that intact coding regions were either unstable or not well represented in the library (R. Firtel, personal communication). Kuspa and Loomis constructed a D. discoideum genomic DNA library in Yeast Artificial Chromosomes (YAC). The library facilitated the effort to map the Dictyostelium genome, but its utility was otherwise limited[6]. With the popularization of Restriction Enzyme Mediated Insertional mutagenesis (REMI)[3], efforts to clone a stable, gene-rich library were largely abandoned.
The issue of cloning genomic DNA was also relevant during the effort to sequence the D. discoideum genome[4]. Eichinger, et al. (2005) reported that the low complexity and AT-richness of the genome made large-insert bacterial clones unstable and presented significant challenges to sequencing and assembly. Researchers resorted to performing shotgun sequencing on electrophoretically separated chromosomes, developing statistical methods to handle these sequences, and framing the assembly with ordered YAC libraries and in-depth HAPPY maps[4,6,7]. An appealing strategy for handling “difficult” DNA was recently employed to clone the genome of a similarly AT-rich organism, Plasmodium berghei[8]. Pfander and colleagues (2011) solved the issue of Plasmodium clone instability using the linear, phage derived pJAZZ vector (Lucigen, Madison, WI)[8,9]. We reasoned that the pJAZZ cloning system might work well for Dictyostelium DNA as well.
As a proof of concept, we wished to build a D. discoideum genomic DNA library in E. coli with a broad chromosomal distribution, high gene content, and stable propagation. We cloned a 4 – 10 kb fraction of RsaI- and SspI-partially digested genomic DNA into the pJAZZ-ok vector. In total we recovered over 200,000 clones, constituting nearly 20 genome equivalents. End-sequencing of 192 inserts revealed an unbiased chromosomal distribution of the coverage, and a PCR screen of 24 randomly selected target genes found that all were represented in the library. We estimate that over 70% of the chromosomally derived inserts span at least one complete open reading frame (ORF). Some inserts encompass gene clusters, while others contain atypically large ORFs. Intergenic regions, known to harbor regulatory elements such as promoter and Kozak motifs, are also well represented in the library. Critically, we showed that individual and pooled genomic DNA library clones replicate stably over many generations and at high cell density. Our library represents a resource for performing molecular biology tasks on D. discoideum DNA in E. coli, demonstrates the potential for cloning dictyostelid DNA for sequencing additional genomes, and may illustrate a path towards a long sought after approach for genetic complementation.
2. RESULTS AND DISCUSSION
2.1 Dictyostelium discoideum genomic DNA library approximates 20-fold coverage
We constructed a ~20 fold coverage genomic DNA library of D. discoideum strain AX4 maintained in E. coli. The library consists of a 4 – 10 kilobase (kb) partial digest fraction cloned in the linear pJAZZ vector in TSA Big Easy cells (Lucigen). Two separate genomic DNA preparations and ligations yielded an estimated 212,752 bacterial colonies, which were collected into 31 pools (Table 1). We end-sequenced the inserts of 192 arbitrarily picked clones, 96 for each round of ligation, and mapped the clones to regions of the genome by BLAST. About 62% of the sequence pairs were unambiguously assigned to a locus among the six chromosomes (Table 1, Additional File 1). Five clones mapped to the duplicated region of chromosome[4]. In keeping with the Dictyostelium community convention for genome annotation and RNA-sequencing mapping, we assigned these clones to the proximal duplicated region[10]. The majority of the non-chromosomal sequences were most similar to the extrachromosomal ribosomal DNA (rDNA) palindrome, which exists in multiple copies and comprises up to 20% of the nuclear DNA[11]. In three instances (~1.6% of clones), the ends mapped to distant coordinates or different chromosomes, suggesting these clones contained concatamers of more than one restriction fragment. The remainder of the unmapped reads were either repetitive or multi-copy, or of such low complexity sequences that they failed to sequence or could not be identified by BLAST. Further analysis focused on the clones derived from and mapped to the chromosomes.
Table 1.
Summary of library colony content and estimated genome coverage
| Ligation Round 1 | Ligation Round 2 | Merged Library | |
|---|---|---|---|
| Pools | 3 | 28 | 31 |
| a Colonies / pool | 5,667 | 6,991 | 6,863 |
| Total colonies | 17,000 | 195,752 | 212,752 |
| b Unique clones | 16,433 | 189,227 | 205,660 |
| c Percent chromosomal | 61 | 62.5 | 62.3 |
| d Average insert size (kb) | 5.2 | 5.2 | 5.2 |
| Genome coverage (fold) | 19.6 | ||
Colony number was extrapolated by counting quadrants of agar plates, and averaging among plates scraped for each pool.
We encountered 2 instances of duplicates among the end-sequenced, mapped clones (4 out of 120, or 96.67% unique). We scaled the total colonies accordingly.
The percent of clones containing DNA from the six chromosomes was determined by mapping end sequences to the genome. Inserts that failed to map to the six chromosomes typically represented the ribosomal palindrome, retro-transposable elements, floating contigs, or concatamerized DNA.
Average insert size was calculated from in silico measurements of the distance between the chromosomal coordinates of mapped end sequences.
Of the 120 mapped clones, only 2 duplicates were encountered, indicating a high degree of clonal diversity (~97%, Additional File 1). The average insert size was estimated to be 5.2 kb based on the coordinates of the end-sequences. We calculated the genomic coverage (Table 1) by the following equation:
Achieving multiple genome equivalents of coverage with inserts of this size could be a powerful enabling technology. This cloning system could be used to finish the genome sequencing of additional dictyostelid species. Sequencing and completing the genomes of D. discoideum and its congeneric D. purpureum required intensive efforts based on whole chromosome shotgun sequencing, YACs and HAPPY maps[4,6,7]. With today’s relatively inexpensive next-generation short read approaches, sequencing new genomes should be quicker and less labor intensive. However, rampant low complexity would preclude the complete assembly of these genomes. Similar libraries could be constructed for those organisms to allow scaffolding of contigs and closure of assembly gaps.
2.2 Library inserts represent an unbiased distribution of chromosomal DNA
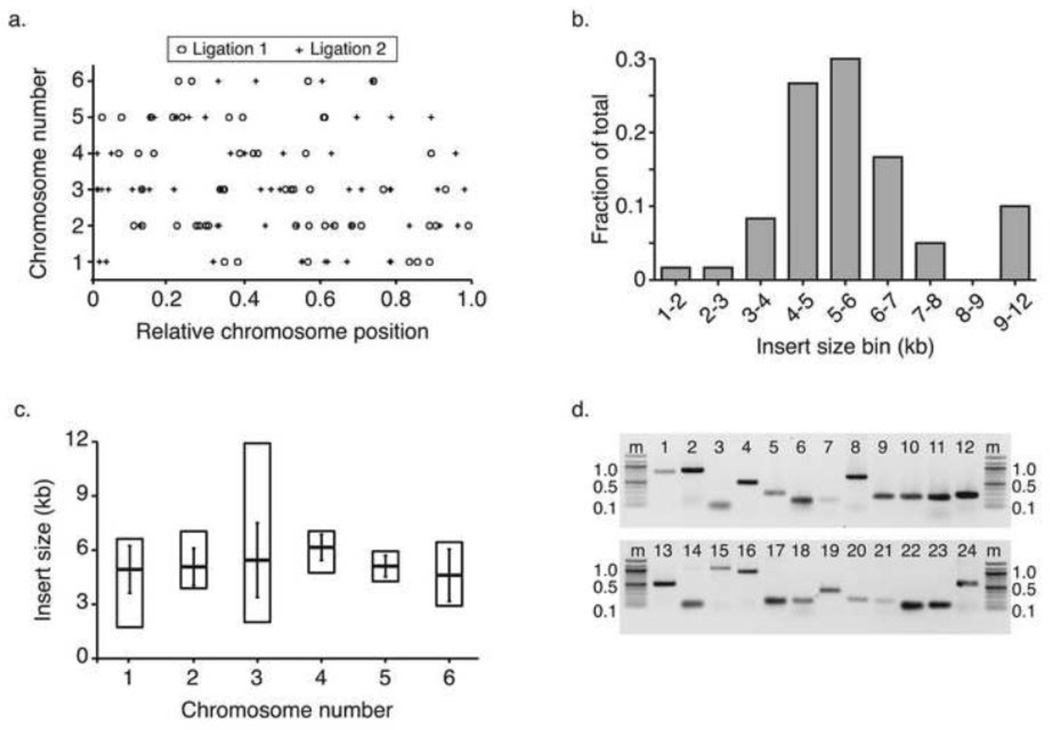
We tested whether the physical distribution of library inserts was representative of the entire genome, or biased in terms of the chromosomal positions of its clones. Plotting the relative chromosomal position of each insert suggested even clone distribution (Figure 1a). The distribution of mapped clones was not statistically different from a random distribution drawn from 10,000 simulations (Two-sample Kolmogorov-Smirnov test, p-value = 0.40). Further, the number of inserts per chromosome was as expected by chance, considering the different sizes of the chromosomes (χ2 p-value = 0.35, d.f. = 5). Next we tested whether the base composition of the cloned inserts reflected the AT-richness of the genomic DNA at large. The average GC content of the end-sequenced inserts was 26%. A simulated, random selection of 10,000 genomic fragments equal to the mean insert size were, on average, 24% GC. This difference was statistically significant (Two-sample Kolmogorov-Smirnov test, p-value < 0.0001), but may result from the exclusion of low complexity clones that failed to sequence or align by BLAST. Alternatively, a slight GC bias in the library might indicate enrichment for gene-containing DNA fragments, which are more GC-rich than intergenic regions.
Figure 1. Unbiased chromosomal origin of Genomic DNA inserts.
We end-sequenced the insert DNA of 96 colonies from each of two separate library preparations (see Table 1). Sequences were compared to the genome using BLAST to identify the genomic origins of the cloned DNA. (a) Clones that mapped to the six chromosomes (y-axis) are displayed as circles (ligation round 1) or crosses (ligation round 2). All chromosome lengths have been scaled from 0 – 1 (x-axis). (b) We determined the lengths of the insert DNA by subtracting the end positions of the genomic coordinates identified by BLAST mapping. Insert sizes (kb) are given on the x-axis while the frequency of those sizes is displayed on the y-axis. (c) The mean insert size (horizontal bars, y-axis) was highly similar for DNA from each chromosome (x-axis). Vertical bars indicate 1 standard deviation from the mean. The range of sizes (vertical boxes) was largest for chromosome 3. (d) PCR reactions targeting 24 genes from around the genome (Additional File 1) were resolved by 0.8% agarose gel electrophoresis. Gels were stained with ethidium bromide and negative images are shown. The marker (m) on either side of each gel image is a 100 base pair ladder (NEB). The sizes of select marker bands are indicated in kilobases on the right and left sides. Numbers above the lanes indicate PCR reaction number.
2.3 The majority of clones contain genes
We aimed to generate a library with cloned fragments sufficiently large to capture full genes and even gene clusters. The D. discoideum genome is gene-rich and compact, with ~13,000 gene models distributed over 34 MB[4,10]. The average gene size is 1.7 kb and the average intergenic segment size is roughly 2.5 kb. The size distribution of the library inserts was mostly consistent with the 4–10 kb DNA fraction purified for cloning (Figure 1b), sufficient to capture at least one gene per insert. The range of sizes was broader among fragments from chromosome three (Figure 1c), but this range could be attributed to two outlier inserts (2 kb and 11 kb), so it is probably not meaningful.
From a functional point of view, we wanted to know whether the library was likely to contain genes of interest. We screened a pool of library DNA by PCR for 24 genes representing all six chromosomes (Additional File 1). Target genes were selected based on existing primer pairs commonly used in the lab for qPCR or other experiments. All 24 targets were successfully amplified from pooled library DNA (Figure 1d), suggesting that most genes one might wish to study are present in the library. We further analyzed the mapped end-sequences to determine the gene content of those clones (Table 2, Additional File 1). Most of the chromosomal inserts (73%) contained at least one complete open reading frame (ORF). Less than four percent of the mapped clones contained no ORFs. Many of the inserts also contained partial ORFs, either exclusively or adjacent to complete genes. The high incidence of complete ORFs is consistent with the high gene density of the D. discoideum genome.
Table 2.
Most chromosomal DNA inserts contain at least one complete gene
| Ligation Round 1 | Ligation Round 2 | Merged Library | |
|---|---|---|---|
| Fraction of clones in library | 0.08 | 0.92 | |
| Percent of inserts with: | |||
| Complete genes | 66 | 74 | 73.4 |
| a Partial genes | 32 | 27 | 27.4 |
| No genes | 2 | 4 | 3.8 |
| Complete genes / insert | 1.0 | 1.5 | 1.5 |
| Partial gene / insert | 1.4 | 1.3 | 1.3 |
Partial genes were not mutually exclusive from complete genes. Often inserts contained both.
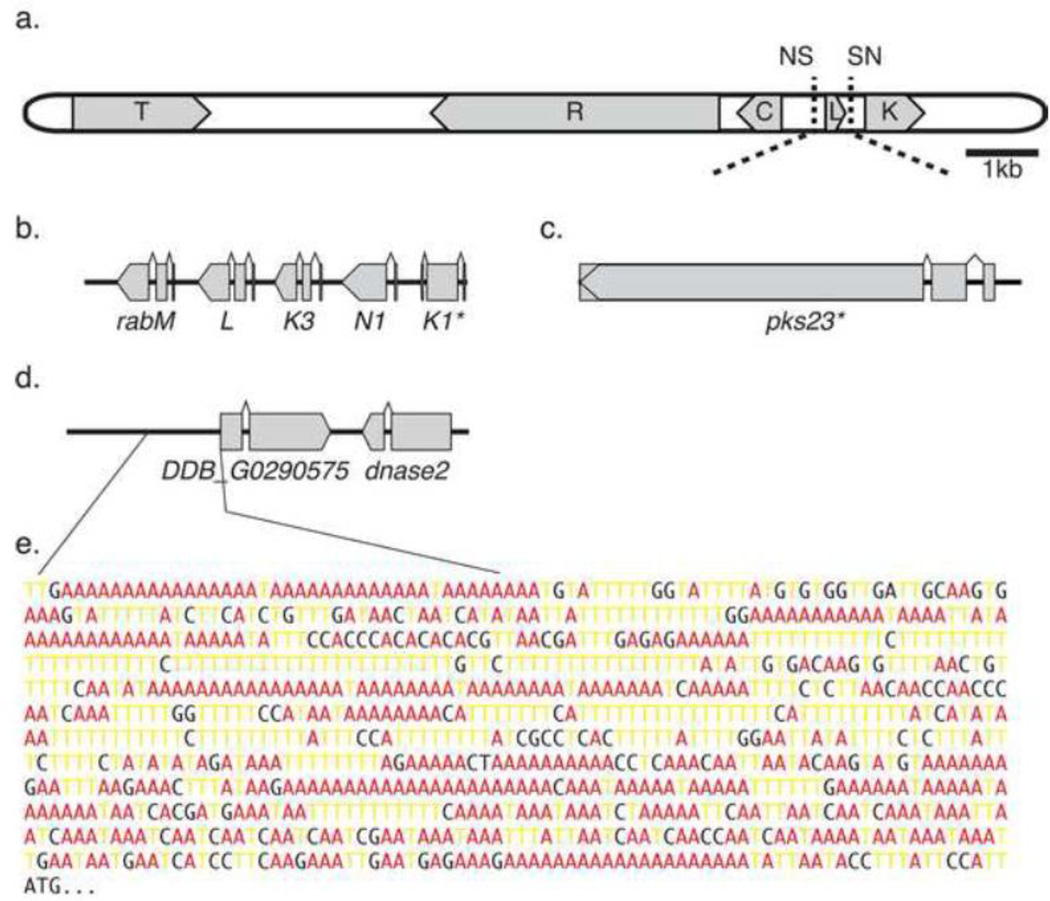
We have illustrated three clones that serve as interesting examples of genic content in the library (Figure 2). For example, clone F03 from the second round of ligation (Additional File 1) contains an insert that includes multiple genes of the rab GTPase family (Figure 2b), demonstrating the potential for identifying cloned gene clusters. Another clone from that set harbors a single nearly complete ORF of the gene polyketide synthase (pks) 23 (Figure 2c). The pks23 locus spans 7.8 kb in the genome, and this clone captured the 5’-most 5.7 kb, and 373 bases of the 5’ upstream region. In general, polyketide synthase genes are large and encode modular proteins involved in biosynthesis of small molecule natural products. Experimental manipulation of this fascinating gene family will benefit greatly from a platform capable of cloning entire pks ORFs. And lastly, clone F04 from the first ligation round (Additional File 1) encompasses two ORFs—dnase2 and the gene model DDB_G0290575, predicted to encode a LIM-family transcription factor (Figure 2d). This insert also contains a relatively large intergenic region 5’ of the DDB_G0290575 ORF.
Figure 2. Most genomic inserts contain genes.
We determined the gene content of each sequenced, mapped genomic DNA insert cloned in the pJAZZ-OK vector. (a) The vector, shown here to scale, is annotated with the following genes or features: T = teln, R = repA, C = cB, L = lacZ, K = kanR, N = NotI site, S = SmaI site. Coding sequences are represented by grey boxes, with the direction of transcription indicated by block arrows. Dotted lines indicate the location of genomic DNA insertion between the SmaI restriction sites. The black bar represents 1 kilobase (kb). Three representative clones are illustrated in (b – d). (b) Some inserts contained numerous open reading frames (ORFs). Clone F03 (of ligation 2) contains five genes from the rab GTPase family (rabK1, rabN1, rabK3, rabL and rabM), spanning positions 4,604,848 to 4,609,616 on chromosome 5. (c) Other inserts contained large genes, such as this nearly complete ORF for pks23, spanning bases 1,324,867 to 1,330,970 on chromosome 4. (d) Clone F04 (of ligation 1) harbors two genes—DDB_G0290575, a predicted LIM family transcription factor; and dnase2— as well as a sizeable intergenic region. (e) The region 1 kb upstream of DDB_G0290575 displays characteristically high AT content and long homopolymer runs. Bases A and T are colored red and yellow, respectively, for emphasis. (b – d) Exons are represented by grey boxes, with the direction of transcription indicated by block arrows. Asterisks indicate partially cloned genes with truncated exons. Thick black lines represent contiguous genomic DNA, while bent lines above the exons indicate splice junctions.
Regulatory elements such as promoters and Kozak motifs are typically found in intergenic segments, and thus can be difficult to clone. PCRs and rote molecular biology methods are made problematic by the extreme AT content found in these stretches of DNA (Figure 2e). The demonstrated ability to clone genes with their adjacent intergenic regions will facilitate studies in which native gene regulation, rather than constitutive expression, is desired. Intergenic regions are often targeted for cloning as “homology arms” for homologous recombination of knockout (KO) cassettes. Library clones could be engineered into KO constructs by inserting a selectable marker (such as Blasticidin S deaminase) into a clone containing the genomic locus of interest, obviating the need for recalcitrant PCRs. In order to be efficient, however, this approach would require an easy means of isolating the desired clone. While colony-lift hybridization is one reliable method for probing libraries, this approach could contribute far more labor cost than it saves. Instead, we propose to use lambda-red mediated recombineering to insert a selectable marker into the target clone within a library pool. This method has been demonstrated for libraries of other organisms[8], but remains to be optimized for the D. discoideum library. Alternatively, we have found that simply using library pool DNA as PCR templates often yields results where genomic DNA templates fail (not shown).
2.4 Dictyostelium discoideum genomic DNA cloned in the pJAZZ vector replicates stably in E. coli
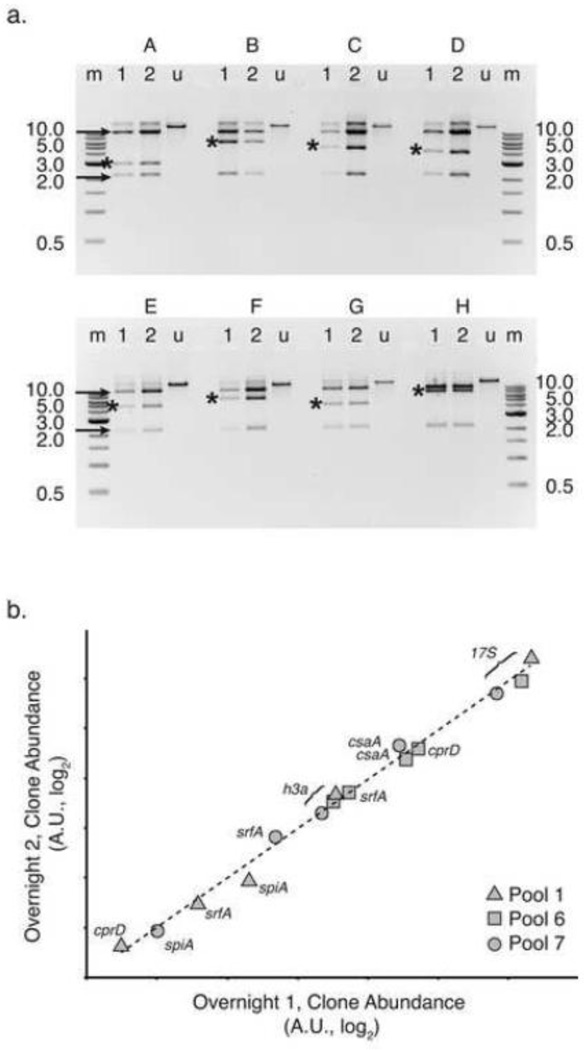
Historically, D. discoideum genomic DNA fragments in this size range have proven unstable when cloned in circular plasmids. Inserts suffer deletions or rearrangements, or fold into cruciform or other toxic structures[4]. We tested dozens of colonies to see whether the genomic DNA inserts were stably replicated and maintained. We inoculated rich media cultures and grew them shaking at 37°C, with added arabinose to induce increased vector copy-number. After 24 hours growth, a 1:1000 dilution of each was used to inoculate a second overnight culture. We harvested the first and second overnight cultures, isolated plasmid DNA, and digested the DNA with the restriction enzyme NotI to release the inserts. Eight representative clones are shown in Figure 3.
Figure 3. Cloned D. discoideum genomic DNA stably replicates in the library.
(a) We digested plasmid DNA purified from haphazardly selected library clones (A – H) with the restriction enzyme NotI. For each clone, plasmid DNA was sampled after growing overnight once (lanes “1”), or after a dilution was grown overnight for a second time (lanes “2”). Undigested plasmid DNA (lanes “u”) from the second overnight culture is shown as well. The 1-kilobase (kb) reference ladders (NEB, lanes “m”) are partially labeled with band sizes to the right and left of the image (0.5 – 10 kb). Black arrows indicate the long and short vector arms, while asterisks mark the insert genomic DNA fragment. The ratio of insert to vector band intensity, as determined by densitometry, is statistically indistinguishable between lanes 1 and 2 for each clone. DNA fragments were resolved by 0.8% agarose gel electrophoresis. Gels were stained with ethidium bromide and negative images are shown. (b) Using qPCR, we amplified marker genes from plasmid DNA isolated from entire library pools grown overnight for 1 or 2 serial dilutions. Cycle threshold (Ct) values were standardized to the kanamycin resistance marker aph, present on all plasmids in all pools. Relative gene abundance, estimated by log2 transformation of the standardized Ct value (2−ΔCt), is plotted for overnight growth 2 (y-axis) versus overnight growth 1 (x-axis). Both axes are log2 scale, with arbitrary units (A.U.). Targets from pool 1 are represented by triangles, pool 6 by squares and pool 7 by circles, as indicated in the legend. Genes h3a, srfA, and rRNA 17S were present in all three pools tested. Gene cprD was present in pools 1 and 6, while spiA was present in pool 1 and 7. The log-log best fit is shown as a dotted line (R2 = 0.99).
We expected to observe three bands by gel electrophoresis, corresponding to the long and short vector arms (10 kb and 2 kb, respectively), and the intact insert (see Figure 2a). The NotI recognition motif occurs only once among the six chromosomes and twice on the extrachromosomal ribosomal palindrome. Thus NotI should rarely cut within cloned genomic DNA. Virtually all clones tested yielded the expected three bands, with the inserts appearing as single, intact fragments (Figure 3a). Most importantly, each clone displayed a consistent digestion pattern for both overnight cultures, indicating that the inserts remain intact over many rounds of replication and cell division, despite the high cell density and the induced copy number. We also observed that the insert bands varied in size between clones, within the range of the genomic DNA fraction, consistent with the clonal diversity realized from end-sequencing. The inserts did not display a “ladder” pattern characteristic of historically deletion- or recombination-prone clones.
To further quantify insert stability, we measured the intensity of all the bands on the gel and calculated the ratio of insert to vector for each sample. We compared these ratios between the first and second overnight preparations for each clone. We found that the proportion of insert to vector varies less than 4% on average between lanes 1 and 2. This difference is statistically insignificant (Student’s paired t-test, p-value = 0.16), and thus we accept the null hypothesis that there is no difference in the quantity of insert relative to vector after multiple overnight dilutions of a given clone.
In addition to asking whether individual clones remain intact, we wished to assess the degree to which the complexity of the library is maintained during replication in E. coli. We used quantitative PCR (qPCR) to measure the relative abundance of various marker genes within entire pools (~7000 clones in each pool, on average) grown overnight in successive serial dilutions. First we identified several pools (numbered 1, 6 and 7) from which we could amplify up to 5 of the marker genes used to survey library diversity in Figure 1d, as well as the rRNA gene 17S (Additional File 1). We then performed qPCR using plasmid DNA isolated from the library pools after one overnight, and after a second outgrowth. Our results showed an extremely high correlation in relative gene abundance (log-log best fit R2=0.99) within a given pool between overnight outgrowths (Figure 3b). The preservation of relative abundance of these six genes, represented in at least two or three different pools, indicates that overall library complexity is robust to biological amplification.
With its demonstrated stability, and the depth and breadth of coverage, this library may prove a critical stepping-stone to a generalized method for haploid genetic complementation. Developing a widely applicable means of complementation has been a white whale of Dictyostelium genetics for generations of researchers[1,5]. Restriction Enzyme Mediated Integration (REMI), the standard for mutagenesis screens for the past two and a half decades, has successfully revealed much of what we know about the genetic basis for development, kin recognition, innate immunity, and other phenomena in D. discoideum[3,12–15]. However, chemical mutagenesis creates a very different spectrum of mutations and expands the scope of phenotypes we can screen. Parasexual genetic crosses can be made to identify complementation groups, but honing in on the actual mutation of interest remains a great challenge[16–18]. The current library is not suitable for complementation studies because it lacks a selectable marker for Dictyostelium transformation. Nevertheless, it demonstrates the feasibility of cloning and sets the stage for future experiments with a resistance-marked version.
3. MATERIALS AND METHODS
3.1 Isolation of high molecular weight genomic DNA by CsCl gradient
We grew Dictyostelium discoideum strain AX4 in 1 L of HL-5 medium to a density of 1×107 cells/mL. Cells were pelleted, washed in double distilled water, and resuspended in 50 mL nuclei buffer (40 mM Tris pH 7.8, 1.5% sucrose, 0.1 mM EDTA, 6 mM MgCl2, 40 mM KCl, 5 mM DTT, 0.4% NP40). After disrupting the plasma membrane by repeated pipetting, we collected the nuclei by centrifugation at ~8×g for 10 minutes. Nuclei were resuspended in 100 mM EDTA and disrupted by the addition of 1.5 volumes 10% sodium lauryl sarcosyl. An equal volume of 4 M ammonium acetate was added, and the debris pelleted by centrifugation (8×g, 15 minutes). The DNA was ethanol-precipitated from the supernatant, then dissolved in 4 mL 1× TE (10 mM Tris-HCl, 1 mM EDTA). Once the DNA re-entered solution, we added 4 g CsCl and 280 µL ethidium bromide (10 mg/mL). The CsCl gradient was formed by ultra-centrifugation at 65K RPM at 18°C for 4 hours. The genomic DNA band was isolated with an 18G1/2 needle under UV light. The DNA fraction was extracted with NaCl-saturated butanol, then ethanol-precipitated and washed with 70% ethanol. The resulting highly pure, high molecular weight genomic DNA pellet was dissolved in 1× TE, and aliquots stored at 4°C.
3.2 Partial restriction digestion and size selection of genomic DNA
We fragmented the genomic DNA by partial digest using the restriction enzymes RsaI and SspI (NEB, Ipswich, MA), which cut the genome every 700±10 base pairs (bp) and 450±45 bp, respectively. We reasoned that varying the enzyme concentrations and incubation times would yield a randomized pool of partially digested DNA fragments. Further, these enzymes create blunt ends ready for ligation without additional processing. Digestions were performed at 37°C using the following enzyme concentrations and sampling times: RsaI, high = 0.0125 Units/µL, sampled at 2, 4, 6, 8, and 10 minutes; RsaI low = 0.00625 U/µL, sampled at 3, 6, 9, and 12 minutes; SspI high = 0.025 U/µL, sampled at 3, 6, 9, and 12 minutes; SspI medium = 0.0125 U/µL, sampled at 5, 10, 15, and 20 minutes; and SspI low = 0.00625 U/µL, sampled at 10, 15, 20, and 25 minutes. Five micrograms of genomic DNA were digested in each enzyme dilution. For each sample aliquot, the reaction was stopped by the addition of EDTA to 20 mM followed by heat inactivation at 70°C for 10 minutes. All samples were pooled, ethanol precipitated, resuspended in 1× TE, and run on a 0.8% agarose gel for size fractionation. A gel slice containing the 4–10 kb fraction was excised and the DNA purified by Qiagen gel extraction kit according to the manufacturer’s recommended protocol.
3.3 Ligation, transformation and pooling of library clones
We preformed two rounds of ligation, each using an independent genomic DNA preparation. Each round included multiple ligation reactions, exhausting the available genomic DNA preparation. As per the Lucigen BigEasy Linear Cloning kit instructions, each reaction included: 1 µL CloneDirect 10× buffer and 1 µL CloneSmart DNA ligase (2 U/µL), 1.5 µL pJAZZ-OK vector arms (100 ng/µL), and 6.5 µL of the genomic DNA preparation. The ligation reactions were incubated at room temperature for 2 – 3 hours, heat inactivated at 70°C for 10 minutes, then transformed into electrocompetent TSA Big Easy E. coli, as per manufacturer’s instructions (Lucigen). Transformed cells were incubated in 1 mL recovery media (Lucigen) for 1 hour at 30°C, and plated on YT-agar plus 30 µg/mL kanamycin, 1 mM IPTG, and 20 µg/mL XGAL. The plates were incubated overnight at 30°C, then stored at 4°C awaiting colony picking or pooling.
3.4 Serial overnight cultures and NotI restriction fingerprinting
From each ligation round, 8–12 colonies were picked into test tubes with 2 mL Terrific Broth (TB) plus 30 µg/mL kanamycin and 0.01% arabinose. The cultures were grown shaking overnight at 37°C. After 24 hours, an aliquot of 2 µL was transferred from each tube to a new vessel with 2 mL of fresh media. The remaining volume was pelleted and stored at −20°C. The dilutions were grown for another 24 hours under identical conditions, then pelleted. Plasmid DNA was extracted from the two pellets by Qiagen miniprep kit, following the manufacturer’s recommended protocol. Approximately 200 ng of each plasmid was digested with 10 units NotI-high fidelity (NEB) for 4–16 hours at 37°C. Digestions were heat inactivated at 70°C for 10 minutes, then analyzed by 0.8% agarose gel electrophoresis.
3.5 End-sequencing and mapping of cloned inserts
From each ligation round, 96 colonies were picked into deep microtiter plates containing 500 µL TB + 30 µg/mL kanamycin per well. The colonies were grown overnight in shaking suspension at 30°C. They were then mixed with sterile glycerol (20% v/v final concentration) and stored at −80°C. Frozen stocks were processed for plasmid preparation and end sequencing with the Big Easy kit primers (SL1: 5'-CAGTCCAGTTACGCTGGAGTC, NZ-revC: 5'- AAATGGTCAGTTAATCAGTTCT).
Sequencing reads were mapped to the D. discoideum genome by BLAST[19]. Reads with ambiguous genomic coordinates were further examined manually. Reads were assigned to chromosomes 1 – 6, the extrachromsomal ribosomal palindrome, or one of the unassembled floating contigs. For the clones that mapped to the chromosomes, we used the end coordinates to estimate the inset size, and to determine which gene models were wholly or partially contained within these limits.
3.6 Simulations of chromosomal insert bias
We tested for bias in the chromosomal origin of the mapped inserts by computational simulation. We generated a sampling distribution by randomly selecting genomic positions at the frequency of chromosome occurrence observed in the library. We repeated this simulation 10,000 times and asked if the relative positional distribution of our inserts was different from the simulated distribution using the Kolmogorov-Smirnov test (R function ks.test)[20,21]. Similarly, to check for base composition bias, we randomly sampled genomic sequences of mean insert size found in the library. We repeated this simulation 10,000 times and compared GC content of the library inserts and the simulated fragments using the Kolmogorov-Smirnov test, as above.
3.7 PCR screen of pooled library DNA for genes of interest
DNA was purified from each of the 31 library pools by Qiagen miniprep kit using cells grown overnight in 2 mL Terrific Broth (TB) plus 30 µg/mL kanamycin. We combined and diluted aliquots of these DNA preparations into a single 1 ng/uL PCR template. PCRs were performed testing for various genes, using primers selected solely for their availability in lab (see Additional File 1 for primer information). A genomic DNA template control was performed in parallel. We varied PCR reaction conditions from gene to gene depending on the melting temperature of the primers and expected amplicon size, in accordance with standard PCR practices[22].
3.8 qPCR of pooled library DNA to measure relative gene abundance
Library pools were screened by PCR with primers for a battery of target genes (see Additional File 1) to identify pools containing numerous markers. PCRs were performed with standard methods, as above (section 3.7). Pools 1, 6, and 7 were selected for further analysis using plasmid DNA isolated from cultures grown in successive overnight dilutions, described in section 3.4.
qPCR was performed using the iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA) and an MJ Research DNA Engine Opticon 2 thermocycler. Each reaction was templated with 1 ng plasmid DNA, and SYBR incorporation measured over 40 thermal cycles. We included reactions to amplify a fragment of the kanamycin resistance marker from the pJAZZ vector backbone as an internal standard. Two technical replicates were amplified for each gene from each pool, and those cycle threshold (Ct) values were averaged. Target gene Ct values were standardized to their corresponding kanamycin Ct value, then log2 transformed (2−ΔCt) to provide an estimate of relative abundance of each gene, from each pool, from each overnight outgrowth.
4. AVAILABILITY
The physical library will be archived at the Dictyostelium stock repository (www.dictybase.org) for availability to the scientific community[23]. The archive will consist of glycerol stocks representing the 31 pools, as well as the 192 clones arrayed for end-sequencing.
Supplementary Material
Highlights.
The genome of Dictyostelium discoideum is very A:T-rich
Previous attempts to generate stable genomic libraries in E. coli have been rather unsuccessful
A library of D. discoideum genomic DNA is constructed in the pJAZZ vector
The library is stable over several rounds of amplification
ACKNOWLEDGEMENTS
We thank Adam Kuspa and Ron Godiska for conversations and advice regarding the library construction, and Balaji Santhanam for help implementing chromosomal distribution simulations and statistical tests. This work was supported by a grant from the National Institute of Child Health and Human Development (P01 HD39691). RDR was supported by a training fellowship from the Keck Center of the Gulf Coast Consortia, on the Training Program in Biomedical Informatics, National Library of Medicine (NLM, T15LM007093-21, PI - G. Anthony Gorry).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional file 1 is an excel workbook consisting of three sheets. Sheets 1 and 2 contain lists of the library clones that were end-sequenced and mapped to the chromosomes. These sheets describe the 96-well plate location of the clone, the genes included in the insert, the chromosomal coordinates of the insert, and whether the genes are completely or partially encompassed by the insert. Sheet 3 is a list of the PCR targets shown in figure 1d, enumerating the gene, gene product description, locus tag, NCBI ID, chromosome number, and forward and reverse primer sequences. Sheet 3 also includes PCR primers used for qPCR in figure 3b.
REFERENCES
- 1.Williams JG. Dictyostelium finds new roles to model. Genetics. 2010;185:717–726. doi: 10.1534/genetics.110.119297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faix J, Kreppel L, Shaulsky G, Schleicher M, Kimmel AR. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 2004;32:e143. doi: 10.1093/nar/gnh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichinger L, Pachebat JA, Glöckner G, Rajandream M-A, Sucgang R, Berriman M, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dynes JL, Firtel RA. Molecular complementation of a genetic marker in Dictyostelium using a genomic DNA library. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7966–7970. doi: 10.1073/pnas.86.20.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuspa A, Loomis WF. Ordered yeast artificial chromosome clones representing the Dictyostelium discoideum genome. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5562–5566. doi: 10.1073/pnas.93.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konfortov BA, Cohen HM, Bankier AT, Dear PH. A high-resolution HAPPY map of Dictyostelium discoideum chromosome 6. Genome Res. 2000;10:1737–1742. doi: 10.1101/gr.141700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfander C, Anar B, Schwach F, Otto TD, Brochet M, Volkmann K, et al. A scalable pipeline for highly effective genetic modification of a malaria parasite. Nat. Methods. 2011;8:1078–1082. doi: 10.1038/nmeth.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godiska R, Mead D, Dhodda V, Wu C, Hochstein R, Karsi A, et al. Linear plasmid vector for cloning of repetitive or unstable sequences in Escherichia coli. Nucleic Acids Res. 2010;38:e88. doi: 10.1093/nar/gkp1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fey P, Gaudet P, Curk T, Zupan B, Just EM, Basu S, et al. dictyBase--a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 2009;37:D515–D519. doi: 10.1093/nar/gkn844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sucgang R, Chen G, Liu W, Lindsay R, Lu J, Muzny D, et al. Sequence and structure of the extrachromosomal palindrome encoding the ribosomal RNA genes in Dictyostelium. Nucleic Acids Res. 2003;31:2361–2368. doi: 10.1093/nar/gkg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dynes JL, Clark AM, Shaulsky G, Kuspa A, Loomis WF, Firtel RA. LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 1994;8:948–958. doi: 10.1101/gad.8.8.948. [DOI] [PubMed] [Google Scholar]
- 13.Artemenko Y, Swaney KF, Devreotes PN. Assessment of development and chemotaxis in Dictyostelium discoideum mutants. Methods Mol. Biol. Clifton NJ. 2011;769:287–309. doi: 10.1007/978-1-61779-207-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuspa A. Restriction enzyme-mediated integration (REMI) mutagenesis. Methods Mol. Biol. Clifton NJ. 2006;346:201–209. doi: 10.1385/1-59745-144-4:201. [DOI] [PubMed] [Google Scholar]
- 15.Santorelli LA, Thompson CRL, Villegas E, Svetz J, Dinh C, Parikh A, et al. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature. 2008;451:1107–1110. doi: 10.1038/nature06558. [DOI] [PubMed] [Google Scholar]
- 16.Katz ER, Sussman M. Parasexual recombination in Dictyostelium discoideum: selection of stable diploid heterozygotes and stable haploid segregants (clones-temperature sensitive-ploidy-fruiting bodies-spore-slime mold) Proc. Natl. Acad. Sci. U. S. A. 1972;69:495–498. doi: 10.1073/pnas.69.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrissey JH, Loomis WF. Parasexual genetic analysis of cell proportioning mutants of Dictyostelium discoideum . Genetics. 1981;99:183–196. doi: 10.1093/genetics/99.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King J, Insall RH. Parasexual genetics of Dictyostelium gene disruptions: identification of a ras pathway using diploids. BMC Genet. 2003;4:12. doi: 10.1186/1471-2156-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Marsaglia G, Tsang WW, Wang J. Evaluating Kolmogorov’s Distribution. J. Stat. Softw. 2003;8:1–4. [Google Scholar]
- 21.Carvalho L. An improved evaluation of Kolmogorov’s distribution. (Submitted.). [Google Scholar]
- 22.New England Biolabs. Guidelines for PCR optimization with thermophilic DNA polymerases. (n.d.). https://www.neb.com/tools-and-resources/usageguidelines/ guidelines-for-pcr-optimization-with-thermophilic-dna-polymerases. [Google Scholar]
- 23.Fey P, Dodson RJ, Basu S, Chisholm RL. One Stop Shop for Everything Dictyostelium: dictyBase and the Dicty Stock Center. In: Eichinger L, Rivero F, editors. Dictyostelium Discoideum Protoc. Humana Press; 2013. [accessed March 18, 2015]. http://link.springer.com/protocol/10.1007%2F978-1-62703-302-2_4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.