Abstract
It is clear that IL-10 plays an essential role in maintaining homeostasis in the gut in response to the microbiome. However, it is unknown whether IL-10 also facilitates immune homeostasis at distal sites. To address this question, we asked whether splenic immune populations were altered in IL-10-deficient (Il10−/−) mice in which differences in animal husbandry history were associated with susceptibility to spontaneous enterocolitis that is microbiome-dependent. The susceptible mice exhibited a significant increase in splenic macrophages, neutrophils and marginal zone (MZ) B cells that was inhibited by IL-10 signaling in myeloid, but not B cells. The increase in macrophages was due to increased proliferation that correlated with a subsequent enhancement in MZ B cell differentiation. Cohousing and antibiotic treatment studies suggested that the alteration in immune homeostasis in the spleen was microbiome-dependent. 16S rRNA sequencing revealed that susceptible mice harbored a different microbiome with a significant increase in the abundance of the bacterial genus Helicobacter. The introduction of H. hepaticus to the gut of nonsusceptible mice was sufficient to drive macrophage expansion and MZ B cell development. Given that myeloid cells and MZ B cells are part of the first line of defense against blood-borne-pathogens, their increase following a breach in the gut epithelial barrier would be protective. Thus IL-10 is an essential gatekeeper that maintains immune homeostasis at distal sites that can become functionally imbalanced upon the introduction of specific pathogenic bacteria to the intestinal track.
Introduction
Interleukin (IL)-10 is an important anti-inflammatory cytokine produced in the gut by a variety of immune cells including B cells, T cells and macrophages, as well as nonhematopoietic cells (1). Genome-wide association studies revealed a linkage between IL-10 polymorphisms and susceptibility to inflammatory bowel disease in humans (2, 3). Humans harboring loss of function mutations in IL10RA and IL10RB, whose gene products encode for the heterodimer IL-10 receptor (R) 1 and IL-10R2 subunits, respectively, succumb to early-onset colitis (4–6). In mice, spontaneous onset of severe intestinal inflammation also occurs in mice deficient in IL-10 or its receptor (7, 8), and is prevented by IL-10 signaling in macrophages (9, 10). Collectively, these studies illuminate the essential role for IL-10 in dampening intestinal inflammation and the maintenance of gut homeostasis.
It has become clear that the gut microbiome plays an essential role in the health of the host and that microbial dysbiosis can contribute to disease (11). For example, increases in Enterobacteriaceae and gram negative anaerobes, including Bacteroides, have been associated with inflammatory bowel disease (IBD) in humans (12–14). A decrease in presumed beneficial bacteria, such as Bifidobacteria species, has also been reported in IBD (14, 15). In mice, spontaneous enterocolitis in Il10−/− mice only occurs in some animal colonies in a microbiome-dependent manner (7, 16, 17). Mice lacking Helicobacter do not succumb to spontaneous colitis in the absence of IL-10 (18). In addition, H. hepaticus-induced colitis susceptibility has been shown to differ between mouse colonies with markedly different microbiomes (16). While the gut microbiome can influence immune responses at distal sites (11, 19), the role of specific bacterial species in systematic inflammation remains only partially understood.
The Il10−/− mouse has not been reported to have defects in hematopoietic lineage development, but at eight weeks an elevation in IgM, IgG1, IgG2b IgG2a and IgA was reported (7). Whether this is due to altered B cell function is not known and a comprehensive analysis of B cell subsets has not been performed in the Il10−/− mouse. While T-dependent immune responses in Il10−/− mice were reported to be normal (7), the slight elevation in IgM is suggestive that T-independent immune responses may be altered. T-independent antibody responses are in part mediated by marginal zone (MZ) B cells that respond very rapidly to blood-borne pathogens due to their localization within the splenic MZ (20). The MZ is situated between the red and white pulp through which most of the blood that enters the spleen flows through before reentering the circulation (20). Thus a breach in the epithelial layer during gut inflammation allowing bacterial translocation would potentially prompt a MZ B cell response. In Il10−/− mice, bacteria were found to penetrate the colon mucus layer and directly contact the epithelial barrier and Enterococcus faecalis, through the production of a metalloprotease, was shown to compromise the barrier (21, 22). These effects on the barrier are thought to contribute to inflammation within the gut, but whether they alter immune cell function and homeostasis at distal sites is unknown.
In this study, we asked whether Il10−/− mice with susceptibility to spontaneous enterocolitis had an imbalance in immune cell homeostasis outside of the gut compared to resistant mice raised in a separate facility. We found that susceptible mice exhibited a significant increase in both MZ B cells and CD11b+ myeloid cells in the spleen that was inhibited by IL-10 signaling in macrophages. Cohousing, antibiotic treatment and 16S rRNA sequencing studies indicated that the immune homeostasis alteration was microbiome-dependent. In addition, introduction of H. hepaticus to nonsusceptible mice was sufficient to drive MZ B cell differentiation and macrophage expansion. These results indicate that introduction of a single bacterial species can produce dysbiosis in the gut and drive a functional imbalance in immune homeostasis in the spleen when the gatekeeper function of IL-10 is compromised.
Materials and Methods
Mice
C57BL/6J and B10.PL (H-2u) WT mice, and B6.129P2-Il10tm1Cgn/J mice (Il10−/−) were purchased from The Jackson Laboratory (Jax) (Bar Harbor, ME). IL-10Rαfl/flCD19Cre+ and IL-10Rαfl/flCD19Cre− mice were provided by Dr. Terrence Geiger, St. Jude’s Children’s Research Hospital, Memphis, TN (23). IL-10Rαfl/flLysMCre+ and IL-10Rαfl/flLysMCre− mice were provided by Dr. Werner Muller, University of Manchester, Manchester, United Kingdom (24). B6.129P2-Il10tm1Cgn/J mice were backcrossed to B10.PL for three generations and then intercrossed to generate B10.PL H-2uxu Il10+/+, Il10+/− and Il10−/− littermates. All animals were housed and/or bred at the Translational Biomedical Research Center of the Medical College of Wisconsin (MCW). All animal protocols were approved by the MCW Institutional Animal Care and Use Committee. At the initiation of all experiments, including cohousing, mice were between 6–8 weeks of age.
Antibodies and Other Reagents
The 2.4G2 antibody was produced locally. Mouse specific CD45R-PE-Texas Red, CD45R-PE, CD5-APC, CD86-V450, Ki-67-FITC, Caspase 3-FITC and CD40 antibodies were purchased from BD Biosciences (San Diego, CA). Mouse specific CD21-eFluor 450, CD23-PE-Cy7, CD23-FITC, CD1d-PE, CD93-biotin, CD93-APC, CD93-PE, TCR-β-FITC, TCR-β-PE, CD4-biotin, CD4-FITC, CD4-APC-eFluor 780, CD8-PE-Cy7, CD11b-biotin, CD11b-eFluor 450 and Foxp3-PE antibodies were purchased from eBioscience (San Diego, CA). Mouse specific CD11b-Alexa Fluor 488, CD45R-Alexa Fluor 594, CD80-PE-Cy5, CD40-Alexa Fluor 647, MHC class II-PE-Cy7, Ly6C-APC, Ly6G-APC-Cy7, Ly6G-Alexa Fluor 647, F4/80-PE-Cy7, CD138-APC, IgM-APC-Cy7, IgD-Pacific Blue, Notch 2-PE, Delta-like 1-Alexa Fluor 647 antibodies and the LEGENDplex multi-analyte flow assay kit were purchased from Biolegend (San Diego, CA). Mouse specific Marco-FITC and MOMA-FITC antibodies were purchased from AbD Serotec (Raleigh, NC). Anti-Bc1-2 and anti-Bcl-xL were purchased from Cell Signaling Technology (Danvers, MA). Anti-mouse IgM-FITC was purchased from SouthernBiotech (Birmingham, AL). Anti-mouse IgM F(ab′)2 was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Streptavidin-PE-Cy5.5 was purchased from eBioscience (San Diego, CA). Anti-BrdU-APC was purchased from BD Biosciences (San Diego, CA). CFSE and DAPI were purchased from Molecular Probes (Eugene, OR). LPS was obtained from Sigma-Aldrich (St. Louis, MO) and CpG from Invivogen (San Diego, CA). Ampicillin and neomycin were purchased from LKT Laboratories, Inc. (St. Paul, MN), and metronidazole and vancomycin were obtained from Sigma-Aldrich (St. Louis, MO).
Cell Isolation, Flow Cytometry and Cell Sorting
Single cell suspensions were prepared from bone marrow, thymus, Peyer’s patches, inguinal lymph nodes and spleens. Peritoneal cavity cells were isolated as previously described (25). 1 × 106 cells were incubated with anti-CD16/CD32 (Fc block) (clone 2.4G2) for 15 min followed by cell surface staining with specific mAb. Intracellular Ki-67 was performed using the anti-mouse/rat Foxp3 staining buffer set from eBioscience (San Diego, CA). Cells were acquired on a LSRII flow cytometer (BD Biosciences) and data were analyzed using FlowJo software (Tree Star, Inc. Ashland, OR). Splenic B cell subsets were characterized as described (26). For in vitro culture and real-time PCR, B cell subsets were sorted using a FACSAria cell-sorter (BD Biosciences) as described (27).
Immunohistology
Spleens from eight week old mice were embedded in Tissue-Tek OCT compound (Sakura Finetek, Torrance, CA) and snap frozen. Seven μm sections were stained with B220-PE and MOMA-FITC and images were captured using a Nikon Eclipse TE200 inverted fluorescent microscope as described (28). Sections stained with B220-Alexa Fluor 594, CD11b-Alexa Fluor 488 and LysG-Alexa Flour 647 were imaged by confocal microscopy with an Olympus Fluoview FV1000 MPE Multiphoton Scanning Microscope.
Detection of chemokines and cytokines in serum and colon tissue
Serum and colon tissue were collected from seven-eight week old naïve mice. Colons were homogenized in PBS containing 0.1% IGEPAL CA-630 (Sigma-Aldrich, St. Louis, MO) and mini protease inhibitor (Roche, Indianapolis, IN) using the gentleMACS Dissociator (Miltenyi Biotec, San Diego, CA). Cytokine and chemokines in serum and colon lysates were determined using LEGENDplex multi-analyte flow cytometry assay kit (Biolegend, San Diego, CA).
Immunization and Detection of Serum Immunoglobulins
Mice were immunized with 30 μg NP-ficoll i.p. and sera were collected 7 and 14 days later. Sera from eight week unimmunized mice were used for detecting resting IgM and IgG3 levels. ELISA was performed to determine NP-specific Ig titers and total resting IgM and IgG3 levels in the serum as described previously (28, 29).
Helicobacter hepaticus Culture
Culture of the MU-94 Helicobacter hepaticus strain utilized in these studies was performed at the University of Missouri in the laboratory of Craig L. Franklin (30, 31). The culture suspension was pipetted into 15 ml conical tubes, wrapped in bubble wrap and placed in a foam shipping container lined with ice packs. A previous experiment demonstrated that H. hepaticus was viable in these conditions for approximately 16 hr (data not shown). Cultures were shipped by overnight courier to ensure arrival in Milwaukee, WI within 16 hr and gavaged immediately upon receipt.
Cohousing and Helicobacter hepaticus Gavage
Il10−/− Jax, or IL-10Rαfl/flCD19Cre+, or IL-10Rαfl/flLysMCre+ mice were cohoused with Il10−/− mice from our colony (B10.PL background) in a 1:1 ratio for four weeks for feces-mediated transfer of the microbiota. Il10−/− Jax mice were orally gavaged with ~5 × 1010 CFU of Helicobacter hepaticus suspended in brucella broth. Control mice received sterile broth only. Cellular analysis of splenic B cell subsets and CD11b+ myeloid cells were performed four weeks after cohousing or bacterial gavage.
Antibiotic Treatment
The following antibiotics were provided ad libitum in drinking water for four weeks: ampicillin trihydrate (1.0 g/L), vancomycin hydrochloride (0.5 g/L), neomycin sulfate (1.0 g/L), and metronidazole (1.0 g/L). This combined antibiotic regimen provides effective broad-spectrum coverage against gram-positive and -negative bacteria (32).
BrdU Incorporation Assay
Mice were i.p. injected with 1 mg BrdU, twice daily, for three days. BrdU+ B cells were tracked 1, 30, and 45 days after the last BrdU injection using the APC BrdU flow kit (BD Biosciences, San Diego, CA). For the B cell turnover assay, mice were provided with BrdU in the drinking water (0.8 mg/ml) for nine days, and the next day BrdU+ B cells were detected using flow cytometry (33, 34).
In Vitro Survival of B Cells
FACS purified B cells (1 × 105) were cultured in vitro in the presence of LPS (1 and 10 μg/ml), CpG (0.1 and 1 μg/ml), goat anti-mouse IgM F(ab′)2 (0.5 and 5 μg/ml) or anti-CD40 (0.1 and 1 μg/ml) for 72 h and cell survival was determined by flow cytometry gating on live DAPI− cells.
RNA Isolation and Real-Time PCR
Total RNA was isolated from FACS purified cells using the RNAqueous -4PCR kit (Ambion, Austin, TX) and first-strand cDNA was synthesized using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA). The quality of the cDNA was assessed by Gapdh PCR. The expression of Gapdh, Hes1, Dtx1, and Rbpj was quantified by real-time PCR using TaqMan primers and probes from Invitrogen and an ABI Prism 7500 PCR machine (Applied Biosystems, Foster City, CA). The amplification cycle was as follows: 10 min hold at 95 °C, and then 40 cycles of 15 s at 95 °C and 60 s at 60 °C. The expression of the Hes1, Dtx1, and Rbpj genes was normalized to the housekeeping gene Gapdh by calculating the ΔCT. The relative expression of each gene is represented as 2(−ΔCT).
Isolation of Microbial DNA, PCR Amplification and Sequencing
Small and large intestines and cecum were isolated from Il10−/− Jax mice that were or were not cohoused with Il10−/− mice from our colony as described (35). The isolated tissues were homogenized and bacterial DNA was isolated using MO BIO Power Analyzer kit (MO BIO Laboratories, Inc., Carlsbad, CA) and PCR was used with unique bar coded primers to amplify the V4 region of the 16S rRNA gene to create an “amplicon library” from individual samples (36, 37). The 5′ primer was: 5′AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCAGCMGCCGGGTAA3′ and the 3′ primer was: 5′CAAGAGAAGACGGCATACGAGATNNNNNNAGTCAGTCAGCCGGACTACHVGGGWTCTAAT3′. The individual PCR reactions were set up as follows: 10 μL of 5x Reaction Buffer, 1.5 μL of dNTPs, 2 μL of the 5′ primer, 2 μl of the 3′ primer, 1.5 μl of the “LongAmp” enzyme kit from New England Biolabs (Ipswich, MA), 30 μL of DNA with the concentration of DNA at 2–5 ng/μl and 3 μl of H2O as described (37). The amplification cycle was as follows: denature at 94 °C for 1 min followed by 32 cycles of 94 °C for 30 s, 50 °C for 1 min, 65 °C for 1 min with a final extension of 65 °C for min. The PCR products were sequenced using NextGen sequencing Illumina GAIIx platform (36). The samples were first quantitated using Pico Green, adjusted to a concentration of 4 nM then used for sequencing (37). Fastq conversion of the raw data files was performed following demultiplexing. Overall reads quality was assessed using the program FASTQC and quality filtering of fastq files was performed using the FASTX toolkit.
Bioinformatics
A total of 11,321,588 sequencing reads of 101 bp length were generated from 12 animals (36 samples total) using Illumina GAIIx sequencing technology. Quality filtering of raw data was performed in two steps: 1) due to low quality of bases toward the 3′ ends of the read, 11 bases were trimmed from the 3′ end for all reads and 2) only high quality reads having >90% bases with a Q Score >30 are retained. The quality filtering step retained 78% of total reads (8,928,469) with average of 248013 reads per sample (range: 127940 – 484143 reads per sample). We clustered those sequences using AbundantOTU+ v.0.93b with the “-abundantonly” option at 97% similarity and generated 824 Operational Taxonomic Units (OTUs) incorporating 99.38% of the total input sequences. We utilized USEARCH (38) (v. 6.1.357) and the Gold reference database to screen for the presence of chimeras in our OTU sequences and a total of 135 OTUs were removed. The remaining 689 OTUs (representing 98.73% of the input sequences) were used for downstream analysis. We assigned taxonomic ranks through the Quantitative Insight into Microbial Ecology (QIIME) (39) (v. 1.8.0) using both uclust consensus taxon assigner and the RDP (40) classifier (v. 2.2) (confidence set to 80%). OTU consensus sequences were then aligned to the GreenGenes core reference alignment (41) using PyNAST (42). We built a phylogenetic tree from those alignments using FastTree (43) (v. 2.1.3) as implemented in the QIIME pipeline (v. 1.8.0). We generated BIOM files form our OTU counts and fed those along with the phylogenetic tree to Phyloseq (44) (v. 1.7.24). Raw OTU counts were normalized and log10 transformed according to the following formula:
The normalized and log10 transformed reads were used in Phyloseq to produce PCoA plots from Bray-Curtis and UniFrac (weighted and un-weighted) (45). Alpha diversity measures (Chao1 richness estimate and Shannon diversity index) were also calculated using Phyloseq after rarefying the raw counts to an even depth of 97,988 reads (the minimum count in all samples). We used a mixed linear model utilizing SAS (v. 9.3) software to analyze the data and accounting for possible effects that may arise from co-housing the mice in the same cage (46).
Statistical Analysis
Data were analyzed using GraphPad prism software and were presented as mean ± SE. Statistical significance was determined using the non-parametric Mann Whitney test or unpaired t-test. p-values <0.05 were considered significant. Our mixed linear model, in which origin and tissue is a fixed effect and cage is a random effect (46), takes the form of:
where Yijkl represents either PCoA axis value OTU log normalized count Shannon or Chao1 value for Origin i, Tissue j, Cage k and replicate l. Oi. is the effect of the ith origin, were origin is set to either Jackson lab animals, Jackson lab animals cohoused with our animals or our Il10−/− animals. Tj is effect from the jth tissue (cecum, small intestine or large intestine). (OT)ij is the interaction effect between Treatment i and tissue j. Ck(i) is the effect from the kth cage that is nested within the ith origin and εijklεijkl denotes the error associated with measuring YijklYijkl. We controlled for the false discovery rate (FDR) by correcting the p-values using Benjamini and Hochberg (BH) approach (47).
Results
MZ B Cells and CD11b+ Myeloid Cells and the T-Independent Immune Response are Significantly Increased in the Spleens of Il10−/− Mice
To determine whether the gut microbiota alters distal immune cell homeostasis in mice susceptible to colitis, we quantitated immune cell populations in Il10−/− mice. For this analysis, we used Il10−/− mice acquired from Jackson Laboratories (Jax) in the year 2000 that were crossed to the B10.PL background (H-2u) and maintained in our colony by heterozygous crossings under SPF conditions. Since our Il10−/− mice are susceptible to spontaneous enterocolitis, we utilized mice at 6–8 weeks of age prior to enterocolitis onset and evidence of T cell activation in the gut (data not shown). First, we compared the total cellularity of Il10+/+ (WT) and Il10−/− littermates and found no difference in the spleen (Fig. 1A), lymph node, thymus, Peyer’s patch or bone marrow (Suppl. Fig. 1A). We next performed a comprehensive immune phenotyping to determine whether the homeostasis of specific immune cell populations was altered in Il10−/− mice. In the spleen (Fig. 1B) and lymph node (Suppl. Fig. 1b), we found that percentage of αβ T cells were significantly reduced, while no difference was observed in the Peyer’s patch or peripheral blood (Suppl. Fig. 1B). We also observed a significant reduction in the absolute number of αβ T cells in the spleen (Fig. 1B). Overall, we did not observe a difference in the ratio of CD4:CD8 T cells in the spleen (Fig. 1C) or in the lymph node, thymus or peripheral blood (Suppl. Fig. 1C, D). For B cells, while the percentage of total B cells was significantly decreased in the spleen of Il10−/− mice the reduction in absolute number did not reach significance (Fig. 1D). Interestingly, the percentage of B cells was significantly increased in the lymph node and decreased in the peripheral blood, while no change was observed in the Peyer’s patch or bone marrow (Suppl. Fig. 1E).
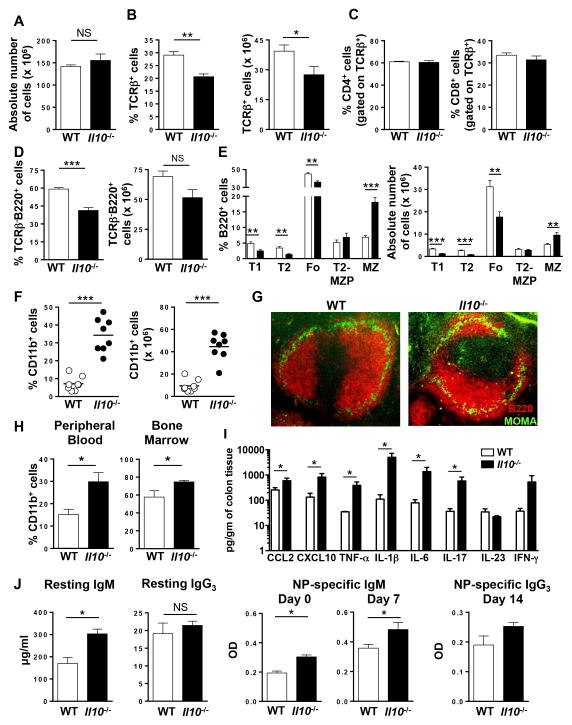
Figure 1. Increased MZ B and CD11b+ cells in the spleen of Il10−/− mice.
(A) The absolute number of cells in the spleen of WT and Il10−/− mice was determined. Data are shown as the mean ± SE of three experiments (n = 8). (B) The percentage (left panel) and absolute number (right panel) of splenic TCRβ+ T cells in WT and Il10−/− mice was determined by flow cytometry. Data are shown as the mean ± SE of three experiments (n = 8). (C) The percentage of TCRβ+CD4+ (left panel) and TCRβ+CD8+ (right panel) T cells in the spleen of WT and Il10−/− mice was determined by flow cytometry. Data are shown as the mean ± SE of two experiments (n = 6). (D) The percentage (left panel) and absolute number (right panel) of splenic TCRβ−B220+ B cells in WT and Il10−/− mice was determined by flow cytometry. Data are shown as the mean ± SE of three experiments (n = 8). (E) The percentage (left panel) and absolute number (right panel) of splenic T1, T2, Fo, T2-MZP and MZ B cell subsets from WT and Il10−/− mice was determined by flow cytometry. Data are shown as the mean ± SE of three experiments (n = 8). (F) Flow cytometric analysis showing the percentage (left panel) and absolute number (right panel) of splenic CD11b+ cells from WT and Il10−/− mice. Each data point represents a single mouse from three independent experiments. (G) Immunofluorescence analysis of spleen sections from WT and Il10−/− mice stained with B220-PE (red) and MOMA-1-FITC (green) were performed. Representative images from one of three mice are shown (200x). (H) The percentage of CD11b+ cells in the peripheral blood (left panel) and bone marrow (right panel) in WT and Il10−/− mice was determined by flow cytometry. Data are shown as the mean ± SE (n = 3). (I) Cytokines and chemokines in the colon of naïve WT and Il10−/− mice at 7–8 weeks of age are shown (n = 4). (J) Resting serum IgM and IgG3 levels in naïve WT and Il10−/− mice were measured by ELISA (n = 6–7) (left panels). NP-specific IgM (day 0 and 7) (middle panels) and IgG3 (day 14) (right panel) serum titers in NP-ficoll immunized WT and Il10−/− mice was determined by ELISA. Data are shown as the mean ± SE (n = 3–4). *p<0.05, **p<0.01, ***p<0.001, NS = not significant.
We then asked whether the homeostasis of specific B cell subsets was altered in Il10−/− mice. Using a gating strategy to identify peritoneal cavity B cell subsets, there was no difference in the absolute number of B2, B1, B1a or B1b cells (Suppl. Fig. 2A). When we gated on splenic B cell subsets (26) (Suppl. Fig. 2B), we found that both the percentage and absolute number of the transitional (T) 1 and T2 precursors and the mature follicular (Fo) populations were significantly decreased in Il10−/− mice (Fig. 1E). There was no difference in the percentage or absolute number of T2-MZ precursors (P), the immediate precursor to MZ B cells (Fig. 1E). However, there was a significant increase in MZ B cells (Fig. 1E), which was confirmed using an alternative gating strategy (Suppl. Fig. 2C) (48, 49). Because MZ B cell retention is regulated in part by macrophages and perhaps neutrophils (50, 51), we quantitated splenic CD11b+ myeloid cells and found that both their percentage and absolute number were significantly increased in Il10−/− mice (Fig. 1F). We next used immunofluorescence to determine whether IL-10 deficiency altered the splenic architecture. By detecting B220+ B cells and MOMA-1+ macrophages to demarcate the MZ, we found an evident increase in the size of the MZ in Il10−/− mice, but no obvious difference in the splenic architecture (Fig. 1G). In measuring CD11b+ myeloid cells at other anatomical sites, we found a similar increase in their percentage in the peripheral blood and bone marrow (Fig. 1H), but not the lymph node (data not shown). These data indicate that the loss of IL-10 has organ specific effects on immune cell homeostasis.
We next determined whether the increase in myeloid cells and MZ B cells was associated with signs of inflammation. To measure local inflammation in the gut, we quantitated the protein expression level of two chemokines and six pro-inflammatory cytokines known to be associated with intestinal inflammation in the colon. As compared to WT mice, Il10−/− mice had significantly increased levels of the chemokines CCL2 and CXCL10 and the cytokines TNF-α, IL-1β, IL-6 and IL-17, but not IL-23 or IFN-γ (Fig. 1I). These data indicate that local inflammation is occurring in Il10−/− mice in our animal colony, which is consistent with their eventual development of spontaneous entercolitis. To determine whether inflammation was detectable systemically, we measured the same panel of chemokines and cytokines in the serum and found no difference between the two cohorts of mice (Table 1). These data suggest that the increase in MZ B cells and myeloid cells in Il10−/− mice is not due to a systemic immune response, but stems from inflammation in the colon.
Table I.
Chemokine and cytokine levels in serum of naïve WT and Il10−/− micea
| IL-23 (pg/ml) | IFN-γ (pg/ml) | TNF-α (pg/ml) | CCL2 (pg/ml) | IL-1β (pg/ml) | IP-10 (pg/ml) | IL-6 (pg/ml) | IL-17 (pg/ml) | |
|---|---|---|---|---|---|---|---|---|
| WT | ||||||||
| 1 | nd | nd | nd | 12.8 | nd | 9.8 | 19.4 | 9.8 |
| 2 | nd | nd | nd | 52.15 | nd | 5.67 | 193.4 | nd |
| 3 | nd | 8.7 | 6.7 | 294.6 | 8.27 | 17.14 | 655.3 | 6.5 |
| 4 | nd | 9.4 | 11.7 | 17.3 | nd | 9.94 | 21.7 | 29.5 |
|
| ||||||||
| Mean ± SE | na | 9.0 ± 0.4 | 9.2 ± 2.5 | 94.2 ± 67.4 | na | 10.6 ± 2.4 | 222.4 ± 149.9 | 15.3 ± 7.2 |
|
| ||||||||
| Il10−/− | ||||||||
| 1 | 6.1 | 49.68 | 15.5 | 15.4 | 16.45 | 33.77 | 257.5 | 52.9 |
| 2 | 6.2 | 25.2 | 17.1 | 19.6 | 21.79 | 21.81 | 61.8 | 20.4 |
| 3 | nd | 39.2 | 11.4 | 13.0 | nd | 25.7 | 253.2 | 31.4 |
| 4 | nd | nd | nd | 630 | nd | 55.4 | 330.4 | nd |
|
| ||||||||
| Mean ± SE | 6.2 ± 0.03 | 38.1 ± 7.1 | 14.6 ± 1.7 | 169.5 ± 153.5 | 19.1 ± 2.7 | 34.2 ± 7.5 | 225.7 ± 57.4 | 34.9 ± 9.5 |
Mice aged 7–8 weeks.
nd: not detected; na: not applicable
We next examined the immunological consequence of an increase in MZ B cells and found that resting levels of IgM, but not IgG3, were significantly elevated in Il10−/− mice (Fig. 1J). To determine whether the increase in MZ B cells impacted the amplitude of the humoral immune response, we immunized with the T-independent antigen NP-ficoll. On day 0, there was a significant increase in NP-specific IgM in Il10−/− mice that was retained on day 7 (Fig. 1J). NP-specific IgG3 was not detectable on day 0 (data not shown) and there was no difference in its generation on day 14 in WT versus Il10−/− mice (Fig. 1J). The increase in NP-specific IgM was not accompanied by an increase in the percentage of plasma cells in the spleen (Suppl. Fig. 2D). However, there was an increase in the absolute number of plasma cells in the spleen of Il10−/− mice (Suppl. Fig. 2D). The increase in IgM was not likely due to B1 cells (52) because we found no difference in the absolute number of peritoneal cavity B cell subsets (Suppl. Fig. 2A). Thus the increase in resting levels of total IgM in Il10−/− mice is likely due to increased numbers of MZ B cells contributing to the IgM pool. Following immunization, the increase in NP-specific IgM is consistent with an increase in the absolute number of plasma cells.
Defects in MZ B Cell and Myeloid Cell Homeostasis is Dependent upon the Housing Environment of the Mice
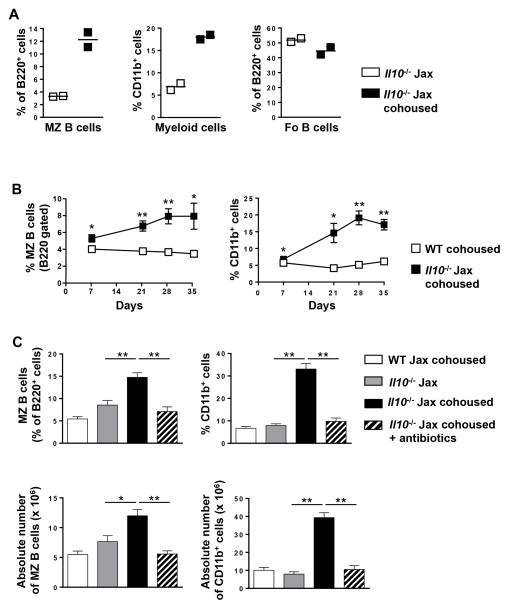
To determine whether the alterations in B cell and myeloid homeostasis in Il10−/− mice was unique to our colony, we performed the same splenic phenotypic analysis on mice purchased directly from Jax and found no significant difference in the absolute number of T cells, B cell subsets and myeloid cells between WT Jax and Il10−/− Jax mice (data not shown). The contrast between mice housed long-term in our colony and mice acquired directly from Jax supports the hypothesis that the microbiome acquired after housing in isolated animal colonies can specifically alter immune cell homeostasis. To evaluate this hypothesis, we determined whether Il10−/− Jax mice cohoused with Il10−/− mice from our colony would acquire the immune homeostasis defect. We found that four weeks of cohousing was sufficient to induce an increase in both MZ B cells and CD11b+ myeloid cells, but not Fo B cells (Fig. 2A). Kinetically, the increase in the percentage of MZ B cells and CD11b+ myeloid cells was evident as early as one week following cohousing, which peaked at four weeks (Fig. 2B). In contrast, the percentage of Fo B cells within the total B cell population did not change (data not shown). To further demonstrate that the acquisition of an imbalance in immune homeostasis was driven by the microbiome, we included a treatment group whereby Il10−/− Jax cohoused mice were provided a combination of non-absorbable broad-spectrum antibiotics in the drinking water (32). The inclusion of antibiotics from the outset of the experiment prevented the increase in the percentage and absolute number of MZ B cells and CD11b+ myeloid cells (Fig. 2C).
Figure 2. Microbiota-mediated increase in splenic MZ B and CD11b+ cells in Il10−/− mice.
(A) The percentage of MZ B cells (gated on B220) (left panel), CD11b+ myeloid cells (middle panel) and Fo B cells (gated on B220) (right panel) was quantitated in Il10−/− Jax housed alone or cohoused with Il10−/− mice from our colony for four weeks. Each data point represents a single mouse. (B) WT and Il10−/− Jax mice were cohoused with Il10−/− mice from our colony and the percentage of MZ B cells (gated on B220) and CD11b+ myeloid cells in the spleen was determined on day 7, 21, 28 and 35. Data are shown as the mean ± SE (n = 3). (C) The percentage (top panels) and absolute number (bottoms panels) of splenic MZ B cells (gated on B220) (left panels) and CD11b+ myeloid cells (right panels) in WT Jax cohoused and Il10−/− Jax and Il10−/− Jax cohoused mice (four weeks) with and without antibiotic treatment was determined by flow cytometry. Data are shown as the mean ± SE of two experiments (n = 6–7). *p<0.05, **p<0.01.
Both Neutrophil and Monocyte/Macrophage Numbers are Specifically Increased in Il10−/− Mice Following Cohousing
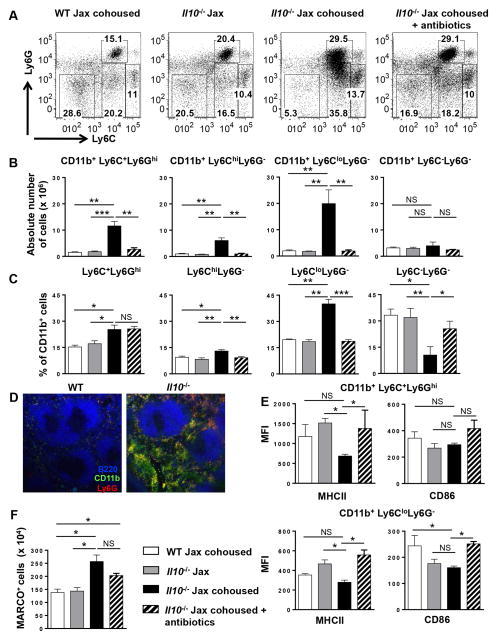
To determine the phenotype of the expanded CD11b+ myeloid population in Il10−/− mice, we subdivided CD11b+ cells by Ly6C and Ly6G expression. Specifically, we quantitated Ly6C+Ly6Ghi neutrophils, Ly6ChiLy6G− inflammatory and Ly6CloLy6G− resident monocyte/macrophages and Ly6C−Ly6G− cells that are likely a combination of resident monocytes/macrophages, dendritic cells and NK cells (Fig. 3A, Suppl. Fig. 2E) (53, 54). When we measured the absolute number of each CD11b+ myeloid cell population in Il10−/− Jax mice following cohousing with Il10−/− mice from our colony, we found a significant increase in the Ly6C+Ly6Ghi neutrophil and in the Ly6ChiLy6G− and Ly6CloLy6G− monocyte/macrophage populations as compared to both WT mice and noncohoused IL10−/− Jax mice (Fig. 3B). No change in the Ly6C−Ly6G− population was observed (Fig. 3B). Antibiotic treatment during the cohousing period prevented the increase in the absolute number of neutrophils and monocyte/macrophages (Fig. 3B). When we measured the relative distribution of each cell type within the CD11b+ population, we observed a significant increase in the percentage of neutrophils and monocytes/macrophages with cohousing (Fig. 3C) that correlated with their increase in absolute number (Fig. 3B). Interestingly, antibiotic treatment did not affect the percentage of neutrophils, but did prevent the increase in the percentage of the monocytes/macrophages (Fig. 3B), which was offset by the reciprocal effect on the Ly6C−Ly6G− population (Fig. 3C). We confirmed that the Ly6ChiLy6G− inflammatory macrophages expressed CCR2, which was not expressed by the Ly6CloLy6G− monocytes/macrophages (data not shown) (55). We next determined the localization of the increased myeloid cells in the spleen by immunofluorescence. As shown in Fig. 3D, the increased numbers of both CD11b+Ly6G− macrophages (green) and CD11b+Ly6G+ neutrophils (yellow) in Il10−/− mice was clearly evident. Interestingly, the myeloid cells were restricted to the red pulp and were completely excluded from the B and T cell zones. We then examined the activation status of the macrophages and neutrophils by flow cytometry staining for MHC class II and CD86, and found that the two cell populations in Il10−/− Jax cohoused mice were not more activated as compared to WT mice and Il10−/− Jax cohoused antibiotic treated mice (Fig. 3E). Surprisingly, Il10−/− Jax cohoused mice expressed lower levels of MHC class II with no difference in CD86 (Fig. 3E). These data indicate that loss of IL-10 signaling outside of the gut does not lead to the over activation of myeloid cells. This finding is consistent with the histology, because unactivated macrophages would not be expected to migrate into the white pulp.
Figure 3. The increase in splenic CD11b+ myeloid cells in Il10−/− mice is due to increased numbers of monocytes/macrophages and neutrophils.
(A) Representative dot plots showing Ly6C and Ly6G expression by splenic CD11b+ myeloid cells from WT Jax cohoused, Il10−/− Jax, Il10−/− Jax cohoused (four weeks) and antibiotic treated (four weeks) Il10−/− Jax cohoused mice. (B) Using the indicated mice in (A) the absolute number of CD11b+Ly6C+Ly6Ghi, CD11b+Ly6ChiLy6G−, CD11b+Ly6CloLy6G−, and CD11b+Ly6C−Ly6G− cells was determined by flow cytometry. Data are shown as the mean ± SE of two experiments (n = 6–7). (C) The percentage of the indicated cell populations in (A) was determined by flow cytometry. (D) Immunofluorescence analysis of spleen sections from WT and Il10−/− mice stained with B220-Alexa Fluor 594 (blue), CD11b-Alexa Fluor 488 (green) and Ly6G-Alexa Fluor 647 (red) were performed. Representative images from one of two mice are shown (100x). (E) The expression of MHCII and CD86 on splenic CD11b+Ly6C+Ly6Ghi (top panels) and CD11b+Ly6CloLy6G− (bottom panels) cells from the mice indicated in (A) was measured by flow cytometry and the mean fluorescence intensity (MFI) is shown. Data are shown as the mean ± SE of two experiments (n = 6). (F) The absolute number of splenic MARCO+ cells (CD11b gated) was determined in the indicated mice in (A). Data are shown as the mean ± SE (n = 3–4). *p<0.05, **p<0.01, ***p<0.001, NS = not significant.
Because MZ B cell retention within the MZ has been reported to be regulated by MARCO+ macrophages resident in the MZ (56), we also determined the absolute number of MARCO+ macrophages and found that their absolute number increased upon cohousing (Fig. 3F). Interestingly, antibiotic treatment did not prevent the increase in MARCO+ macrophages (Fig. 3F), suggesting that their increase alone is not responsible for the increase in MZ B cells.
The Increase in MZ B Cells in Il10−/− Mice is Due to Enhanced MZ B Cell Development
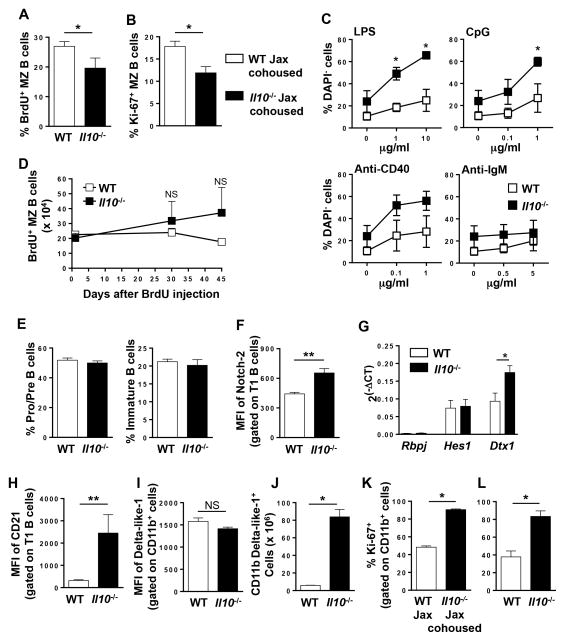
Next, we investigated the mechanism driving the immune homeostasis defect. First, we examined proliferation and found that total B cells from Il10−/− mice exhibited a slight proliferative advantage over WT when stimulated with LPS, CpG or anti-CD40, but not anti-IgM (data not shown). However, this did not translate into increased steady state proliferation of Il10−/− MZ B cells in vivo as measured by BrdU incorporation (Fig. 4A) nor did cohousing increase the number of Ki-67+ MZ B cells (Fig. 4B). Interestingly, MZ B cells were proliferating less in the Il10−/− mice (Fig. 4A, B). These findings are consistent with similar expression of the activation markers CD80, CD86, MHC class II and CD40 by WT and Il10−/− MZ B cells (data not shown) (57). To determine whether Il10−/− MZ B cells have a survival advantage, we repeated the in vitro culture using FACS purified MZ B cells and found that in the absence of stimulation Il10−/− MZ B cells had a higher level of survival after three days in culture (Fig. 4C). Following stimulation with LPS or CpG both the WT and Il10−/− MZ B cells expanded with a similar dose-dependent kinetics that was proportional to the number of surviving cells without simulation, which resulted in a significant increase in the number of Il10−/− MZ B cells (Fig. 4C). Although anti-CD40 induced expansion of both WT and Il10−/− MZ B cells there was not a significant difference in their cell number (Fig. 4C). Anti-IgM only led to a marginal expansion of both populations (Fig. 4C). The survival advantage was marginally evident following a BrdU pulse chase assay after 30 and 45 days (Fig. 4D). This finding is consistent with the similar expression levels of the cell survival proteins Bcl-2 and Bcl-xL in MZ B cells from WT and Il10−/− mice (data not shown). In addition, Il10−/− MZ B cells were not undergoing reduced levels of apoptosis (data now shown). These cumulative data indicate that the increased numbers of MZ B cells in Il10−/− mice is not due to increased cell survival or proliferation. Thus we examined whether B cell differentiation is altered in Il10−/− mice. In the bone marrow, we found no alteration in the proportion of pro/pre and immature B cells in Il10−/− mice, indicating that IL-10 is not required for early B cell differentiation (Fig. 4E). To specifically examine whether enhanced MZ lineage differentiation is occurring in the spleen, we measured Notch-2 expression in the T1 subset because it is required to drive precursor maturation into the MZ B cell lineage (58, 59). Using flow cytometry, we found that Notch-2 expression was significantly increased on Il10−/− T1 cells (Fig. 4F). However, Notch-2 expression was not altered in the mature Fo and MZ subsets (data not shown). We then determined if the increase in Notch-2 expression led to increased signaling by measuring the transcript levels of its downstream effector molecules (60, 61). While we saw no difference in mRNA levels of Rbpj and Hes1, we found that Deltex 1 (Dtx1) transcripts were significantly increased in Il10−/− T1 B cells (Fig. 4G). In addition, the cell surface expression of CD21, another Notch-2 regulated protein (59), was significantly increased on T1 B cells in Il10−/− mice (Fig. 4H). Finally, we asked whether the expression of the ligand for Notch-2, Delta-like 1, was increased on CD11b+ myeloid cells in Il10−/− mice. While we did not detect a difference in the expression level of Delta-like 1 on myeloid cells (Fig. 4I) or in the percentage of CD11b+ cells that express Delta-like 1 (data not shown), there was a ~15 fold-increase in the absolute number of CD11b+Delta-like 1+ cells in the Il10−/− mice (Fig. 4J). When we examined the proliferation of Il10−/− CD11b+ myeloid cells in the spleen in the steady state, we found that in Il10−/− Jax cohoused mice they underwent a significantly higher rate of proliferation as compared to WT Jax mice (Fig. 4K). Identical results were obtained in WT and Il10−/− mice from the home colony (Fig. 4L). These cumulative data suggest that the increase in MZ B cells in Il10−/− mice is due to increased differentiation of the T1 subset into the MZ lineage via upregulation of Notch-2 and increased availability of its ligand Delta-like 1, while the increase in myeloid cells is due to increased proliferation.
Figure 4. Enhanced MZ B cell survival and increased Notch-2 signaling in B cells from Il10−/− mice.
(A) BrdU (0.8 mg/ml) was administered in the drinking water for nine days and the percentage of BrdU+ MZ B cells were determined by flow cytometry. Data are shown as the mean ± SE of two experiments (n = 4). (B) The percentage of proliferating (Ki-67+) MZ B cells from WT Jax cohoused and IL-10−/− Jax cohoused (four weeks) mice was determined by flow cytometry. Data are shown as the mean ± SE of two experiments (n = 3–4). (C) MZ B cells were FACS purified from WT and Il10−/− mice and cultured in medium alone or stimulated with the indicated concentrations of LPS, CpG, anti-CD40 or anti-IgM. Four days later, the percentage of DAPI− viable cells was determined by flow cytometry. Data are shown as the mean ± SE of three experiments. (D) WT and Il10−/− mice were i.p. injected with BrdU for three days and the absolute number of BrdU+ MZ B cells was determined by flow cytometry 1, 30, and 45 days later. Data shown are the mean ± SE from one representative experiment of two (n = 3). (E) The percentage of Pro/Pre (B220+IgM−) (left panel) and immature (B220loIgM+) (right panel) B220-gated B cells in the bone marrow of WT and Il10−/− mice was determined by flow cytometry. Data are shown as the mean ± SE (n = 3). (F) The mean fluorescence intensity (MFI) of Notch2 expression on splenic T1 B cells (B220+CD21lo/−IgMhiCD23−CD93+) from WT and Il10−/− mice was determined by flow cytometry. Data are shown as the mean ± SE of two experiments (n = 5–6). (G) The relative expression of Notch2 target genes was determined by real-time PCR in splenic T1 B cells from WT and Il10−/− mice. Data shown is normalized to Gapdh from two experiments (n = 6). (H) The MFI of CD21 on splenic T1 B cells from WT and Il10−/− mice was measured by flow cytometry. Data are shown as the mean ± SE of two experiments (n = 6). (I, J) The MFI of Delta-like 1 expression on CD11b+ cells (I) and the absolute number of splenic CD11b+Delta-like-1+ cells (J) in WT and Il10−/− mice was determined by flow cytometry. Data shown are the mean ± SE of one representative experiment of two (n = 3). (K, L) Proliferation of splenic CD11b+ cells from WT Jax and Il10−/− Jax cohoused (four weeks) (K) and WT and Il10−/− (L) mice was determined by Ki-67 staining using flow cytometry. Data are shown as the mean ± SE of two experiments (n = 8, K), (n = 4, L). *p<0.05, **p<0.01, NS = not significant.
The Increase in MZ B and Myeloid Cells in Il10−/− Mice is Macrophage, but not B Cell, Intrinsic
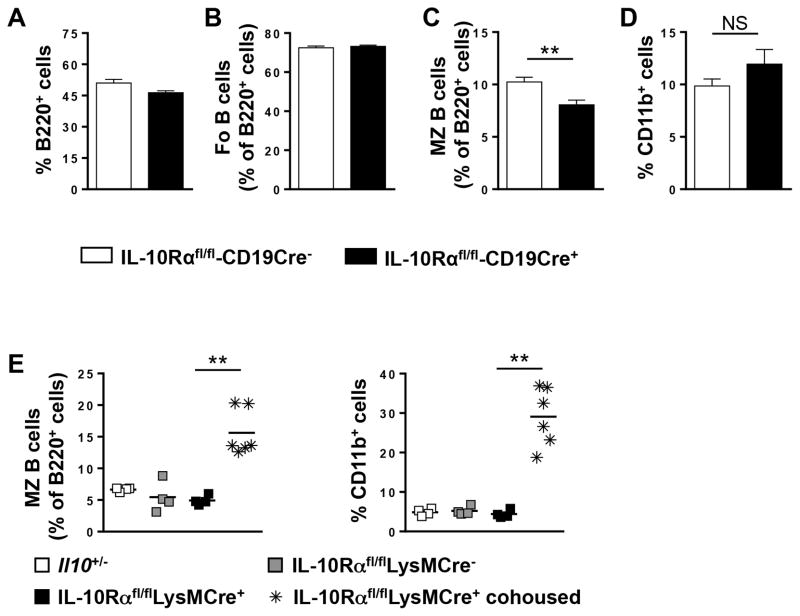
We next determined whether the increase in MZ B cells and myeloid cells was due to a loss of IL-10 signaling in one or both cell types. To examine the necessity for IL-10 signaling in B cells, we utilized IL-10Rfl/flCD19Cre+ mice and found that upon cohousing these mice exhibited no alteration in the percentage of total (Fig. 5A) or Fo B cells (Fig. 5B) as compared to IL-10Rfl/flLysMCre negative control mice. Interestingly, the percentage of MZ B cells was significantly decreased (Fig. 5C). In addition, the percentage of CD11b+ cells was not altered (Fig. 5D). To render monocytes/macrophages and other myeloid subsets deficient in IL-10R signaling, we utilized IL-10Rfl/flLysMCre+ mice and found that upon cohousing these mice had an increase in both the percentage and absolute number (data not shown) of both MZ B cells and CD11b+ cells at a level similar to Il10−/− mice from our colony (Fig. 5E). WT levels of both populations were observed in the IL10Rfl/flLysMCre− and IL10Rfl/flLysMCre+ non-cohoused controls (Fig. 5E). These cumulative data indicate that IL-10R signaling in monocytes/macrophages, but not B cells, is required to regulate the steady state homeostasis of MZ B cells and CD11b+ myeloid cells in the spleen.
Figure 5. The increase in CD11b+ myeloid cells Il10−/− mice is due to enhanced proliferation in a cell-intrinsic manner.
The percentage of total splenic B cells (B220+) (A) and Fo (B) and MZ (C) B cells within the B220 gate and CD11b+ (D) cells was determined by flow cytometry from IL-10Rαfl/flCD19Cre+ and IL-10Rαfl/flCD19Cre− mice cohoused with mice from our colony for four weeks. Data are shown as the mean ± SE of two experiments (n = 6). (E) The percentage of splenic MZ B cells (left panel) and CD11b+ myeloid cells (right panel) from Il10+/−, IL-10Rαfl/flLysMCre−, IL-10Rαfl/flLysMCre+, and IL-10Rαfl/flLysMCre+ cohoused (four weeks) mice was determined by flow cytometry. Each data point represents a single mouse from three experiments. **p<0.01, NS = not significant.
Il10−/− Jax Mice Acquire A Distinct Microbial Community After Cohousing
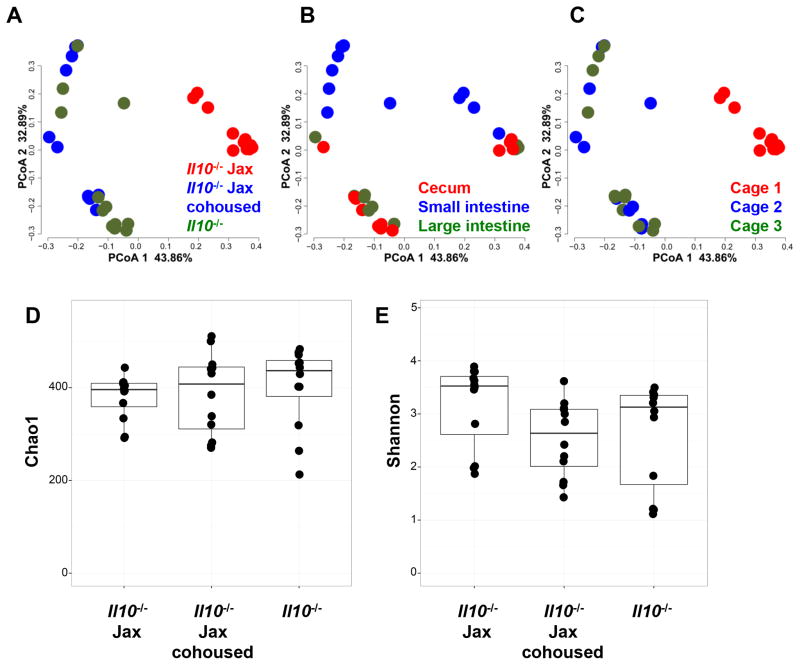
In order to identify the microbial community associated with the transition from Jackson Labs, we performed Illumina sequencing targeting the V4 region of the 16S rRNA gene, after quality filtering 8,928,469 sequences of length=90 from 12 animals (four in each group sampled from three different tissues). Those sequences were then clustered into Operational Taxonomic Units (OTUs), groups of sequences with an average percent identity of 97%. A Principal Coordinate Analysis (PCoA) analysis of these OTUs revealed that Il10−/− Jax mice upon immediate receipt from Jax showed distinct clustering from Il10−/− mice housed in our facility and Il10−/− Jax mice cohoused with our animals (Fig. 6A). The FDR was p<0.05 for Il10−/− Jax mice compared to IL10−/− mice in our facility and co-housed Il10−/− Jax mice.
Figure 6. Distinct and significant clusters are formed between Il10−/− Jax mice as compared to Il10−/− Jax co-housed and Il10−/− from our colony.
(A) Bray-Curtis Principal Coordinate Analysis (PCoA) plots generated from the operational taxonomic unit (OTU) colored by origin (A) were generated comparing Il10−/− Jax mice analyzed immediately upon receipt from Jax (n=4) to Il10−/− mice in our facility (n=4) and to Il10−/− Jax mice cohoused with our mice for four weeks (n=4). (B) PCoA was conducted as for (A) colored by tissue source. (C) PCoA was conducted as for (A) colored by cage. Alpha diversity analysis using Chao1 richness estimate (D) and Shannon diversity index (E) between the three groups is shown.
In order to separate the effect of the Jax microbiota from the tissue type from which we sampled (Fig. 6B) and the cage (Fig. 6C) in which the animals were housed in, we utilized a mixed linear model as described previously (46) with the terms: origin indicating Il10−/− Jax mice, Il10−/− Jax mice cohoused with our mice or our Il10−/− mice, tissue indicating the source tissue of the sample (cecum, small intestine or large intestine) and cage indicating the cage at which the animals were housed. The results from this analysis have been deposited at http://metagenomics.anl.gov/linkin.cgi?project=14383. Among the first five PCoA axes, we observed many highly significant effects for tissue, tissue×origin and origin. These results are consistent with the Il10−/− Jax mice having a distinct microbiome that shifts over time and with cohousing in our animal facility. It is interesting to note that the differences between the Jax mice gut microbiome and our facility’s mice gut microbiome (Fig. 6A) appear to be larger than the differences that are attributable to the tissue that is sampled (Fig. 6B). This emphasizes the profound influence of the animal facility environment over the gut microbiome. Analysis of these data utilizing alternative algorithms (for example utilizing UNIFRAC distance as opposed to Bray-Curtis or assigning OTUs through the open-reference algorithm in QIIME) yielded nearly identical patterns of clustering. (Suppl. Fig. 3A–I). Our conclusion of a distinct microbial community in the Il10−/− Jax mice is therefore unlikely to be the result of particular analysis choice we made in our pipeline. However, alpha diversity analysis (Fig. 6D and E) showed no significant differences in terms of richness or diversity between the three groups.
MZ B Cell and Myeloid Cell Homeostasis is Dysregulated by Introduction of a Single Bacterial Strain, Helicobacter Hepaticus
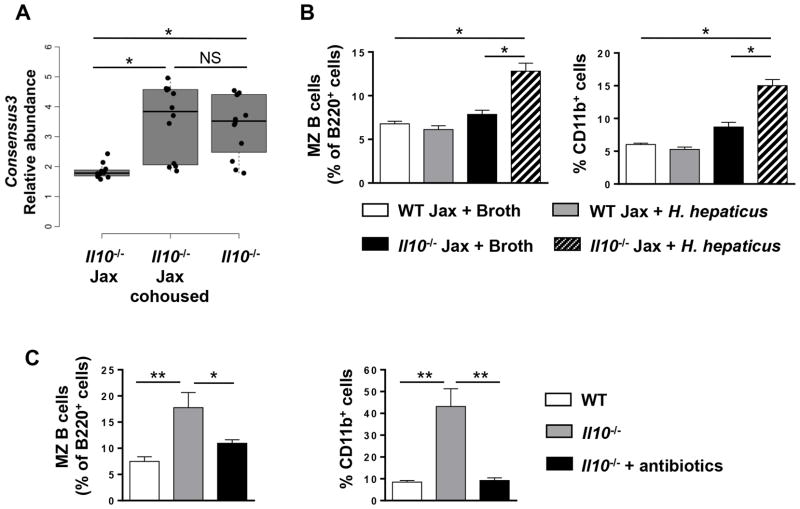
In order to determine the biological impact of differences between Jax mice and mice in our facility, we focused on Helicobacter because of its known association with colitis (11, 18). In addition, our Il10−/− mice harbor Helicobacter and succumb to spontaneous colitis while the Il10−/− Jax mice do not. We found that Consensus3 classified to genus level as Helicobacter by both Ribosomal Database project (RDP) Classifier (confidence score > 0.8) and uclust was significantly lower in Il10−/− Jax mice FDR-p< 0.05) (Fig. 7A). To determine whether the presence of Helicobacter was responsible for the immune homeostasis defect in Il10−/− mice, we inoculated Il10−/− Jax mice once with Helicobacter hepaticus by gavage. We found that Il10−/− Jax mice gavaged with H. hepaticus had a significant increase in both MZ B cells and CD11b+ myeloid cells as compared to Il10−/− mice that were gavaged broth (Fig. 7B). WT Jax mice gavaged with H. hepaticus did not exhibit an increase in either MZ B cells or CD11b+ myeloid cells (Fig. 7B). We next asked whether the immune homeostasis defect was permanent and found that antibiotic treatment of our Il10−/− mice reversed the increase in MZ B and CD11b+ myeloid cells (Fig. 7C). Together, these results indicate that a single bacterial strain upon introduction to the intestinal track can modulate immune cell homeostasis at distal sites in addition to contributing to a disease state in immunocompromised individuals.
Figure 7. Il10−/− Jax mice as compared to Il10−/− Jax co-housed and Il10−/− from our colony had an increased abundance of Helicobacter that when introduced to Il10−/− Jax mice was sufficient to drive an increase in MZ B and CD11b+ cells.
(A) A box and whisker plot shows the minimum, first quartile, median, third quartile and maximum relative abundance of Consensus3 (Helicobacter spp.) in Il10−/− Jax with and without cohousing and Il10−/− from our colony (n = 4). (B) The percentage of MZ B (left panel) and CD11b+ (right panel) splenic cells in H. hepaticus or control broth gavaged WT Jax and Il10−/− Jax mice was determined by flow cytometry. Data are shown as the mean ± SE from one representative experiment of two (n = 4). (C) The percentage of splenic MZ B cells (left panel) and CD11b+ cells (right panel) from WT, Il10−/− and antibiotic treated (four weeks) Il10−/− mice was determined by flow cytometry. Data are shown as the mean ± SE of two independent experiments (n = 5–6). *p<0.05, **p<0.01, NS = not significant.
Discussion
We have demonstrated that differences in the microbiota between genetically similar mice can vary profoundly depending on the animal husbandry history leading to an imbalance in immune cell development and homeostasis at distal sites outside of the intestinal track. Specifically, we showed that MZ B and CD11b+ myeloid cell numbers were significantly increased in the spleen of Il10−/− mice from our colony as compared to WT mice from our colony and Il10−/− Jax mice. Sequencing of the microbiome revealed that our mice were colonized with a distinct bacterial microbiota, which was transferred to Il10−/− Jax mice upon cohousing with our mice in conjunction with the acquisition of the homeostasis defect in MZ B and myeloid cells. We also found that the transfer of a single bacterial species was sufficient to drive the imbalance in immune homeostasis seen in our colony. These results have profound implications for the reproducibility of experimental data when animals with deficiencies in immune regulatory mediators are used and emphasize the importance of controlling the composition of the microbial community.
When genetically altered mice are phenotyped for deficiencies in immune cell populations, it is very uncommon for specific B cell subsets to be examined. Thus even profound differences in the relatively rare population of MZ B cells (4–8% of splenic B cells) can be missed. This is clearly demonstrated in our study where we found that total B cells were decreased in Il10−/− mice from our colony (Fig. 1D). This reduction was largely due to a decrease in Fo B cells (Fig. 1E), and because Fo B cells are the major B cell subset in the spleen, their decrease was able to offset the increase in MZ B cells in the total B cell pool (Fig. 1E). We found that in Il10−/− mice from our colony there were increased levels of resting IgM and antigen-specific IgM following immunization with a T-independent antigen (Fig. 1J). While we attribute this finding to an increase in MZ B cells, if the increase in MZ B cells had not been observed an alternate explanation would be that IL-10 directly regulates IgM production. If the same experiment had been performed with Il10−/− Jax mice, no alteration in IgM levels would have been observed, which would be interpreted as IL-10 playing no role in IgM production. Thus two laboratories doing the same experiment using the same mouse strain would come to very different conclusions. In support of the importance of phenotyping B cell subsets, in previous studies in mice deficient in the cannabinoid receptor 2, we found that while the percentage of Fo B cells was not altered in the spleen, the percentage of MZ B cells was significantly reduced (28). These data indicate that assuming B cell development and homeostasis is normal based on using global B cell markers such as B220 and CD19 is not sufficient when phenotyping genetically modified mice.
Using two different gating strategies (26, 48), we were able to definitively demonstrate that MZ B cell numbers are increased in the spleen of Il10−/− Jax mice as early as seven days after cohousing with mice from our colony (Fig. 2B). Using a variety of approaches, we determined that the increase in MZ B cells in Il10−/− mice was not due to increased levels of proliferation or enhanced cell survival (Fig. 4). However, we cannot rule out a role for either increased expression of B cell survival factors or increased MZ B cell responsiveness to survival factors such as BAFF in Il10−/− mice. We do not favor the increased survival factor hypothesis because it was previously shown that serum levels of the B cell survival factor BAFF were not increased in Il10−/− mice (62). Given the requirement for Notch-2 signaling for the differentiation of the MZ lineage (59), our data demonstrating an increase in Notch-2 expression by the T1 precursors (Fig. 4F) suggests that the increase in MZ B cells is due to increased lineage commitment to the MZ lineage. This would mean that fewer progenitors would be available to commit to the Fo lineage, which is what we observed (Fig. 1E). The increase in the mRNA expression of the Notch2 target gene Dtx1 (Fig. 4G) and surface expression of CD21 (Fig. 4H) indicates that the increase in Notch2 protein has a functional outcome likely due to enhanced signaling. Little is known regarding how Notch2 expression is regulated in MZ B cells, but its function is dependent upon binding to its ligand Delta-like 1 (20, 58). Delta-like 1 expression by splenic non-hematopoietic cells, likely a stromal cell, is sufficient to drive MZ B cell differentiation (63). The site of MZ commitment is unknown, but it likely occurs outside of the MZ. Our finding that Delta-like 1 is expressed by monocytes/macrophages in the spleen (Fig. 4I) is consistent with the literature (64). We found that neither the MFI of Delta-like 1 on CD11b+ cells (Fig. 4I) nor their percentages expressing Delta-like 1 (data not shown) were altered in Il10−/− mice. However, the increase in total myeloid cells in the Il10−/− spleen resulted in an increase in the total number of Delta-like 1+ cells by ~15-fold (Fig. 4J). Thus while in the steady state hematopoietic cells do not promote MZ B cell differentiation, we propose that during inflammation the increase in CD11b+Delta-like 1+ cells in the spleen increases Notch2 ligand availability transiently increasing the number of MZ B cells likely by both promoting their differentiation and retention. This speculative hypothesis is supported by our finding that antibiotic treatment of Il10−/− mice, presumably dampening the inflammatory response, was sufficient to reduce the number of MZ B cells and CD11b+ myeloid cells in the spleen (Fig. 7C). Further support of this hypothesis is the finding that intradermal injection of Borrelia burgdorferi, the causative agent of Lyme disease, led to a transient increase in MZ B cells in mice that correlated with the first appearance of the spirochete in other tissues (65).
While we favor the hypothesis that the increase in MZ B cells in Il10−/− mice is due to enhanced differentiation, we cannot rule out the possibility that MZ B cells are being retained within the spleen. Using a Delta-like 1 blocking antibody, it was shown that Notch2 engagement with its ligand was required for retention of the pre-established long-lived MZ B cell pool within the spleen in adult mice (64). In addition, MARCO+ macrophages within the MZ are also thought to contribute to MZ B cell retention (56). However, the increase in MARCO+ macrophages in cohoused Il10−/− mice without and without antibiotics suggests that retention is not likely the major mechanism that drives the increase in MZ B cells (Fig. 3F). An unanswered question is the myeloid cell type that potentially drives MZ B cell differentiation. By flow cytometry phenotyping, we found that both neutrophils and monocytes/macrophages are significantly increased in Il10−/− Jax mice upon cohousing with mice from our colony (Fig. 3B, C). Based on the composition of the myeloid cells, we favor the hypothesis that the Ly6CloLy6G− monocytes/macrophages are the required cell. Cohousing of Il10−/− Jax mice significantly increased the absolute number of neutrophils (Ly6C+Ly6Ghi), inflammatory macrophages (Ly6ChiLy6G−) and the Ly6CloLy6G− monocytes/macrophages, the expansion of which was prevented by the inclusion of antibiotics (Fig. 3B). However, when the composition of the myeloid cell population was determined, the percentage of neutrophils within the CD11b+ population was identical in the cohoused with and without antibiotic groups (Fig. 3A, C). The most dramatic change in composition was in the Ly6CloLy6G− population, which was increased by over 50% after cohousing. We cannot rule out a role for neutrophils given that Puga and colleagues recently demonstrated that they are located peri-MZ in the spleen (51). That elegant study largely conducted in humans further demonstrated that neutrophils provide a helper function to MZ B cells by producing factors promoting isotype class switching, somatic hypermutation and immunoglobulin production (51). Interestingly, a broad group of neutropenic patients was shown to have reduced numbers of circulating MZ B cells with a poorly developed MZ (51). However, the study was not designed to determine whether neutrophils are required for development of the MZ and/or lineage commitment of MZ B cells.
In our kinetic study, upon cohousing MZ B cells and CD11b+ cells increased in parallel in the Il10−/− Jax mice with a significant increase evident as early as day 7 post-cohousing (Fig. 2B). From these data it was not clear if the expansion of the two populations is linked or are independent events. We investigated this question by conditionally knocking out the IL-10R in B cells or macrophages. For B cells, we used CD19-Cre that achieves close to 100% depletion in splenic B cells (66). For macrophages, we utilized LysM-Cre that largely targets macrophages with a reported depletion rate of the IL-10R in 90% of monocytes/macrophages (24). Depletion in neutrophils has also been reported (24). Loss of IL-10R signaling in B cells had no impact on B cell or myeloid cell homeostasis (Fig. 5A–D) and supports our findings in global Il10-deficient mice that IL-10 has no major intrinsic role in B cell development, function or homeostasis. In contrast, loss of IL-10 signaling in myeloid cells was sufficient to drive an increase in both MZ B cells and myeloid cells further supporting our hypothesis that macrophages and/or neutrophils can promote MZ B cell differentiation (Fig. 5E). The expansion in splenic myeloid cells was due to increased proliferation with up to 90% of the cells proliferating in the absence of IL-10 (Fig. 4K, L). Since peripheral neutrophils do not divide and they make up 10% of the myeloid population in the Il10−/− mice, this indicates that 100% of the monocytes/macrophages were undergoing cell division. This remarkable finding indicates that IL-10 is a master regulator of monocyte/macrophage proliferation. The increase in CD11b+ cells in the bone marrow (Fig. 1H) indicates that IL-10 may exert this control even during myeloid cell development. In support of this hypothesis are studies in the Rag1−/− CD4+CD25−CD45RBhi T cell transfer model of colitis where increased numbers of both macrophages and neutrophils were observed in the spleen and bone marrow. This study concluded that the increase in myeloid cells was largely due to an increase in proliferative hematopoietic stem cells and skewed differentiation towards granulocyte-monocyte progenitors resulting in a reduction in the percentages of both lymphoid and erythroid progenitors (67). Of importance to our study, is the finding that H. hepaticus-induced colitis in Rag1−/− mice also resulted in the accumulation of myeloid cells in the spleen and BM (67). The increase in myeloid cells was shown to be IFN-γ-dependent (67). While we did not specifically examine myeloid cell commitment in the BM, it is possible that the combination of IL-10-deficiency and H. hepaticus also results in skewed myeloid commitment in the BM leading to increased number of neutrophils in the spleen and peripheral blood. It is not clear whether this would be the result of altered production of pro-inflammatory cytokines, such as IFN-γ, due to the loss of IL-10 immunosuppression. However, we do not favor this hypothesis because it has been shown that serum levels of a large number of cytokines were not increased in Il10−/− mice as compared to WT (62), which we further confirmed in mice from our colony (Table I). Although, there were no signs of systemic inflammation (Table I), levels of a number of pro-inflammatory cytokines and chemokines were significantly elevated in the colon of Il10−/− mice (Fig. 1I). This suggests that local colonic inflammation drives the expansion of myeloid cells in Il10−/− mice.
Our finding that the introduction of H. hepaticus to the microbiome of normal Il10−/− mice can drive myeloid cell expansion (Fig. 7B) indicates that the introduction of a single bacterial species to immune compromised individuals has the potential to alter their immune status in essentially every tissue. For infectious disease, overcoming the IL-10 negative feedback loop would be beneficial with increased numbers of myeloid cells better able to clear infections. However, in autoimmunity, such as in multiple sclerosis, or in colitis where macrophages are thought to be drivers of disease, increased numbers of myeloid cells that cannot be negatively regulated would be detrimental. Indeed, it was recently shown that loss of IL-10 signaling in macrophages was sufficient to lead to spontaneous colitis in mice (9, 10). The ability of one bacterial species that is seemingly harmless upon introduction to alter the host immune status is likely why it has been difficult to link particular micro-organisms with the onset of autoimmunity and other diseases.
While the phenomenon of irreproducibility is not new to science or immunology here we clearly show that it can be affected by the introduction of a single bacterial species (Fig. 7B). Another clear example of a single bacterial species altering immune homeostasis is segmented filamentous bacteria (SFB) that when introduced to the gut induced lamina propria CD4 T cells to produce IL-17 (68, 69). Introduction of SFB into germ free K/BxN mice that exhibit attenuated arthritis was sufficient to induce lamina propria Th17 cells and the onset of arthritis (70). While SFP is linked to pathogenesis, both Bacteroides and Clostridium species have been implicated in immunoprotection (71, 72). While the mechanisms whereby particular bacteria alter immune homeostasis and function are largely unknown, B. fragiles immune regulatory activity is dependent upon expression of the capsular polysaccharide A (PSA) (72). In experimental autoimmune encephalomyelitis (EAE), B. fragiles expressing PSA could not restore susceptibility in antibiotic-treated mice, while mice that received PSA− bacteria exhibited EAE similar to controls (72). The protection was dependent upon Foxp3+ T regulatory cells (Treg), suggesting that PSA plays a role in Treg conversion (72). Likewise, Clostridium likely via the induction of Treg in the lamina propria can provide protection against colitis (71).
In this study, we demonstrated that the introduction of a single bacterial species to the intestinal track altered immune cell homeostasis at a distal site in the spleen. This is the first time that introduction of a single taxa to the microbiome has been shown to effect MZ B cell homeostasis in the spleen. Moreover, our results have profound implications for scientific reproducibility as we have demonstrated that differences in the history of animal husbandry can directly impact measurements of the immune system via differences in the microbial community. Failure to explicitly consider these differences will likely lead to results that are difficult to reproduce. While we have demonstrated a specific role for an IL-10 regulatory loop in controlling activation of macrophages and subsequent MZ B cell differentiation by the microbiota, similar regulatory pathways likely exist for other cytokines and inflammatory mediators. MZ B cells are poised to rapidly respond to blood-borne pathogens given their localization within the MZ in the spleen and their partially activated phenotype (57). Thus a specific increase in MZ B cells would facilitate faster clearance of potential pathogens that transverse the gut endothelium and enter the blood during inflammation. Future work will undoubtedly continue to define the role of gut bacteria in signaling to the immune system via breaches in the gut barrier.
Supplementary Material
Acknowledgments
The authors would like to thank Shelley Morris for help with the animal studies, Nichole Miller and Michael Hayward for technical assistance, Dr. Sridhar Rao for help with the design and implementation of the microbiome studies and Dr. Monica Mann for her initial suggestion to more carefully examine the Il10−/− mouse.
This work was supported by National Institutes of Health grants R01 AI069358-01A2 (BD), R56 AI106672-01 (BD), R21 AI097619 (NHS), R01 GM099526 (NHS), R01 DK088831 (NHS), R01 AI056153 (TLG) and the Blood Center Research Foundation (BD). The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical Translational Science (UL1TR000165) and Heflin Center.
References
- 1.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 2.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D, Sina C, Onnie CM, Weersma RK, Stokkers PC, Wijmenga C, Gazouli M, Strachan D, McArdle WL, Vermeire S, Rutgeerts P, Rosenstiel P, Krawczak M, Vatn MH, Mathew CG, Schreiber S. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 3.Moran CJ, Walters TD, Guo CH, Kugathasan S, Klein C, Turner D, Wolters VM, Bandsma RH, Mouzaki M, Zachos M, Langer JC, Cutz E, Benseler SM, Roifman CM, Silverberg MS, Griffiths AM, Snapper SB, Muise AM. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2013;19:115–123. doi: 10.1002/ibd.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hatscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, Hugot JP, Daussy C, Verkarre V, Pigneur B, Fischer A, Klein C, Cerf-Bensussan N, Ruemmele FM. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Amer J Gastroenterol. 2011;106:1544–1555. doi: 10.1038/ajg.2011.112. [DOI] [PubMed] [Google Scholar]
- 6.Kotlarz D, Beier R, Murugan D, Diestelhorst J, Jensen O, Boztug K, Pfeifer D, Kreipe H, Pfister ED, Baumann U, Puchalka J, Bohne J, Egritas O, Dalgic B, Kolho KL, Sauerbrey A, Buderus S, Gungor T, Enninger A, Koda YK, Guariso G, Weiss B, Corbacioglu S, Socha P, Uslu N, Metin A, Wahbeh GT, Husain K, Ramadan D, Al-Herz W, Grimbacher B, Sauer M, Sykora KW, Koletzko S, Klein C. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterol. 2012;143:347–355. doi: 10.1053/j.gastro.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 8.Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, Sordat B, Gibbs VC, Aguet M. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187:571–578. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim KW, Brenner O, Krauthgamer R, Varol C, Muller W, Jung S. Macrophage-Restricted Interleukin-10 Receptor Deficiency, but Not IL-10 Deficiency, Causes Severe Spontaneous Colitis. Immunity. 2014;40:720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Shouval DS, Biswas A, Goettel JA, McCann K, Conaway E, Redhu NS, Mascanfroni ID, Al Adham Z, Lavoie S, Ibourk M, Nguyen DD, Samsom JN, Escher JC, Somech R, Weiss B, Beier R, Conklin LS, Ebens CL, Santos FG, Ferreira AR, Sherlock M, Bhan AK, Muller W, Mora JR, Quintana FJ, Klein C, Muise AM, Horwitz BH, Snapper SB. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40:706–719. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 12.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veenbergen S, Samsom JN. Maintenance of small intestinal and colonic tolerance by IL-10-producing regulatory T cell subsets. Curr Opin Immunol. 2012;24:269–276. doi: 10.1016/j.coi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004;53:1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Favier C, Neut C, Mizon C, Cortot A, Colombel JF, Mizon J. Fecal β-D-galactosidase production and Bifidobacteria are decreased in Crohn’s disease. Dig Dis Sci. 1997;42:817–822. doi: 10.1023/a:1018876400528. [DOI] [PubMed] [Google Scholar]
- 16.Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG, Josenhans C, Suerbaum S. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One. 2013;8:e70783. doi: 10.1371/journal.pone.0070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakoff-Nahoum S, Hao L, Medzhitov R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25:319–329. doi: 10.1016/j.immuni.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66:5157–5166. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molloy MJ, Bouladoux N, Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Semin Immunol. 2012;24:58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- 21.Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, Hansson GC. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, Tonkonogy SL, Mair K, Krueger D, Pruteanu M, Shanahan F, Vogelmann R, Schemann M, Kuster B, Sartor RB, Haller D. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterol. 2011;141:959–971. doi: 10.1053/j.gastro.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Alli R, Steeves M, Nguyen P, Vogel P, Geiger TL. The T cell response to IL-10 alters cellular dynamics and paradoxically promotes central nervous system autoimmunity. J Immunol. 2012;189:669–678. doi: 10.4049/jimmunol.1200607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pils MC, Pisano F, Fasnacht N, Heinrich JM, Groebe L, Schippers A, Rozell B, Jack RS, Muller W. Monocytes/macrophages and/or neutrophils are the target of IL-10 in the LPS endotoxemia model. Eur J Immunol. 2010;40:443–448. doi: 10.1002/eji.200939592. [DOI] [PubMed] [Google Scholar]
- 25.Ray A, Dittel BN. Isolation of mouse peritoneal cavity cells. J Vis Exp. 2010;35:e1488. doi: 10.3791/1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava B, Quinn WJ, 3rd, Hazard K, Erikson J, Allman D. Characterization of marginal zone B cell precursors. J Exp Med. 2005;202:1225–1234. doi: 10.1084/jem.20051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu S, Campbell HM, Dittel BN, Ray A. Purification of specific cell population by fluorescence activated cell sorting (FACS) J Vis Exp. 2010:e1546. doi: 10.3791/1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu S, Ray A, Dittel BN. Cannabinoid receptor 2 is critical for the homing and retention of marginal zone B lineage cells and for efficient T-independent immune responses. J Immunol. 2011;187:5720–5732. doi: 10.4049/jimmunol.1102195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu S, Ray A, Dittel BN. Cannabinoid Receptor 2 (CB2) Plays a Role in the Generation of Germinal Center and Memory B Cells, but Not in the Production of Antigen-Specific IgG and IgM, in Response to T-dependent Antigens. PLoS One. 2013;8:e67587. doi: 10.1371/journal.pone.0067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livingston RS, Myles MH, Livingston BA, Criley JM, Franklin CL. Sex influence on chronic intestinal inflammation in Helicobacter hepaticus-infected A/JCr mice. Comp Med. 2004;54:301–308. [PubMed] [Google Scholar]
- 31.Myles MH, Dieckgraefe BK, Criley JM, Franklin CL. Characterization of cecal gene expression in a differentially susceptible mouse model of bacterial-induced inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:822–836. doi: 10.1002/ibd.20138. [DOI] [PubMed] [Google Scholar]
- 32.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray A, Basu S, Williams CB, Salzman NH, Dittel BN. A novel IL-10-independent regulatory role for B cells in suppressing autoimmunity by maintenance of regulatory T cells via GITR ligand. J Immunol. 2012;188:3188–3198. doi: 10.4049/jimmunol.1103354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ray A, Basu S, Miller NM, Chan AM, Dittel BN. An Increase in Tolerogenic Dendritic Cell and Natural Regulatory T Cell Numbers during Experimental Autoimmune Encephalomyelitis in Rras−/− Mice Results in Attenuated Disease. J Immunol. 2014;192:5109–5117. doi: 10.4049/jimmunol.1302254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman NH, de Jong H, Paterson Y, Harmsen HJ, Welling GW, Bos NA. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiol. 2002;148:3651–3660. doi: 10.1099/00221287-148-11-3651. [DOI] [PubMed] [Google Scholar]
- 36.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar R, Eipers P, Little RB, Crowley M, Crossman DK, Lefkowitz EJ, Morrow CD. Getting started with microbiome analysis: sample acquisition to bioinformatics. Curr Protoc Hum Genet. 2014;82:18 18 11–18 18 29. doi: 10.1002/0471142905.hg1808s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 39.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCafferty J, Muhlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–2125. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y, Hochberg Y IBSEN study group. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Soc Ser. 1995;57:289–300. [Google Scholar]
- 48.Roark JH, Park SH, Jayawardena J, Kavita U, Shannon M, Bendelac A. CD1.1 expression by mouse antigen-presenting cells and marginal zone B cells. J Immunol. 1998;160:3121–3127. [PubMed] [Google Scholar]
- 49.Martin F, Kearney JF. Marginal-zone B cells. Nat Rev Immunol. 2002;2:323–335. doi: 10.1038/nri799. [DOI] [PubMed] [Google Scholar]
- 50.den Haan JM, Kraal G. Innate immune functions of macrophage subpopulations in the spleen. J Innate Immun. 2012;4:437–445. doi: 10.1159/000335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puga I, Cols M, Barra CM, He B, Cassis L, Gentile M, Comerma L, Chorny A, Shan M, Xu W, Magri G, Knowles DM, Tam W, Chiu A, Bussel JB, Serrano S, Lorente JA, Bellosillo B, Lloreta J, Juanpere N, Alameda F, Baro T, de Heredia CD, Toran N, Catala A, Torrebadell M, Fortuny C, Cusi V, Carreras C, Diaz GA, Blander JM, Farber CM, Silvestri G, Cunningham-Rundles C, Calvillo M, Dufour C, Notarangelo LD, Lougaris V, Plebani A, Casanova JL, Ganal SC, Diefenbach A, Arostegui JI, Juan M, Yague J, Mahlaoui N, Donadieu J, Chen K, Cerutti A. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat Immunol. 2012;13:170–180. doi: 10.1038/ni.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehrenstein MR, Notley CA. The importance of natural IgM: scavenger, protector and regulator. Nat Rev Immunol. 2010;10:778–786. doi: 10.1038/nri2849. [DOI] [PubMed] [Google Scholar]
- 53.Rose S, Misharin A, Perlman H. A novel Ly6C/Ly6G-based strategy to analyze the mouse splenic myeloid compartment. Cytometry A. 2012;81:343–350. doi: 10.1002/cyto.a.22012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 55.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 56.Karlsson MC, Guinamard R, Bolland S, Sankala M, Steinman RM, Ravetch JV. Macrophages control the retention and trafficking of B lymphocytes in the splenic marginal zone. J Exp Med. 2003;198:333–340. doi: 10.1084/jem.20030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oliver AM, Martin F, Gartland GL, Carter RH, Kearney JF. Marginal zone B cells exhibit unique activation, proliferative and immunoglobulin secretory responses. Eur J Immunol. 1997;27:2366–2374. doi: 10.1002/eji.1830270935. [DOI] [PubMed] [Google Scholar]
- 58.Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, Tamaoki N, Mailhos C, Ish-Horowicz D, Habu S, Owen MJ. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 59.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, Nakagami-Yamaguchi E, Hamada Y, Aizawa S, Hirai H. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 60.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 61.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 62.Scapini P, Lamagna C, Hu Y, Lee K, Tang Q, DeFranco AL, Lowell CA. B cell-derived IL-10 suppresses inflammatory disease in Lyn-deficient mice. Proc Natl Acad Sci USA. 2011;108:E823–832. doi: 10.1073/pnas.1107913108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheng Y, Yahata T, Negishi N, Nakano Y, Habu S, Hozumi K, Ando K. Expression of Delta-like 1 in the splenic non-hematopoietic cells is essential for marginal zone B cell development. Immunol Lett. 2008;121:33–37. doi: 10.1016/j.imlet.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 64.Moriyama Y, Sekine C, Koyanagi A, Koyama N, Ogata H, Chiba S, Hirose S, Okumura K, Yagita H. Delta-like 1 is essential for the maintenance of marginal zone B cells in normal mice but not in autoimmune mice. Int Immunol. 2008;20:763–773. doi: 10.1093/intimm/dxn034. [DOI] [PubMed] [Google Scholar]
- 65.Malkiel S, Kuhlow CJ, Mena P, Benach JL. The loss and gain of marginal zone and peritoneal B cells is different in response to relapsing fever and Lyme disease Borrelia. J Immunol. 2009;182:498–506. doi: 10.4049/jimmunol.182.1.498. [DOI] [PubMed] [Google Scholar]
- 66.Li B, Alli R, Vogel P, Geiger TL. IL-10 modulates DSS-induced colitis through a macrophage-ROS-NO axis. Mucosal Immunol. 2013;7:869–878. doi: 10.1038/mi.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griseri T, McKenzie BS, Schiering C, Powrie F. Dysregulated hematopoietic stem and progenitor cell activity promotes interleukin-23-driven chronic intestinal inflammation. Immunity. 2012;37:1116–1129. doi: 10.1016/j.immuni.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 70.Wu HJ, Ivanov, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.