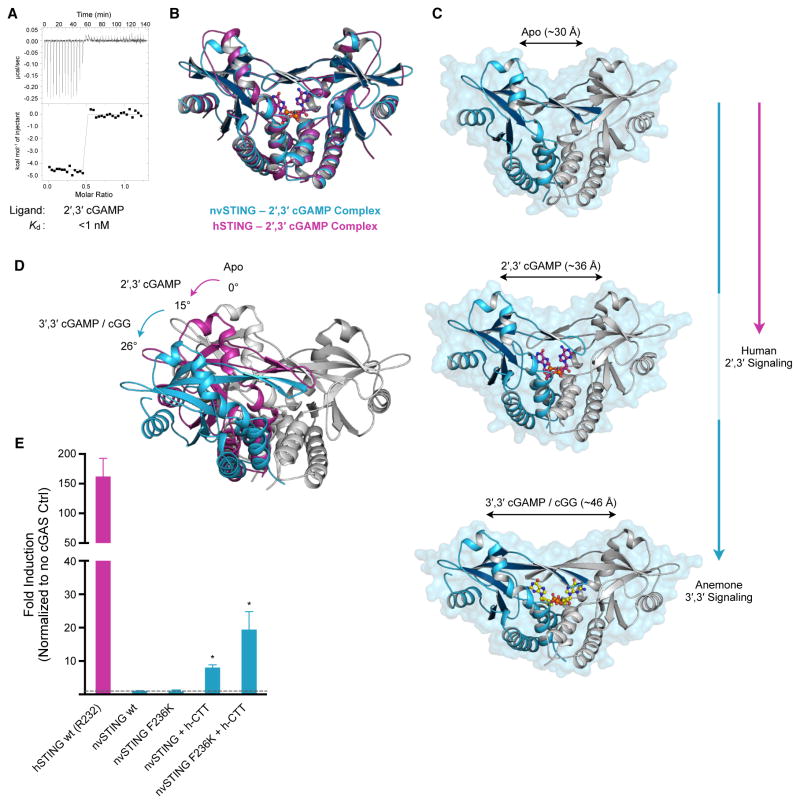
Summary
In humans, the cGAS-STING immunity pathway signals in response to cytosolic DNA via 2′,3′ cGAMP, a cyclic dinucleotide (CDN) second messenger containing mixed 2′–5′ and 3′–5′ phosphodiester bonds. Prokaryotes also produce CDNs, but these are exclusively 3′ linked, and thus the evolutionary origins of human 2′,3′ cGAMP signaling are unknown. Here we illuminate the ancient origins human cGAMP signaling by discovery of a functional cGAS-STING pathway in Nematostella vectensis, an anemone species >500 million years diverged from humans. Anemone cGAS appears to produce a 3′,3′ CDN that anemone STING recognizes through nucleobase-specific contacts not observed in human STING. Nevertheless, anemone STING binds mixed-linkage 2′,3′ cGAMP indistinguishably from human STING, trapping a unique structural conformation not induced by 3′,3′ CDNs. These results reveal that human mixed-linkage cGAMP achieves universal signaling by exploiting a deeply conserved STING conformational intermediate, providing critical insight for therapeutic targeting of the STING pathway.
Graphical abstract
Introduction
Cyclic dinucleotides (CDNs) are critical second messenger signaling molecules widely dispersed throughout prokaryotes (Danilchanka and Mekalanos, 2013). Until recently, CDNs were not thought to function in animals. However, the discovery of an innate immunity pathway responsible for surveillance of cytosolic DNA has revealed a prominent role for CDN signaling in vertebrates (Sun et al., 2013). In this pathway, cytosolic double-stranded DNA is detected after it binds directly to an enzyme called cyclic GMP–AMP synthase (cGAS). Upon dsDNA binding, cGAS is allosterically activated to produce a CDN second messenger called cyclic GMP–AMP (cGAMP). cGAMP binds directly to an endoplasmic reticulum receptor protein called STING (Stimulator of IFN Genes), thereby activating a signaling pathway that induces transcription of type I interferon and other co-regulated genes (Burdette et al., 2011; Ishikawa and Barber, 2008; Jin et al., 2008; Sun et al., 2013; Sun et al., 2009; Zhong et al., 2008). Whereas all currently known prokaryotic CDNs have two canonical 3′–5′ phosphodiester linkages, the human cGAS product contains a unique 2′–5′ bond resulting in a mixed linkage cyclic GMP–AMP molecule, denoted as 2′,3′ cGAMP (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013a; Zhang et al., 2013). The bacterium Vibrio cholerae encodes an enzyme called DncV that is a structural homolog of cGAS and synthesizes a related second messenger with canonical 3′–5′ bonds (3′,3′ cGAMP), suggesting that human CDN signaling may have deep ancestral roots (Davies et al., 2012; Kranzusch et al., 2014).
In mammals, the ER-resident receptor protein STING is essential for interpreting CDN signals (Jin et al., 2011a; Sauer et al., 2011). The carboxy-terminal domain of STING forms a homodimer that creates a recessed binding pocket for endogenous 2′,3′ cGAMP or prokaryotic 3′,3′ CDNs released by intracellular pathogens (Burdette et al., 2011; Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012; Woodward et al., 2010; Yin et al., 2012). Upon its activation, STING recruits the downstream TBK1 kinase to phosphorylate the transcription factor IRF3. Activated IRF3 enters the nucleus where it mediates transcription of interferon β and other co-regulated genes (Blaauboer et al., 2014; Ishikawa et al., 2009; Liu et al., 2015; Tanaka and Chen, 2012). Additionally, STING signaling can lead to STAT6 phosphorylation and NF-κB activation (Chen et al., 2011; McWhirter et al., 2009). Endogenous 2′,3′ cGAMP is a significantly more potent agonist of human STING than are prokaryotic 3′,3′ CDNs, and triggers a structural rearrangement in the STING beta-strand lid domain that is thought to be responsible for establishing a signaling competent state (Gao et al., 2013b; Zhang et al., 2013). It is not known how these structural rearrangements result in recruitment and activation of downstream signaling molecules, and thus it remains unclear why 2′,3′ cGAMP is a particularly potent STING agonist.
Recent analyses of cGAS and STING deficient mice have shown that the cGAS-STING pathway is essential for cytosolic immune detection of DNA and is critical for immune responses against numerous viral and bacterial pathogens (Gall et al., 2012; Ishikawa et al., 2009; Li et al., 2013; Sauer et al., 2011; Schoggins et al., 2014; Zhang et al., 2014). The small-molecule nature of 2′,3′ cGAMP and the ability of STING to initiate potent immune responses in diverse cell types has spurred interest in using STING agonists as novel vaccine adjuvants or immunotherapeutics. In particular, recent data highlight the potency of CDNs as anti-tumor immunotherapy agents (Corrales et al., 2015; Deng et al., 2014; Fu et al., 2015; Woo et al., 2014). However, current understanding of the molecular principles underlying the unique ability of 2′,3′ cGAMP to broadly activate various STING alleles remains incomplete. We rationalized that analyzing the evolutionary origins of the human cGAS-STING pathway would help establish the principles underlying universal 2′,3′ cGAMP signaling.
Prior bioinformatics analyses have identified candidate STING homologs throughout diverse metazoans (Wu and Chen, 2014; Wu et al., 2014). However, only mammalian STING proteins have been functionally characterized and shown to bind CDNs. Thus it remains possible that ancestral STING had other functions and that the ability to bind CDNs is a recent innovation that arose in vertebrates alongside modern innate immunity. In contrast to this view, we show here that binding to CDNs is a deeply evolutionarily conserved function of STING. Using a paleo-biochemical approach, we tracked the function of STING through diverse animal lineages and identified functional cGAS and STING homologs in the sea anemone Nematostella vectensis, an animal divergent from humans by >500 million years of evolution. These results reveal the surprisingly ancient evolutionary origins of cGAS-STING signaling and demonstrate that the CDN second messenger pathway in animals predates the emergence of interferon signaling and modern innate immunity. We show that anemone cGAS activates human STING, and specific reporter and CDN phosphodiesterase assays suggest that the anemone cGAS product is 3′,3′ cGAMP. Moreover, we determined a complete series of seven crystal structures of anemone STING, encompassing two unbound structures and five ligand-bound structures. These structures comprise the only complete set of STING structures in any species and reveal an unexpected nucleobase-specific recognition of 3′,3′ cGAMP by anemone STING. Most importantly, however, our results illustrate that a unique property of 2′,3′ cGAMP is its ability to trap a distinct and deeply evolutionarily conserved STING conformation not seen with 3′,3′ CDNs. We propose that the cGAS-STING pathway has exploited a unique mode of evolution where the protein components remain structurally conserved, while chemical changes in the interconnecting second messenger drive functional innovation. These results illuminate the evolution and ancestral origins of a critical second-messenger signaling pathway in human immunity, and outline the conserved functionality required for design of broadly active STING chemotherapeutics.
Results
STING cyclic dinucleotide recognition predates modern innate immunity
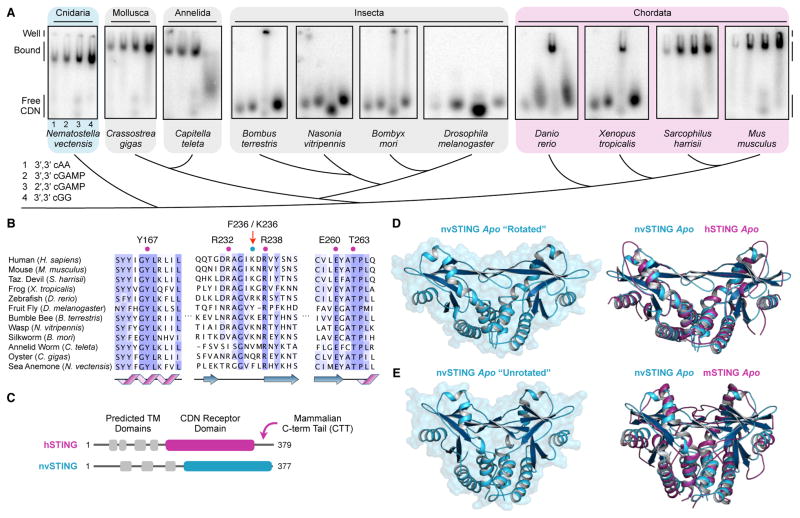
To probe STING function in diverse animal lineages, candidate STING genes were identified by protein homology using a hidden Markov model to search published animal genomes. The C-terminal STING domains of eleven different STING orthologs were expressed and purified from E. coli, and tested for their ability to interact with 32P-radiolabelled CDNs using an electrophoretic mobility gel-shift assay. All four known naturally occurring CDNs were tested: 3′,3′ c-di-AMP (cAA), 3′,3′ cGAMP, 2′,3′ cGAMP and 3′,3′ c-di-GMP (cGG) (Figure 1A). The vertebrate STING orthologs we analyzed readily interacted with the endogenous product 2′,3′ cGAMP. While all mammalian STING proteins tested also bound to 3′,3′ CDNs, we did not detect robust 3′,3′ CDN binding with STING proteins from more diverse vertebrates including D. rerio (zebrafish) and X. tropicalis (western clawed frog). In support of preservation of a functional cGAS-cGAMP-STING pathway across vertebrate evolution, mammalian, Xenopus and Danio genomes contain readily identifiable cGAS homologs with an intact active site and Zn-ribbon insertion critical for human cGAS function (Ablasser et al., 2013; Kranzusch et al., 2013; Wu et al., 2014).
Figure 1. STING Cyclic Dinucleotide Recognition Predates Modern Innate Immunity.
(A) Gel-shift assay measuring complex formation between recombinant STING proteins and radiolabeled CDNs. Each panel shows reactions in order: (1) 3′,3′ cAA, (2) 3′,3′ cGAMP, (3) 2′,3′ cGAMP and (4) 3′,3′ cGG. (B) Phylogenetic alignment of animal STING proteins colored by amino-acid conservation. Key CDN interacting residues are indicated. (C) Cartoon schematic of human and anemone STING proteins. Predicted transmembrane domains are indicated in grey, and the crystallized CDN receptor domain is indicated in magenta or blue. Notably, the cterminal tail required for hSTING interferon signaling is absent in nvSTING. (D,E) Crystal structure of apo nvSTING CDN receptor domain in “rotated” and “unrotated” states. Overlaid structures compare the apo nvSTING structures (blue) and apo hSTING (PDB 4F9E) or apo mouse STING (PDB 4KC0) (magenta) revealing conformational dynamics and an ancestrally conserved STING fold. See also Figure S1.
In contrast to vertebrate STING proteins, we observed no interaction between the insect Drosophila STING and any of the four naturally occurring CDNs. To ascertain the generality of this result, we tested three additional STING alleles from insect genomes (bumble bee, B. terrestris; wasp, N. vitripennis; and silkworm, B. mori), and again were unable to detect CDN interaction other than nonspecific well-shifts. A search for cGAS homologs in insect genomes reveals possible nucleotidyltransferase superfamily related genes but no obvious homologs of human cGAS that contain both an enzymatic active site and a Zn-ribbon domain for dsDNA-specific recognition (Civril et al., 2013; Hancks et al., 2015; Kranzusch et al., 2013; Wu et al., 2014). These results suggest that insect STING proteins may have lost or decreased the ability to bind to CDNs, or may only do so with additional co-factors missing from our assay.
Amongst invertebrates, we characterized putative STING homologs within phyla Annelida, Mollusca and Cnidaria. Despite <30% amino acid identity with vertebrate STING proteins, the invertebrate STING homologs we tested exhibited a robust ability to bind both 3′,3′ CDNs and 2′,3′ cGAMP (Figure 1A). The most divergent STING homolog examined was from the starlet sea anemone (Nematostella vectensis), an animal with >500 million years of evolutionary divergence from humans (Putnam et al., 2007). Consistent with our biochemical findings, phylogenetic alignment of human STING (hSTING) and anemone STING (nvSTING) indicates that despite low sequence identity (29%) the key residues previously implicated in hSTING CDN recognition are conserved in ancestrally divergent STING homologs (Figure 1B,C, Figure S1).
STING architecture is conserved in anemone
To confirm that invertebrate STING homologs exhibit evolutionary conservation of key structural features, we determined crystal structures of the C-terminal CDN receptor domain of nvSTING using selenomethionine-derivatized crystals for experimental phase determination (Figure 1D,E, Table 1). Apo nvSTING crystallized in two distinct crystal forms, an open “rotated” wing conformation (2.1 Å) and a closed “unrotated” wing conformation (2.9 Å). In each case, the structures reveal a dimeric protein receptor with clear structural homology to human and mouse STING (Chin et al., 2013; Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012; Yin et al., 2012). Our two distinct “rotated” and “unrotated” apo nvSTING structures correlate with previously observed apo structures of human and mouse STING, respectively, indicating that these two forms likely represent alternatively sampled STING conformations, and not species-specific differences between the human and mouse proteins (Figure 1D,E). Importantly, the structures of apo nvSTING reveal the conserved presence of a beta-strand lid domain poised to arch over a deep central ligand-binding pocket (Figure 1D,E). Taken together, our results provide strong evidence that the overall STING fold, and its ability to bind CDNs, is deeply conserved throughout metazoans, predating the clear emergence of interferons and ‘modern’ innate immunity (Zhang et al., 2015).
Table 1.
Crystallographic Statistics
| nvSTING–cGG c(G[3′–5′]pG[3′–5′]p) |
nvSTING–3′,3′
cGAMP c(G[3′–5′]pA[3′–5′]p) |
nvSTING–cAA c(A[3′–5′]pA[3′–5′]p) |
nvSTING
Apo “Rotated” |
nvSTING–cGG
(SeMet) c(G[3′–5′]pG[3′–5′]p) |
|
|---|---|---|---|---|---|
| Data Collection | |||||
| Resolution (Å) | 48.54–1.84 (1.90–1.84) | 40.63–1.99 (2.04–1.99) | 40.83–2.95 (3.06–2.95) | 41.15–2.10 (2.16–2.10) | 48.66–2.39 (2.48–2.39) |
| Wavelength (Å) | 1.11587 | 1.11587 | 1.11587 | 1.11587 | 0.97967 |
| Space group | P 31 | P 31 | P 31 | P 31 | P 31 |
| Unit cell: a, b, c (Å) | 81.11, 81.11, 97.07 | 81.27, 81.27, 99.12 | 81.66, 81.66, 98.76 | 82.30, 82.30, 100.35 | 80.77, 80.77, 97.33 |
| Unit cell: α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Molecules per ASU | 2 | 2 | 2 | 2 | 2 |
| No. reflections: total | 693819 | 473676 | 89635 | 494252 | 323356 |
| No. reflections: unique | 61587 | 49839 | 15258 | 43684 | 27919 |
| Completeness (%) | 98.7 (82.5) | 99.5 (93.9) | 98.3 (85.5) | 98.4 (81.1) | 99.2 (92.7) |
| Multiplicity | 11.3 (9.3) | 9.5 (8.2) | 5.8 (5.5) | 11.3 (8.1) | 11.6 (10.9) |
| I/σI | 10.3 (1.5) | 13.4 (1.8) | 6.1 (1.0) | 9.4 (1.0) | 17.1 (5.2) |
| CC(1/2) (%) | 99.8 (50.8) | 99.9 (63.4) | 95.9 (34.4) | 99.8 (34.0) | 99.8 (91.5) |
| Rpim (%) | 3.6 (69.9) | 3.3 (73.9) | 10.5 (89.2) | 4.4 (88.2) | 3.7 (16.2) |
| No. sites | 8 | ||||
| Refinement | |||||
| Resolution (Å) | 48.54–1.84 | 40.63–1.99 | 40.83–2.95 | 41.15–2.10 | |
| Free reflections (%) | 5 | 5 | 5 | 5 | |
| R-factor / R-free | 17.2 / 20.9 | 16.9 / 19.0 | 22.1 / 24.6 | 17.2 / 20.2 | |
| Bond distance (RMS Å) | 0.003 | 0.008 | 0.003 | 0.011 | |
| Bond angles (RMS °) | 0.740 | 1.053 | 0.944 | 1.322 | |
| Structure/Stereochemistry | |||||
| No. atoms: protein | 3060 | 3060 | 3063 | 3006 | |
| No. atoms: ligand | 46 | 45 | 88 | 0 | |
| No. atoms: water | 392 | 256 | 0 | 209 | |
| Average B-factor: protein | 37.6 | 51.3 | 74.4 | 54.2 | |
| Average B-factor: ligand | 24.7 | 33.5 | 85.9 | - | |
| Average B-factor: solvent | 47.6 | 52.7 | - | 55.8 | |
| Ramachandran plot: favored | 98.4% | 97.8% | 98.6% | 97.8 | |
| Ramachandran plot: allowed | 1.6% | 2.2% | 1.4% | 2.2 | |
| Ramachandran plot: outliers | 0% | 0% | 0% | 0% | |
| Rotamer outliers: | 0.9% | 0.9% | 1.5% | 1.2% | |
| MolProbity score | 1.06 | 1.29 | 1.49 | 1.19 | |
| Protein Data Bank ID | 5CFL | 5CFM | 5CFN | 5CFO | |
| nvSTING–cGG
F276K c(G[3′–5′]pG[3′–5′]p) |
nvSTING–2′,3′
cGAMP c(G[2′–5′]pA[3′–5′]p) |
nvSTING
Apo “Unrotated” |
|
|---|---|---|---|
| Data Collection | |||
| Resolution (Å) | 40.93–2.07 (2.12–2.07) | 42.49–2.10 (2.16–2.10) | 43.04–2.85 (3.00–2.85) |
| Wavelength (Å) | 1.11588 | 1.11587 | 1.11587 |
| Space group | P 31 | P 21 21 21 | P 21 21 21 |
| Unit cell: a, b, c (Å) | 81.39, 81.39, 100.55 | 40.50, 77.97, 101.35 | 43.04, 43.04, 170.94 |
| Unit cell: α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Molecules per ASU | 2 | 2 | 2 |
| No. reflections: total | 290406 | 283149 | 31005 |
| No. reflections: unique | 45536 | 19434 | 7901 |
| Completeness (%) | 99.6 (94.6) | 99.6 (95.6) | 99.4 (97.5) |
| Multiplicity | 6.4 (6.0) | 14.6 (6.6) | 3.9 (4.0) |
| I/σI | 8.0 (1.1) | 16.2 (1.6) | 12.6 (3.2) |
| CC(1/2) (%) | 99.5 (31.3) | 99.9 (62.1) | 99.7 (97.5) |
| Rpim (%) | 5.2 (90.1) | 2.4 (54.1) | 3.8 (23.1) |
| Refinement | |||
| Resolution (Å) | 40.93–2.07 | 42.49–2.10 | 43.04–2.85 |
| Free reflections (%) | 5 | 5 | 5 |
| R-factor / R-free | 17.2 / 20.6 | 24.2 / 25.9 | 25.7 / 27.8 |
| Bond distance (RMS Å) | 0.009 | 0.003 | 0.003 |
| Bond angles (RMS °) | 1.298 | 0.733 | 0.805 |
| Structure/Stereochemistry | |||
| No. atoms: protein | 2986 | 2963 | 2844 |
| No. atoms: ligand/ion | 92 | 45 | 4 (Calcium) |
| No. atoms: water | 241 | 56 | 3 |
| Average B-factor: protein | 48.7 | 67.4 | 64.7 |
| Average B-factor: ligand/ion | 54.4 | 31.1 | 76.5 |
| Average B-factor: water | 53.5 | 51.1 | 14.2 |
| Ramachandran plot: favored | 98.0% | 96.1% | 94.6% |
| Ramachandran plot: allowed | 2.0% | 3.9% | 5.4% |
| Ramachandran plot: outliers | 0% | 0% | 0% |
| Rotamer outliers: | 1.2% | 0.6% | 4.8% |
| MolProbity score | 1.35 | 1.72 | 2.28 |
| Protein Data Bank ID | 5CFP | 5CFQ | 5CFR |
The cGAS-STING signaling pathway is conserved in sea anemone
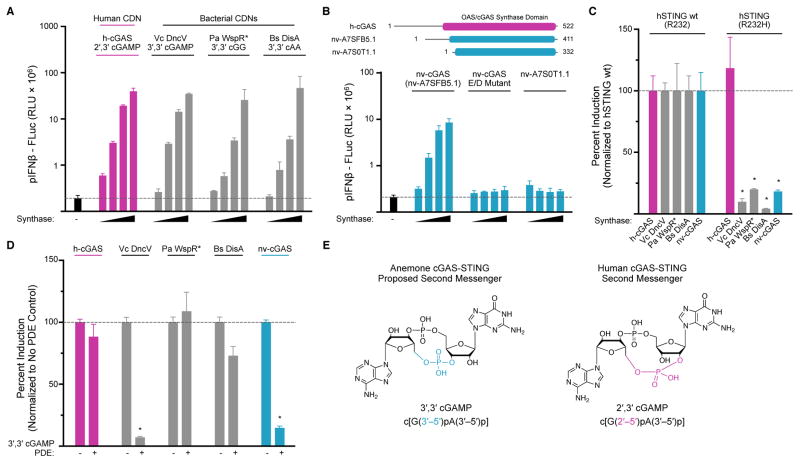
To determine if the entire cGAS-STING pathway is evolutionarily conserved, we next asked if invertebrate genomes encode a functional cGAS-like enzyme that is able to produce an endogenous CDN second messenger. We focused our efforts on the sea anemone N. vectensis, as this was the most divergent species found to encode a functional STING ortholog. We cloned all 12 N. vectensis genes encoding putative cGAS-like proteins with predicted functional active sites. We screened these enzymes for CDN synthase activity by transiently transfecting them into human STING-interferon β luciferase reporter cells that respond to all known naturally occurring CDNs and CDN synthases from bacteria and humans (Diner et al., 2013) (Figure 2A). We found that expression of only a single anemone gene, nv-A7SFB5.1 (Ensembl accession A7SFB5.1), induced robust hSTING-dependent interferon β signaling in human cells (Figure 2B and Figure S2). Overexpression of all other candidate N. vectensis genes, including the closely related protein nv-A7S0T1.1, did not result in any detectable STING activation (Figure 2B and Figure S2). The other candidate genes are inactive in this assay either because they are functionally diverse nucleotidyltransferases unrelated to CDN synthases or possibly because essential co-factors are missing from the cellular assay. Alignment of nv-A7SFB5.1 and human cGAS reveals a shared predicted catalytic active site (Figure S2), and disruption by amino-acid substitution of the predicted magnesium ion coordination motif in nv-A7SFB5.1 (E131A/D133A) abolishes all signal induction in human cells. Identification of nv-A7SFB5.1 (now renamed nv-cGAS) extends the occurrence of CDN synthases to invertebrate animals, and demonstrates that cGAS and STING are ancestrally conserved signaling components in animal biology.
Figure 2. Anemone cGAS Signals Via a 3′,3′-Linked Cyclic Dinucleotide.
(A) Cell culture assay using wildtype hSTING (R232) and an interferon β luciferase reporter to measure CDN synthase activity in transfected human cells. Titrated synthases include human cGAS (h-cGAS), Vibrio cholerae DncV (Vc DncV), Pseudomonas aeruginosa WspR D70E (Pa WspR*), Bacillus subtilis DisA (Bs DisA) and the product CDNs are indicated. (B) Cartoon schematic of candidate N. vectensis cGAS-like enzymes, and screen for CDN synthase activity using interferon β assay as in A. Activity is detected with gene nv-cGAS (nv-A7SFB5.1), but not in a catalytic inactive nv-cGAS control (E/D mutant) or in the closely related candidate gene nv-A7S0T1.1. (C) Cell interferon β luciferase as in A, using either a wildtype hSTING (R232) allele responsive to all CDNs, or a mutant hSTING (R232H) allele highly selective for 2′,3′ cGAMP. Production of 2′,3′ cGAMP by human cGAS activates both STING alleles, while 3′,3′ CDN production by bacterial synthases and nv-cGAS only activates wildtype hSTING. (D) Cell interferon β luciferase as in A, with supplementation of a 3′,3′ cGAMP specific phosphodiesterase (PDE) (Vibrio cholerae VCA0681). Vc DncV and nv-cGAS 3′,3′ cGAMP production is ablated by 3′,3′ cGAMP PDE co-expression, while other synthases are not significantly affected. (E) Cartoon schematic of proposed anemone 3′,3′ cGAMP and human 2′,3′ cGAMP second messengers. Data in C and D are normalized to hSTING wt and synthase control. Error bars represent the SE of the mean of at least three independent experiments (asterisk denotes p <0.002). See also Figures S2 and S3.
Anemone cGAS signals via a 3′,3′-linked cyclic dinucleotide
Human cGAS produces a cyclic GMP–AMP second messenger containing a mixed 2′–5′ and 3′–5′ phosphodiester linkage (2′,3′) distinct from all previously characterized bacterial 3′,3′ CDNs. To determine the nv-cGAS product identity and linkage specificity we first attempted to reconstitute nv-cGAS enzymatic activity in vitro using purified components. In contrast to the robust in vitro activity of recombinant human cGAS, nv-cGAS activity is not detected in vitro upon stimulation in the presence of dsDNA (Figure S3). These results suggest that an alternative ligand present within the complex cellular environment may be responsible for controlling nv-cGAS activity.
As the activating ligand for nv-cGAS is currently unknown, we instead further characterized nv-cGAS activity and product identity in transfected cells, where we observed the enzyme to be active and able to stimulate STING signaling (Figure 2B). We took advantage of polymorphic hSTING alleles that exhibit differing specificities for 2′,3′ and 3′,3′ CDNs: hSTING R232 (the wild-type allele), which responds to all CDNs, and hSTING R232H which responds preferentially to 2′,3′ cGAMP (Ablasser et al., 2013; Corrales et al., 2015; Diner et al., 2013; Jin et al., 2011b; Kranzusch et al., 2014).
To confirm that the hSTING alleles behave as expected, we tested their responsiveness to transfection of well-characterized CDN synthases. Human cGAS produces 2′,3′ cGAMP and thus activates both hSTING variants (Figure 2C). In contrast, the Vibrio cholerae synthase DncV, which produces a 3′,3′ cGAMP dinucleotide, is unable to activate the hSTING R232H allele (Diner et al., 2013; Kranzusch et al., 2014). Similarly, overexpression of nv-cGAS was unable to activate the hSTING R232H allele (Figure 2C). Collectively, these data indicate that nv-cGAS produces a 3′,3′ linked CDN distinct from human 2′,3′ cGAMP.
We next sought to determine the dinucleotide base identity produced by nv-cGAS. Vibrio cholerae DncV is a close structural homolog of human cGAS (Kato et al., 2015; Kranzusch et al., 2014; Zhu et al., 2014), and the enzyme active sites of DncV and cGAS are similar enough that a single amino acid substitution converts human cGAS into a DncV-like 3′,3′ cGAMP synthase (Kranzusch et al., 2014). Given that the nv-cGAS sequence is predicted to have similar changes to catalytic pocket amino acids important for substrate positioning (Figure S2), we hypothesized that the 3′,3′ CDN nv-cGAS product is most likely to be 3′,3′ cGAMP. To test this hypothesis, we took advantage of a recently characterized phosphodiesterase that is highly specific for degradation of 3′,3′ cGAMP (Gao et al., 2015). Vibrio cholerae cGAP1 efficiently hydrolyzes 3′,3′ cGAMP, but has no activity on any other CDN product (Gao et al., 2015). We confirmed this specificity in our reporter system by showing that co-expression of cGAP1 ablated DncV 3′,3′ cGAMP signaling but only had a minimal (<2-fold) impact on STING activation by bacterial 3′,3′ cGG (Pseudomonas WspR) or 3′,3′ cAA (Bacillus DisA) synthases (Figure 2D). Importantly, cGAP1 abolished the ability of nv-cGAS to induce STING signaling (Figure 2D), consistent with nv-cGAS producing 3′,3′ cGAMP (Figure 2E). These results demonstrate that nv-cGAS produces a 3′,3′ CDN and suggest that the mixed phosphodiester-linkage chemistry found in human 2′,3′ cGAMP is an evolutionarily recent immune second messenger adaptation.
nvSTING specifically recognizes guanine bases in 3′,3′ second messengers
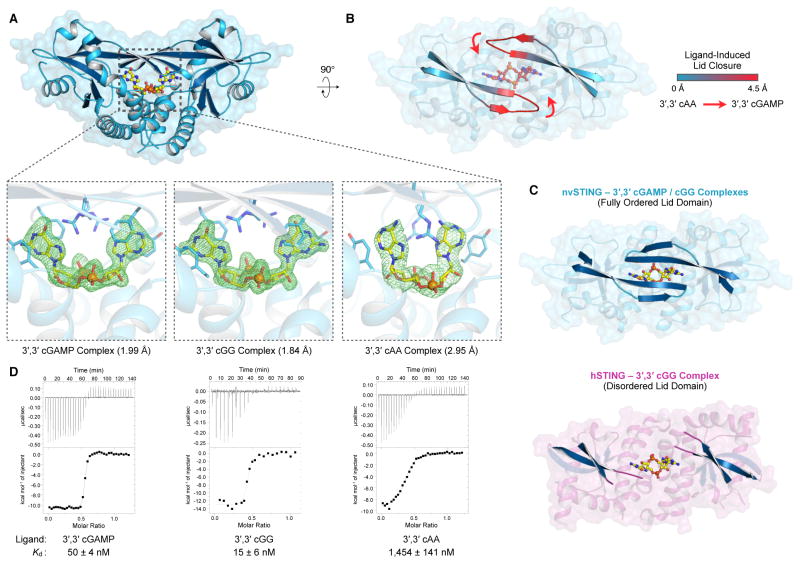
In order to characterize the interaction of nvSTING with CDN ligands, we determined crystal structures of nvSTING bound to 3′,3′ cGAMP (2.0 Å), 3′,3′ cGG (1.8 Å) and 3′,3′ cAA (3.0 Å) (Figure 3A). Interestingly, this series of structures uncovered a nucleobase-specific recognition of guanine not previously seen in mammalian STING structures. Similar to the previously reported beta-strand lid observed to form in hSTING in response to its agonist ligand, the nvSTING beta-strand lid domain undergoes an ~4 Å movement to close tightly over the top of 3′,3′ cGAMP and 3′,3′ cGG ligands, but is notably splayed open and more loosely organized when nvSTING is bound to 3′,3′ cAA (Figure 3B). In line with the contrasting activities of 3′,3′ vs. 2′,3′ second messengers, the ability of the nvSTING beta-strand lid to specifically reorganize upon 3′,3′ cGAMP or 3′,3′ cGG binding is in stark contrast to previous crystal structures of hSTING bound to 3′,3′ cGG, where the hSTING beta-strand lid remained disordered (Figure 3C) (Ouyang et al., 2012; Yin et al., 2012). These results suggest that recognition of the guanine nucleobase is a unique property of nvSTING.
Figure 3. nvSTING Specifically Recognizes Guanine Bases in 3′,3′ Second Messengers.
(A) Crystal structure of nvSTING in complex with 3′,3′ cGAMP, 3′,3′ cGG or 3′,3′ cAA as indicated. Zoomed-in cutaway includes simulated-annealing Fo–Fc omit maps of ligand density contoured to 5.0 σ (3′,3′ cGAMP, cGG) or 4.0 σ (cAA). (B) Top-down view of nvSTING–3′,3′ cAA crystal structure. The beta-strand lid domains are highlighted and colored according to range of movement compared to the closed nvSTING–3′,3′ cGAMP / cGG crystal structures. Red arrows denote the direction of beta-strand lid closure upon 3′,3′ cGAMP / cGG binding. (C) Crystal structures of nvSTING–3′,3′ cGAMP / cGG (blue) and hSTING–3′,3′ cGG (PDBs 45FY [R232] and 4F9G [H232]) (magenta) complexes. The beta-strand lid domain is highlighted illustrating the fully closed nvSTING lid domain compared with the disordered and loosely organized hSTING lid domain. (D) ITC measurements of nvSTING affinity for specific 3′,3′ CDN ligands as indicated. ITC data is representative of at least three independent experiments. See also Figure S4 and Table S1.
To validate the base-specific contacts observed in the nvSTING 3′,3′ cGAMP and 3′,3′ cGG structures, we measured ligand affinity using isothermal titration calorimetry (ITC). nvSTING exhibited a remarkably high affinity of for 3′,3′ cGAMP and cGG ligands (50 and 15 nM respectively) compared to the previously described affinity of hSTING for 3′,3′ cGG (~1,000–4,400 nM) (Figure 3D, Table S1) (Ouyang et al., 2012; Yin et al., 2012; Zhang et al., 2013). In further confirmation of the selective recognition of guanine nucleobases, the affinity of nvSTING for 3′,3′ cAA is two orders of magnitude lower (~1,500 nM) and in closer agreement to the previously measured low-micromolar affinities of hSTING and mouse STING for 3′,3′ CDNs (Figure 3D).
Structural basis of nvSTING guanine-specific cyclic dinucleotide recognition
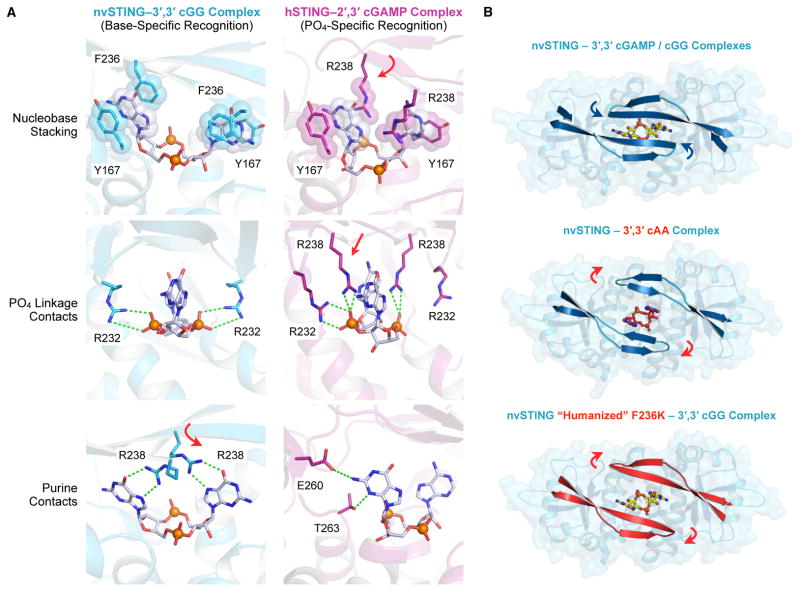
We next sought to explain the structural basis for specific recognition of guanine bases by nvSTING. In our nvSTING–CDN crystal structures we observe three main types of protein–CDN contacts; for clarity, these contacts are described in relation to the hSTING equivalent amino-acid positions (Figure 4A). First, a nucleobase stacking interaction sandwiches the purine CDN bases in place through hydrophobic base-stacking between nvSTING Y167 and F236. Second, R232 residues reach down from the organized beta-strand lid to interact with the phosphates linkage on either side of the CDN. Similar contacts are also seen between Y167 and R232 of hSTING and 2′,3′ cGAMP. However, nvSTING has additional contacts between the beta-strand lid R238 and the Hoogsteen edge of the guanine base resulting in sequence specific interactions not possible with an adenine base (Figure 4A). Interestingly, the R238 residue is also conserved in hSTING at the identical amino acid position, but in hSTING, R238 contacts the phosphate linkage rather than making guanine-specific contacts. This key difference is explained by the unique presence of phenylalanine at position 236 in nvSTING. This residue generates an additional base-stacking interaction that is not provided by the corresponding K236 in hSTING. hSTING compensates for the lack of F236 by positioning R238 over the CDN nucleobase face. In nvSTING, the presence of F236 frees R238 to make base-specific interactions.
Figure 4. Structural Basis of nvSTING Guanine-Specific Cyclic Dinucleotide Recognition.
(A) Detailed STING–CDN interactions in nvSTING–3′,3′ cGG (blue) and hSTING–2′,3′ cGAMP (magenta) complexes as described in the Results. nvSTING exhibits guanine base-specific ligand recognition while hSTING exhibits 2′,3′ phosphodiester linkage-specific ligand recognition. Variance between nvSTING and hSTING at position F236/K236 induces repositioning of R238 to support either guanine-specific contacts (nvSTING, red arrow) or phosphodiester-linkage contacts (hSTING, red arrows). For clarity, nvSTING amino acids are numbered according to hSTING sequence. (B) Top-down view of nvSTING–3′,3′ CDN crystal complexes and relative positioning of the beta-strand lid domain. The wildtype nvSTING lid domain (blue) is tightly closed over 3′,3′ cGAMP / cGG ligands (blue arrows) and in contrast the lid domain remains open in the 3′,3′ cAA bound structure (red cAA ligand and indicating red arrows). A humanizing F236K mutation in nvSTING prevents guanine-recognition and the nvSTING(F236K) lid domain (red) remains open when bound to 3′,3′ cGG. See also Figure S4.
To confirm these results and the specific importance of nvSTING F236, we created a humanized version of nvSTING with an F236K mutation and re-crystallized this mutant in complex with 3′,3′ cGG (2.1 Å). The humanized nvSTING(F236K)–3′,3′ cGG structure reveals an open beta-strand lid in the identical confirmation as the nvSTING–3′,3′ cAA structure (Figure 4B and Figure S4). Likewise, when bound to the humanized nvSTING(F236K) protein, the 3′,3′ cGG base now adopts the less rigid nvSTING–3′,3′ cAA conformation and ITC measurements demonstrate greatly reduced guanine-specific affinity (Figure S4B,C, Table S1). Together, these results demonstrate that subtle changes in the STING lid domain have a cascading affect, allowing adaptation of STING from base-specific recognition in anemone to that of the human phosphodiester-linkage specific system.
2′,3′ cGAMP traps a unique and conserved STING conformation
Collectively, our data demonstrate that the N. vectensis cGAS-STING signaling pathway relies on endogenous production and base-specific recognition of a 3′,3′ second messenger. However, our initial phylogenetic survey of STING alleles by gel-shift assay demonstrated that all functional ancestrally-related STING alleles exhibited the ability to complex with the endogenous human second messenger 2′,3′ cGAMP (Figure 1). Indeed, ITC measurements with nvSTING confirmed a specific high-affinity interaction with 2′,3′ cGAMP (<1 nM) (Figure 5A, Table S1). We hypothesized that understanding the evolutionary origins of the STING–2′,3′ cGAMP interaction might reveal the conserved structural principles underlying specific recognition of 2′,3′ cGAMP. Therefore, we determined the crystal structure of the nvSTING–2′,3′ cGAMP complex. Crystals of nvSTING–2′,3′ cGAMP grew in a unique crystal form distinct from each of the other six nvSTING structures, and we determined the structure by molecular replacement using a monomer core derived from the nvSTING–3′,3′ cGAMP structure as an initial search model.
Figure 5. 2′,3′ cGAMP Traps a Unique and Conserved STING Conformation.
(A) ITC measurements of nvSTING affinity for 2′,3′ cGAMP. (B) Structural overlay of nvSTING–2′,3′ cGAMP complex (blue) and hSTING–2′,3′ cGAMP complex (magenta) (PDB 4KSY) demonstrating the monomer wing rotation and core CDN-interacting portion of nvSTING and hSTING are unchanged. (C) Structural comparison of nvSTING structures in various ligandbound complexes. CDN ligands lock STING in alternative conformations as measured by the distance between the apical monomer wing domains (monomer 1 in blue, monomer 2 in grey). nvSTING–3′,3′ CDN interactions result in complete monomer rotation (blue vertical line) while primary hSTING–2′,3′ cGAMP signaling traps a partially rotated intermediate structure (magenta line). (D) Endogenous anemone 3′,3′ second messengers trigger an ~26° rotation in monomer wing domains from the apo state (apo grey, 3′,3′-bound in blue), while human 2′,3′ cGAMP traps an ~15° rotated structural intermediate in the nvST ING and hSTING structures (2′,3′-bound in magenta). (E) Cell interferon β luciferase as in Figure 2A, using indicated STING plasmids stimulated with human cGAS overexpression. Error bars represent the SE of the mean of at least three independent experiments (asterisk denotes p <0.002). ITC data is representative of at least three independent experiments. See also Figure S5 and Table S1.
Surprisingly, the structure of nvSTING–2′,3′ cGAMP (2.1 Å) reveals a partially rotated conformational state distinct from all previous nvSTING–3′,3′ CDN structures. In this partially rotated 2′,3′ cGAMP-bound state, the apical wings of each STING monomer are rotated by ~15° from the apo positions, in comparison to the ~26° of rotation observed in the 3′,3′ CDN-bound structures (Figure 5B,C,D). Remarkably, the partially rotated state observed in the nvSTING–2′,3′ cGAMP structure is essentially identical to the rotation observed in the hSTING–2′,3′ cGAMP structures (Figure 5B, Figure S5) (Gao et al., 2013b; Zhang et al., 2013). Although nvSTING and hSTING are only 29% identical at the amino acid level, and exhibit significant structural differences when bound to 3′,3′ CDNs, the 2′,3′ cGAMP bound structures are nearly superimposable (1.3 Å main-chain RMSD). This observation indicates that the unique chemical nature of 2′,3′ cGAMP, rather than the protein allele itself, dictates the wing pitch conformation of STING.
2′,3′ cGAMP appears to be a recent vertebrate innovation. Yet, despite this, our results demonstrate that the unique partially rotated conformation adopted by hSTING when bound to 2′,3′ cGAMP is conserved in a distant STING ortholog. To test whether such structural conservation is accompanied by functional conservation, we asked whether nvSTING could successfully replace hSTING in a human cell setting. As noted above, nvSTING lacks the long unstructured carboxy-terminal tail (CTT) previously shown to be required for hSTING to recruit the critical downstream TBK1 and IRF3 signaling components (Liu et al., 2015; Tanaka and Chen, 2012). Consistent with its lack of a CTT, wt nvSTING is unable to activate interferon β luciferase signaling upon stimulation by human cGAS (Figure 5E). Remarkably, however, appending the hSTING CTT (residues 341–379) to nvSTING is sufficient to permit low-level interferon β stimulation in human cells. The ability of nvSTING to functionally replace hSTING in human cells is further enhanced by the addition of the humanizing F236K mutation, where chimeric nvSTING(F236K) signaling is now boosted to ~20–25-fold over background levels (Figure 5E). Together these results demonstrate the minor changes required to adapt a STING allele to modern 2′,3′ cGAMP interferon signaling, and reveal a possible path for cGAS-STING functional evolution.
Discussion
In vertebrates, the cGAS-STING signaling pathway enables cells to initiate interferon-mediated gene expression in response to foreign DNA from viruses, bacteria and tumors. Although STING homologs are present in diverse animals, it has been unclear whether activation by CDNs is an evolutionarily conserved function of STING, and if so, whether STING first evolved to respond to exogenous CDNs produced by bacteria, or endogenous CDNs produced as second messengers. To determine the evolutionary origins and conserved signaling principles of cGAS-STING immunity, we investigated the pathway in divergent metazoan species. By surveying phylogenetically distant STING variants, we found that CDN binding is a deeply conserved function of STING that was likely present >500 million years ago in the common ancestor of humans and cnidaria. This observation implies that the ability to respond to CDNs via STING may have been a conserved property of the common metazoan ancestor. Furthermore, our data suggest that most features of modern innate immunity, including the evolution of interferons, followed the evolution of the STING–CDN interaction. In addition to interferon signaling, STING–CDN recognition causes downstream activation of the NF-κB pathway (McWhirter et al., 2009), and ancestral NF-kB signaling (Wolenski et al., 2011) may explain evolutionary conservation of the cGAS-STING pathway prior to interferon-based immunity.
To investigate whether cGAS, like STING, is evolutionarily ancient, we focused on the most divergent species in which we observed a robust STING–CDN interaction, the sea anemone N. vectensis. Although N. vectensis contains several candidate cGAS-like genes, these enzymes all lack defining features of human cGAS, including the Zn-ribbon that is critical for binding to dsDNA. Thus, signaling assays were essential to identify a true functional ortholog of cGAS capable of activating human STING. Our identification of nv-cGAS implies that the entire cGAS-STING signaling network was also already present >500 million years ago. Although it remains possible that STING first evolved to detect exogenous bacterial CDNs, our data suggest that the likely function of STING in the primitive metazoan ancestor was to detect an endogenous second messenger produced by a cGAS-like enzyme. Recombinant purified nv-cGAS is not active in vitro and the biochemical trigger that activates nv-cGAS remains unknown. However, the ability of nv-cGAS to signal in transfected human cells suggests that, analogous to DNA-dependent activation of human cGAS, an additional cellular co-factor or activator is able to induce nv-cGAS enzymatic activity.
Anemone cGAS appears to differ from its human ortholog in the regiochemistry of its CDN product. By taking advantage of human STING variants that respond specifically to 2′,3′ cGAMP, as well as a 3′,3′ cGAMP-specific phosphodiesterase from V. cholerae, we propose that nv-cGAS produces 3′,3′ cGAMP. This product is distinct from the 2′,3′ cGAMP produced by vertebrate cGAS enzymes, and suggests that 2′,3′ cGAMP is a recent vertebrate innovation, though we cannot formally rule out the possibility that uncharacterized enzymes in anemone or other invertebrates might also make a 2′,3′-linked CDN. Nevertheless, consistent with the apparent production of 3′,3′ cGAMP in the anemone system, crystal structures and biochemical analysis demonstrate intimate contacts within the high affinity nvSTING–3′,3′ cGAMP complex. The ability of nvSTING to specifically recognize guanine bases is particularly striking, and is a capability not seen in hSTING.
Evolution-based model of STING signaling
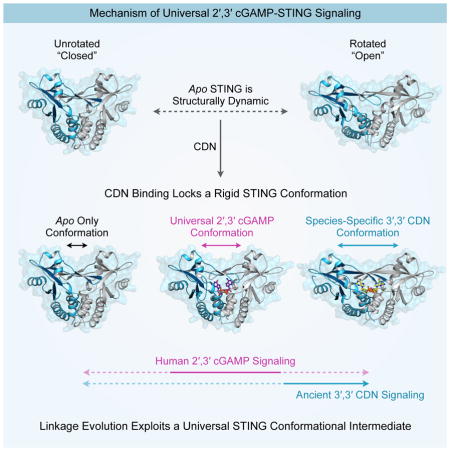
Our coherent series of seven nvSTING structural states now allow collective placement of available hSTING and mouse STING structures to illustrate a dynamic movement of STING monomers rotating against each other, analogous to the opening and closing of a butterfly’s wings. Our crystallographic studies captured two distinct apo forms of nvSTING. In one of these forms, apo nvSTING adopts an unrotated “closed” conformation similar to a previously published mouse STING apo structure (Chin et al., 2013). The other apo nvSTING structure has a rotated “open” wing pitch and exhibits a loosely organized lid, similar to published apo hSTING structures (Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012; Yin et al., 2012). The most parsimonious explanation of these results is that inactive apo STING in all species samples diverse conformations. In contrast to the existing model of hSTING activation, in which ligand binding induces a transition from a rotated-apo to a compact-bound conformation, we propose that apo STING is structurally dynamic and that the primary function of CDN binding is to stabilize a specific STING conformation (Figure 6, Movie S1).
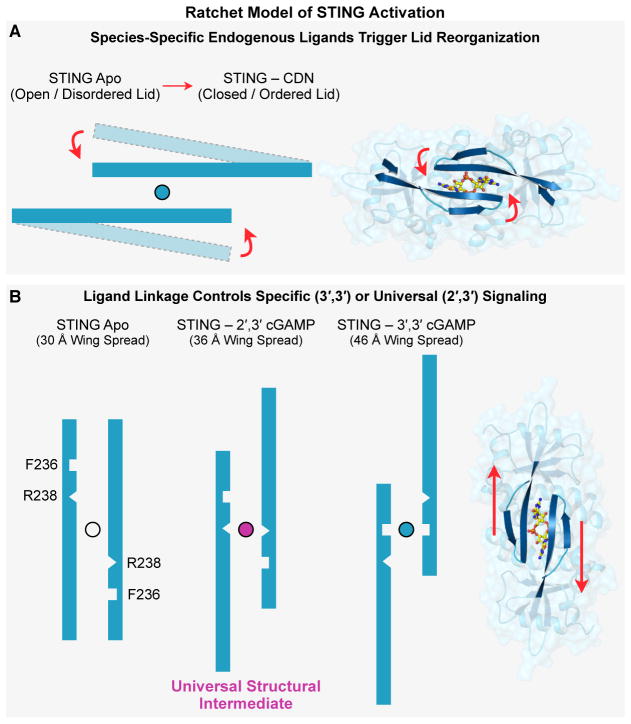
Figure 6. Ratchet Model of STING Activation.
(A,B) Ratchet model of CDN-induced conformational change leading to STING activation as described in Results. Specific lid domain interactions allow STING to discriminate endogenously produced second-messengers in a species-specific manner. Correct CDN engagement transduces reorganization of the STING lid to create large conformation changes in the apical wing domains. The unique ability of 2′,3′ cGAMP to capture a conserved structural intermediate allows universal signaling independent of STING allele diversity. See also Movie S1.
The ability of hSTING to induce downstream signaling is believed to require the formation of a beta-strand lid that covers the CDN binding pocket (Gao et al., 2013b; Zhang et al., 2013). hSTING appears to form this lid preferentially in response to 2′,3′ cGAMP. In contrast, we found that nvSTING readily forms an ordered beta-strand lid in response to multiple CDNs, including 2′,3′ cGAMP, 3′,3′ cGAMP and 3′,3′ cGG. Lid formation thus appears to be a conserved feature of STING. However, 3′,3′ CDN binding to nvSTING results in a significantly more rotated wing pitch than is observed in the hSTING–2′,3′ cGAMP structure. We conclude that key species-specific positions within the lid regulate CDN recognition to induce a ratchet-like movement that controls overall pitch of STING monomers. High affinity nvSTING–3′,3′ CDN interactions are made possible by a phenylalanine at the hSTING position 236, allowing a type of nucleobase stacking not possible with mammalian STING alleles. The uniquely compact regiochemical conformation of 2′,3′ cGAMP (Shi et al., 2015) allows it to target a more intermediately rotated state of STING where R238, a key residue involved in base-specific nvSTING–3′,3′ CDN, now completes the nucleobase stacking (Figure 6). It is possible that subsequent to the evolution of 2′,3′ cGAMP, F236 was lost to enhance discrimination of endogenous mammalian 2′,3′ cGAMP from bacterial 3′,3′ CDNs. Indeed, we observed that a humanizing F236K mutation specifically reduces 3′,3′ CDN binding while retaining 2′,3′ cGAMP recognition.
A unique feature of 2′,3′ cGAMP thus appears to be its ability to stabilize an intermediate, partially-rotated form that is not captured by canonical 3′,3′ CDNs (Figure 6). Importantly, our results demonstrate that this specific structural state, and thus the associated ability of STING to distinguish bacterial (3′,3′) from mixed-linkage (2′,3′) CDNs, is not a recent adaptation of hSTING, but is instead a fundamental and deeply evolutionarily conserved feature of the STING architecture, offering an explanation for the universal signaling potential of 2′,3′ cGAMP.
STING agonists are being developed as novel immunotherapeutics and vaccine adjuvants (Corrales et al., 2015; Deng et al., 2014; Fu et al., 2015; Woo et al., 2014). A previous small molecule STING agonist, DMXAA, showed considerable promise in pre-clinical mouse models as a potent anti-tumor agent. However, DMXAA failed in a phase III clinical trial because subtle variation in hSTING versus mouse STING affects DMXAA binding (Gao et al., 2014; Lara et al., 2011; Prantner et al., 2012). This experience suggests that a more detailed molecular understanding of how CDN binding translates into STING activation is critical for the development of therapeutics targeting STING. Our work has produced a complete set of STING structures encompassing its unbound conformational states and its conformations when bound to each of the four naturally occurring CDNs. These results identify the partially rotated 2′,3′ cGAMP-bound structure as a unique evolutionarily conserved state that provides a molecular template that we propose should be targeted in future efforts to develop effective STING agonists.
Experimental Procedures
STING–CDN Complex Gel-Shift Assay
Recombinant STING proteins were expressed and purified from E. coli and radiolabelled CDNs were enzymatically prepared according to previously developed conditions (Kranzusch et al., 2014). STING–CDN reaction mixtures were separated on a native acrylamide gel. See also supplemental experimental procedures.
Structure Determination
nvSTING was crystallized in apo form or in complex with CDN ligands by hanging drop vapor diffusion, and structures were determined using single anomalous dispersion methods. See also supplemental experimental procedures.
Cell-Based Interferon β Luciferase Assay
Synthase assays, and the N. vectensis screen for cGAS-like enzymes, were conducted in 293T human kidney cells as previously described (Diner et al., 2013; Kranzusch et al., 2014). Briefly, cells were transfected using Lipofectamine 2000 (Invitrogen) in a 96-well format as indicated, and at 24 h post-transfection control Renilla and interferon β Firefly luciferase values were assayed as previously described (Lee et al., 2015). Data were combined from multiple experiments and analyzed by an unpaired, two-tailed t test in Prism.
Supplementary Material
Highlights.
Binding of CDNs is an evolutionarily ancient STING function, predating interferons
cGAS-STING function is conserved in anemone, >500 million years diverged from humans
Anemone cGAS produces a canonical 3′,3′ linked CDN similar to those in bacteria
Vertebrate 2′,3′ cGAMP signaling exploits a deeply conserved STING conformation
Acknowledgments
X-ray data were collected at the Lawrence Berkeley National Lab Advanced Light Source (beamline 8.3.1). The authors are grateful to D. Burdette, E. Diner, and M. Raulet for assistance with initial identification of animal STING homologs; A. Whiteley for phosphodiesterase advice; J. Holton, G. Meigs, and A. Gonzalez for technical assistance with data collection and processing; R. Wilson for advice on ITC experiments; and members of the Berger, Doudna and Vance labs for helpful comments and discussion. This work was funded by HHMI (R.E.V. and J.A.D.), NIH P01 AI063302 (R.E.V), G. Harold and Leila Y. Mathers Foundation (J.M.B.), and NIGMS Center for RNA Systems Biology (A.S.Y.L. and J.A.D.). P.J.K. is supported as an HHMI Fellow of the Life Sciences Research Foundation, and A.S.Y.L. is supported as an American Cancer Society Fellow (PF-14-108-01-RMC). J.A.D. and R.E.V. are HHMI Investigators.
Footnotes
Accession Numbers
Coordinates of nvSTING cGG, 3′,3′ cGAMP, cAA, apo “rotated”, (F276K) cGG, 2′,3′ cGAMP and apo “unrotated” have been deposited in the RCSB Protein Data Bank under accession numbers 5CFL, 5CFM, 5CFN, 5CFO, 5CFP, 5CFQ and 5CFR.
Supplemental information includes five figures, one movie, one table and supplemental experimental procedures.
Author Contributions
Experiments were designed by P.J.K., S.C.W., J.A.D. and R.E.V. in consultation with J.M.B. All structural and biochemical experiments were conducted by P.J.K. and S.C.W. A.S.Y.L. performed cell-signaling assays with assistance from P.J.K. and S.C.W. The manuscript was written by P.J.K., S.C.W., J.A.D., and R.E.V., and all authors contributed to editing the manuscript and support the conclusions.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaauboer SM, Gabrielle VD, Jin L. MPYS/STING-mediated TNF-alpha, not type I IFN, is essential for the mucosal adjuvant activity of (3′-5′)-cyclic-di-guanosine-monophosphate in vivo. Journal of immunology. 2014;192:492–502. doi: 10.4049/jimmunol.1301812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Chin KH, Tu ZL, Su YC, Yu YJ, Chen HC, Lo YC, Chen CP, Barber GN, Chuah ML, Liang ZX, et al. Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING. Acta crystallographica Section D, Biological crystallography. 2013;69:352–366. doi: 10.1107/S0907444912047269. [DOI] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo SR, Lemmens E, Banda T, Leong JJ, et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell reports. 2015 doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154:962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, Mechette K, Leong JJ, Lauer P, Liu W, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Science translational medicine. 2015;7:283ra252. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Tao J, Liang W, Zhao M, Du X, Cui S, Duan H, Kan B, Su X, Jiang Z. Identification and characterization of phosphodiesterases that specifically degrade 3′3′-cyclic GMP-AMP. Cell research. 2015 doi: 10.1038/cr.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013a;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013b;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Zillinger T, Wang W, Ascano M, Dai P, Hartmann G, Tuschl T, Deng L, Barchet W, Patel DJ. Binding-pocket and lid-region substitutions render human STING sensitive to the species-specific drug DMXAA. Cell reports. 2014;8:1668–1676. doi: 10.1016/j.celrep.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Hartley MK, Hagan C, Clark NL, Elde NC. Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape. PLoS genetics. 2015;11:e1005203. doi: 10.1371/journal.pgen.1005203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature structural & molecular biology. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. Journal of immunology. 2011a;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Molecular and cellular biology. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, Cambier JC. Identification and characterization of a loss-of-function human MPYS variant. Genes and immunity. 2011b;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Ishii R, Hirano S, Ishitani R, Nureki O. Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase. Structure. 2015 doi: 10.1016/j.str.2015.01.023. [DOI] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell reports. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Wilson SC, Solovykh MS, Vance RE, Berger JM, Doudna JA. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell. 2014;158:1011–1021. doi: 10.1016/j.cell.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara PN, Jr, Douillard JY, Nakagawa K, von Pawel J, McKeage MJ, Albert I, Losonczy G, Reck M, Heo DS, Fan X, et al. Randomized phase III placebo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent vadimezan (ASA404) in advanced non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:2965–2971. doi: 10.1200/JCO.2011.35.0660. [DOI] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. The Journal of experimental medicine. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner D, Perkins DJ, Lai W, Williams MS, Sharma S, Fitzgerald KA, Vogel SN. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates stimulator of interferon gene (STING)-dependent innate immune pathways and is regulated by mitochondrial membrane potential. The Journal of biological chemistry. 2012;287:39776–39788. doi: 10.1074/jbc.M112.382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infection and immunity. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nature structural & molecular biology. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- Shi H, Wu J, Chen ZJ, Chen C. Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1507317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nature structural & molecular biology. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science signaling. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolenski FS, Garbati MR, Lubinski TJ, Traylor-Knowles N, Dresselhaus E, Stefanik DJ, Goucher H, Finnerty JR, Gilmore TD. Characterization of the core elements of the NF-kappaB signaling pathway of the sea anemone Nematostella vectensis. Molecular and cellular biology. 2011;31:1076–1087. doi: 10.1128/MCB.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, Duggan R, Wang Y, Barber GN, Fitzgerald KA, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annual review of immunology. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- Wu X, Wu FH, Wang X, Wang L, Siedow JN, Zhang W, Pei ZM. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic acids research. 2014;42:8243–8257. doi: 10.1093/nar/gku569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Molecular cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li L, Guo X, Litman GW, Dishaw LJ, Zhang G. Massive expansion and functional divergence of innate immune genes in a protostome. Scientific reports. 2015;5:8693. doi: 10.1038/srep08693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. Journal of immunology. 2014;193:2394–2404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Zhu D, Wang L, Shang G, Liu X, Zhu J, Lu D, Wang L, Kan B, Zhang JR, Xiang Y. Structural biochemistry of a Vibrio cholerae dinucleotide cyclase reveals cyclase activity regulation by folates. Molecular cell. 2014;55:931–937. doi: 10.1016/j.molcel.2014.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.