Abstract
T follicular regulatory cells (TFR) are a suppressive CD4+ T cell subset that migrates to germinal centers (GC) during antigen presentation by up-regulating the chemokine receptor CXCR5. In the GC, TFR control T follicular helper cells (TFH) expansion and modulate the development of high-affinity antigen specific responses. Here we identified and characterized TFR as CXCR5+ CCR7−-“follicular” T regulatory cells (TREG) in lymphoid tissues of healthy rhesus macaques, and we studied their dynamic throughout infection in a well-defined animal model of HIV pathogenesis. TFR were infected by SIVmac251 and had comparable levels of SIV-DNA to CXCR5− CCR7+-“T-zone” TREG and TFH. Contrary to the SIV-associated TFH expansion in the chronic phase of infection, we observed an apparent reduction of TFR frequency in cell suspension, as well as a decrease of CD3+ Foxp3+ cells in the GC of intact lymph nodes. TFR frequency was inversely associated with the percentage of TFH and, interestingly, with the avidity of the antibodies that recognize the SIV-gp120 envelope protein. Our findings show changes in the TFH/TFR ratio during chronic infection and suggest possible mechanisms for the unchecked expansion of TFH cells in HIV/SIV infection.
Introduction
The generation of long-lived plasma cells and high affinity antibodies is largely dependent on T-B-cell interaction in the B-follicles of secondary lymphoid organs (1) (2) (3). Antigen-activated B cells making contact with a specialized subset of CD4+ T cells, called T follicular helper cells (TFH), can enter the germinal centers (GCs) to undergo to somatic hypermutation and affinity maturation (4). TFH home to B follicles and GC (5) (6, 7) (8, 9) by up-regulating the chemokine (C-X-C motif) receptor 5 (CXCR5) and down-regulating the chemokine (C-C motif) receptor 7 (CCR7) (10)(5)(6). TFH express high levels of programmed death 1 (PD-1), inducible co-stimulator (ICOS), and Bcl-6, a master transcriptional regulator that orchestrates TFH differentiation (11)(12)(13). In the GC, TFH provide signals for B-cell survival and differentiation (10)(5) via IL-21 production and CD40L expression, and they promote the generation of antibodies with high affinity (11)(12)(13, 14)(15)(16).
GC reactions are tightly regulated to prevent the emergence of B-cell clones that are specific or cross-reactive against self-antigens, while selecting for high affinity antibodies to microbes (17)(18). The maintenance of the appropriate number of TFH is crucial (19); the absence of TFH has a negative impact in the generation of the GC (20)(21), while their excessive accumulation leads to increased GC reactions and the onset of some autoimmune diseases (4)(22)(23)(24).
CD4+ T follicular regulatory cells (TFR) contain TFH numbers and in doing so, they control the magnitude of GC responses (25) (26). Similarly to TFH, TFR migrate to the GC by expressing CXCR5 and down regulating CCR7 during T-cell activation (6)(27)(28)(29)(25). TFR differentiate from natural CXCR5− Foxp3+ CD25+-TREG and express high levels of the typical TREG markers (i.e. Foxp3, CD25, CTLA-4) and TFH canonical markers such as ICOS, PD-1 and Bcl-6 (25) (26). While Bcl-6 is essential for CXCR5 expression on B- and TFH cells and for their localization to the GC (25)(26), TFR co-express Blimp-1, which is known to repress CXCR5 expression (25)(30). Ablation of the activated T-cell nuclear factor (NFAT)-2 in mice results in reduced expression of CXCR5 on TFR, but not on TFH, suggesting that this transcriptional factor may enable the proper localization of TFR within B-cell follicles, possibly by inhibiting Blimp-1–mediated repression of CXCR5 expression (31). TFR restrict TFH numbers, and help to maintain a steady ratio of IgM+ to IgM− (switched) B cells (32) via IL-10 production (29); in vivo depletion of CD4+ T cells with suppressive activity including TFR, or in vivo blockade of IL-10 or transforming grow factor –β (TGF-β) receptors results in TFH expansion, loss of normal proportion of IgM− B cells and in increased levels of high affinity antibodies (26) (29)(33).
A hallmark of HIV and SIV infection is the immune dysfunction of humoral responses characterized by loss of memory B cells and hypergammaglobulinemia (34) (35). TFH frequency is significantly increased in the lymph nodes of HIV infected individuals and chronically SIVmac251 infected macaques (8)(36). Production of the IL-21 cytokine by TFH is significantly reduced during HIV/SIV infection, possibly affecting GC homeostasis and the development of effective humoral responses to the virus (37). The HIV/SIV associated changes in TFH number and function may contribute to the impairment of B-cell responses (9)(36)(38), however other studies have found associations between the levels of functional TFH and broadly neutralizing antibodies in chronic HIV patients (39). While the relative role of TFH in HIV pathogenesis needs further investigation, it would be important to understand the molecular and cellular mechanisms that regulate TFH expansion.
TFR dynamic in HIV-infection has not been investigated yet. We identified TFR as CXCR5+-TREG in the lymph nodes of rhesus macaques, a well-established model of HIV infection. We show that 1) TFR are infected by SIVmac251, 2) there is an apparent decrease in TFR levels, particularly during chronic infection, 3) TFR levels are associated with the levels of TFH and the total frequency of IgG+ B cells, and 4) TFR levels are inversely correlated with the avidity of antibodies to SIV-gp120 protein. Taken together these findings suggest a potential role for TFR in modulating humoral responses against HIV/SIV.
Materials and Methods
Animals and challenge
All of the animals used in this study were colony-bred rhesus macaques (Macaca mulatta) obtained from Covance Research Products (Alice, TX). The animals were housed and maintained in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All surgery was performed under general anesthesia, and all efforts were made to minimize suffering. All macaques were negative for simian retrovirus, simian T-cell leukemia virus type 1, and herpesvirus B. Macaques were infected with a single high dose (6300 TCID50) (40) or with 10 repeated low doses (120 TCID50) of SIVmac251 given rectally (41) (Table I).
Table I.
Serological data, Acute and Chronic animals
| Acute | Challenge | Weeks p.i | VL | CD4 count |
|---|---|---|---|---|
| P061 | High dose I.R | 2 | 5806601 | 748 |
| P066 | High dose I.R | 2 | 25556960 | 489 |
| P074 | High dose I.R | 2 | 30925780 | 333 |
| P136 | High dose I.R | 2 | 39844500 | 382 |
| P273 | High dose I.R | 2 | 34856014 | 524 |
| P251 | Low dose I.R | 3 | 115008398 | 647 |
| P434 | Low dose I.R | 3 | 215693077 | 517 |
| P650 | Low dose I.R | 3 | 23469446 | 489 |
| P716 | Low dose I.R | 3 | 108209623 | 454 |
| P712 | Low dose I.R | 3 | 49568224 | 757 |
| Chronic | Challenge | Weeks p.i | VL | CD4 count |
|---|---|---|---|---|
| P015 | High dose I.R | 14 | 121368 | 485 |
| P061 | High dose I.R | 14 | 368663 | 928 |
| P074 | High dose I.R | 14 | 11883300 | 437 |
| P134 | High dose I.R | 14 | 16963740 | 260 |
| P136 | High dose I.R | 14 | 19165270 | 340 |
| P266 | High dose I.R | 15 | 172649 | 265 |
| P273 | High dose I.R | 15 | 163587 | 360 |
| P274 | High dose I.R | 15 | 1297398 | 265 |
| E060 | Low dose I.R | 14 | 4957329 | nd |
| E074 | Low dose I.R | 14 | nd | nd |
| P632 | Low dose I.R | 12 | 7825190 | 111 |
| P761 | Low dose I.R | 15 | 31347 | 1330 |
| P758 | Low dose I.R | 15 | 24934 | 1431 |
| P760 | Low dose I.R | 15 | 1425169 | 426 |
| R158 | Low dose I.R | 15 | 1440103 | 820 |
| P733 | Low dose I.R | 15 | 1358565 | 212 |
| P746 | Low dose I.R | 15 | 143386 | 618 |
| P754 | Low dose I.R | 15 | 2168071 | 734 |
| P698 | Low dose I.R | 15 | 21182252 | 572 |
| P609 | Low dose I.R | 15 | 5462462 | 239 |
| P251 | Low dose I.R | 15 | 111656 | 406 |
| P633 | Low dose I.R | 15 | 1104050 | 514 |
| P761 | Low dose I.R | 15 | 147114 | 1330 |
Viral load: SIV/RNA copies/ml plasma Absolute CD4+ T number/mm2 of blood
I.R= intrarectal; p.i. post infection; VL= viral load
Cell isolation from lymph nodes and mucosa
Cells were isolated from blood, lymph nodes and spleen by density-gradient centrifugation. Tissues from the rectum, jejunum and colon were treated with 1 mM Ultra Pure DTT (Invitrogen Life Technologies) for 30 min followed by incubation in calcium/magnesium-free HBSS (Invitrogen Life Technologies) for 60 min with stirring at room temperature to remove the epithelial layer. Lamina propria lymphocytes were separated by cutting the tissue into small pieces and incubating in 10% FBS IMDM (Invitrogen Life Technologies) with collagenase D (400 U/ml; Boehringer Mannheim) and DNase (1 μg/ml; Invitrogen Life Technologies) for 2.5 h at 37°C. Mononuclear cells were placed over 42% Percoll (General Electric Healthcare) and centrifuged at 800 × g for 25 min at 4°C. Lamina propria lymphocytes were collected from the cell pellet (42).
Antibodies & staining
We used the following antibodies: CD3-Alexa700 (SP34-2), CD4-PerCP-Cy5.5 (L200), CD95-PeCy5 (DX2), CD197-PeCy7 (CCR7, clone 3D12), CD25-APCcy7 (M-A251), CD195-PE (CCR5, clone 3A9), CD14-Alexa700 (M5E2), CD16-Alexa700 (3G8), CD56-Alexa700 (B159), IgM-PerCP-Cy5.5 (G20-127), IgG-Qdot605 (G18-145), Ki67-PE (B56), CD21-PECy7 (B-ly4), all from BD Biosciences; Bcl-6-PE (IG191E/A8); CD278-PacBlue (ICOS, clone C398.4A), CD25-PacBlue (BC96), PD-1-APC (EH12.2H7), CD39-BV421 (MOCP-21), CD39 (A1) from BioLegend; Foxp3-FITC (PCH101), CXCR5-PE-eFluor610 (MU5UBEE), CD20-Qdot650 (2H7) all from eBioscience; CD103-FITC (αE integrin, clone 2G5), CD19-PE-Cy5 (J3-119), CD127-PE (eBioRDR5) from Beckman Coulter. The α4β7 antibody (Act-1) was obtained through the NIH Nonhuman Primate Reagent Resource Program (AIDS Reagent Program, Division of AIDS, NIAID, NIH). Vivid-amine-reactive dye was used to discriminate live/dead cells (Invitrogen). IgA-Texas Red (polyclonal) was obtained from SouthernBiotech, and CD38-FITC (clone AT-1) from StemCell.
For phenotypic characterization of CD4+ T cells subsets, cells were stained with surface markers CD3, CD4, CD95, CD25, CCR7, CXCR5, ICOS and Vivid. Cells were then fixed and permeabilized according to eBioscience’s instructions and stained with anti-Foxp3 and anti-Bcl-6 for 30 min. The appropriate isotype-matched control Ab was used to define positivity. TFR cells were gated as live CD3+ CD4+ CD95+ T cells, and their percentage was calculated as the frequency of CXCR5+ & CCR7− within Foxp3+ CD25+ cells (% of TREG) or within CD95+ CD4+ T cells (% of memory CD4+ T cells) (26). Similarly, Double Positive were CXCR5+ & CCR7+ cells and CCR7+-TREG were CXCR5− & CCR7+. Finally, TFH cells were gated as CXCR5+ PD-1hi cells within the Foxp3 negative region or the memory CD4+ T cells population (Figure 1A).
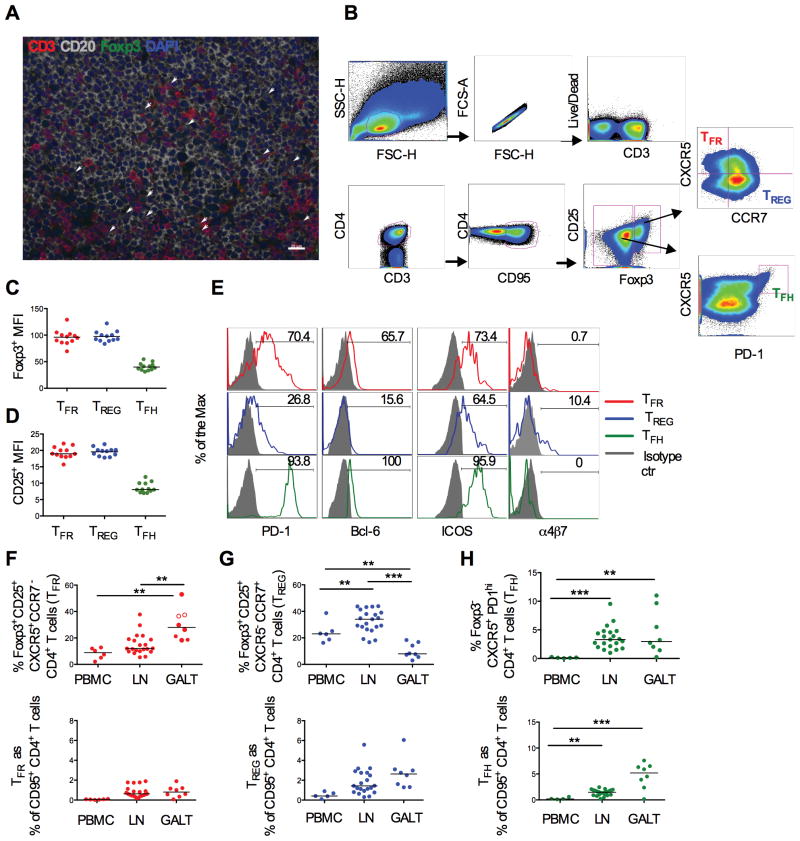
Figure 1. Characterization and distribution of TFR in SIV-uninfected macaques.
(A) Double positive CD3+ (red) and Foxp3+ cells (green) are present in B-follicles (CD20 in grey) in lymph nodes from a naïve macaque (blue =DAPI; scale bar: 20μm). (B) Representative flow cytometry plots showing the gating strategy for TFR, CCR7+-TREG and TFH cells in lymph nodes. All the subsets were gated on singlet/live/CD3+ CD4+ CD95+. TFR and CCR7+-TREG were identified as Foxp3+ CD25+ and CXCR5+ CCR7− or CXCR5− CCR7+, respectively; TFH cells as Foxp3− CXCR5+ PD-1high. (C) Geometric mean (MFI) of Foxp3 and (D) CD25 expression. (E) Cell surface expression of PD-1, Bcl-6 ICOS and α4β7 in TFR (red), TREG (blue) and TFH cells (green). Isotype controls are in grey. Frequency of (F) CXCR5+ & CCR7− cells and (G) CXCR5− & CCR7+ cells within Foxp3+ CD25+ CD4+ T (upper panel) or within memory CD4+ T cells (corresponding lower panels) in blood, peripheral lymph nodes and in the GALT (colon, jejunum and rectal mucosa) of naïve animals. The median is shown. (H) TFH frequency on Foxp3 negative (upper panel) and memory CD4+ T cells (lower panel) in different tissues.
B cells were gated as live/lineage negative (linneg: CD3− CD14− CD16− CD56−) positive for CD20 and or CD19 markers. For plasmablasts, cells were stained with lineage markers, CD20, CD38, CD39, IgM, IgG and IgA. Cells were treated with Cytofix/Cytoperm (BD Biosciences) and stained with Ki67. Plasmablasts (PBs) were gated as lineage negative (linneg CD20+ and/or CD19+ CD21− Ki67+ CD38+ CD39+ (43). Marker expression was analyzed with a LSRII flow cytometer using FACSDiva software (BD Biosciences). FACS analysis was performed using FlowJo software (Tree Star, Inc., Ashland, OR). A minimum of 10,000 cells per tube was analyzed.
CD4+ T cells counts
The absolute number of CD4+ T cells was calculated as previously described (44).
Sorting
To determine the RNA levels for IL-10, TGF-β and SIV-DNA, cells from lymph nodes were stained with Vivid, CD3, CD4, CD25, CCR7 and CXCR5. TFR cells were defined as live CD3+ CD4+ CD25+ CXCR5+ & CCR7−, TREGS as CXCR5− & CCR7+. For proliferation, CD25+ CD4+ T cells (live CD3+ CD4+ CD25+) were sorted. Sorting was performed on a FACS ARIA (BD Biosciences).
Migration assay
Sorted live CD3+ CD4+ CD25+ cells were migrated for 1 hour to 1μg/ml CXCL13 (R&D Systems, Cat no. 801-CX/CF) using 5μm Pore Polycarbonate Membrane inserts (Millipore).
Proliferation
Cell proliferation was determined by dilution of CFSE (Life Technologies). Briefly, CD25-depleted (CD3+ CD4+) cells were stained with CFSE for 10 minutes and were then placed in a 24-well plate in the presence or absence of CD3 (10 μg/ml; clone FN18) with soluble αCD28 (1 μg/ml; clone CD28.2), in the presence or absence of autologous CD3+ CD4+ CD25+ cells migrated to CXCL13 (10:1 ratio), for 4 days. Cells were then stained and analyzed by FACS as described above.
Cyclosporin A in vitro treatment
Cells from lymph nodes were incubated 30h with 50ug of cyclosporine A (Sigma) and incubated for 6h with or without PMA and in the presence of brefeldin A (Golgi-Plug; BD Biosciences).
RT-PCR
Total RNA was extracted from whole tissue with RNeasy Plus (Qiagen) and reverse transcribed with the High-capacity cDNA reverse transcription kit (Applied Byosistems). After reverse transcription the relative amounts of transcripts were determined by real-time PCR with the SYBR Green qPCR Master Mix (Promega) using 0.2 μM of PCR primers for IL-10 (5′-AGAACCACGACCCAGACATC-3′ (forward), 5′-GGCCTTGCTCTTGTTTTCAC-3′ (reverse)), and transforming growth factor β (TGF-β). The TGF-β was described elsewhere (45). Quantification of cDNA was normalized in each reaction according to the internal β-actin control (5′-GGCACCCAGCACAATGAAG-3′ (forward), and 5′-GCTGATCCACATCTGCTGG-3′ (reverse)). A real-time nucleic acid sequence-based amplification (NASBA) assay was used to quantitative SIV-RNA in plasma (46). SIV-DNA was quantified as previously described (40).
Immunohistochemistry in lymph nodes
The primary antibodies included Anti-Foxp3 (Abcam, Rabbit), Anti-CD20 (DAKO, mouse Ig2a) and Anti-CD3 (UCD Rat). Tris-buffered saline with 0.05% Tween 20 was used for all washes. Antibody diluent (Dako, Inc., Carpinteria CA) was used for all antibody dilutions. For all primary antibodies, slides were subjected to an antigen retrieval step consisting of incubation in AR10 (Biogenex Inc, San Ramon, CA) for 2 min at 125 °C in the Digital Decloaking Chamber (Biocare Medical, Concord, CA) followed by cooling to 90 °C before rinsing in water. Primary antibodies were replaced by normal rabbit IgG, mouse IgG (Invitrogen, Grand Island, NY) and rat IgG (Vector, Burlingame, CA) were included with each staining series as the negative control. Nonspecific binding sites were blocked with 10% goat serum and 5% bovine serum albumin (BSA, Jackson ImmunoResearch, West Grove, PA). Binding of the primary antibodies was detected simultaneously using Alexafluor Goat anti Rat 568, Alex fluor Goat anti Mouse IgG2a 647, and Alex fluor Goat anti Rabbit 488. All slides were coverslipped using ProLong Gold with 4′, 6-diamidino-2-phenylindole dihydrochloride hydrate (DAPI, Molecular probes, Grand Island, NY) to stain nuclei. All the control experiments gave appropriate results with minimal nonspecific staining.
Slides were visualized with epi-fluorescent illumination using a Zeiss Axioplan 2 microscope (Carl Zeiss Inc., Thornwood, NY) and appropriate filters. Digital images were captured and analyzed by using Openlab software (Inprovision, Waltham, MA). Alex flour 647 was captured in black and white channel while other fluorescence dyes were pictured in color channels. Five high-power (40x) microscope fields were randomly chosen and captured digitally with the system described above. Each captured field includes an area of approximately 0.04 mm2. Only clearly positive cells with distinctly labeled nuclei (DAPI) and bright staining were considered positive. Individual positive cells in the five captured high-power microscope fields of the immunohistochemical stained tissue sections were counted manually by a single observer. The numbers of positive cells are presented as cells per square millimeter.
Avidity assay
Avidity was analyzed as previously described (41). Briefly, recombinant SIV gp120 protein made from codon-optimized SIVmac239 gp120 fused to the C-terminal tag of HIV-1 gp120 was used as an antigen for the capture ELISA to detect SIV Abs against conformational epitope. Antibody avidity was determined by parallel ELISA. Heat-inactivated plasma samples were serially diluted and applied to a 96-well plate capturing SIVmac239 gp120. After 1 h of incubation, the plate was washed and half the samples were treated with Tris-buffered saline (TBS), while the paired samples were treated with 1.5 M sodium thiocyanate (NaSCN; Sigma-Aldrich) for 10 min at room temperature. The plate was washed and a goat anti-monkey IgG-detecting Ab (Fitzgerald) was used. The avidity index (%) was calculated by taking the ratio of the NaSCN-treated plasma dilution giving an OD of 0.5 to the TBS-treated plasma dilution giving an OD of 0.5 and multiplying by 100. Plasma of uninfected normal macaques served as negative controls.
Statistical analysis
Tests of two groups of animals for differences between cell types, tissues or stages of infection were performed using the exact Wilcoxon rank sum test. Differences before and after infection within the same group of animals were assessed using the Wilcoxon signed rank test. Differences across three stages of infection were modeled using repeated measures analysis of variance when distributional assumptions were met. Correlation analyses were performed using the exact Spearman rank correlation method. Trends across three groups were assessed by the Jonckheere-Terpstra test. Due to the exploratory nature of this study, the p values reported were not corrected for multiple comparisons. Values of p ≤ 0.05 were labeled statistically significant, and we note that for outcomes where all pairwise comparisons of three groups are possible, values of p ≤ 0.02 remain significant after correction for the multiple tests.
Results
Characterization and tissue distribution of TFR in naïve Rhesus macaques
TFR localize in the GC of mice and humans (29)(25)(26)(47). We confirmed the presence of Foxp3+ CD3+ T cells in healthy macaque’s B-cell follicles of lymph nodes, by immunohistochemistry (Figure 1A). We then characterized TFR in cell suspension by flow cytometry, and compared their frequency, phenotype and localization to that of CCR7+-TREG and TFH cells. The gating strategy used to define these 3 cell subsets is shown in Figure 1B. TFR and TREG were gated within the live Foxp3+ CD25+ CD95+ CD4+ T cells, and defined as CXCR5 positive and CCR7 negative, consistent with GC location, and as CXCR5 negative and CCR7 positive, consistent with T-zone location, respectively (Figure 1B). Of note, the TREG population identified by this strategy only includes a specific subset based on the expression or the lack thereof of the two considered chemokines. We will refer to this subset as CCR7+-TREG or TREG for simplicity.
In agreement with Sage et al. and Linterman et al., we used the Foxp3 marker to distinguish between TFH and TFR subsets because both populations express CXCR5, PD-1, ICOS and Bcl-6 (48)(25). TFH were defined as Foxp3neg CXCR5+ PD-1high CD4+ T cells (Figure 1B).
Consistent with their mouse counterparts, macaque TFR expressed comparable levels of Foxp3 (Figure 1C), equal intensity and frequency of CD25, and frequency of CD39 to TREG (Figure 1D and Supplemental Figure 1A and B) (25)(26). TFR were also negative for CD127, the marker for the IL-7 receptor, and expressed common TFH markers, such as PD-1, Bcl-6 and ICOS (Supplemental Figure 1C and Figure 1E) (28). Only a subset of TREG but not of TFR or TFH cells was positive for the αE (CD103) and α4β7 integrin (25)(26) (Figure 1E and data not shown).
Within the Foxp3+ CD25+ population we identified an additional CXCR5+ CCR7+ double positive (DP) cell subset (Figure 1B). DP cells expressed intermediate levels of PD-1 as compared to TFR and TREG and had equal levels of Bcl-6 to TFR cells (Supplemental Figure 1D).
We looked at the distribution of TFR, TREG and TFH in blood, peripheral lymph nodes, and in the gut-associated lymphoid tissue (GALT; colon, jejunum and rectal mucosa) obtained from 6, 21 and 8 naïve macaques, respectively (Figure 1F–H). Representative flow plots obtained from blood, lymph node and rectal mucosa tissue from one healthy animal are shown in Supplemental Figure 1E. TFR were mainly in the GALT and lymph nodes, and only a few were detected in blood (Figure 1F and Supplemental Figure 1E). Within the Foxp3+ CD25+ population, cells that expressed only CXCR5 were less frequent in lymph nodes than in the GALT (p<0.0001 by the Wilcoxon rank sum test; Figure 1F, upper panel), and the opposite was observed for cells that expressed only CCR7 (p<0.0001 by the Wilcoxon rank sum test; Figure 1G, upper panel). The apparent difference in tissue distribution in lymph nodes and GALT was lost when we looked at the frequency of TFR and TREG within the memory CD4+ T cell population (lower panels Figure 1F and G). DP cells were equally distributed among all the tissues analyzed, including the blood (Supplemental Figure 1F). Finally TFH frequency was significantly higher in lymph nodes and in the GALT than in the blood, as previously described (PBMCs vs. LN, p<0.0001; PBMCs vs. GALT, p=0.0031) (49) (Figure 1H).
Because we could not determine whether DP cells home exclusively to the B-zone, we excluded them from the rest of the analysis.
Macaque TFR suppress CD4+ T cells and TFH proliferation of in vitro
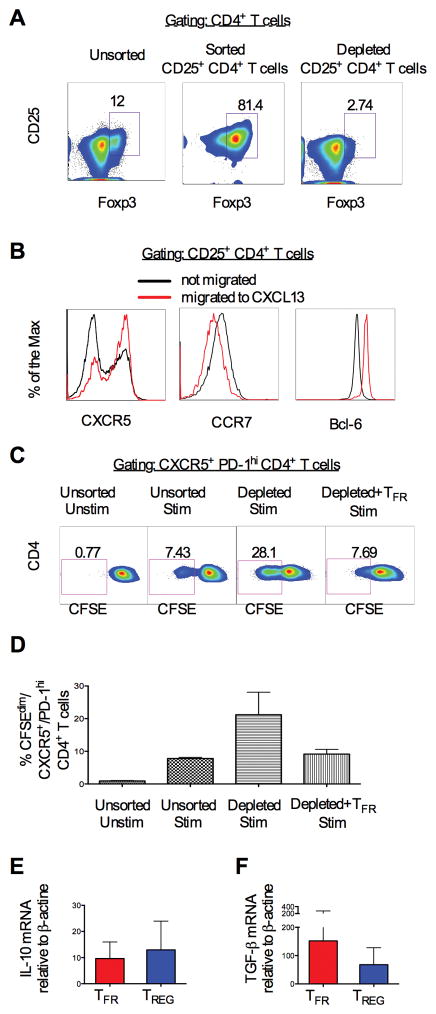
In mice, TFR control TFH numbers and decrease their proliferation in vivo and in vitro (26). We studied if macaque’s lymph nodes also contained a suppressive CD4+ T cell population that homes to the B follicles. We sorted CD4+ CD25+ live cells from the lymph nodes of 2 naïve animals and isolated those capable of migrating in response to CXCL13, the ligand for CXCR5 (Figure 2). While we could not use Foxp3, an intracellular marker, to discriminate suppressor CD4+ T cells, sorted CD25+ CD4+ T cells from lymph nodes consisted primarily of Foxp3+ CD4+ T cells (Figure 2A). Regulatory CD4+ T cells that migrated to CXCL13 had higher levels of CXCR5, lower levels of CCR7 and expressed higher levels of Bcl-6 than those that did not migrate (Figure 2B). This strategy allowed us to obtain a population of CD4+ T cells, highly enriched for TFR, which could be used in downstream functional assays. Unsorted and sorted cells were stimulated with or without CD3 and CD28, in the presence or absence of migrated CD25+ CD4+ T cells (follicular-TREG-enriched population). We assessed proliferation (% of CFSEdim cells) within CXCR5+ PD-1high CD4+ T cells (TFH gated cells) by FACS analysis. Representative plots of the gated CXCR5+ CD4+ T cells (Gate 1) are shown in Supplemental Figure 2A. Interestingly, in both the two lymph nodes used for this analysis, TFH proliferated less in the presence of TFR-enriched cells following stimulation than in the absence of CD25+ cells, as shown in the representative plots in Figure 2C and graphically in Figure 2D. As expected, TFR also reduced non-TFH CD4+ T cells subsets proliferation (Gate 2), Supplemental Figure 2B).
Figure 2. TFR suppress in vitro TFH cell proliferation.
(A) Representative density plot showing unsorted (upper panel), sorted CD25+ CD4+ T cells (central panel) and CD25+ CD4+ T-depleted cells (lower panel) from a lymph node of a naïve macaque. (B) Cell surface expression of CXCR5, CCR7 and Bcl-6 on CD4+ T cells that migrated (red line) or did not migrate (black line) to CXCL13. (C) Representative density plot showing proliferation (CFSEdim) of stimulated (CD3, CD28) unsorted, CD25+ depleted CD4+ T cells alone or co-cultured with CXCL13 migrated-CD25+ CD4+ T cells. (D) Percentage of proliferating CXCR5+ PD1high CD4+ T cells in all conditions. The bars represent the mean ± standard error from 3 lymph nodes. (E) IL-10 and (F) TGF-β mRNA measured by RT-PCR from TFR and CCR7+-TREG sorted from lymph nodes of 4 naïve animals. The bars represent the mean ± standard error.
Consistent with their regulatory function and similar to TREG, TFR produce IL-10 and TGF-β, which together suppress the proliferative potential and function of CD4+ T cells in mice (50) (51). Thus, we measured the levels of IL-10 and TGF-β in enriched populations of TFR (CD3+ CD4+ CD25+ CXCR5+ CCR7−) and CCR7+-TREG (CD3+ CD4+ CD25+ CXCR5− CCR7+) obtained from peripheral lymph nodes of 3 naïve macaques. TFR had equivalent IL-10 and TGF-β mRNA levels, by RT-PCR, than CCR7+-TREG (Figure 2E and F).
SIVmac251 infection and TFR cell frequency
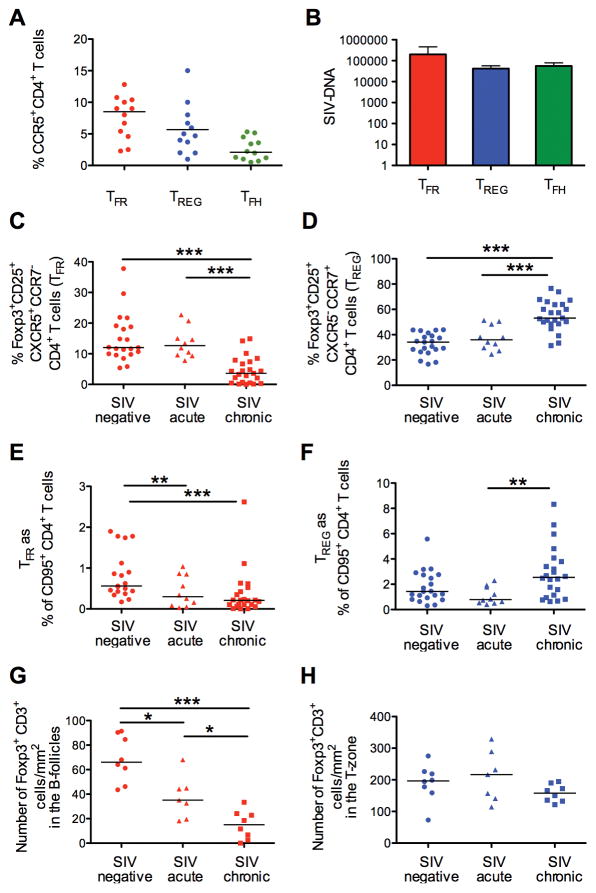
Sorted TFR and TREG from lymph nodes of 4 chronically infected macaques expressed comparable levels of CCR5 (Figure 3A), and harbored equivalent levels of SIV-DNA (Figure 3B). Similar SIV-DNA levels were also found in TFH cells.
Figure 3. TFR susceptibility and dynamics during SIV chronic infection.
(A) Percentage of CCR5+ TFR, CCR7+-TREG and TFH. (B) SIV-DNA levels in sorted TFR CCR7+-TREG and TFH by PCR. (C) Frequency of TFR and (D) CCR7+-TREG within Foxp3+ CD25+ cells, and (E) TFR and (F) CCR7+-TREG within memory CD4+ T cells in lymph nodes of naïve, acutely and chronically SIV infected macaques. (G) Number of Foxp3+ CD3+ cells in the B-follicles (TFR) and (H) in the T-zone (TREG) of intact lymph nodes from naïve, acutely and chronically infected macaques.
We analyzed the effect of SIVmac251 infection on the frequency of TFR and CCR7+-TREG in the lymph nodes of 10 acutely (2 or 3 weeks after infection) and 23 chronically infected (12–15 weeks after infection) macaques (Figure 3C and D, Table I). Representative dot-plots for 2 macaques before and after infection are shown in Supplemental Figure 3A. Cells expressing only CXCR5+ were significantly reduced within the Foxp3+ CD25+ population during chronic infection (chronic vs. negative p< 0.0001; chronic vs. acute p< 0.0001) (Figure 3C), while CCR7-single positive cells simultaneously increased (chronic vs. negative p<0.0001; chronic vs. acute p= 0.0003) when compared to non-infected and chronically infected animals (Figure 3D).
We looked at the levels of TFR and CCR7+-TREG with respect to the memory CD95+ CD4+ T population. TFR showed a downward trend from negative to acute to chronic (p=0.0005 by the Jonckheere-Terpstra test for trend), with marginal differences in acute (negative vs. acute: p=0.049), and a significant decrease in chronic infection (negative vs. chronic p=0.0003) (Figure 3E). A trend for increase in CCR7+-TREG levels was also observed and significance was reached between the values detected in acute and in chronic phase (p=0.0018) (Figure 3F).
We then performed immunohistochemistry in lymph nodes, at 3 and 12 weeks after infection from 7 and 8 animals, respectively (Figure 3G and H). The numbers of CD3+ cells expressing Foxp3 in the B-cell follicles was significantly reduced in acute infection (p=0.0022, n=7) and contracted even further in chronic infection (p<0.0001, n=8), when compared to naïve (n=8) animals (Figure 3G and Supplemental Figure 3B). The number of Foxp3+ CD3+ cells in the T-zone did not change, as described by others (Figure 3H) (52).
We did not see any significant correlation between the frequency of TFR or TREG and the SIV-RNA plasma levels (data not shown).
To further explore possible mechanisms for the SIV-associated decrease of TFR, we looked at markers of immune activation. We could not find any association with the frequency of Ki67+ CD4+ T cells in lymph nodes of 10 chronic animals (data not shown).
In mice NFAT-2 is critical for the up-regulation of CXCR5 on TFR (31), hence for their migration to the GC. Thus NFAT-2 may be involved in the reduction of TFR during chronic infection. To determine whether CXCR5 expression on macaque TFR was also dependent on NFAT activity, we treated lymph nodes cells from 2 naïve animals with cyclosporin A (CsA), and measured changes in the CXCR5 and CCR7 levels within Foxp3+ CD25+ CD4+ T cells (Supplemental Figure 3C and D). While CsA treatment had no effect on CCR7, it decreased CXCR5 expression levels on Foxp3+ CD25+ CD4+ T cells.
Decreased TFR and increased TFH frequency during SIVmac251 infection
The decrease of CXCR5+ regulatory T-cells may be associated with TFH expansion during SIVmac251 infection. We measured the frequency of TFH in lymph nodes of infected macaques as shown in Figure 1B. The percentage of TFH cells within the memory CD4+ T cell population did not change during acute infection but this population significantly expanded during chronic infection (week 12–15 after infection), as described by others (8) (negative vs. chronic and acute vs. chronic p<0.0001) (Figure 4A and B). Of note, we found a significant inverse correlation between the levels of TFR and TFH on memory CD4+ T cells during acute and chronic infection (R=−0.82, p=0.0058 and R=−0.69, p= 0.0010 by the Spearman Rank test) (Figure 4B and 4C).
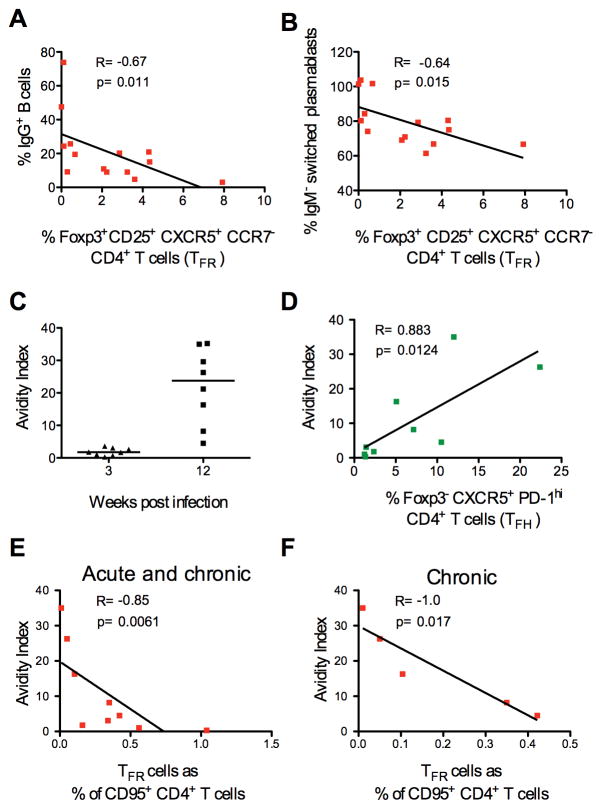
Figure 4. Association between TFR and TFH levels in SIV infection.
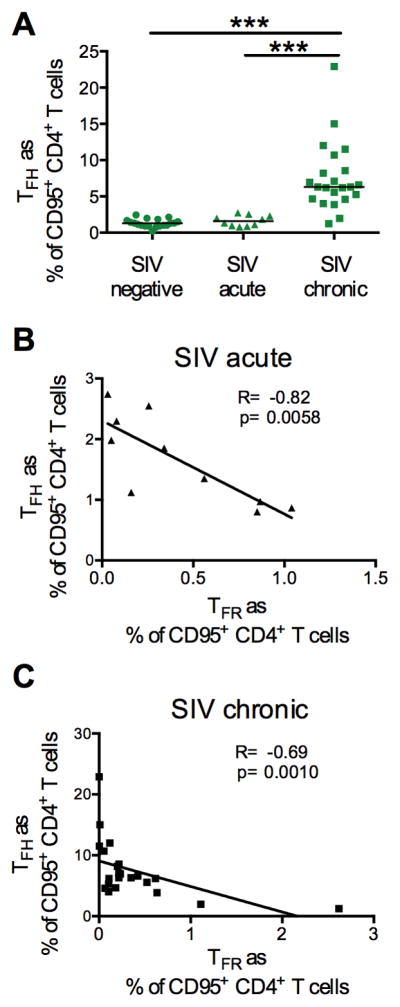
(A) Frequency of TFH on memory CD4+ T cells in lymph nodes of naïve, acutely and chronic SIV-infected macaques. (B) Correlation between the percentage of TFR and TFH on memory CD4+ T cells in acute and (C) chronic infection.
TFR frequency correlates with decreased avidity of antibodies to the gp120
In mice, TFR cells play a role in reducing plasma-cell differentiation (26). We measured the frequency of IgM+, IgG+ or IgA+ CD20+ B cells and plasmablasts, defined as lineage− (CD3− CD14− CD16− CD56−) CD20+ and/or CD19+ and CD21− Ki67+ CD38+ CD39+ in lymph nodes of SIVmac251 chronically infected animals (n=14) by flow cytometry. While we did not find any associations with the frequency of total memory B cells or plasmablasts measured in lymph nodes, we found a weak negative correlation between the frequency of IgG+ B cells and the frequency of IgM-switched plasmablasts (IgG+ and IgA+) and the percentage of TFR within Foxp3+ CD25+ cells (Figure 5A and B).
Figure 5. TFR levels and SIV specific antibody avidity.
(A) Correlation between the frequency of TFR cells on Foxp3+ CD25+ cells and IgG+ B cells or (B) switched IgM− plasmablasts in lymph nodes of chronically infected macaques. (C) Avidity index of plasma SIV-gp120 IgG measured in acutely and chronically infected macaques (the median is shown). (D) Correlation between the frequency of TFH or (E) TFR cells and the avidity index in plasma of all the SIV infected macaques and (F) in chronically infected macaques.
TFH are associated with the avidity to influenza virus and SIV (8)(53). In SIVmac251 infected macaques, avidity to the gp120 is low during acute infection and increases during chronic infection (Figure 5C). We confirmed that TFH were positively associated with gp120 avidity when all the infected macaques were considered (R=0.88; p=0.0031; Figure 5D), but not when the acute and chronic phases values were looked at separately. Importantly, TFR levels on memory CD4+ T cells were associated with a reduction of binding high avidity antibodies to SIV-gp120 in all the infected animals (R=−0.85; p=0.0061, and in chronic phase (R=−1.0 and p=0.017), but not during the acute phase (R=−0.80; p=0.33) (Figure 5E and F and data not shown).
Discussion
In this study we identified TFR in lymphoid tissues of healthy non-vaccinated rhesus macaques. We confirmed the presence of Foxp3+ T cells in the B-zone of intact lymph nodes of macaques. Because TFR share markers of TFH and TREG, they have been isolated and functionally characterized in mice as TFH cells positive for Foxp3 (25), or alternatively, as “follicular” TREG expressing CXCR5 (26)(47). We opted for the latter identification strategy to characterize macaque TFR in lymph nodes. In addition, we identified a subset of CCR7-expressing TREG that are CXCR5 negative and therefore, in principle, unable to enter the GC, and another subset of CXCR5 & CCR7-positive TREG (DP). Because it is possible that DP may localize at the T-B borders/mantle zone, following gradients of CXCL13 (B-cell follicles-GC) and of CCL21 and CCL19 (T-zone), we excluded this population from our analysis (7).
In accordance with their mouse counterpart, macaque TFR expressed high levels of PD-1, ICOS and Bcl-6, they were CD39+ and CD127−, and mainly resided in the GALT and lymph nodes. In a few macaques, TFR had similar levels of IL-10 and TGF-β mRNA as CCR7+-TREG (25, 26). It is possible that IL-10 and TGF-β may play a role in the TFR-mediated control of TFH proliferation, as shown in mice. We did not directly assess the suppressive ability of sorted CD25+ CXCR5+ CCR7− cells, however lymph nodes of healthy non-infected macaques contained a CXCR5high CCR7low Bcl-6hi CD25+ CD4+ population displaying in vitro chemotaxis toward CXCL13, and suppressive activity on CD4+ T cells and TFH proliferation. Further characterization is needed to confirm IL-10 and TGF-β production by macaque TFR and their role in the apparent suppression.
We took advantage of the established similarities between SIV-infection of macaques and HIV-1 infection of humans to study and TFR susceptibility and dynamic during infection in comparison to CCR7+-TREG. Previous studies on CD4+ T susceptibility have shown that CD25+ Foxp3+ cells are less susceptible than other subsets to HIV/SIV infection, due to their anergic nature and to the Foxp3-mediated inhibition of HIV-1-LTR activation (54)(55)(56)(57). Therefore the relative frequency of CD25+ Foxp3+ CD4+ T increases in acute and chronic HIV/SIV infection, while their absolute number remains the same (58)(59)(56). Similarly, we observed a trend for increased frequency of CCR7+-TREG within the memory CD4+ population and no differences in the number of CD3+Foxp3+ cells in the T-zone of intact lymph nodes in chronic infection.
TFR from naïve macaques expressed comparable levels of surface CCR5 to TREG, and had equivalent levels of SIV-DNA following infection. Conversely, we saw a reduction in TFR frequency and a decrease of CD3+ Foxp3+ cells in the B-follicles of infected animals, in particular during the chronic phase of infection. While we could not discern between CD8+ T cells and CD4+ T cells in our immunohistochemical analysis, some chronically infected animals had negligible numbers of Foxp3+ T cells in the GC indicating an overall reduction of regulatory cells, likely including TFR.
We could not determine whether TFR are more susceptible to SIVmac251 infection than TREG, and we did not observe any association between the TFR levels and viral replication levels or with immune activation.
The reduction in Foxp3+ cells in the GC may be driven by SIV-associated changes in the homing patterns of these cells. NFAT-2 is an essential transcriptional factor for CXCR5 expression on mice TFR (31). HIV-envelope induces NFAT-2 translocation to the nucleus, where it binds to multiple sites within the HIV-LTR (60)(61). HIV/SIV may therefore alter NFAT-dependent expression of CXCR5. We showed that cyclosporin A, a calcineurin inhibitor that blocks NFAT dephosphorylation, decreases CXCR5 levels on macaque CD4+CD25+Foxp3+ cells, but we were unable to test this intriguing hypothesis in our model due to the lack of reagents that cross-react with macaque NFAT proteins.
TFH numbers expand in acute and chronic viral infections, such as in Influenza A virus (IAV) infection in mice, and in chronic Hepatitis B and HIV/SIV in humans and macaques (62)(63, 64). In particular during acute IAV infection, a temporary TFH expansion occurs 3 days after challenge (62). Differently, in HIV/SIV infection, a sustained expansion of TFH is seen in chronic, but not during the acute phase of infection, as we also observed in our study (8).
We found an association between the levels of TFR and TFH in acutely and chronic macaques infected with SIVmac251. It is possible that a reduction or lack of expansion of TFR may contribute to the increased TFH number in chronic infection. Alternatively, the persistence of the antigen may lead to the increase in TFH cells, resulting in higher levels of PD-1 in the GC and in TFR reduction (47). Additionally, changes in TFR function, other regulatory subsets in the GC (CD8+ and NK T-cells), imbalanced cytokine milieu (i.e. increased IL-6), and immune activation are likely to participate to the HIV-associated increase in TFH numbers (8)(19).
To our knowledge our study is the first to describe TFR dynamics and changes in the TFH/TFR ratio during SIV infection, together with the accompanied study by Chowdhury and colleagues reporting similar findings in SIVsmE660 infected macaques.
By suppressing TFH numbers and proliferation, TFR modulate B-cell responses in mice (26)(29)(47). No correlation was found with the frequency of plasmablasts or with the IgA+ B cells. We found an association between the relative frequency of CXCR5+ cells within the CD25+ Foxp3+ population and the frequency of total IgG+ B cells and of overall switched IgM− (IgA+ IgG+) B-cells and plasmablasts in lymph nodes (32).
The accumulation of TFH in chronic SIV infection is associated with increased titers of higher avidity SIV-specific immunoglobulins (8). Interestingly, we observed an antithetic role of TFR and TFH in the avidity of antibodies to the SIV-gp120 protein throughout the infection, and only TFR levels were strongly correlated with the increased in avidity during chronic infection. It has been proposed that TFH accumulation, together with HIV-associated changes in cell function, may lead to a reduction in affinity maturation due to a lowered competition for B-cell selection. However, so far, the role of TFH cells in HIV/SIV pathogenesis has been studied without making a clear distinction between TFR and TFH. Thus, the relative contribution of TFH and TFR in the impairment in B-cell selection during HIV infection remains to be determined.
In summary, we identified a population of macaques CD4+ T cells with a phenotype, function and location consistent with TFR cells, and we revealed SIV-associated changes in the TFR and TFH ratio, adding to the complexity of humoral immunity to HIV.
Supplementary Material
Acknowledgments
We thank James Arthos and Claudia Cicala for helpful discussion, and Namal P. Liyanage and Dallas P. Brown and Veronica Galli for critical reading of the manuscript.
This work was entirely supported by the Intramural Research Program at the National Cancer Institute, National Institutes of Health.
Nonstandard abbreviations
- TREGS
T regulatory cells
- TFH
T follicular helper
- TFR
T follicular regulatory cells
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Garside P, Ingulli E, Merica RR, Johnson JG, Noelle RJ, Jenkins MK. Visualization of specific B and T lymphocyte interactions in the lymph node. Science. 1998;281:96–99. doi: 10.1126/science.281.5373.96. [DOI] [PubMed] [Google Scholar]
- 2.Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. European journal of immunology. 1991;21:2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- 3.Jacob J, Kelsoe G, Rajewsky K, Weiss U. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991;354:389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 4.Schwickert TA, Victora GD, Fooksman DR, Kamphorst AO, Mugnier MR, Gitlin AD, Dustin ML, Nussenzweig MC. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. The Journal of experimental medicine. 2011;208:1243–1252. doi: 10.1084/jem.20102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. The Journal of experimental medicine. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, Cyster JG. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. The Journal of experimental medicine. 1999;190:1123–1134. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. The Journal of experimental medicine. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, Pan L, Poholek A, Rao SS, Brenchley JM, Alam SM, Tomaras GD, Roederer M, Douek DC, Seder RA, Germain RN, Haddad EK, Koup RA. CD4 T follicular helper cell dynamics during SIV infection. The Journal of clinical investigation. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong JJ, Amancha PK, Rogers KA, Courtney CL, Havenar-Daughton C, Crotty S, Ansari AA, Villinger F. Early lymphoid responses and germinal center formation correlate with lower viral load set points and better prognosis of simian immunodeficiency virus infection. Journal of immunology. 2014;193:797–806. doi: 10.4049/jimmunol.1400749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. The Journal of experimental medicine. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crotty S. Follicular helper CD4 T cells (TFH) Annual review of immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 12.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. Journal of immunology. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 15.Vinuesa CG, Linterman MA, Goodnow CC, Randall KL. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunological reviews. 2010;237:72–89. doi: 10.1111/j.1600-065X.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt N, Bustamante J, Bourdery L, Bentebibel SE, Boisson-Dupuis S, Hamlin F, Tran MV, Blankenship D, Pascual V, Savino DA, Banchereau J, Casanova JL, Ueno H. IL-12 receptor beta1 deficiency alters in vivo T follicular helper cell response in humans. Blood. 2013;121:3375–3385. doi: 10.1182/blood-2012-08-448902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinuesa CG, Sanz I, Cook MC. Dysregulation of germinal centres in autoimmune disease. Nature reviews Immunology. 2009;9:845–857. doi: 10.1038/nri2637. [DOI] [PubMed] [Google Scholar]
- 18.Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nature immunology. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 19.Pratama A, Vinuesa CG. Control of TFH cell numbers: why and how? Immunology and cell biology. 2014;92:40–48. doi: 10.1038/icb.2013.69. [DOI] [PubMed] [Google Scholar]
- 20.de Vinuesa CG, Cook MC, Ball J, Drew M, Sunners Y, Cascalho M, Wabl M, Klaus GG, MacLennan IC. Germinal centers without T cells. The Journal of experimental medicine. 2000;191:485–494. doi: 10.1084/jem.191.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warnatz K, Bossaller L, Salzer U, Skrabl-Baumgartner A, Schwinger W, van der Burg M, van Dongen JJ, Orlowska-Volk M, Knoth R, Durandy A, Draeger R, Schlesier M, Peter HH, Grimbacher B. Human ICOS deficiency abrogates the germinal center reaction and provides a monogenic model for common variable immunodeficiency. Blood. 2006;107:3045–3052. doi: 10.1182/blood-2005-07-2955. [DOI] [PubMed] [Google Scholar]
- 22.King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annual review of immunology. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 23.Park HJ, Kim DH, Lim SH, Kim WJ, Youn J, Choi YS, Choi JM. Insights into the role of follicular helper T cells in autoimmunity. Immune network. 2014;14:21–29. doi: 10.4110/in.2014.14.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG. Follicular helper T cells are required for systemic autoimmunity. The Journal of experimental medicine. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, Fagarasan S, Liston A, Smith KG, Vinuesa CG. Foxp3+ follicular regulatory T cells control the germinal center response. Nature medicine. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nature medicine. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim HW, Hillsamer P, Banham AH, Kim CH. Cutting edge: direct suppression of B cells by CD4+ CD25+ regulatory T cells. Journal of immunology. 2005;175:4180–4183. doi: 10.4049/jimmunol.175.7.4180. [DOI] [PubMed] [Google Scholar]
- 28.Lim HW, Hillsamer P, Kim CH. Regulatory T cells can migrate to follicles upon T cell activation and suppress GC-Th cells and GC-Th cell-driven B cell responses. The Journal of clinical investigation. 2004;114:1640–1649. doi: 10.1172/JCI22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander CM, Tygrett LT, Boyden AW, Wolniak KL, Legge KL, Waldschmidt TJ. T regulatory cells participate in the control of germinal centre reactions. Immunology. 2011;133:452–468. doi: 10.1111/j.1365-2567.2011.03456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nature immunology. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaeth M, Muller G, Stauss D, Dietz L, Klein-Hessling S, Serfling E, Lipp M, Berberich I, Berberich-Siebelt F. Follicular regulatory T cells control humoral autoimmunity via NFAT2-regulated CXCR5 expression. The Journal of experimental medicine. 2014;211:545–561. doi: 10.1084/jem.20130604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolniak KL, Shinall SM, Waldschmidt TJ. The germinal center response. Critical reviews in immunology. 2004;24:39–65. doi: 10.1615/critrevimmunol.v24.i1.20. [DOI] [PubMed] [Google Scholar]
- 33.Mqadmi A, Zheng X, Yazdanbakhsh K. CD4+CD25+ regulatory T cells control induction of autoimmune hemolytic anemia. Blood. 2005;105:3746–3748. doi: 10.1182/blood-2004-12-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moir S, Malaspina A, Ogwaro KM, Donoghue ET, Hallahan CW, Ehler LA, Liu S, Adelsberger J, Lapointe R, Hwu P, Baseler M, Orenstein JM, Chun TW, Mican JA, Fauci AS. HIV-1 induces phenotypic and functional perturbations of B cells in chronically infected individuals. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10362–10367. doi: 10.1073/pnas.181347898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Milito A, Nilsson A, Titanji K, Thorstensson R, Reizenstein E, Narita M, Grutzmeier S, Sonnerborg A, Chiodi F. Mechanisms of hypergammaglobulinemia and impaired antigen-specific humoral immunity in HIV-1 infection. Blood. 2004;103:2180–2186. doi: 10.1182/blood-2003-07-2375. [DOI] [PubMed] [Google Scholar]
- 36.Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, Davis I, Farber D, Hartjen P, Haag F, Alter G, Schulze zur Wiesch J, Streeck H. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. The Journal of clinical investigation. 2012;122:3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cubas RA, Mudd JC, Savoye AL, Perreau M, van Grevenynghe J, Metcalf T, Connick E, Meditz A, Freeman GJ, Abesada-Terk G, Jr, Jacobson JM, Brooks AD, Crotty S, Estes JD, Pantaleo G, Lederman MM, Haddad EK. Inadequate T follicular cell help impairs B cell immunity during HIV infection. Nature medicine. 2013;19:494–499. doi: 10.1038/nm.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong JJ, Amancha PK, Rogers K, Ansari AA, Villinger F. Spatial alterations between CD4(+) T follicular helper, B, and CD8(+) T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. Journal of immunology. 2012;188:3247–3256. doi: 10.4049/jimmunol.1103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, Poignard P, Crotty S A. V. I. P. C. P. I. International. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaccari M, Keele BF, Bosinger SE, Doster MN, Ma ZM, Pollara J, Hryniewicz A, Ferrari G, Guan Y, Forthal DN, Venzon D, Fenizia C, Morgan T, Montefiori D, Lifson JD, Miller CJ, Silvestri G, Rosati M, Felber BK, Pavlakis GN, Tartaglia J, Franchini G. Protection afforded by an HIV vaccine candidate in macaques depends on the dose of SIVmac251 at challenge exposure. Journal of virology. 2013;87:3538–3548. doi: 10.1128/JVI.02863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pegu P, Vaccari M, Gordon S, Keele BF, Doster M, Guan Y, Ferrari G, Pal R, Ferrari MG, Whitney S, Hudacik L, Billings E, Rao M, Montefiori D, Tomaras G, Alam SM, Fenizia C, Lifson JD, Stablein D, Tartaglia J, Michael N, Kim J, Venzon D, Franchini G. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. Journal of virology. 2013;87:1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cecchinato V, Tryniszewska E, Ma ZM, Vaccari M, Boasso A, Tsai WP, Petrovas C, Fuchs D, Heraud JM, Venzon D, Shearer GM, Koup RA, Lowy I, Miller CJ, Franchini G. Immune activation driven by CTLA-4 blockade augments viral replication at mucosal sites in simian immunodeficiency virus infection. Journal of immunology. 2008;180:5439–5447. doi: 10.4049/jimmunol.180.8.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Vos J, Hose D, Reme T, Tarte K, Moreaux J, Mahtouk K, Jourdan M, Goldschmidt H, Rossi JF, Cremer FW, Klein B. Microarray-based understanding of normal and malignant plasma cells. Immunological reviews. 2006;210:86–104. doi: 10.1111/j.0105-2896.2006.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaccari M, Boasso A, Ma ZM, Cecchinato V, Venzon D, Doster MN, Tsai WP, Shearer GM, Fuchs D, Felber BK, Pavlakis GN, Miller CJ, Franchini G. CD4+ T-cell loss and delayed expression of modulators of immune responses at mucosal sites of vaccinated macaques following SIV(mac251) infection. Mucosal immunology. 2008;1:497–507. doi: 10.1038/mi.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmann-Lehmann R, Swenerton RK, Liska V, Leutenegger CM, Lutz H, McClure HM, Ruprecht RM. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one-versus two-enzyme systems. AIDS research and human retroviruses. 2000;16:1247–1257. doi: 10.1089/08892220050117014. [DOI] [PubMed] [Google Scholar]
- 46.Romano JW, Williams KG, Shurtliff RN, Ginocchio C, Kaplan M. NASBA technology: isothermal RNA amplification in qualitative and quantitative diagnostics. Immunological investigations. 1997;26:15–28. doi: 10.3109/08820139709048912. [DOI] [PubMed] [Google Scholar]
- 47.Wollenberg I, Agua-Doce A, Hernandez A, Almeida C, Oliveira VG, Faro J, Graca L. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. Journal of immunology. 2011;187:4553–4560. doi: 10.4049/jimmunol.1101328. [DOI] [PubMed] [Google Scholar]
- 48.Sage PT, Francisco LM, Carman CV, Sharpe AH. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nature immunology. 2013;14:152–161. doi: 10.1038/ni.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu H, Wang X, Lackner AA, Veazey RS. PD-1(HIGH) Follicular CD4 T Helper Cell Subsets Residing in Lymph Node Germinal Centers Correlate with B Cell Maturation and IgG Production in Rhesus Macaques. Frontiers in immunology. 2014;5:85. doi: 10.3389/fimmu.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanjabi S, Zenewicz LA, Kamanaka M, Flavell RA. Anti-inflammatory and pro-inflammatory roles of TGF-beta, IL-10, and IL-22 in immunity and autoimmunity. Current opinion in pharmacology. 2009;9:447–453. doi: 10.1016/j.coph.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arce J, Levin M, Xie Q, Albanese J, Ratech H. T-regulatory cells in lymph nodes: correlation with sex and HIV status. American journal of clinical pathology. 2011;136:35–42. doi: 10.1309/AJCPAR39BFWNHWRO. [DOI] [PubMed] [Google Scholar]
- 53.Leon B, Bradley JE, Lund FE, Randall TD, Ballesteros-Tato A. FoxP3+ regulatory T cells promote influenza-specific Tfh responses by controlling IL-2 availability. Nature communications. 2014;5:3495. doi: 10.1038/ncomms4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreno-Fernandez ME, Zapata W, Blackard JT, Franchini G, Chougnet CA. Human regulatory T cells are targets for human immunodeficiency Virus (HIV) infection, and their susceptibility differs depending on the HIV type 1 strain. Journal of virology. 2009;83:12925–12933. doi: 10.1128/JVI.01352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Gowans EJ, Chougnet C, Plebanski M, Dittmer U. Natural regulatory T cells and persistent viral infection. Journal of virology. 2008;82:21–30. doi: 10.1128/JVI.01768-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran TA, de Goer de Herve MG, Hendel-Chavez H, Dembele B, Le Nevot E, Abbed K, Pallier C, Goujard C, Gasnault J, Delfraissy JF, Balazuc AM, Taoufik Y. Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PloS one. 2008;3:e3305. doi: 10.1371/journal.pone.0003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grant C, Oh U, Fugo K, Takenouchi N, Griffith C, Yao K, Newhook TE, Ratner L, Jacobson S. Foxp3 represses retroviral transcription by targeting both NF-kappaB and CREB pathways. PLoS pathogens. 2006;2:e33. doi: 10.1371/journal.ppat.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chevalier MF, Weiss L. The split personality of regulatory T cells in HIV infection. Blood. 2013;121:29–37. doi: 10.1182/blood-2012-07-409755. [DOI] [PubMed] [Google Scholar]
- 59.Boasso A, Vaccari M, Hryniewicz A, Fuchs D, Nacsa J, Cecchinato V, Andersson J, Franchini G, Shearer GM, Chougnet C. Regulatory T-cell markers, indoleamine 2,3-dioxygenase, and virus levels in spleen and gut during progressive simian immunodeficiency virus infection. Journal of virology. 2007;81:11593–11603. doi: 10.1128/JVI.00760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cicala C, Arthos J, Censoplano N, Cruz C, Chung E, Martinelli E, Lempicki RA, Natarajan V, VanRyk D, Daucher M, Fauci AS. HIV-1 gp120 induces NFAT nuclear translocation in resting CD4+ T-cells. Virology. 2006;345:105–114. doi: 10.1016/j.virol.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 61.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annual review of immunology. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 62.Boyden AW, Legge KL, Waldschmidt TJ. Pulmonary infection with influenza A virus induces site-specific germinal center and T follicular helper cell responses. PloS one. 2012;7:e40733. doi: 10.1371/journal.pone.0040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu TT, Song XF, Lei Y, Hu HD, Ren H, Hu P. Expansion of circulating TFH cells and their associated molecules: involvement in the immune landscape in patients with chronic HBV infection. Virology journal. 2014;11:54. doi: 10.1186/1743-422X-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng J, Lu L, Hua C, Qin L, Zhao P, Wang J, Wang Y, Li W, Shi X, Jiang Y. High frequency of CD4+ CXCR5+ TFH cells in patients with immune-active chronic hepatitis B. PloS one. 2011;6:e21698. doi: 10.1371/journal.pone.0021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.