SUMMARY
Endosomal protein recycling is a fundamental cellular process important for cellular homeostasis, signaling, and fate determination that is implicated in several diseases. WASH is an actin nucleating protein essential for this process and its activity is controlled through K63-linked ubiquitination by the MAGE-L2-TRIM27 ubiquitin ligase. Here, we show that the USP7 deubiquitinating enzyme is an integral component of the MAGE-L2-TRIM27 ligase and is essential for WASH-mediated endosomal actin assembly and protein recycling. Mechanistically, USP7 acts as a molecular rheostat to precisely fine-tune endosomal F-actin levels by counteracting TRIM27 auto-ubiquitination/degradation and preventing overactivation of WASH through directly deubiquitinating it. Importantly, we identify de novo heterozygous loss-of-function mutations of USP7 in individuals with a neurodevelopmental disorder, featuring intellectual disability and autism spectrum disorder. These results provide unanticipated insights into endosomal trafficking, illuminate the cooperativity between a ubiquitin ligase and a deubiquitinating enzyme, and establish a role for USP7 in human neurodevelopmental disease.
INTRODUCTION
Post-translational modification of proteins by ubiquitin serves as an important cellular regulatory event to spatially and temporally control the stability and activity of proteins (Komander and Rape, 2012). Ubiquitination is highly regulated and controlled by E3 ubiquitin ligases (Deshaies and Joazeiro, 2009). However, equally important is the counterbalance by deubiquitinating enzymes that reverse the action of ubiquitin ligases (Komander et al., 2009; Reyes-Turcu et al., 2009). How these two seemingly opposing activities work in concert to allow precise control of cellular signaling and biological processes is not fully resolved. Ubiquitin-specific protease 7 (USP7; also known as herpesvirus-associated ubiquitin-specific protease, HAUSP) is a deubiquitinating enzyme in the ubiquitin-specific protease family that can cleave multiple chain linkages, including short K48-linked ubiquitin chains and longer K63-linked ubiquitin chains (Li et al., 2002; Nicholson and Suresh Kumar, 2011; Schaefer and Morgan, 2011). USP7 regulates the ubiquitination of many proteins, including the MDM2-p53 pathway, that impacts a number of diverse cellular and pathophysiological processes, such as DNA repair, transcription, immune responses, viral replication, and cancer (Li et al., 2002; Nicholson and Suresh Kumar, 2011). Knockout of Usp7 in mice results in early embryonic lethality that cannot be rescued by p53 knockout, suggesting additional roles of Usp7 (Kon et al., 2010).
MAGE-L2 is part of the melanoma antigen gene (MAGE) family of E3 ubiquitin ligase regulators (Doyle et al., 2010). MAGE-L2 binds to the TRIM27 ubiquitin ligase and mediates its recruitment to endosomes where it facilitates retromer-dependent recycling of proteins from endosomes back to the trans-Golgi network (retrograde; such as CI-M6PR) or to the plasma membrane (such as integrins) (Burd and Cullen, 2014; Hao et al., 2013). The MAGE-L2-TRIM27 ubiquitin ligase promotes endosomal protein recycling by K63-linked poly-ubiquitination of WASH, an actin nucleation promoting factor essential for this fundamental recycling pathway (Derivery et al., 2009; Gomez and Billadeau, 2009; Hao et al., 2013; Seaman et al., 2013). Non-degradative ubiquitination of WASH leads to its activation through unlocking an auto-inhibited state, resulting in endosomal actin assembly and protein recycling (Hao et al., 2013; Jia et al., 2010). Importantly, MAGEL2 is one of the protein-coding genes typically deleted in Prader-Willi syndrome (PWS), a human neurodevelopmental disease (Boccaccio et al., 1999; Lee et al., 2000). Furthermore, truncating mutations of MAGEL2 result in Schaaf-Yang syndrome (MIM 615547) with marked clinical overlap to PWS, highlighted by hypotonia, hypogonadism, intellectual disability, and autism spectrum disorder (Schaaf et al., 2013). The Magel2 knockout mice recapitulate several of these phenotypes (Bischof et al., 2007; Kozlov et al., 2007; Mercer et al., 2009; Mercer and Wevrick, 2009; Tennese and Wevrick, 2011). This suggests that the regulation of WASH-mediated endosomal actin assembly and protein recycling by the MAGE-L2-TRIM27 ubiquitin ligase plays an important role in neurodevelopment.
RESULTS
USP7 is an integral component of the MAGE-L2-TRIM27 ubiquitin ligase complex
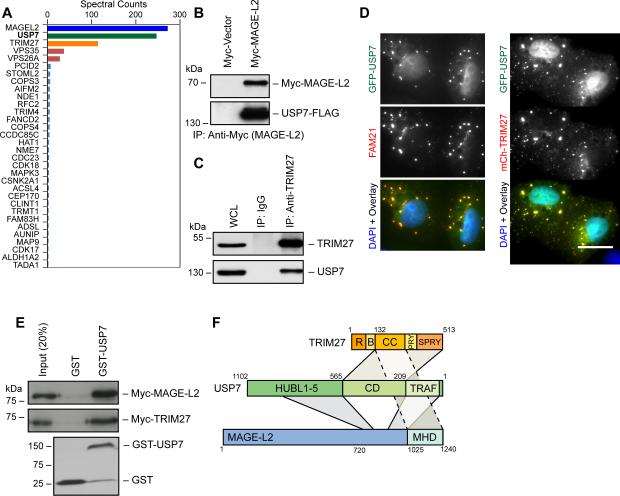
To identify MAGE-L2-TRIM27 regulatory factors, we performed tandem affinity purification of MAGE-L2 followed by LC-MS/MS. As expected, MAGE-L2 co-purified with the E3 ubiquitin ligase TRIM27 (Figure 1A). Strikingly, we unexpectedly found USP7 co-purifying with MAGE-L2 (Figure 1A). The robust interaction of USP7 with MAGE-L2 (and TRIM27) was confirmed by co-expression and endogenous immunoprecipitation experiments (Figure 1B-C). Expression of GFP-tagged USP7 in U2OS cells revealed co-localization with mCherry-TRIM27 cytoplasmic puncta and the WASH complex (FAM21), although the majority of GFP-USP7 localizes to the nucleus (Figure 1D). In vitro experiments using recombinant proteins revealed that USP7 not only directly bound MAGE-L2, but also directly bound TRIM27 (Figure 1E), consistent with previous results (Zaman et al., 2013). Through a series of in vitro binding studies and cellular co-immunoprecipitation experiments, we find that MAGE-L2 interacts with USP7 through two distinct interfaces, most prominently with the USP7 N-terminal TRAF domain and more weakly with the C-terminal HUBL1-3 regulatory domains (Faesen et al., 2011) (Figure S1A-D and summarized in Figure 1F). In contrast, TRIM27's C-terminal domains interact with the catalytic domain of USP7 (Figure S1A-D and summarized in Figure 1F). Consistent with the importance of USP7 in proper assembly of the complex, attempts to purify the MAGE-L2-TRIM27 complex failed in the absence of co-expression and purification with USP7 in insect cells (Figure S1E). However, co-expression of all three subunits of the complex yielded stoichiometric amounts of MAGE-L2, TRIM27, and USP7 (Figure S1F). These results suggest that MAGE-L2, TRIM27, and USP7 form an intricate and stable protein complex.
Figure 1. USP7 is an integral component of the MAGE-L2-TRIM27 ubiquitin ligase complex and is required for WASH-mediated endosomal protein recycling.
(A) Tandem affinity purification of MAGE-L2 followed by LC-MS/MS identified USP7 as a strong binding partner of MAGE-L2. Spectral counts normalized to control pulldown are shown.
(B) Co-expressed Myc-MAGE-L2 and USP7-FLAG co-immunoprecipitate. Cells were transfected with indicated constructs for 48 hrs before anti-Myc (MAGE-L2) immunoprecipitation and immunoblotting.
(C) Endogenous TRIM27 and USP7 co-immunoprecipitate. Cell lysates were immunoprecipitated with the indicated antibodies followed by immunoblotting.
(D) GFP-tagged USP7 was expressed in U2OS cells and co-localization was determined with mCherry-tagged TRIM27 or anti-FAM21. Co-localization was quantitated by Pearson's correlation (Rr) and is shown. Scale bar represent 20 μm.
(E) Both recombinant MAGE-L2 and TRIM27 bind purified GST-USP7, but not GST, in vitro. GST pulldown assays were performed with the indicated proteins followed by immunoblotting.
(F) Summary of results determining the binding and architecture of the MAGE-L2-TRIM27-USP7 complex.
See also Figure S1.
USP7 is required for WASH-mediated endosomal actin accumulation and protein recycling
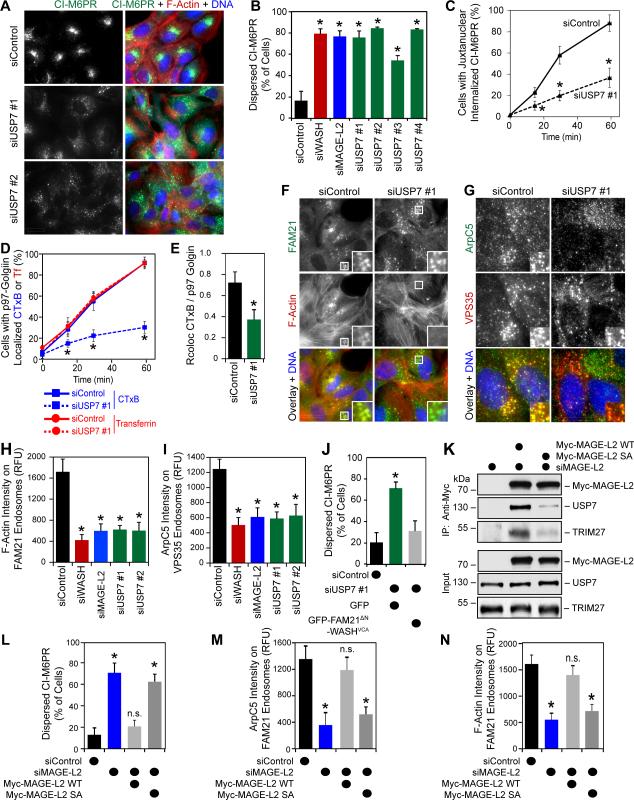
Given that MAGE-L2-TRIM27 promotes WASH activation, endosomal actin assembly, and protein recycling through WASH ubiquitination (Hao et al., 2013), we hypothesized that USP7 may inhibit these processes by counteracting MAGE-L2-TRIM27's activity. Surprisingly, knockdown of USP7 had the opposite effect, resulting in phenotypes that mimicked MAGE-L2, TRIM27, or WASH knockdown, including impairment of CI-M6PR protein recycling (Figure 2A-B and S2A) that resulted in endosomal accumulation of CI-M6PR (Figure S2B). Importantly, these phenotypes were evident in both p53-proficient (U2OS, Figure 2A-B) and deficient (HeLa, Figure S2C-D) cells and were not due to defects in the general trans-Golgi network architecture (Figure S2E). Additionally, knockdown of USP7 impaired retrograde trafficking of other WASH/retromer-dependent proteins, including cell surface CI-M6PR (Figure 2C), cholera toxin subunit B (CTxB; Figure 2D-E and S2F), and TGN46 (Figure S2G). Depletion of USP7 also impacted WASH-dependent recycling of Integrin α5 to the plasma membrane (Figure S2H). However, other WASH-independent endosomal recycling pathways, such as trafficking of transferrin, were unaffected by USP7 knockdown (Figure 2D and S2F). Consistent with the impaired trafficking of WASH/retromer-dependent proteins due to alterations in WASH activity, knockdown of USP7 decreased levels of F-actin and the Arp2/3 subunit ArpC5 (Figure 2F-I) on FAM21/VPS35-postive endosomes to a degree similar to knockdown of MAGE-L2 or WASH. Knockdown of USP7 had no general effect on total cellular levels of ArpC5 (Figure 3A). Importantly, defects in CI-M6PR retrograde transport in USP7-RNAi cells could be rescued by elevating endosomal F-actin levels using an endosomal-targeted active WASH construct we previously characterized (Figure 2J) (Hao et al., 2013).
Figure 2. USP7 is required for WASH-mediated endosomal protein recycling.
(A and B) Knockdown of USP7 disrupts CI-M6PR recycling. U2OS cells were treated with the indicated siRNAs for 72 hrs followed by immunostaining (A) and quantification (B).
(C) Trafficking of cell surface labeled CI-M6PR is impaired upon knockdown of USP7. U2OS cells were treated with indicated siRNAs for 72 hrs before cell surface CI-M6PR was labeled with anti-CI-M6PR for 30 mins. Cells were fixed at the indicated time points after labeling, the internalized CI-M6PR was detected, and the percentage of cells with juxtanuclear internalized CI-M6PR was determined.
(D and E) Depletion of USP7 impairs cholera toxin subunit B (CTxB), but not transferrin (Tf) trafficking. U2OS cells were treated with the indicated siRNAs for 72 hrs before fluorescently labeled CTxB and Tf were added to cells for 15 mins. Cells were fixed at the indicated time points post-labeling and the percentage of cells with CTxB and Tf localized with p97-golgin was determined by counting (D) or Pearson correlation (E).
(F-I) Knockdown of USP7 decreases endosomal F-actin and Arp2/3 levels. U2OS cells were treated with the indicated siRNAs for 72 hrs followed by staining for F-actin (Phalloidin) and endosomal marker FAM21 (F) and quantification (G) or ArpC5 and endosomal marker VPS35 (H) and quantitation (I).
(J) USP7-RNAi induced defects in CI-M6PR recycling are rescued by expression of endosomal-targeted WASHVCA. Cells were transfected with the indicated siRNAs for 24 hrs followed by expression of the indicated plasmids for an additional 48 hrs before immunostaining and quantitation for CI-M6PR localization in GFP-positive transfected cells.
(K) MAGE-L2 SA mutant has reduced USP7 affinity. HeLa cells were treated siMAGE-L2 for 24 hrs followed by expression of the indicated constructs for 48 hrs before anti-Myc (MAGE-L2) immunoprecipitation and immunoblotting.
(L) USP7 interaction with MAGE-L2 is critical for CI-M6PR endosomal protein recycling. HeLa cells were treated with the indicated siRNAs for 24 hrs followed by expression of the indicated constructs for 48 hrs before anti-CI-M6PR immunostaining. Quantitation was performed only on transfected cells.
(M and N) HeLa cells expressing MAGE-L2 SA mutant have reduced ArpC5 (M) and F-actin (N) on FAM21-positive endosomes. HeLa cells were treated with the indicated siRNAs for 24 hrs followed by expression of the indicated constructs for 48 hrs before anti-ArpC5 immunostaining (K) or F-actin staining (Phalloidin; L). Quantitation was performed only on transfected cells.
Results are representative of at least three replicate experiments. More than 100 cells were quantitated. Values shown are mean + SD. Asterisks indicates p<0.05. Scale bars represent 20 μm. See also Figure S2.
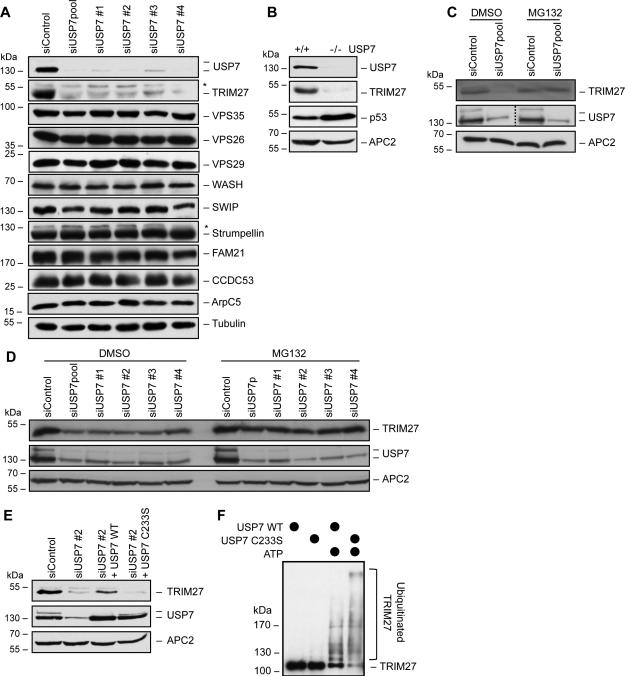
Figure 3. USP7 protects TRIM27 from auto-ubiquitination-induced degradation.
(A) Knockdown of USP7 dramatically reduces TRIM27 levels, without affecting WASH and retromer complex levels. HeLa cells were treated with the indicted siRNAs for 72 hrs before immunoblotting for the indicted proteins. Asterisks indicate non-specific bands.
(B) USP7 knockout (USP7−/−) cells have reduced TRIM27 protein levels. Cell lysates from the indicated HCT116 cells were immunoblotted.
(C and D) Proteasome inhibitor (MG132) rescues TRIM27 protein levels upon USP7 knockdown. p53-proficient U2OS (C) and p53-deficient HeLa (D) cells were treated with the indicated siRNAs for 72 hrs, incubated with DMSO (vehicle) or MG132 for 4 hrs, and immunoblotted.
(E) USP7 deubiquitinating enzymatic activity is critical for maintenance of TRIM27 protein levels. HeLa cells were treated with the indicated siRNAs for 24 hrs before expression of the indicated constructs for 48 hrs followed by immunoblotting for the indicated proteins.
(F) USP7 deubiquitinates TRIM27 auto-ubiquitination in vitro. Purified proteins were used in in vitro ubiquitination reactions before samples were separated by SDS-PAGE and anti-TRIM27 immunoblotting was performed.
Results are representative of at least three replicate experiments. See also Figure S3.
USP7 is known to deubiquitinate and regulate several proteins. To determine whether its role in endosomal actin assembly and WASH-dependent protein trafficking was through its interaction with MAGE-L2-TRIM27, we identified a MAGE-L2 mutant that disrupted USP7 interaction. MAGE-L2 (amino acids 820-1034) interacts with the USP7 TRAF domain (Figure 1F and S1), which has previously been shown to bind to a conserved motif A/P-X-X-S (Sheng et al., 2006). Within this portion of MAGE-L2, six TRAF binding motifs were identified (Figure S2I). Ser to Ala mutation of these motifs (MAGE-L2 SA) blocked USP7 interaction (Figure 2K). MAGE-L2 SA mutant also showed decreased binding to TRIM27, suggesting that USP7 facilitates MAGE-L2-TRIM27 complex formation in cells (Figure 2K). Importantly, knockdown of endogenous MAGE-L2 and expression of MAGE-L2 SA disrupted CI-M6PR endosomal protein recycling (Figure 2L and S2J), Arp2/3 localization to FAM21-positive endosomes (Figure 2M), and F-actin accumulation on FAM21-postive endosomes (Figure 2N). Taken together, these findings suggest that the USP7 deubiquitinating enzyme forms a stable complex with the MAGE-L2-TRIM27 E3 ubiquitin ligase and that all three proteins work in concert to control WASH-mediated actin assembly and protein recycling.
USP7 prevents auto-ubiquitination and degradation of TRIM27
To investigate how USP7 supports MAGE-L2-TRIM27 and WASH-mediated protein recycling, we investigated whether USP7 controls the steady state levels or localization of any of the known components of the endosomal protein recycling pathway, including the important WASH actin assembly and retromer coat-like complexes. WASH and retromer complexes localized properly on endosomes (Figure S3) and their protein levels were unaffected by USP7 knockdown (Figure 3A). However, TRIM27 protein levels were dramatically decreased (Figure 3A). This result was confirmed to be an on-target effect using multiple siRNAs targeting USP7 and knockout of USP7 in HCT116 cells (USP7−/−) (Cummins et al., 2004) (Figure 3A-B). Importantly, these effects were independent of p53 status and could be rescued by the MG132 proteasome inhibitor (Figure 3C-D), suggesting that USP7 prevents ubiquitin-mediated degradation of TRIM27 by the proteasome. To determine whether USP7 stabilizes TRIM27 through its deubiquitinating activity, we suppressed endogenous USP7 and re-expressed wild-type or a USP7 mutant (C233S) that lacks deubiquitinating enzymatic activity. Whereas expression of wild-type USP7 was competent to rescue TRIM27 protein levels, expression of USP7 C233S was not (Figure 3E). To confirm that USP7 directly controls TRIM27 auto-ubiquitination levels, we performed in vitro ubiquitination studies in which recombinant TRIM27 auto-ubiquitination activity was determined in the presence of wild-type USP7 or USP7 C233S inactive mutant. Consistent with our cellular data, wild-type USP7 restrained TRIM27 auto-ubiquitination in vitro when compared to auto-ubiquitination of TRIM27 in the presence of USP7 C233S (Figure 3F and S1F). These results suggest that USP7 is an integral component of the MAGE-L2-TRIM27 ubiquitin ligase complex that prevents auto-ubiquitination-induced degradation of TRIM27.
USP7 acts as a molecular rheostat to control WASH activity and endosomal actin assembly through deubiquitination of WASH
Next, we examined whether USP7's sole role in endosomal protein recycling was stabilization of TRIM27. To do so, we identified a non-ubiquitinatable mutant of TRIM27 that was stable even in the absence of USP7. First, we mutated all lysines in TRIM27 to arginine (termed, all K-R) and examined whether this TRIM27 mutant was resistant to USP7 knockdown. This was indeed the case (Figure 4A and S4A). To further refine which lysines in TRIM27 contribute to its auto-ubiquitination and instability in the absence of USP7, we mapped its ubiquitination sites by anti-diGly affinity proteomics which revealed five ubiquitination sites (K79, K292, K304, K380, and K382) (data not shown). Mutation of these individually or in various combinations ultimately identified a TRIM27 3KR mutant (K304R, K380R, and K382R) that was stable even in the absence of USP7 (Figure 4A, S4A, and data not shown). Importantly, TRIM27 3KR incorporated into the MAGE-L2-TRIM27-USP7 complex (Figure 4B and S4B) and is fully functional in supporting CI-M6PR recycling (Figure 4C). Thus, TRIM27 3KR allowed us to uncouple USP7's activity on stabilizing TRIM27 from any other potentially unknown activity of USP7 on this pathway. We determined whether cells reconstituted with TRIM27 3KR mutant are resistant to USP7-RNAi-induced defects in CI-M6PR recycling, as would be expected if USP7's only role was stabilization of TRIM27. However, this was not the case as knockdown of USP7 in TRIM27 3KR cells still impaired CI-M6PR recycling similarly to TRIM27 wild-type cells (Figure 4D). Analysis of these cells revealed that knockdown of USP7 in TRIM27 3KR cells significantly increased Arp2/3 and F-actin levels on FAM21-positive endosomes (Figure 4E-F). This lead us to hypothesize that in addition to preventing TRIM27 auto-ubiquitination and degradation, USP7 also prevents over-activation of WASH and improper CI-M6PR recycling by precisely tuning WASH ubiquitination and endosomal actin levels. Consistent with this hypothesis, the level of ubiquitinated WASH is increased upon knockdown of USP7 in TRIM27 3KR cells (Figure 4G). Furthermore, ubiquitination of WASH in vitro by MAGE-L2-TRIM27 was restrained by wild-type USP7 when compared to inactive USP7 C233S mutant (Figure 4H). Taken together, these results suggest that USP7 limits overproduction of endosomal F-actin by WASH through its ability to deubiquitinate WASH (Figure 4I) and that precise endosomal F-actin levels are critical for proper endosomal protein recycling. Consistent with this, artificial elevation of endosomal F-actin (Figure 4J and S4C) by overexpression of the WASH K220D mutant that mimics the activation of WASH by ubiquitination (Hao et al., 2013) causes defects in CI-M6PR recycling (Figure 4K and S4D).
Figure 4. USP7 buffers MAGE-L2-TRIM27-induced WASH ubiquitination by deubiquitinating WASH.
(A) TRIM27 3KR mutant is stable in the absence of USP7. HeLa cells were treated with the indicated siRNAs for 24 hrs before the indicated constructs were expressed for 48 hrs followed by immunoblotting.
(B) TRIM27 3KR binds MAGE-L2. The indicated proteins were expressed in cells for 48 hrs before anti-Myc immunoprecipitation was performed and co-purifying proteins were detected by immunoblotting.
(C) TRIM27 3KR mutant is still functional and can rescue TRIM27-RNAi-induced CI-M6PR mistrafficking. HeLa cells were treated with the indicated siRNAs for 24 hrs before expression of the indicated constructs for 48 hrs followed by anti-CI-M6PR immunostaining and quantification.
(D) Knockdown of USP7 in cells reconstituted with TRIM27 3KR still abrogates CI-M6PR recycling, suggesting another critical activity of USP7. HeLa cells were treated with the indicated siRNAs for 24 hrs before expression of the indicated constructs for 48 hrs followed by anti-CI-M6PR immunostaining and quantification. Quantitation was performed only on transfected cells as marked by anti-HA (TRIM27) staining.
(E and F) USP7 limits WASH-mediated endosomal Arp2/3 localization (E) and endosomal F-actin (F). HeLa cells were treated with the indicated siRNAs for 24 hrs before expression of the indicated constructs for 48 hrs followed by immunostaining and quantification. Quantitation was performed only on transfected cells as marked by anti-HA (TRIM27) staining. Endosomes were marked with anti-FAM21.
(G) Knockdown of USP7 in TRIM27 3KR cells increases WASH ubiquitination levels. HeLa cells were treated with the indicated siRNAs for 24 hrs before expression of the indicated constructs for 48 hrs followed by anti-Myc (ubiquitin) immunoprecipitation and immunoblotting.
(H) USP7 wild-type, but not inactive USP7 C233S, limits MAGE-L2-TRIM27-induced WASH ubiquitination (and TRIM27 auto-ubiquitination) in vitro. In vitro ubiquitination reactions were performed as indicated. Samples were separated by SDS-PAGE and immunoblotted for TRIM27 (left) or WASH (right).
(I) Model for dual activities of USP7 in preventing TRIM27 from auto-ubiquitination-induced degradation, but also limiting WASH ubiquitination levels and activity through its deubiquitination.
(J and K) U2OS cells were transiently transfected with the indicated constructs for 48 hrs before immunostaining and quantification of F-actin on FAM21-positive endosomes in GFP-positive cells (J) or quantification of CI-M6PR localization in GFP-positive cells (K).
Results are representative of at least three replicate experiments. More than 100 cells were quantitated. Values shown are mean + SD. Asterisks indicates p<0.05. n.s. indicates not significant (p>0.05). See also Figure S4.
Disruption of USP7 in humans is associated with a neurodevelopmental disorder, featuring intellectual disability and autism spectrum disorder
Given our identification of USP7 as a critical component of the MAGE-L2-TRIM27 ligase complex that regulates WASH-mediated endosomal protein recycling and our previous identification of truncating MAGEL2 mutations in children with Schaaf-Yang syndrome (Schaaf et al., 2013), we hypothesized that mutations in USP7 may lead to similar clinical phenotypes. We queried clinical databases of chromosome microarray analyses on approximately 94,000 individuals (Baylor College of Medicine Molecular Genetics Laboratory and Signature Genomic Laboratories, February 2014), the publically available DECIPHER (DatabasE of genomiC varIation and Phenotype in Humans using Ensembl Resources) database, and the database of clinical whole exome sequencing at the Whole Genome Laboratory of Baylor College of Medicine (approximately 1,500 cases at the time of query, i.e. February 2014). We identified six cases with heterozygous chromosomal microdeletions (see Figure 5A and Table S1 for genomic coordinates), and one case with a heterozygous nonsense mutation in USP7 (Figure 5A and Table 1). Subsequent testing revealed that all seven mutations were de novo (p<0.0001, two-tailed Chi-square analysis with Yates’ correction and Bonferroni correction for multiple comparisons, see supplemental experimental procedures). In a genotype-to-phenotype approach, we obtained detailed clinical information on the affected individuals (Table 1 and supplemental clinical notes), which revealed striking overlap with the phenotypes observed in individuals with MAGEL2 loss-of-function mutations. In summary, the most prevalent phenotypes were intellectual disability (100%), autism spectrum disorder (83%), epilepsy (71%), aggressive behavior (57%), hypotonia (57%), and hypogonadism (80%). These results suggest haploinsufficiency of USP7 as a mechanism for pathogenesis in human neurodevelopment.
Figure 5. Mutation or deletion of USP7 impairs WASH-mediated protein recycling and occurs in children with a neurodevelopmental disorder.
(A) Schematic illustrating USP7 genomic locus deletions or mutations in seven subjects with intellectual disability, autism spectrum disorder, and other neurodevelopmental phenotypes (see Table 1).
(B) Partial knockdown of USP7 decreases TRIM27 protein levels (bottom) and impairs CI-M6PR recycling (top). HeLa cells were treated with the indicated siRNA concentrations for 72 hrs before anti-CI-M6PR immunostaining and quantification (top) or cell lysates were immunoblotted (bottom). Asterisks indicates non-specific band (bottom).
(C and D) USP7+/− HCT116 cells have defects in CI-M6PR recycling. The indicated HCT116 cells were subjected to anti-CI-M6PR immunostaining (C) and quantification (D).
(E and F) USP7+/− cells have decreased Arp2/3 (E) and F-actin (F) on VPS35-positive endosomes. The indicated HCT116 cells were subjected to immunostaining and quantification.
(G) Other activities of USP7 are not significantly altered in USP7+/− cells, such as alteration in p53, p21, or Bub3 levels. Cell lysates from the indicated HCT116 cells were separated by SDS-PAGE and immunoblotted.
(H-J) Expression of subject #6 USP7 Y143X in HCT116 USP7+/− cells does not rescue defects in CI-M6PR recycling (H) or accumulation of Arp2/3 (I) or F-actin (J) on FAM21-positive endosomes. The indicated HCT116 cells were transfected with GFP and wild-type (WT) or Y143X USP7 for 72 hrs before immunostaining and quantitation of GFP-positive cells.
Results are representative of at least three replicate experiments. More than 100 cells were quantitated. Values shown are mean + SD. Asterisks indicates p<0.05. n.s. indicates not significant (p>0.05). Scale bar represents 20 μm. See also Figure S5.
Table 1.
Mutation summary and clinical phenotypes of 7 individuals with USP7 mutations. See also Table S1
| Subject | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Summary |
|---|---|---|---|---|---|---|---|---|
| Sex | M | M | F | M | M | F | M | 5 M; 2 F |
| Age (years) | 9 | 10 | 9 | 8 | 5 | 13 | 8 | 8.9 ± 2.4 (SD) |
| Mutation Type | Del | Del | Del | Del | Del | Nonsense c.429C>G (p.Y143X) | Del | |
| Inheritance | De novo | De novo | De novo | De novo | De novo | De novo | De novo | |
| Genomic Size | 4.74 Mb | 91.6 kb | 3.27 Mb | 3.72 Mb | 267 kb | 1 b | 280 kb | |
| Genes affected (#) | 17 | 2 | 8 | 9 | 2 | 1 | 5 | |
| Symptoms | ||||||||
| DD/ID | + | + | + | + | + | + | + | 7/7 |
| Autism spectrum disorder | + | + | + | + | + | - | o | 5/6 |
| Seizures | + | - | + | + | + | + | - | 5/7 |
| Cryptorchidism/micropenis | + | + | N/A | - | + | N/A | + | 4/5 |
| Hypotonia | + | + | - | - | + | + | - | 4/7 |
| Aggressive Behavior | - | + | + | + | + | - | - | 4/7 |
Abbreviations: Del, deletion; DD/ID, developmental delay/intellectual disability; F, female; M, male; N/A, not applicable; +, present; -, not present; o, unknown; b, base; kb, kilobases; Mb, megabases; #, number; SD, standard deviation.
USP7 is haploinsufficient for its specific function in endosomal protein recycling
Given these findings, we next investigated whether USP7 is haploinsufficient for its role in regulating TRIM27 protein levels and WASH-mediated endosomal protein recycling on a molecular basis. Partial knockdown of USP7 resulted in a significant decrease in TRIM27 protein levels and impairment of CI-M6PR recycling (Figure 5B and S5A). Furthermore, knockout of one USP7 allele in HCT116 cells (USP7+/−) resulted in impaired CI-M6PR recycling (Figure 5C-D), decreased Arp2/3 localization to VPS35-positive endosomes (Figure 5E and S5B), and decreased F-actin accumulation on VPS35-positive endosomes (Figure 5F and S5C). Importantly and as reported previously (Cummins and Vogelstein, 2004; Kon et al., 2010), many of the other functions of USP7, including regulation of the p53 pathway, were not dramatically altered in USP7+/− cells as compared to USP7−/− cells (Figure 5G). Furthermore, specific abrogation of the p53 regulatory pathway by knockdown of HDM2 or HDMX did not impair localization of Arp2/3 or accumulation of F-actin on FAM21-positive endosomes (Figure S5D-F). Consistent with these findings, knockdown of MAGE-L2 or TRIM27 did not impact other functions of USP7, such as regulation of the MDM2-p53 pathway (Figure S5G). Additionally, reconstitution of USP7+/− cells with USP7 Y143X found in subject #6 failed to rescue CI-M6PR recycling or rescue Arp2/3 and F-actin levels on FAM21-positive endosomes (Figure 5H-J). These results, in combination with the similarities to clinical manifestations of children with MAGEL2 mutation, suggest that disruption of the MAGE-L2-TRIM27-USP7 WASH regulatory pathway is associated with Schaaf-Yang syndrome phenotypes in humans, prominently featuring intellectual disability and autism spectrum disorder.
MAGE-L2-TRIM27-USP7 plays an important role in hypothalamic neurons
To begin to understand how disruption of the MAGE-L2-TRIM27-USP7 regulatory axis controlling retromer and WASH-mediated endosomal protein recycling may contribute to disease, we investigated which anatomical tissues may be critically dependent on this regulatory pathway by examining if MAGE-L2, TRIM27, or USP7 are enriched in specific tissues. Unlike the ubiquitous TRIM27 and USP7 genes (Figure S6A-B) that have functions independent of regulation of WASH, MAGE-L2 is highly enriched in the brain of both mice and humans (Figure 6A-C). Detailed analysis of the various regions of the mouse brain revealed that MAGE-L2 is highly enriched in the hypothalamus (Figure 6A-B). Intriguingly, the hypothalamus is strongly implicated in several of the clinical symptoms associated with MAGEL2 and USP7 deletion/mutation, including PWS and autism (Cassidy et al., 2012; Jacobson, 2014; Tennese and Wevrick, 2011; Wetsel et al., 1991). Therefore, we examined whether MAGE-L2-TRIM27-USP7 plays an important role in regulation of WASH in hypothalamic neurons that endogenously express MAGE-L2 (Miller et al., 2009). Partial knockdown of USP7 resulted in reduction in TRIM27 protein levels (Figure 6D) and impairment in Arp2/3 localization (Figure 6E-F) and F-actin assembly (Figure 6E and 6G) on FAM21-positive endosomes. Knockdown of other genes within the deletion regions around the USP7 genomic locus (Figure 5A) did not decrease TRIM27 protein levels (Figure S6C) or levels of Arp2/3 on FAM21-positive endosomes (Figure S6D). Furthermore, CRISPR/Cas9 knockout of USP7 in hypothalamic neurons resulted in similar phenotypes, including reduced TRIM27 protein levels (Figure 6H) and diminished the level of F-actin on FAM21-positive endosomes (Figure 6I-J). Therefore, MAGE-L2-TRIM27-USP7 may be critically important for proper hypothalamic function, and disruption of this pathway may contribute to the development of Schaaf-Yang syndrome with its various phenotypic manifestations.
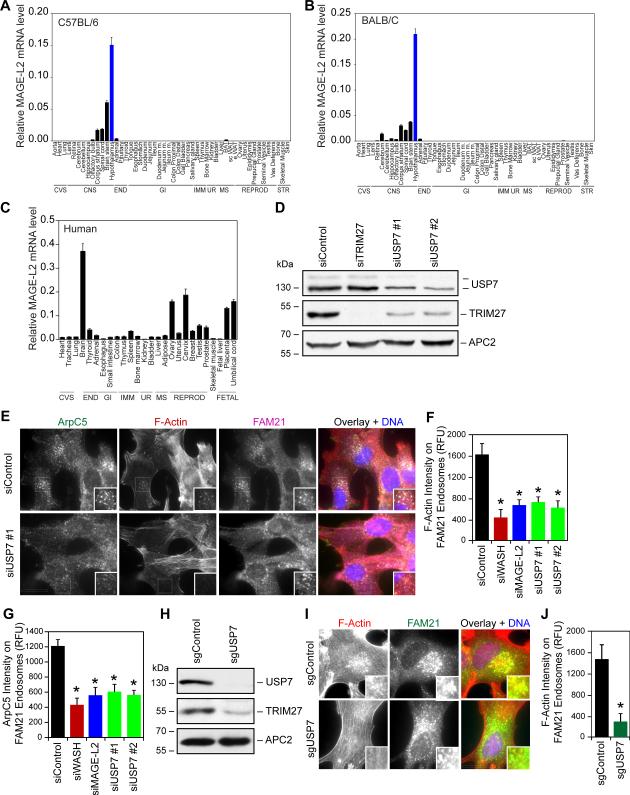
Figure 6. MAGE-L2-TRIM27-USP7 plays critical role in the hypothalamus.
(A and B) RT-QPCR analysis reveals that MAGE-L2 is highly enriched in the hypothalamus of both C57BL/6 (A) and BALB/c (B) mice. The indicated tissues were collected from six mice, pooled, and RT-QPCR analysis of MAGE-L2 expression was performed and normalized to 18S rRNA.
(C) Expression of human MAGE-L2 is enriched in the brain. RNA from the indicated tissues were obtained and RT-QPCR analysis of MAGE-L2 expression was performed and normalized to 18S rRNA.
(D) Knockdown of USP7 in hypothalamic neurons significantly decreases TRIM27 protein levels. Gt1-7 hypothalamic neurons were treated with the indicated siRNAs for 72 hrs before cell lysates were immunoblotted for the indicated proteins.
(E-G) Knockdown of MAGE-L2 or USP7 in hypothalamic neurons impairs F-actin accumulation (E and F) and Arp2/3 endosomal localization (E and G). Gt1-7 hypothalamic neurons were treated with the indicated siRNAs for 72 hrs before immunostaining (E) and quantification (F and G).
(H-J) CRISPR/Cas9 knockout of USP7 decreases TRIM27 protein levels (H) and levels of F-actin on FAM21-positive endosomes (I-J) levels. Gt1-7 hypothalamic neurons were transfected with GFP-Cas9 and the indicated gRNAs for 48 hrs before GFP-positive cells were collected by flow cytometry and analyzed by immunoblotting (H) and immunostaining (I-J) after culturing. Results are representative of at least three replicate experiments. More than 100 cells were quantitated. Values shown are mean + SD. Asterisks indicates p<0.05. Scale bar represents 20 μm. See also Figure S6.
DISCUSSION
Ubiquitination plays many important regulatory roles during the complex process of endosomal protein transport, including endocytosis, sorting and targeting of proteins to multi-vesicular bodies, and regulation of recycling pathways. Previous studies have implicated multiple deubiquitinating enzymes in these processes, including USP8 that is required for retrograde transport of CI-M6PR (MacDonald et al., 2014). We provide evidence that USP7 functions in the endosomal protein recycling pathway through its incorporation into the MAGE-L2-TRIM27 ubiquitin ligase complex. Intriguingly, USP7 has dual activities: 1) promoting WASH ubiquitination by preventing TRIM27 auto-ubiquitination and degradation through its deubiquitinating activity, 2) limiting WASH ubiquitination through direct deubiquitination of WASH. These seemingly opposing activities of USP7 allow for precision control and tuning of WASH activity. This is critically important because endosomal F-actin follows the ‘Goldilocks principle’ in which too little or too much is detrimental for retromer-dependent protein recycling (Gomez and Billadeau, 2009; Hao et al., 2013) (Figure 4). In this way, USP7 acts to buffer WASH ubiquitination to maintain optimal activity and endosomal F-actin levels. This level of cooperativity observed between USP7 and MAGE-L2-TRIM27 may extend to other DUB-E3 ligase pairs.
In addition to defining a role for USP7 in cellular protein trafficking and its mechanisms of action, we report a genetic disorder due to disruption of USP7. We show that heterozygous deletion or mutation of USP7 results in a number of neurological and behavioral phenotypes. Importantly, there is marked clinical overlap with Schaaf-Yang syndrome caused by MAGE-L2 loss-of-function, such as hypotonia, developmental delay/intellectual disability, autism spectrum disorder, and hypogonadism. Other phenotypes appear to be more distinct to USP7 haploinsufficiency, i.e. seizures and aggressive behaviors. Conversely, some phenotypes characteristic of PWS were not associated with deletion or mutation of USP7, namely infantile feeding difficulties, excessive weight gain, hyperphagia, and distinct craniofacial features. This suggests a spectrum of genotype-phenotype correlations with classic PWS on one side, MAGEL2-associated Schaaf-Yang syndrome, and now USP7-associated disease at the other end of the spectrum. Consistent with our findings, a genome-wide association study implicated TRIM27 in autism spectrum disorder (St Pourcain et al., 2013). Additionally, genomic duplications involving USP7 were reported in individuals with autism spectrum disorder (Sanders et al., 2011), further supporting the idea of dosage sensitivity, with both too little and too much USP7 causing imbalances of neuronal homeostasis. Furthermore, conditional knockout of USP7 in the mouse brain results in neonatal lethality that cannot be fully rescued by p53 knockout (Kon et al., 2011). This is consistent with our failure to identify individuals homozygous null for USP7 and our findings suggesting that USP7 has critical p53-independent functions in the brain.
Our results indicate that the disruption of USP7-mediated regulation of MAGE-L2-TRIM27 and WASH impairs endosomal protein recycling and F-actin assembly. This cellular function of USP7 may contribute to the described USP7-associated disease, although we cannot exclude that some of the phenotypes may be attributed to other nuclear functions of USP7 or TRIM27, such as regulation of TNFα-induced cell death through deubiquitination of RIP1 (Zaman et al., 2013). Similarly to the linkage of endocytic trafficking to neurodegenerative disorders, including mutation of the retromer subunit VPS35 in Parkinson disease (Vilarino-Guell et al., 2011), our findings imply that alterations in recycling of membrane proteins in the brain may contribute to impaired neurological and cognitive functions. Consistently, a role for endosomal trafficking proteins in a neurodevelopmental disorder of the mouse was recently reported (Watson et al., 2015). Finally, our results suggest that chemically activating WASH in these patients may have therapeutic potential.
EXPERIMENTAL PROCEDURES
Cell culture, transfections, CRISPR/Cas9, and antibodies
GT1-7 hypothalamic neuronal cells (kindly provided by Pamela Mellon, UCSD)(Wetsel et al., 1991), HCT116 USP7+/+, USP7+/−, USP7−/− cells (kindly provided by Bert Vogelstein, Johns Hopkins University) (Cummins et al., 2004), HEK293 (ATCC), HeLa Tet-ON (Clontech), or U2OS (ATCC) cells were grown in DMEM (Invitrogen) supplemented with 10% FBS (Hyclone), 2 mM L-Glutamine (Invitrogen), 100 units/ml penicillin (Invitrogen), 100 mg/ml streptomycin (Invitrogen), and 0.25 mg/ml amphotericin B (Invitrogen). Plasmid and siRNA transfection were performed for 48-96 hours with Effectene (QIAGEN) or Lipofectamine RNAiMAX (Invitrogen), respectively, according to the manufacturer's protocol.
siRNAs (GE Dharmacon or Sigma) and antibodies used in this study are listed in the supplemental experimental procedures. USP7 knockout GT1-7 cells were generated by cotransfection of plasmids encoding GFP-Cas9 and a gRNA plasmid targeting USP7 (5’-GGTTGCCTCGGAGCGCCAAC-3’). Forty-eight hours after transfection, cells were flow sorted for GFP-positive cells. USP7 disruption was confirmed by western blotting.
Tandem affinity purification, immunoprecipitation, and immunoblotting
Tandem affinity purification (TAP) was performed using 293/TAP-Vector or 293/TAP-MAGEL2 stable cell lines as described previously (Doyle et al., 2010) and detailed in extended supplemental procedures. Immunoprecipitation and immunoblotting were performed as described previously (Potts and Yu, 2005) and in extended supplemental procedures.
In vitro binding assays
Proteins were purified from bacterial or SF9 cells using standard protocols and described in detail in supplemental experimental procedures. In vitro binding assays were performed as described previously (Doyle et al., 2010) with details in supplemental experimental procedures.
Immunofluorescence, microscopy, and quantitative measurements
Immunofluorescence was performed essentially as described previously (Hao et al., 2013; Potts and Yu, 2007) and detailed in the supplemental experimental procedures. Retrograde transport assays measuring trafficking of internalized CI-M6PR, CTxB, and transferrin were performed as previously described (Hao et al., 2013). Cells were imaged with a 63X or 100X objective on a DeltaVision or Nikon TiU inverted fluorescence microscope. Images were acquired with a CoolSnap HQ2 charge-coupled device camera (Photometrics) at 0.3-μm intervals, deconvolved using the nearest neighbor algorithm, and stacked to better resolve endosome structures. Intensity measurements were performed using ImageJ Software. For those experiments specifically quantitating the amount of endosomal localized F-Actin or ArpC5, images (>100 cells over multiple independent experiments) were thresholded in ImageJ to create a mask (or region of interest) for the relevant endosomes (anti-FAM21 or anti-VPS35 signal) and the relative fluorescent units intensity of F-Actin (phalloidin) or anti-ArpC5 signal overlapping with FAM21 or VPS35 in the thresholded (masked) region was determined. The threshold for endosomal markers was kept constant across all images analyzed to prevent bias. Quantitation of CI-M6PR trafficking was performed by blind analysis of the percentage of cells showing compact juxtanuclear (normal) or dispersed (abnormal) CI-M6PR staining. For colocalization analysis, Pearson correlation measurements were performed using ImageJ colocalization package. At least 100 cells were counted for each condition in each experiment. Statistical analysis was performed using two-tailed unpaired student's t-test.
In vitro ubiquitination assay
In vitro ubiquitination assays were performed as described previously (Doyle et al., 2010). Assays were performed using 100 nM His-Ube1 (Biomol), 2.5 μM E2 (His-UbcH5c), 150 nM E3 (His-MAGE-L2/MBP-TRIM27/MBP-USP7 WT or C223S mutant), 2.5 μM ubiquitin (Biomol), 25 nM substrate (GST-WASH), 0.5 mM Mg-ATP (Biomol), 20 units/ml inorganic pyrophosphatase (Sigma), and 1 mM DTT in 1x ubiquitylation buffer (Biomol). Negative control reactions were set up as described above except Mg-ATP was replaced with 5 mM EDTA. Reactions were stopped by the addition of SDS sample buffer after incubation at 30°C for 2 hours and subjected to SDS-PAGE and immunoblotting for auto-ubiquitination of TRIM27 and ubiquitination of WASH.
Human subjects
Clinical chromosome microarray databases (Baylor College of Medicine Molecular Genetics Laboratory, BCM-MGL, and Signature Genomic Laboratories), the publically available DECIPHER (DatabasE of genomiC varIation and Phenotype in Humans using Ensembl Resources) database, and the database of clinical whole exome sequencing at the Whole Genome Laboratory (WGL) of Baylor College of Medicine were queried in February 2014. The Baylor and Signature Genomic chromosome microarray databases contained samples of 94,242 individuals combined at that time, while the DECIPHER database contained approximately 10,000 individuals at the time of query. We limited our search on those smaller than 5 Mb in size. Seven cases were identified, of which six were successfully enrolled in this study (Signature Genomics: subject 1; DECIPHER: subjects 2 and 7; BCM-MGL: subjects 3, 4, and 5). The WGL database of exome sequencing encompassed approximately 1,500 cases at the time of query. Importantly, these are consecutive, unrelated samples without prescreening criteria. Samples are submitted as a clinical test, on the referring physician's determination that the patient likely has a genetic change that has led to genetic disease.
As part of a study approved by the Institutional Review Board of Baylor College of Medicine, the referring physicians of subjects 1, 3, 4, 5, and 6 were re-contacted, and families granted permission to publish genomic and phenotypic information. For subjects 2 and 7, patient consent was obtained by the attending geneticist for public and collaborative group data sharing, giving access to genomic and phenotypic data and patient reports. Following informed consent, we performed a comprehensive chart review of medical records. As well, providers were asked to fill out a clinical questionnaire. See Supplemental Clinical Notes for details on subjects.
RNA preparation and cDNA synthesis
Specifics regarding tissue collection from animals are described in supplemental experimental procedures. RNA was extracted using RNAStat60 (TelTest) according to the manufacturer's directions. Total RNA was pooled in equal quantities for each tissue (n = 6). Genomic DNA contamination was eliminated by DNase I (Roche) treatment in 4.5 mM MgCl2. cDNA for qPCR assays was prepared from 4 μg DNase-treated RNA using High Capacity cDNA Reverse Transcription kit (Life Technologies). Gene expression levels were measured as described in supplemental experimental procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Potts lab, Michael Rosen, Daniel Billadeau, and Ezra Burstein for helpful discussions and critical reading of the manuscript. We also thank Pamela Mellon and Bert Vogelstein for critical reagents, Genevera I. Allen for statistical support and discussion, and Pawel Stankiewicz for his invaluable contributions on patient data. This work was supported by Michael L. Rosenberg Scholar in Medical Research fund (PRP), CPRIT R1117 (PRP), WELCH Foundation I-1821 (PRP), NIH R01GM111332 (PRP), the Joan and Stanford Alexander Family (CPS), and the Foundation for Prader-Willi Research (CPS and PRP).
We are indebted to the patients and their families for their willingness to participate in our study. This study makes use of data generated by the DECIPHER Consortium. A full list of centers that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust. Those who carried out the original analysis and collection of the data for DECIPHER bear no responsibility for the further analysis or interpretation of it by our group or the registered users of DECIPHER.
The project was supported in part by IDDRC grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. Cores: translational core.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization – PRP; Investigation – YH, KFT, PRP; Formal analysis – PRP, YH, KFT, CPS, MDF; Resources – FX, WB, SLK, AP, JAR, CLC, BI, IDK, SEN, JPP, TMM, RM, RCP, MSS; Data curation – MDF, CPS; Writing – Original draft – PRP, YH; Writing – Review and edit – PRP, CPS, YH, KFT, MDF, JAR, BI; Visualization – PRP, CPS, YH, KFT, MDF; Supervision – PRP, CPS; Project administration – PRP, CPS; Funding acquisition – PRP, CPS.
REFERENCES
- Bischof JM, Stewart CL, Wevrick R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum Mol Genet. 2007;16:2713–2719. doi: 10.1093/hmg/ddm225. [DOI] [PubMed] [Google Scholar]
- Boccaccio I, Glatt-Deeley H, Watrin F, Roeckel N, Lalande M, Muscatelli F. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum Mol Genet. 1999;8:2497–2505. doi: 10.1093/hmg/8.13.2497. [DOI] [PubMed] [Google Scholar]
- Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harbor perspectives in biology. 2014;6 doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genetics in medicine : official journal of the American College of Medical Genetics. 2012;14:10–26. doi: 10.1038/gim.0b013e31822bead0. [DOI] [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1–486. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell cycle. 2004;3:689–692. [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YH, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell. 2013;152:1051–1064. doi: 10.1016/j.cell.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson L. Hypothalamic-pituitary-adrenocortical axis: neuropsychiatric aspects. Comprehensive Physiology. 2014;4:715–738. doi: 10.1002/cphy.c130036. [DOI] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci U S A. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nature reviews Molecular cell biology. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Zhong J, Kobayashi Y, Li M, Szabolcs M, Ludwig T, Canoll PD, Gu W. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ. 2011;18:1366–1375. doi: 10.1038/cdd.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, Hogenesch JB, Muglia LJ, Van Gelder RN, Herzog ED, Stewart CL. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39:1266–1272. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- Lee S, Kozlov S, Hernandez L, Chamberlain SJ, Brannan CI, Stewart CL, Wevrick R. Expression and imprinting of MAGEL2 suggest a role in Prader-willi syndrome and the homologous murine imprinting phenotype. Hum Mol Genet. 2000;9:1813–1819. doi: 10.1093/hmg/9.12.1813. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- MacDonald E, Urbe S, Clague MJ. USP8 controls the trafficking and sorting of lysosomal enzymes. Traffic. 2014;15:879–888. doi: 10.1111/tra.12180. [DOI] [PubMed] [Google Scholar]
- Mercer RE, Kwolek EM, Bischof JM, van Eede M, Henkelman RM, Wevrick R. Regionally reduced brain volume, altered serotonin neurochemistry, and abnormal behavior in mice null for the circadian rhythm output gene Magel2. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:1085–1099. doi: 10.1002/ajmg.b.30934. [DOI] [PubMed] [Google Scholar]
- Mercer RE, Wevrick R. Loss of magel2, a candidate gene for features of Prader-Willi syndrome, impairs reproductive function in mice. PLoS One. 2009;4:e4291. doi: 10.1371/journal.pone.0004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NL, Wevrick R, Mellon PL. Necdin, a Prader-Willi syndrome candidate gene, regulates gonadotropin-releasing hormone neurons during development. Human molecular genetics. 2009;18:248–260. doi: 10.1093/hmg/ddn344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell biochemistry and biophysics. 2011;60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CP, Gonzalez-Garay ML, Xia F, Potocki L, Gripp KW, Zhang B, Peters BA, McElwain MA, Drmanac R, Beaudet AL, et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet. 2013;45:1405–1408. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer JB, Morgan DO. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J Biol Chem. 2011;286:45186–45196. doi: 10.1074/jbc.M111.310094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Gautreau A, Billadeau DD. Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- St Pourcain B, Whitehouse AJ, Ang WQ, Warrington NM, Glessner JT, Wang K, Timpson NJ, Evans DM, Kemp JP, Ring SM, et al. Common variation contributes to the genetic architecture of social communication traits. Molecular autism. 2013;4:34. doi: 10.1186/2040-2392-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennese AA, Wevrick R. Impaired hypothalamic regulation of endocrine function and delayed counterregulatory response to hypoglycemia in Magel2-null mice. Endocrinology. 2011;152:967–978. doi: 10.1210/en.2010-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JA, Bhattacharyya BJ, Vaden JH, Wilson JA, Icyuz M, Howard AD, Phillips E, DeSilva TM, Siegal GP, Bean AJ, et al. Motor and Sensory Deficits in the teetering Mice Result from Mutation of the ESCRT Component HGS. PLoS Genet. 2015;11:e1005290. doi: 10.1371/journal.pgen.1005290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetsel WC, Mellon PL, Weiner RI, Negro-Vilar A. Metabolism of pro luteinizing hormone-releasing hormone in immortalized hypothalamic neurons. Endocrinology. 1991;129:1584–1595. doi: 10.1210/endo-129-3-1584. [DOI] [PubMed] [Google Scholar]
- Zaman MM, Nomura T, Takagi T, Okamura T, Jin W, Shinagawa T, Tanaka Y, Ishii S. Ubiquitination-deubiquitination by the TRIM27-USP7 complex regulates tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol. 2013;33:4971–4984. doi: 10.1128/MCB.00465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.