Abstract
The ATR-Chk1 pathway is critical for DNA damage responses and cell cycle progression. Chk1 inhibition is more deleterious to cycling cells than ATR inhibition, raising questions about ATR and Chk1 functions in the absence of extrinsic replication stress. Here, we show that a key role of ATR in S phase is to coordinate RRM2 accumulation and origin firing. ATR inhibitor (ATRi) induces massive ssDNA accumulation and replication catastrophe in a fraction of early S-phase cells. In other S-phase cells, however, ATRi induces moderate ssDNA and triggers a DNA-PK and Chk1-mediated backup pathway to suppress origin firing. The backup pathway creates a threshold such that ATRi selectively kills cells under high replication stress, whereas Chk1 inhibitor induces cell death at a lower threshold. The levels of ATRi-induced ssDNA correlate with ATRi sensitivity in a panel of cell lines, suggesting that ATRi-induced ssDNA could be predictive of ATRi sensitivity in cancer cells.
Introduction
The ATR (ATM [ataxia telangiectasia-mutated]- and rad3-related) kinase is a crucial safeguard of the genome (Ciccia and Elledge, 2010; Cimprich and Cortez, 2008; Flynn and Zou, 2011; Marechal and Zou, 2013). In response to a wide range of intrinsic and extrinsic genotoxic stress, ATR acts as a master regulator of DNA damage signaling to orchestrate DNA repair, DNA replication, and cell cycle progression. In particular, when DNA replication is compromised, ATR plays a key role in stabilizing the genome (Zeman and Cimprich, 2014). Several recent studies have shed light on how ATR functions in cells challenged with replication inhibitors, such as hydroxurea (HU) and aphidicolin (APH) (Couch et al., 2013; Ragland et al., 2013; Toledo et al., 2013). In the presence of high levels of extrinsic replication stress, ATR is critical for preventing excessive cleavage of replication forks by nucleases and replication catastrophe, a state in which replicating chromosomes undergo severe fragmentation (Couch et al., 2013; Ragland et al., 2013; Toledo et al., 2013). Although these studies provided important insights into one of the pivotal roles of ATR, how ATR functions under physiological and pathological conditions in the absence of extrinsic replication stress is yet to be elucidated.
ATR and its homologues in a number of organisms are critical for the survival of proliferating cells. In budding yeast, the ATR homologue Mec1 is essential for viability unless Sml1, a repressor of ribonucleotide reductase, is deleted (Zhao et al., 1998). In mouse and C. elegans, loss of ATR leads to embryonic lethality (Brown and Baltimore, 2000; Garcia-Muse and Boulton, 2005). Conditional deletion of ATR from the human colon cancer cell line HCT116 also leads to cell death (Cortez et al., 2001). However, ATR homologs in some other organisms, such as fission yeast and Drosophila, are not essential for viability (Enoch et al., 1992; Laurencon et al., 2003). Interestingly, in mouse, the effects of ATR loss on proliferating cells are not uniform in cell populations. For example, deletion of ATR in cells from blastocyosts resulted in different levels of genomic instability, arranging from a few DNA breaks to severe chromosomal fragmentation (Brown and Baltimore, 2000). Deletion of ATR during nervous system development induced cell death only in specific progenitor cells (Lee et al., 2012). These observations raise an important question as to why some proliferating cells are more dependent on ATR than others.
How ATR functions during S phase is still poorly understood. During the response to DNA damage or replication stress, ATR phosphorylates and activates its effector kinase Chk1 (Liu et al., 2000). It has been long believed that ATR and Chk1 function as a kinase cascade. Like ATR, Chk1 is critical for genomic stability during DNA replication (Forment et al., 2011; Petermann et al., 2008; Petermann et al., 2010; Syljuasen et al., 2005). Both ATR and Chk1 have been implicated in the regulation of origin firing even in the absence of extrinsic stress (Couch et al., 2013; Eykelenboom et al., 2013; Maya-Mendoza et al., 2007; Petermann et al., 2010; Shechter et al., 2004). However, a recent study reported unexpected differences between the effects of ATR inhibitor (ATRi) and Chk1 inhibitor (Chk1i) on cycling cells (Toledo et al., 2011). Whereas Chk1i induced massive γH2AX accumulation in a large fraction of U2OS cells, ATRi only induced γH2AX in a small fraction of the cells. This result raises the possibility that ATR and Chk1 may not always function as a linear pathway, and it posts a question about how ATR and Chk1 function in concert during DNA replication.
While both ATR and Chk1 are important in cycling cells, the nature of the intrinsic stress that they deal with remains enigmatic. Interestingly, certain proliferation-promoting oncogenic events, such as Ras activation and Myc overexpression, render cancer cells sensitive to ATR suppression (Gilad et al., 2010; Murga et al., 2011; Schoppy et al., 2012), leading to the hypothesis that ATR is important for countering the replication stress in cancer cells. Nevertheless, how replication stress can be measured in normal and cancer cells remains elusive. Because multiple ATRi and Chk1i are being tested in clinical trials for cancer therapy (Foote et al., 2013; Josse et al., 2014; Karp et al., 2012; Ma et al., 2013; Sausville et al., 2014; Seto et al., 2013), understanding the mechanisms of action and unique properties of these inhibitors may help to guide their applications in clinical settings.
In this study, we used multiple inhibitors and siRNAs to interrogate the functions of ATR and Chk1 during S phase. Unexpectedly, we found that acute inactivation of ATR in S-phase cells led to two distinct outcomes. Upon ATRi treatment, a fraction of S-phase cells accumulated high levels of ssDNA and underwent replication catastrophe. In contrast, other S-phase cells initially acquired moderate levels of ssDNA but subsequently recovered from the “ATRi shock” through a Chk1-mediated mechanism. The critical role of ATR in suppressing replication catastrophe was traced to its functions in promoting RRM2 (ribonucleotide reductase M2) accumulation and limiting replication origin firing in early S phase. In the ATRi-treated cells escaping from replication catastrophe, ATRi triggered a DNA-PK and Chk1-mediated backup pathway to suppress origin firing. Importantly, the Chk1-mediated backup pathway in ATRi-treated cells creates a threshold of tolerable replication stress, allowing ATRi to selectively kill cells under high replication stress. In contrast to ATRi, Chk1i disrupted the backup pathway and induced cell death even when replication stress was moderate. Notably, the levels of ATRi-induced ssDNA correlated with ATRi-induced cell death in a panel of cell lines, suggesting that ATRi-induced ssDNA is a quantitative indicator of replication stress that could be used to predict the ATRi sensitivity of cancer cells.
Results
Acute ATR inactivation leads to two distinct outcomes in S-phase cells
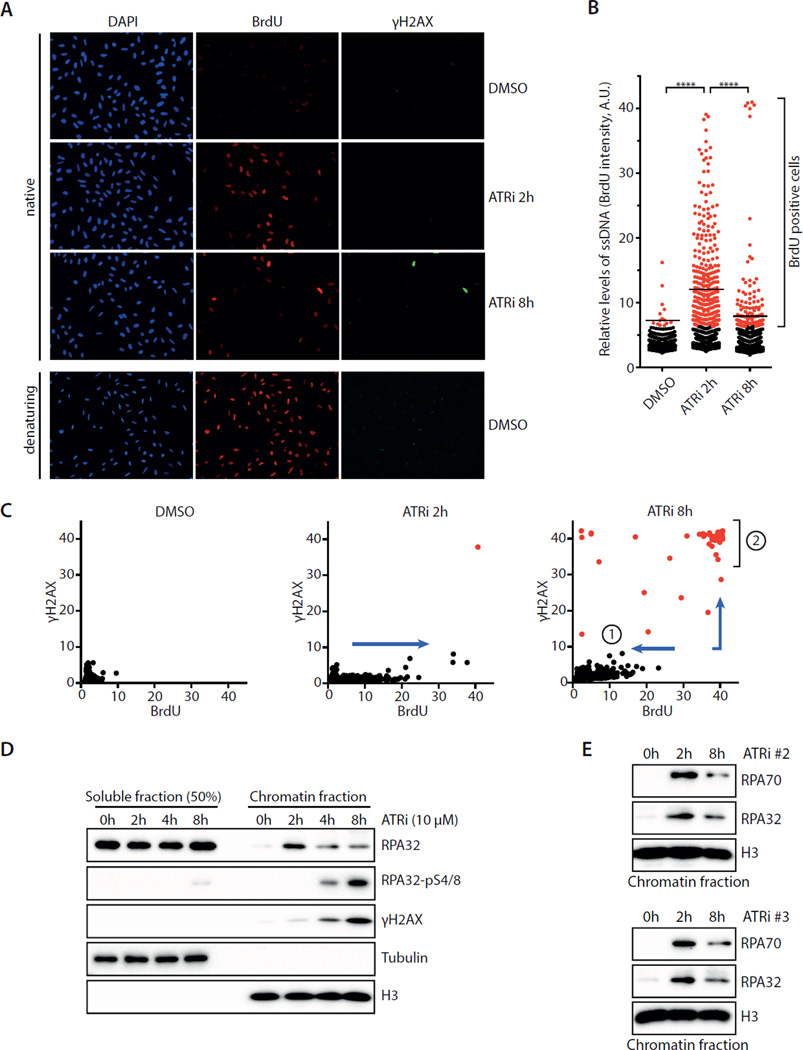
To investigate how ATR functions in cycling cells, we acutely inactivated ATR in U2OS cells with the ATR inhibitor VE-821 and followed the effects over time (Fig. S1A–B) (Reaper et al., 2011). To visualize ssDNA, DNA was labeled with BrdU and analyzed by native BrdU staining. An increase of ssDNA was detected in S-phase cells 2 hours after ATRi treatment (Fig. 1A–C, S1C). At 8 hour after ATRi treatment, a fraction (∼5%) of S-phase cells displayed very high levels of ssDNA and became strongly positive for γH2AX and TUNEL staining (Fig. 1A–C, S1D–E), indicating that they were undergoing replication catastrophe (Toledo et al., 2013). Surprisingly, however, the majority of ATRi-treated cells displayed less ssDNA at 8 hour than at 2 hour (Fig. 1A–C). This reduction in ssDNA was not due to loss of S-phase cells (Fig. S1F). Consistent with the induction of ssDNA at 2 hour, increased amounts of RPA were detected on chromatin by fractionation and immunostaining (Fig. 1D, S1G). Subsequently, the levels of RPA on chromatin gradually declined. Similar decline of chromatin-bound RPA was also observed in cells treated with two other ATR inhibitors, AZ20 and EPT-46464 (Fig. 1E) (Foote et al., 2013; Toledo et al., 2011). Despite the overall decline of chromatin-bound RPA, phosphorylated RPA32 and gH2AX gradually accumulated on chromatin in a fraction of cells after 2 hours (Fig. 1A–D, S1H). Thus, while a subpopulation of ATRi-treated cells acquired high levels of ssDNA and DNA damage, a distinct cell subpopulation gradually recovered. These results raise an important question as to how the distinct effects of ATRi are manifested in S-phase cells.
Fig. 1. Acute ATR inhibition exerts two distinct effects on S-phase cells.
A. U2OS cells were cultured in BrdU for 36 h, treated with DMSO or ATRi (10 μM VE-821), and analyzed for BrdU and γH2AX by immunestaining. B. Quantification of the BrdU intensity of 1,000 U2OS cells treated with DMSO or ATRi. Black lines indicate median BrdU intensities of BrdU-positive cells in various cell populations. C. Quantification of the BrdU and γH2AX intensities of 1,200 U2OS cells treated with DMSO or ATRi. Cell subpopulation 1 displayed less ssDNA at 8 h than at 2h. Cell subpopulation 2 displayed very high levels of ssDNA and became γH2AX-positive at 8h. D. Levels of RPA32, pRPA32, and γH2AX in the soluble and chromatin fractions of ATRi-treated cells were analyzed by Western blot. E. The levels of chromatin-bound RPA32 and pRPA32 were analyzed in cells treated with ATRi#2 (AZ20) and ATRi#3 (EPT-46464). See also Fig. S1.
ATR suppresses ssDNA accumulation in early S phase
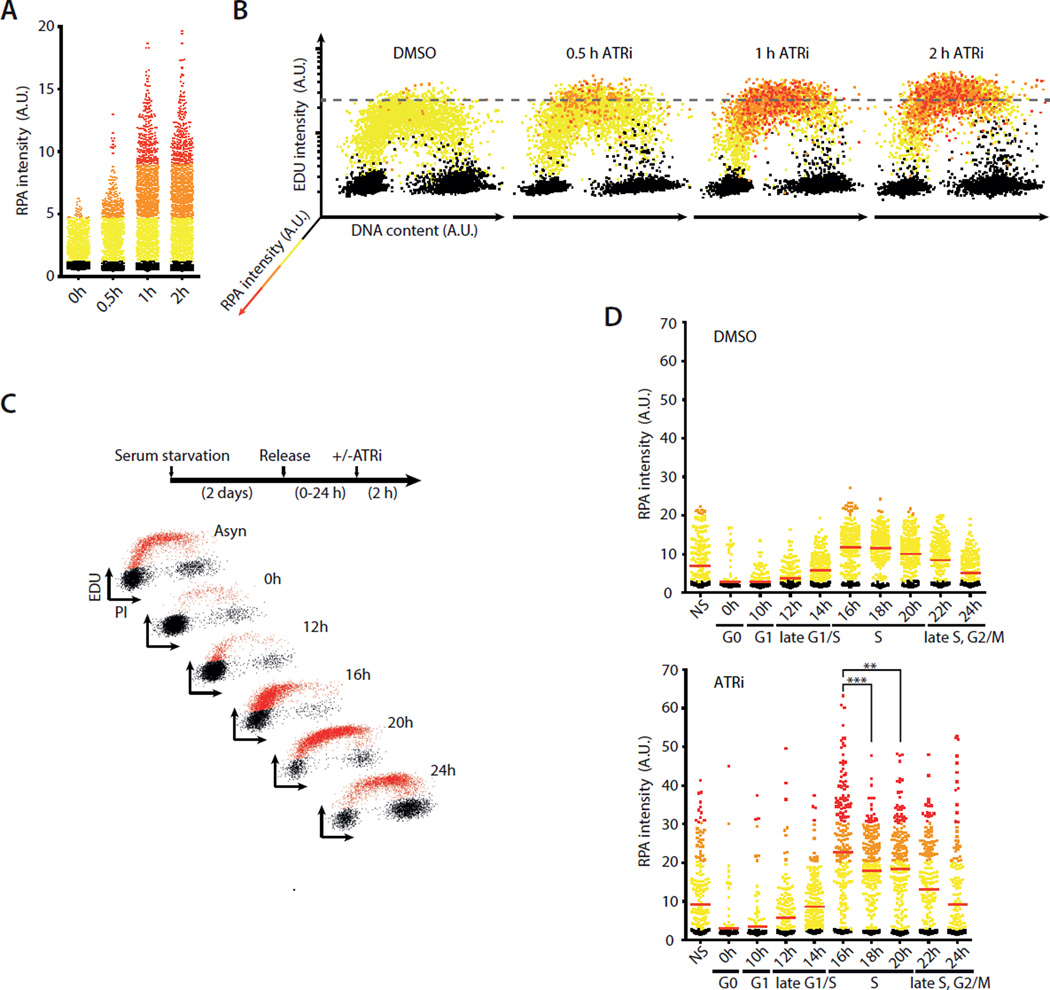
The distinct effects of ATRi on S-phase cells prompted us to investigate whether a fraction of replicating cells are particularly vulnerable to ATR inactivation. Consistent with the induction of ssDNA by ATRi, the staining of chromatin-bound RPA in individual cells gradually increased during the first 2 hours (Fig. 2A). When cells were sorted according to EdU incorporation, DNA content, and RPA staining, it was evident that the chromatin binding of RPA occurred most efficiently in a fraction of cells in early-to-mid S phase (Fig. 2B).
Fig. 2. ATR suppresses ssDNA accumulation in early S phase.
A-B. Quantification of chromatin-bound RPA, EdU incorporation, and DNA contents of 5,000 U2OS cells treated with DMSO or ATRi (10 μM VE-821). Cells were color-coded according to the intensity of RPA staining as shown in the left panel. C. T98G cells were synchronously released from G0 and analyzed for EdU incorporation at the indicated times. D. Staining intensity of chromatin-bound RPA was analyzed at different stages of the cell cycle after ATRi or DMSO treatment. Red lines indicate mean RPA intensities in various cell populations. **, P<0.01; ***, P<0.001. See also Fig. S2.
To test more directly if early or mid S-phase cells are most vulnerable to ATR inactivation, we treated synchronously growing T98G cells with ATRi in different stages of the cell cycle (Fig. 2C, S2). T98G cells were synchronized in G0 by serum starvation and then released into the cell cycle. Even in the absence of ATRi, low levels of ssDNA were detected in replicating cells (Fig. 2D). Interestingly, the basal levels of ssDNA in replicating cells peaked in early S phase, suggesting that cells in this cell-cycle window are facing relatively high levels of intrinsic replication stress. Furthermore, ATRi induced higher levels of ssDNA in early S-phase cells than in mid or late S-phase cells (Fig. 2D), suggesting that ATR is particularly important for the suppression of ssDNA in early S phase.
ATR suppresses DNA damage by promoting RRM2 accumulation and limiting origin firing
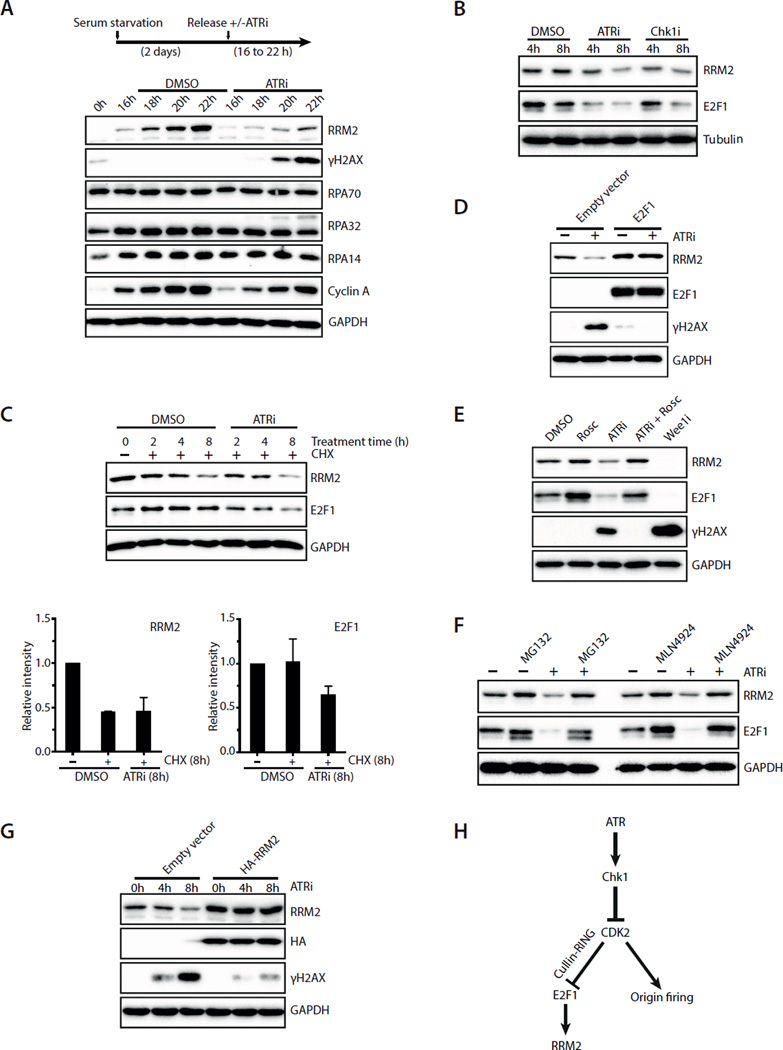
To understand why early S-phase cells are vulnerable to ATR inactivation, we tested if certain DNA replication factor(s) is limiting during this period. In the presence of HU, ATRi induces excessive firing of replication origins and a massive increase of stalled replication forks, which ultimately leads to exhaustion of RPA and replication catastrophe (Toledo et al., 2013). Even in the absence of HU, ATRi induced a surge of origin firing in 2 hours (Fig. S3A). To test if RPA is limiting in early S phase, we monitored the levels of all three RPA subunits in synchronously growing T98G cells (Fig. 3A). The levels of RPA70, RPA32, and RPA14 were constant throughout the cell cycle and not affected by ATRi, ruling out RPA as the limiting factor in early S phase.
Fig. 3. ATRi suppresses DNA damage by promoting RRM2 accumulation and limiting origin firing.
A. T98G cells were synchronously released from G0 in the presence or absence of ATRi (10 μM VE-821). Levels of RRM2, γH2AX, RPA70, RPA32, RPA14 and Cyclin A were analyzed during the time course. B. Asynchronously growing U2OS cells were treated with ATRi (10 μM VE-821) or Chk1i (2 μM MK-8776). Levels of RRM2 and E2F1 were analyzed at the indicated times. C. Levels of RRM2 and E2F1 were analyzed in U2OS cells treated with DMSO or ATRi in the presence of cycloheximide (CHX). Relative levels of RRM2 and E2F1 were quantified from 3 blots (n=3). Error bars: S.D. D. U2OS cells transfected with empty vector or E2F1-expressing plasmids were treated with ATRi for 8 h. Levels of RRM2, E2F1, and γH2AX were analyzed. E. U2OS cells were treated with the indicated inhibitors for 8 h. Levels of RRM2, E2F1, and γH2AX were analyzed. F. U2OS cells were treated with DMSO or ATRi for 8 h in the presence or absence of MG132 or MLN4924. G. U2OS cells infected with HA-RRM2-expressing retrovirus or control virus were treated with ATRi for 8 h. Levels of RRM2 and γH2AX were analyzed at the indicated times. H. A model in which ATR coordinates RRM2 accumulation and origin firing in early S phase. See also Fig. S3.
In contrast to RPA, RRM2, a cell cycle-regulated subunit of the ribonucleotide reductase, gradually accumulated in early S phase (Fig. 3A) (Chabes et al., 2003; D’Angiolella et al., 2012). Notably, ATRi attenuated the accumulation of RRM2 in S phase (Fig. 3A). Even in asynchronous U2OS cells, ATRi and Chk1i (MK-8776) reduced the levels of RRM2 (Fig. 3B, S3B), suggesting that the ATR-Chk1 pathway promotes RRM2 accumulation in cycling cells. Surprisingly, although RRM2 is an unstable protein, its degradation was not enhanced by ATRi in cells treated with cycloheximide (CHX) (Fig. 3C). Knockdown of Cyclin F, the F-box protein required for RRM2 ubiquitylation in G2 (D’Angiolella et al., 2012), did not suppress the reduction of RRM2 in ATRi-treated cells (Fig. S3C). In contrast to RRM2, E2F1, the transcription activator of the RRM2 gene (DeGregori et al., 1995; Zhang et al., 2009), was increasingly degraded in ATRi-treated cells in the presence of CHX (Fig. 3C). Similar to RRM2, E2F1 was reduced in ATRi and Chk1i–treated cells (Fig. 3B, S3B). Concomitant with the reduction of E2F1, RRM2 mRNA levels declined (Fig. S3D). Importantly, overexpression of E2F1 completely suppressed the reduction of RRM2 in ATRi-treated cells (Fig. 3D), suggesting that E2F1 degradation is responsible for the reduction of RRM2 by ATRi. Together, these results show that the ATR-Chk1 pathway promotes RRM2 accumulation by stabilizing E2F1.
The ATRi-induced reduction in E2F1 and RRM2 was suppressed by the CDK inhibitor roscovitine, the proteasome inhibitor MG132, and the Nedd8-activating enzyme inhibitor MLN4924 (Fig. 3E–F) (Soucy et al., 2009). In contrast, Wee1 inhibitor, which activates CDK1/2 (Beck et al., 2012; Hughes et al., 2013), drastically reduced E2F1 and RRM2 levels even in the absence of ATRi (Fig. 3E). These results suggest that the ATR-Chk1 pathway promotes RRM2 accumulation by antagonizing a CDK1/2, Cullin-RING ubiquitin ligase and proteasome-mediated mechanism that degrades E2F1 (see Fig. 3H).
In ATRi-treated cells, roscovitine not only elevated RRM2 levels but also reduced the induction of γH2AX (Fig. 3E). Furthermore, a CDK2-specific inhibitor also reduced γH2AX (Fig. S3E). These results suggest that a reduction in CDK2 activity may suppress ATRi-induced DNA damage by increasing RRM2 levels. Indeed, expression of RRM2 significantly reduced the γH2AX induced by ATRi or Chk1i (Fig. 3G, S3F). In addition to its effects on RRM2, roscovitine also decreased origin firing in ATRi-treated cells (Fig. S3A). To test if suppression of origin firing reduces ATRi-induced DNA damage, we used siRNA to knock down CDC7, a key regulator of replication initiation (Labib, 2010). Knockdown of CDC7 indeed reduced ATRi-induced γH2AX (Fig. S3G). Thus, ATR, through restricting CDK2 activity, promotes RRM2 accumulation and limits origin firing in S phase (Fig. 3H). Both of these effects of ATR contribute to the suppression of DNA damage in replicating cells (Fig. 3H). How ATR is activated during unperturbed S phase and how E2F1 is suppressed by CDK2 and Cullin ligases remain to be investigated. The transient accumulation of ssDNA in S-phase cells may trigger limited ATR activation, thereby coordinating RRM2 accumulation and origin firing. Interestingly, the budding yeast ATR homolog Mec1 is required for priming the Mcm2–7 helicase for phosphorylation by Cdc7 (Randell et al., 2010). The limited ATR activation during S phase may promote origin firing but also restrict it to a tolerable level, preventing ssDNA from accumulating to a high level that triggers replication catastrophe (see Fig. 7A).
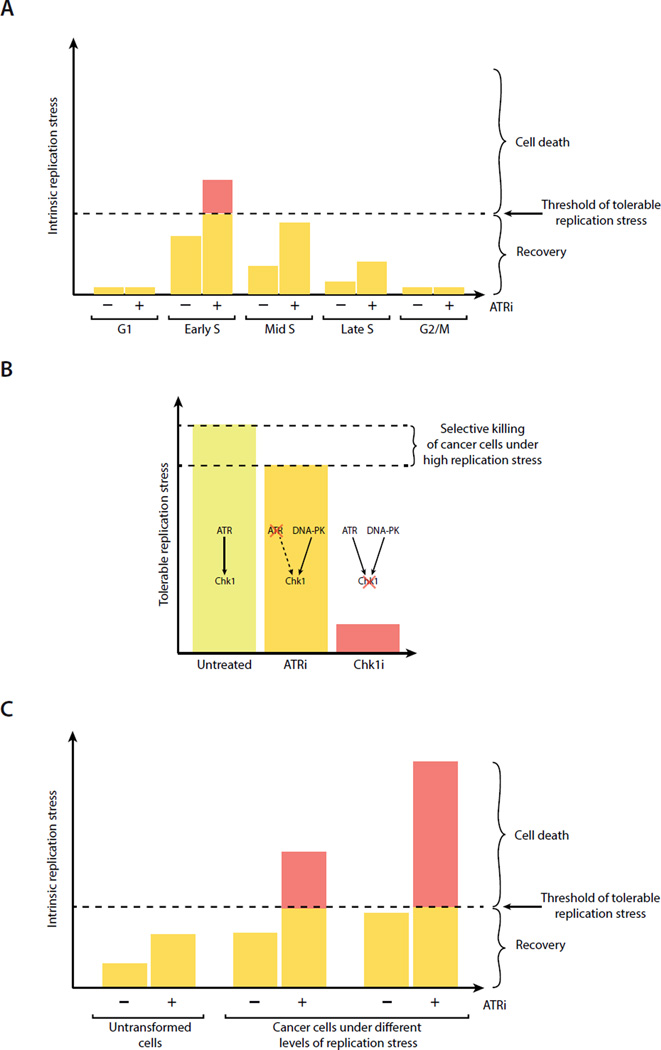
Fig. 7. Modeling the roles for ATR, DNA-PK, and Chk1 in countering replication stress.
A. A fraction of early S-phase cells are particularly vulnerable to ATR inactivation. B. ATRi selectively kills cells under high replication stress, whereas Chk1i induces cell death even in cells in which replication stress is moderate. C. ATRi-induced ssDNA is an indicator of replication stress that may predict the ATRi sensitivity of cancer cells.
Recovery of ATRi-treated cells via a Chk1-mediated mechanism
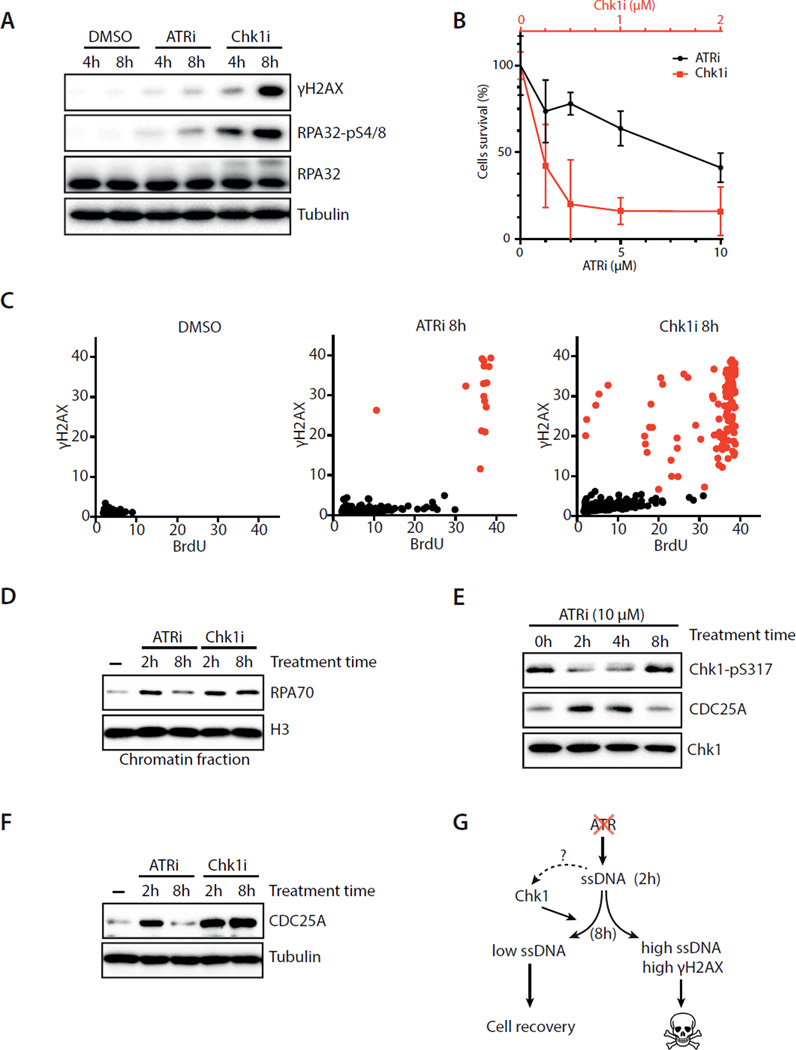
The correlations of high ssDNA with replication catastrophe and moderate ssDNA with recovery suggest that the fate of ATRi-treated cells may be dictated by a threshold of ssDNA (see Fig. 7A). To investigate the underlying mechanism of recovery, we first tested if ATRi gradually looses its potency during this process. Even after 8 hours of incubation with cells, ATRi still effectively blocked the CPT-induced Chk1 phosphorylation (Fig. S4A). Although ATRi remained active at 8 hour, its effects were significantly different from those of Chk1i. Using Chk1 autophosphorylation at S296 as a functional readout (Okita et al., 2012), we carefully titrated ATRi and Chk1i to determine their respective concentrations required for Chk1 inactivation (Fig. S1A, S4B–C). We found that 10 μM ATRi and 1–2 μM Chk1i similarly inhibited CPT-induced Chk1 autophosphorylation (Fig. S4B). Furthermore, CDC25A, which is degraded in a Chk1-dependent manner (Busino et al., 2003; Jin et al., 2003), was similarly stabilized by 10 μM ATRi and 1–2 μM Chk1i in unperturbed cycling cells (Fig. S4C). At functionally equivalent concentrations, Chk1i induced much more γH2AX-positive cells than did ATRi (Fig. 4A). Similar observations were made using different ATRi and Chk1i (Fig. S4D–E), as well as multiple independent ATR and Chk1 siRNAs (Fig. S4F–G). Importantly, cells were significantly more sensitive to Chk1i than ATRi (Fig. 4B, S4H), showing that Chki is indeed more cytotoxic than ATRi. These results confirm and extend the observation by Toledo et al. (Toledo et al., 2011), prompting us to further investigate why ATRi and Chk1i exert different effects.
Fig. 4. ATRi-treated cells recover via a Chk1-mediated mechanism.
A. U2OS cells were treated with DMSO, ATRi (10 μM VE-821), or Chk1i (2 μM MK-8776). Levels of RPA32, pRPA32, and γH2AX were analyzed at the indicated times. B. U2OS cells were treated with increasing concentrations of ATRi or Chk1i for 24 h and then cultured in inhibitor-free media. Cell survival was analyzed 4 days after treatment. Error bar: S.D. (n=3). C. U2OS cells were treated with DMSO, ATRi, or Chk1i for 8 h. BrdU and γH2AX intensities were quantified in 1,200 cells at the indicated times. D. U2OS cells were treated with ATRi or Chk1i. Levels of chromatin-bound RPA were analyzed at the indicated times. E. U2OS cells were treated with ATRi, and levels of pChk1 and CDC25A were analyzed at the indicated times. F. Levels of CDC25A in U2OS cells treated with ATRi or Chk1i were compared at the indicated times. G. Amodel in which Chk1 promotes the recovery of ATRi-treated cells with moderate ssDNA. See also Fig. S4.
We next compared the effects of ATRi and Chk1i on cycling cells at different time points. ATRi and Chk1i induced similar levels of ssDNA at 2 hour (Fig. S4I). However, at 8 hour, ssDNA was reduced in the majority of ATRi-treated cells but increased in Chk1i–treated cells (Fig. 4C, S4J). The fraction of Chk1i–treated cells displaying high levels of ssDNA was also positive for γH2AX (Fig. 4C), suggesting that they were undergoing replication catastrophe. Consistent with the ssDNA results, the levels of chromatin-bound RPA declined from 2 to 8 hour only in ATRi-treated cells, but not in Chk1i–treated cells (Fig. 4D). These results show that the recovery observed in ATRi-treated cells does not occur in Chk1i–treated cells, raising the possibility that Chk1 is involved in recovery.
Since Chk1 is a downstream effector of ATR in the DNA damage response, it is surprising that Chk1 may function in recovery independently of ATR. To follow the function of Chk1 during recovery, we analyzed the phosphorylation of Chk1 at S317 and Chk1-mediated CDC25A destabilization in ATRi-treated cells (Busino et al., 2003; Jin et al., 2003; Zhao and Piwnica-Worms, 2001). Before ATRi treatment, a basal level of pChk1 was detected (Fig. 4E). At 2 hour after ATRi treatment, the level of pChk1 was reduced, whereas the level of CDC25A was elevated. Subsequently, pChk1 gradually reappeared, and CDC25A gradually declined. At 8 hour, the levels of pChk1 and CDC25A became similar to those before ATRi treatment. Importantly, CDC25A levels remained high at 8 hour in Chk1i–treated cells (Fig. 4F). Even at low concentrations that only partially stabilized CDC25A at 2 hour, Chk1i prevented the decline of CDC25A at 8 hour (Fig. S4K). Consequently, even 0.25–0.5 μM Chk1i induced higher levels of γH2AX than 10 μM ATRi at late time points (Fig. S4L). Thus, during the recovery of ATRi-treated cells, Chk1 regains its basal phosphorylation on S317 and its function in destabilizing CDC25A. These results suggest that a large fraction of S-phase cells recover from the initial ATRi response via a Chk1-mediated mechanism (Fig. 4G).
DNA-PK phosphorylates Chk1 to suppress origin firing and promote recovery in ATRi-treated cells
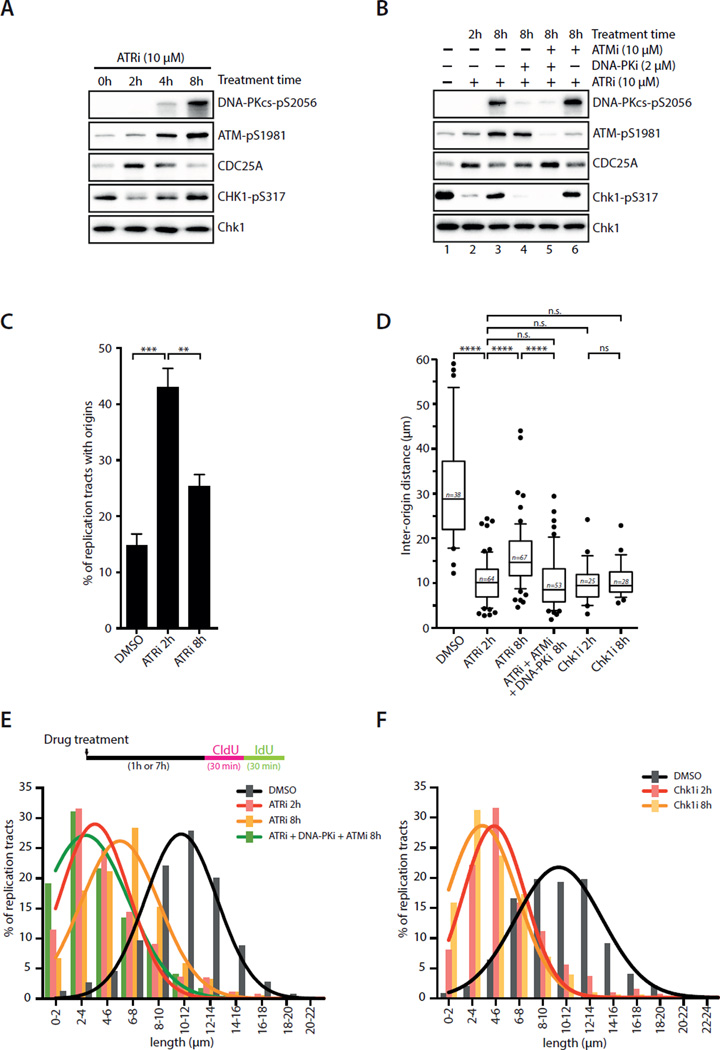
To understand the regulation of Chk1 during recovery, we investigated how Chk1 is phosphorylated at S317 at 8 hour after ATRi treatment. Even in cells that were repeatedly treated with ATRi, pChk1 still reappeared (Fig. S5A), excluding ATR as the kinase that phosphorylates Chk1. Both ATM and DNA-PK were autophosphorylated 8 hours after ATRi treatment (Fig. 5A). Surprisingly, the ATRi-induced phosphorylation of Chk1, as well as the phosphorylation of H2AX and RPA32, was dependent on DNA-PK but not ATM (Fig. 5B lanes 4, 6; S5B–D). The induction of γH2AX and pRPA32 was reduced by knockdown of KU70, a DSB (DNA double-stranded break) sensor for DNA-PK, as well as SLX4 and MUS81, two components of a nuclease complex that processes aberrant replication forks (Fig. S5E–F) (Couch et al., 2013; Forment et al., 2011; Ragland et al., 2013). These results suggest that the aberrant replication forks induced by ATRi are processed by SLX4 and MUS81, leading to KU-dependent DNA-PK activation. When both DNA-PK and ATM were inhibited, the reappearance of pChk1 was eliminated and CDC25A was stabilized at 8 hour (Fig. 5B lane 5; Fig. S5B). Thus, the resurgence of Chk1 function during recovery is primarily driven by DNA-PK, but ATM also has a secondary role in this process.
Fig. 5. Regulation and function of Chk1 during recovery.
A. Levels of pDNA-PK, pATM, pChk1 and CDC25A were analyzed in ATRi-treated U2OS cells at the indicated times. B. U2OS cells were treated with ATRi, ATMi, DNA-PKi, or the combinations of these inhibitors. Levels of pDNA-PK, pATM, pChk1 and CDC25A were analyzed 8 h after treatment. C. The percentage of replication tracts containing fired origins was determined in RPE1 cells treated with DMSO or ATRi at the indicated times. Error bars: S.D. (n=3 experiments). **P<0.01; ***P<0.001. D. RPE1 cells were treated with DMSO or various inhibitors as indicated. The inter-origin distance was analyzed using DNA fiber assay at the indicated times. Error bars: S.E.M. (n=25 to 67 as indicated). ****, P<0.0001; n.s., not significant. E-F. RPE1 cells were treated with DMSO or various inhibitors as indicated. The length of continuous replication tracts was determined using DNA fiber assay at the indicated times (>600 forks per condition, n=3 experiments). See also Fig. S5.
Our finding that DNA-PK phosphorylates Chk1 in ATRi-treated cells raised a question as to whether this alternative pathway involves the known regulators of Chk1 phosphorylation in the ATR-Chk1 pathway. Knockdown of Rad17 and Clapsin, a sensor and a mediator of the ATR-Chk1 pathway, did not prevent Chk1 from regaining S317 phosphorylation in ATRi-treated cells (Fig. S5G–H). In contrast, knockdown of TopBP1, an activator of the ATR kinase and a mediator of the ATR-Chk1 pathway, abolished pChk1 8 hours after ATRi treatment (Fig. S5I). Although the role of TopBP1 in the DNA-PK-Chk1 pathway is still unclear, a recent study suggested that DNA-PK phosphorylates TopBP1 to promote Chk1 phosphorylation (Vidal-Eychenie et al., 2013). Our results suggest that the sensing and signaling mechanisms of the DNA-PK-Chk1 pathway are distinct from those of the ATR-Chk1 pathway.
To gain mechanistic insights into how Chk1 promotes recovery, we used DNA fiber assay to monitor origin firing and fork progression in ATRi-treated cells. At 2 hour after ATRi treatment, origin firing was elevated, reducing in the inter-origin distance and replication fork speed (Fig. 5C–E). At 8 hour, origin firing has clearly declined, whereas the inter-origin distance and fork speed have partially recovered (Fig. 5C–E). As in ATRi-treated cells, in Chk1i–treated cells the inter-origin distance was reduced and fork speed was decreased at 2 hour (Fig. 5D, 5F) (Maya-Mendoza et al., 2007; Petermann et al., 2006; Petermann et al., 2010). However, neither the inter-origin distance nor fork speed changed from 2 to 8 hour in Chk1i–treated cells (Fig. 5D, 5F), suggesting that Chk1 is needed to suppress origin firing during recovery. Consistently, when both DNA-PK and ATM were inhibited in ATRi-treated cells, the inter-origin distance did not increase and fork speed did not recover from 2 to 8 hour (Fig. 5D–E). The regain of Chk1 function in destabilizing CDC25A during recovery suggests that Chk1 suppresses origin firing by inhibiting CDK2. Indeed, inhibition of CDK in Chk1i–treated cells by roscovitine 2 hours after Chk1i treatment enabled the cells to recover (Fig. S5J). Furthermore, activation of CDK in ATRi-treated cells by Wee1i 2 hours after ATRi treatment prevented recovery (Fig. S5K).
ATRi but not Chk1i selectively kills cells under high replication stress
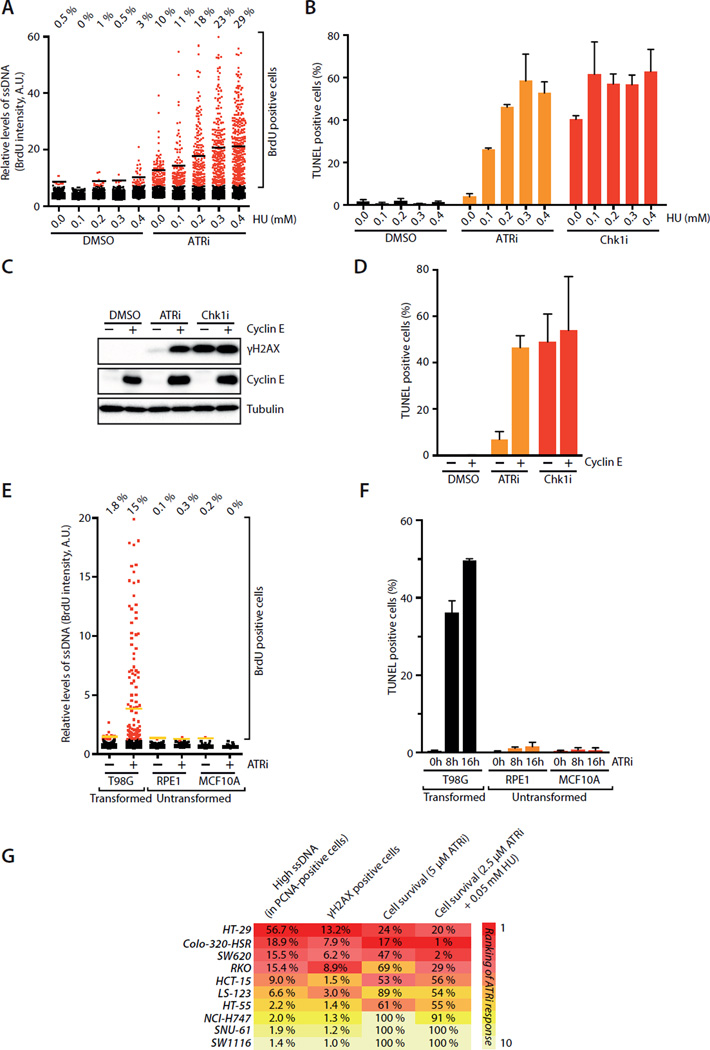
If the Chk1-mediated backup pathway promotes the recovery of ATRi-treated cells, it may create a threshold of tolerable replication stress. Consequently, ATRi may selectively kill cells under high replication stress, whereas Chk1i may kill cells even when replication stress is moderate (see Fig. 7B). To test these possibilities, we treated U2OS cells with ATRi in the presence of increasing concentrations of HU. As HU concentration rose, ATRi induced increasing levels of ssDNA (Fig. 6A), showing that the level of ATRi-induced ssDNA is an indicator of replication stress. Importantly, as the level of ssDNA rose, increasing fractions of ATRi-treated cells underwent replication catastrophe (Fig. 6B), confirming that ATRi selectively kills cells under high replication stress. In the presence of HU, Chk1i induced slightly more ssDNA than ATRi at 2 hour (Fig. S6A). At a later time point, Chk1i induced much more ssDNA than ATRi regardless of the presence or absence of HU (Fig. S6B). These results suggest that recovery occurs more efficiently in ATRi-treated cells than in Chk1i–treated cells independently of the levels of replication stress. In contrast to ATRi-treated cells, large fractions of Chk1i–treated cells underwent replication catastrophe even in the absence of HU (Fig. 6B), supporting the notion that disruption of the Chk1-mediated backup pathway kills replicating cells even if they are under moderate replication stress.
Fig. 6. ATRi selectively kills cells under high replication stress.
A. U2OS cells were treated with ATRi for 2 h in the presence of increasing concentrations of HU. The BrdU intensity of 1,000 U2OS cells was quantified. Black lines indicate mean BrdU intensities of BrdU-positive cells in various populations. B. U2OS cells were treated with ATRi or Chk1i for 16 h in the presence of increasing concentrations of HU. Cell death was measured by the TUNEL assay. Error bars: S.D. (n=3). C-D. U2OS cells were induced to overexpress Cyclin E or left uninduced, and treated with ATRi or Chk1i for 16h. Levels of Cyclin E and γH2AX were analyzed by Western (C). Fractions of TUNEL-positive cells were quantified (D). Error bars: S.D. (n=3). E-F. T98G, RPE1, and MCF10A cells were treated with DMSO or ATRi. Levels of ssDNA were analyzed by native BrdU staining 2 h after ATRi treatment (E). Yellow lines indicate mean BrdU intensities of BrdU-positive cells in various populations. Levels of cell death were measured by TUNEL assay at the indicated times (F). Error bars: S.D. (n=3). G. Quantification of ssDNA, γH2AX, and cell survival of 10 colorectal cell lines after ATRi treatment. Levels of ssDNA and γH2AX were analyzed 2 h and 16 h after the indicated treatments, respectively. Fractions of ATRi-treated cells displaying stronger BrdU staining than untreated cells were determined (see Supplemental Methods). Cell survival was analyzed 6 days after the indicated treatments using CellTiter-Glo. See also Fig. S6.
Replication stress arises from not only extrinsic but also intrinsic sources, such as the oncogenic events in cancer cells (Bartkova et al., 2005; Gorgoulis et al., 2005). Cyclin E is commonly overexpressed in cancer cells, and it interferes with DNA replication (Bartkova et al., 2005; Neelsen et al., 2013). ATRi induced ssDNA, γH2AX and replication catastrophe more efficiently in Cyclin E-overexpressing U2OS cells than in control cells (Fig. 6C–D, S6C) (Toledo et al., 2011). Loss of the tumor suppressor Rb also impairs DNA replication (Manning et al., 2014). Again, ATRi induced γH2AX more efficiently in Rb-depleted cells than in control cells (Fig. S6D). In marked contrast to ATRi, Chk1i induced γH2AX and replication catastrophe indiscriminately of the levels of replication stress (Fig. 6C–D, S6D). Importantly, ATRi induced much higher levels of ssDNA and replication catastrophe in two cancer cell lines, U2OS and T98G, than in two untransformed cell lines, RPE1 and MCF10A (Fig. 6E–F, S6E–G), suggesting that ATRi selectively kills cancer cells under high replication stress (see Fig. 7C). We recently showed that cancer cells dependent on the alternative telomere-lengthening (ALT) pathway are hypersensitive to ATRi (Flynn et al., 2015). Interestingly, whereas U2OS cells are ALT-positive, T98G cells express active telomerase (Sano et al., 1998). Thus, even in telomerase-positive cancer cells under high replication stress, ATRi induces massive ssDNA accumulation and replication catastrophe.
ATRi-induced ssDNA is an indicator of replication stress in cancer cells
ATRi induced higher levels of ssDNA and replication catastrophe in cancer cells than in non-transformed cells, suggesting that ATRi-induced ssDNA may be an indicator of replication stress in cancer cells. To test this possibility, we treated a panel of 10 colorectal cancer cell lines with ATRi and analyzed its effects on ssDNA (at 2 hour), γH2AX (at 16 hour), and cell survival (in 6 days) (Fig. S6H–K). Cell lines of colorectal cancer were selected for this experiment because the oncogenic drivers in this context have been well characterized (Kinzler and Vogelstein, 1996). Furthermore, colorectal cancer is not associated with ALT (Heaphy et al., 2011), giving us an opportunity to test if ATRi-induced ssDNA predicts ATRi sensitivity in non-ALT cells. All the cell lines replicated DNA efficiently (Fig. S6L), and they displayed a range of ATRi sensitivity (Fig. S6J–K). Remarkably, when the cell lines were ranked according to the ATRi-induced ssDNA accumulation, a correlation of ATRi-induced ssDNA and ATRi sensitivity was clearly discernable (Fig. 6G). This correlation was even more evident when a low concentration of HU was used to sensitize the ATRi response (Fig. 6G, S6J–K). These results suggest that the levels of ATRi-induced ssDNA in the colorectal cancer cells are indeed reflective of intrinsic replication stress and predictive of ATRi sensitivity. In contrast to ATRi-induced ssDNA, none of the common mutations of colorectal cancer, either individually or in combinations, are predictive of ATRi sensitivity (Fig. S6M) (Kinzler and Vogelstein, 1996). Furthermore, the microsatellite instability, CpG island methylation phenotype, and chromosomal instability of these cell lines did not correlate with ATRi sensitivity (Fig. S6N) (Ahmed et al., 2013). Therefore, ATRi-induced ssDNA in cancer cells is a unique indicator of replication stress that predicts ATRi sensitivity.
Discussion
A key function of ATR in unperturbed S phase
While recent studies have shed light on the function of ATR in the response to replication inhibition, how ATR acts during the unperturbed cell cycle is still poorly understood. In this study, we found that ssDNA accumulates in the genome even during unperturbed DNA replication, especially in early S phase. Furthermore, acute ATR inactivation leads to increased ssDNA accumulation, particularly in early S-phase cells. Due to the massive ssDNA accumulation resulting from ATR inactivation, a fraction of S-phase cells undergo replication catastrophe. These results suggest that a key function of ATR in unperturbed S phase is to protect early S-phase cells by suppressing ssDNA accumulation. Even during unperturbed DNA replication, cells face a threshold of intrinsic replication stress (Fig. 7A). If the level of intrinsic replication stress in an individual cell goes above the threshold, ssDNA would trigger replication catastrophe in this cell. In a population of cycling cells, early S-phase cells have the highest probability to cross the threshold. During unperturbed S phase, ATR functions to limit ssDNA and protect cells from this threshold. When ATR is lost, the surge of ssDNA drives a fraction of cells over the threshold, killing them through replication catastrophe (Fig. 7A). This model explains the critical function of ATR in unperturbed S phase, and it clarifies why the effects of ATR loss are not uniform in replicating cell populations.
Why are early S-phase cells particularly vulnerable to ATR loss? As cells enter S phase, a large number of early replication origins are activated by CDK2 and CDC7 (Rhind and Gilbert, 2013). This surge of DNA synthesis creates a large demand on dNTPs. Despite the demand on dNTPs, RRM2, a critical factor for dNTP synthesis, is still low in early S phase. This conflict between dNTP consumption and production in early S phase may create a situation resembling the depletion of dNTPs by HU. In this study, we found that ATR promotes RRM2 accumulation by antagonizing a CDK2, Cullin-RING and proteasome-mediated mechanism that down regulates E2F1. Furthermore, the ATR-Chk1 pathway limits origin firing by restricting CDK2 activity. Through these two distinct effects, ATR promotes the synthesis of dNTPs and limits their consumption in early S phase, thereby suppressing ssDNA accumulation and replication catastrophe. Consistent with our findings, a recent study reported that overexpression of RRM2 suppressed the genomic instability in ATR-deficient mice (Lopez-Contreras et al., 2015). Similar to human ATR, the budding yeast ATR homolog Mec1 also promotes dNTP synthesis and prevents inappropriate firing of dormant origins (Huang and Elledge, 1997; Santocanale et al., 1999). It is tempting to speculate that the synthesis and consumption of dNTPs during DNA replication vary in different cellular and organismal contexts, which may explain why ATR is essential in some situations but not others.
The functions of ATR in S phase share similarities to its functions in the DNA damage response. When activated by DNA damage or replication inhibition, ATR suppresses origin firing (Costanzo et al., 2003; Toledo et al., 2013). ATR/ATM and Chk1 regulate E2F1 stability and RRM2 levels after DNA damage (Lin et al., 2001; Zhang et al., 2009). Despite these similarities, the roles of ATR in the absence or presence of extrinsic stress are not identical. While ATR limits CDK2 activity and origin firing in S phase, it does not trigger a cell-cycle arrest. The levels of Chk1 phosphorylation in unperturbed cycling cells are significantly lower than those induced by DNA damage, suggesting that some of ATR functions may be controlled by signaling thresholds. Interestingly, a recent study showed that yeast Mec1 phosphorylates different substrates in the absence or presence of DNA damage (Bastos de Oliveira et al., 2015), raising the possibility that ATR has distinct substrate specificity in S phase and the DNA damage response. Further studies are needed to reveal how ATR is elicited and tuned in these distinct functional modes.
A Chk1-mediated backup pathway for ATR
In this study, we have made an unexpected observation that a large fraction of S-phase cells were able to recover from their initial response to ATRi. During the initial response to ATRi, many S-phase cells acquired moderate levels of ssDNA. Presumably because the levels of ssDNA in these cells have not reached the threshold, they did not undergo replication catastrophe. Instead, the ssDNA in these cells triggered a DNA-PK and Chk1-mediated backup pathway to destabilize CDC25A, reduce CDK2 activity, and suppress origin firing. Consequently, ssDNA is gradually reduced in these cells, allowing them to recover from the “ATRi shock”. Both ATM and DNA-PK have been implicated in the response to extrinsic replication stress when ATR is absent (Chanoux et al., 2009). Interestingly, DNA-PK but not ATM is the main upstream kinase of Chk1 in ATRi-treated cells during recovery, although ATM may have a secondary role in this process. The activation of DNA-PK requires ssDNA accumulation, the SLX4-associated nuclease complex, and KU70. These results suggest that the accumulation of ssDNA at replication forks may give rise to aberrant DNA structures susceptible to SLX4-mediated nucleolytic processing (Couch et al., 2013; Forment et al., 2011; Ragland et al., 2013), generating DSBs that activate DNA-PK. Why DNA-PK but not ATM is preferentially used in this backup pathway is still unclear. Although the activation of this backup pathway may require the transient presence of a few DSBs, it allows a large fraction of S-phase cells to avoid replication catastrophe and continue to proliferate in the absence of ATR.
The discovery of the DNA-PK and Chk1-mediated backup pathway explains why Chk1i is more cytotoxic in cycling cells than ATRi (Fig. 7B). Owning to the backup pathway, ATRi-treated cells can still cope with a substantial level of replication stress. This explains why ATRi selectively kills cells under high replication stress. On the other hand, Chk1i disrupts not only the canonical ATR-Chk1 cascade but also the backup pathway, thereby lowering the threshold of tolerable replication stress drastically. As a result, Chk1i kills replicating cells even when they are under moderate replication stress, explaining its high cytotoxicity. It should be noted that although many ATRi-treated cells can recover from the initial ATRi response, some of these cells are only partially recovered. Consistent with a previous study (Couch et al., 2013), we found that prolonged ATRi treatment increased cell death (Fig. 4B, S4H). Nonetheless, the cytotoxicity of ATRi is lower than Chk1i in a wide range of concentrations. Our findings suggest that ATRi is more selective than Chk1i toward cells under high replication stress, revealing an important distinction between these inhibitors that could guide their applications in cancer therapy.
ATRi-induced ssDNA as an indicator of replication stress
Although ATR is long known to be a master regulator of cellular responses to replication stress, whether and how replication stress can be quantified remains elusive. A quantitative understanding of replication stress is crucial for explaining the functions of ATR in specific oncogenic, developmental, aging and therapeutic contexts (Brown and Baltimore, 2000; Flynn et al., 2015; Gilad et al., 2010; Lee et al., 2012; Murga et al., 2009; Murga et al., 2011; Reaper et al., 2011; Ruzankina et al., 2007). In this study, we found that the levels of ATRi-induced ssDNA vary in individual cells and in different stages of S phase. In HU-treated cells, the levels of ATRi-induced ssDNA rise with HU concentrations, suggesting that ATRi-induced ssDNA reflects replication stress quantitatively. Furthermore, in a panel of cancer cell lines, the levels of ATRi-induced ssDNA correlate with ATRi-induced cell death. These results suggest that ATRi-induced ssDNA is also an indicator of intrinsic replication stress, and it is predictive of ATRi sensitivity in cancer cells (Fig. 7C). Although replication stress could arise from many different sources, induction of ssDNA may be a common effect (Flynn and Zou, 2011). Elicited by ssDNA (Zou and Elledge, 2003), ATR acts to counter replication stress by suppressing ssDNA accumulation (Toledo et al., 2013). When ATR is lost, replication stress is unleashed to drive further ssDNA formation, generating a quantifiable product reflecting its own strength. We propose that ATRi-induced ssDNA is a quantitative indicator of both extrinsic and intrinsic replication stress. The use of ATRi-induced ssDNA to measure replication stress may help to explain the roles of ATR in different functional contexts and predict the outcomes of ATR inhibition, providing a quantitative view of the interplay between replication stress and the ATR checkpoint.
Methods
Cell culture
U2OS, T98G and RPE1-hTERT cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin. U2OS cells expressing HA-RRM2 were generated by retroviral infection (pBabe-HA-RRM2) and puromycin selection. U2OS-derived cells carrying inducible Cyclin E were cultured in DMEM supplemented with 10% FBS and 4 µg/ml tetracycline (Bartkova et al., 2006). Cyclin E expression was induced by washing off tetracycline 24 h before drugs treatment. MCF-10A cells were cultured in DMEM/F12 supplemented with 5 % Horse Serum, 2 ng/ml EGF, 0.5 µg/ml hydrocortizone, 100 ng/ml cholera toxin, 10 µg/ml Insulin and 1% penicillin/streptomycin. HT-29, SNU-61, NCI-H747, HCT-15 and Colo-320-HSR were cultured in Roswell Park Memorial Institute 1640 Medium (RPMI 1640) GlutaMAX™-I supplemented with 10% FBS, 1% penicillin/streptomycin, 1% Glucose and 1% Sodium Pyruvate. SW1116, SW620, HT-55, RKO and LS-123 were cultured in DMEM/F12 supplemented with 10% FBS and 1% penicillin/streptomycin.
Inhibitors
The kinase inhibitors used in this study are: ATRi (10 µM VE-821), ATRi#2 (1 µM AZ-20), ATRi#3 (10 µM EPT-46464), Chk1i (2 µM MK-8776), Chk1i#2 (0.3 µM UCN-01), Roscovitine (25 µM), DNA-PKi (2 µM NU7441), ATMi (10 µM KU-55933), and Wee1i (0.25 µM MK-1775). When various inhibitors were used in combination with ATRi or in comparison with ATRi, they were added to cell cultures at the same time as ATRi unless indicated otherwise.
Supplementary Material
Acknowledgement
We thank J. Bartek, C. Bakkenist, B. Chen, N. Dyson, M. Lopes, S. Maheswaran, M. Pagano and W. Wei for reagents, S. Yazinski and V. Comaills for advice on DNA fiber and RT-qPCR assays, E. Hardy for technical assistance and members of the Zou lab for helpful discussions. R.B. is partly supported by a Tosteson Postdoctoral Award, a Marsha Rivkin Scientific Scholar Award and a fellowship from the Phillips Foundation. L.Z. is a Jim & Ann Orr Massachusetts General Hospital (MGH) Research Scholar. This work is supported by grants from the US National Institutes of Health (GM076388) and the Federal Share of the MGH Proton Program to L.Z., and a grant from the Wellcome Trust (102696) to C.H.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknaes M, Hektoen M, Lind GE, Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. doi: 10.1038/oncsis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bastos de Oliveira FM, Kim D, Cussiol JR, Das J, Jeong MC, Doerfler L, Schmidt KH, Yu H, Smolka MB. Phosphoproteomics Reveals Distinct Modes of Mec1/ATR Signaling during DNA Replication. Mol Cell. 2015;57:1124–1132. doi: 10.1016/j.molcel.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck H, Nahse-Kumpf V, Larsen MS, O’Hanlon KA, Patzke S, Holmberg C, Mejlvang J, Groth A, Nielsen O, Syljuasen RG, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol. 2012;32:4226–4236. doi: 10.1128/MCB.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci U S A. 2003;100:3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem. 2009;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Betous R, Carroll CM, Jung SY, Qin J, Cimprich KA, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch T, Carr AM, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Eykelenboom JK, Harte EC, Canavan L, Pastor-Peidro A, Calvo-Asensio I, Llorens-Agost M, Lowndes NF. ATR activates the S-M checkpoint during unperturbed growth to ensure sufficient replication prior to mitotic onset. Cell Rep. 2013;5:1095–1107. doi: 10.1016/j.celrep.2013.10.027. [DOI] [PubMed] [Google Scholar]
- Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suva ML, Benes CH, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–140. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote KM, Blades K, Cronin A, Fillery S, Guichard SS, Hassall L, Hickson I, Jacq X, Jewsbury PJ, McGuire TM, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H–indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J Med Chem. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- Forment JV, Blasius M, Guerini I, Jackson SP. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS One. 2011;6:23517. doi: 10.1371/journal.pone.0023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. Embo J. 2005;24:4345–4355. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, Brown EJ. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–9702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Elledge SJ. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes BT, Sidorova J, Swanger J, Monnat RJ, Jr, Clurman BE. Essential role for Cdk2 inhibitory phosphorylation during replication stress revealed by a human Cdk2 knockin mutation. Proc Natl Acad Sci U S A. 2013;110:8954–8959. doi: 10.1073/pnas.1302927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse R, Martin SE, Guha R, Ormanoglu P, Pfister TD, Reaper PM, Barnes CS, Jones J, Charlton P, Pollard JR, et al. ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase i inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 2014;74:6968–6979. doi: 10.1158/0008-5472.CAN-13-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp JE, Thomas BM, Greer JM, Sorge C, Gore SD, Pratz KW, Smith BD, Flatten KS, Peterson K, Schneider P, et al. Phase I and pharmacologic trial of cytosine arabinoside with the selective checkpoint 1 inhibitor Sch 900776 in refractory acute leukemias. Clin Cancer Res. 2012;18:6723–6731. doi: 10.1158/1078-0432.CCR-12-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencon A, Purdy A, Sekelsky J, Hawley RS, Su TT. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics. 2003;164:589–601. doi: 10.1093/genetics/164.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Shull ER, Frappart PO, Katyal S, Enriquez-Rios V, Zhao J, Russell HR, Brown EJ, McKinnon PJ. ATR maintains select progenitors during nervous system development. Embo J. 2012;31:1177–1189. doi: 10.1038/emboj.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Lopez-Contreras AJ, Specks J, Barlow JH, Ambrogio C, Desler C, Vikingsson S, Rodrigo-Perez S, Green H, Rasmussen LJ, Murga M, et al. Increased Rrm2 gene dosage reduces fragile site breakage and prolongs survival of ATR mutant mice. Genes Dev. 2015;29:690–695. doi: 10.1101/gad.256958.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CX, Ellis MJ, Petroni GR, Guo Z, Cai SR, Ryan CE, Craig Lockhart A, Naughton MJ, Pluard TJ, Brenin CM, et al. A phase II study of UCN-01 in combination with irinotecan in patients with metastatic triple negative breast cancer. Breast Cancer Res Treat. 2013;137:483–492. doi: 10.1007/s10549-012-2378-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Yazinski SA, Nicolay B, Bryll A, Zou L, Dyson NJ. Suppression of genome instability in pRB-deficient cells by enhancement of chromosome cohesion. Mol Cell. 2014;53:993–1004. doi: 10.1016/j.molcel.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. Embo J. 2007;26:2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, D’Artista L, Schleker T, Guerra C, Garcia E, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen KJ, Zanini IM, Herrador R, Lopes M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol. 2013;200:699–708. doi: 10.1083/jcb.201212058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita N, Minato S, Ohmi E, Tanuma S, Higami Y. DNA damage-induced CHK1 autophosphorylation at Ser296 is regulated by an intramolecular mechanism. FEBS Lett. 2012;586:3974–3979. doi: 10.1016/j.febslet.2012.09.048. [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T, Caldecott KW. Claspin promotes normal replication fork rates in human cells. Mol Biol Cell. 2008;19:2373–2378. doi: 10.1091/mbc.E07-10-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland RL, Patel S, Rivard RS, Smith K, Peters AA, Bielinsky AK, Brown EJ. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013;27:2259–2273. doi: 10.1101/gad.223180.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP. Mec1 is one of multiple kinases that prime the Mcm2–7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40:353–363. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5:010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Asai A, Mishima K, Fujimaki T, Kirino T. Telomerase activity in 144 brain tumours. Br J Cancer. 1998;77:1633–1637. doi: 10.1038/bjc.1998.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Sharma K, Diffley JF. Activation of dormant origins of DNA replication in budding yeast. Genes Dev. 1999;13:2360–2364. doi: 10.1101/gad.13.18.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausville E, Lorusso P, Carducci M, Carter J, Quinn MF, Malburg L, Azad N, Cosgrove D, Knight R, Barker P, et al. Phase I dose-escalation study of AZD7762, a checkpoint kinase inhibitor, in combination with gemcitabine in US patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;73:539–549. doi: 10.1007/s00280-014-2380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, Fernandez-Capetillo O, Diehl JA, Brown EJ. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest. 2012;122:241–252. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto T, Esaki T, Hirai F, Arita S, Nosaki K, Makiyama A, Kometani T, Fujimoto C, Hamatake M, Takeoka H, et al. Phase I, dose-escalation study of AZD7762 alone and in combination with gemcitabine in Japanese patients with advanced solid tumours. Cancer Chemother Pharmacol. 2013;72:619–627. doi: 10.1007/s00280-013-2234-6. [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Eychenie S, Decaillet C, Basbous J, Constantinou A. DNA structure-specific priming of ATR activation by DNA-PKcs. J Cell Biol. 2013;202:421–429. doi: 10.1083/jcb.201304139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Jones TL, Martin SE, Caplen NJ, Pommier Y. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J Biol Chem. 2009;284:18085–18095. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.