Abstract
Routes to carbon-13 enrichment of bacterially expressed proteins include achieving uniform or positionally selective (e.g. ILV-Me, or 13C′, etc.) enrichment. We consider the potential for biosynthetically directed fractional enrichment (e.g. carbon-13 incorporation in the protein less than 100%) for performing routine n-(D)dimensional NMR spectroscopy of proteins. First, we demonstrate an approach to fractional isotope addition where the initial growth media containing natural abundance glucose is replenished at induction with a small amount (e.g. 10%w/w u-13C-glucose) of enriched nutrient. The approach considered here is to add 10% (e.g. 200 mg for a 2 g/L culture) u-13C-glucose at the induction time (OD600=0.8), resulting in a protein with enhanced 13C incorporation that gives almost the same NMR signal levels as an exact 20% 13C sample. Second, whereas fractional enrichment is used for obtaining stereospecific methyl assignments, we find that 13C incorporation levels no greater than 20%w/w yield 13C and 13C-13C spin pair incorporation sufficient to conduct typical 3D-bioNMR backbone experiments on moderate instrumentation (600 MHz, RT probe). Typical 3D-bioNMR experiments of a fractionally enriched protein yield expected backbone connectivities, and did not show amino acid biases in this work, with one exception. When adding 10% u-13C glucose to expression media at induction, there is poor preservation of 13Cα-13Cβ spin pairs in the amino acids ILV, leading to the absence of Cβ signals in HNCACB spectra for ILV, a potentially useful editing effect. Enhanced fractional carbon-13 enrichment provides lower-cost routes to high throughput protein NMR studies, and makes modern protein NMR more cost-accessible.
Introduction
An excess of signal still exists in routine 3D-NMR experiments of liquid phase protein samples, even when employing gradient coherence selection and non-traditional sampling (e.g. non-uniform sampling, NUS) to minimize experimental times.1 Bacterial host expression protocols continue to improve, aiding researchers in achieving soluble 0.5–1 mM protein samples, 2–6 while steady progress continues to be made in improving sensitivity through new probe designs (e.g. 77 K and 20 K cryogenic probes, reduced coil diameters), improvements in console electronics, and increasing static field strengths. It would be desirable to identify means to realize value from situations of excess NMR signal in liquid phase protein NMR. Cost savings could also be paired with automation of data analysis to aid high throughput structural studies by NMR.7–16 In principle, 13C incorporation into proteins at percentages less than 100%, conserving adjacent spin labels without amino acid bias, would reduce dependence on stable isotopes in many 3D NMR experiments on proteins, such as those for performing backbone assignments that normally require expensive isotopic enrichment. The practice of 10%w/w carbon-13 biosynthetically directed fractional (herein denoted 10%w/w-BDF) enrichment has been well developed for obtaining stereospecific methyl side chain assignments.17–20 Further, there is strong fundamental support that carbon-13 spin pairs and triples will be highly conserved in amino acid synthesis and thus in proteins produced by 10%w/w-BDF.18, 21, 22 The 13C-BDF method should therefore have high utility for enabling broader 3D-NMR experiments on proteins, but does not appear to have been investigated for this use to date. This work investigates two questions relating to fractional 13C incorporation into biosynthetically expressed proteins.
First, we considered if it would be possible to enhance the incorporation of the 13C isotope into expressed proteins by adding the fractional amount of u-13C-glucose at the time of induction. We find that the addition of 10% u-13C-glucose by mass into cultures at the time of induction results in about 18% carbon-13 incorporation into the expressed protein, and have confirmed this effect under several conditions including different induction temperatures and durations, and also different glucose concentrations. We denote such an approach opt-10%w/w-BDF. Second, we investigated whether fractional enrichment of proteins on the scale of 10–20% is suitable for producing proteins that are amenable to typical 3D-NMR backbone assignment experiments. We find that no more than 20% carbon-13 incorporation is needed for acquiring even challenging 3D-bioNMR experiments (e.g. 3D-HN(CA)CO) on moderate instrumentation (600 MHz, RT probe), while less incorporation easily provides enough signal for others (e.g. 3D-HNCO).
Uniform, fractional and specific isotopic enrichment for protein NMR structural studies have been well developed. 2, 17, 19, 23–28 Protocols for specific amino acid enrichment include selective ILV methyl enrichment,29 selective alanine methyl enrichment,30, 31 selective carbon enrichment of the protein backbone,32 and amino-acid selective enrichment at the carbonyl position.33 These and other targeted labeling approaches 25, 34–36 enable the study of very large proteins, but their efficacy is dependent on the complicated biosynthetic and catabolic pathways that they rely upon. Broadly, uniform enrichment approaches focus on the use of gram quantities of u-13C-glucose, or other 13C-enriched metabolites, in the growth media. Although costly, u-13C-glucose is still among the least expensive sources of 13C-enriched carbon suitable for E. coli metabolism. Due to the low cost of 15NH4Cl, protein samples are produced with 100% 15N, which will be assumed for the remainder of this report.
As a result of these considerations, u-13C-glucose and 100% 15NH4Cl enriched amino acids and their precursors, have gained widespread adoption in bacterial host protein expression of samples for NMR study. Although there are more methods of bacterial protein production than can be reviewed here, three general bacterial growth protocols for isotopic incorporation can be identified. The most common is to provide the specifically or uniformly enriched metabolites in the base media prior to growth. 6, 28, 37–39 A second strategy introduces the enriched metabolites into resuspended cell media during the expression phase once a certain optical density (OD600) has been achieved; specifically, the culture is grown with natural abundance nutrients, centrifugally pelleted, and then resuspended into media containing enriched metabolites.40 However, the weak centrifugation followed by resuspension has the potential to stress cellular membranes. Finally, pre-induction isotope addition, guided by monitoring dissolved oxygen as a function of glucose consumption, 32, 33 avoids the stresses of centrifugation and resuspension, but has been aimed at achieving ~100% incorporation instead of at identifying the minimum incorporation required for n-D NMR protein structural studies, and also relies on oxygen sensing.24 In principle, cell-free expression also offers routes to employing lower quantities of enriched metabolites in the production of uniformly enriched biomolecules; the application of cell-free protein expression for obtaining large-scale and enriched samples 41, 42 is promising but appears to still be in early phases of adoption and development.
A general feature of pre-induction isotope addition is the benefit of using somewhat reduced amounts of isotopically labeled metabolites, where the goal to date has been to achieve uniform carbon-13 incorporation (e.g. 100% 13C). Alternately, the question may be posed, what level of incorporation of isotopes into proteins can still facilitate the acquisition of typical 3D-NMR experiments that require 13C-13C spin pairs? Prior detailed work by Szyperski demonstrated that 10%w/w-BDF with u-13C-glucose in the initial growth media will result in produced proteins that retain 13C-13C spin pairs and triples,18, 43 which suggests that such samples could be suitable for broader NMR analysis. We show that the density of spin pairs in 10%w/w-BDF enriched protein samples facilitates all typical 3D-NMR backbone assignment experiments on moderate instrumentation (600 MHz, RT probe), and introduce a simple method to enhance the carbon-13 BDF isotope incorporation into proteins. Specifically, introducing the 10%w/w u-13C-glucose into the base media at the time of induction (OD600 = 0.7–0.8 for 2 g/L glucose, OD600 = 1.0–1.1 for 4 g/L glucose) produces samples that give spectra consistent with ca. 20%w/w incorporation, and which also does not depend on the use of internal glucose monitoring. This work suggests that BDF and opt-BDF enrichment strategies will significantly lower the cost per sample of protein NMR studies and enable employing NMR more broadly in high throughput structural studies. This methodology targets routine NMR structural investigations of well-behaved proteins and has implications for high throughput work. For NMR structural studies at the leading edge that target intrinsically difficult systems (high MW, low solubility, complexes, dark states, disordered proteins, etc…), uniform enrichment at the highest possible sample concentrations are normally required.
In other words, isotope incorporation is recast as a minimization problem to identify the least amount of u-13C-glucose subject to the constraint of enabling all typical 13C-dependent 3D-NMR backbone experiments of small to moderate size proteins.
Experimental
Yeast ubiquitin (8.6 kDa) was chosen as a model protein. A plasmid containing the yeast ubiquitin sequence under control of the lac operon (Courtesy Prof. J. Morgan, University of California San Francisco) was transformed into E. coli BL21 competent cells. All samples of yeast ubiquitin were prepared from 1 L cultures and induced at OD600 = 0.8 unless otherwise indicated. Identical cell growth conditions were enforced, including 100–105 rpm shaking in Fernbach flasks at 37°C in M9 media (40 mM Na2HPO4, 20 mM KH2PO4, 8 mM NaCl, 2 mM MgSO4, 0.1 mM CaCl2, 5 μg/ml thiamine, 1.0 g/L NH4Cl) and containing 1.0 g 15NH4Cl, followed by a 48–64 hr induction at 18 °C or 5.5 hr at 37 °C (see Table 1). The samples of yeast ubiquitin tested two possible parameters: fractional u-13C-glucose in the initial media at inoculation, or timing the addition of u-13C-glucose to the culture to occur at induction. All expression tests used 2.0 g/L or 4.0 g/L of glucose in M9 buffer (2.0 mM MgSO4, 0.1 mM CaCl2, 5 μg/mL thiamine, 100 μg/ml Kanamycin) supplemented with a commercial vitamin mix (BME, B6891 Sigma Aldrich) and a cocktail of supplemental metals (Fe, Ca, Mn, Co, Cu, Zn, Mo, Mg), whose primary component is iron. For cold induction, the incubation chamber was equilibrated to 18°C for 20 minutes before adding IPTG.
Table 1.
Enhanced fractional enrichment of carbon-13 incorporation into yeast ubiquitin as measured by comparing the 1D-HNCO integral values (range: 9.5 to 7.0 ppm) via and optimized BDF methods. Each trial is a pair of the control (exact 10%w/w-BDF) and enhance (opt-10%w/w-BDF samples, and is independent of all other trials. Thus this table summarizes data from 12 distinct samples.
| Trial | Glucose (g) (u-13C/na) | Induction Temp | Induction OD600 (ctrl/opt) | Induction Duration | Incorporation# | |
|---|---|---|---|---|---|---|
| 1 | 0.20/1.80 | 18 | 0.8/0.8 | 64 hr | 18% | Fig1a |
| 2 | 18 | 0.8/0.8 | 64 hr | 18% | Fig1b | |
| 3 | 18 | 0.74/0.74 | 48 hr | 17% | Fig2a | |
| 4 | 37 | 0.79/0.74 | 5.5 hr | 20% | Fig2b | |
| 5 | 0.40/3.60 | 18 | 1.00/0.99 | 60 hr | 17% | Fig3a |
| 6 | 18 | 1.10/1.10 | 64 hr | 18% | Fig3b |
Estimated error 1%.
Number of transient recorded for the 1D-HNCO experiments are 1: 256, 2: 128, 3: 512, 4: 512, 5: 512, 6, 4096.
This work reports results from over twenty independent preparations of yeast ubiquitin (Tables 1–2, Figures 1–5, Figure 9, Figure S.3). Trials 1–9 comprise 18 of these samples. Each trial consists of a control sample that yields a known isotope incorporation (e.g. 10%w/w-BDF, 20%w/w-BDF) and an enhanced fractionally enriched sample (e.g. opt-10%w/w-BDF, opt-20%w/w-BDF) that were treated as a pair with respect to all experimental procedures (i.e. same incubator, same dialysis, sharing a centrifugal rotor, etc…). Each control sample was prepared with exactly 10%w/w or 20%w/w of u-13C-glucose in the initial M9 growth media (e.g. 200 mg u-13C glucose + 1.8 g na-glucose, or 400 mg u-13C glucose + 3.6 g na-glucose). Each enhanced fractionally enriched sample was prepared by growing cultures with na-glucose initially, and then adding the fractional amount of u-13C-glucose at induction. The induction period was followed by centrifugation (4 °C) of the media and freezing of the resulting cell pellet (−20 °C). The pellet was thawed and resuspended in a phosphate buffer (50 mM NaPO4, 0.5 M NaCl, 10 mM Imidazole, and 2%[m/v] glycerol), and membranes were disrupted through sonication (5 x 30 s, 4 °C). The resulting cell debris was discarded after centrifugation. Affinity chromatography (His-bind resin, Novagen), followed by size exclusion chromatography (FPLC, GE Healthcare AKTApurifier and G-75 16/60 sepharose column) was performed. Each sample was then dialyzed into a de-gassed phosphate buffer (25 mM [Pi], 25 mM NaCl, pH=5.2) and was spin-concentrated (GE Healthcare Vivaspin 20, 3 kDa) to reduce the total volume. Final samples contained 10% D2O.
Table 2.
Several conditions bearing the common theme that induction occurred on or after the onset of stationary phase all result in a much lower enhancement of carbon-13 incorporation. Contrasting the results in Tables 1 and 2 supports that enhanced carbon-13 incorporation is higher when the fractional quantity of u-13C-glucose is added near the end of log phase growth, prior to stationary phase. Number of transient recorded for the 1D-HNCO experiments are 7: 512, 8: 384, 9, 64.
Figure 1.
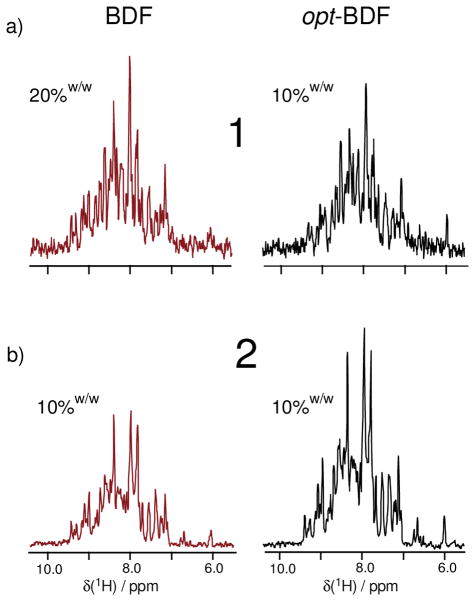
Comparison of 1D-HNCO slices which illustrate differences in carbon-13 isotope incorporation (100% 15N in all samples) as a result of implementing (left column) exact 10%w/w or 20%w/w fractional carbon-13 enrichment versus (right column) enhanced fractional enrichment, both using 200 mg u-13C-glucose (10%w/w). These data are also summarized in Table 1. (a) The 1D-HNCO slices (256 transients each) indicate that opt-10%w/w-BDF samples produce SNR similar to the exact 20%w/w sample. An independent trial (b) (128 transients each) contrasts an opt-10%w/w-BDF sample with an exact 10%w/w sample, where the almost two fold improvement is observed.
Figure 5.
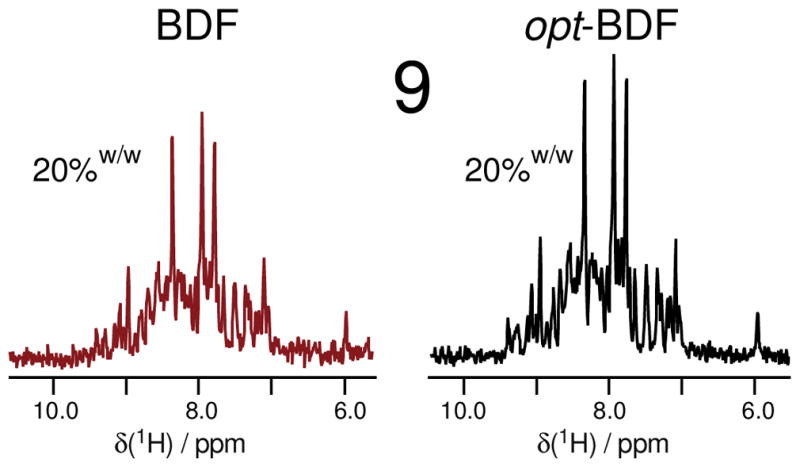
Comparison of the 1D-HNCO (64 transients each) slices of exact 20% and opt-20%w/w-BDF samples shows modest SNR improvement which is attributed to a poor growth rate at the time of induction due to limited nutrient availability in the initial media (which contained just 1.6 g na-glucose). (see also Table 2)
Figure 9.
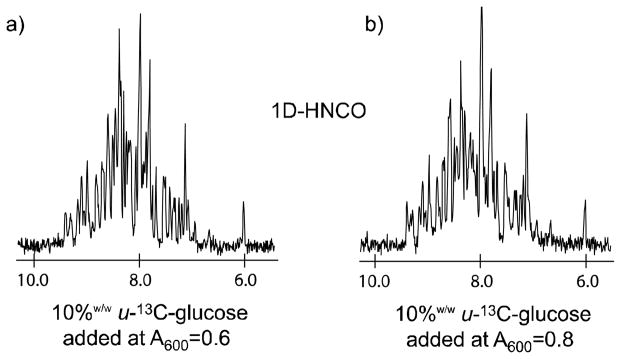
Complex factors control the level of enhanced isotope incorporation as a function of timing the addition of the fractional amount of u-13C-glucose, as shown by comparing 1D-HNCO (1024 transients each) slices which received the 0.2 g u-13 C-glucose (a) one hour prior to induction (OD600=0.6) and (b) at induction (OD600=0.8). The samples were prepared to identical concentrations (0.57 mM) in equal volumes as described in the Experimental section of the text. The degree of isotope incorporation is seen to be nearly identical in both trials, although a slight advantage can be ascribed to (b) which followed the opt-10%w/w-BDF protocol.
Final samples to be compared by NMR spectroscopy were carefully standardized using UV absorption (A280) to have identical or similar concentrations; furthermore, identical volumes were pipetted into 5 mm Shigemi NMR susceptibility matched tubes (Allison Park, PA) in which the locations of the glass plugs were set to identical positions. All spectra were recorded on a 4-channel, 600MHz spectrometer (Oxford AS 600/51, Varian Inc., DirectDrive [tm] vnmrs) equipped with a triple resonance 5 mm 1H[13C,15N] PFG room-temperature probe.
Incorporation of 13C into the C′ position of expressed ubiquitin was measured by taking integrals over the entire signal region of the first slice of 1D-HNCO experiments (Figures 1–5, 9, Tables 1–2). In order to obtain accurate results, we utilized very large numbers of transients (e.g. 256-4k) that far exceed the number that would be necessary in experimentation. Spectra were baseline corrected and line broadening on the order of 2–4 Hz was employed to further improve the accuracies of the integrals. We obtained both integrals and RMS noise levels of these spectra using the Rowland NMR Toolkit. The enhancement is then obtained by taking the ratio of the SNR values, where we note that RMS noise was essentially identical among spectra to be compared. When slight differences in protein concentration occurred, the integrals were standardized by the relative integral values.
Results and Discussion
The first question to be considered is if it is possible to enhance the fractional incorporation of carbon-13 into an expressed protein by adding the fractional u-13C-glucose at the time of induction. In other words, when using 10%w/w u-13C-glucose, is it possible to enrich the protein at a level greater than 10%? A standard suite of 3D-NMR experiments was conducted on a 600 MHz, 4-channel spectrometer using a room temperature probe and employing both uniform and non-uniform sampling44 on fractionally carbon-13 enriched yeast ubiquitin samples. To begin, the first slice of a 3D-HNCO spectrum (aka 1D-HNCO) is taken to be a measure of 13C incorporation on the protein backbone. Figure 1 illustrates real-world signal-to-noise comparisons corresponding to trials 1 and 2 in Table 1, when 10%w/w mass equivalence of u-13C-glucose was added to the E. coli growth media either at inoculation (BDF) or at induction (opt-BDF). Specifically, when the 10%w/w or 20%w/w quantity of u-13C-glucose is added at the initiation of growth, then exact 10%w/w or 20%w/w carbon-13 incorporation into the protein occurs (left column of Figure 1). On the other hand, the spectra in the right column of Figure 1 clearly demonstrate signals greater than 10% carbon-13 incorporation in two independently produced samples, where the enriched 10%w/w amounts of u-13C-glucose were added at induction (e.g. simultaneously with IPTG when OD600=0.8) and the initial growth media contained 90%w/w (1.8 g) of natural abundance glucose. In each row of Figure 1, care was taken to compare samples under identical conditions (see experimental details) including concentration and volume, for example. Further, we also tested dilute conditions in one case (Figure 1a) and concentrated conditions in the other (Figure 1b) in the unlikely event that instrument-based biases could occur as a function of signal intensity. Numerical results for trials 1 and 2 are summarized in Table 1, wherein we conclude that the signal to noise ratio (SNR) of the first slice of the 3D-HNCO is increased by almost two-fold simply by withholding the u-13C-glucose until IPTG was added (opt-10%w/w-BDF). For example, in Figure 1a, the real-world SNR that is observed from an opt-10%w/w-BDF sample is seen to be comparable with an exact 20%w/w-BDF sample. Further, in Figure 1b, an independently prepared opt-10%w/w-BDF sample is seen to have about twice the SNR of a 10%w/w-BDF sample.
The initial trials given in Figure 1 demonstrate that enhanced fractional isotope incorporation can be achieved by adding the fractional amount of u-13C-glucose at the time of induction. In trials 1 and 2, the carbon-13 incorporation is measured to be 18%, based on measuring signal area relative to RMS noise in a signal-free region. The accuracy of these measurements can be appreciated also by contrasting the opt-BDF samples in the right column of Figure 1 to exact fractional samples in the left column.
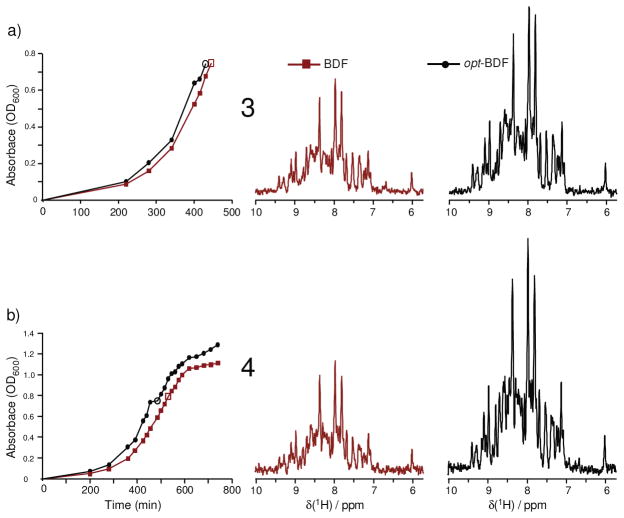
A next step is to characterize variations in typical growth and induction parameters. Trials 1 and 2 considered long induction periods at lower temperature (e.g. 64 hrs at 18 °C). Trial 3 finds 17 % carbon-13 incorporation for opt-BDF using a somewhat shorter 48 hr induction, while trial 4 finds a 20% carbon-13 incorporation for opt-BDF using just 5.5 hr of induction at 37 °C. The 1D-HNCO slices for trials 3 and 4 are illustrated in Figure 2, which also shows the respective growth curves to see that induction does occur during exponential phase. In trial 3, the growth was monitored up to induction and can clearly be seen to be strongly exponential. In trial 4, OD600 data were recorded throughout induction and are illustrated in Figure 2b for about the first five hours of induction, where it is seen that the cells decrease their rates of proliferation shortly after induction. These data from Trials 1–4 repeatedly obtain about 18% carbon-13 incorporation when 200 mg u-13C-glucose is added to a culture that has grown to OD600 ~ 0.8, and which was initially prepared with 1.8 g na-glucose.
Figure 2.
Comparison of signal intensities relative to noise in 1D-HNCO (512 transients each) slices. Enhanced fractional carbon-13 enrichment can be reproduced under several conditions, including (a) a different induction time (48 hr at 18 °C) in trial 3 and (b) short induction at 37 °C in trial 4, resulting in 17% and 20% carbon-13 incorporation respectively. (Table 1) Open symbols on the cell density data indicate the time of induction.
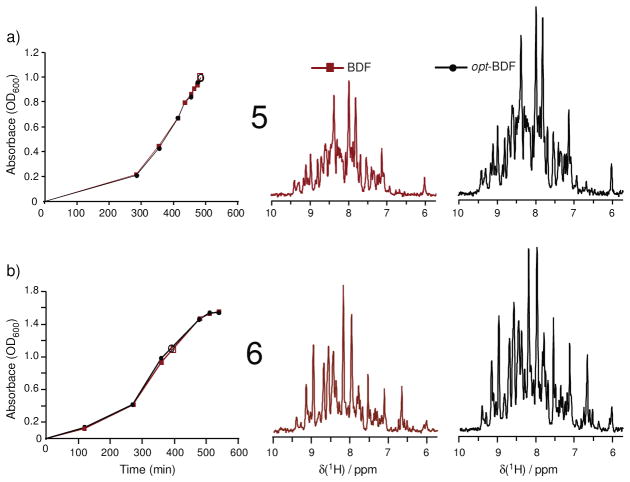
It is common when performing carbon-13 enrichment to use a total quantity of 2 g/L of glucose on the basis of cost, which has been the focus of this work. However, the potential to obtain opt-BDF fractional isotope enhancements in minimal media that use 4 g/L of glucose was tested in trials 5–6 (Table 1, Figure 3). Again, on the basis of integrated signal areas relative to noise, typical incorporations of 17–18% are found. In particular, it is important to recognize that the induction should occur at a higher cell density since the high 4 g/L glucose levels permit greater proliferation prior to stationary phase. A careful study of the growth of cultures utilizing 4 g/L glucose relative to the time of induction was conducted to identify that OD600= 1.0–1.1 is optimal for adding the fractional u-13C-glucose (Figure S.1, supporting information) to obtain the opt-BDF enhancement.
Figure 3.
Comparison of signal intensities relative to noise in 1D-HNCO slices. Enhanced fractional carbon-13 isotope incorporation is demonstrated in two independent trials that employed glucose at a higher concentration of 4 g/L (3.6 g na-glucose + 0.4 g u-13C-glucose added at induction). Panels (a) and (b) are trials 5 (512 transients per HNCO 1D slice) and 6 (4096 transients per HNCO 1D slice), respectively (Table 1). A comment should be added on the enhancement in panel (b): line widths in the opt-BDF spectrum of (b) were broader due to less efficient decoupling, but integrated signal areas confirm the 18% enhancement in (b). Open symbols indicate the time of induction.
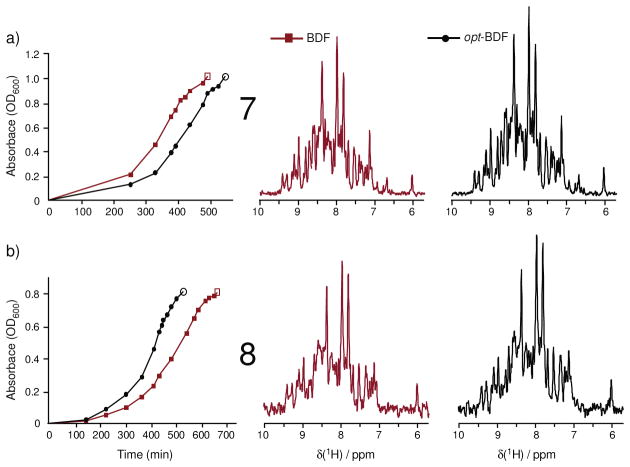
Many variables can be considered in pursuing the opt-BDF methodology, and some cases are presented in Table 2 (Figures 4–5) that resulted in less enhancement of isotope incorporation. Trials 7 and 8 (Figure 4) both considered how the opt-BDF strategy performs if cultures are induced at or shortly after the onset of stationary phase, and both find incorporation levels of just 12–13% based on 10%w/w u-13C-glucose. These findings agree with the general understanding that the onset of stationary phase is associated with starvation and decreased protein production and therefore decreased use of glucose.
Figure 4.
Comparison of signal intensities relative to noise in 1D-HNCO slices. The importance of adding the fractional quantity of u-13C-glucose during log phase growth is supported by two independent trials in which the cultures were each induced at a stage that exhibited the onset of rollover (trials 7 (512 transients) and 8 (384 transients), Table 1). These trials yielded just 12–13% incorporation using 10%w/w enriched glucose. Open symbols indicate the time of induction.
Next, trial 9 tested the use of 20%w/w u-13C-glucose in the opt-BDF protocol (Figure 5). However, a culture initially containing 80%w/w (1.6 g na-glucose) of the glucose at inoculation cannot reach satisfactory cell densities before transitioning into stationary phase growth. Glucose starvation of E. coli when starting the culture with just 1.6 g glucose forces the culture into stationary phase at a low cell density, which is undesirable for protein production since non-essential biosynthetic pathways are down regulated and rates of cell death increase. In stationary phase, gluconeogenic catabolic processes that can degrade expressed proteins are up regulated.22 Indeed the opt-20%w/w–BDF sample achieved poorer enhancement of isotope incorporation (Table 2, Figure 5) relative to opt-10%w/w–BDF samples. Furthermore, the opt-20%w/w–BDF trial also showed decreased net protein yield compared to typical trials using 2.0 g/L natural abundance glucose. In contrast, cultures that followed the opt-10%w/w–BDF protocol were unaffected by withholding 10% glucose in the base media (e.g. Figure 2–3, growth remained exponential until induction) and produced net amounts of ubiquitin effectively identical to samples prepared with media that contained 2.0 g/L glucose at inoculation. In this work, slight variations in growth rates were more strongly dependent on factors such as the cell density of the overnight cultures, flask location in the shaking incubator, and the frequency of accessing the cultures.
Ubiquitin prepared by opt-10%w/w–BDF reproducibly gave almost double the carbon-13 incorporation in the produced protein (Table 1). We conclude that the strategy of withholding 20%w/w u-13C-glucose in the initial media prevents reaching cell densities of OD600=0.8, and produces too little relative enhancement of the carbon-13 incorporation (23% vs. 20%). At first, the relative enhancements obtained with the opt-20%w/w-BDF and opt-10%w/w–BDF samples seem counterintuitive as we might have expected the 20% opt-BDF approach to experience less dilution of the u-13C-glucose when added at induction. However in hindsight the low enhancement (23%) could have been expected as a consequence of extreme nutrient starvation and the well-known stationary phase growth characteristic of E. coli cultures (e.g. trials 7 and 8).
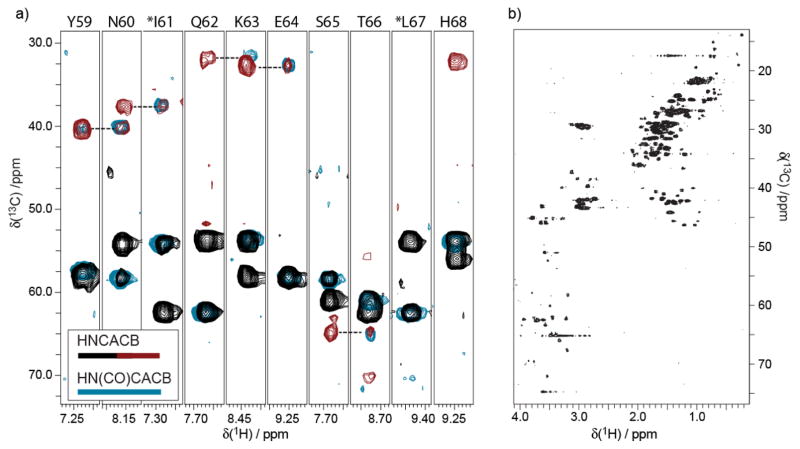
The second question to be considered is if it is feasible to use samples prepared by biosynthetically directed fractional 13C-isotope incorporation for backbone assignments of soluble proteins? It appears this question has not been widely considered, particularly as a minimization problem. The utility of many 3D-NMR experiments depends on the presence of adjacent carbon-13 nuclei in the protein backbone and side chain. The natural question then is whether fractional enrichment places special restrictions on experiments that exploit 13C-13C connectivities? A 0.4 mM opt-10%w/w–BDF sample was employed in 3D-HNCA and 3D-HN(CO)CA experiments using 2.4 hrs and 19 hrs respectively, and high SNR is obtained in both experiments (Figure S.2 Supporting Information). Achieving high SNR with 3D-HNCA on fractionally enriched samples is expected, as this is an efficient experiment, and furthermore relies only upon incorporating carbon-13 in one position on the backbone. However, the observation of high SNR in the less sensitive 3D-HN(CO)CA experiment with just a 0.4 mM opt-10%w/w–BDF sample strongly supports the finding of preservation of 13C-13C spin pairs in the protein backbone as predicted by analyses of digests of sparsely enriched protein samples.18 Next, a typical strip analysis of the HNCACB/HN(CO)CACB pair of experiments (10 hrs and 19.6 hrs respectively) is shown in Figure 6. The HNCACB was applied to a 0.8 mM opt-10%w/w-BDF sample, and the HN(CO)CACB to the 1.5 mM opt-20%w/w-BDF sample of yeast ubiquitin. Good incorporation of 13C within single amino acids is well demonstrated, particularly for the Cα backbone walk (again, see Figure S.2 for example strips from the HNCA/HN(CO)CA pair). It is seen in Figure 6 that the Cβ peaks are weak in some instances, and in a few cases some are missing, although many Cβ connectivities are observed.
Figure 6.
(a) The use of opt-BDF samples for HNCACB/HN(CO)CACB experiments tests the presence of 13C-13C spin labels. The HN(CO)CACB (light blue contour lines) is superimposed with the HNCACB (black/red contour lines) experiment in order to verify a typical backbone walk pattern. The HNCACB data were acquired on a 0.8 mM sample produced with the opt-10%-BDF method, while the HN(CO)CACB employed a 1.5 mM sample produced following opt-20%w/w-BDF. The 3D-HNCACB (4 scans per increment nonuniformly spanning a 48[13C] x 24[15N] matrix, sw13C=80 ppm, sw15N=35 ppm, 2.0 second recycle time, 150 ms acquisition) was acquired in just 10 hrs, while the HN(CO)CACB was acquired in 19.6 hrs (4 scans per increment for a 64[13C] x 32[15N] matrix, sw13C=80 ppm, sw15N=35ppm, 2.0 second recycle time, 150 ms acquisition), both processed via FFT, including linear prediction in the 15N dimension. (b) The inclusion of 13C throughout the expressed protein is illustrated by a representative 13C-HSQC (512 increment, 8 scans per increment, 1.5 s recycle time) of the same sample used for the HNCACB in panel (a).
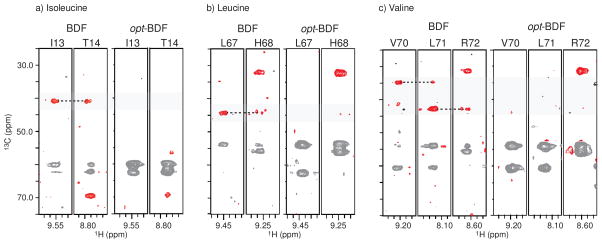
The phenomenon of missing CB peaks in CACB correlation spectra of opt-BDF samples requires a closer look, as it appears in Figure 6 that the missing peaks may be localized to isoleucine (I61) and leucine (L67). Indeed, in bacterial biosynthesis of ILV, the CACB pair is not preserved, suggesting that opt-BDF samples may systematically break CACB spin pairs. This hypothesis is confirmed by viewing selected ILV strips, Figure 7, from 10%-BDF and opt-10%w/w-BDF samples, where priming the 13C source prior to induction with 10%-BDF results in more CACB spin pairs than adding the 10% carbon-13 source at the time of induction. This selective ‘knockout’ of ILV signal in CACB spectra is potentially useful in facilitating assignments. Although the Cβ peak for E64 is missing, this appears to be pathological as Cβ peaks are observed for other glutamate residues in ubiquitin.
Figure 7.
Representative strips comparing the same opt-10%w/w-BDF HNCACB experiment in Figure 6 to an exact 10%w/w-BDF sample demonstrate that the opt-BDF protocol disrupts the CACB connectivity in ILV residues of ubiquitin, but 10% enrichment at the beginning of growth does not. The 10%-BDF sample was prepared to about 1 mM and the HNCACB acquired for it consumed 22 hrs (8 scans per increment for a 64[13C] x 32[15N] matrix, sw13C=80 ppm, sw15N=35 ppm, 1.2 second recycle time, 150 ms acquisition).
Although we observe good Cβ connectivities in spectra obtained by both 10%-BDF and opt-10%w/w-BDF, these spectra can be weak. Since these spectra were obtain on ~ 1mM sample and an RT probe at 600 MHz, we note that the use of higher fields and cryogenic probes will yield substantially higher sensitivity. Overall, these results resolve the question of whether the use of fractional u-13C-glucose incorporation yields a level of backbone incorporation of bonded 13C-13C spin pairs within given amino acids as well as at least the β-carbon position of side chains18, 43 that is sufficient for a broader suite of nD-NMR protein experiments.
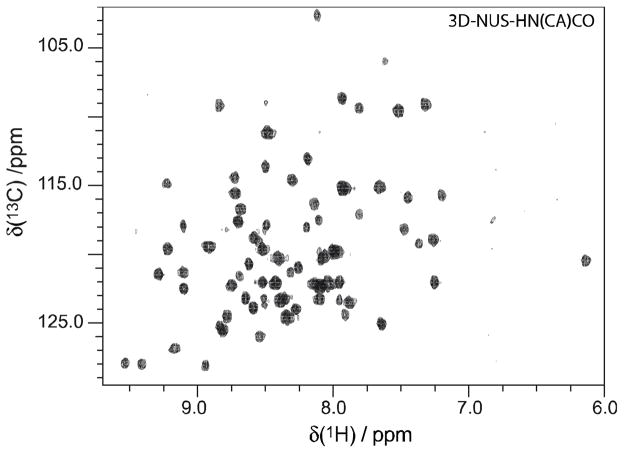
Complete backbone assignments have been verified solely with these data (not shown), indicating that all amino acids are detected without major biases, although an interesting question for future study is to characterize the possibility of amino acid dependent variations in carbon-13 incorporation in fractionally versus uniformly enriched samples. Thus these results support prior work demonstrating that positional isotopic incorporation through fractional backbone enrichment (1,3-13C-pyruvate) through the TCA cycle is conserved from the primary carbon source to the resulting protein.32 In other words, since glycolysis is simply the conversion of glucose (6-C molecule) to pyruvate (3-C molecule), it is reasonable to predict that u-13C-glucose incorporation into each amino acid will also be conserved. As further validation, the projection of all planes of the challenging 3D-HN(CA)CO experiment is illustrated in Figure 8, acquired in 38 hrs with just a 0.4 mM opt-10%w/w-BDF sample. This 3D-HNCACO spectrum was obtained with the assistance of non-uniform sampling, which has been shown to result in equal or higher intrinsic sensitivity to uniform sampling.45 However, the sensitivity gains by NUS for protein bioNMR are modest since no enhancement is possible in constant-time dimensions (e.g. 15N), while the rather short evolution times in decaying dimensions are evolved to short timescales that limit the NUS enhancement.46, 47 The major role of NUS in Figure 8 is to enable sufficient resolution in the indirect dimensions in a reasonable total experimental time without sacrificing sensitivity. Again, it is clear that significant incorporation of 13C-13C′ pairs is obtained (Figure 8), and that sensitivity is high given the concentration. Overall, empirical evidence indicates that a 20%w/w level of fractional incorporation (or its equivalent by opt-10%w/w-BDF) enables the acquisition of even the most challenging 3D-NMR experiments on moderate instrumentation (600 MHz, RT probe) and is seen to be a conservatively high limit.
Figure 8.
Adjacent 13C-13C spin labels in the backbone of proteins prepared by opt-10%w/w-BDF methods are demonstrated by the projection of all planes of a 3D-HN(CA)CO acquired on a 0.40 mM opt-10%w/w-BDF sample non-uniformly acquired (32 scans per increment nonuniformly spanning a 64[13C] x 40[15N] matrix with 500 samples, sw13C=20 ppm, sw15N=35 ppm, 2.0 second recycle time, 150 ms acquisition) in 38 hrs. This and all other experiments were performed on a 600 MHz spectrometer using a room-temperature probe.
A number of variations on the opt-BDF protocol for 13C enrichment developed here can be envisioned. We briefly report on two variant opt-BDF protocols that were also attempted in this work. First, we considered the question of when the fractional amount of u-13C-glucose most profitably can be added to optimize the incorporation of carbon-13 into the produced protein. For example, Neri and coworkers added the fractional amount of u-13C-glucose approximately one hour before the cell cultures reached OD600=0.8,17, partially charging biosynthetic pathways with 13C just prior to induction. We therefore performed a trial to compare the carbon-13 incorporation when the fractional amount of enriched glucose is added at different times: one culture received 0.2 g of u-13C-glucose at OD600=0.6, and the other at OD600=0.8, where both were induced at OD600=0.8. There is little difference in the signal intensities of the one-dimensional HNCO slices recorded for these two samples (Figure 9). On closer inspection, it can be seen that the sample following the opt-10%w/w-BDF protocol does have a slight SNR advantage. Adding the enriched nutrient about one hour prior to induction can provide a head start for isotopic incorporation in to amino acid synthesis prior to the addition of IPTG, but it also entails adding the fractional amount of u-13C-glucose at a time when there is a larger background of na-glucose. We interpret the findings of Figure 9 to imply that it is more favorable to minimize the na-glucose background at the time of induction rather than to isotopically charge the biosynthetic pathways ahead of induction. Second, we considered a variant in which thiamine is supplemented at the time of induction to promote glucose metabolism at the same time that the 10%-13C-glucose addition takes place. These data can be found in the Supplemental Information (Figure S.3), where the opt-BDF sample with supplemental thiamine (1mL/L of 0.1% thiamine added at induction) still demonstrated 17% 13C incorporation, consistent with incorporation levels of previous opt-BDF samples (Table 1). Thiamine is an essential vitamin for glycolysis and, while this test did not show an effect, it is possible that variations in added thiamine could alter the isotope incorporation.
One of the trials (trial 4, Table 1, Figure 2b) considered here resulted in 20% carbon-13 incorporation with just 10%w/w u-13C-glucose, the highest enhancement that has been obtained in this work to date. It is encouraging to see that this level of enhancement is possible, and it suggests in particular that shorter induction periods could be favorable in preventing scrambling of isotopes through catabolic degradation processes.
The need to add the fractional u-13C-glucose during log phase growth is clearly established by contrasting the enhancements reported in Figures 2–4 as a function of the time at which the cultures were induced. It may be noted that while the 12–13% carbon-13 incorporation for cultures that are induced in stationary phase is viewed as disappointing, this modest enhancement still has utility since even a 13% 13C enriched sample can be used to acquire spectra in almost half the time that would be needed for equivalent spectra with a 10% 13C sample.
The straightforward protocol described here for enhanced fractional enrichment of bacterially synthesized proteins appears to provide a few advantages over related methods. It is well known that traditional monitoring of OD600 reports on glucose-dependent progression through cell cycle phases, as long as glucose is the growth-limiting factor (and not pH, nitrogen source, etc.). This work finds that OD600 values can be used as the sole criterion for adding a fractional quantity of u-13C-glucose at induction just prior to stationary phase in a simple shaking, incubation chamber. The enhanced BDF procedure of adding sub-gram level amounts of enriched metabolites at induction should be transferable to the growing suite of protocols that use high levels of glucose in M9 media to reach very high cell densities,2, 27, 40 as evidenced by the results found here also for cultures employing 4 g/L glucose (Figure 3).
In principle, preparing a uniformly 13C enriched sample at much lower concentration could be an alternative to the methodology proposed here. However the present opt-BDF scheme represents a transparent modification to existing liter-scale bacterial host expression protocols and does not require efforts to change the scale of the expression that might involve, for example, specialized fermentation equipment. Regardless, variations on opt-BDF at different expression scales, or even in cell-free expression systems may be compelling alternatives to explore.
Conclusion
We demonstrate the ability to enhance the biosynthetically directed fractional incorporation of carbon-13 into expressed proteins if the fractional quantity of enriched glucose is added at induction. Specifically, this work arrives at a recommendation of a tenfold reduction in u-13C-glucose utilization (opt-10%w/w–BDF), which consistently yields samples whose spectra are nearly identical to those prepared with exact 20% carbon-13 incorporation. To obtain carbon-13 incorporation into the final expressed protein of about 18%, we have identified that 10%w/w fractional quantities of u-13C-glucose should be added at OD600 = 0.7–0.8 for cultures using 2 g/L glucose, or at OD600 = 1.0–1.1 for cultures using 4 g/L glucose, with either cold (18 °C) or native (37 °C) induction temperatures (Table 1). These conditions are widely employed in liter-scale bacterial host over-expression of proteins.
Further, this work finds that even on moderate instrumentation (600MHz, RT-probe), and adhering to the experiment acquisition times afforded by practices such as non-uniform sampling, fractional carbon-13 enrichment at exact 10% and 20% levels lead to sensitive 3D-bioNMR spectra for backbone assignments of proteins. To obtain the more difficult 3D-NMR spectra (e.g. HN(CO)CACB and HN(CA)CO) we have shown that about 20% carbon-13 incorporation is required for the moderate concentrations (0.5–1.5 mM) and instruments (600 MHz, RT-probe) used herein that were intended to mimic real-world conditions. This work also revealed an editing effect in which ILV residues did not give detectable Cβ peaks in HNCACB spectra.
Since it is possible to use a 10%w/w quantity of u-13C-glucose to produce samples that behave spectroscopically as if they were nearly 20%w/w carbon-13 enriched, doubling the value of the u-13C-glucose used, this work suggests that further efforts should be devoted to optimizing isotope incorporation. It can also be recognized that when higher fields and cryogenic probes are used, even lower levels of fractional enrichment than reported here can be employed since the key principle is that biosynthetic pathways preserve 13C-13C spin pairs and triples in the backbones of expressed products.
Supplementary Material
Highlights.
Biosynthetic fractional enrichment of proteins at 10–20% levels of carbon-13 incorporation into the protein facilitate all typical 3D-bioNMR experiments for performing backbone assignments, confirming that 13C-13C spin pairs are conserved in the protein backbone during fractional enrichment.
The incorporation of carbon-13 into the expressed protein can be enhanced by adding the fractional quantity of u-13C-glucose at the time of induction.
An ‘editing’ effect in which ILV residues are knocked out of HNCACB experiments performed on enhanced fractionally enriched samples may be useful in performing assignments.
The enhancement of fractional carbon-13 incorporation is sensitive to cell life cycle changes and is highest (e.g. 18% 13C incorporation for 10%w/w u-13C-glucose) when the fractional quantity of glucose is added during log phase just prior to the onset of stationary phase.
The enhancement of fractional carbon-13 incorporation using the simple protocols described here is obtained under a variety of popular expression conditions including cold (18 °C) and native (37 °C) induction temperatures, different induction durations, and both 2 g/L and 4 g/L net total glucose concentrations.
Acknowledgments
We thank NSF (MRI - 0521108) and Bucknell University for the acquisition of the 600 MHz spectrometer. We also thank Charles Clapp Ph.D.(BU), Thomas Selby Ph.D.(BU), Brian Breczinski, and the Bucknell Chemistry Department for their contributions, as well as Jeff Hoch and Alan Stern for the Rowland NMR Toolkit (UCONN, Rowland Institute-RNMTK). R.E.S. thanks the Drs. Anthony and Joyce D. Kales fund for support. We thank Prof. Thomas Szyperski for valuable feedback and Prof. Jeff Peng for helpful suggestions. This work was supported by NIH 1R15GM084443. The vector expressing yeast ubiquitin was kindly supplied by the J. Morgan laboratory (Univ. California San Francisco).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Cited Literature
- 1.Kim S, Szyperski T. GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. Journal of the American Chemical Society. 2003;125(5):1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- 2.Berthold DA, Jeisy VJ, Sasser TL, Shea JJ, Frericks HL, Shah G, Rienstra CM. Top Ten Tips for Producing 13C, 15N Protein in Abundance. Cambridge Isotopes Laboratories Research Literature Inc; 2007. (Application Note 15) [Google Scholar]
- 3.Liu GH, Shen Y, Atreya HS, Parish D, Shao Y, Sukumaran DK, Xiao R, Yee A, Lemak A, Bhattacharya A, Acton TA, Arrowsmith CH, Montelione GT, Szyperski T. NMR data collection and analysis protocol for high-throughput protein structure determination. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(30):10487–10492. doi: 10.1073/pnas.0504338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montelione GT. The Protein Structure Initiative: achievements and visions for the future. F1000 Biol Rep. 2012;4:7. doi: 10.3410/B4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivashanmugam A, Murray V, Cui C, Zhang Y, Wang J, Li Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 2009;18(5):936–48. doi: 10.1002/pro.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expression and Purification. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Billeter M, Wagner G, Wuthrich K. Solution NMR structure determination of proteins revisited. Journal of Biomolecular NMR. 2008;42(3):155–158. doi: 10.1007/s10858-008-9277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crippen GM, Rousaki A, Revington M, Zhang YB, Zuiderweg ERP. SAGA: rapid automatic mainchain NMR assignment for large proteins. Journal of Biomolecular NMR. 2010;46(4):281–298. doi: 10.1007/s10858-010-9403-2. [DOI] [PubMed] [Google Scholar]
- 9.Hyberts SG, Wagner G. IBIS - A tool for automated sequential assignment of protein spectra from triple resonance experiments. Journal of Biomolecular NMR. 2003;26(4):335–344. doi: 10.1023/a:1024078926886. [DOI] [PubMed] [Google Scholar]
- 10.Jung YS, Zweckstetter M. Mars -- robust automatic backbone assignment of proteins. J Biomol NMR. 2004;30(1):11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 11.Moseley HNB, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Nuclear Magnetic Resonance of Biological Macromolecules, Pt B. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 12.Williamson MP, Craven CJ. Automated protein structure calculation from NMR data. Journal of Biomolecular NMR. 2009;43(3):131–143. doi: 10.1007/s10858-008-9295-6. [DOI] [PubMed] [Google Scholar]
- 13.Zimmerman DE, Kulikowski CA, Huang YP, Feng WQ, Tashiro M, Shimotakahara S, Chien CY, Powers R, Montelione GT. Automated analysis of protein NMR assignments using methods from artificial intelligence. Journal of Molecular Biology. 1997;269(4):592–610. doi: 10.1006/jmbi.1997.1052. [DOI] [PubMed] [Google Scholar]
- 14.Lee W, Bahrami A, Markley JL. ADAPT-NMR Enhancer: complete packagte for reduced dimensionality in protein NMR spectroscopy. Bioinformatics. 2013;29(4):515–517. doi: 10.1093/bioinformatics/bts692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee W, Kim JH, Westler WM, Markley JL. PONDEROSA, an automated 3D-NOESY peak picking program, enables automated protein structure determination. Bioinformatics. 2011;27(12):1727–1728. doi: 10.1093/bioinformatics/btr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montelione GT, Arrowsmith C, Girvin ME, Kennedy MA, Markley JL, Powers R, Prestegard JH, Szyperski T. Unique opportunities for NMR methods in structural genomics. J Struct Funct Genomics. 2009;10(2):101–6. doi: 10.1007/s10969-009-9064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific Nuclear Magnetic-Resonance Assignments of the Methyl-Groups of Valine and Leucine in the DNA-Binding Domain of the 434-Repressor by Biosynthetically Directed Fractional C-13 Labeling. Biochemistry. 1989;28(19):7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 18.Szyperski T. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids. An efficient analytical tool to investigate intermediary metabolism. Eur J Biochem. 1995;232(2):433–48. doi: 10.1111/j.1432-1033.1995.tb20829.x. [DOI] [PubMed] [Google Scholar]
- 19.Szyperski T, Neri D, Leiting B, Otting G, Wuthrich K. Support of H-1-Nmr Assignments in Proteins by Biosynthetically Directed Fractional C-13-Labeling. Journal of Biomolecular NMR. 1992;2(4):323–334. doi: 10.1007/BF01874811. [DOI] [PubMed] [Google Scholar]
- 20.Hilty C, Wider G, Fernandez C, Wuthrich K. Stereospecific assignments of the isopropyl methyl groups of the membrane protein OmpX in DHPC micelles. Journal of Biomolecular NMR. 2003;27(4):377–382. doi: 10.1023/a:1025877326533. [DOI] [PubMed] [Google Scholar]
- 21.Umbarger HE. Amino-Acid Biosynthesis and Its Regulation. Annual Review of Biochemistry. 1978;47:533–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 22.Neidhardt FC, editor. Escherichia Coli Salmonella Typhimurium. 1–2. American Society for Microbiology; Washington DC: 1987. [Google Scholar]
- 23.Baleja JD, Cherry JJ, Liu Z, Gao H, Nicklaus MC, Voigt JH, Chen JJ, Androphy EJ. Identification of inhibitors to papillomavirus type 16 E6 protein based on three-dimensional structures of interacting proteins. Antiviral Res. 2006;72(1):49–59. doi: 10.1016/j.antiviral.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai ML, Huang Y, Sakaguchi K, Clore GM, Gronenborn AM, Craigie R. An efficient and cost-effective isotope labeling protocol for proteins expressed in Escherichia coli. Journal of Biomolecular NMR. 1998;11(1):97–102. doi: 10.1023/a:1008222131470. [DOI] [PubMed] [Google Scholar]
- 25.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. Uniform and residue-specific N-15-labeling of proteins on a highly deuterated background. Journal of Biomolecular NMR. 2004;29(3):289–297. doi: 10.1023/B:JNMR.0000032523.00554.38. [DOI] [PubMed] [Google Scholar]
- 26.Gardner KH, Kay LE. The use of H-2, C-13, N-15 multidimensional NMR to study the structure and dynamics of proteins. Annual Review of Biophysics and Biomolecular Structure. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 27.Lian LY, Middleton DA. Labelling approaches for protein structural studies by solution-state and solid-state NMR. Progress in Nuclear Magnetic Resonance Spectroscopy. 2001;39(3):171–190. [Google Scholar]
- 28.McIntosh LP, Dahlquist FW. Biosynthetic incorporation of 15N and 13C for assignment and interpretation of nuclear magnetic resonance spectra of proteins. Q Rev Biophys. 1990;23(1):1–38. doi: 10.1017/s0033583500005400. [DOI] [PubMed] [Google Scholar]
- 29.Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated N-15-, C-13-, H-2-labeled proteins. Journal of Biomolecular NMR. 1999;13(4):369–374. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 30.Sheppard D, Guo CY, Tugarinov V. Methyl-detected ‘out-and-back’ NMR experiments for simultaneous assignments of Ala beta and Ile gamma 2 methyl groups in large proteins. Journal of Biomolecular NMR. 2009;43(4):229–238. doi: 10.1007/s10858-009-9305-3. [DOI] [PubMed] [Google Scholar]
- 31.Sheppard D, Guo CY, Tugarinov V. 4D H-1-C-13 NMR Spectroscopy for Assignments of Alanine Methyls in Large and Complex Protein Structures. Journal of the American Chemical Society. 2009;131(4):1364. doi: 10.1021/ja808202q. [DOI] [PubMed] [Google Scholar]
- 32.Guo CY, Geng C, Tugarinov V. Selective backbone labeling of proteins using {1,2-C-13(2)}-pyruvate as carbon source. Journal of Biomolecular NMR. 2009;44(3):167–173. doi: 10.1007/s10858-009-9326-y. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi K, Ng E, Malia TJ, Wagner G. 1-C-13 amino acid selective labeling in a (HN)-H-2-N-15 background for NMR studies of large proteins. Journal of Biomolecular NMR. 2007;38(1):89–98. doi: 10.1007/s10858-007-9152-z. [DOI] [PubMed] [Google Scholar]
- 34.Waugh DS. Genetic tools for selective labeling of proteins with alpha-N-15-amino acids. Journal of Biomolecular NMR. 1996;8(2):184–192. doi: 10.1007/BF00211164. [DOI] [PubMed] [Google Scholar]
- 35.Tong KI, Yamamoto M, Tanaka T. A simple method for amino acid selective isotope labeling of recombinant proteins in E-coli. Journal of Biomolecular NMR. 2008;42(1):59–67. doi: 10.1007/s10858-008-9264-0. [DOI] [PubMed] [Google Scholar]
- 36.Lin MT, Sperling LJ, Schmidt HLF, Tang M, Samoilova RI, Kumasaka T, Iwasaki T, Dikanov SA, Rienstra CM, Gennis RB. A rapid and robust method for selective isotope labeling of proteins. Methods. 2011;55(4):370–378. doi: 10.1016/j.ymeth.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemaster DM. Isotope Labeling in Solution Protein Assignment and Structural-Analysis. Progress in Nuclear Magnetic Resonance Spectroscopy. 1994;26:371–419. [Google Scholar]
- 38.Clore GM, Gronenborn AM. Structures of Larger Proteins in Solution - 3-Dimensional and 4-Dimensional Heteronuclear Nmr-Spectroscopy. Science. 1991;252(5011):1390–1399. doi: 10.1126/science.2047852. [DOI] [PubMed] [Google Scholar]
- 39.Bax A. Multidimensional Nuclear-Magnetic-Resonance Methods for Protein Studies. Current Opinion in Structural Biology. 1994;4(5):738–744. [Google Scholar]
- 40.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. Journal of Biomolecular NMR. 2001;20(1):71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 41.Kigawa T, Yabuki T, Yokoyama S. Large-scale protein preparation using the cell-free synthesis. Tanpakushitsu Kakusan Koso. 1999;44(4 Suppl):598–605. [PubMed] [Google Scholar]
- 42.Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T, Yokoyama S. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999;442(1):15–9. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- 43.Szyperski T, Bailey JE, Wuthrich K. Detecting and dissecting metabolic fluxes using biosynthetic fractional C-13 labeling and two-dimensional NMR spectroscopy. Trends in Biotechnology. 1996;14(12):453–459. [Google Scholar]
- 44.Rovnyak D, Frueh DP, Sastry M, Sun ZYJ, Stern AS, Hoch JC, Wagner G. Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. Journal of Magnetic Resonance. 2004;170(1):15–21. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Palmer MR, Suiter CL, Henry GE, Rovnyak J, Hoch JC, Polenova T, Rovnyak D. Sensitivity of Nonuniform Sampling NMR. Journal of Physical Chemistry B. 2015;119(22):6502–6515. doi: 10.1021/jp5126415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rovnyak D, Sarcone M, Jiang Z. Sensitivity enhancement for maximally resolved two-dimensional NMR by nonuniform sampling. Magnetic Resonance in Chemistry. 2011;49(8):483–491. doi: 10.1002/mrc.2775. [DOI] [PubMed] [Google Scholar]
- 47.Rovnyak D, Hoch JC, Stern AS, Wagner G. Resolution and sensitivity of high field nuclear magnetic resonance spectroscopy. Journal of Biomolecular NMR. 2004;30(1):1–10. doi: 10.1023/B:JNMR.0000042946.04002.19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.