Abstract
Purpose
A no b-wave (nob) electroretinography (ERG) phenotype arose spontaneously in a colony of C3H mice and was named nob5. A mutation was identified in the Gpr179 gene in homozygous nob5 mice. There is a concern that this mutation is also present in additional C3H sublines and may compromise retinal research performed using these lines. In this report, therefore, we provide a phenotype and genotype survey of nob5 in six C3H substrains present at the Jackson Laboratory.
Methods
Fundus changes were evaluated in the six C3H substrains with image-guided optical coherence tomography (OCT), and retinal function was assessed with ERG. The substrains were genotyped with PCR using appropriate primers for the nob5 mutation. Additionally, the genomic sequences of C3H/HeJ, available from the Jackson Laboratory, and C3H/HeH, available from the Wellcome Trust Sanger Institute, were examined for the Gpr179nob5 mutation.
Results
Two C3H congenic strains, C3Sn.BLiA-Pde6b+/DnJ and C3A.BLiA-Pde6b+/J, wild-type for Pde6b, used as the sighted control strains and had normal fundi, OCT, and ERG responses. Four C3H strains C3H/HeJ, C3HeB/FeJ, C3H/HeOuJ, and C3H/HeSnJ bearing the Pde6brd1 allele exhibited a grainy fundus appearance, retinal degeneration on OCT, and no rod and cone ERG responses. The nob5 mutation was not observed in the six C3H strains assessed with PCR genotyping. Further, the genomic sequences of C3H/HeJ and C3H/HeH did not contain the nob5 mutation.
Conclusions
The Gpr179nob5 allele is not present in C3H substrains at the Jackson Laboratory. Therefore, the usefulness of these C3H strains as commonly used models to study the effects of photoreceptor degeneration is not compromised.
Introduction
The rd1 retinal degeneration model was described by Dr. Clyde E. Keeler more than 80 years ago [1]. It was subsequently identified as a disruption in phosphodiesterase 6 beta (Pde6brd1, formerly rd1, rd, identical to Keeler rodless retina, r) [2-4]. This mutation has been found among many common laboratory inbred strains, as well as wild-derived inbred strains including all C3H substrains [4-6].
The nob5 mutant bears a mutation in the Gpr179 gene (Gpr179nob5) and has a normal a-wave but no b-wave (nob) ERG phenotype. The mutation arose as a spontaneous mutation in a colony of C3H mice and was identified with electroretinography (ERG) when this line was outcrossed to a line of C3H mice that lack the rd1 mutation [7]. About 1 year later, Balmer et al. [8] identified the same allele of Gpr179nob5 in a C3H-derived transgenic mouse line named tg21.
In an interesting article, published on January 23, 2015, Nishiguchi et al. [9] reported that a Gpr179nob5 mutation occurring in their C3H/HeN inbred mice interfered with restoration of functional b-wave, but not a-wave, responses in mice undergoing gene therapy for the Pde6brd1 mutation. This is valuable information for the research community. However, in their discussion, Nishiguchi et al. stated that “C3H mouse lines with the rd1 mutation are widely distributed for research purposes by most major animal suppliers worldwide. Indeed, a review of over 130 research articles, where the rd1 background was stated, shows that 67% of these studies use the C3H line. Our investigations suggest that the Gpr179 mutation may be present in many C3H lines, including those provided by the Jackson Laboratory, Charles River and Harlan Laboratories.” The researchers outlined the pedigreed relationship between C3H sublines. The founders of the C3H/HeJ substrain were received at The Jackson Laboratory (Bar Harbor, ME) from Heston in 1948, and the founders of the C3H/HeN substrain were sent to the National Institutes of Health (NIH; Bethesda, MD) by Heston in 1951 and subsequently sent to the Charles River (Wilmington, MA) Laboratory from the NIH in 1974. The authors theorized that “the Gpr179 mutation was likely already present at the time the line was propagated to the Jackson Laboratory by Heston in 1948. This would indicate that most, if not all, of the studies using rd1-C3H mice as a model of retinal degeneration published within the past 65 years (since 1948) probably used the double mutants unknowingly and that the conclusions, based on the erroneous assumption that these mice are a simple model of photoreceptor degeneration, should be interpreted with caution.” We reviewed the premise that the Gpr179nob5 mutation is carried in many C3H strains, including those at the Jackson Laboratory, and found that the conclusion drawn by the authors [9] is incorrect.
Methods
Mice
The mice screened in this study were bred and maintained in standardized conditions of the Production and Research Animal Facilities at the JAX. The strains used were C3H/HeJ (Stock #659), C3HeB/FeJ (Stock #658), C3H/HeOuJ (Stock #635), C3H/HeSnJ (Stock #661), C3A.BLiA-Pde6b+/J (Stock #1912-background is C3H/HeA), and C3Sn.BLiA-Pde6b+/DnJ (Stock #3648-background is C3H/HeSnJ). They were maintained on a NIH31 6% fat chow diet and provided acidified water, in a pathogen-free vivarium environment with a 14 h:10 h light-dark cycle. One pair of retired breeders from the Jackson Laboratory Mouse Repository was screened for each strain. All experiments were approved by the Institutional Animal Care and Use Committee and were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Clinical and functional retinal evaluation
Mouse eyes, dilated with 1% atropine (Alcon Laboratories, Inc., Fort Worth, TX), were evaluated with image-guided (Micron III) optical coherence tomography (OCT) from Phoenix Research Laboratories (Pleasanton, CA).
For electroretinographic evaluation, following 2 h dark adaptation, the mice were anesthetized with an intraperitoneal injection of xylazine (80 mg/kg) and ketamine (16 mg/kg) in normal saline. Additional anesthetic was given if akinesia was inadequate. Briefly, dark-adapted, rod-mediated ERGs were recorded with the responses to short-wavelength flashes over 4.0-log units to the maximum intensity by the photopic stimulator. Cone-mediated ERGs were recorded with white flashes after 10 min of complete light adaptation. The signals were sampled at 0.8 msec intervals and averaged.
Genotyping protocols
Genomic DNA for genotyping mice was prepared from the tail tips by the rapid, hot sodium hydroxide and Tris (Hot SHOT) procedure [10], and 1 µl of the DNA supernatant was used in a 10 µl PCR reaction. The PCR condition were: initial denaturation for 3 minutes at 94 °C followed by 35 cycles of denaturation for 15 s at 94 °C, annealing for 1 min at 60 °C, extension for 1 min at 72 °C, and a final extension step for 7 min at 72 °C. The Gpr179nob5 control DNA samples were generously provided by Dr. Ronald G. Gregg at the Department of Biochemistry, University of Louisville, Louisville, KY (from two nob5 mutant strains, C3H.Pde6b+ and tg21) [8]. After electrophoretic separation on a 2% agarose gel, amplicons were visualized with ethidium bromide under UV illumination. The Gpr179nob5 mutation is associated with a transposon insertion of the ERV2 retroviral sequence in the first intron of the gene [7]. For genotyping the Gpr179nob5 allele, two protocols were used. The first protocol [9] used a pair of primers, GPR179F1 (5′-CTG GCT GGC TAG TCA CAG TCT C-3′) and GPR179R3 (5′-CAC AGC TGT CAG CGC CAT CTT GTA AC-3′), that generate a 537 bp product from the nob5 mutant allele, and primer GPR179F1 paired with primer GPR179R4 (5′-CTT AGC AAT GAC GCT GGG GAT TCA G-3′) generated a larger product (782 bp) from the wild-type control DNA. The second protocol [8] used a pair of primers, Gpr179 fwd (5′-TGT GCC TGG GTA TCT GTT GA-3′) and Gpr179 rev (5′-GCT TAC ACA CTT ACA CAC AGA TAG AT-3′), that generate a 100 bp product from wild-type control DNA, while primer Gpr179 fwd paired with primer Gpr179 insert (5′-GCA TGT GCC AAG GGT ATC TT-3′) generated a 400 bp larger product from the nob5 mutant allele.
Results
Retinal phenotypes in C3H substrains
Two C3H congenic strains, C3Sn.BLiA-Pde6b+/DnJ and C3A.BLiA-Pde6b+/J, wild-type for Pde6b, were used as sighted control strains as they are the wild-type for the Pde6b mutation. These strains had a normal fundus appearance (Figure 1A) and normal retinal structure as assessed with OCT (Figure 1B) at 6 weeks of age. The other four C3H substrains bearing the Pde6brd1 mutation exhibited severe, early onset retinal degeneration at 6 weeks of age. Retinal vessels were attenuated, and areas of depigmentation were observed on the fundus examination (Figure 1C). Loss of photoreceptors was observed with OCT (Figure 1D).
Figure 1.
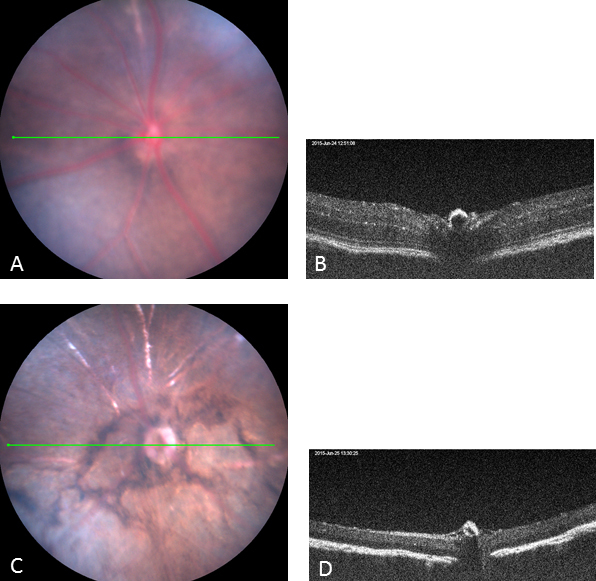
The clinical retinal phenotype of two C3H congenic strains wild-type for Pde6b (used as the sighted control strains) and four C3H rd1 substrains. A: Normal fundus. B: Normal retinal structure by image-guided optical coherence tomography (OCT) at 6 weeks of age (representative strain is C3A.BLiA-Pde6b+/J). C: Retinal vessels are attenuated, and areas of retinal depigmentation are observed. D: Retinal degeneration with image-guided OCT at 6 weeks of age (the representative strain is C3H/HeJ). The green line across the fundus image indicates the location of the B-scan.
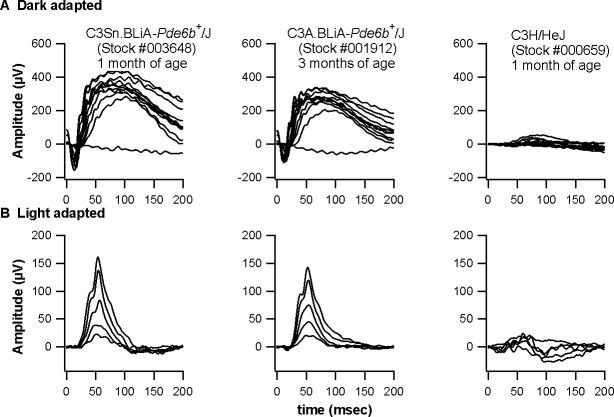
The C3Sn.BLiA-Pde6b+/DnJ and C3A.BLiA-Pde6b+/J mice exhibited a normal ERG response (Figure 2, left two columns). This result indicates that these two C3H congenic strains wild-type for Pde6b gene do not bear the Grp179nob5 mutation. As expected, the four remaining C3H substrains bearing the Pde6brd1 mutation showed significantly reduced ERG responses (Figure 2, right column).
Figure 2.
Electroretinograms of two C3H congenic strains wild-type for Pde6b (used as the sighted control strains) and four C3H rd1 substrains (the representative strain is C3H/HeJ). A: Dark-adapted conditions. B: Light-adapted conditions. Dark-adapted responses were recorded after overnight dark adaptation, while light-adapted responses were recorded with a background light of 1.46 log cd/m2 following 10 min exposure to the same intensity.
The Gpr179nob5 mutation is not present in C3H substrains at the Jackson Laboratory
The Mouse Genome Project has whole-genome sequences available for 36 key laboratory mouse strains, including two C3H substrains, C3H/HeH and C3H/HeJ. The genomic sequences of these strains were examined, and they did not contain the Gpr179nob5 mutation.
We subsequently genotyped for the presence of the Gpr179nob5 allele in the six C3H substrains available at the Jackson Laboratory using the genotyping protocol provided in Nishiguchi et al. [9] (Figure 3A) and Balmer et al. [8] (Figure 3B). No Gpr179nob5 mutation was observed in the C3H substrains assessed: C3H/HeJ (Stock #659), C3HeB/FeJ (Stock #658), C3H/HeOuJ (Stock #635), C3H/HeSnJ (Stock #661), C3A.BLiA-Pde6b+/J (Stock #1912-background is C3H/HeA), and C3Sn.BLiA-Pde6b+/DnJ (Stock #3648-background is C3H/HeSnJ).
Figure 3.
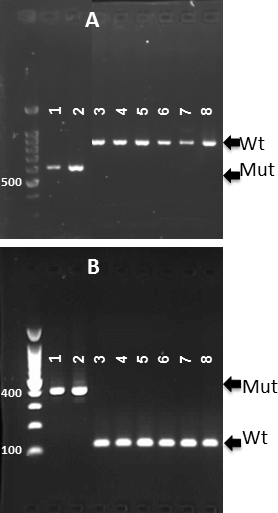
Genotyping for the Gpr179nob5 allele in C3H substrains at the Jackson Laboratory: 1 and 2 are the positive control DNA samples provided by Dr. Ronald G. Gregg at Department of Biochemistry, University of Louisville, Louisville, KY; from two nob5 mutant strains, C3H.Pde6b+ and tg21); 3. C3H/HeJ (Stock #659); 4. C3HeB/FeJ (Stock #658); 5. C3H/HeOuJ (Stock #635) and 6. C3H/HeSnJ (Stock #661) are from the production colony at The Jackson Laboratory; 7. C3A.BLiA-Pde6b+/J (Stock #1912-background is C3H/HeA); 8. C3Sn.BLiA-Pde6b+/DnJ (Stock #3648-background is C3H/HeSnJ).
Discussion
The nob5 mouse mutant arose as a result of a spontaneous mutation in a colony of C3H mice. The nob5 mouse model was characterized by the absence of the ERG b-wave, a component that reflects depolarizing bipolar cell (DBC) function. GPR179 is required for depolarizing bipolar cell function and is mutated in nob5 mice [7]. The Gpr179nob5 mutation was rediscovered in a C3H-derived transgenic mouse line (tg21) [8] and, again, in a C3H/HeN strain used for adenoassociated virus (AAV)–mediated gene supplementation of the rd1 mutation [9]. Pde6b AAV-mediated gene supplementation of the C3H/HeN-rd1 mice resulted only in the structural preservation of photoreceptors and the restoration of the photoreceptor-mediated a-wave but not in restoration of the bipolar cell-mediated b-wave because C3H/HeN harbors the nob5 mutation [9]. Taken together, these results suggest that nob5 could be a founder mutation that exists in C3H substrains. However, this study demonstrated that C3H/HeJ (Stock #659), C3HeB/FeJ (Stock #658), C3H/HeOuJ (Stock #635), C3H/HeSnJ (Stock #661), C3A.BLiA-Pde6b+/J (Stock #1912-background is C3H/HeA), and C3Sn.BLiA-Pde6b+/DnJ (Stock #3648-background is C3H/HeSnJ) do not have the Gpr179nob5 mutation, suggesting nob5 arose in the C3H strain after 1948, when it was originally sent to the Jackson Laboratory.
The records related to C3H/He found in the earliest issues of Mouse News Letter show that Strong gave C3H mice to Andervont in 1930, which Andervont later passed to Heston in 1941 [11]. The National Institutes of Health (C3H/HeN) and the Netherlands Cancer Institute (C3H/HeA) each obtained C3H/He mice from Heston in 1951 [12,13], several years after Heston gave C3H/He to the Jackson Laboratory (C3H/HeJ). The February 1955 Harwell entry in the Mouse News Letter [14] reported that C3H/H arrived at Harwell in 1954 from Edinburgh where C3H mice had arrived in 1953 from W.L. Russell at Oak Ridge. The absence of the nob5 mutation from C3H/HeJ and its sublines C3H/HeOuJ, C3H/HeSnJ, and C3HeB/FeJ is expected since a recent subline of C3H/HeA also lacks the nob5 mutation that is found in C3H/HeN. Confirming the probability that the nob5 mutation arose after 1951 when C3H/HeA and C3H/HeN separated is the report by Balmer et al. [8] that C3H/HeH also does not have the nob5 mutation.
Table 1 lists the C3H substrains with information on the presence or absence of the Gpr179nob5 and Pde6brd1 mutation as well as the availability of these C3H substrains. Nishiguchi et al. [9] purchased their rd1 mice from Harlan Laboratories (Blackthorn, UK) and Charles River (Margate, UK), and the strain name was C3H/HeN. By looking at the C3H substrains from these two suppliers, the strain C3H/HeNHsd (Harlan Laboratories) was derived from a nucleus colony obtained from the National Institutes of Health (with no date), and the other strain C3H/HeNCrl (Charles River) was also obtained from the NIH in 1974. It is possible that the origin of the nob5 mutation is from the C3H/HeN strain. Investigators should genotype for the presence of the Gpr179nob5 mutation in their mouse lines derived from the C3H/HeN strain.
Table 1. The presence of the Gpr179nob5 and Pde6brd1 mutation in C3H substrains.
| Strain | Stock # or Stain code | Gpr179nob5 | Pde6brd1 | References and/or resource |
|---|---|---|---|---|
| C3H.Pde6b+ |
|
Yes |
No |
[7,8] |
| tg21 |
|
Yes |
No |
[8] |
| C3H/HeH |
|
No |
Yes |
[8] |
| C3H/HeN or C3H/HeNHsd |
|
Yes |
Yes |
[9]/Harlan Laboratories |
| C3H/HeN or C3H/HeNCrl |
025 |
Yes |
Yes |
[9]/Charles River |
| C3H/HeJ |
000659 |
No |
Yes |
[5]/The Jackson Laboratory |
| C3HeB/FeJ |
000658 |
No |
Yes |
[5] /The Jackson Laboratory |
| C3H/HeOuJ |
000635 |
No |
Yes |
[5] /The Jackson Laboratory |
| C3H/HeSnJ |
000661 |
No |
Yes |
[5] /The Jackson Laboratory |
| C3A.BLiA-Pde6b+/J |
001912 |
No |
No |
The Jackson Laboratory |
| C3Sn.BLiA-Pde6b+/DnJ | 003648 | No | No | The Jackson Laboratory |
Acknowledgments
I thank Dr. Ronald G. Gregg at Department of Biochemistry, University of Louisville, Louisville, KY, for providing DNA samples for the two Gpr179nob5 strains, and Drs. Patsy Nishina, Jürgen Naggert, and Melissa Berry for careful review of the manuscript. I am grateful to Marge Strobel and Belinda Harris for providing C3H substrains. This work has been supported by the National Eye Institute Grants R01EY019943, R01EY011996 and R01EY016501.
References
- 1.Keeler CE. The inheritance of a retinal abnormality in white mice. Proc Natl Acad Sci USA. 1924;10:329. doi: 10.1073/pnas.10.7.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeler C. Retinal degeneration in the mouse is rodless retina. J Hered. 1966;57:47–50. doi: 10.1093/oxfordjournals.jhered.a107462. [DOI] [PubMed] [Google Scholar]
- 3.Pittler SJ, Keeler CE, Sidman RL, Baehr W. PCR analysis of DNA from 70-year-old sections of rodless retina demonstrates identity with the mouse rd defect. Proc Natl Acad Sci USA. 1993;90:9616–9. doi: 10.1073/pnas.90.20.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowes C, Li T, Frankel WN, Danciger M, Coffin JM, Applebury ML, Farber DB. Localization of a retroviral element within the rd gene coding for the beta subunit of cGMP phosphodiesterase. Proc Natl Acad Sci USA. 1993;90:2955–9. doi: 10.1073/pnas.90.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. Vision Res. 2002;42:517–25. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 6.Chang B, Hurd RE, Wang J, Nishina PM. Survey of Common Eye Diseases in Laboratory Mouse Strains. Invest Ophthalmol Vis Sci. 2013;54:4974–81. doi: 10.1167/iovs.13-12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peachey NS, Ray TA, Florijn R, Rowe LB, Sjoerdsma T, Contreras-Alcantara S, Baba K, Tosini G, Pozdeyev N, Iuvone PM, Bojang P, Jr, Pearring JN, Simonsz HJ, van Genderen M, Birch DG, Traboulsi EI, Dorfman A, Lopez I, Ren H, Goldberg AF, Nishina PM, Lachapelle P, McCall MA, Koenekoop RK, Bergen AA, Kamermans M, Gregg RG. GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2012;90:331–9. doi: 10.1016/j.ajhg.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balmer J, Ji R, Ray TA, Selber F, Gassmann M, Peachey NS, Gregg RG, Enzmann V. Presence of the Gpr179(nob5) allele in a C3H-derived transgenic mouse. Mol Vis. 2013;19:2615–25. [PMC free article] [PubMed] [Google Scholar]
- 9.Nishiguchi KM, Carvalho LS, Rizzi M, Powell K, Holthaus SM, Azam SA, Duran Y, Ribeiro J, Luhmann UF, Bainbridge JW, Smith AJ, Ali RR. Gene therapy restores vision in rd1 mice after removal of a confounding mutation in Gpr179. Nat Commun. 2015;6:6006. doi: 10.1038/ncomms7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques. 2000;29:52–4. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 11.Borges P. Standard Stocks, Jackson Laboratory. Mouse News Letter. 1951;5:27. [Google Scholar]
- 12.Jay GE., Jr Strains maintained by the inbred nucleus of the Laboratory Aids Branch, National Institutes of Health. Mouse News Letter. 1951;5:46. [Google Scholar]
- 13.Mühlbock O. Inbred Strains, The Netherlands Cancer Institute. Mouse News Letter. 1952; 6:22. [Google Scholar]
- 14.Barnes DWH. Inbred Strains, Harwell. Mouse News Letter. 1955;12:25. [Google Scholar]