Abstract
Purpose
Retinoblastoma (RB) is a rare intraocular malignant tumor of the developing retina with an estimated incidence of 1:20,000 live births in children under the age of 5 years. In addition to the abnormal whitish appearance of the pupil or leukocoria, strabismus has also been reported as a clinical symptom of the disease. RB1 is the first cloned tumor suppressor gene, and mutational inactivation of this gene is responsible for the development of RB during early childhood. The purpose of this study was to identify mutational alterations in the RB1 gene in Pakistani patients with RB.
Methods
During this study, 70 clinically evaluated patients with RB were recruited from different regions of Pakistan. The cases included 23 sporadic bilateral (32.9%), 34 sporadic unilateral (48.6%), nine familial bilateral (12.8%), and four familial unilateral (5.7%) cases. Constitutional causative mutations in the RB1 gene were screened via direct sequencing of all RB1 exons and their flanking regions.
Results
In this report, genetic testing resulted in the identification of 18 mutations in 25 patients with RB including six novel RB1 mutations. Of the total mutations identified, 13 (72.22%) were found to be null mutations caused by nine nonsense, three deletions, and one insertion. Two (11.11%) missense, two (11.11%) splice site mutations, and one (5.55%) base substitution in the promoter region were also found. Moreover, ten intronic variants were identified, one of which is novel.
Conclusions
Molecular screening and identification of these mutations in Pakistani patients with RB provide the mutational variants of the RB1 gene in the Pakistani population. The detection of oncogenic mutations in patients with RB and genetically predisposed individuals is a major step in clinical management, prognosis, follow-up care, accurate genetic counseling, and presymptomatic diagnosis of RB.
Introduction
Retinoblastoma (RB; MIM# 180200) is a rare intraocular malignant tumor of the developing retina. RB was first described as a tumor of the eye, with enucleation as a suggested treatment [1] while later it was claimed that the tumor proliferates from embryonic retinal cells, and thus, the term “retinoblastoma” was used [2]. The most common clinical sign of RB, which usually alerts parents or pediatricians, is the abnormal whitish appearance of the pupil or leukocoria [3]. RB has an estimated incidence of 1:20,000 live births and usually occurs in children younger than 5 years [4]. The average age of onset is reported as 18 months, which in unilateral and bilateral cases is around 24 and 15 months, respectively. Genetic basis of this tumor can be heritable (10%) or non-heritable mutations (90%). In 40% of patients, the laterality is reported as bilateral, and 60% are unilateral [5]. The causative RB gene, RB1 (GenBank accession number L11910), is located on chromosome 13q14.2 and was identified and cloned as the first tumor suppressor gene [6]. This gene extends over 178.143 kb genomic DNA and consists of 27 exons that transcribe 4.772 kb mRNA. The 2.787 kb extended CDS encode the 928-amino-acid-long p105RB nuclear phosphoprotein (pRB) that contains three domains: N-terminal, A/B pocket, and C-terminal domain [7]. After modification by cell cycle–regulated hyperphosphorylation, pRB plays an important role in regulating cell cycle progression by crossing the G1/S checkpoint [8]. The first mutation in hereditary RB is usually inherited as an autosomal dominant pattern in germ cells, but the progression toward malignancy requires a second mutation at the cellular level (tumor) in an adjacent allele as proposed by Knudson in his two-hit hypothesis [9] while in non-hereditary RB cases, both mutational events in adjacent alleles occur in the somatic (retinal) cells that lead to the development of a tumor. Neither mutation is present in the DNA of the constitutional cells [10,11]. Patients with heritable RB mostly have bilateral, multifocal, and early onset tumors while the non-heritable form is usually unilateral, lacks family history, and has no risk of transmission to the next generation [5]. Mutations in the RB1 gene are mostly scattered in coding exons, splice site junctions, the promoter region, and even in deep intronic regions. Mutations that result in early stop codons are usually associated with high penetrant RB while missense mutations, small in-frame insertions, and deletion and splicing mutations are associated with low expressivity and incomplete penetrance of the disease phenotype [12]. During this study, a total of 70 clinically evaluated patients with RB, including 38 unilateral and 32 bilateral RB cases, were recruited from different regions of Pakistan. Using the direct sequencing method, six novel and 12 reported RB1 mutations were found. This study provides alterations in RB1 in Pakistani patients with RB and increases the spectrum of known variations in the RB1 gene.
Methods
Patients
A total of 70 patients were recruited after detailed clinical evaluation by team of senior ophthalmologists and oncologists. All these patients were recruited from the outdoor clinic of the Department of Ophthalmology, Lahore General Hospital. At the time of recruitment most of the patients were presented with Leukocoria, as clinical symptom of the disease. By having 44 (62.8%) male and 26 (37.2%) female patients, the male to female sex ratio was found to be 1.7:1. The age at the time of diagnosis was documented from 2-84 months with overall mean as 18 months while the mean onset age of the patients with unilateral and bilateral Rb was found to be 22 months and 13 months respectively (Appendix 1). The treatment modalities included enucleation with a long part of the optic nerve in unilateral cases and at least one eye in bilateral cases along with chemotherapy. After primary enucleation, the diagnosis was confirmed with a histopathological examination, and patients were further evaluated for any additional signs of metastasis. The study was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained for molecular analysis from each parent/guardian. This study adhered to the ARVO statement on human subjects and was approved by the Institutional Review Board (IRB) of the University of Veterinary and Animal Sciences and by Lahore General Hospital, Lahore, Pakistan.
DNA extraction
Peripheral blood samples were collected from 70 RB probands. In addition to the patient samples, the available relatives of hereditary patients with RB and blood samples from 100 normal Pakistani individuals used as control were collected. DNA was extracted with the phenol/chloroform method [13]. Genomic DNA was dissolved in a low Tris-EDTA solution and was quantified by using a microvolume spectrophotometer (NanoDrop 2000, UV-Vis Spectrophotometer, San Diego, CA).
Detection of mutations in RB1 gene using Sanger sequencing
Primers for amplification of all 27 exons, their flanking region, and the promoter of the RB1 gene were designed by using Primer Blast software (Appendix 2). Genomic DNA was amplified with the thermal cycler ABI 2700 (Applied Biosystems, Foster City, CA) in a total volume of 25 µl containing 25–50 ng of genomic DNA, 1 pmol of forward and reverse primer, 1.5 mM MgCl2, 1x PCR reaction buffer, 2.5 mM dNTPs, and 1–1.5 U Taq polymerase (AmpliTaq Gold 360 Master Mix; Applied Biosystems, Foster City, CA). PCR reactions were performed at initial denaturation at 94 °C for 3 min; ten cycles of 94 °C (45 s), annealing at 67–57 °C with descending one degree in each cycle (45 s), and extension at 72 °C (1 min). Later annealing was performed at 57 °C for 25 cycles (45 s), and a final extension step at 72 °C for 10 min was added. Screening of germline mutations in all DNA samples was performed with direct Sanger sequencing. The unincorporated deoxynucleoside triphosphates and primers were removed by precipitation initially with 75% and then with 70% ethanol. PCR products were sequenced using the ABI Big Dye Terminator v 3.1 Sequencing Standard Kit (Applied Biosystems) and run on an ABI 3130XL genetic analyzer (Applied Biosystems). The sequence data were analyzed by comparison with the consensus sequence of the RB1 gene (GenBank L11910.1) using BioEdit Sequence Alignment Editor software. The sequences were analyzed manually and with Nucleotide-Nucleotide BLAST (Blastn) or BLAST 2 Sequences (2.2.10) on the NCBI web [14]. Additional information about RB1 gene mutations and polymorphism were confirmed from the following databases including the RB1 variation database: rb1-lsdb, The Human Gene Mutation Database (HGMD), 1000 Genome Project, ClinVar, Exome Aggregation Consortium, and Single Nucleotide Polymorphism database (dbSNP).
Results
A total of 70 Pakistani patients with RB were genetically tested to detect germline RB1 mutations. The patients were 38 unilateral (54.3%) and 32 bilateral (45.7%) cases that included 23 bilateral/non-heritable (32.9%), 34 unilateral/non-heritable (48.6%), nine bilateral/heritable (12.8%), and four unilateral/heritable (5.7%) cases. The patients included 26 girls and 44 boys (sex ratio of 1.7) with age at diagnosis ranging from 2 to 84 months. The overall mean of the age of onset was recorded as 18 months while the mean age of onset of the patients with unilateral and bilateral RB was 22 months and 13 months, respectively. Six siblings who were predisposed as carriers but younger than 5 years were also screened.
RB1 screening
Eighteen distinct germline mutations were found in 25 patients with RB with Sanger sequencing, of which 13 (72.22%) were null mutations caused by nine nonsense mutations, three deletions, and one insertion. Two (11.11%) missense, two (11.11%) splice site mutations, and one (5.55%) base substitution in the promoter region were also found to be likely pathogenic when analyzed with MutationTaster and Polymorphism Phenotyping-2 (PolyPhen-2). These disease-causing alterations included six novel and 12 reported mutations. The novel mutations include two small deletions (c.1116_1119delCACT in exon 11 and c.1436_1437delAC in exon 16) and four substitutions (c.148G>T in exon 2, c.610G>T in exon 7, c.947A>T in exon 10, and g.1991G>C in promoter region). Twelve reported mutations in 18 patients include nine substitutions (c.160G>T in exon 2, c.289G>T in exon 3, c.751C>T in exon 8, c.967G>T in exon 10, c.1072C>T in exon 11, c.1654C>T in exon 17, c.2063T>C in exon 20, and c.2359C>T in exon 23), one deletion (c.772_776del in exon 8), one insertion (c.2060_2061insTCATT in exon 20), and two altered splice site (c.380+1G>T in intron 3 and c.1215+1G>A in intron 12) mutations. Details of the identified mutant variants are presented in Table 1.
Table 1. Mutations in the RB1 gene identified in a cohort of 70 Pakistani RB patients.
| Patient ID |
*U/B |
Exon/ |
g.Position |
c.Position |
Protein change |
Type of mutation |
Remarks |
|---|---|---|---|---|---|---|---|
| **H/NH/MC/FC |
Intron |
||||||
| No. | |||||||
| RB35 |
U/NH |
Promoter |
g.1991G>C |
- |
- |
Substitution |
Novel |
| RB37 |
U/NH |
Exon 2 |
g.5434G>T |
c.148G>T |
p.Glu50* |
Nonsense |
Novel |
| RB23 |
B/NH |
Exon 2 |
g.5446G>T |
c.160G>T |
p.Glu54* |
Nonsense |
*** |
| rb1-lsdb |
|||||||
| RB51a |
B/H |
Exon 3 |
g.39470G>T |
c.289G>T |
p.Glu97* |
Nonsense |
rb1-lsdb |
| RB51b |
B/H |
||||||
| RB49 |
U/NH |
Intron 3 |
g.39562G>T |
c.380+1G>T |
- |
Splicing |
rb1-lsdb |
| RB28 |
U/NH |
Exon 7 |
g.56855G>T |
c.610G>T |
p.Glu204* |
Nonsense |
Novel |
| RB40 |
B/H |
Exon 8 |
g.59683C>T |
c.751C>T |
p.Arg251* |
Nonsense |
rb1-lsdb |
| RB40 |
MC |
||||||
| RB67 |
B/NH |
||||||
| RB20 |
B/NH |
Exon 8 |
g.59704_59708delAACAG |
c.772_776 |
p.Asn258 |
Deletion |
rb1-lsdb |
| RB44a |
B/H |
delAACAG |
Glufs*11 |
||||
| RB44b |
B/H |
||||||
| RB44c |
B/H |
||||||
| RB44 |
MC |
||||||
| RB38 |
U/NH |
Exon 10 |
g.64337A>T |
c.947A>T |
p.Asn316Ile |
Missense |
Novel |
| RB22a |
U/H |
Exon 10 |
g.64357G>T |
c.967G>T |
p.Glu323* |
Nonsense |
rb1-lsdb |
| RB22b |
U/H |
||||||
| RB22 |
FC |
||||||
| RB18 |
B/NH |
Exon 11 |
g.65386C>T |
c.1072C>T |
p.Arg358* |
Nonsense |
rb1-lsdb |
| RB65 |
B/NH |
Exon 11 |
g.65430_65433delCACT |
c.1116_1119 |
p.Thr373 |
Deletion |
Novel |
| delCACT |
Glnfs*6 |
||||||
| RB03 | B/NH | Intron 12 | g.70330G>A | c.1215+1G>A | - | Splicing | rb1-lsdb |
Mutation name is based on full length RB1 transcript (NM_000321.1) and encoded protein (NP_000312). Nucleotide numbering commenced with the A of the ATG translation initiation codon as +1. *U/B=Unilateral/Bilateral **H/NH/MC/FC=Hereditary/Non-Hereditary/Father Carrier/Mother Carrier ***rb1-lsdb=RB1 variation database.
In addition to novel and reported mutant variants in the RB1 gene, one novel and several reported variants were also found in the Pakistani population as c.380+45C>T in intron 3, c.501–77G>A in intron 4, c.920C>T in exon 9, c.940–64C>T in intron 9 (novel), c.1128–72T>G in intron 11, c.1695+99A>T in intron 17, c.1695–1696delAA in intron 17, c.1815–104A>G in intron 18, c.1961–10T>C in intron 19, c.2663+33T>C in intron 25, and c.2664–10T>A in intron 25. The detailed mutational changes and polymorphism of the RB1 gene in Pakistani patients with RB are given in Appendix 1.
Discussion
The detection of constitutional causative mutations in patients with RB and genetically predisposed individuals is a major step in clinical management, information for prognosis, treatment planning, follow-up care, genetic counseling, and presymptomatic diagnosis of RB. It also eradicates the need for expensive and time-consuming tests for family members who are not carriers [15]. Despite the importance of finding the molecular basis of RB in its clinical management, only a few clinical reports have been reported in Pakistani patients with RB [16-18] while the prognostic significance of molecular screening in 51 Pakistani patients with RB has been reported [19]. This is the second molecular-based screening of germline RB1 gene mutations in 70 Pakistani individuals with RB. In this study, in these patients, boys showed predominance tumor burden compared to girls (26 girls and 44 boys; sex ratio of 1:1.7), which might be due to gender bias in receiving healthcare treatment and follow-up as decided by their parents and might have thus influenced the uneven sex ratio seen here. The proportion of bilateral cases (32/70; 45.7%) versus unilateral cases (38/70; 54.3%) in our cohort of patients is higher than in previous reports. The frequency of bilateral and unilateral tumors is 26.7% and 71.9% in the United States [20], 35.4% and 64.51% in Pakistan [21], and 32% and 68% in China, respectively [19]. Overall, the total mutation detection rate was 35.7% (25/70) in the index patients. The mutation rate in the patients with bilateral and unilateral RB was 56.3% (18/32) and 18.4% (7/38), respectively. A similar detection rate (18%) for germline mutations in unilateral non-heritable cases was previously reported [22]. Out of 70 patients with RB, 18 mutations were found in 25 patients; thus, the mutation detection rate was 77.8% (7/9) in familial/bilateral patients, 50% (2/4) in familial/unilateral, 47.8% (11/23) in non-familial/bilateral, and 14.7% (5/34) in non-familial/unilateral patients.
Probably pathogenic null and missense mutations
Null mutations due to nonsense and frameshift mutations are mostly reported as pathogenic due to nonsense mediated decay (NMD) [23]. Previously, the possibility of the occurrence of 420 different nonsense mutations in the RB1 gene was figured out [24] while out of a total of 46 arginine codons in the RB1 gene, 14 are encoded by CGA/CGG from which 12 codons have been found to undergo recurrent C>T transition leading to early stop codons [25].
In our 20 patients, 13 different null mutations were found, including four novel and nine previously reported mutations. Four known mutations (p.Arg251*, p.Arg358*, p.Arg552*, and p.Arg787*) in exon 8, 11, 17, and 23 were identified. The premature stop codon is generated due to the C>T transition possibly by deamination of 5-methyl-cytosine at CpG dinucleotides. Further, p.Glu50* (exon 2; novel), p.Glu54* (exon 2), p.Glu97* (exon 3), p.Glu204* (exon 7; novel), and p.Glu323* (exon 10) resulted from the G>T transversion. Both of our patients with c.148G>T (p.Glu50*) and c.160G>T (p.Glu54*) had late onset up to 30–36 months, possibly due to less expressivity (Appendix 1). Other frameshift mutations include novel deletions p.Thr373Glnfs*6 (exon 11) and p.Asp479Glufs*13 (exon 16), a known deletion p.Asn258Glufs*11 (exon 8), and a reported insertion p.Leu688Hisfs*10 (exon 20). Three of the known mutations (p.Arg251*, p.Glu323*, and p.Asn258Glufs*11) were not restricted to the patients with RB but were also found in three asymptomatic carrier parents of the familial patients possibly due to incomplete penetrance. These null mutations result in premature termination codons (PTCs), coding aberrant mRNAs that are subjected to NMD. Even if these mRNAs containing PTCs escape the conserved surveillance mechanism of NMD, they will encode a nonfunctional truncated protein that lacks a N-terminus, A/B pocket, or C-terminus of pRB.
One known missense mutation p.Leu688Pro was found in a bilateral patient, while one novel missense mutation p.Asn316Ile (exon 10) was found in a unilateral non-hereditary patient. The genotyping of parents for this variant was not performed due to the unavailability of their samples. The change of leucine to proline at the 688 position caused by the c.2063T>C transition is located within a stretch of residues annotated in Uniprot as a special region “Pocket Domain B” (IPR002719). MutationTaster and PolyPhen-2 analysis regarded this change as “disease causing” and “possibly damaging.” The novel missense mutation p.Asn316Ile was also analyzed with Project HOPE and MutationTaster. Although the wild-type residue asparagine is not conserved at this position, the mutant residue isoleucine was not found in the other homologous sequences. Isoleucine is smaller and more hydrophobic; thus, it might lead to loss of interaction and disturbance in the correct folding, rendering p.Asn316Ile possibly damaging to the protein.
The RB1 promoter region contains binding sites for retinoblastoma binding factor (RBF1), specificity protein 1 (SP1), and activation transcription factor (ATF). In previous studies, mutations in SP1 (g.1858–1863 and g.1905–1910), RBF-1 (g.1859–1867), and ATF (g.1866–1872) were reported [26]. One novel alteration (G>C) in the promoter region at the g.1991 position was also found in this study. Although no functional studies were performed, MutationTaster analysis and absence of this variant in 100 ethnically matched controls and the dbSNP indicate that this change is most likely a disease-causing variant. This mutation at g.1991 might disrupt the regulatory sequence in the RB1 promoter and thus might alter the control of expression of the RB1 gene.
Probably pathogenic intronic substitution
Two previously reported splice site mutations, c.380+1G>T and c.1215+1G>A in intron 3 and 12, were found in two patients with RB. Analysis with Human Splicing Finder software [27] showed that the consensus values of the wild-type splice donor site were decreased by 31%. Splice site mutations might result in interruption of normal splicing sites, activation of cryptic splice sites, exon skipping, creation of a pseudo-exon within an intron, or intron retention. c.380+1G>T was found in a 3-year-old patient with bilateral, non-familial RB with onset at 2 months while c.1215+1G>A was found in a 3-year-old patient with unilateral, non-familial RB. Regarding, mRNA splicing, c.1215+1G>A is known to cause skipping of exon 12 in mature mRNA [28].
Probably nonpathogenic substitution
Although polymorphisms in RB1 have been reported, limited and fragmentary information on the incidence among Asians is available. A recurrent substitution p.Thr307Ile was found in two bilateral patients with RB. The wild-type residue (threonine) is not conserved in the homologous sequences of other species. This variant is listed as a mutation in the HGMD (CM030499) and rb1-lsdb database, but it is predicted to be “benign” by PolyPhen-2 and “polymorphism” by MutationTaster. In addition, this variant has been frequently reported as a variant in South Asian populations (13-48939088-C-T). Although p.Thr307Ile is a known polymorphism (rs183898408), we did not find any pathogenic variant in these two patients (Appendix 1). Kadam-Pai et al. reported the ethnic variations of RB susceptibility gene polymorphism A>G SNP at g.153104A>G (c.1815–104A>G, rs4151580) in intron 18 in eight Asian populations including Pakistani [29]. This polymorphism was also identified in our ten RB samples. A better understanding of the distribution of RB1 polymorphisms among Asian populations can facilitate the design of specifically tailored strategies for studying RB1 inheritance by pedigree segregation analysis in different Asian ethnicities. One novel intronic variant c.940–64C>T found in intron 9 was probably nonpathogenic or of unknown significance while other nine intronic substitutions found outside splicing sites were previously reported as polymorphisms.
Genotype phenotype correlation, penetrance, and expressivity of RB1 alterations
As a tumor suppresser gene, RB1 is a negative regulator of the cell cycle. pRB is a major regulatory unit in the late G1/S checkpoint of cell cycle progression. Changes in different regions of RB1 might cause pRB dysfunction that affects interaction with other proteins, regulation of protein encoding genes involved in replication, cell differentiation. and apoptosis [30]. This ultimately leads to the development of a tumor as domains A and B of pRB act as a repressor motif for E2F to inhibit transcription and ultimately pRB synthesis. Germline carriers usually develop bilateral or multifocal tumors; however, some rare families exhibit low penetrance, variability in the age of onset, number of tumors, and variable expressivity of the disease due to which bilaterally affected, unilaterally affected, and unaffected mutation carriers are known to coexist [31]. In such situations, the existence of MDM2 (OMIM 614401) and MDM4 (OMIM 602704) as modifier genes appears highly probable and must be screened [32].
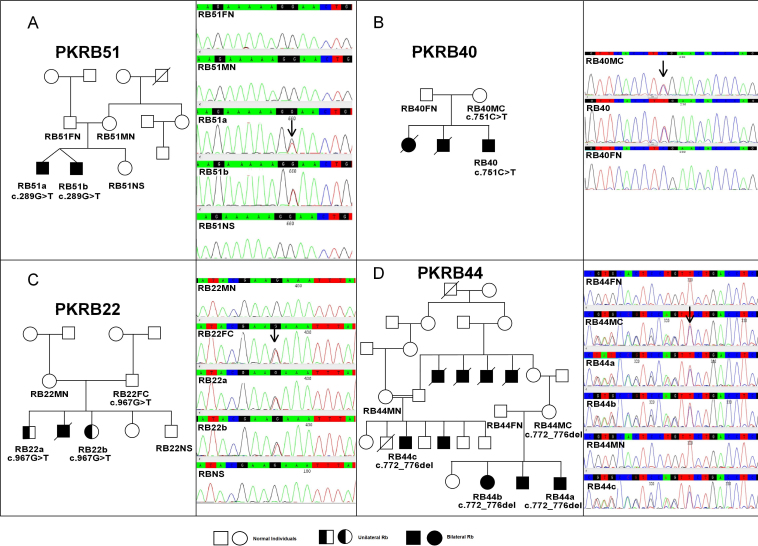
Mutations in familial RB show variable expressivity and phenotypic variability. Due to low penetrance, a germline carrier might not develop RB in some cases, while decreased expressivity may result in the development of unilateral RB or retinoma instead of bilateral RB [31]. The major reasons for this low penetrance and less expressivity could be a changed translation start site, a mutation at the C-terminus, or non-truncating mutations in low penetrant alleles that partly inactivate pRB [33]. The same low or incomplete penetrance was observed in three asymptomatic carrier (normal) parents, RB44MC, RB40MC, and RB22FC, who transmitted likely pathogenic variants p.Asn258Glufs*11, p.Arg251*, and p.Glu323*, respectively, to their offspring (Figure 1). In family PKRB22, both familial patients with RB (RB22a and RB22b) inherited p.Glu323* from their normal carrier father and developed unilateral RB. Thus, in this family p.Glu323* showed low penetrance and low expressivity while two other mutations p.Asn258Glufs*11 and p.Arg251* showed high expressivity in terms of the bilateral phenotype in familial patients with RB (RB44a, RB44b, RB44c, and RB40) but low penetrance in normal carrier parents (RB44MC and RB40MC). These results suggested that some germline mutations may not be directly responsible for malignant or bilateral tumor development in familial patients, but the cell maturation stage at the time of the second non-constitutional mutation may also be involved [34]. There could be a possibility of mutational mosaicism that might be a potential reason for the lack of tumor development in asymptomatic carrier parents. In PKRB51, twin brothers with RB carried p.Glu97*, but none of the parents and a normal sibling had this mutation as carriers. Thus, it might be a de novo mutation (Figure 1).
Figure 1.
Sequencing results of the constitutive causative heterozygous mutations of the RB1 gene. A: c.289G>T, p.Glu97* in pedigree PKRB51. B: c.751C>T, p.Arg251* in pedigree PKRB 40. C: c.967G>T, p.Glu323* in pedigree PKRB22. D: c.772_776del, p.Asn258Glufs*10 in pedigree PKRB44. Abbreviations: FN represents father normal, MN represents other normal, FC represents father carrier, MC mother carrier, NS represents normal sibling
Increased understanding and advancement in technology have led to a high mutation detection rate. Patients with no mutations detected in this study might have a large deletion, a mutation in deep non-coding sequences, or low-level mosaicism, which cannot be detected with direct sequencing. Allele-specific PCR and exome sequencing with next-generation sequencing were also reported to detect low levels of mosaicism [35].
This study has reported the molecular genetic analysis and frequency of RB1 mutations in Pakistani patients with RB. Molecular screening of the RB1 gene is a standard diagnostic in developed countries; thus, our preliminary results will be helpful in the future for designing and implementing screening strategies for patients with RB in Pakistan. Genetic testing also allows early detection of cancer and can guide treatment options. In addition, this will ultimately lead to appropriate genetic counseling, help parents make better choices in family planning, and lead to better clinical management of patients with RB and relatives at risk.
Acknowledgments
The authors thank all the RB affected patients and their families for their participation and cooperation in term of their valuable time and precious blood samples. We would like to extend our thanks and gratitude to the University of Veterinary and Animal Sciences for the financial support of this project. We sincerely acknowledge Prof. Dr. Syed Ali Haider (late), senior surgeon and ophthalmologist at Lahore General Hospital, for his assistance and valuable help throughout the progress of this research.
Appendix 1. Detailed description of collected RB patients.
Mutation name is based on full length RB1 transcript (NM_000321.1) and encoded protein (NP_000312). Nucleotide numbering commenced with the A of the ATG translation initiation codon as +1. *U/B = Unilateral/Bilateral **H/NH = Hereditary/Non-hereditary ***rb1-lsdb = RB1 variation Database, (http://rb1-lovd.d-lohmann.de/home.php?select_db=RB1) ****ExAC = Exome Aggregation Consortium, http://exac.broadinstitute.org/variant/13-48939088-C-T. To access the data, click or select the words “Appendix 1.”
Appendix 2. Sequence of primers used to amplify exons of RB1 gene in Pakistani RB patients.
To access the data, click or select the words “Appendix 2.”
References
- 1.Wardrop J. Observations on Fungus Haematodes or Soft Cancer: In Several of the Most Important Organs of the Human Body: Ramsay; 1809. [Google Scholar]
- 2.Verhoeff F, Jackson E. Minutes of the proceedings. In: 62nd annual meeting. Trans Am Ophthalmol Soc 1926; 38–43. [Google Scholar]
- 3.Shields JA, Augsburger JJ. Current approaches to the diagnosis and management of retinoblastoma. Surv Ophthalmol. 1981;25:347–72. doi: 10.1016/0039-6257(81)90072-2. [DOI] [PubMed] [Google Scholar]
- 4.Kivelä T. The epidemiological challenge of the most frequent eye cancer: retinoblastoma, an issue of birth and death. Br J Ophthalmol. 2009;93:1129–31. doi: 10.1136/bjo.2008.150292. [DOI] [PubMed] [Google Scholar]
- 5.Lohmann DR, Brenda GL. Retinoblastoma: Gene Reviews 2013. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1452/
- 6.Lee WH, Bookstein R, Hong F, Young LJ, Shew JY, Lee E. Human retinoblastoma susceptibility gene: cloning, identification, and sequence. Science. 1987;235:1394–9. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- 7.Saijo M, Kishino T, Niikawa N, Matsuura Y, Morino K, Tamai K, Taya Y. Molecular cloning of a human protein that binds to the retinoblastoma protein and chromosomal mapping. Genomics. 1995;27:511–9. doi: 10.1006/geno.1995.1084. [DOI] [PubMed] [Google Scholar]
- 8.Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–42. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- 9.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klutz M, Horsthemke B, Lohmann DR. RB1 gene mutations in peripheral blood DNA of patients with isolated unilateral retinoblastoma. Am J Hum Genet. 1999;64:667–8. doi: 10.1086/302254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohmann DR, Gallie BL. Retinoblastoma: revisiting the model prototype of inherited cancer. Am J Med Genet C Semin Med Genet. 2004;129C:23–8. doi: 10.1002/ajmg.c.30024. [DOI] [PubMed] [Google Scholar]
- 12.Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013;132:1077–130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Press; pp 1659–1662; 1989. [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Dhar SU, Chintagumpala M, Noll C, Chévez BP, Paysse EA, Plon SE. Outcomes of integrating genetics in management of patients with retinoblastoma. Arch Ophthalmol. 2011;129:1428–34. doi: 10.1001/archophthalmol.2011.292. [DOI] [PubMed] [Google Scholar]
- 16.Arif M, Iqbal Z. Zia ul I. Retinoblastoma in NWFP, Pakistan. J Ayub Med Coll Abbottabad. 2009;21:60–2. [PubMed] [Google Scholar]
- 17.Islam F, Zafar SN, Siddiqui SN, Khan A. Clinical course of retinoblastoma. J Coll Physicians Surg Pak. 2013;23:566–9. [PubMed] [Google Scholar]
- 18.Noorani SAJ, Shaikh ZA. Retinoblastoma: clinical picture and grouping at the time of first presentation. Pak J Med Sci. 2011;27:1055–9. [Google Scholar]
- 19.Khan AA, Mehboob R, Bukhari MH. Prognostic significance of retinoblastoma gene mutation in retinoblastoma eye with respect to pathological risk factors. Natural Science. 2013;5:411–8. [Google Scholar]
- 20.Broaddus E, Topham A, Singh AD. Incidence of retinoblastoma in the USA: 1975–2004. Br J Ophthalmol. 2009;93:21–3. doi: 10.1136/bjo.2008.138750. [DOI] [PubMed] [Google Scholar]
- 21.Arif M, Islam Z. Retinoblastoma: postenucleation orbital recurrence. Can J Ophthalmol. 2010;45:606–9. doi: 10.3129/i10-059. [DOI] [PubMed] [Google Scholar]
- 22.Rushlow D, Piovesan B, Zhang K, Prigoda‐Lee NL, Marchong MN, Clark RD, Gallie BL. Detection of mosaic RB1 mutations in families with retinoblastoma. Hum Mutat. 2009;30:842–51. doi: 10.1002/humu.20940. [DOI] [PubMed] [Google Scholar]
- 23.Zhang K, Nowak I, Rushlow D, Gallie BL, Lohmann DR. Patterns of missplicing caused by RB1 gene mutations in patients with retinoblastoma and association with phenotypic expression. Hum Mutat. 2008;29:475–84. doi: 10.1002/humu.20664. [DOI] [PubMed] [Google Scholar]
- 24.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037–47. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 25.Cowell JK, Smith T, Bia B. Frequent constitutional C to T mutations in CGA-arginine codons in the RB1 gene produce premature stop codons in patients with bilateral (hereditary) retinoblastoma. Eur J Hum Genet. 1994;2:281–90. doi: 10.1159/000472372. [DOI] [PubMed] [Google Scholar]
- 26.Cowell JK, Bia B, Akoulitchev A. A novel mutation in the promotor region in a family with a mild form of retinoblastoma indicates the location of a new regulatory domain for the RB1 gene. Oncogene. 1996;12:431–6. [PubMed] [Google Scholar]
- 27.Desmet FO, Hamroun D, Lalande M, Collod Béroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn JM, Phillips R, Zhu X, Becker A, Gallie B. Mutations in the RB1 gene and their effects on transcription. Mol Cell Biol. 1989;9:4596–604. doi: 10.1128/mcb.9.11.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadam-Pai P, Su XY, Miranda JJ, Soemantri A, Saha N, Heng CK, Lai PS. Ethnic variations of a retinoblastoma susceptibility gene (RB1) polymorphism in eight Asian populations. J Genet. 2003;82:33–7. doi: 10.1007/BF02715879. [DOI] [PubMed] [Google Scholar]
- 30.Herwig S, Strauss M. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 31.Harbour JW. Molecular basis of low-penetrance retinoblastoma. Arch Ophthalmol. 2001;119:1699–704. doi: 10.1001/archopht.119.11.1699. [DOI] [PubMed] [Google Scholar]
- 32.Castéra L, Sabbagh A, Dehainault C, Michaux D, Mansuet-Lupo A, Patillon B, Lamar E, Aerts I, Lumbroso Le Rouic L, Couturier J, Stoppa Lyonnet D, Gauthier Villars M, Houdayer C. MDM2 as a modifier gene in retinoblastoma. J Natl Cancer Inst. 2010;102:1805–8. doi: 10.1093/jnci/djq416. [DOI] [PubMed] [Google Scholar]
- 33.Abouzeid H, Schorderet DF, Balmer A, Munier FL. Germline mutations in retinoma patients: relevance to low-penetrance and low-expressivity molecular basis. Mol Vis. 2009;15:771–7. [PMC free article] [PubMed] [Google Scholar]
- 34.Gallie BL, Dunn J, Chan H, Hamel P, Phillips R. The genetics of retinoblastoma. Relevance to the patient. Pediatr Clin North Am. 1991;38:299–315. doi: 10.1016/s0031-3955(16)38079-8. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Moran K, Richards-Yutz J, Toorens E, Gerhart D, Ganguly T, Shields CL, Ganguly A. Enhanced sensitivity for detection of low-level germline mosaic RB1 mutations in sporadic retinoblastoma cases using deep semiconductor sequencing. Hum Mutat. 2014;35:384–91. doi: 10.1002/humu.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]