SUMMARY
The selective inhibition of bacterial β-glucuronidases was recently shown to alleviate drug-induced gastrointestinal toxicity in mice, including the damage caused by the widely used anticancer drug irinotecan. Here, we report crystal structures of representative β-glucuronidases from the Firmicutes Streptococcus agalactiae and Clostridium perfringens and the Proteobacterium Escherichia coli, and the characterization of a β-glucuronidase from the Bacteroidetes Bacteroides fragilis. While largely similar in structure, these enzymes exhibit marked differences in catalytic properties and propensities for inhibition, indicating that the microbiome maintains functional diversity in orthologous enzymes. Small changes in the structure of designed inhibitors can induce significant conformational changes in the β-glucuronidase active site. Finally, we establish that β-glucuronidase inhibition does not alter the serum pharmacokinetics of irinotecan or its metabolites in mice. Together, the data presented advance our in vitro and in vivo understanding of the microbial β-glucuronidases, a promising new set of targets for controlling drug-induced gastrointestinal toxicity.
Graphical Abstract
INTRODUCTION
It is increasingly clear that the bacteria that live on and within the human body are involved in a range of physiological processes. Changes to the microbiota have been found to accompany several disease states, including diabetes, atherosclerosis, obesity, malnutrition, cancer, gastrointestinal (GI) inflammatory disorders, and neurological diseases (Cho et al., 2012; Hsiao et al., 2013; Methé et al., 2012; The Human Microbiome Project Consortium, 2012; Morgan et al., 2012; Nicholson et al., 2012; Plottel and Blaser, 2011; Smith et al., 2013; Turnbaugh et al., 2006; Wang et al., 2011). Ongoing chemical interactions between mammalian tissues and their colonizing microbiota appear to play important roles in the symbiosis that sustains both domains of life (Mani et al., 2014; Redinbo, 2014). A key player in this interplay is glucuronic acid. This small sugar is conjugated, typically in the liver, to drugs and endobiotics marked for excretion, commonly via the bile duct to the GI tract (Holmes et al., 2011). In their glucuronidated states, most chemicals are inactive. The GI microbiota, however, contain β-glucuronidase enzymes that scavenge this sugar as a source of carbon. Removal of the glucuronide moiety produces aglycones that are often still eliminated, although they can be reabsorbed into the plasma and return to the liver as part of normal enterohepatic recycling. Importantly, reactivated aglycones can also be highly toxic to the GI tissues, and, as outlined below, there is evidence that such compounds can lead to serious side effects in human patients (Wallace and Redinbo, 2013).
Irinotecan (CPT-11) is a potent anticancer drug used to treat solid tumors and other malignancies (Cunningham et al., 1998; Douillard et al., 2000; Hurwitz et al., 2004). It is an essential component of FOLFIRI (where IRI indicates irinotecan), which is widely used for late-stage colorectal cancer (Kelly and Goldberg, 2005), and FOLFIRINOX, which was recently recommended as the first-line regimen for pancreatic cancer (Conroy et al., 2011). Irinotecan causes severe diarrhea in a significant fraction of patients, commonly leading to dose reductions or treatment termination (Chabot, 1996; Ma and McLeod, 2003; Mathijssen et al., 2001). The irinotecan prodrug is activated to the human topoisomerase I poison SN-38 in patients; this active metabolite is further processed by UDP-glucuronosyltransferases to become the inactivated glucuronide conjugate SN-38-G, which is sent to the GI tract for elimination (Chabot, 1996; Ma and McLeod, 2003) (Figure S1). In the GI tract, bacterial β-glucuronidase enzymes remove the glucuronic acid and reactivate the topoisomerase I poison SN-38, which causes GI damage and the diarrhea that is dose-limiting for irinotecan (Dranitsaris et al., 2005; Gupta et al., 1994; Rothenberg et al., 2001; Stein et al., 2010) (Figure S1). According to the Food and Drug Administration package insert for irinotecan, up to 88% of patients experience diarrhea and 31% show grade 3–4 diarrhea, which requires significant medical intervention. Thus, the alleviation of the chemotherapy-induced diarrhea caused by irinotecan is an unmet medical need.
A low-potency β-glucuronidase inhibitor showed promise in 2004 in reducing the GI toxicity associated with irinotecan in rats (Fittkau et al., 2004). We reported more potent (<μM) and novel inhibitors of bacterial β-glucuronidases in 2010, and showed that they significantly reduced the GI damage caused by irinotecan in mice (Wallace et al., 2010). We showed that these inhibitors were non-toxic to both bacterial and mammalian cells, and demonstrated using crystal structures and other data that the inhibitors were selective for the bacterial β-glucuronidases by more than 1,000-fold over the human enzyme ortholog (Roberts et al., 2013; Wallace et al., 2010). We pinpointed the basis of the selectivity to a loop present in bacterial β-glucuronidases that is missing from the human enzyme ortholog; the inhibitors bind to this loop, making them ineffective against mammalian β-glucuronidases (Wallace et al., 2010).
We have since demonstrated that the same approach can be applied to the non-steroidal anti-inflammatory drugs (NSAIDs), some of the world's most widely used therapeutics (Brune and Hinz, 2004). NSAIDs cause serious lower GI ulcers that have been less appreciated than the gastric side effects more commonly associated with these compounds (Laine et al., 2003; Lanas, 2010). Seventy percent of long-term NSAID users have chronic lower GI inflammation, increased permeability, and malabsorption, 40% have ulcers, and 30% suffer from GI bleeding and anemia (Lanas and Sopena, 2009). Like SN-38, NSAIDs are inactivated by glucuronidation and reactivated in the GI tract back to the parent drugs, which can then cause significant enteropathy largely in the small intestine via mechanisms that are not completely understood (Boelsterli et al., 2012). In mouse models of GI toxicity caused by the NSAIDs diclofenac, ketoprofen, or indomethacin, we showed that oral dosing of bacterial β-glucuronidase inhibitors significantly reduced the numbers of small-intestinal ulcers observed (LoGuidice et al., 2012; Mani et al., 2014; Saitta et al., 2014). We also demonstrated that inhibition of bacterial β-glucuronidases did not alter serum levels of diclofenac in mice (LoGuidice et al., 2012). Thus, modulating the activity of enzymes encoded by the symbiotic bacteria in the mammalian GI tract can improve tolerance to two classes of drugs, irinotecan and NSAIDs.
To date, these investigations have focused on the β-glucuronidase from Escherichia coli (Roberts et al., 2013; Wallace et al., 2010). While the Proteobacteria, including E. coli, are present in the mammalian GI tract, other phyla are more significantly predominant, particularly the Firmicutes and the Bacteroidetes (The Human Microbiome Project Consortium, 2012). Thus, to expand our understanding of the structure, function, and in vitro inhibition of bacterial β-glucuronidases, we cloned, overexpressed, purified, characterized, and determined the crystal structures of representative β-glucuronidases from the Firmicutes species Streptococcus agalactiae and Clostridium perfringens, each of which is present in the human GI tract. We also characterized a β-glucuronidase from Bacteroides fragilis, a GI microbe representative of the Bacteroidetes phylum. We demonstrate that a small chemical change to an inhibitor affects its efficacy, pharmacokinetics, and binding to a bacterial β-glucuronidase. Finally, we show that the inhibition of GI bacterial β-glucuronidases in mice does not alter the serum levels of irinotecan or its key metabolites SN-38 and SN-38-G. Together, these data extend our understanding of the microbial β-glucuronidases, which might serve as targets to reduce the toxicity and improve the efficacy of irinotecan and NSAIDs.
RESULTS
Crystal Structures of Firmicutes β-Glucuronidases
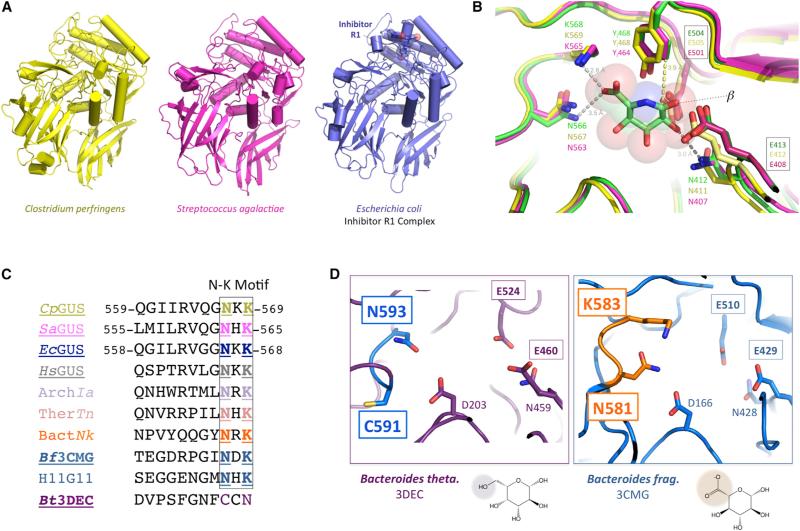
To advance our understanding of the structure and function of bacterial β-glucuronidase, the β-glucuronidases from two Firmicutes species associated with the mammalian GI microbiota, C. perfringens (CpGUS) and S. agalactiae (SaGUS), were cloned, overexpressed, and purified, and the structures of each were determined to 2.3 Å resolution (Figure 1A and Table S1). The structure of the β-glucuronidase from the Proteobacterial species E. coli (EcGUS), also present in the mammalian GI tract, was also determined in complex with a novel inhibitor, R1, to 2.4 Å resolution (Figure 1A and Table S1). The structures of the two Firmicutes enzymes are the first from this phylum. The CpGUS structure (PDB: 4JKM) shares 0.77 Å root-mean-square deviation (rmsd) over 453 equivalent Cα positions with the structure of apo EcGUS (PDB: 3K46) and 48% sequence identity (Wallace et al., 2010). The P21212 (PDB: 4JKL) and I222 (PDB: 4JKK) SaGUS structures share 0.91 and 0.93 Å rmsd, respectively, over 432 and 431 equivalent Cα positions with the structure of E. coli β-glucuronidase. SaGUS and EcGUS share 42% sequence identity. The two Firmicutes enzymes, CpGUS and SaGUS, share 0.55 Å (P21212) and 0.56 Å (I222) rmsd over 440 and 436 equivalent Cα positions and 53% sequence identity. The two SaGUS structures share 0.22 Å rmsd over 527 equivalent Cα positions. The three microbial enzymes, CpGUS, SaGUS (P21212), and EcGUS, share 0.63, 0.85, and 0.72 Å rmsd, respectively, with the structure of human β-glucuronidase (HsGUS, PDB: 3HN3) (Jain et al., 1996) (Hassan et al., 2013) over 457, 424, and 441 equivalent Cα positions, and 46%, 38%, and 48% sequence identities, respectively. Thus, the Firmicutes enzymes reported here exhibit overall structures similar to the EcGUS and HsGUS structures reported previously, and all are members of the glycosyl hydrolase 2 (GH2) family of enzymes.
Figure 1. Firmicutes and Bacteroidetes β-Glucuronidase Crystal Structures.
(A) Novel crystal structures of bacterial β-glucuronidases, two from the Firmicutes phylum, Clostridium perfringens (yellow; CpGUS) and Streptococcus agalactiae (magenta; SaGUS), and one from the Proteobacteria phylum, Escherichia coli (EcGUS) in complex with designed Inhibitor R1 (blue).
(B) Superposition of the active sites the β-glucuronidases from C. perfringens (yellow) and S. agalactiae (magenta) on that of E. coli β-glucuronidase (green) in complex with the glucuronic acid mimic glucaro-δ-lactam, showing the binding of the ligand's carboxylate moiety (left) and indicating that the proximity of the conserved tyrosine indicated (YI468/464) favors the alignment of a β linkage (right) for catalysis. The labels for the two catalytic glutamic acid residues of each enzyme are boxed.
(C) The Asn (N) and Lys (K) residues create the N-K loop conserved in β-glucuronidases but not in the putative β-galactosidase 3DEC from Bacteroides thetaiotaomicron (Bt3DEC, purple). β-Glucuronidases listed include those presented here (e.g., CpGUS, SaGUS), as well as those from mammalian (Homo sapiens, HsGUS), Archaeal (ArchIa), Thermatogae (ThermTn), and other sources. The enzymes for which a crystal structure is available are underlined. ArchIa, Archaea Ignisphaera aggregans, WP_013302863; TherTn, Thermotogae Thermotoga naphthophila, ADA67771; BactNk, Bacteroidetes Niastella koreensis, AEV98753; H11G11, 745-residue protein from uncultured bacterium.
(D) The glycosyl hydrolase from B. thetaiotaomicron (PDB: 3DEC; Bt3DEC; left) lacks the Lys and Asn residues necessary to bind a glucuronic acid sugar for catalysis; thus, this enzyme appears to be a β-galactosidase. By contrast, the glycosyl hydrolase from Bacteroides fragilis (PDB: 3CMG; Bf3CMG; right) maintains the active-site residues capable of glucuronic acid binding. The labels for the two catalytic glutamic acid residues of each enzyme are boxed.
β-Glucuronidase N-K Fingerprint
Using these β-glucuronidase crystal structures as a guide, we next sought to define functionally relevant sequence motifs. β-Glucuronidases align ether-linked glucuronides for hydrolysis using hydrophobic, hydrogen bonding, and electrostatic interactions with the glucuronic acid sugar moiety (Figure 1B). The asparagine and lysine residues that contact the carboxylic acid group unique to glucuronic acid relative to galactose are particularly critical (Figure 1B). In an alignment of β-glucuronidase sequences obtained from the NCBI, the asparagine and lysine (N-K) residues that contact the carboxylic acid are completely conserved, including in the three structures reported here (CpGUS, SaGUS, and EcGUS), as well as in the HsGUS enzyme (Figure 1C). This motif is present in more distantly related β-glucuronidases from Archaeal and Thermotoga sources, as well as Bacteroides species and a gene obtained from uncultured sample whose gene product, H11G11, was demonstrated to have β-glucuronidase activity (Gloux et al., 2011) (Figure 1C). In addition to N-K, a tyrosine residue (Y468 or Y464 in Figure 1B) located ~4 Å from the glucuronic acid binding site appears to ensure by steric occlusion that only β-linked substrates are processed by the two catalytic glutamic acid residues of β-glucuronidases; an α linkage would clash with the aromatic ring at this position (Figure 1B). We examined the N-K motif region in the structures of GH2 family enzymes from B. fragilis (RCSB: 3CMG) and Bacteroides thetaiotaomicron (RCSB: 3DEC) that were deposited in the Research Collaboratory for Structural Bioinformatics in 2012. As shown in Figures 1C and 1D, 3CMG maintains the N-K motif associated β-glucuronidases, while the 3DEC glycosyl hydrolase lacks the N-K motif and instead contains Cys and Asn residues in these locations (Figures 1C and 1D). Thus, 3CMG would be expected to be a β-glucuronidase, and 3DEC, because it lacks the N-K motif, would be expected to act on distinct substrates, perhaps as a β-galactosidase. As shown below, 3CMG is a β-glucuronidase. Thus, the N-K sequence motif can be employed as a fingerprint to identify β-glucuronidases from the large GH2 family of proteins.
Bacterial and M Loops of Microbial β-Glucuronidases
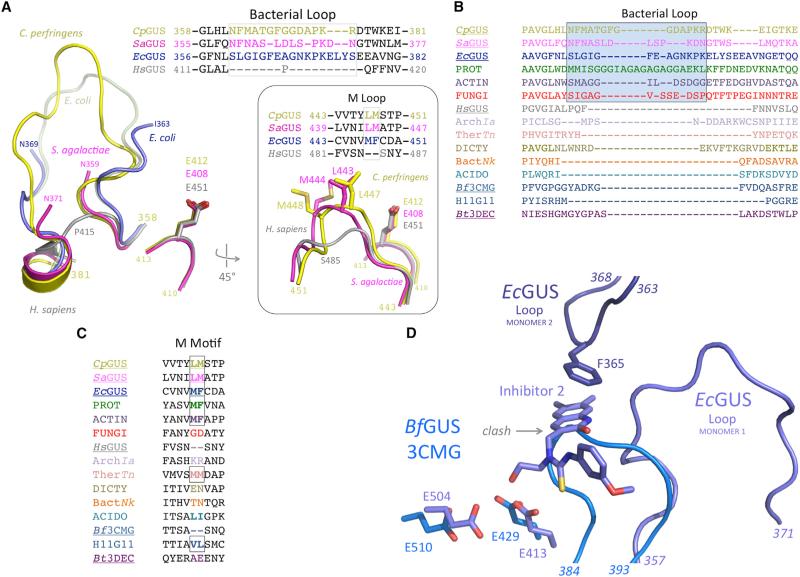
The bacterial β-glucuronidases presented here contain two additional motifs unique relative to the human β-glucuronidase structure. First, many contain a ‘‘bacterial loop’’ that folds over the active sites of microbial enzymes, which processes small substrates (Wallace et al., 2010; Figure 2A). By contrast, human β-glucuronidase, which processes larger glucosaminoglycan-glucuronide substrates, lacks this loop. In the CpGUS and SaGUS structures presented here, this loop is 14 and 13 residues in length, respectively, is visualized in its entirety in CpGUS, and is partially ordered in SaGUS and in the EcGUS-R1 complex (where it is 17 residues in length; Figures 2A and S2). The EcGUS-Inh2 structure reported previously contained a fully ordered loop, and a comparison of this loop and the ordered loop from CpGUS presented here demonstrates that this region is structurally flexible (Figures 2A and S2). Bacterial loops are also divergent in sequence, sharing only 11%–36% identity between the EcGUS, CpGUS, and SaGUS. In HsGUS, this region is replaced by a single residue, P415 (Figure 2A).
Figure 2. Bacterial- and Mammalian-Type β-Glucuronidases Exhibit Distinct Bacterial and M Loops.
(A) The three novel bacterial β-glucuronidase structures presented here (yellow, magenta, blue) superimposed on the structure of human β-glucuronidase (gray) reveals both the lack of a bacterial loop in the mammalian enzyme, as well as the lack of an M loop in the mammalian enzyme (inset).
(B) The bacterial loop, boxed, is maintained in the L-GUS enzymes, including those from several fungi (red), but is missing from the no-loop β-glucuronidases (NL-GUS) and the B. thetaiotaomicron (Bt3DEC) β-galactosidase. The enzymes for which a crystal structure is available are underlined. PROT, Proteobacteria Vibrio harveyi, WP_005434141; ACTIN, Actinobacteria Corynebacterium massiliense, WP_022863751; FUNGI, Eukaryota Aspergillus niger, CAK36851; ArchIa, Archaea Ignisphaera aggregans, WP_013302863; TherTn, Thermotogae Thermotoga naphthophila, ADA67771; DICTY, Dictyoglomi Dictyoglomus turgidum, ACK42813; BactNk, Bacteroidetes Niastella koreensis, AEV98753; ACIDO, Acidobacteriua Terriglobus roseus, WP_01478374; H11G11, 745-residue protein from uncultured bacterium.
(C) A hydrophobic M loop is missing from several no-loop β-glucuronidases (NL-GUS), such as HsGUS and Bf3CMG, but is present in others, and is conserved in the bacterial loop-containing β-glucuronidases (L-GUS). The enzymes for which a crystal structure is available are underlined.
(D) Active-site superposition of EcGUS-Inh2 complex (dark blue) on BfGUS (3CMG; light blue) showing the catalytic residues and bacterial loop of EcGUS. The 384–393 region of BfGUS, which is significantly shorter than the bacterial loop of EcGUS (357–371), would clash with the binding site of compounds such as Inhibitor 2. In addition, in the EcGUS tetramer, loops from different monomers (e.g., 1 and 2 shown) participate in binding contacts with Inhibitor 2. BfGUS does not form an analogous tetramer and cannot form a similar inhibitor binding site.
Sequence analysis of bacterial loops in a set of microbial β-glucuronidase enzymes from the NCBI database reveals that some of the enzymes contain the loop and some do not (Figure 2B). Phyla that encode a β-glucuronidase with an intact bacterial loop include Firmicutes, Proteobacteria, and Actinobacteria, as well as representatives from the fungi such as Aspergillus niger (Figure 2B). Phyla with β-glucuronidases lacking the bacterial loop include the Bacteroidetes, Dictyoglomi, and Acidobacteria. The β-glucuronidase from B. fragilis, 3CMG, lacks the bacterial loop. Representative β-glucuronidases in Archaea, such as Ignisphaera aggregans, and in Thermatogae, such as Thermotoga naphthophila (Figure 2B), also lack the bacterial loop. All sequences in this alignment contain the conserved catalytic β-glucuronidase residues outlined in Figure 1B, as well as the N-K motif (see Figure 1C), and are therefore considered to be representative of the range of β-glucuronidases present in sequenced microbiota.
Second, a minor structural feature unique to the microbial β-glucuronidases is the presence of a dipeptide within the active site composed of Leu-Met in Cp and SaGUS, and Met-Phe in EcGUS (Figure 2A). This motif is missing from HsGUS but is present in most, but not all, L-GUS and NL-GUS enzymes from NCBI sequences (Figure 2C). Due to the prevalence of Met residues in this short region, we refer to it as the M loop. In the Cp, Sa, and EcGUS structures, this region is located ~7.5 Å from catalytic residue E413 and contacts bound inhibitors, suggesting that it may play a role in substrate binding and catalytic function.
Based on these sequence and structural considerations, we propose that microbial β-glucuronidases can be divided into two major groups: the L-GUS enzymes (loop-β-glucuronidases), resembling the initially characterized bacterial enzymes and containing a bacterial loop, and the NL-GUS (no-loop) enzymes, which, as shown here, maintain all major features of β-glucuronidases but lack the bacterial loop. As shown in more detail below, these groupings provide a useful tool in understanding the β-glucuronidases of known sequence.
Microbial β-Glucuronidases Are Distinct in Catalytic Activity and Inhibition
We compared the kcat, KM, and catalytic efficiencies (kcat/KM) of the microbial β-glucuronidases examined here using the standard p-nitrophenyl glucuronide β-glucuronidase assay substrate. The B. fragilis β-glucuronidase 3CMG, a representative GUS from the Bacteroidetes phylum, was overexpressed in E. coli and purified for kinetic characterization. Marked differences in all three parameters were observed (Table 1), with EcGUS exhibiting the best activity (kcat = 120 s–1, KM = 0.13 mM, and kcat/KM = 920 s–1 mM–1), SaGUS showing moderate activity (kcat = 80 s–1, KM = 0.36 mM, and kcat/KM = 224 s–1 mM–1), and CpGUS demonstrating the slowest kcat (2.6 s–1), the weakest KM (1.1 mM), and the least catalytic efficiency kcat/KM (2.4 s–1 mM–1). The B. fragilis β-glucuronidase 3CMG (BfGUS) exhibited a poor KM of only 1.9 mM, but a kcat (18 s–1) roughly three times higher than CpGUS, and thus showed a catalytic efficiency nearly five times higher than CpGUS. These data establish for the first time that BfGUS is a β-glucuronidase, and that the four enzymes from three representative GI bacterial phyla exhibit a range of catalytic efficiencies with this standard β-glucuronidase assay substrate.
Table 1.
Catalytic Activity of β-Glucuronidases
| Enzyme | kcat (s−1) | KM (mM) | kcat/KM (s−1 mM−1) |
|---|---|---|---|
| EcGUS | 120 ± 12 | 0.13 ± 0.01 | 920 |
| SaGUS | 80 ±2 | 0.36 ± 0.03 | 224 |
| CpGUS | 2.6 ± 0.6 | 1.1 ± 0.2 | 2.4 |
| BfGUS | 18 ± 1 | 1.9 ± 0.3 | 9.5 |
Data are presented as the average over ±3 experiments ± SEM for Escherichia coli (EcGUS), Clostridium perfringens (CpGUS), Streptococcus agalactiae (SaGUS), and Bacteroides fragilis (BfGUS) β-glucuronidases. kcat, catalytic rate; KM, Michaelis constant; kcat/KM, catalytic efficiency.
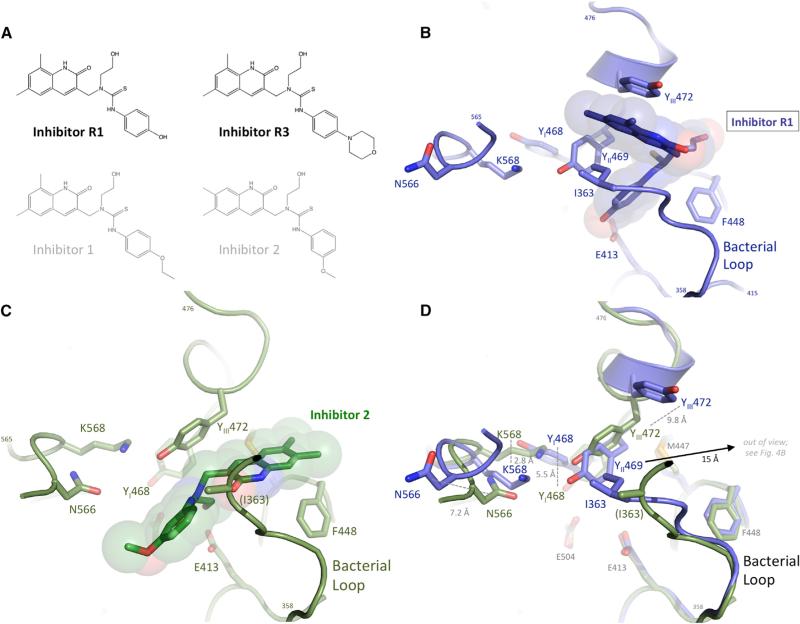
We also examined the ability of the Proteobacteria and Firmicutes enzymes to be inhibited by Inhibitors 1–8 previously identified by a 10,000-compound high-throughput screen using EcGUS (Roberts et al., 2013; Wallace et al., 2010; Figure S3). These compounds all inhibit EcGUS, with all Ki values below 2 μM (Table S2). Two novel compounds, R1 and R3, which are designed analogs of Inhibitor 1 and are shown in Figure 3A, were also synthesized and exhibited Ki values of 1.9 and 0.61 μM, respectively, toward EcGUS, and EC50 values of 4.5 and 3.1 μM, respectively, in cultured E. coli cells (Tables S2 and S3). In previous studies, Inhibitor 1 demonstrated an EC50 of 17 nM for cultured E. coli.
Figure 3. Novel Bacterial β-Glucuronidase Inhibitors.
(A) Inhibitors R1 and R3 were designed, based on Inhibitor 1 (gray), to reduce relative GI absorption. Inhibitor 2, for which a crystal structure bound to E. coli β-glucuronidase is available, is also shown (gray).
(B) Inhibitor R1 bound to the active site of E. coli β-glucuronidase, making contacts with the bacterial loop, as well as two active-site tyrosines, 469 and 472. The catalytic residue E413 is also shown, as are the β-glucuronidase fingerprint residues N566 and K568.
(C) Binding of Inhibitor 2 to the active site of E. coli β-glucuronidase (Wallace et al., 2010), making distinct contacts, relative to Inhibitor R1, with the bacterial loop and one active-site tyrosine, 472.
(D) Superposition of the active sites of E. coli β-glucuronidase bound to Inhibitors R1 (blue) and 2 (green) but with the ligands removed. The relative shifts in five active-site residues are shown, including a 15-Å movement of Y469 and a shift in the positions of the β-glucuronidase fingerprint residues N566 and K568.
Interestingly, all ten compounds exhibit markedly differential effects toward the two novel microbial β-glucuronidases, CpGUS and SaGUS, presented here (Table S2). For example, Inhibitors R1, R3, 7, and 8 show no inhibition of the Cp and Sa β-glucuronidases, but are 0.61–2 μM inhibitors of the E. coli enzyme. Similarly, Inhibitors 2, 3, and 6 are 0.2–0.7 μM inhibitors of EcGUS, but are up to 16-fold weaker (e.g., 11 μM for Inhibitor 3 toward Sa versus 0.68 μM for Ec) toward the Firmicute enzymes from SaGUS and CpGUS. Thus, distinct chemotypes can exert ortholog-specific effects on the activities of purified microbial β-glucuronidases.
We next examined whether a no-loop (NL) β-glucuronidase from the Bacteroidetes phylum can be inhibited by the compounds we have in hand. We found that BfGUS is not subject to inhibition by Inhibitors 1–8 at up to 100-μM inhibitor concentrations (Table S2). This result is reminiscent of the data collected previously on the mammalian GUS from bovine, which was also not subject to inhibition by Inhibitors 1–8 (Roberts et al., 2013; Wallace et al., 2010). Both bovine GUS and BfGUS are NL-GUS enzymes, in contrast to the Firmicutes and Proteobacteria enzymes, which are L-GUS enzymes with bacterial loops. Superimposing the structure of EcGUS in complex with Inhibitor 2 with BfGUS from crystal structure 3CMG reveals that the residues 384–393 of BfGUS would clash with the inhibitor binding site (Figure 2D). Furthermore, while both enzymes are shown here to form tetramers by multi-angle light scattering in solution, by structural analysis the tetramers they form are distinct (Figure S4). Indeed, BfGUS fails to maintain the dimer contacts shown previously for EcGUS to be involved in inhibitor binding (Wallace et al., 2010) (Figure 2D). Taken together, these observations provide molecular explanations for the lack of inhibition of BfGUS by Inhibitors 1–8. Thus, we conclude the NL-GUS enzymes will not be subject to inhibition by the reagents that potently inhibit the L-GUS enzymes.
Reducing Serum Exposure of Inhibitors
We next hypothesized that inhibitors intended to target enzymes in the GI lumen would be improved by limiting their serum exposure. We have reported previously that Inhibitor 1 exhibits 21% oral bioavailability (F) in mice, but which increases to 99% when the animals are given the pan-cytochrome P450 inhibitor ABT (LoGuidice et al., 2012). This suggests that Inhibitor 1 is absorbed from the GI tract and is subject to metabolism by P450 enzymes. Thus, we sought to reduce the serum exposure of Inhibitor 1 by creating two novel analogs, R1 and R3 (Figures 3A, S5, and S6). The O-ethyl group from Inhibitor 1, expected from crystal structural analysis to accommodate derivatization, was replaced with a hydroxyl (R1) or morpholine (R3) group, each hypothesized to reduce GI absorption. As noted above, inhibitors R1 and R3 are 0.6–2 μM inhibitors of EcGUS, but do not inhibit SaGUS and CpGUS at up to 100 μM. We performed preliminary pharmacokinetic (PK) studies on mice treated orally with Inhibitor 1 and Inhibitors R1 and R3 to determine plasma levels of these compounds over 24 hr, with or without the cytochrome P450 inhibitor ABT. We found that the R1 compound exhibited significantly reduced oral bioavailability (%F) alone or with ABT relative to Inhibitor 1 or R3 (Table 2). Inhibitor R1 also showed poor plasma exposure, as measured by maximum concentration and area under the curve, relative to Inhibitors 1 and R3. Thus, R1 is either metabolized quickly or remains in the GI tract when given orally to mice. The ability of Inhibitor R1 to alleviate CPT-11-induced GI damage is examined below.
Table 2.
Oral Bioavailability of β-Glucuronidase Inhibitors
| Tmax (hr) | Cmax (ng/ml) | AUC (ng/ml/hr) | %F | |
|---|---|---|---|---|
| Inh-1 | 0.5 | 510 ± 200 | 960 | 21 |
| Inh-1 + ABT | 4.0 | 740 ± 20 | 4,900 | 99 |
| R1 | 0.3 | 13 ± 0.4 | 24 | 4 |
| R1 + ABT | 0.3 | 27 ± 20 | 75 | 12 |
| R3 | 0.3 | 82 ± 10 | 140 | 6 |
| R3 + ABT | 1.0 | 780 ± 200 | 2,800 | 100 |
Oral bioavailability (F) with and without treatment with the pan-cytochrome P450 inhibitor ABT. Tmax, maximum time; Cmax, maximum concentration; AUC, area under the curve.
E. coli β-Glucuronidase-Inhibitor R1 Complex Crystal Structure
We determined the 2.4-Å resolution crystal structure of EcGUS in complex with Inhibitor R1. This complex revealed a surprising level of structural variability in the active-site and bacterial-loop regions of this microbial β-glucuronidase enzyme. Inhibitor R1 was found to bind in a unique manner to EcGUS when compared with its close chemical homolog, Inhibitor 2 (Wallace et al., 2010). Inhibitor R1 forms a hydrogen bond between catalytic residue E413 and this compound's phenyl ring hydroxyl (Figure 3B). In contrast, Inhibitor 2 forms a hydrogen bond between its ethoxy group and E413 (Figure 3C), similar to that observed in the EcGUS-Inhibitor 3 complex (Wallace et al., 2010). Inhibitors 2 and R1 are rotated ~120° about the horizontal axis with respect to one another in the EcGUS active site, as shown in Figures 3B and 3C. We observed a significant conformational change associated with Inhibitor R1 binding to EcGUS. N566 and K568 of the N-K loop, and tyrosines 468, 469, and 472 all shift in position from 2.8 to 15Å relative to the Inhibitor 2 bound structure (Figure 3D).
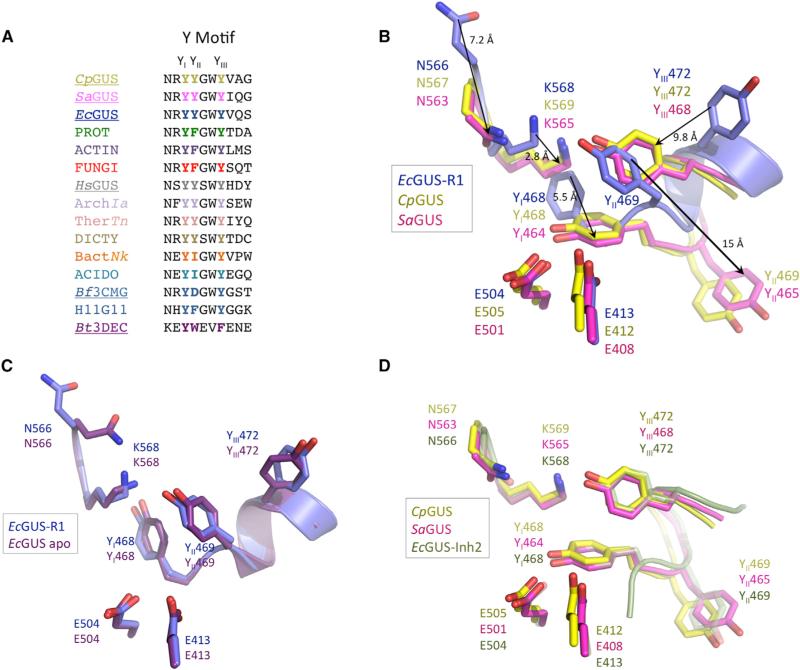
These observations define a mobile Y loop within the active site of microbial β-glucuronidases (Figure 4A). As shown in Figures 3D and 4B, Y468, Y469, and Y472 in the EcGUS-R1 complex shift in position by 5.5, 9.8, and 15Å, respectively, relative to their locations in the Inhibitor 2-EcGUS complex. These three tyrosines are conserved in the sequences of the CpGUS and SaGUS enzymes examined here (Figure 4A), and can be labeled as YI (Y468, Y468, and Y464 in Ec, Cp, and Sa, respectively), YII (Y469, Y469, and Y465), and YIII (Y472, Y472, and Y468). The Y loop is capable of adopting two different positions in the crystal structures of microbial β-glucuronidases resolved to date, as noted previously (Wallace et al., 2010) and extended here. The Y-loop residues in the EcGUS-R1 complex are positioned nearly identically to that observed in the apo structure of EcGUS reported previously, and can be considered to be “inactive” because the position of YI sterically blocks the Lys in the N-K loop from forming a salt bridge with the carboxylate unique to glucuronic acid (Figure 4C). The Y-loop residues in the EcGUS-Inhibitor 2 and 3 structures (Wallace et al., 2010), by contrast, are aligned well with those in the CpGUS and SaGUS structures reported here, and the Lys of the N-K loop is positioned to contact a bound glucuronic acid sugar (Figure 4D; see also Figure 1B). Taken together, these data highlight the structural plasticity present in the active sites of GUS enzymes, particularly in the bacterial-loop and Y-loop regions, and indicate that these shifts can affect the position of other regions of the enzyme, such as the N-K loop.
Figure 4. Bacterial β-Glucuronidases Exhibit Dynamic Y Loops.
(A) The first (YI), second (YII), and third (YIII) tyrosines of the Y loop are conserved in both the bacterial- and mammalian-type β-glucuronidases. See the legend of Figure 2 for labels.
(B) Superposition of the active sites of C. perfringens (yellow; CpGUS), S. agalactiae (magenta; SaGUS), and the E. coli Inhibitor R1 complex (blue; EcGUS-R1) showing the dramatic shifts in position in tyrosine residues YI (Tyr 464 or 468), YII (Tyr 465 or 469), and YIII (Tyr 468 or 472) of the Y loop, and in the β-glucuronidase fingerprint sequence of the N-K loop. The view is rotated approximately 45° about the horizontal axis relative to Figure 3, and the two catalytic Glu residues of each enzyme are shown.
(C) The E. coli β-glucuronidase Inhibitor R1 complex (blue; EcGUS-R1) superimposes well on the active site of the unliganded structure of the enzyme (purple; EcGUS apo) (Wallace et al., 2010).
(D) In contrast to (C), the unliganded C. perfringens (yellow; CpGUS) and S. agalactiae (magenta; SaGUS) β-glucuronidase structures superimpose well in this region with the liganded E. coli b-glucuronidase Inhibitor 2 complex (green; EcGUS-Inh2) (Wallace et al., 2010).
Irinotecan Toxicity and Pharmacokinetics in Mice
Finally, we sought to determine how inhibition of microbial β-glucuronidase activity would affect irinotecan pharmacokinetics and GI damage in mice. First, we examined Inhibitor R1 for its ability to alleviate irinotecan (CPT-11)-induced diarrhea in mice. Without inhibitor, mice (n = 9 per group) that receive 50 mg/kg CPT-11 intraperitoneally (i.p.) develop bloody diarrhea after 8–10 days, with ~25% of treated mice showing this acute side effect at day 8, ~60% at day 9, and 100% at day 10 (Figure 5A). Inhibitors 1 and R1 were provided to the same number of mice twice daily by oral gavage at 10 μg per dose, for a total of 20 μg per day. As a positive control, the previously described Inhibitor 1 was found to reduce the number of mice with this side effect to 20% at day 9 and 40% at day 10. Inhibitor R1, which demonstrates ~10-fold reduced in vitro inhibition of EcGUS and no inhibition of the Firmicutes enzymes (Table S2), showed reduced alleviation of CPT-11-induced diarrhea, to 20% of mice at day 8, 40% at day 9, and 60% at day 10 (Figure 5A). Thus, while Inhibitor R1 showed a potentially beneficial lack of serum exposure in mice (Table 2), it is deficient relative to Inhibitor 1 in its alleviation of CPT-11-induced injury.
Figure 5. Microbial β-Glucuronidase Inhibitors in Mice Treated with CPT-11.
(A) CPT-11 (irinotecan) produced delayed diarrhea in 25%, 60%, and 100% of mice in 8, 9, and 10 days, respectively. Oral delivery of Inhibitor R1, a 1.9-μM inhibitor of E. coli β-glucuronidase, reduces the number of mice that experience diarrhea at days 9 and 10 to 40% and 60%, respectively, but is less effective than Inhibitor 1, a 0.16-μM inhibitor, on which it was based. N = 9 mice per group.
(B) The circulating plasma levels of CPT-11, its active metabolite SN-38, or its inactive glucuronide conjugate SN-38-G are unaffected by the oral delivery of the microbial β-glucuronidase Inhibitor 1 (Inh1).
Second, we examined the potential impact of Inhibitor 1 on the serum levels of CPT-11 and its key metabolites SN-38 and SN-38-G in mice. To date, it has remained unclear whether the reabsorption of the SN-38 created by GI microbial β-glucuronidases plays a role in circulating levels of this active metabolite of CPT-11. Using the same dosing scheme as above, plasma levels of CPT-11, SN-38, and SN-38-G were determined using mass spectrometry. As shown in Figures 5B and Table S4, the presence of Inhibitor 1 appears not to alter significantly the levels of CPT-11, SN-38, and SN-38-G. These data support the conclusion that inhibiting the reactivation of SN-38 from SN-38-G in the GI tract would not reduce serum in the SN-38 capable of reaching tumors via the serum.
DISCUSSION
Firmicutes and Bacteroidetes are the predominant microbial phyla in the mammalian GI tract (The Human Microbiome Project Consortium, 2012); here, we examine representative β-glucuronidases from each to advance our understanding of the structure and function of these enzymes, which are involved in drug-induced GI toxicity and its alleviation (LoGuidice et al., 2012; Mani et al., 2014; Roberts et al., 2013; Saitta et al., 2014; Wallace et al., 2010). We find that purified β-glucuronidases from the Firmicutes, Proteobacteria, and Bacteroidetes phyla exhibit 10-, 50-, and 100-fold differences with respect to one another in substrate binding, reaction rate, and catalytic efficiency, respectively (Table 1). Furthermore, we find that some are amenable to inhibition while others are not inhibited by a range of chemotypes (Table S2). These observations support the conclusion that the GI microbiome maintains an array of enzyme orthologs capable of scavenging glucuronic acid from a range of chemically distinct substrates. Diverse endobiotic, food, and xenobiotic glucuronides are expected to be regularly delivered to the GI tract, and thus the microbial species therein are poised to utilize these diverse sources of carbon. It will be of interest to determine in future studies whether specific symbiotic bacteria are responsible for reactivating specific drug- or endobiotic glucuronides. For example, a particular set of bacteria may be most efficient at reactivating SN-38G while another efficiently processes NSAID glucuronides. Such knowledge may enable the development of more tailored inhibitors for specific drug-induced GI enteropathies.
At the structure-function level, we find that β-glucuronidases exhibit conformational flexibility within their active sites, and can adopt active and inactive states (Figures 3 and 4). We also demonstrate that β-glucuronidases contain Asn and Lys residues (an N-K motif) in their active site that coordinates the carboxylate group unique to glucuronic acid relative to galactose (Figure 1). This observation was anticipated by the work of Matsumura and Ellington (2001), who evolved glucuronidase activity from galactosidase enzymes and noted that the N-K motif was essential. The aromatic-rich Y motif is also conserved across glucuronidases of known sequence; these tyrosines make contact with substrate and inhibitor ligands and, in the case of EcGUS, are observed to adopt distinct conformations depending on the nature of the ligand bound. Thus, we propose that the N-K and Y motifs should join the catalytic glutamic acid residues as structural features considered essential for β-glucuronidase activity. A B. fragilis glycosyl hydrolase, the structure of which was reported in 2008 (RCSB: 3CMG), was annotated at that time as a β-galactosidase, an understandable conclusion given its significant similarity to β-galactosidases from other Bacteroides species (e.g., 3DEC, 3BGA). However, unlike such related β-galactosidases, 3CMG contains an N-K motif, and here we confirm in in vitro assays that it is a β-glucuronidase. This result highlights the potential for using the N-K motif to identify the β-glucuronidases present in the GI microbiome.
We conducted a phylogenetic analysis of microbial and mammalian β-glucuronidase sequences from a taxonomically diverse group of cultured organisms. The resulting phylogenetic tree of β-glucuronidases from organisms in all three kingdoms indicates the tremendous diversity of β-glucuronidase sequences and the broad distribution of this gene across the tree of life (Figure S7). This analysis further supports the conclusion that the β-glucuronidases can be effectively divided into those that maintain a loop of their active sites (L-GUS) and those that do not (NL-GUS). The majority of loop-positive bacteria are allocated into two phyla, Firmicutes and Actinobacteria, both prominent in the human gut. Sequenced representatives of the Bacteroidetes, also a prominent GI phylum, are loop-negative, as are the mammalian β-glucuronidases. While closely related organisms generally share similar loop phenotypes, this not a universal feature of the tree. For example, within the Firmicutes (Figure S7, blue branches), some loop-positive sequences cluster with loop-negative sequences. We also find loop-positive fungal (Figure S7, orange), archaeal Crenarchaeota, and even Euryarchaeota β-glucuronidases (Figure S7, cyan and light blue, respectively), in spite of the fact that the mammalian β-glucuronidases are loop-negative. The presence is likely linked to the propensity for organisms to utilize smaller glucuronides rather than larger ones as sources of carbon that can be scavenged. However, phylogenetic and biochemical analysis of Ascomycete fungi suggested previously that the β-glucuronidase gene was horizontally transferred from bacteria, potentially enabling fungi to make use of glucuronic acids as a carbon source in the context of a vertebrate host (Wenzl et al., 2005).
We find that the presence of the active-site loop confers sensitivity to inhibition. We demonstrate that the NL β-glucuronidase from B. fragilis (BfGUS) is inhibited neither by the eight microbial β-glucuronidase inhibitors reported previously (Wallace et al., 2010; Roberts et al., 2013) nor by Inhibitors R1 and R3 presented here. It was previously shown that deleting the loop from EcGUS creates an enzyme that retains activity but is no longer subject to inhibition (Wallace et al., 2010). Together, these results define a crucial functional difference between L and NL β-glucuronidases. We predict that NL-GUS enzymes will be unable to utilize glucuronide conjugates such as SN-38-G as well as the L-GUS enzymes, as NL-GUS proteins lack the active-site loop predicted to be necessary to close over these molecules. By contrast, L-GUS enzymes have been shown to utilize small glucuronides such as SN-38-G and diclofenac-G as substrates (Wallace et al., 2010; Roberts et al., 2013). Thus, the ability to selectively inhibit L-GUS enzymes over their NL-GUS counterparts appears particularly useful in the prevention of drug-induced GI toxicity.
We hypothesized that limiting the serum exposure of inhibitors would be beneficial to their efficacy, as such compounds would potentially spend more time in the GI tract. The designed Inhibitor 1 analog R1 did show reduced serum exposure after oral dosing in mice. However, it was found to be less efficacious than Inhibitor 1 in alleviating diarrhea. There may be several explanations for this result. First, R1 shows reduced potency against EcGUS, and complete lack of inhibition of SaGUS or CpGUS. Thus, it is relatively weak as an inhibitor in vitro, which may correspond to weak efficacy in vivo. Second, the reduction of Inhibitor R1 levels in the plasma could be caused by its rapid metabolism and elimination and not by its reduced absorption from the GI tract. Examination of intestinal contents for intact inhibitors, and detailed inhibitor metabolism studies, will be required to understand the pharmacokinetics and pharmacodynamics of the microbiota-targeted compounds in vivo.
Importantly, we find that inhibition of microbial GI β-glucuronidases does not alter the serum levels of CPT-11 and its key metabolites in mice. Controversy has existed over the route by which SN-38, the active metabolite of CPT-11, reaches colon tumors. Particularly, it is not clear whether the reabsorption of SN-38 reactivated by the GI microbiota from the GI lumen plays a significant role in colon tumor exposure, and subsequent efficacy. Recent data using transgenic mice have suggested that, like most chemotherapies, SN-38 delivery to the tumors would be expected to occur through the systemic vasculature (Chen et al., 2013). Furthermore, SN-38 produced in the GI lumen is subject to epithelial metabolism back to SN-38-G and export back to the lumen (Chen et al., 2013). The data presented here support the conclusion that reabsorption of reactivated SN-38 from the GI tract does not play a role in plasma levels of this compound, CPT-11, or SN-38-G. We therefore hypothesize that inhibition of GI bacterial β-glucuronidases would not negatively affect tumor response to CPT-11. Recent data from another group validates this hypothesis and demonstrates that Inhibitor 1 and other β-glucuronidase inhibitors improve tumor regression in mice receiving CPT-11 (Kong et al., 2014). Thus, by reducing the GI side effects of CPT-11, bacterial β-glucuronidase inhibitors might also improve outcomes in human patients receiving CPT-11 as part of the FOLFIRI or FOLFIRINOX regimen for colon and pancreas cancers, respectively.
SIGNIFICANCE
Conjugating glucuronic acid to chemicals marked for excretion is fundamental to xenobiotic and endobiotic metabolism. Glucuronide conjugates are reactivated by glucuronide-scavenging β-glucuronidases in the GI microbiota, and such reactivated compounds can significantly damage the GI tract. This effect has been established for the chemotherapeutic irinotecan and for NSAIDs. Selective, potent, and non-lethal inhibition of bacterial β-glucuronidases was recently shown to alleviate drug-induced GI damage. Here we elucidate the structure and function of β-glucuronidases from the mammalian GI microbial phyla Firmicutes and Bacteroidetes. We show that β-glucuronidases exhibit marked differences in their catalytic activities and propensities for inhibition, and that small changes in inhibitor structure can alter β-glucuronidase active-site conformation. A phylogeny of extant β-glucuronidases establishes two major enzyme clusters: L-GUS enzymes with active-site loops, and NL-GUS enzymes without loops. GI bacterial β-glucuronidase inhibition in mice does not alter the serum pharmacokinetics of irinotecan, its active metabolite SN-38, or its inactive glucuronide SN-38-G. Furthermore, reducing inhibitor serum exposure does not enhance alleviation of drug-induced GI toxicity, suggesting that increasing GI residence of inhibitors is not essential to efficacy. Finally, unlike the Proteobacteria and Firmicutes enzymes, the B. fragilis β-glucuronidase is not subject to inhibition by our selective reagents. Together, these data advance our understanding of the structure, function, and inhibition of the microbial β-glucuronidases, a promising set of new therapeutic drug targets for controlling drug-induced GI toxicity and the first targets identified in the microbiome.
EXPERIMENTAL PROCEDURES
Protein Purification, Crystallization, and Structure Determination
Genomic DNA was derived from S. agalactiae 2603V/R (ATCC #BAA-611D-5), and was graciously provided by Dr. Bruce McClane (University of Pittsburgh School of Medicine) for C. perfringens str. 13. Enzymes were cloned by PCR into pLIC-His (S. agalactiae) and pLIC-MBP (C. perfringens) ligation independent cloning (LIC) vectors. The pLIC-His vector contains an N-terminal 6x-histidine tag, and the pLIC-MBP plasmid incorporates an N-terminal MBP-fusion and 6x-histidine tag. Proteins were expressed in E. coli BL21-DE3 AI competent cells (Invitrogen) and purified by column chromatography.
Similar methods were employed for BfGUS. His-SaGUS crystals were optimized for quality in 0.1–0.3 M potassium thiocyanate and 20%–28% polyethylene glycol (PEG) 3350 at 16°C. Crystals of MBP-CpGUS were grown at 16°C in 0.1 M 2-(N-morpholino)ethanesulfonic acid (pH 6.0–6.5) and 28%–36% PEG 400. The E. coli β-glucuronidase (EcGUS)-R1 complex crystals were obtained as first described by Wallace et al. (2010) with modifications for co-crystallization as described by Roberts et al. (2013). Crystals were flash-frozen in liquid nitrogen in preparation for X-ray data collection. Diffraction data were collected on the 23-ID beamline at GM/CA-CAT (Advanced Photon Source, Argonne National Laboratory). Data were processed using standard methods and structures determined by molecular replacement using the EcGUS structure as a search model (PDB: 3K46). Structures were refined using standard methods. See Supplemental Information for more details. Coordinates and structure factors can be found at PDB: 4JKM, 4JKK, 4JKL, 5CZK.
Enzyme Assays
Assays were performed using the His-SaGUS, MBP-CpGUS, BfGUS, and His-EcGUS enzymes. Kinetic assays used p-nitrophenyl glucuronide as the absorbance substrate. Reactions were conducted in 96-well, clear-bottom assay plates (Costar) at 37°C in a 50-μl total volume. The reaction was performed at 37°C and product formation was measured using a PHERAstar Plus microplate reader (BMG Labtech). Initial velocities were determined for multiple substrate and enzyme concentrations, and Michaelis-Menten kinetics were used to calculate KM, kcat, and catalytic efficiency. Inhibition assays were performed using the same enzymes to assess the efficiency of Inhibitors 1–8, R1, and R3 to disrupt enzymatic activity from various bacterial species. Cell-based assays to calculate EC50 were performed as described by Roberts et al. (2013) and Wallace et al. (2010). High-performance liquid chromatography (HPLC) assays of SN-38-G (Santa Cruz Biotechnology) conversion to SN-38 (Santa Cruz) were conducted as described by Wallace et al. (2010). Structures were refined using standard methods. See Supplemental Information for more details.
CPT-11 and Inhibitor Studies in Mice
Animal experiments were performed according to the Institutional Animal Care and Use guidelines approved by the Institutional Animal Care and Use Committee (IACUC nos. 20070715 and 20100711) of the Albert Einstein College of Medicine, Bronx, NY.
Alleviation of CPT-11-induced toxicity studies were performed as described previously (Roberts et al., 2013). Inhibitor R1 and R3 pharmacokinetic analyses were performed identically to those described previously (LoGuidice et al., 2012). CPT-11, SN-38, and SN-38-G pharmacokinetic analysis was performed as follows. CPT-11 was administered at 50 mg/kg i.p. × 1 with or without 20 μg of Inhibitor 1 per day (oral gavage of 10 mg twice daily for 3 days prior to CPT-11 administration and once at CPT-11 administration) in Balb/cJ wild-type mice. Analytes were separated using a Shimadzu LC-20 HPLC instrument equipped with a Waters XBridge C18 column (2.5 mm, 2.1 × 50 mm). Quantitative analysis was performed by a Thermo Exactive Orbitrap mass spectrometer equipped with a heated electrospray ionization source, operating in positive mode with enhanced resolution and optimized for high dynamic range. Structures were refined using standard methods. See Supplemental Information for more details.
Supplementary Material
Highlights.
Microbiome drug targets are examined from Firmicutes and Bacteroides
Marked differences seen in catalytic activities and propensities for inhibition
Inhibition does not alter serum pharmacokinetics of irinotecan or its metabolites
Phylogeny defines major enzyme groups guided by structural features
ACKNOWLEDGMENTS
Supported by the University Cancer Research Fund of North Carolina (M.R.R.) and by NIH grants CA98468 (M.R.R.) and CA161879 (S.M.). J.D.I. is an employee of Symberix Pharmaceuticals, which is developing microbiome-targeted reagents to treat drug-induced GI toxicity. M.R.R. is a Founder of Symberix Pharmaceuticals and a member of its Scientific Advisory Board.
Footnotes
AUTHOR CONTRIBUTIONS
B.D.W., M.D., E.F., and M.R.R. determined the structures of the Firmicutes GUS enzymes. A.B.R. and M.R.R. determined the structure of EcGUS in complex with R1. R.M.P., J.D.I., K.A.B., S.J.P., S.J.R., S.R.McS., B.D.W., A.B.R., and M.R.R. determined the activities of the GUS enzymes and their propensities for inhibition. M.K.V. and S.M. determined the effects of R1 on CPT-11-induced GI toxicity in mice. L.G., C.P.-R., and L.K. performed the phylogenetic analysis of GUS sequences. S.K.O. and W.C.Z. examined the serum pharmacokinetics of CPT-11 and key metabolites in mouse plasma. J.J., S.V.F., and M.R.R. synthesized R1 and R3 and examined their pharmacokinetics in mice. All authors participated in analyzing the data and writing the manuscript.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2015.08.005.
REFERENCES
- Boelsterli UA, Redinbo MR, Saitta K. Multiple NSAID-induced hits injure the small intestine: underlying mechanisms and novel strategies. Toxicol. Sci. 2012;131:654–667. doi: 10.1093/toxsci/kfs310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune K, Hinz B. The discovery and development of antiinflammatory drugs. Arthritis Rheum. 2004;50:2391–2399. doi: 10.1002/art.20424. [DOI] [PubMed] [Google Scholar]
- Chabot GG. Clinical pharmacology and pharmacodynamics of irinotecan. Ann. N. Y. Acad. Sci. 1996;803:164–172. doi: 10.1111/j.1749-6632.1996.tb26385.x. [DOI] [PubMed] [Google Scholar]
- Chen S, Yueh M-F, Bigo C, Barbier O, Wang K, Karin M, Nguyen N, Tukey RH. Intestinal glucuronidation protects against chemo-therapy-induced toxicity by irinotecan (CPT-11). Proc. Natl. Acad. Sci. USA. 2013;110:19143–19148. doi: 10.1073/pnas.1319123110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, Adenis A, Raoul J-L, Gourgou-Bourgade S, de la Fouchardière C, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Pyrhönen S, James RD, Punt CJ, Hickish TF, Heikkila R, Johannesen TB, Starkhammar H, Topham CA, Awad L, et al. Randomised trial of irinotecan plus supportive care versus supportive care alone after fluorouracil failure for patients with metastatic colorectal cancer. Lancet. 1998;352:1413–1418. doi: 10.1016/S0140-6736(98)02309-5. [DOI] [PubMed] [Google Scholar]
- Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- Dranitsaris G, Maroun J, Shah A. Severe chemotherapy-induced diarrhea in patients with colorectal cancer: a cost of illness analysis. Support Care Cancer. 2005;13:318–324. doi: 10.1007/s00520-004-0738-7. [DOI] [PubMed] [Google Scholar]
- Fittkau M, Voigt W, Holzhausen H-J, Schmoll H-J. Saccharic acid 1.4-lactone protects against CPT-11-induced mucosa damage in rats. J. Cancer Res. Clin. Oncol. 2004;130:388–394. doi: 10.1007/s00432-004-0557-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloux K, Berteau O, Oumami El H, Béguet F, Leclerc M, Doré J. A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4539–4546. doi: 10.1073/pnas.1000066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta E, Lestingi TM, Mick R, Ramirez J, Vokes EE, Ratain MJ. Metabolic fate of irinotecan in humans: correlation of glucuronidation with diarrhea. Cancer Res. 1994;54:3723–3725. [PubMed] [Google Scholar]
- Hassan MI, Waheed A, Grubb JH, Klei HE, Korolev S, Sly WS. High resolution crystal structure of human beta-glucuronidase reveals structural basis of lysosome targeting. PLoS One. 2013;8:e79687. doi: 10.1371/journal.pone.0079687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- Jain S, Drendel WB, Chen ZW, Mathews FS, Sly WS, Grubb JH. Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs. Nat. Struct. Biol. 1996;3:375–381. doi: 10.1038/nsb0496-375. [DOI] [PubMed] [Google Scholar]
- Kelly H, Goldberg RM. Systemic therapy for metastatic colorectal cancer: current options, current evidence. J. Clin. Oncol. 2005;23:4553–4560. doi: 10.1200/JCO.2005.17.749. [DOI] [PubMed] [Google Scholar]
- Kong R, Liu T, Zhu X, Ahmad S, Williams AL, Phan AT, Zhao H, Scott JE, Yeh L-A, Wong STC. Old drug new use—amoxapine and its metabolites as potent bacterial β-glucuronidase inhibitors for alleviating cancer drug toxicity. Clin. Cancer Res. 2014;20:3521–3530. doi: 10.1158/1078-0432.CCR-14-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine L, Connors LG, Reicin A, Hawkey CJ, Burgos-Vargas R, Schnitzer TJ, Yu QF, Bombardier C. Serious lower gastrointestinal clinical events with nonselective NSAID or coxib use. Gastroenterology. 2003;124:288–292. doi: 10.1053/gast.2003.50054. [DOI] [PubMed] [Google Scholar]
- Lanas A. A review of the gastrointestinal safety data—a gastroenterologist's perspective. Rheumatology. 2010;49:ii3–ii10. doi: 10.1093/rheumatology/keq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanas A, Sopena F. Nonsteroidal anti-inflammatory drugs and lower gastrointestinal complications. Gastroenterol. Clin. North Am. 2009;38:333–352. doi: 10.1016/j.gtc.2009.03.007. [DOI] [PubMed] [Google Scholar]
- LoGuidice A, Wallace BD, Bendel L, Redinbo MR, Boelsterli UA. Pharmacologic targeting of bacterial β-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J. Pharmacol. Exp. Ther. 2012;341:447–454. doi: 10.1124/jpet.111.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, McLeod H. Lessons learned from the irinotecan metabolic pathway. Curr. Med. Chem. 2003;10:41–49. doi: 10.2174/0929867033368619. [DOI] [PubMed] [Google Scholar]
- Mani S, Boelsterli UA, Redinbo MR. Interrogating and modulating mammalian-microbial communication for improved health. Annu. Rev. Pharmacol. Toxicol. 2014;55:559–580. doi: 10.1146/annurev-pharmtox-011613-140007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathijssen RHJ, van Alphen RJ, Verweij J, Loos WJ, Nooter K, Stoter G, Sparreboom A. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11). Clin. Cancer Res. 2001;7:2182–2194. [PubMed] [Google Scholar]
- Matsumura I, Ellington AD. In vitro evolution of beta-glucuronidase into a beta-galactosidase proceeds through non-specific intermediates. J. Mol. Biol. 2001;305:331–339. doi: 10.1006/jmbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- Methé BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, Gevers D, Petrosino JF, Abubucker S, Badger JH, et al. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redinbo MR. The Microbiota, chemical symbiosis, and human disease. J. Mol. Biol. 2014;426:3877–3891. doi: 10.1016/j.jmb.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Wallace BD, Venkatesh MK, Mani S, Redinbo MR. Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 2013;84:208–217. doi: 10.1124/mol.113.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg ML, Meropol NJ, Poplin EA, Van Cutsem E, Wadler S. Mortality associated with irinotecan plus bolus fluorouracil/leucovorin: summary findings of an independent panel. J. Clin. Oncol. 2001;19:3801–3807. doi: 10.1200/JCO.2001.19.18.3801. [DOI] [PubMed] [Google Scholar]
- Saitta KS, Zhang C, Lee KK, Fujimoto K, Redinbo MR, Boelsterli UA. Bacterial beta-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen, or diclofenac: mode of action and pharmacokinetics. Xenobiotica. 2014;44:28–35. doi: 10.3109/00498254.2013.811314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Voigt W, Jordan K. Chemotherapy-induced diarrhea: pathophysiology, frequency and guideline-based management. Ther. Adv. Med. Oncol. 2010;2:51–63. doi: 10.1177/1758834009355164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Wallace BD, Redinbo MR. The human microbiome is a source of therapeutic drug targets. Curr. Opin. Chem. Biol. 2013;17:379–384. doi: 10.1016/j.cbpa.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh L-A, Mani S, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl P, Wong L, Kwang-won K, Jefferson RA. A functional screen identifies lateral transfer of beta-glucuronidase (GUS) from bacteria to fungi. Mol. Biol. Evol. 2005;22:308–316. doi: 10.1093/molbev/msi018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.