Abstract
Hypertonicity stimulates Nuclear Factor of Activated T-cells 5 (NFAT5) nuclear localization and transactivating activity. Many transcription factors are known to contain intrinsically disordered regions (IDRs) which become more structured with local environmental changes such as osmolality, temperature and tonicity. The transactivating domain of NFAT5 is predicted to be intrinsically disordered under normal tonicity, and under high NaCl, the activity of this domain is increased. To study the binding of co-regulatory proteins at IDRs a cDNA construct expressing the NFAT5 TAD was created and transformed into E.coli cells. Transformed E.coli cells were mass produced by fermentation and extracted by cell lysis to release the NFAT5 TAD. The NFAT5 TAD was subsequently purified using a His-tag column, cation exchange chromatography as well as hydrophobic interaction chromatography and then characterized by mass spectrometry (MS).
Keywords: Intrinsically disordered proteins, Transcription factors, Hypertonicity, Fermentation
Introduction
Nuclear Factor of Activated T-cells 5 (NFAT5) is an osmoregulatory transcription factor that is bidirectionally regulated by change in tonicity, such as variation in NaCl (2; 14). Hypertonicity activates NFAT5 by increasing its RNA and protein abundance (9; 10), its nuclear localization (9; 10) and its transactivating activity (6). Hypertonic activation of NFAT5 increases the RNA and protein abundance of its target genes including those proteins that synthesize or transport multiple organic osmolytes, stabilize other proteins, or function in water balance (2). The result is protection of cells from the deleterious effects of hypertonicity found, in particular, in the normal functioning of the kidney medulla. NFAT5 also has far reaching roles in immunity (3), carcinoma invasion (4), and atherosclerosis (8) that apparently, are independent of its response to tonicity.
High NaCl increases both the activity and phosphorylation of the transactivation domain (TAD) located between amino acids 872-1271 of NFAT5 (6). As is characteristic of all transcription factors, the TAD of NFAT5 is the region where co-regulatory proteins, co-activators or -repressors, would be expected to bind and as such the TAD is critical to NFAT5 function.
Transcription factors contain intrinsically disordered (ID) regions where binding interactions that control transcription occur. Binding of co-regulatory proteins at ID regions accompanies transition from disorder to order and is critical to initiate and sustain transcriptional activity (5; 7; 12). The TAD region of NFAT5 is predicted to be intrinsically disordered (GlobPlot 2 Version 2.3 (http://globplot.embl.de/) (Algorithm: Lusselle/Linding definition) but, currently, there is no published crystal structure. This prompted our purification of the NFAT5 TAD for structural and binding interaction studies.
A cDNA construct expressing the NFAT5 TAD was created and transformed into E.coli cells. Transformed E.coli cells were mass produced by fermentation and extracted by cell lysis to release the NFAT TAD. The NFAT5 TAD was subsequently purified using a his-tag column, cation exchange chromatography as well as hydrophobic interaction chromatography and then characterized by mass spectrometry (MS).
Materials and Methods
Plasmids, DNA constructs and oligonucleotides
pET-41b was purchased from EMD Millipore and modified to create pET-41b-MBP-AcTEV-872-1271 F1005W AcTEV (pET-MBP-872-1271 and Figure 1). Human NFAT5 cDNA clone KIAA0827 was a gift from Dr. Takahiro Nagase (Kazusa DNA Research Institute, Chiba, Japan). All cDNA inserts used in cloning were generated using GeneArt Gene Synthesis (Life Technologies, Carlsbad, CA).
Figure 1.

Sequence map for pET-MBP-872-1271.
Sequence coding for maltose binding protein (MBP, pMAL-p5E, New England Biolabs) was cloned into pET-41b using the NdeI and SpeI restriction sites. MBP insertion replaced sequence coding for glutathione-S-transferase (GST) but retained that coding for a 6x His tag. Then, sequence coding for AcTEV protease binding site followed by amino acids 872-1271 of NFAT5 protein (KIAA0827) with an F1005W mutation (TTT to TGG), and a second AcTEV protease binding site was inserted between the EcoRV and XhoI restriction sites. A second His tag (8X) was retained as found in pET-41b between the XhoI and AvrII restriction sites. All constructs were generated by using standard cloning procedures and verified by restriction enzyme digestion and DNA sequencing (GeneWiz, Frederick, MD).
Bacterial expression and Fermentation
pET-MBP-872-1271 was transformed into HMS174 E. coli competent cells (EMD4Biosciences, USA). 20 ng of plasmid was added to 50 µL competent cells in a 1.5 mL micro-tube and incubated on ice for 10 mins. The cell mix was heat-pulsed in a 42 °C water bath for 90 seconds and plated on LB agar with 50 mg/L kanamycin. A single colony was chosen from the plate after overnight incubation at 37 °C, and inoculated into 100 mL modified LB* with 50 mg/L kanamycin in a 500 ml baffled shake flask. Following incubation at 37 °C 220 rpm overnight, the culture was used to inoculate the large fermenter.
A 14 L BioFlo 110 fermenter (New Brunswick Scientific, Edison, NJ) with a working volume of 10 L was used for large-scale fermentation. The fermenter was batched with 10 g/L tryptone, 5 g/L YE, 5 g/L NaCl, 3.5 g/L glucose, 0.5 g/L MgSO4·7H2O, 12 g/L K2HPO4, 50 mg/L kanamycin, pH 7.0. Temperature was controlled at 37 °C, pH was controlled at 7.0 with 30% NH4OH, and dissolved oxygen (DO) was controlled at 30%. BioCommand Plus software from NBS was used for data collection. Cells were induced with 0.5 mM IPTG at 37 °C when OD600 reached above 3.0. Addition of 2.4 g/L of glucose was added at the time of induction and one hour later. 2–3 hour after induction, cells were harvested using a Sharple continuous centrifuge at a flow rate of 250 mL/min. Cell pellets were stored at −80 °C until protein purification.
Cell lysis of pET-MBP-872-1271
Frozen pellets of E. coli cells (10 g) were suspended in 100 mL 25 mM NaPi, pH 7.4, 150 mM NaCl, 1 mM EDTA buffer containing a cocktail of protease inhibitors (Complete, EDTA, Roche, USA) and lysed using a microfluidizer at 17,000 psi (2 cycles) at 4 °C. The cell lysate was centrifuged (19,000 rpm for 35 min) and the supernatant was discarded. The pellets were then re-suspended with 25 mM NaPi, pH 8.0, and 1 mM EDTA buffer (native buffer) and lysed again using a microfluidizer at 17,000 psi. The lysate was centrifuged (19,000 rpm for 35 min) to remove debris and the resulting supernatant was retained for cleavage by protease AcTEV. Each step of lysis was analyzed using SDS-PAGE.
Proteolytic Cleavage of pET-MBP-872-1271
The enzymatic cleavage of pET-MBP-872-1271 was completed with 1 unit of AcTEV per 10 units (29 mg) of pET-MBP-872-1271 in 25 mM NaPi, pH 8.0, 1 mM EDTA buffer, containing a cocktail of protease inhibitors (Complete, EDTA, Roche, USA) and incubated at 4 °C overnight with rocking to give NFAT5 872-1271 (Figure 2).
Figure 2.
Amino acid sequence of NFAT5 872-1271.
Purification of 872-1271
Digested samples were passed through a 0.22 µm filter and mixed with pre-equilibrated Ni Sepharose Excel resin (GE Health Care Life Sciences, Pittsburgh, PA) to remove the free pET-MBP-6xHis-tag and 8xHis-tag, and shaken (180 rpm) for 2 hours at 6 °C. The resin was first washed with 10 column volumes (CVs) of 25 mM NaPi, pH 7.4, 150 mM NaCl, 1 mM EDTA, 10 mM Imidazole buffer (W1), followed by 10 CVs of 25 mM NaPi, pH 7.4, 150 mM NaCl, 1 mM EDTA, 20 mM Imidazole buffer (W2). The His-tag was eluted with 5 CVs of 25 mM NaPi, pH 7.4, 150 mM NaCl, 1 mM EDTA, 250 mM Imidazole buffer. All fractions were analyzed using SDS-PAGE.
Fractions containing 872-1271 (W1) were dialyzed into 25mM NaPi, pH 7.4, 1 mM EDTA, filtered through a 0.22 µm filter and loaded onto a pre-equilibrated chromatography column containing DEAE resin (TOSOH, TSK-gel, 13 µm, 21.5 mm × 15 cm) to remove remaining MBP-His-tag as well as remaining impurities with a flow rate of 2.0 mL/min and a 0–600 mM NaCl gradient over 60 min. The flow through was concentrated (30×) using a 10 kDa centrifugal filter unit (Amicon Ultra-15, Merck Millipore, Billerica, MA). 600 mM NaCl was added to the 30× concentrated protein and the soluble protein was loaded onto a chromatography column containing Phenyl-5PW resin (TSK, 10 µm, 7.5 mm × 7.5 cm) to complete purification with a flow rate of 0.5 mL/min and a 0.6-0 mM NaCl over 60 min and 0.5 mL fractions were collected and analyzed with SDS-PAGE. Fractions containing NFAT5 were buffered (25 mM NaPi, pH 7.4, 1 mM EDTA), concentrated (10×) using a 10 kDa centrifugal filter unit (Amicon Ultra-0.5 Merck Millipore, Billerica, MA) and dialyzed into 25 mM NaPi, pH 7.4, 1 mM EDTA for spectrophotometric measurements of changes in structure using fluorescence.
HPLC-Mass spectrometry for Intact Protein
Proteins were injected into a reverse phase HPLC (Agilent 1100 series HPLC, Agilent Technologies, Santa Clara, CA) with a Vydac C18 (218TP5205, 2.1 × 50mm, 5:M, Grace Vydac) and introduced into the mass spectrometer as described (1; 13) Positive ion Electrospray Ionization (ESI) mass spectra for intact protein were obtained with an Agilent 6224 mass spectrometer equipped with an ESI interface and a time-of-flight (TOF) mass detector (Agilent Technologies, Santa Clara, CA). Mass spectra were analyzed and deconvoluted as described (11), using an Agilent software, MassHunter version B.04.00 (Agilent Technologies, Santa Clara, CA).
Results and Discussion
Expression Hosts and Induction Temperature
There are various E.coli competent cells available for recombinant protein expression. Some contain additional tRNA transferases to recognize codons rarely used in E. coli, such as Rosetta2. Some provide extra mRNA stability, such as One Shot® BL21 Star™. Some contain chaperones to assist protein folding, such as ArcticExpress. In order to select the host that expresses the greatest amount of pET-MBP-872-1271, pET-MBP-872-1271 plasmids were transformed into four hosts, and the expression level of pET-MBP-872-1271 was determined.
Conditions affecting recombinant protein production during fermentation include pH, dissolved oxygen level, and temperature for growth and induction. During cell growth, pH is maintained at neutral pH (~7) and dissolved oxygen is kept above 30%, which ensures E. coli cells are under aerobic conditions. The optimal growth temperature of E.coli is 37 °C however, the optimal induction temperature can vary. Therefore, two induction temperatures, 16 °C and at 37 °C were studied on a small scale to determine optimal conditions to be used during the final fermentation.
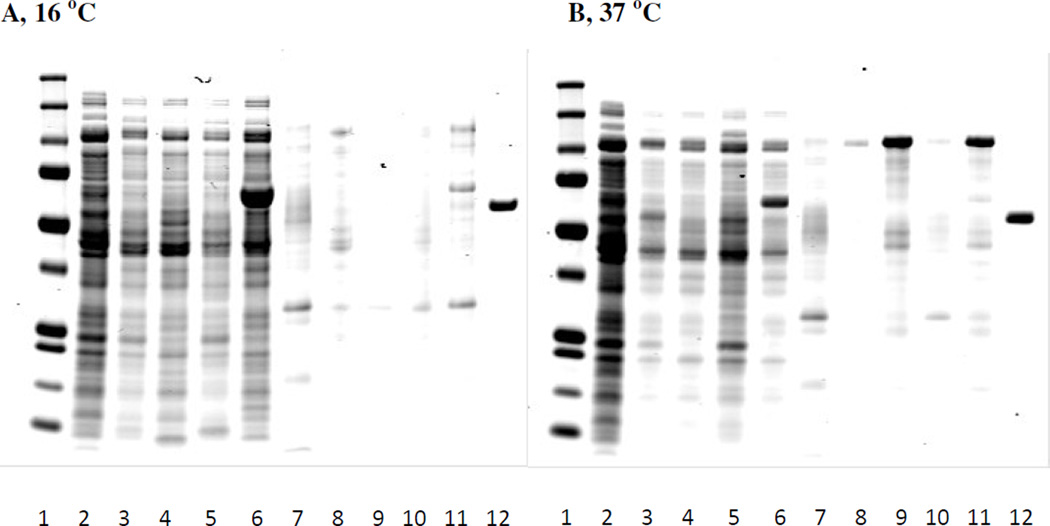
Four E. coli host cells, Rosetta2pLysS, HMS174, One Shot® BL21 Star™ (DE3) pLysS and ArcticExpress were grown at 37 °C until 1.0 OD600. Each host cell culture was induced at either 16 °C overnight or at 37 °C for 3 hours. Cell pellets were collected and re-suspended with HEPES buffer before cell lysis. Figure 3 is a Coomassie stained gel showing the expression of pET-MBP-872-1271 from the cell growths. Induction at 16 °C gave some pET-MBP-872-1271 expressed as insoluble protein in the pellets. However, the soluble (supernatant) pET-MBP-872-1271 is less at 16 °C when compared to induction at 37 °C. Among the four cell types induced at 37 °C, HMS174 produced comparable amount of soluble protein to the other three hosts, but the most insoluble protein. Since this insoluble protein could be re-solubilized, HMS174 was chosen as the host. Though 37 °C was chosen as induction temperature, the length of induction was increased based on analysis of the samples taken from fermenter in a later experiment.
Figure 3.
pET-MBP-872-1271 expression by different E. coli hosts. Panel A: E. coli cells were induced with IPTG at 16 °C for overnight. Panel B: E. coli cells were induced with IPTG at 37 °C for 3 hours. Lane 1: MW marker. Lane 2: before induction supernatant. Lane 3: Rosetta2 pLysS after induction, supernatant. Lane 4: HMS174 after induction, supernatant. Lane 5: One Shot® BL21 Star™ (DE3) pLysS after induction, supernatant. Lane 6: ArcticExpress after induction, supernatant. Lane7: before induction pellets. Lane 8: Rosseta2 pLysS after induction, pellets. Lane 9: HMS174 after induction, pellets. Lane 10: One Shot® BL21 Star™ (DE3) pLysS after induction, pellets. Lane11: ArcticExpress after induction, pellets. Lane 12: BSA std, 0.1 mg/ml.
10 L Fermentation
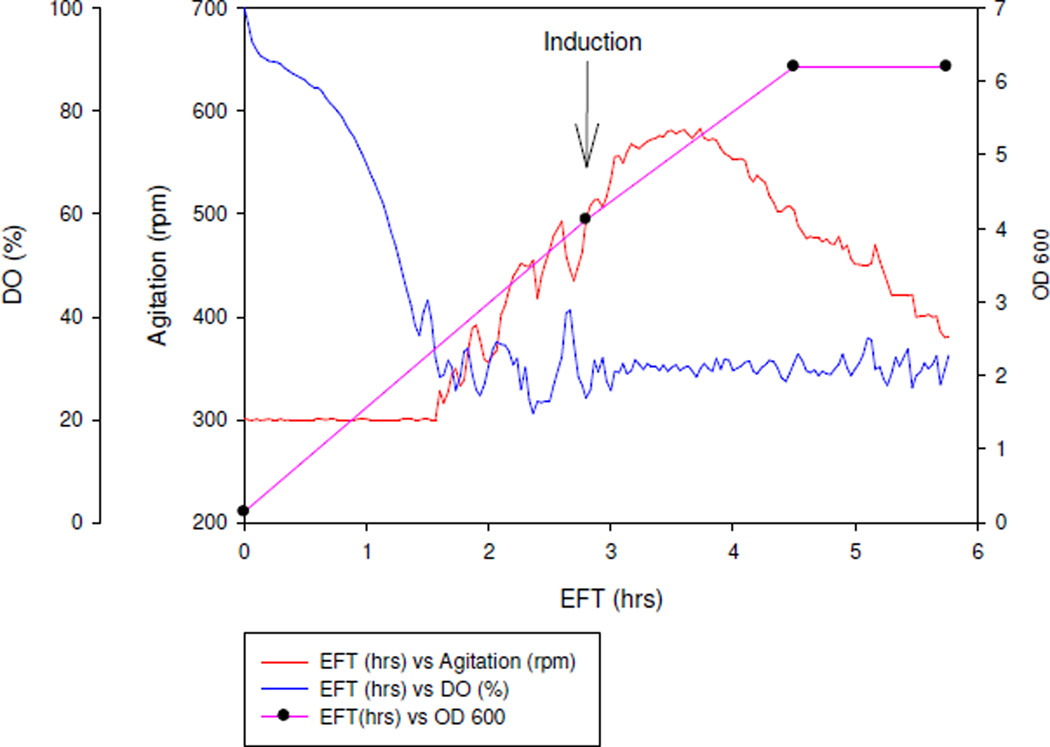
Dissolved oxygen (DO) in the fermenter was controlled at 30% by adjusting agitation using the built-in controller of the fermenter. The higher the oxygen demand, the higher the agitation, since higher agitation increases the fermenter’s ability to deliver more oxygen by breaking down air bubbles to smaller bubbles. As elapsed fermentation time (EFT) increased, agitation increased due to cell growth. Recombinant E.coli cells were induced after reaching 3 OD600. As shown in Figure 4, agitation slowed down 0.5 hrs into IPTG induction, which indicated the burden of recombinant protein synthesis to the E.coli native system. One hour after induction, agitation started to decrease, which continued until the end of the fermentation, three hours after induction. This type of drastic decrease on oxygen demand after induction normally coincides with inclusion body formation and toxicity to the cells. Plateau of the cell density at the end of fermentation also indicated that recombinant E.coli was under stress.
Figure 4.
Online data and OD600 profile from 10 L fermentation.
*EFT: elapsed fermentation time
* DO: dissolved oxygen
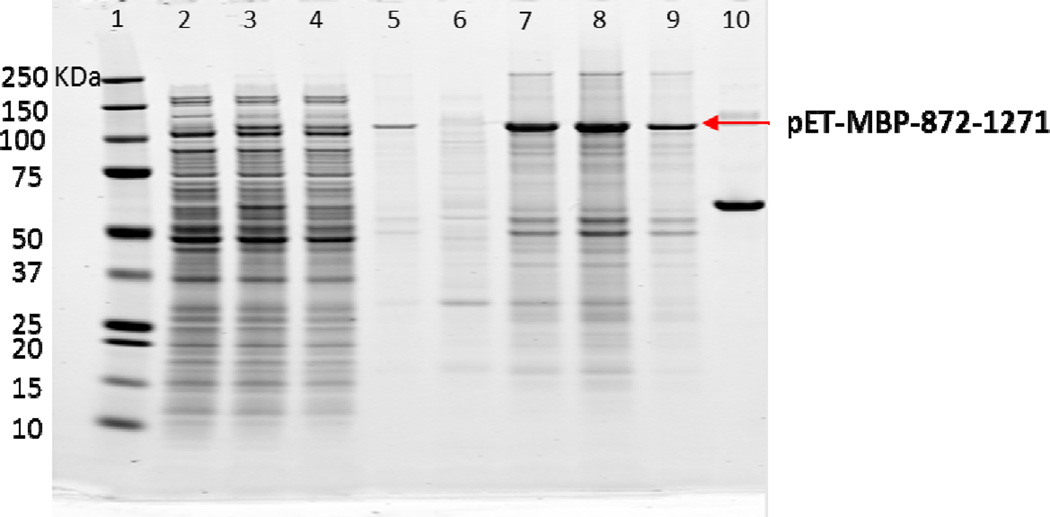
Figure 5 is a Coomassie stained gel of the expression of pET-MBP-872-1271 from various samples. Though the calculated mass of pET-MBP-872-1271 is 95 kDa, there was clearly a band positioned at 110 kDa appearing after induction. Even at 1.7 hours post induction, the majority of the pET-MBP-872-1271 was expressed as inclusion bodies (Lane 7) and less than 20% of pET-MBP-872-1271 expressed was soluble (Lane 3). As time progressed to 3 hours post induction, more pET-MBP-872-1271 was expressed as inclusion bodies (Lane 8) and less as soluble protein (Lane 4). The inclusion bodies from 3 hours post induction were re-suspended with cell lysis buffer and sonicated twice before the sample was centrifuged again. Surprisingly, a portion of the inclusion body sample could be re-solubilized by native buffer and appeared in the supernatant after centrifugation, as shown in lane 5. When a larger volume of buffer was used to re-suspend the pellets, more pET-MBP-872-1271 was re-solubilized. Therefore, the pET-MBP-872-1271 that appeared in the pellets after cell homogenization was not mis-folded protein. pET-MBP-872-1271 was in the pellets was due to low solubility in native buffer. However, this provided a simple clean up strategy to obtain pure pET-MBP-872-1271. Instead of using supernatant after cell homogenization, the pellets were suspended with native buffer and the very clean re-solubilized protein was further purified.
Figure 5.
Coomassie stained gel of fermentation samples. Lane 1: marker. Lane 2: before induction supernatant. Lane 3: 1.7 hour after induction, supernatant. Lane 4: 3 hour after induction, supernatant. Lane 5: supernatant after re-suspending pellets in lane 8. Lane 6: before induction pellets. Lane 7: 1.7 hour after induction, pellets. Lane 8: 3 hour after induction, pellets. Lane 9: pellets after re-suspending pellets in lane 8. Lane 10: BSA standard (0.1 mg/ml)
Purification and confirmation of pET-MBP-872-1271
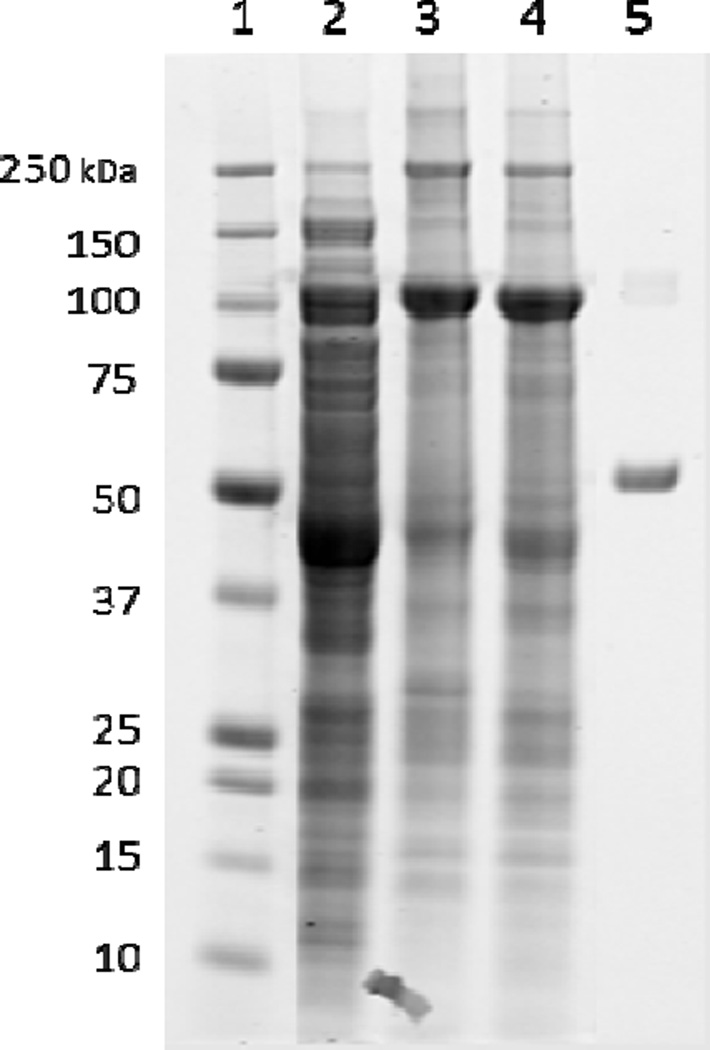
Cell lysis using a microfluidizer followed by centrifugation provided a cell pellet that was washed with Na-Pi buffer to remove the cell debris and other proteins which are not of interest. A subsequent lysis using the microfluidizer and centrifugation yielded the majority of soluble protein (29 mg) in the supernatant at 95 kDa (Figure 6). Previous attempts of cell lysis of constructs not containing MBP proved it to be crucial for protein solubility and stability. Constructs that did not contain MBP did not express, therefore MBP increased the stability of the protein and it did not degrade in the presence of MBP.
Figure 6.
Cell lysate and washes of pET-MBP-872-1271. Lane 1: protein standards (kDa). Lane 2: supernatant after centrifuging cell lysate containing cell debris. Lane 3: pellet after centrifuging cell lysate. Lane 4: soluble protein, pET-MBP-872-1271 at 95 kDa. Lane 5: Bovine serum albumin (BSA) standard (0.1mg/mL).
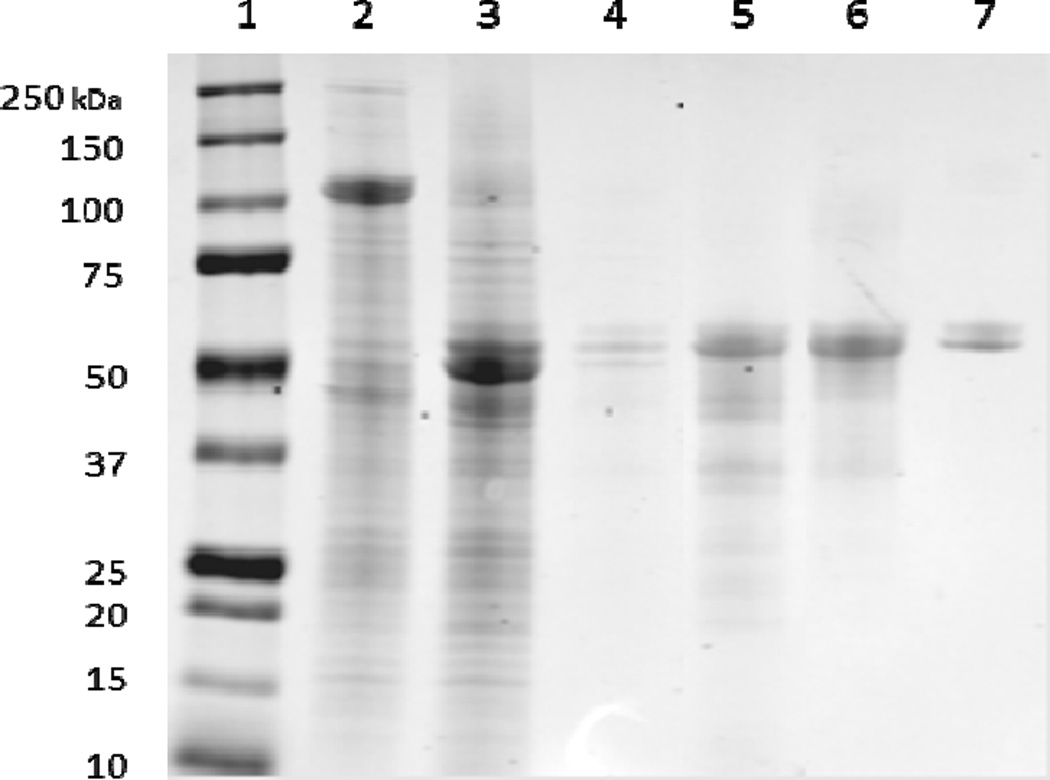
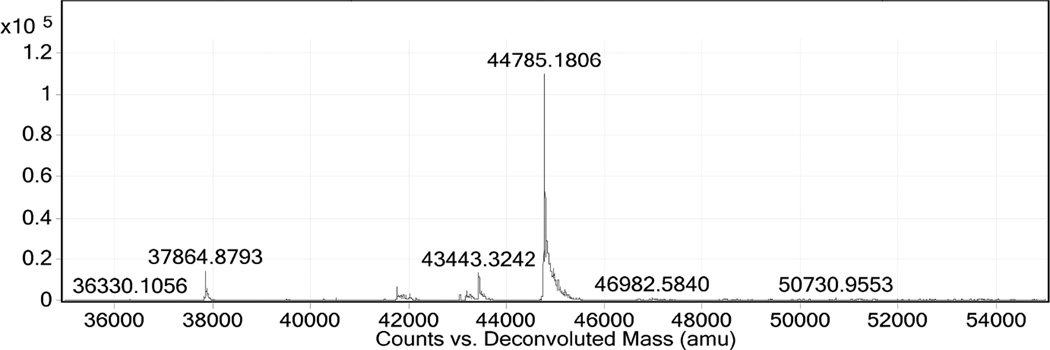
10 units of pET-MBP-872-1271 per 1 unit of AcTEV protease was digested to completion after overnight incubation at 4 °C as shown in Figure 7. Following digestion, the protein was filtered and mixed with Ni-Sepharose excel resin for 2 hours at 6 °C for purification of 872-1271. The protein of interest was expected to be present in the flow through due to the his-tag cleavage from the protease since both his-tags were cleaved from 872-1271 by AcTEV, but instead eluted (9 mg) with 10 mM imidazole due to possible non-specific binding (Figure 7, Lane 4). The protein of interest ran higher on the SDS-PAGE gel than the MBP portion (51 kDa) of the construct, which may be due to a conformational difference between the two cleaved proteins. Identification of the cleaved MBP was verified by MS (data not shown). After MBP cleavage, the band never runs tightly on the gel, potentially due to the disorder of the protein. The predicted pI of 872-1271 is 4.3. While in NaPi buffer with a pH of 7.4, the protein would be expected to have net negative charge and therefore bind to the anion exchange DEAE column. However, the protein eluted on the flow through when loaded onto the DEAE resin (Figure 7, Lane 5). The DEAE column bound the cleaved MBP and therefore increased the purity of the target protein. Since the protein did not display anionic character, a hydrophobic interaction column (HIC) was used to explore separation based on hydrophobicity. Separation of 0.2 mg/mL 872-1271 was achieved with the HIC Phenyl-5w resin with elution of the protein at the end of the NaCl gradient with 0 NaCl mM (Figure 7, Lane 6). The protein could not be concentrated beyond 0.2 mg/mL due to insolubility at higher concentrations. Detergent, Triton-X, was employed to increase solubility, but could not be easily removed via dialysis for further fluorescence studies. Figure 8 shows HPLC-MS confirmation of the protein 872-1271 with a mass of 44785 Da. The calculated mass is 44786.9 Da. The protein contains two cysteine residues and the mass identified is the protein containing a disulfide bond. After incubation with DTT, the mass of the sample was measured at 44787 (data not shown).
Figure 7.
Digestion of pET-MBP-872-1271. Lane 1: protein standards (kDa). Lane 2: pET-MBP-872-1271 at 95 kDa post cell lysis and wash. Lane 3: Protein 872-1271 at approximately 60 kDa post digestion. Lane 4: 872-1271 elution in wash 1 with 10mM imidazole on Ni-Sepharose excel His-tag affinity column. Lane 5: 30× concentrated flow through of 872-1271 from DEAE column. Lane 6: 10× concentrated 0 mM NaCl elution of 872-1271 from HIC column. Lane 7: BSA standard (0.1 mg/mL).
Figure 8.
HPLC-MS identification of 872-1271 at 44785 kDa.
Conclusions
We have demonstrated the construct pET-MBP-872-1271 can be expressed in E.coli and grown through fermentation. Solubility and stability of the protein proved to be crucial in the purification steps. Low solubility in native buffer allowed for simple clean up through multiple washes before proteolytic cleavage. Successful specific cleavage using AcTEV followed by a His-tag affinity column further purified 872-1271. Continued purification with a DEAE column followed by an HIC column was necessary to complete purification of 872-1271, which was confirmed with HPLC-MS. The protein could be concentrated to 0.2 mg/mL which is sufficient to study its intrinsic disorder using fluorescence.
Highlights.
construct pET-MBP-872-1271 can be expressed in E.coli and grown through fermentation
low solubility in native buffer allowed for simple clean up through multiple washes before proteolytic cleavage
specific cleavage using AcTEV followed by a His-tag affinity column further purified 872-1271
continued purification with a DEAE column followed by an HIC column was necessary to complete purification of 872-1271
concentrated to 0.2 mg/mL; sufficient to study its intrinsic disorder using fluorescence
protein mass confirmed with HPLC-MS
Acknowledgments
We would like to thank the by the NIH NHLBI Biochemistry Core for their support of this work acquiring the HPLC-MS data and analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
formula for modified LB: 10 g/L tryptone, 5 g/L YE, 5 g/L NaCl, 3.5 g/L glucose, 0.5 g/L MgSO4·7H2O, 12 g/L K2HPO4, pH 7.0
Contributor Information
Jenna F. DuMond, Email: jenna.dumond@nih.gov.
Yi He, Email: hey4@nhlbi.nih.gov.
Maurice B. Burg, Email: burgm@mail.nih.gov.
Joan D. Ferraris, Email: ferraris@nhlbi.nih.gov.
Reference List
- 1.Apffel A, Fischer S, Goldberg G, Goodley PC, Kuhlmann FE. Enhanced sensitivity for peptide mapping with electrospray liquid chromatography-mass spectrometry in the presence of signal suppression due to trifluoroacetic acid-containing mobile phases. J Chromatogr A. 1995;712:177–190. doi: 10.1016/0021-9673(95)00175-m. [DOI] [PubMed] [Google Scholar]
- 2.Burg MB, Ferraris JD, Dmitrieva NI. Cellular response to hyperosmotic stresses. Physiol Rev. 2007;87:1441–1474. doi: 10.1152/physrev.00056.2006. [DOI] [PubMed] [Google Scholar]
- 3.Buxade M, Lunazzi G, Minguillon J, Iborra S, Berga-Bolanos R, Del VM, Aramburu J, Lopez-Rodriguez C. Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J Exp Med. 2012;209:379–393. doi: 10.1084/jem.20111569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen M, Sinha M, Luxon BA, Bresnick AR, O'Connor KL. Integrin alpha6beta4 controls the expression of genes associated with cell motility, invasion, and metastasis, including S100A4/metastasin. J Biol Chem. 2009;284:1484–1494. doi: 10.1074/jbc.M803997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 6.Ferraris JD, Williams CK, Persaud P, Zhang Z, Chen Y, Burg MB. Activity of the TonEBP/OREBP transactivation domain varies directly with extracellular NaCl concentration. Proc Natl Acad Sci U S A. 2002;99:739–744. doi: 10.1073/pnas.241637298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garza AS, Ahmad N, Kumar R. Role of intrinsically disordered protein regions/domains in transcriptional regulation. Life Sci. 2009;84:189–193. doi: 10.1016/j.lfs.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Halterman JA, Kwon HM, Zargham R, Bortz PD, Wamhoff BR. Nuclear factor of activated T cells 5 regulates vascular smooth muscle cell phenotypic modulation. Arterioscler Thromb Vasc Biol. 2011;31:2287–2296. doi: 10.1161/ATVBAHA.111.232165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko BC, Turck CW, Lee KW, Yang Y, Chung SS. Purification, identification, and characterization of an osmotic response element binding protein. Biochem Biophys Res Commun. 2000;270:52–61. doi: 10.1006/bbrc.2000.2376. [DOI] [PubMed] [Google Scholar]
- 10.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a Rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci U S A. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 12.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 13.Taggart C, Cervantes-Laurean D, Kim G, McElvaney NG, Wehr N, Moss J, Levine RL. Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J Biol Chem. 2000;275:27258–27265. doi: 10.1074/jbc.M004850200. [DOI] [PubMed] [Google Scholar]
- 14.Woo SK, Dahl SC, Handler JS, Kwon HM. Bidirectional regulation of tonicity-responsive enhancer binding protein in response to changes in tonicity. Am J Physiol Renal Physiol. 2000;278:F1006–F1012. doi: 10.1152/ajprenal.2000.278.6.F1006. [DOI] [PubMed] [Google Scholar]