Abstract
The DNA alkylating agent busulfan is used to ‘precondition’ patients with leukemia, lymphomas and other hematological disorders prior to hematopoietic stem cell transplants. Busulfan is metabolized via conjugation with glutathione (GSH) followed by intramolecular rearrangement to the GSH analog γ–glutamyl-dehydroalanyl-glycine (EdAG). EdAG contains the electrophilic dehydroalanine, which is expected to react with protein nucleophiles, particularly proteins with GSH binding sites such as glutaredoxins (Grx’s). Incubation of EdAG with human Grx-1 or Grx-2 results in facile adduction of cys-23 and cys-77, respectively, as determined by ESI-MS/MS. The resulting modified proteins are catalytically inactive. In contrast, the glutathione transferase A1-1 includes a GSH binding site with a potentially reactive tyrosinate (Tyr-9) but it does not react with EdAG. Similarly, Cys-112 of GSTA1-1, which lies outside the active site and is known to form disulfides with GSH, does not react with EdAG. The results provide the first demonstration of the reactivity of any busulfan metabolites with intact proteins, and they suggest that GSH-binding sites containing thiolates are most susceptible. The adduction of Grx’s by EdAG suggests the possible alteration of proteins that are normally regulated via Grx-dependent reversible glutathionylation or deglutathionylation. Dysregulation of Grx-dependent processes could contribute to cellular toxicity of busulfan.
Keywords: busulfan, glutathionylation, drug metabolism, redox regulation, mass spectrometry
The myleoablative agent busulfan is the ‘drug of choice’ to precondition patients with leukemia, lymphomas, or nonmalignant blood diseases prior to bone marrow transplant, or hematopoietic stem cell transplant (HSTC). Although busulfan has clear advantages over total body irradiation for preconditioning, and it has become a standard component of HSTC, it has a narrow therapeutic index that makes it difficult to dose optimally (1–3). Toxicity associated with busulfan treatment includes pulmonary injury, seizures, cataracts and hepatic sinusoidal obstruction syndrome (SOS), which is frequently fatal in patients in whom it develops (4–11). Many very recent studies have aimed to improve busulfan safety by varying dosing regimens or to establish predictive pharmacokinetic biomarkers, and these efforts emphasize the urgent need for more complete understanding of busulfan system pharmacology (12,13). The recent high level of interest in pharmacokinetic strategies for busulfan dosing provides a striking contrast to the paucity of information concerning the molecular mechanisms for its toxicity. Increased understanding of the mechanisms of busulfan toxicity could lead to improved safety of HSTC protocols.
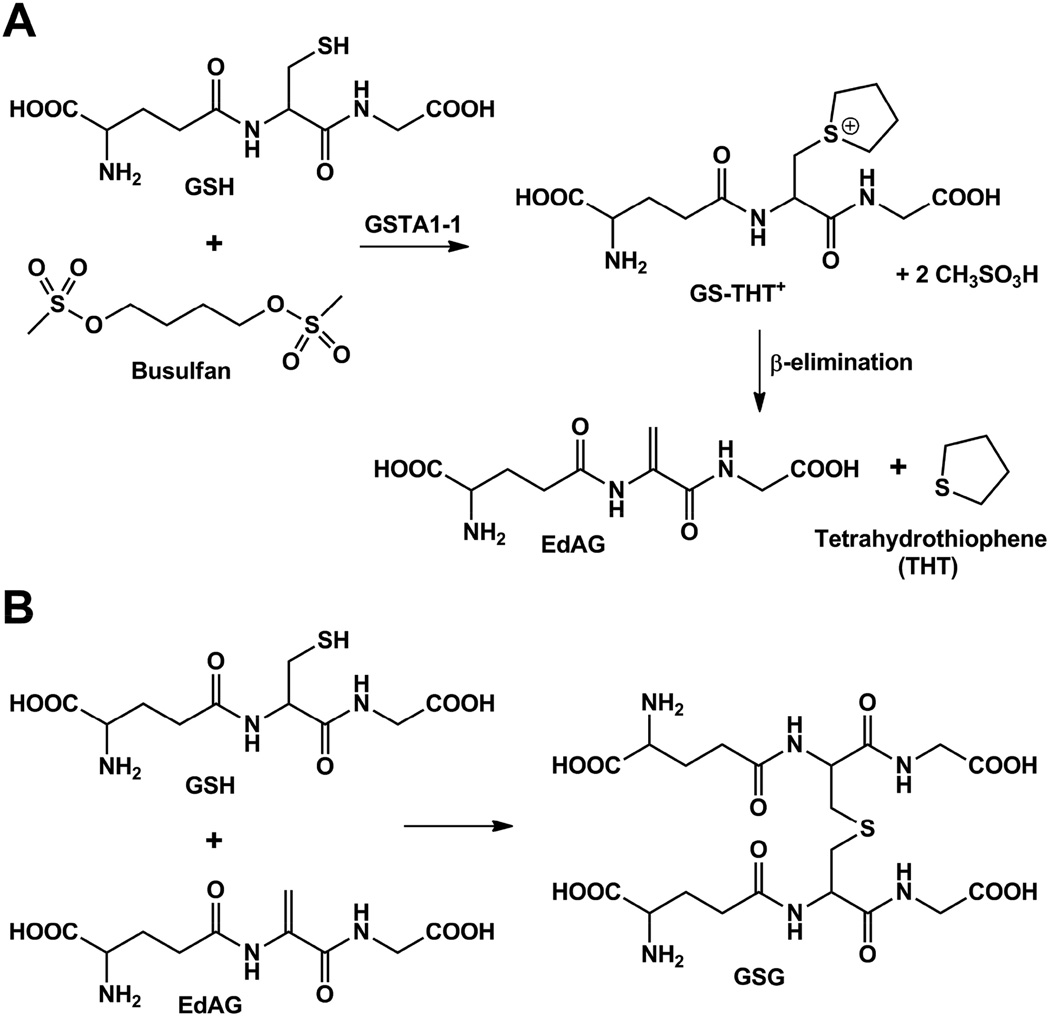
Metabolites of busulfan have been considered as potential sources of toxicity, and the enzymes responsible for the metabolism have been considered as genetic factors in toxicity, but no definitive mechanisms have been demonstrated (14–16). The metabolite pool of busulfan is derived from its initial Glutathione Transferase (GST)-catalyzed reaction with GSH and includes tetrahydrothiophene (THT+) and oxidized THT+ variants (17,18). In addition, the dehydroalanine analog of GSH, or EdAG (γ-glutamyl-dehydroalanyl-glycine), is formed, as shown in Figure 1A. EdAG demands consideration as a source of toxicity due to its chemical reactivity. EdAG is known to react with additional GSH to form the nonreducible lanthionine GSG (Figure 1B), and it has been speculated to react with protein thiols to yield lanthionine linkages in irreversibly glutathionylated proteins (19,20). This reaction has not been experimentally demonstrated, however.
Figure 1.
Glutathione-dependent metabolites of busulfan. A) The GSH adduct of busulfan rearranges to the GS-THT+, which subsequently undergoes β-elimination to EdAG. B) EdAG undergoes additional reaction with GSH to form GSG.
The possibility of irreversible glutathionylation has important implications for the mechanism of busulfan toxicity, because it could lead to dysregulation of many proteins that are normally regulated by reversible glutathionylation. Proteins that contribute to many different cellular functions are regulated via reversible glutathionylation in response to the redox status of the cell, including transcriptional activators, cytoskeletal proteins, mitochondrial electron transfer components, signaling proteins, and enzymes involved in intermediary metabolism (21–23). Normally, such regulation relies on formation and reduction of mixed disulfides of GSH with protein cys residues in response to redox status. Therefore, EdAG could globally impair cellular homeostasis if it forms irreversible lanthionine linkages with proteins, or if it formed such a linkage with enzymes that control reversible glutathionylation.
Hypothetically, proteins that contain reactive cys residues within GSH binding sites would be particularly susceptible to EdAG covalent binding. Because EdAG is a direct structural analog of GSH, it would likely have affinity for GSH-binding sites including those in glutaredoxins (Grx’s), Thioredoxins (Trx’s) or other GSH-binding proteins. Grx motifs include a conserved GSH binding site, or Grx fold, that occurs in many proteins (24–27). In many cases Grx domains include a nucleophilic cys that forms disulfide bonds with GSH during enzymatic catalysis (26). Glutaredoxin-1 (Grx-1), for example, utilizes cys-23 to form a mixed disulfide with GSH (Grx-S-SG) to catalyze the glutathionylation of proteins at their free thiols, or it can reduce protein-S-SG disulfides to result in their deglutathionylation with intermediate formation of Grx-S-SG. Grx-2 utilizes cys-77 for a similar, but mechanistically distinct, reaction. Thus, Grx-1 and Grx-2 are critically important enzymes in the regulation of the ‘glutathiome’, or the set of proteins normally regulated via reversible glutathionylation.
Here, we demonstrate that exposure of EdAG to cytosolic Grx-1 or mitochondrial Grx-2 leads to facile formation of a lanthionine linkage with their active site cys residues. Formation of covalent linkage leads to the expected inhibition of function. This combination of effects has significant implications for the mechanism of busulfan toxicity. Most importantly, the results establish the chemical competence of EdAG for reacting with cys residues in folded proteins.
Materials and Methods
Chemicals
All chemicals were purchase from Sigma-Aldrich unless otherwise stated.
Synthesis of γ-Glutamyl-dehydroalanyl-glycine (EdAG)
EdAG was synthesized via elimination of 2,4-dinitrothiophenolate from S-(2,4-dinitrophenyl)glutathione as previously described by Asquith et al. (28). Specifically, the intermediate S-(2,4-Dinitrophenyl)glutathione was synthesized as described by Younis (29). Briefly, a solution of 1-chloro-2,4-dinitrobenzene (10 mmol in 20 ml of methanol) was added drop wise to a solution of reduced glutathione-(10 mmol in 40 ml of 1 N NaHCO3) and the mixture was then stirred at room temperature. After one hour the reaction was complete, as determined by 1H NMR. The reaction mixture was then filtered, and the product precipitated by acidification with 1 M HCl. The yellow precipitate was collected by vacuum filtration, recrystallized from hot water, desiccated at 60 °C for 48 hours and characterized by 1H NMR [DMSO-d6: δ 1.79 (m, 1H), 1.94 (m, 1H), 2.24–2.37 (m, 2H), 3.27 (m, 1H), 3.34 (m, 1H), 3.64 (dd, 1H), 3.67–3.79 (m, 2H), 4.60 (td, J = 9.3, 4.1 Hz, 1H), 7.97 (d, J = 9.1 Hz, 1H), 8.47 (dd, J = 8.9, 2.5 Hz, 1H), 8.68 (d, J = 8.7 Hz, 1H), 8.87 (d, J = 2.5 Hz, 1H), and 8.98 (t, J = 5.8 Hz, 1H)] and ESI-MS (positive ion mode, [MH]+ = 474.1 m/z). The yield of S-(2,4-dinitrophenyl)glutathione was 75%.
γ-Glutamyl-dehydroalanyl-glycine (EdAG) was synthesized from this product. Two mmol of S-(2,4-dinitrophenyl)glutathione were dissolved in 50 ml of 0.5 M NaOH, and the solution was stirred at room temperature. After 45 min the reaction was complete, as determined by 1H NMR. The solution was then extracted with n-butanol (6 × 20 ml) and the organic layers (containing the 2,4-dinitrothiophenolate) were discarded. To remove sodium ions, the orange aqueous layer (pH ~ 12) was titrated with Dowex 50W-X8 cation exchange resin (hydrogen form, Biorad, 20–50 mesh, pre-equilibrated with n–butanol saturated water) until the pH approached a value of ~ 4.0. The resulting pale yellow aqueous solution was filtered and concentrated to a small volume (~ 0.5 ml) by a rotary vacuum evaporator. The EdAG was then precipitated by addition of 100 ml of cold (-20 °C) ethanol (200 proof, Decon Labs., Inc.). Upon centrifugation (4100 rpm for 5 min at 4 °C), a light brown solid was recovered and subsequently dried at room temperature and under atmospheric pressure. The product was then re-dissolved in water containing 0.1% Formic acid (F.A.) and purified by C18 RP-HPLC (Syncronis aQ, 250 × 10 mm, 5 μm particle size, Thermo Scientific). A water + 0.1% F. A. mobile phase was employed and the flow rate was set to 3 ml/min. EdAG eluted at 9.39 min. The fractions containing EdAG were pooled and lyophilized, yielding a white solid with purity > 98-% as determined by 1H NMR. The product was fully characterized by 1H, 13C and 1H-13C NMR (30). 1H NMR (D2O, pH ~ 3): δ 2.17 (q, J = 7.3 Hz, 2H), 2.58 (td, J = 7.3, 4.5 Hz, 2H), 3.82 (t, J = 6.4 Hz, 1H), 4.00 (s, 2H), 5.68 ppm (s, 1H), 5.74 ppm (s, 1H). ESI-MS (positive ion mode, [MH]+ = 274.1 m/z). The yield of EdAG from starting S-(2,4-dinitrophenyl)glutathione was 60%. A scheme depicting the overall two step synthesis is shown in the accompanying reference 30 along with detailed methods and spectral data.
Recombinant human Grx-1 and Grx-2(41-164) expression and purification
The plasmids for expression in E.coli of human Grx-1 and Grx-2(41-164) [this engineered truncation is referred as Grx-2 for the remainder of the manuscript] were kindly donated by Prof. Caryn E. Outten (Department of Chemistry and Biochemistry, University of South Carolina, U.S.A.).
Expression and purification of Grx-1 and Grx-2 were accomplished as previously described (31, 32), although tris(2-carboxyehtyl)phosphine (TCEP) was used instead of DTT. Both proteins were obtained at a purity > 90% as determined by SDS-PAGE. In all instances, protein concentrations were determined by UV-Vis absorbance using molar extinction coefficients (ε280 nm) of 3160 and 6400 M−1cm−1 for Grx-1 and Grx-2(41-164), respectively. MS characterization indicates that Grx-1 expressed in E. coli is obtained without the N-terminal Met.
Determination of glutaredoxin activity (HED assay)
The catalytic activities of purified hGrx-1 and hGrx-2 were determined using a standard coupled enzyme assay and monitoring reduction of the model substrate 2-hydroxyethyl disulfide (HED), as previously described (33). Briefly, 0.7 mM HED was added to a mixture containing 100 mM Tris•HCl, pH 7.9, 1 mM GSH, 0.2 mM NADPH, 2 mM EDTA, 0.1 mg/ml bovine serum albumin and 6 μg/ml (~ 3 units/ml) baker’s yeast glutathione reductase (Sigma-Aldrich, 500 units/mg protein). After 3 min incubation glutaredoxin was added to the sample cuvette (Grx-1 at 10 nM or Grx-2 at 100 nM), and an equal amount of buffer to the reference cuvette. All the aforementioned concentrations are intended as the final values in the cuvette. The decrease in absorbance at 340 nm (due to the NAPDH oxidation) was followed using a Varian Cary 3E UV-Vis and the data were fitted to a linear regression using GraphPad Prism version 4.00 (GraphPad Software, San Diego, CA, USA). The activity was expressed in units/mg protein (i.e. micromoles of NADPH oxidized/min/mg protein) using a molar extinction coefficient of 6220 M−1cm−1 and was determined after subtracting the spontaneous reduction rate observed in the absence of glutaredoxin. The specific activities were 220 and 25 units/mg for Grx-1 and Grx-2, respectively.
Incubation of glutaredoxins with EdAG
All incubations were performed in phosphate-buffered saline (PBS) at pH 7.4, containing 250 μM TCEP, under agitation at 120 rpm and at a temperature of 37 °C, unless otherwise stated. The initial EdAG/protein molar ratio was invariably set to 20 (i.e. 50 μM Glutaredoxin and 1 mM EdAG).
For the reaction time course experiments, aliquots were taken at variable time points and immediately stored at −80 °C. Each aliquot was then thawed immediately before mass spectrometry analysis.
Reaction aliquots subjected to glutaredoxin activity determination were not, in any case, frozen before analysis. In these instances, a small portion of the time point aliquot was taken and immediately tested for activity.
Glutaredoxins tryptic digestion
hGrx-1, hGrx-2(41-164), and their EdAG-ylated counterparts were proteolyzed with trypsin for mass spectrometry analysis. Briefly, aliquots containing ~ 5 μg protein at ~ 500 μg/ml were denatured at room temperature by adding urea at a final concentration of 8 M. After addition of Tris•HCl, pH 8.0, to a final concentration of 50 mM, dithiothreitol (DTT) was added to a final concentration of ~ 10 mM and the mixture was left for 1 hour at room temperature. Subsequently, iodoacetamide (IAM) was added to a final concentration of ~ 20 mM and the mixture kept in the dark and at room temperature for one hour, after which fresh DTT was added to a final concentration of ~ 10 mM to quench the unreacted IAM. The mixture was then diluted 80 fold with 50 mM ammonium bicarbonate, pH 8.0, 1 mM CaCl2, and added with sequencing grade trypsin to a 1:20 trypsin/protein mass ratio and incubated overnight at 37 °C. Acetonitrile was then added to a final concentration of 5% and the pH decreased to ~ 2.5 by addition of formic acid 88%.
Mass Spectrometry
All mass analyses were performed on a SYNAPT G2-Si quadrupole time of flight spectrometer (Waters, Milford, MA). To ensure high mass accuracy throughout an analysis, a lock mass (leucine enkephalin, [M+H]+ = 556.2771 Da) was sampled every 60 seconds during the run.
For the intact protein analysis, ~ 5 μg were applied to a POROS-R1 column (150 × 2.1 mm, 10 μm particle size, Applied Biosystem) and subjected to a binary mobile phase linear gradient (A = 0.1% F.A.; B = ACN + 0.1% F.A.) from 10% to 95% B over the course of 17 min, at a flow rate of 0.3 ml/min. The MS spectra acquisition was done in positive mode, scanning through a m/z range of 200–3000 Da.
In all instances where it was necessary to estimate the EdAG-protein/protein molar ratio in a protein/EdAG reaction mixture, smoothed peaks of identical charge states were integrated and the fractional area (i.e. A+x/(A+x + B+x), where A+x and B+x are intended as the computed integrals of the EdAG-ylated protein and the unmodified protein, respectively) calculated. This approach is based on the assumption/approximation that both the native and the EdAG-ylated proteins shared equivalent ionization efficiency.
For the tryptic digestion LC-MS/MS analysis, ~ 350 ng digested protein were resolved on an UPLC BEH C18 column (100 × 1.0 mm, 1.7 μm particle size, Waters) and subjected to a linear gradient (A = 0.1% F.A.; B = ACN + 0.1% F.A.) from 5% to 50% B over the course of 24 min, at a flow rate of 0.08 ml/min.
The MS spectra were acquired with data dependent acquisition (DDA), with a survey scan of 1 second through a m/z range of 50–2000 Da, and a subsequent MS/MS scan from 50 to 1200 Da for 1 second with the trap collision energy of 30 eV. The mass error was found, in all instances, to be under 5 ppm.
All data acquisition, processing and visualization were performed using MassLynx (Waters). Peptides were manually assigned using exact mass and MS/MS spectra with the aid of protein prospector (prospector.ucsf.edu).
Results
Characterization of EdAG
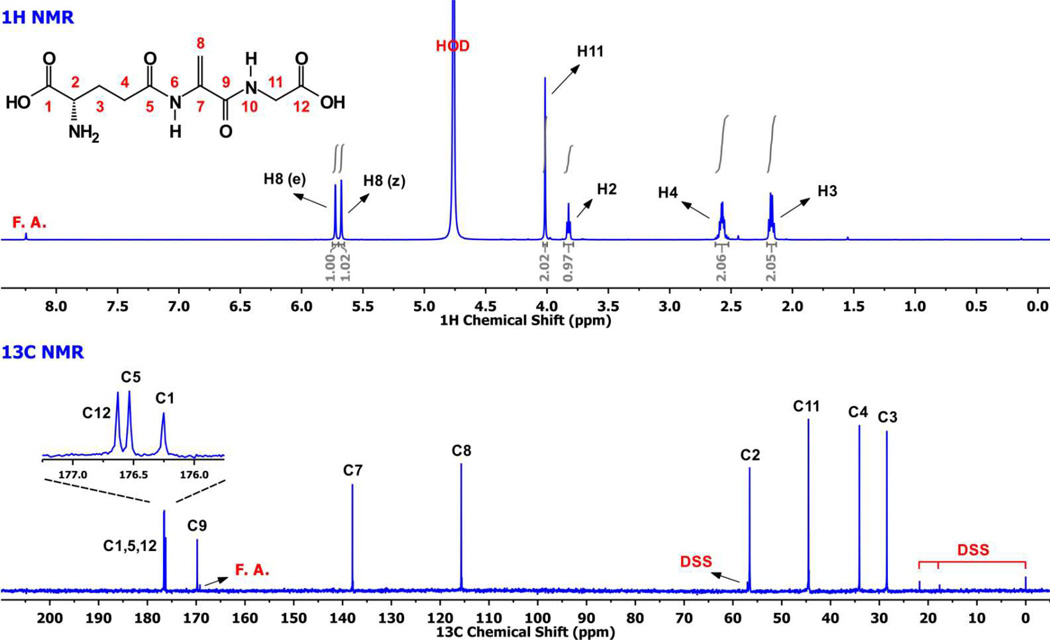
Although the synthesis of EdAG has been reported (17, 29), no complete characterization is published. Therefore, we performed a full NMR characterization as a measure of purity and as a confirmation of structure. The simple 1-dimensional 1H and 13C NMR spectra are shown in Figure 2, which indicates that the purity of our final product is at least 99%. The full characterization via DQF-COSY, TOCSY, NOESY, 1H-13C HSQC and 1H-13C HMBC is provided elsewhere (30).
Figure 2.
1H-NMR spectrum of ~ 5 mM EdAG in unbuffered D2O, pH ~ 3 and 13C-NMR spectrum of ~ 40 mM EdAG in unbuffered H2O/D2O 90:10 at pH ~ 3. The spectra provided a criterion for purity, which was estimated at >98%. ‘FA’ is formic acid and ’DSS’ is 2,2-dimethyl-2-silapentane-5-sulfonate.
Grx’s form covalent adducts with EdAG
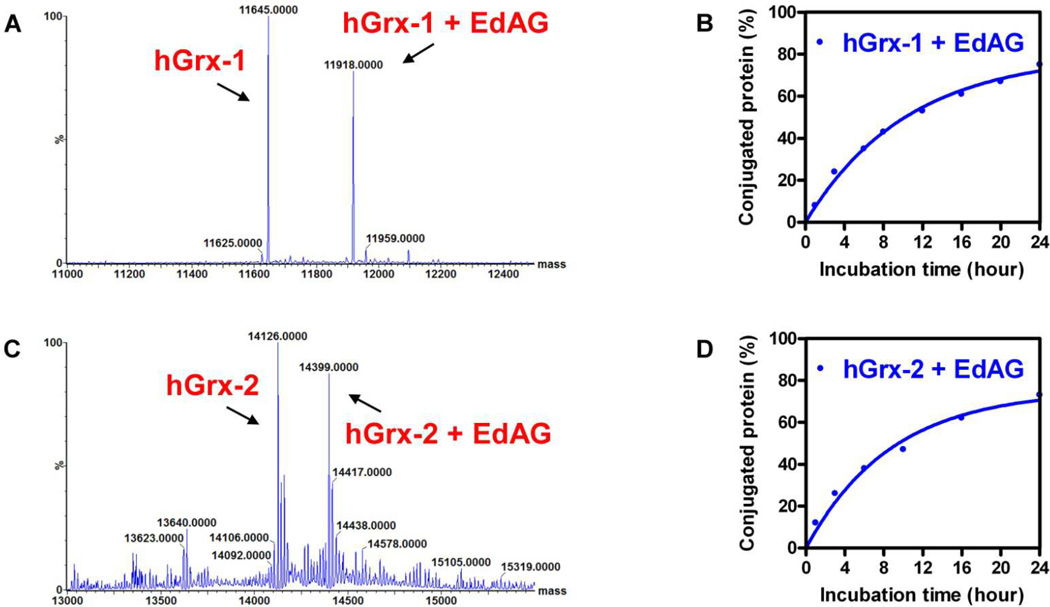
In order to test the hypothesis that EdAG efficiently reacts with Grx’s, it was incubated separately with purified human Grx-1 or Grx-2, which represent cytosolic and mitochondrial isoforms respectively. Both isoforms play critical roles in cellular redox regulation and control reversible glutathionylation of a wide range of proteins (24,27). The incubations included 50 μM protein with a 20-fold excess of EdAG, at physiologic pH at 37 °C, where the majority of protein thiols are protonated, and not highly reactive. The reaction was monitored with ESI-MS at varying times after mixing. Both the intact, unmodified protein and the expected adduct were monitored. Figure 3 shows the time course of the appearance of the adduct expressed as percent of the total protein for Grx-1 and Grx-2. Both proteins show a similar time course under these conditions wherein 50% of the protein is adducted in ~ 10 hrs.
Figure 3.
A, C) Deconvoluted ESI-MS spectra of the reaction mixture obtained from 1 mM EdAG and 50 μM Grx-1 or Grx-2 at 8 and 10 hrs, respectively. B, D) Time course of the EdAG adduct formation with Grx-1 and Grx-2, respectively. The reactions were conducted at 37 °C in PBS, pH 7.4, and in presence of 250 μM TCEP. The percentage of adducted protein was calculated from the fractional area of the most intense charge state (i.e. [M+11H]+11 for Grx-1/EdAG-Grx-1 and [M+16H]+16 for Grx-2/EdAG-Grx-2) in the MS spectrum.
Identification of the site of modification
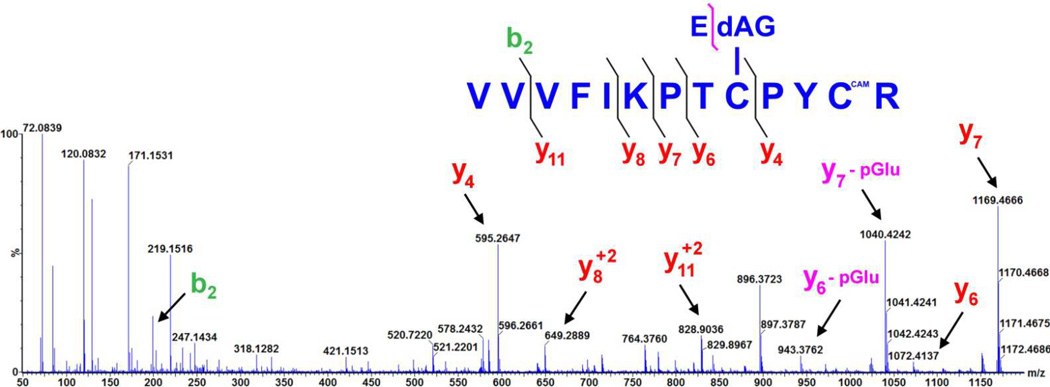
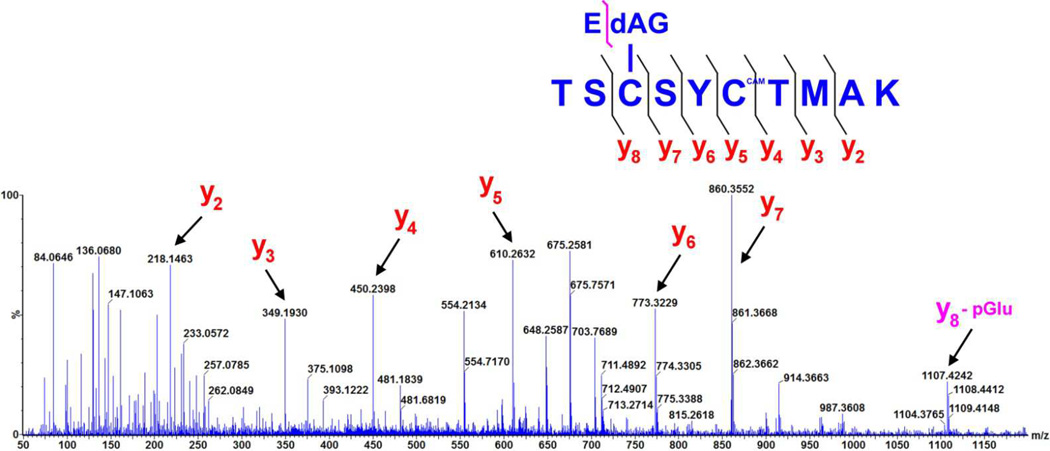
In order to determine the site of modification, ESI-MS/MS was performed with trypsin digested samples of either Grx-1 or Grx-2 incubated with EdAG. The results for Grx-1 and Grx-2 are shown in Figure 4 and Figure 5 respectively. Fragmentation of the peptide corresponding to VVVFIKPTCPYCR clearly reveals EdAG adduction at cys-23 (bold underlined) for Grx-1. In addition to the ions resulting from fragmentation at the peptide backbone, a neutral loss of the pyroglutamic acid (loss of 129 m/z) is observed for many fragments, consistent with the γ-glutamyl linkage on EdAG, which is a well-established fragmentation for GSH adducts (34, 35). The ions corresponding to loss of pyroglutamic acid from the EdAG-adducted peptide are labeled with ‘yx - pGlu’ in Figure 4. Similar results are observed for Grx-2 incubated with EdAG. Here cys-77 is clearly adducted in the peptide TSCSYCTMAK, where the adducted cys-77 is bold and underlined.
Figure 4.
High mass accuracy MS/MS spectrum of the Grx-1 tryptic peptide with EdAG adducted to cys23, derived from a precursor ion with mass 618.98 Da (i.e. [M+3H]3+). Selected fragment ions, including those corresponding to the neutral loss of 129 m/z which is diagnostic for the loss of pyroglutamic acid, are highlighted. The y4 and y7 ions identify cys23 as the site of EdAG adduction. The symbol ‘CAM’ in the peptide sequence indicates that the specific cys residue was the carbamidomethyl derivative, resulting from reaction with iodoacetamide.
Figure 5.
High mass accuracy MS/MS spectrum of the Grx-2 tryptic peptide with EdAG adducted to cys77, derived from a precursor ion with mass 712.78 Da (i.e [M+2H]2+). Selected fragment ions, including one corresponding to the neutral loss of 129 m/z which is diagnostic for the loss of pyroglutamic acid, are highlighted. The y5–7 and y8 ions unequivocally identify in cys77 the site of EdAG-ylation.
Each of the other cys-containing peptides showed no adduction as determined by MS. Specifically, the peptides containing cys-8, cys-79 and cys-83 of Grx-1 and cys-68 and cys-153 of Grx-2 were found to be unmodified, and the data are shown elsewhere (30). Interestingly, the two non-active site cys residues in Grx-2, as well as the two active site cys residues were found to slowly form disulfides during the incubation time in the presence or absence of EdAG (30), by the progressive mass shift of the arbitrarily selected [M+11H]11+ charge species during the incubation. These species were easily reduced by TCEP, but no cysteines other than Cys-23 of Grx-1 or Cys-77 of Grx-2 were ever adducted by EdAG. However, this disulfide heterogeneity made it difficult to quantify catalytic activity of the individual species present, as described below.
Catalytic activity of EdAG-adducted Grx
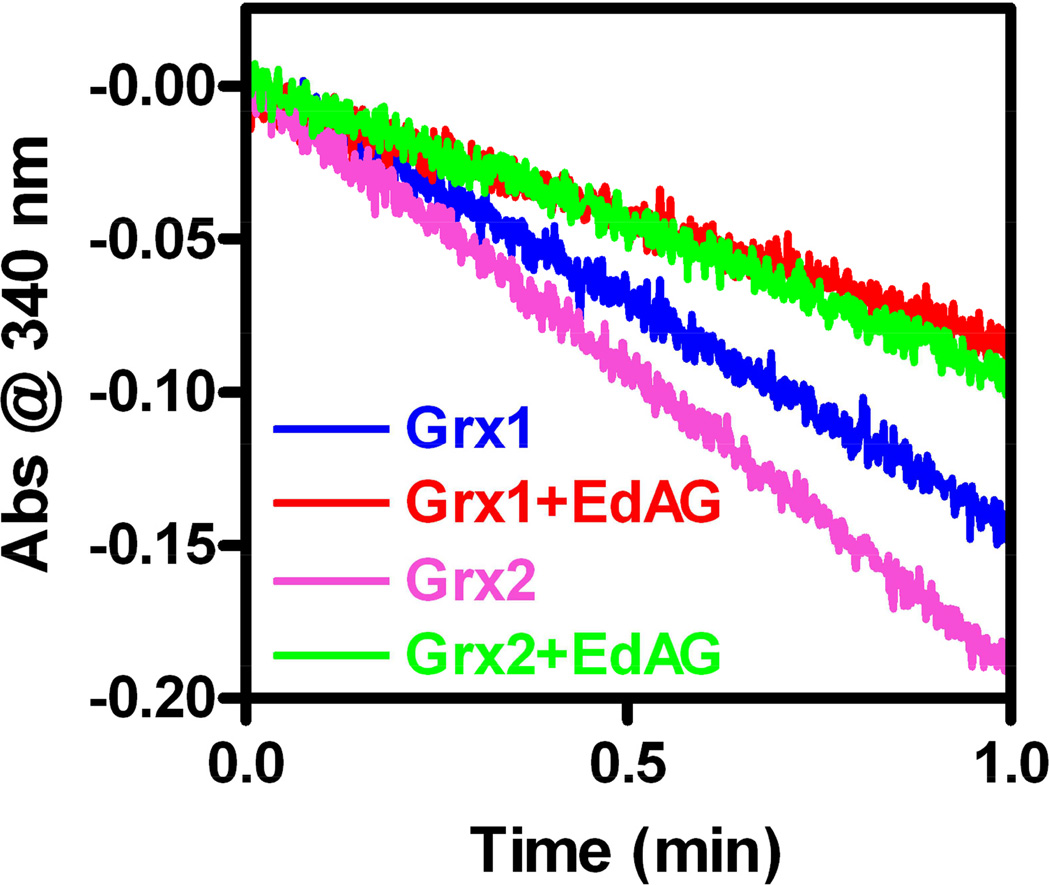
The formation of a lanthionine linkage at the catalytic cys residues would be expected to inactivate Grx-1 and Grx-2. To demonstrate this, the standard hyroxyethyl disulfide (HED) assay was used to compare the catalytic activities with native Grx’s. In this assay the HED disulfide is reduced and a mixed disulfide is formed between hydroxyl ethyl thiol and GSH. This disulfide is reduced by Grx to yield a mixed disulfide between the active site cys and GSH, which in turn is reduced by GSH to yield GSSG. The GSSG is reduced by Grx utilizing NADPH as a source of reducing equivalents. The GSH-dependent oxidation of NADPH was monitored spectrophotometrically (33). For both Grx-1 and Grx-2 some catalytic activity was lost during the time of incubation with EdAG, even in the absence of EdAG. This was particularly evident with the Grx-2 construct, which is a deletion of the wild type. Also, as noted above, during the incubation we observed formation of disulfides between non-active site cysteines, and separately between active site cysteines, in the intact Grx-2 in the absence and presence of EdAG, resulting in a heterogeneous mixture of reduced, partially oxidized, and fully oxidized species (shown in ref. 30). We presume that these disulfides were formed during the incubation, rather than being present before the incubation, because the sample included 250 μM TCEP at the start. With the accumulation of these disulfide-bonded forms it was not possible to determine the catalytic activity of the differentially oxidized Grx species. It was also difficult to determine whether any disulfide bonding between nonactive site cys residues had an impact on the EdAG adduction of the active site cys-77 as we could not resolve the reduced, partially oxidized or fully oxidized protein, so the percent of protein adducted with EdAG was based on total protein, and not fully reduced protein. In addition, due to uncertainty about the ionization efficiency of unmodified vs. EdAG-modified proteins in the mass spectrometer, quantitation of the amount of protein adducted is only semi-quantitative. As a result of these complexities, it was difficult to quantitatively correlate the loss of catalytic activity with EdAG adduction. This made it very difficult to determine KI and kinact parameters for this active site directed inhibitor. Qualitatively, however, EdAG adduction clearly correlated with loss of activity. For partially adducted samples of Grx-1, the enzyme that was estimated to be 32% adducted at cys-23, based on mass spectrometry, lost 47% of the catalytic activity of a control sample that was not treated with EdAG. An essentially identical result was obtained with Grx-2. Thus both Grx-1 and Grx-2 are nearly equally susceptible to EdAG adduction and the resulting loss of catalytic activity. The greater loss of activity compared to extend of adduction may reflect multiple pathways of inactivation, such as the disulfide bond formation described above and some time-dependent unfolding. Data demonstrating the loss of activity for partially adducted Grx-1 and Grx-2 are shown in Figure 6.
Figure 6.
Catalytic activity of Grx’s after reaction with EdAG. Samples of 50 μM Grx’s in PBS pH 7.4 were incubated at room temperature (Grx-1) or at 4 °C (Grx-2) in presence or in the absence of 1 mM EdAG. After 24 hours, HED activity assays were conducted as described in the material and methods section. With partial (~35%) adduction by EdAG, Grx catalytic activity is decreased ~47%. The loss of activity above the level of adduction is likely due to oxidation and denaturation.
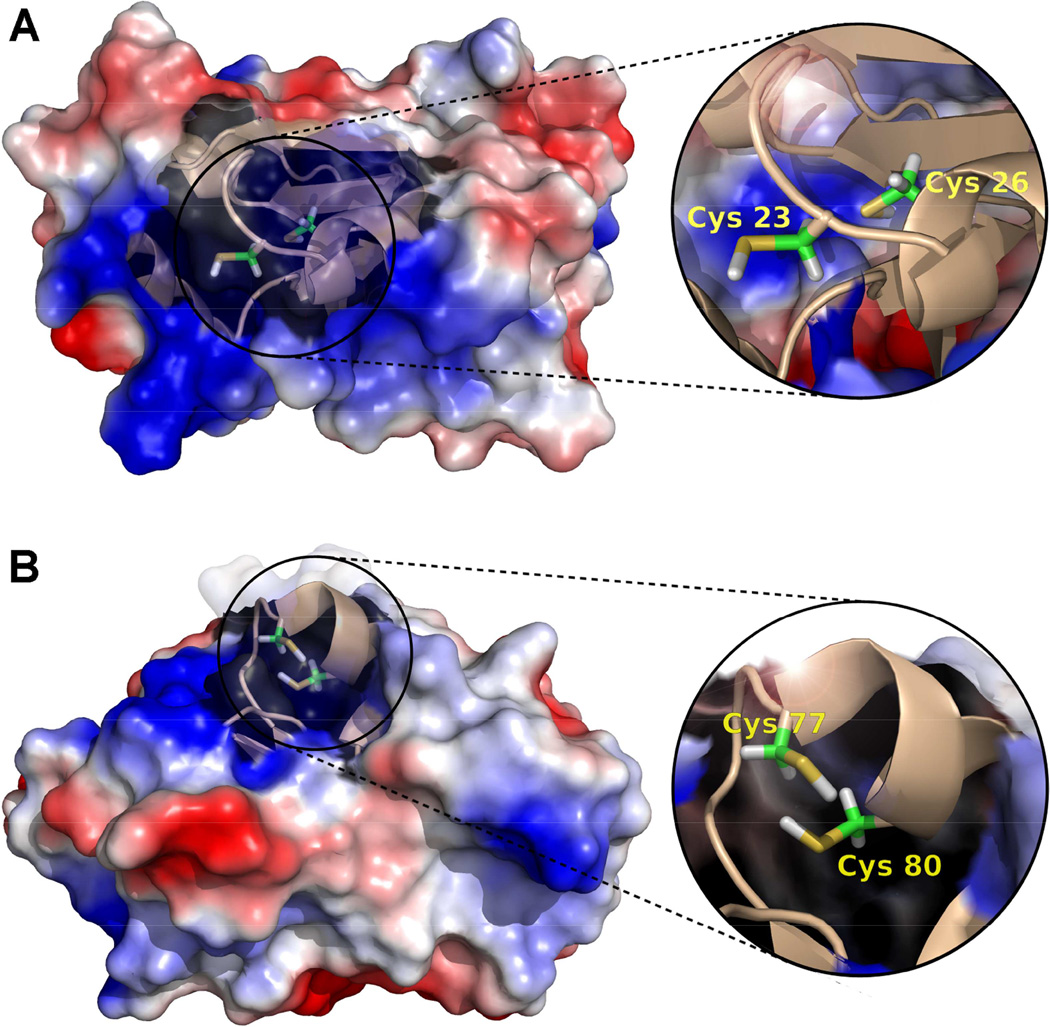
Molecular models of Grx-1 and Grx-2 were generated from available crystal structures (36, 37), and the location of the adducted cys residues is shown in Figure 7. The active sites of the two isoforms are similar, and it is not surprising that they would be similarly sensitive to EdAG. The structures emphasize that the adducted cys residues are not exposed on the surface of the proteins, but are within the GSH binding sites.
Figure 7.
Electrostatic potential surface and cartoon representations of A) hGrx-1 (adapted from pdb: 1JHB, model 8), and B) hGrx-2 (adapted from pdb: 2FLS). The close ups show the structural details of the catalytic cys residues (23, 26 and 77, 80 for Grx-1 and Grx-2, respectively). Images were generated using the PyMol Molecular Graphics System, Version 1.2r1, Schrödinger, LLC.
EdAG does not react with glutathione transferase
For comparison, hGSTA1-1 was incubated with EdAG and monitored for adducts corresponding to glutathionylated protein. GSTA1-1 is the predominant hepatic isoform that catalyzes the conjugation of GSH to many electrophilic drugs, including busulfan. GSTA1-1 provides a useful comparison because it contains a glutaredoxin motif that binds GSH in the active site and has been shown qualitatively to have some affinity for EdAG (17). Although there is no reactive cysteine in the active site of GSTA1-1, the active site residue Tyr-9 is partially ionized at pH 7.4 due to a low pKa and the tyrosinate potentially has some nucleophilic character (38). Therefore, reaction of tyrosinate-9 with EdAG was considered as a possibility. Moreover, Cys 112 of GSTA1-1 lies far from the active site but is known to form a disulfide with GSH (39). Therefore, the possibility of adduct formation of EdAG at Cys-112 was considered. In effect, GSTA1-1 provides a test of the reactivity of EdAG in GSH binding sites with nucleophiles other than cys and a test of the ability of GSH-reactive thiols on proteins to adduct EdAG. In contrast to Grx-1 and Grx-2, GSTA1-1- does not react with EdAG. We observe no adduction at either Cys-112 or Tyr-9. MS data for the intact protein are shown in reference 30.
Discussion
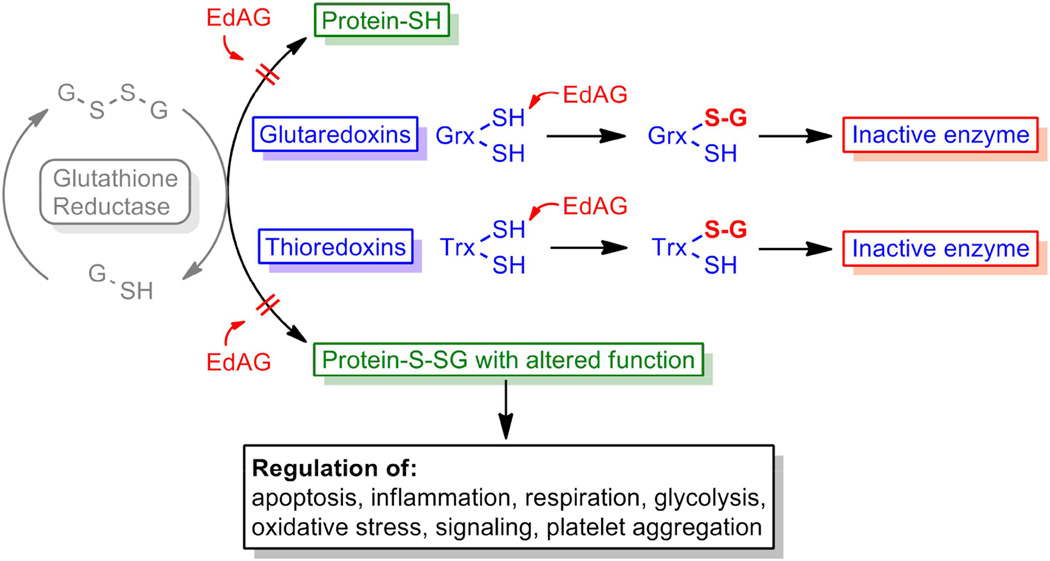
The results are the first to demonstrate the ability of EdAG to form irreversible lanthionine linkages with proteins. The reaction of EdAG with Grx-1 and Grx-2 has significant implications for the cellular toxicity of busulfan. Many proteins spanning a wide range of function are regulated by reversible glutathionylation and Grx’s play a critical role in regulating their glutathionylation in response to redox status. These processes are schematized in Figure 8. The fact that both the cytosolic Grx-1 and the mitochondrial Grx-2 are irreversibly inactivated suggests that a wide range of cellular functions would be dysregulated in response to sufficiently high concentrations of EdAG. Presumably, the highest concentrations of EdAG achieved in vivo would be in hepatocytes which contain the highest concentrations of GSH and GSTs. It is reasonable to speculate that EdAG contributes to the hepatic sinusoidal obstruction syndrome, or other toxicities, associated with busulfan treatment. Of course, toxicity in other organs would be possible if EdAG reaches the systemic circulation. For example, the mechanism of action of busulfan as a myeloablative agent might include its effects on glutathionylation status. Speculatively, the ability of busulfan to affect cancerous bone marrow cells may not be limited to its activity as a DNA alkylator. Possibly the effects of EdAG on bone marrow cells could contribute to its therapeutic effects, if it were generated in those cells or if it reached the bone marrow after being generated in the liver.
Figure 8.
Possible effects of EdAG on the Glutathiome. EdAG could alter the glutathiome directly by inhibiting Grx’s, as shown by the results presented here or, hypothetically, by inhibiting Trx’s. In addition EdAG could form irreversible adducts with cys residues on proteins that are normally activated or inhibited by glutathionylation. Adduction by EdAG could either activate or inhibit such proteins.
As depicted in Figure 8, in principle EdAG may also form lanthionine linkages with cys residues that are not in GSH binding sites. The major determinants of reactivity would be the pKa of the cys and its steric accessibility. This likely explains the differential reactivity of the cys residues in Grx’s, which have pKa’s near 4 and are nucleophilic (26, 27) vs. the surface cys-112 of GSTA1-1, which likely has a pKa near 9.5. Figure 7 includes the speculation that thioredoxins, which have nucleophilic cys residues with low pKas, also would react with EdAG, although we have not explicitly looked for that in this initial demonstration of EdAG reactivity. With regard to cys residues with ‘normal’ pKas, here we have only sampled cys-112 of GSTA1-1, which is known to form disulfides with GSH (39), and found that EdAG does not react with it, indicating that differential reactivity of EdAG with cys residues across the proteome is likely. An additional in vivo parameter that would be important determinant of reactivity would be concentration. Proteins that are more highly expressed might be adducted preferentially over proteins with more accessible cys residues or more reactive cys residues. In light of this, it would be useful to perform proteomic experiments aimed to identify any additional proteins that react with EdAG in vivo. If EdAG reacts with cys residues that are reversibly glutathionylated as part of a regulatory mechanism, then this could also dysregulate the glutathiome. Thus, EdAG could alter the glutathiome by reacting with cys residues that normally participate in regulation via reversible glutathionylation and by its inactivation of Grx’s or Trx’s that control the process of glutathionylation/deglutathionylation. In addition to Grx’s and Trx’s, sulfiredoxins utilize GSH to catalyze deglutathionylation (40) and GST omega may play a role in GSH-independent deglutathionylation processes (41). Thus, these enzymes are potential targets of EdAG as well. We have limited Figure 8 to Grx’s and Trx’s because they appear to be the major determinants of glutathionylation status but these other proteins could also be affected in as much as they have reactive cys residues in potential GSH binding sites. Similarly, GSTP1-1 has been suggested to mediate protein glutathionylation, but the only reactive cys (Cys-47) is not part of the canonical GSH binding site. Still, GSTP1-1 could be an additional interesting target for EdAG.
The results are novel from a biochemical perspective. We are unaware of other examples of irreversible protein glutathionylation caused by any drug or drug metabolite. Thus, any effects of busulfan on the glutathiome, might represent a novel drug-dependent mechanism of toxicity. In addition it is interesting to emphasize that the EdAG reaction with proteins represents the only example of which we are aware, of a drug (busulfan) creating a ‘reactive metabolite’ from an endogenous peptide (GSH). Note that, as depicted in Figure 1, none of the atoms originally found in busulfan are maintained in the ultimate GSH adduct with EdAG, and this is confirmed by the results with protein adducts. The irreversible protein adduct with EdAG contains only atoms from GSH and none from busulfan. This is an intriguing variation on the more common situation in which a drug is metabolized to a reactive species that results in incorporation of the drug scaffold into the protein. In addition, lanthionine linkages have been observed in antibiotic natural products and the enzymes that catalyze their formation represent potential tools for incorporation of novel lanthionines in engineered proteins (42). The reaction with EdAG represents an interesting source of ‘non-natural’ lanthionines. Taken together, the results demonstrate that the frequently used drug busulfan can participate in interesting chemical reactivity with proteins by novel mechanisms.
Highlights.
Busulfan is used in hematopoietic stem cell transplants, but its system pharmacology has not been described.
The busulfan metabolite EdAG irreversibly gluathionylates active site cysteines of glutaredoxins in vitro.
Busulfan metabolism leads to inactivation of glutaredoxins via covalent modification by EdAG.
EdAG may lead to dysregulation of the ‘glutathiome’ or the set of proteins normally regulated by reversible glutathionylation.
Acknowledgements
This work was supported by The Department of Medicinal Chemistry, University of Washington. The authors acknowledge Professor Caryn Outten Department of Chemistry and Biochemistry, University of South Carolina for supplying constructs for the expression of GRx-1 and Grx-2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author Contributions
WMA conceived and coordinated the study and wrote portions of the paper. MS designed, performed and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
References
- 1.McCune JS, Holmberg LA. Busulfan in hematopoietic stem cell transplant setting. Expert Opin Drug Metab Toxicol. 2009;5:957–969. doi: 10.1517/17425250903107764. [DOI] [PubMed] [Google Scholar]
- 2.Reece D, Song K, LeBlanc R, Mezzi K, Olujohungbe A, White D, Zaman F, Belch A. Efficacy and safety of busulfan-based conditioning regimens for multiple myeloma. Oncologist. 2013;18:611–618. doi: 10.1634/theoncologist.2012-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferry C, Socie G. Busulfan-cyclophosphamide versus total body irradiation-cyclophosphamide as preparative regimen before allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia: what have we learned? Exp Hematol. 2003;31:1182–1186. doi: 10.1016/j.exphem.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Barker CC, Butzner JD, Anderson RA, Brant R, Sauve RS. Incidence, survival and risk factors for the development of veno-occlusive disease in pediatric hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2003;32:79–87. doi: 10.1038/sj.bmt.1704069. [DOI] [PubMed] [Google Scholar]
- 5.Reiss U, Cowan M, McMillan A, Horn B. Hepatic venoocclusive disease in blood and bone marrow transplantation in children and young adults: incidence, risk factors, and outcome in a cohort of 241 patients. J Pediatr Hematol Oncol. 2002;24:746–750. doi: 10.1097/00043426-200212000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Cesaro S, Pillon M, Talenti E, Toffolutti T, Calore E, Tridello G, Strugo L, Destro R, Gazzola MV, Varotto S, Errigo G, Carli M, Zanesco L, Messina C. A prospective survey on incidence, risk factors and therapy of hepatic veno-occlusive disease in children after hematopoietic stem cell transplantation. Haematologica. 2005;90:1396–1404. [PubMed] [Google Scholar]
- 7.Jones RJ, Lee KS, Beschorner WE, Vogel VG, Grochow LB, Braine HG, Vogelsang GB, Sensenbrenner LL, Santos GW, Saral R. Venoocclusive disease of the liver following bone marrow transplantation. Transplantation. 1987;44:778–783. doi: 10.1097/00007890-198712000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Carreras E, Bertz H, Arcese W, Vernant JP, Tomas JF, Hagglund H, Bandini G, Esperou H, Russell J, de la Rubia J, Di Girolamo G, Demuynck H, Hartmann O, Clausen J, Ruutu T, Leblond V, Iriondo A, Bosi A, Ben-Bassat I, Koza V, Gratwohl A, Apperley JF. Incidence and outcome of hepatic veno-occlusive disease after blood or marrow transplantation: a prospective cohort study of the European Group for Blood and Marrow Transplantation. European Group for Blood and Marrow Transplantation Chronic Leukemia Working Party. Blood. 1998;92:3599–3604. [PubMed] [Google Scholar]
- 9.Bruno B, Souillet G, Bertrand Y, Werck-Gallois MC, So Satta A, Bellon G. Effects of allogeneic bone marrow transplantation on pulmonary function in 80 children in a single paediatric centre. Bone Marrow Transplant. 2004;34:143–147. doi: 10.1038/sj.bmt.1704549. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Tripathi RC, Tripathi BJ. Drug-induced ocular disorders. Drug Saf. 2008;31:127–141. doi: 10.2165/00002018-200831020-00003. [DOI] [PubMed] [Google Scholar]
- 11.Eberly AL, Anderson GD, Bubalo JS, McCune JS. Optimal prevention of seizures induced by high-dose busulfan. Pharmacotherapy. 2008;28:1502–1510. doi: 10.1592/phco.28.12.1502. [DOI] [PubMed] [Google Scholar]
- 12.Krivoy N, Hoffer E, Lurie Y, Bentur Y, Rowe JM. Busulfan use in hematopoietic stem cell transplantation: pharmacology, dose adjustment, safety and efficacy in adults and children. Curr Drug Saf. 2008;3:60–66. doi: 10.2174/157488608783333899. [DOI] [PubMed] [Google Scholar]
- 13.Diestelhorst C, Boos J, McCune JS, Russell J, Kangarloo SB, Hempel G. Physiologically based pharmacokinetic modelling of Busulfan: a new approach to describe and predict the pharmacokinetics in adults. Cancer Chemother Pharmacol. 2013;72:991–1000. doi: 10.1007/s00280-013-2275-x. [DOI] [PubMed] [Google Scholar]
- 14.Uppugunduri CR, Rezgui MA, Diaz PH, Tyagi AK, Rousseau J, Daali Y, Duval M, Bittencourt H, Krajinovic M, Ansari M. The association of cytochrome P450 genetic polymorphisms with sulfolane formation and the efficacy of a busulfan-based conditioning regimen in pediatric patients undergoing hematopoietic stem cell transplantation. Pharmacogenomics J. 2014;14:263–271. doi: 10.1038/tpj.2013.38. [DOI] [PubMed] [Google Scholar]
- 15.El-Serafi I, Terelius Y, Twelkmeyer B, Hagbjork AL, Hassan Z, Hassan M. Gas chromatographic-mass spectrometry method for the detection of busulphan and its metabolites in plasma and urine. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;913–914:98–105. doi: 10.1016/j.jchromb.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava A, Poonkuzhali B, Shaji RV, George B, Mathews V, Chandy M, Krishnamoorthy R. Glutathione S-transferase M1 polymorphism: a risk factor for hepatic venoocclusive disease in bone marrow transplantation. Blood. 2004;104:1574–1577. doi: 10.1182/blood-2003-11-3778. [DOI] [PubMed] [Google Scholar]
- 17.Younis IR, Elliott M, Peer CJ, Cooper AJ, Pinto JT, Konat GW, Kraszpulski M, Petros WP, Callery PS. Dehydroalanine analog of glutathione: an electrophilic busulfan metabolite that binds to human glutathione S-transferase A1-1. J Pharmacol Exp Ther. 2008;327:770–776. doi: 10.1124/jpet.108.142208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czerwinski M, Gibbs JP, Slattery JT. Busulfan conjugation by glutathione S-transferases alpha, mu, and pi. Drug Metab Dispos. 1996;24:1015–1019. [PubMed] [Google Scholar]
- 19.Cooper AJ, Pinto JT, Callery PS. Reversible and irreversible protein glutathionylation: biological and clinical aspects. Expert Opin Drug Metab Toxicol. 2011;7:891–910. doi: 10.1517/17425255.2011.577738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Townsend DM, Lushchak VI, Cooper AJ. A comparison of reversible versus irreversible protein glutathionylation. Adv Cancer Res. 2014;122:177–198. doi: 10.1016/B978-0-12-420117-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mailloux RJ, Willmore WG. S-glutathionylation reactions in mitochondrial function and disease. Front Cell Dev Biol. 2014;2:68. doi: 10.3389/fcell.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Stroher E, Millar AH. The biological roles of glutaredoxins. Biochem J. 2012;446:333–348. doi: 10.1042/BJ20112131. [DOI] [PubMed] [Google Scholar]
- 25.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 26.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta. 2013;1830:3217–3266. doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Hanschmann EM, Godoy JR, Berndt C, Hudemann C, Lillig CH. Thioredoxins, glutaredoxins, and peroxiredoxins--molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid Redox Signal. 2013;19:1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asquith RS, Carthew P. The preparation and subsequent identification of a dehydroalanyl peptide from alkali-treated oxidised glutathione. Biochim Biophys Acta. 1972;285:346–351. doi: 10.1016/0005-2795(72)90319-4. [DOI] [PubMed] [Google Scholar]
- 29.Younis IR. Ph.D. thesis. West Virginia University; 2008. In vitro elucidation of the metabolic fate of the anticancer drug busulfan. [Google Scholar]
- 30.Scian M, Atkins WM. Supporting Data for Characterization of the Busulfan Metabolite EdAG and the Glutaredoxins that it Adducts. Data in Brief. doi: 10.1016/j.dib.2015.09.002. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouldin SD, Darch MA, Hart PJ, Outten CE. Redox properties of the disulfide bond of human Cu,Zn superoxide dismutase and the effects of human glutaredoxin 1. Biochem J. 2012;446:59–67. doi: 10.1042/BJ20120075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem. 2008;283:8868–8876. doi: 10.1074/jbc.M709567200. [DOI] [PubMed] [Google Scholar]
- 34.Xie C, Zhong D, Chen X. A fragmentation-based method for the differentiation of glutathione conjugates by high-resolution mass spectrometry with electrospray ionization. Anal Chim Acta. 2013;788:89–98. doi: 10.1016/j.aca.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Jian W, Liu HF, Zhao W, Jones E, Zhu M. Simultaneous screening of glutathione and cyanide adducts using precursor ion and neutral loss scans-dependent product ion spectral acquisition and data mining tools. J Am Soc Mass Spectrom. 2012;23:964–976. doi: 10.1007/s13361-012-0354-6. [DOI] [PubMed] [Google Scholar]
- 36.Sun C, Berardi MJ, Bushweller JH. The NMR solution structure of human glutaredoxin in the fully reduced form. J Mol Biol. 1998;280:687–701. doi: 10.1006/jmbi.1998.1913. [DOI] [PubMed] [Google Scholar]
- 37.Johansson C, Kavanagh KL, Gileadi O, Oppermann U. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J Biol Chem. 2007;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- 38.Atkins WM, Wang RW, Bird AW, Newton DJ, Lu AY. The catalytic mechanism of glutathione S-transferase (GST). Spectroscopic determination of the pKa of Tyr-9 in rat alpha 1-1 GST. J Biol Chem. 1993;268:19188–19191. [PubMed] [Google Scholar]
- 39.Lyon RP, Atkins WM. Kinetic characterization of native and cysteine 112-modified glutathione S-transferase A1-1: reassessment of nonsubstrate ligand binding. Biochemistry. 2002;41:10920–10927. doi: 10.1021/bi0262810. [DOI] [PubMed] [Google Scholar]
- 40.Findlay VJ, Tapiero H, Townsend DM. Sulfiredoxin: a potential therapeutic agent? Biomed Pharmacother. 2005;59:374–379. doi: 10.1016/j.biopha.2005.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menon D, Board PGJ. A role for glutathione transferase Omega 1 (GSTO1-1) in the glutathionylation cycle. J Biol Chem. 2013;288:25769–25779. doi: 10.1074/jbc.M113.487785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]