Summary
The ERG gene is fused to TMPRSS2 in approximately 50% of prostate cancers (PrCa) resulting in its overexpression. However, whether this is the sole mechanism underlying ERG elevation in PrCa is currently unclear. Here we report that ERG ubiquitination and degradation is governed by the Cullin 3-based ubiquitin ligase SPOP and that deficiency in this pathway leads to aberrant elevation of the ERG oncoprotein. Specifically, we find that truncated ERG (ΔERG), encoded by the ERG fusion gene, is stabilized by evading SPOP-mediated destruction, while prostate cancer-associated SPOP mutants are also deficient in promoting ERG ubiquitination. Furthermore, we show that SPOP/ERG interaction is modulated by CKI-mediated phosphorylation. Importantly, we demonstrate that DNA damage drug, topoisomerase inhibitors, can trigger CKI activation to restore the SPOP/ΔERG interaction and its consequent degradation. Thus SPOP functions as a tumor suppressor to negatively regulate the stability of the ERG oncoprotein in prostate cancer.
Introduction
Prostate cancer (PrCa) is the second leading cause of cancer death for men in western countries (Siegel et al., 2013). Extensive genomic studies revealed that PrCa is driven by accumulation of genetic alterations including PTEN loss (Li et al., 1997) and gene fusions (Tomlins et al., 2007). Gene fusion products, by juxtaposing two separate genes, may result in a chimeric protein with different functions, such as the BCR–ABL1 gene fusion in chronic myeloid leukemia (CML) (Ren, 2005). Alternatively, a proto-oncogene fusing to a strong promoter/enhancer can result in up-regulation of mRNA levels. For example, the IgH-Myc fusion in lymphoma (Adams et al., 1985) and the E26 transformation-specific (ETS) family of transcription factor fusions in prostate cancer (Kumar-Sinha et al., 2008).
The most common ETS gene fusion is TMPRSS2-ERG, which occurs in approximately 50% of prostate cancers (Kumar-Sinha et al., 2008; Tomlins et al., 2005). On the other hand, overexpression of other ETS genes such as ETV1, ETV4 and ETV5 due to gene fusion, have been reported in only 5–10% of prostate cancers (Rubin et al., 2011). Fusion of ERG to TMPRSS2 results in increased mRNA levels of ERG and expression of N-terminally truncated ERG protein under the control of androgen-regulated TMPRSS2 promoter (Tomlins et al., 2005). Recent studies have demonstrated that overexpression of ERG fusion proteins facilitate prostate cancer development largely by promoting cell migration and invasion (Carver et al., 2009b; Tomlins et al., 2008), thereby functioning as an oncogene. Moreover, the TMPRSS2-ERG fusion has been found in the prostate cancer precursor high-grade prostatic intraepithelial neoplasia (HGPIN), indicating that it is an early molecular event associated with invasion in prostate cancer (Perner et al., 2007). Furthermore, the deubiquitinase USP9X interacts with and stabilizes ERG to promote prostate cancer (Wang et al., 2014). However, little is known about how ERG protein stability is physiologically governed by E3 ligase(s) in vivo and aberrantly regulated in prostate cancer.
Recently, systematic sequencing studies further revealed that recurrent somatic mutation is another key feature of prostate cancer (Barbieri et al., 2012; Berger et al., 2011). Notably, the most frequently mutated gene is SPOP (speckle-type POZ protein), which encodes a Cullin 3-based E3 ubiquitin ligase, with recurrent mutation in 6–15% of primary human prostate cancers (Barbieri et al., 2012; Lindberg et al., 2013). Structurally, SPOP contains two conserved domains: an N-terminal MATH domain that recruits substrates, and a C-terminal BTB domain that binds Cullin 3 (Zhuang et al., 2009). Several SPOP substrates have been identified in the context of prostate, including the androgen receptor (AR) (An et al., 2014; Geng et al., 2014), steroid receptor coactivator 3 (SRC-3) (Li et al., 2011), DEK and TRIM24 (Theurillat et al., 2014). Furthermore, prostate cancer-associated SPOP mutants are deficient in binding with and promoting the degradation of substrates leading to increased prostate cancer cell proliferation and invasion (An et al., 2014; Barbieri et al., 2012), indicating the loss-of-function of SPOP mutations and the tumor suppressive role of SPOP in prostate cancer. Therefore, identification of additional SPOP substrates would benefit prostate cancer clinical diagnosis and therapy.
Interestingly, these two common genetic alterations (SPOP somatic mutations and ERG fusions) appear to be mutually exclusive in prostate cancer (Barbieri et al., 2012), but the reason for the segregation of these two genetic events remains largely unknown. Since both SPOP mutations and ERG fusion are tightly associated with prostate cancer development, they might affect similar downstream pathways or targets to facilitate prostate cancer progression.
Results
SPOP specifically interacts with and promotes ubiquitination and degradation of ERG
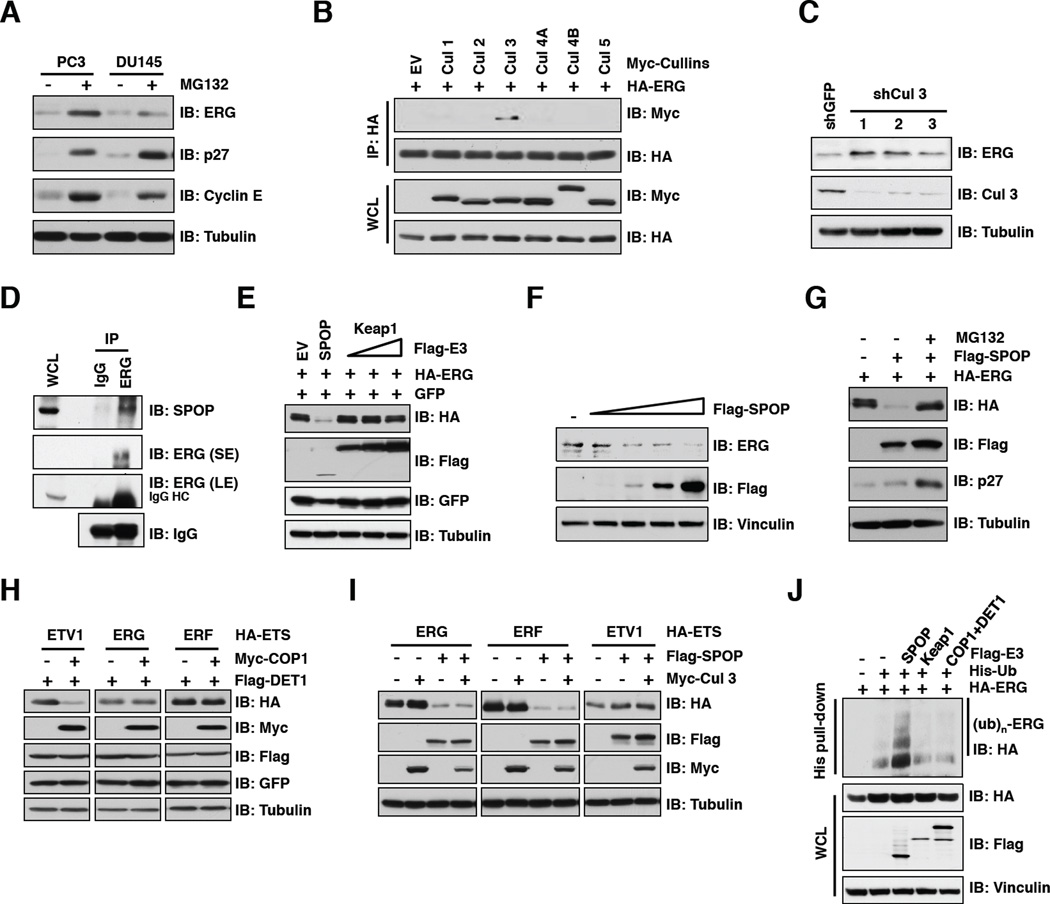
Given the prevalence and the critical role of ERG in prostate cancer progression (Carver et al., 2009a; Tomlins et al., 2008), it is important to understand how ERG protein stability is governed in vivo and whether it is aberrantly regulated in prostate cancer. To this end, we observed that the abundance of endogenous ERG protein levels were significantly increased upon treating PC3 and DU145 prostate cancer cells, which express very low protein levels of wild-type ERG, with the proteasome inhibitor MG132 (Figure 1A). Importantly, ERG protein abundance detected by western blot analysis was markedly reduced by multiple shERG vectors in PC3 cells, confirming that the ERG antibody specifically recognizes endogenous ERG in prostate cancer cells (Figure S1A). As the multi-subunit Cullin-Ring complexes are the largest known class of E3 ubiquitin ligases (Petroski and Deshaies, 2005), we next examined whether a specific Cullin-Ring complex contributes to ERG destruction. We found that ERG specifically interacted with Cullin 3, but not other members of the Cullin family (Figure 1B). Consistent with this finding, depletion of endogenous Cullin 3 led to an increase in ERG abundance (Figure 1C), indicating that the Cullin 3 pathway is involved in controlling ERG stability. Previous studies have established that Cullin 3 exerts its E3 ubiquitin ligase activity by recruiting various BTB/POZ domain-containing proteins as substrate-specific adaptors, including Keap1 and SPOP (Genschik et al., 2013). Notably, we observed that both ERG and ERF specifically interacted with SPOP, but not Keap1 or COP1, a Cullin 4-based E3 ligase substrate adaptor protein with a tumor suppressive role in prostate cancer (Vitari et al., 2011), in vivo and in vitro (Figures 1D and S1B–D). Consistent with this finding, we demonstrated that ectopic expression of SPOP, but not Keap1, decreased expression of ERG in a dose-dependent manner (Figures 1E, 1F and S1E). More importantly, SPOP-mediated destruction of ERG could be blocked by MG132 (Figure 1G).
Figure 1. SPOP specifically interacts with the Cullin 3/SPOP E3 ubiquitin ligase.
A. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from PC3 and DU145 cells. Where indicated, cells were treated with 10 µM MG132 for 10 hr before harvesting.
B. IB analysis of WCL and immunoprecipitates (IP) derived from 293T cells transfected with indicated constructs. Cells were treated with 10 µM MG132 for 10 hr before harvesting.
C. IB analysis of WCL derived from PC3 cells infected with the indicated lentiviral shRNA vectors.
D. IB analysis of LNCaP WCL and anti-ERG IP. Rabbit IgG was used as a negative control for the IP. Cells were treated with 10 µM MG132 for 10 hr before harvesting.
E-I. IB analysis of WCL derived from 293 cells (E, G-I) or PC3 cells (F) transfected with indicated plasmids. Where indicated, 10 µM MG132 was added for 10 hr before harvesting.
J. IB analysis of WCL and His pull-down products derived from PC3 cells transfected with plasmids expressing the indicated proteins.
See also Figure S1
Next, in keeping with a previous study (Vitari et al., 2011), we found that Cullin 4A/COP1 promoted the destruction of ETV1, but not ERG or ERF (Figure 1H). On the other hand, SPOP specifically promoted ERG and ERF, but not ETV1, turnover (Figure 1I). We further confirmed that SPOP-WT, but not Keap1, COP1 or the E3 ligase activity-deficient mutant form of SPOP (SPOP-ΔBTB), could promote ERG ubiquitination in vivo (Figures 1J and S1F). These data together support the notion that the Cullin 3/SPOP E3 ligase complex specifically regulates ERG protein stability.
SPOP negatively regulates ERG-mediated cell migration and invasion
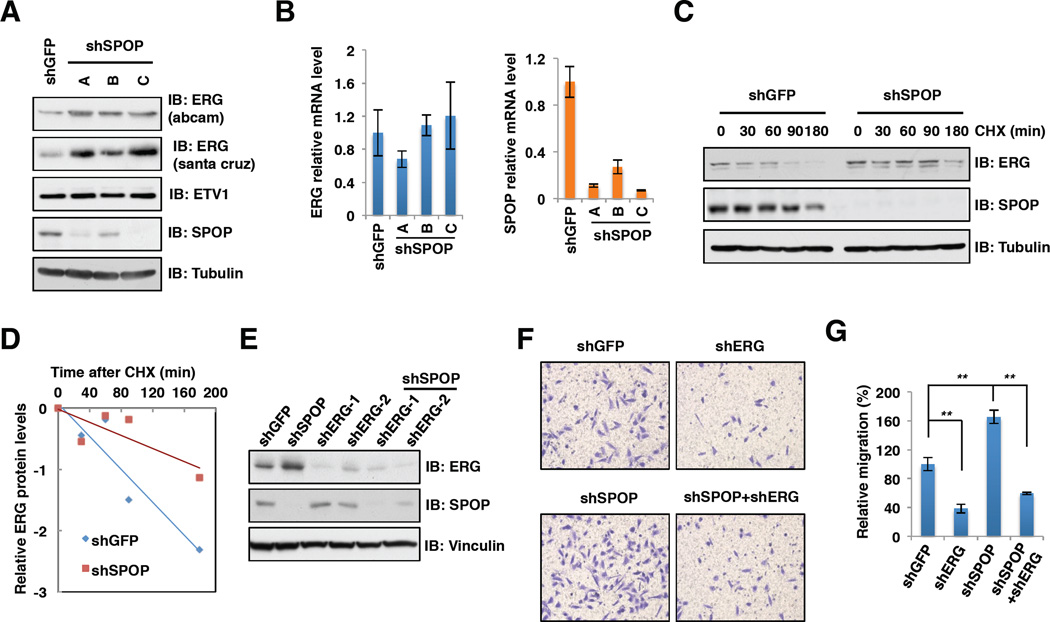
Consistent with a critical role for SPOP in regulating ERG stability, depletion of endogenous SPOP by multiple shRNA vectors led to a noticeable accumulation in protein abundance of ERG, but not ETV1 or ERF, with minimal changes in ERG mRNA levels in prostate cancer cells (Figures 2A, 2B, S2A–C and Table S1). Hence, in the remainder of the study, we primarily focus on elucidating how SPOP controls the stability of the ERG protein. Importantly, the half-life of endogenous ERG protein was extended after depleting SPOP (Figures 2C and 2D), suggesting that SPOP controls ERG expression largely through a post-translational mechanism.
Figure 2. Depletion of SPOP leads to increased ERG protein levels and cell migration and invasion.
A. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from PC3 cells infected with the indicated lentiviral shRNA vectors.
B. Real-time PCR analysis to examine the ERG and SPOP mRNA levels after depletion of SPOP. Data was shown as mean ± SD for three independent experiments.
C-E. IB analysis of PC3 cells infected with the indicated lentiviral shRNA constructs (C, E). Where indicated, 100 µg/ml cycloheximide (CHX) was added and cells were harvested at indicated time points. ERG protein abundance in (C) was quantified by ImageJ and plotted as indicated (D).
F-G. Representative images of migrated PC3 cells infected with indicated lentiviral shRNA constructs in migration assay (F) and quantification of migrated cells (G). Data was shown as mean + SD for three independent experiments. **p < 0.001, t-test.
See also Figure S2
To explore the critical biological function of SPOP targeting ERG for degradation, we next examined the effects of SPOP depletion on cell proliferation, migration and invasion. In agreement with previous studies (Carver et al., 2009b; Tomlins et al., 2007; Tomlins et al., 2008), we observed that depletion of SPOP or ERG has minor effects on cell growth and apoptosis in PC3 cells (Figures S2D–F). However, we found that depletion of ERG decreased cell migration (Figures 2E–G) and invasion (Figures S2G and S2H). In contrast, depletion of SPOP enhanced the invasive ability of the cells. More importantly, simultaneous depletion of SPOP and ERG reduced cell migration and invasion compared with SPOP single knockdown, arguing that SPOP modulates cell migration and invasion largely through governing ERG protein abundance (Figures 2E-2G, S2G and S2H). Consistent with these results, ERG target genes including ADAMTS1, CXCR4, OPN and MMP9, all of which play important roles in promoting cell migration and invasion (Carver et al., 2009b; Egeblad and Werb, 2002; Thalmann et al., 1999), were found to be upregulated at both the mRNA and protein levels upon SPOP depletion (Figures S2I, S2J and Table S1). Therefore, SPOP functions as a tumor suppressor in prostate by targeting the major prostate cancer driver ERG for ubiquitination and degradation. Together, these results suggest that SPOP is the physiological E3 ligase that promotes ERG ubiquitination and destruction in prostate cancer (Figure S2K).
Prostate cancer-associated SPOP mutants fail to interact with ERG to promote ERG destruction
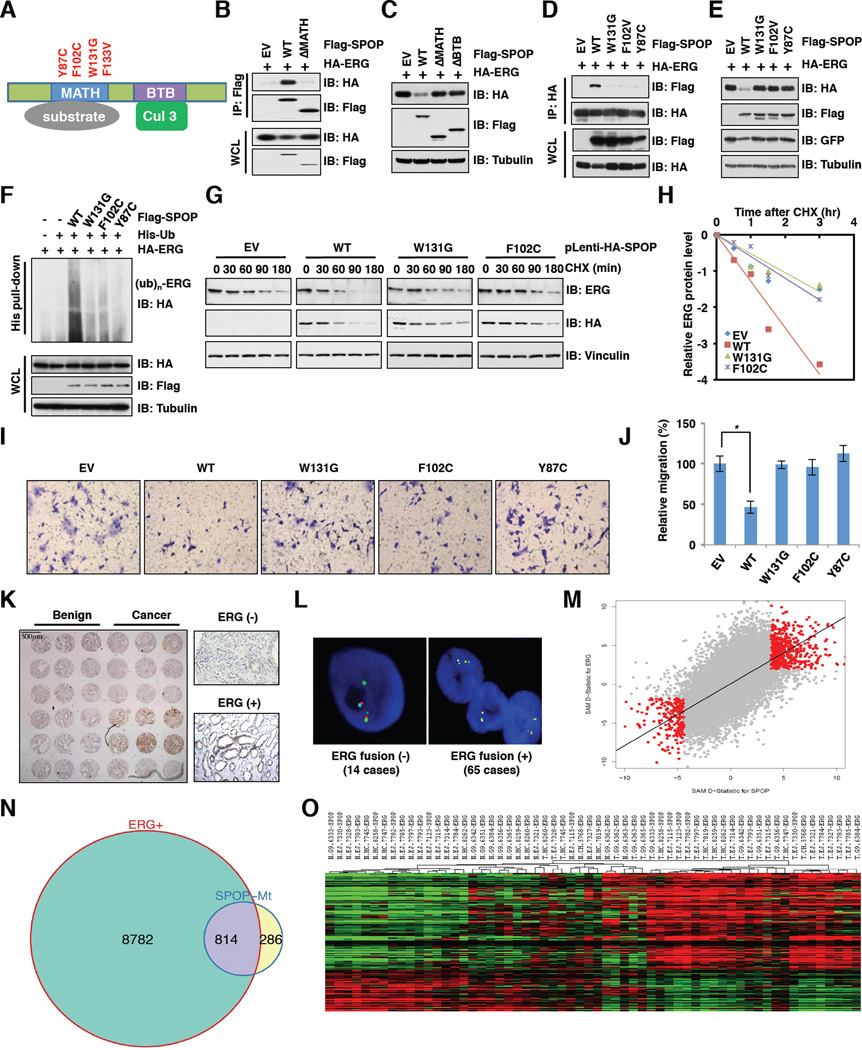
All SPOP somatic mutations identified in prostate cancers, such as Y87C, F102C, W131G and F133V, are clustered in the MATH domain (Figure 3A) and display impaired substrate binding (Geng et al., 2013; Zhuang et al., 2009). To examine whether these SPOP mutations affect ERG stability, we first determined that deletion of the MATH domain abrogated SPOP binding to ERG (Figure 3B), while loss of either the MATH domain or the BTB domain inhibited SPOP-mediated ERG degradation (Figure 3C). Next, we found that various prostate cancer-associated SPOP mutants failed to interact with ERG (Figures 3D and S3A) to promote ERG ubiquitination and destruction (Figures 3E and 3F). On the other hand, ectopic expression of SPOP-WT, but not SPOP-mutants, resulted in a marked reduction of the half-life of endogenous or ectopically expressed ERG (Figures 3G, 3H and S3B, S3C). In keeping with a possible loss-of-function phenotype associated with SPOP mutants in promoting ERG destruction, only cells expressing wild-type, but not mutated, SPOP reduced endogenous ERG protein levels that subsequently led to decreased cell migration (Figures 3I, 3J and S3D–G).
Figure 3. Prostate cancer-associated SPOP mutants fail to regulate ERG stability.
A. Schematic of SPOP domains and prostate cancer-associated mutations.
B. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with indicated constructs. Cells were treated with 10 µM MG132 for 10 hr before harvesting.
C. IB analysis of 293T cells transfected with the indicated plasmids.
D. IB analysis of WCL and IP derived from 293T cells transfected with indicated constructs. Cells were treated with 10 µM MG132 for 10 hr before harvesting.
E. IB analysis of 293T cells transfected with the indicated plasmids.
F. IB analysis of WCL and His pull-down products derived from PC3 cells transfected with plasmids expressing the indicated proteins.
G. IB analysis of WCL derived from DU145 cells stably expressing SPOP-WT or mutants. Cells were treated with 100 µg/ml cycloheximide (CHX) for the indicated time period before harvesting.
H. ERG protein abundance in (G) was quantified by ImageJ and plotted as indicated.
I. Representative images of migrated DU145 cells infected with the indicated lentiviral shRNA constructs in migration assays.
J. Quantification of migrated cells in (I). Data was shown as mean ± SD for three independent experiments. *p < 0.05, t-test.
K. Protein levels of ERG were up-regulated in human prostate cancer samples but not in comparable benign prostate tissue. The tissue microarray slide was stained with anti-ERG antibody (left). High power views of negative and positive nuclear staining of ERG were shown (right).
L. Identification of TMPRSS2-ERG status by FISH in cases with positive staining in (K). The green signal (ERG) is separated or split from the red-aqua signal pair (TMPRSS2-ERG fusion) in ERG fusion (+) samples but not ERG fusion (−) samples.
M. Gene expression changes were positive correlation in SPOP mutation and ERG fusion clinical specimens. Please refer to supplemental information for details.
N. SPOP mutation and ERG fusion share common gene signature. Venn diagram showing the overlap of genes significantly differentially expressed in SPOP mutation and ERG fusion samples from TCGA.
O. Heatmap of the common genes associated with SPOP mutation and ERG fusion. N represents matched normal samples from ERG fusion or SPOP mutation patients; T represents matched tumor samples from ERG fusion or SPOP mutation patients; the number represents TCGA patient ID.
See also Figure S3
It has been previously reported that in prostate cancer cells, SPOP mutants are also deficient in promoting ubiquitination and subsequent degradation of NCOA3 (Geng et al., 2013), DEK and TRIM24 (Theurillat et al., 2014). To further examine the contribution of ERG or these above-mentioned SPOP substrates in mediating the tumorigenesis phenotypes in cells harboring SPOP mutations, we depleted each of these genes or ERG in DU145 cells stably expressing the well-characterized SPOP-F102C mutant (Zhuang et al., 2009). Notably, we found that depletion of each of these proteins leads to decreased cell migration, with depletion of ERG exhibiting the most significantly suppressive effects (Figures S3H–J). Furthermore, prostate cancer data sets from the cBio database (http://www.cbioportal.org/public-portal) (Cerami et al., 2012) showed that ERG fusion occurs in approximately 50% of prostate cancers, whereas the frequency of aberrant NCOA3, DEK or TRIM24 expression is only 2–3% in the prostate cancer setting (Figure S3K). Hence, these results together indicate that ERG may be the major disease-relevant driver of prostate cancer. As such, SPOP mutations disrupt its ability to target ERG for ubiquitination, which may lead to aberrant elevation of ERG oncoprotein abundance to facilitate prostate cancer progression.
SPOP mutations contribute to elevated ERG protein levels and share common gene signatures with ERG fusion in clinical specimens
Having demonstrated SPOP as a physiological upstream E3 ligase for ERG, we next explored whether loss-of-function SPOP mutations correlated with elevated ERG protein levels in pathological conditions such as human prostate cancer. It is noteworthy that SPOP mutations are mutually exclusive with TMPRSS2-ERG fusion that leads to increased ERG expression at both the mRNA and protein levels (Barbieri et al., 2012; Clark and Cooper, 2009). In keeping with this notion, we analyzed all of TCGA prostate cancer samples (236 patients) and found that SPOP mutation and ERG fusion are mutually exclusive (Figures S3L and S3M).
However, some PrCa cases with moderate ERG protein levels did not harbor any ERG fusion (Park et al., 2010). We thus hypothesized that in ERG expression-positive but ERG fusion-negative cases, SPOP mutations may contribute to increased ERG protein levels. To test this notion, we generated a tissue microarray from 239 PrCa cases and identified 79 ERG positive cases by IHC (Figure 3K). Using fluorescence in situ hybridization (FISH) analysis, we identified 14 of the 79 ERG IHC positive samples to be negative for TMPRSS2-ERG fusion (Figure 3L). Sanger sequencing analysis demonstrated that 5 of the 14 cases harbored SPOP mutations (Figure S3N). Interestingly, besides the well-characterized F133V mutation (Barbieri et al., 2012), we identified a novel SPOP mutation (R139K) located in the MATH domain and further validated that the R139K mutation was also deficient in promoting ERG degradation (Figure S3O). These results indicate that SPOP mutations likely contribute to the moderate ERG elevation in those prostate cancer cases without known ERG fusions, largely through stabilization of the ERG protein (Figure S3P).
To further understand the correlation between SPOP mutation and aberrant ERG expression in PrCa pathophysiology, we analyzed the gene signature of ERG fusion versus SPOP mutation in clinical specimens. By using two-class paired significance analysis of microarray data in tumor and normal samples within SPOP mutation cases and the TMPRSS2-ERG fusion cases in TCGA, we found a significant correlation between the expression changes observed in tumors with SPOP mutations and in tumors with ERG fusion (Rho=0.64, P<2.2e-16) (Figure 3M). Compared with normal samples, 1100 and 9596 genes showed significant differential expression in SPOP mutation and ERG fusion prostate tumor samples, respectively (Figure 3N). More importantly, there are 814 genes that are seemingly co-regulated by SPOP mutation and ERG fusion events with 574 up-regulated genes and 240 down-regulated genes (Figures 3N and 3O). These results support the notion that SPOP mutation and ERG fusion share common gene signatures. Therefore, SPOP mutation leads to increased expression of ERG protein and its targets to favor PrCa progression, which is similar to how the ERG fusion protein functions in PrCa.
SPOP-mediated ubiquitination and destruction of ERG depends on the SPOP binding motif
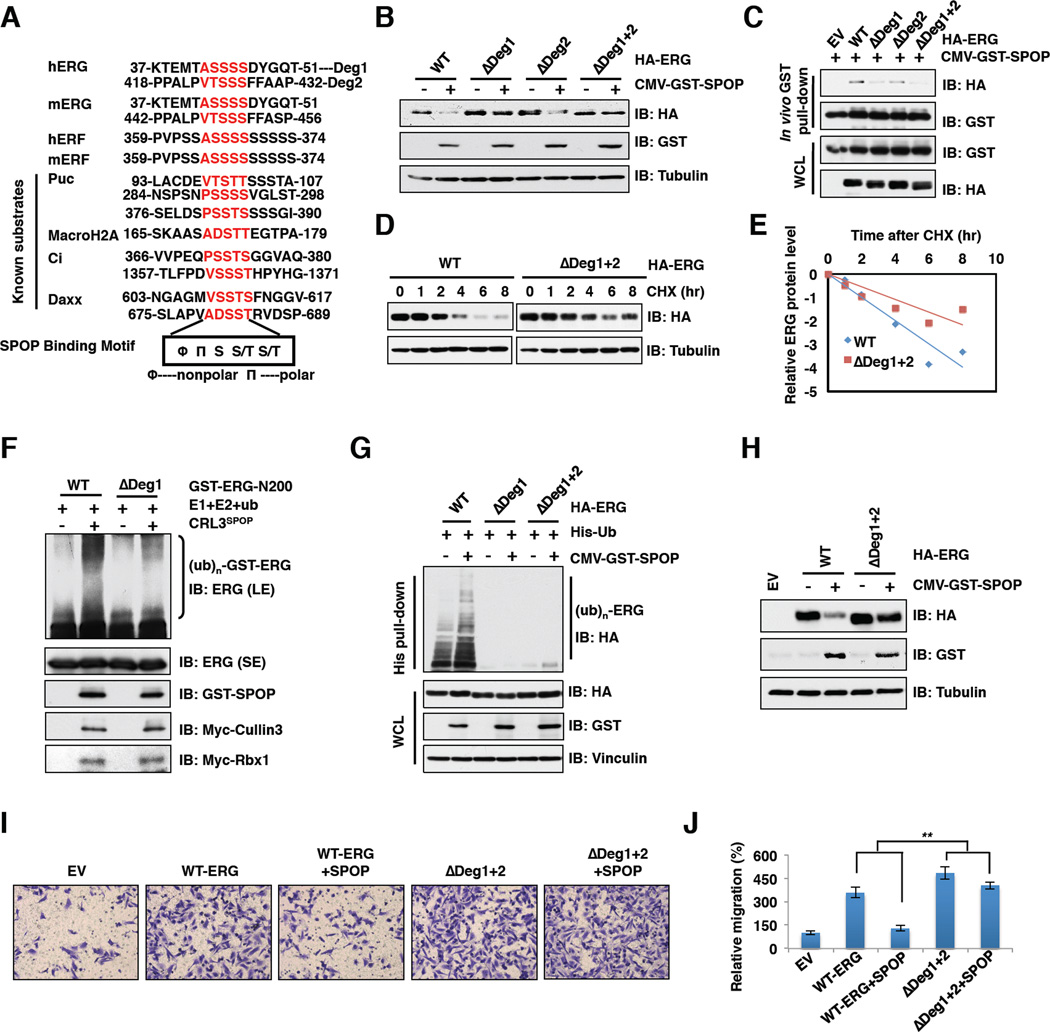
Consistent with previous reports that known SPOP substrates share a SPOP-binding consensus motif Φ-Π-S-S/T-S/T (Φ-nonpolar; Π-polar) (Zhuang et al., 2009), we found that ERG contains two putative SPOP binding motifs or “degrons” located in exon 4 and exon 11 (Figures 4A and 6A). Notably, deletion of degron 1 (ΔDeg1) but not degron 2 (ΔDeg2) largely blocked SPOP-mediated ERG degradation, while ERG was no longer subjected to degradation by SPOP when both degrons 1 and 2 were deleted (ΔDeg1+2) (Figures 4B and S4A). These data suggest that degron 1 is the major SPOP binding site, while degron 2 plays a dispensable role in SPOP-mediated ERG destruction. Consistently, compared with wild-type, deletion of degron 2 only moderately reduced, while deletion of degron 1 or both degrons dramatically decreased ERG interaction with SPOP in vivo and in vitro (Figures 4C and S4B). These results confirm degron 1 as the major functional degron in conferring SPOP-mediated destruction of ERG, thus ΔDeg1+2 and ΔDeg1 mutants behaved similarly in most assays we examined (Figures 4B, 4C, 4G and S4A, S4B). However, to be on the conservative side eliminating any possible contribution of degron 2 to ERG stability, ΔDeg1+2 mutant was used as the non-degradable version of ERG for biochemical and cellular assays hereafter. Moreover, in support of the critical role of identified degrons in mediating SPOP-dependent degradation of ERG, the half-life of ΔDeg1+2 was significantly extended compared with wild-type (Figures 4D and 4E), and the ubiquitination of degron-deleted ERG was reduced in vitro and in vivo (Figures 4F and 4G). Next, we examined whether SPOP-mediated ERG degradation plays any physiological role in prostate cancer. Notably, ectopic expression of either ERG-WT or ΔDeg1+2 led to elevated mRNA levels of ERG targets (Figure S4C and Table S1) and as a result, significantly increased cell migration (Figures 4I and 4J). More importantly, co-expression with SPOP could suppress ERG-WT, but not ΔDeg1+2-mediated enhancement of cell migration (Figures 4I and 4J), which may be explained by the observation that ectopic expression of SPOP led to a significant reduction in the expression levels of ERG-WT, but not ΔDeg1+2 in vivo (Figure 4H).
Figure 4. SPOP promotes ERG ubiquitination and destruction depending on the degron motif.
A. Sequence alignment of ERG with the SPOP binding motif (degron) in known SPOP substrates.
B. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with indicated constructs.
C. IB analysis of WCL and GST pull down products derived from 293T cells transfected with indicated constructs. Cells were treated with 10 µM MG132 for 10 hr before harvesting.
D. IB analysis of WCL derived from 293T cells transfected with indicated HA-ERG plasmids. Cells were treated with 100 µg/ml cycloheximide (CHX) for the indicated time period before harvesting.
E. ERG protein abundance in (D) was quantified by ImageJ and plotted as indicated.
F. The SPOP/Cullin 3 complex promotes ERG ubiquitination in vitro. Please refer to supplemental information for experimental details.
G. IB analysis of WCL and His tag pull-down products derived from PC3 cells transfected with plasmids expressing the indicated proteins.
H. IB analysis of PC3 cells transfected with the indicated constructs.
I-J. Representative images of migrated PC3 cells transfected with indicated constructs in migration assay (I) and quantification of migrated cells (J). Data was shown as mean ± SD for three independent experiments. **p < 0.001, t-test.
See also Figure S4
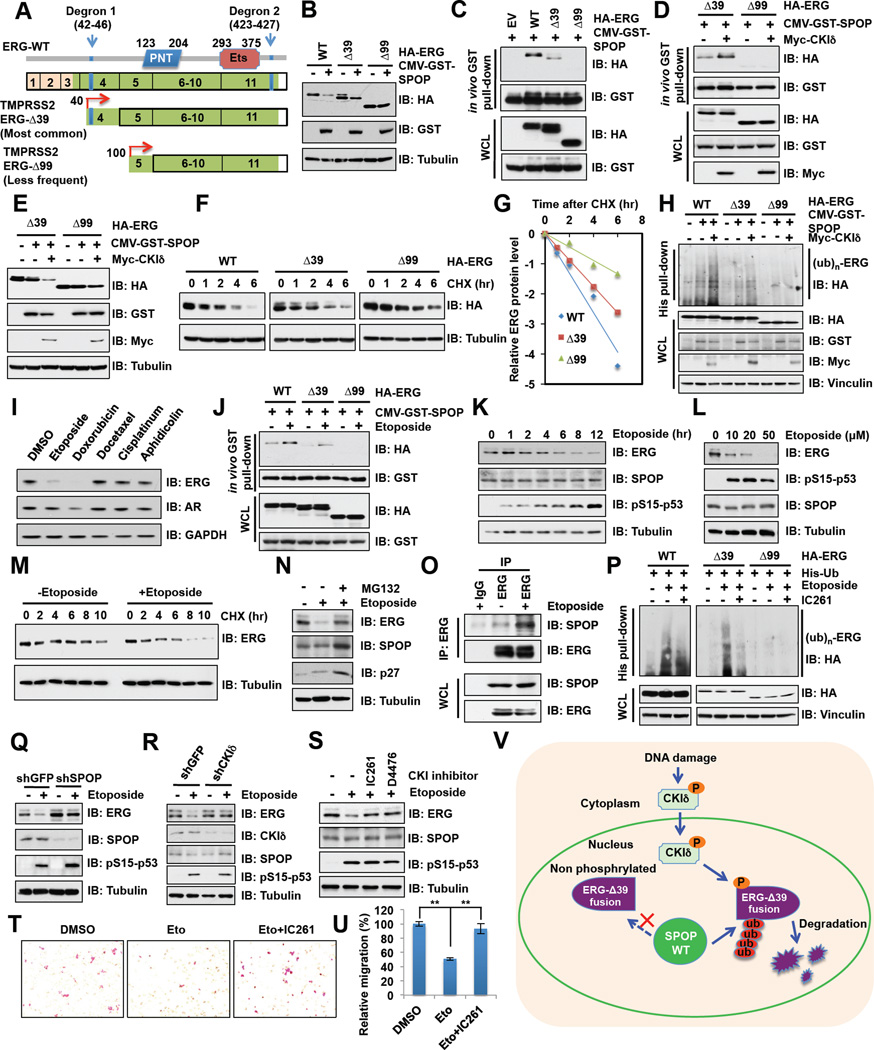
Figure 6. Etoposide-induced degradation of TMPRSS2-ERG fusion proteins is dependent on SPOP and CKIδ.
A. Schematic of major putative TMPRSS2-ERG fusion protein products and the degrons.
B. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with the indicated constructs.
C-D. IB analysis of WCL and GST pull down products derived from 293T cells transfected with indicated constructs. Cells were treated with 10 µM MG132 for 10 hr before harvesting.
E-G. IB analysis of WCL derived from 293T cells transfected with the indicated constructs demonstrate that CKIδ triggers SPOP-mediated degradation of the ERG-Δ39 fusion product (E) by shortening its half-life measured by CHX chase assay (F-G).
H. In vivo ubiquitination assay analysis of WCL and His tag pull-down products derived from PC3 cells transfected with the indicated plasmids.
I. IB analysis of VCaP cells treated with various DNA damage drugs for 12 hr.
J. In vivo GST-pull down analysis to demonstrate that etoposide treatment restore SPOP interaction with the ERG-Δ39 fusion product.
K-L. IB analysis of WCL derived from PC3 cells treated with 20 µM etoposide for indicated time (K) or treated with etoposide for 12 hr (L) before harvesting.
M. IB analysis of VCaP cells treated with etoposide for 12 hr before performing CHX chase analysis.
N. IB analysis of VCaP cells treated with etoposide or together with MG132 for 12 hr before harvesting.
O. IB analysis of WCL and anti-ERG IP derived from VCaP cells.
P. IB analysis of WCL and His tag pull-down products derived from PC3 cells transfected with the indicated plasmids.
Q-R. IB analysis of WCL derived from VCaP cells infected with the indicated lentiviral shRNA constructs. Cells were treated with 20 µM etoposide for 12 hr before harvesting.
S. IB analysis of VCaP cells treated with etoposide and CKI inhibitors IC261 (50 µM) and D4476 (20 µM) for 12 hr before harvesting.
T-U. Representative images of migrated VCaP cells treated with etoposide (Eto) or together with CKI inhibitor (IC261) in migration assay and quantification of migrated cells in (T). Data was shown as mean ± SD for three independent experiments. **p < 0.001, t-test.
V. A schematic illustration of the proposed model of mechanistically how DNA damaging agents including etoposide could promote nuclear translocation of CKIδ, thereby triggering CKI-dependent phosphorylation of the otherwise non-recognizable degron present in the ERG-Δ39 fusion product, restoring its interaction with SPOP and subsequent ubiquitination and degradation by the Cullin 3/SPOP E3 ligase.
See also Figure S6
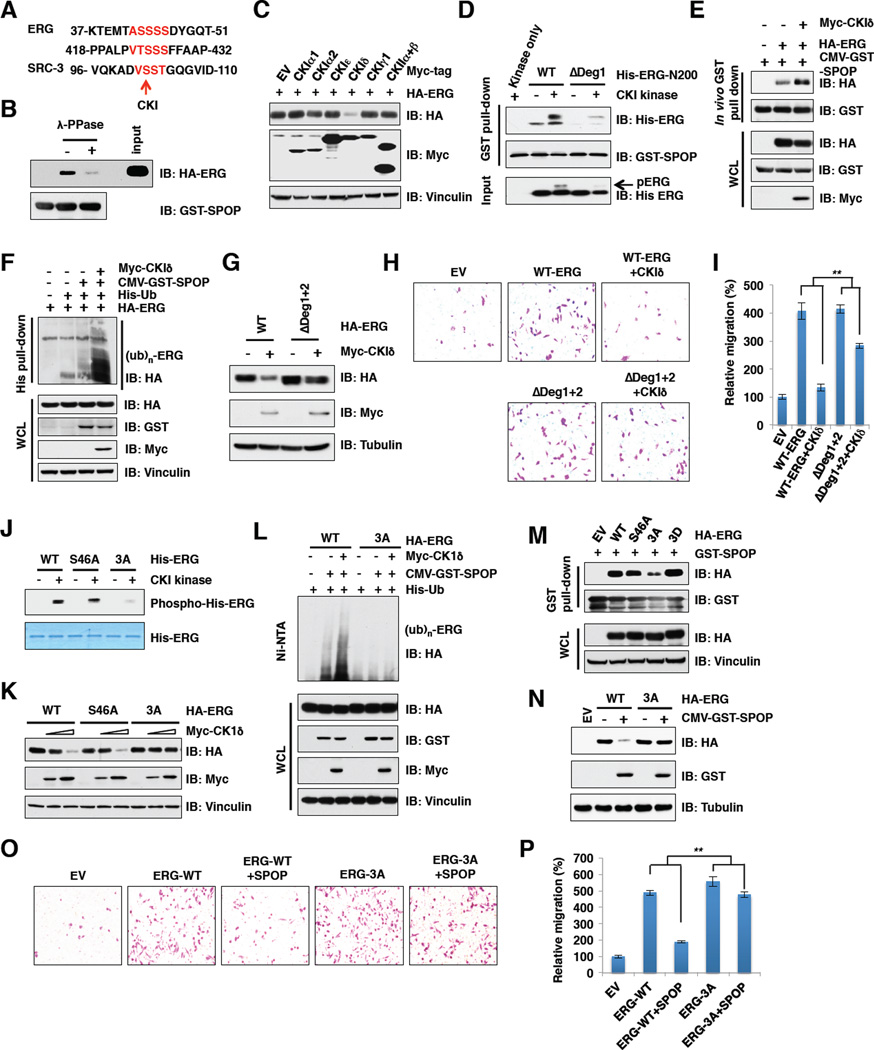
Casein kinase I phosphorylates ERG in the degron 1 to trigger SPOP/ERG interaction and promote degradation of ERG
Although proper substrate phosphorylation is required for recognition by many well-studied SCF type of E3 ubiquitin ligases, such as β-TRCP (Cardozo and Pagano, 2004) and FBW7 (Welcker and Clurman, 2008), it is unclear whether similar modifications are needed for Cullin 3-based SPOP E3 ligase recognition of its substrates. In support of this notion, a recent study identified that SPOP promoted SRC-3 degradation in a CKIe-dependent manner (Li et al., 2011). In keeping with the fact that ERG and SRC-3 share a similar degron sequence with a stretch of Ser/Thr residues (Figure 5A), we found that the interaction of ERG with SPOP was significantly reduced upon lambda protein phosphatase (λ-PPase) treatment (Figure 5B), indicating that phosphorylation of ERG may facilitate ERG and SPOP interaction. Next, we attempted to identify the kinase(s) responsible for ERG phosphorylation. The Scansite program (http://scansite.mit.edu) predicted that the Ser/Thr residues in the ERG degrons are likely CKI or CKII sites. Interestingly, only CKId, but not other CKI isoforms or CKII could promote ERG degradation under ectopic expression conditions (Figure 5C). In contrast, we found that the CKI inhibitor IC261 or D4476 treatment resulted in accumulation of ERG (Figure S5A). Importantly, using in vitro kinase assays, we further demonstrated that deletion of degron 1 largely abolished CKI-mediated phosphorylation of ERG (Figure S5B), indicating that serine residues within degron 1 are the major CKI phosphorylation sites.
Figure 5. CKI triggers the SPOP and ERG interaction to promote ERG degradation.
A. Sequence alignment of ERG with the phospho-degron in SRC-3, a known SPOP substrate.
B. In vitro GST pull-down assay demonstrate that SPOP/ERG interaction is phosphorylation dependent.
C. Immunoblot (IB) analysis of 293T cells transfected with indicated constructs.
D. GST-SPOP proteins purified from 293T cells were pulled-down with bacterially purified His-ERG-N200 prior treated with or without CKI kinase for 30 min. The samples were subjected to IB analysis.
E. IB analysis of WCL and IP derived from 293T cells transfected with indicated plasmids. Cells were treated with 10 µM MG132 for 10 hr before harvesting.
F. IB analysis of WCL and His tag pull-down products derived from PC3 cells transfected with the indicated plasmids.
G. IB analysis of 293T cells transfected with the indicated constructs.
H-I. Representative images of migrated PC3 cells transfected with indicated constructs in migration assay (H) and quantification of migrated cells (I). Data was shown as mean ± SD for three independent experiments. **p < 0.001, t-test.
J. In vitro kinase assays to demonstrate that ERG-3A mutant cannot be phosphorylated by CKI.
K. IB analysis of 293T cells transfected with the indicated constructs.
L. IB analysis of WCL and His tag pull-down products derived from PC3 cells transfected with the indicated plasmids.
M. In vitro GST-pull down assays to demonstrate a decreased interaction between GST-SPOP and the ERG-3A mutant.
N. IB analysis of 293T cells transfected with the indicated constructs.
O-P. Representative images of migrated PC3 cells transfected with indicated constructs in migration assays (O) and quantification of migrated cells (P). Data was shown as mean + SD for three independent experiments. **p < 0.001, t-test.
See also Figure S5
Consistent with these results, CKI inhibitor treatment significantly decreased SPOP and ERG interaction (Figures S5C and S5D), whereas phosphorylation of ERG within degron 1 by CKI in vitro or co-overexpression of CKIδ in cells enhanced the association of ERG with SPOP to promote ERG ubiquitination (Figures 5D-5F). Importantly, ΔDeg1+2 exhibited significant resistance to CKIδ-mediated ERG degradation (Figure 5G). These results indicate that phosphorylation of ERG by CKIδ within the SPOP-recognition degron triggers its interaction with SPOP to promote ERG destruction (Figure S5M). More importantly, when expressed at comparable levels, CKIδ could suppress ERG-WT, but not ΔDeg1+2-mediated enhancement of cell migration (Figures 5G-5I). Moreover, cell migration was significantly enhanced by inactivating CKIδ via either depleting endogenous CKIδ (Figures S5E–G) or the use of CKI inhibitors (Figures S5H and S5I). On the other hand, overexpression of CKIδ could conversely, inhibit cell migration in a dose-dependent manner (Figures S5J–L).
In support of serine residues in degron 1 being major CKI-mediated phosphorylation sites, in vivo Ser46 phosphorylation is detected by high-resolution tandem mass spectrometry (LC-MS/MS) analysis (Figure S5N). However, mutating S46 to an alanine alone could not diminish CKI-mediated ERG phosphorylation in vitro (Figure 5J), indicating that other serine residues within degron 1 may be CKI phosphorylation sites. In support of this notion, mutating S44, S45 and S46 to alanines (ERG-3A) largely abolished CKI-mediated phosphorylation of ERG in vitro (Figure 5J). Consistently, the ERG-3A mutant displayed a marked elevation of resistance to CKIδ-mediated ERG degradation (Figure 5K) and deficiency in SPOP/CKI-mediated ubiquitination in cells (Figure 5L). As a result, ERG-3A displayed significantly reduced interaction, whereas the phospho-mimetic mutant, ERG-3D, exhibited relatively enhanced interaction, with SPOP in cells (Figure 5M). As a consequence, SPOP could efficiently suppress cell migration induced by ERG-WT, but not ERG-3A (Figures 5N–P). These results coherently suggest that CKI functions as the upstream modifying kinase that phosphorylates multiple serine residues within degron 1 and subsequently enhances SPOP-mediated degradation of ERG to govern its biological functions.
Impaired association between SPOP and TMPRSS2-ERG fusion proteins can be restored by CKI-mediated phosphorylation
The most frequently detected ERG fusion transcripts are the TMPRSS2 exon 1 fused to ERG exon 4 or exon 5, with exon 4 fusion being the predominant form (Clark et al., 2007). Both fusions encode N-terminally truncated ERG proteins, which lack the first 39 or 99 amino acids (designated ERG-Δ39 and ERG- Δ99, respectively) (Figure 6A). As a result, the TMPRSS2-ERG fusion results in androgen-induced overexpression of ERG at both mRNA and protein levels, which facilitates prostate cancer progression largely by promoting cell migration and invasion (Carver et al., 2009b; Tomlins et al., 2008).
Given that the ETV1 fusion protein is insensitive to COP1-mediated degradation (Vitari et al., 2011), we next examined whether these ERG fusion proteins are resistant to SPOP-mediated degradation. Notably, in keeping with our observation that degron 1 is the major degron while degron 2 is largely nonfunctional in mediating SPOP-dependent ERG degradation (Figures 4A-4C), ERG- Δ99 that lacks degron 1 could not be degraded by SPOP (Figure 6B). However, although ERG- Δ39 contains both degrons, it displayed significant resistance to SPOP-mediated degradation (Figure 6B). Mechanistically, ERG-D39 displayed significantly reduced capability, while ERG- Δ99 completely failed, to interact with SPOP both in vivo and in vitro (Figures 6C and S6A). As the identified degron 1 (aa 42–46) is in close proximity to the fusion break point, it is possible that deletion of the first 39 amino acids may cause a conformational change, leading to the masking of the otherwise recognizable degron 1. This led us to hypothesize that unlike ERG-Δ99 that lacks degron 1, the interaction of ERG-Δ39 with SPOP might be restored upon the re-exposure of degron 1. In keeping with this notion, we found that CKI-dependent phosphorylation of degron 1, which could enhance SPOP interaction with WT-ERG (Figures 5D and 5E), could also facilitate the interaction between SPOP and ERG-Δ39 (Figure 6D). Furthermore, ectopic expression of CKIδ could trigger SPOP-dependent degradation (Figure 6E) and ubiquitination of ERG-Δ39, but not ERG-Δ99 (Figure 6H). Consistent with these results, the half-life of ERG-Δ39 and ERG-Δ99 was significantly extended (Figures 6F and 6G). Clinically, this is of significant importance, as fusion between TMPRSS2 and ERG exon 4 comprises the largest population of fusion-positive prostate cancer cases and restoration of SPOP-mediated degradation of this ERG fusion protein could therefore inhibit tumorigenesis promoted by such a gene fusion in a large patient population.
Etoposide promotes the degradation of ERG fusion proteins in a SPOP and CKIδ dependent manner
However, due to the current lack of CKI agonists, we went on to explore whether DNA damaging agents, which have been reported to activate CKIδ in part by triggering its nuclear localization (Alsheich-Bartok et al., 2008; Wang et al., 2012), could also promote SPOP-mediated destruction of TMPRSS2-ERG protein. Indeed, we found that DNA damaging drugs, mainly topoisomerase inhibitors including etoposide and doxorubicin, could trigger CKIδ nuclear translocation (Figure S6B) and significantly reduced the protein levels of TMPRSS2-ERG fusion in VCaP cells (Figure 6I), in part by restoring the interaction between SPOP and TMPRSS2-ERG (Figure 6J). On the other hand, docetaxel, a clinically well-established anti-mitotic chemotherapy drug for prostate cancer (Tannock et al., 2004), had no significant effect on the abundance of TMPRSS2-ERG fusion protein (Figure 6I). Since androgen receptor protein levels were also moderately changed upon doxorubicin, but not etoposide treatment (Figure 6I), which might affect ERG mRNA levels, we chose to focus on etoposide to further study how DNA damage might govern ERG stability in the reminder of the study. Notably, we found that etoposide down-regulated TMPRSS2-ERG protein levels in VCaP cells in both a time and dose-dependent manner (Figures 6K and 6L) largely through shortening its protein half-life (Figures 6M and S6C). More importantly, etoposide-induced ERG reduction could be blocked by MG132 (Figure 6N), indicating that etoposide regulates ERG expression largely in an ubiquitination-dependent post-translational manner. Consistently, we found that etoposide treatment led to an enhanced association of TMPRSS2-ERG with endogenous SPOP (Figure 6O), which subsequently resulted in an elevated ubiquitination of TMPRSS2-ERG that could be reduced by treatment with the CKI inhibitor, IC261 (Figure 6P).
Consistent with our finding of SPOP as an E3 ubiquitin ligase in controlling ERG stability, we found that depletion of endogenous SPOP largely abolished etoposide-triggered degradation of both TMPRSS2-ERG (Figure 6Q) and WT-ERG (Figure S6D). Notably, although inhibiting CKI in prostate cancer cell lines with WT-ERG background resulted in accumulation of ERG-WT (Figure S5A) and enhancement of cell migration (Figures S5H and S5I), CKI inhibitors did not significantly affect TMPRSS2-ERG protein levels and cell migration in VCaP cells (Figures S6E–F). This is in keeping with the notion that TMPRSS2-ERG fusion escapes the SPOP/CKI degradation unless cells are challenged with etoposide to restore SPOP/CKI-mediated ERG ubiquitination (Figure 6V). Consistently, inactivation of CKIδ by either depletion of CKIδ (Figure 6R) or the use of CKI inhibitors IC261 and D4476 (Figure 6S) efficiently blocked etoposide-induced TMPRSS2-ERG degradation.
Moreover, consistent with the observation that CKI-dependent phosphorylation facilitates SPOP-mediated degradation of WT-ERG, we found that etoposide also triggered the degradation of endogenous WT-ERG in both PC3 and LNCaP cells (Figures S6G and S6H), a process that could be blocked by treatment with CKI inhibitor (Figures S6I and S6J). As a result, in VCaP cells harboring ERG fusion (Figures 6T and 6U), as well as in WT-ERG expressing PC3 (Figures S6K and S6L) and LNCaP cells (Figures S6M and S6N), cell migration ability was significantly decreased upon etoposide treatment, which could be further restored by CKI inhibitor. Moreover, depleting SPOP attenuated the effects of etoposide in suppressing prostate cancer cell migration (Figures S6O and S6P), illustrating a critical physiological role for SPOP in mediating etoposide-induced destruction of ERG. Altogether, these results suggest that etoposide could suppress prostate cancer cell migration by promoting the degradation of both ERG-WT and ERG fusion proteins in a SPOP and CKIδ dependent manner.
Discussion
The recurrent ERG fusion with TMPRSS2 and other partners discovered by the Chinnaiyan group have been widely considered as one of the most important molecular findings in prostate cancer in the past several decades (Tomlins et al., 2005). Importantly, it is an early and frequent event (over 50% of prostate cancer). Thus, it is well accepted that ERG overexpression plays a pivotal role in promoting prostate cancer progression (Rubin et al., 2011). Here we provide experimental evidence demonstrating that the E3 ubiquitin ligase SPOP plays a critical tumor suppressive role in prostate cancer by controlling ERG oncoprotein stability.
Although previous studies and our results (Figure S6Q) indicated that SPOP could indirectly regulate the expression of ERG by targeting AR ubiquitination and degradation (An et al., 2014; Geng et al., 2014), our work provides evidence that SPOP could also directly control the protein levels of ERG in a post-translational manner. Consistently, mutations at the E3 ubiquitin ligase (SPOP) level to disrupt SPOP/ERG interaction, or fusions at the substrate (ERG) level to impair the degron, can prevent SPOP-mediated destruction of the ERG oncoprotein, leading to stabilization of ERG (Figure S2K). Moreover, it is widely accepted that ERG fusion events are predominant in prostate cancer and most ERG fusions lost the first 3 or 4 exons (Clark and Cooper, 2009). Thus, our current study provides a possible mechanism to explain why ERG fusion proteins are more stable, in part by evading SPOP-mediated degradation.
Furthermore, we found that SPOP mutations and ERG fusion share common gene signatures in clinical specimens (Figures 3M-3O). These results further support the notion that aberrant activation of ERG signaling, either by genetic fusion events to shed off the degron sequences in ERG, or by mutating the upstream SPOP E3 ubiquitin ligase, will lead to the activation of a cohort of common substrates to favor prostate cancer development (Figure S3P). Therefore, our study provides a possible molecular mechanism underlying the mutual exclusivity of SPOP mutation and ERG fusion.
However, as the TMPRSS2-ERG gene product alone is not sufficient to drive prostate tumorigenesis (Carver et al., 2009b; Tomlins et al., 2008), it is possible that SPOP mutation might affect ERG and other oncogenic pathways, or cooperates with other genetic alterations such as PTEN loss to facilitate prostate tumorigenesis (Carver et al., 2009b; King et al., 2009). Nonetheless, given the prevalence of TMPRSS2-ERG fusion in prostate cancer and its role in prostate cancer progression, it is of significance that DNA damaging drugs such as etoposide facilitates the degradation of ERG fusion proteins in part by promoting nuclear accumulation of CKIδ to trigger SPOP-mediated degradation of ERG fusion proteins (Figure 6V). Importantly, as ERG fusions have been reported to be mutually exclusive with SPOP mutations (Barbieri et al., 2012), the preferable presence of WT-SPOP in most ERG-fusion prostate cancer cases makes it possible to restore SPOP-mediated ubiquitination and degradation of TMPRSS2-ERG as a prostate cancer treatment strategy. Moreover, we identified that cells stably expressing SPOP mutants or the non-degradable ERG-Δ99 fusion protein, but not ERG-WT or ERG-Δ39, displayed resistance to etoposide-induced suppression of cell migration (Figures S6R–V). These results indicate that in clinical settings, deficiencies in SPOP-mediated ERG degradation in patients with either SPOP mutation or ERG-Δ99 fusion may prevent a desirable clinical outcome. To this end, optimal treatment strategy based on genetic status may provide better and personalized clinical treatments for individual prostate cancer patient.
Experimental Procedures
In vivo ubiquitination assay
PC3 cells were transfected with His-ub and the desired constructs. Thirty-six hours post-transfection, cells were treated with 20 µM MG132 for 6 hr. Cells were lysed in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 µM imidazole, pH 8.0) and sonicated. The lysates were incubated with Ni-NTA matrices (Qiagen) for 3 hr at room temperature. The His pull-down products were washed twice with buffer A, twice with buffer A/TI (1 volume buffer A and 3 volume buffer TI) and one time with buffer TI (25 mM Tris-HCl, 20 mM imidazole, pH 6.8). The pull-down proteins were resolved by SDS-PAGE for immunoblotting.
Migration and invasion assay
For cell migration, 2×104 to 2×105 cells were plated in an 8.0 µm 24-well plate chamber insert (Corning, 3422) with serum-free medium on the top of insert and 3T3 conditional medium containing 10% FBS at the bottom of the insert. Cells were incubated for 24 hr and fixed with 4% paraformaldehyde for 15 min. After washed with PBS, cells on the top of the insert were scraped with a cotton swab. Cells adherent to the bottom were stained with 0.5% crystal violet blue for 15 min and then washed with ddH2O. The positive staining cells were examined under microscope. For cell invasion assay, Corning BioCoat GFR Matrigel Invasion Chambers (354483) were used instead of chamber insert used in migration assay. The following steps were performed as migration assay described above.
In vitro kinase assay
In vitro kinase assays were adapted from described previously (Inuzuka et al., 2010). Briefly, about 2 µg of bacterially purified ERG was incubated with CKI kinase in the presence of 1 µM ATP and kinase reaction buffer (10 µM Tris-HCl, pH7.5, 10 mM MgCl2, 0.1 mM EDTA and 2 mM DTT) at 30°C for 30 min.
Real-time RT-PCR analysis
In vitro kinase assays were performed according to the protocol as described (Inuzuka et al., 2010) with minor modifications.
Tissue microarray (TMA), Immunohistochemistry (IHC) and FISH
The construction of tissue microarray (TMA) from cases of prostatectomy has been described previously (Huang et al., 2005). Briefly, cases of prostatectomy (n=239) were reviewed and the areas of PrCa and benign prostate tissue circled separately. Three cores of PrCa and benign prostate were taken from each case and transferred to two recipient blocks to construct the TMAs. A 5 µm section was cut from each of the TMA blocks and used for immunohistochemical (IHC) study. Anti-ERG antibody was purchased from Epitomics (AC-0105, clone EP111), and the IHC was performed as described previously (Park et al., 2010). Detection of TMPRSS2-ERG gene fusion by Fluorescence In-Situ Hybridization (FISH) has been described previously (Schelling et al., 2013).
Supplementary Material
Highlights.
The E3 ubiquitin ligase SPOP promotes ERG degradation in a CKI-dependent manner
Prostate cancer-associated SPOP mutants fail to promote ERG destruction
ERG fusion proteins evade SPOP-mediated degradation and could be restored by CKI
Etoposide-induced ERG fusion protein degradation depends on SPOP and CKI
Acknowledgments
We thank, Brian North, Alan W Lau, Lixin Wan and Jianping Guo for critical reading of the manuscript, Pengbo Zhou for providing valuable reagents, Min Yuan and Susanne Breitkopf for help with mass spectrometry experiments. W. W. is an ACS research scholar. This work was supported in part by the NIH grants (W.W., GM094777 and CA177910) and 1S10OD010612 (J.M.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
W.G., X.D., A.L. and Z.L. designed and performed most of the experiments with assistance from H.I., P.L., S.V., J.Z., L.C. and Y.S. J.A. performed mass spectrometry analysis. A.B. performed the bioinformatics analysis. W.W., P.P.P. and J.H. guided and supervised the study. W.G. and W.W. wrote the manuscript. All authors commented on the manuscript.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Alsheich-Bartok O, Haupt S, Alkalay-Snir I, Saito S, Appella E, Haupt Y. PML enhances the regulation of p53 by CK1 in response to DNA damage. Oncogene. 2008;27:3653–3661. doi: 10.1038/sj.onc.1211036. [DOI] [PubMed] [Google Scholar]
- An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–669. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Carver BS, Tran J, Chen Z, Carracedo-Perez A, Alimonti A, Nardella C, Gopalan A, Scardino PT, Cordon-Cardo C, Gerald W, et al. ETS rearrangements and prostate cancer initiation. Nature. 2009a;457:E1. doi: 10.1038/nature07738. discussion E2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009b;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, Eeles R, Martin FL, Phillips DH, Crundwell M, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–439. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, Zimmermann M, Bond R, Shou J, Li C, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng C, Rajapakshe K, Shah SS, Shou J, Eedunuri VK, Foley C, Fiskus W, Rajendran M, Chew SA, Zimmermann M, et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 2014;74:5631–5643. doi: 10.1158/0008-5472.CAN-14-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 2013;32:2307–2320. doi: 10.1038/emboj.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Yao JL, Zhang L, Bourne PA, Quinn AM, di Sant’Agnese PA, Reeder JE. Differential expression of interleukin-8 and its receptors in the neuroendocrine and non-neuroendocrine compartments of prostate cancer. The American journal of pathology. 2005;166:1807–1815. doi: 10.1016/S0002-9440(10)62490-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, O’Malley BW. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Lindberg J, Klevebring D, Liu W, Neiman M, Xu J, Wiklund P, Wiklund F, Mills IG, Egevad L, Gronberg H. Exome sequencing of prostate cancer supports the hypothesis of independent tumour origins. Eur Urol. 2013;63:347–353. doi: 10.1016/j.eururo.2012.03.050. [DOI] [PubMed] [Google Scholar]
- Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. The American journal of surgical pathology. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelling LA, Williamson SR, Zhang S, Yao JL, Wang M, Huang J, Montironi R, Lopez-Beltran A, Emerson RE, Idrees MT, et al. Frequent TMPRSS2-ERG rearrangement in prostatic small cell carcinoma detected by fluorescence in situ hybridization: the superiority of fluorescence in situ hybridization over ERG immunohistochemistry. Human pathology. 2013;44:2227–2233. doi: 10.1016/j.humpath.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, Oudard S, Theodore C, James ND, Turesson I, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- Thalmann GN, Sikes RA, Devoll RE, Kiefer JA, Markwalder R, Klima I, Farach-Carson CM, Studer UE, Chung LW. Osteopontin: possible role in prostate cancer progression. Clin Cancer Res. 1999;5:2271–2277. [PubMed] [Google Scholar]
- Theurillat JP, Udeshi ND, Errington WJ, Svinkina T, Baca SC, Pop M, Wild PJ, Blattner M, Groner AC, Rubin MA, et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346:85–89. doi: 10.1126/science.1250255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Vitari AC, Leong KG, Newton K, Yee C, O’Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- Wang S, Kollipara RK, Srivastava N, Li R, Ravindranathan P, Hernandez E, Freeman E, Humphries CG, Kapur P, Lotan Y, et al. Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4251–4256. doi: 10.1073/pnas.1322198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Inuzuka H, Zhong J, Fukushima H, Wan L, Liu P, Wei W. DNA damage-induced activation of ATM promotes beta-TRCP-mediated Mdm2 ubiquitination and destruction. Oncotarget. 2012;3:1026–1035. doi: 10.18632/oncotarget.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.