Abstract
A carbapenem-resistant Enterobacter cloacae strain, WCHECl-14653, causing a fatal bloodstream infection, was characterized by genome sequencing and conjugation experiments. The strain carried two carbapenemase genes, blaNDM-1 and blaKPC-2, on separate IncF plasmids. The coexistence of blaNDM-1 and blaKPC-2 conferred slightly higher-level carbapenem resistance compared with that of blaNDM-1 or blaKPC-2 alone, and the coexistence of two IncF plasmids may generate new platforms for spreading carbapenemase genes.
TEXT
Carbapenem-resistant Enterobacteriaceae (CRE) are a serious challenge for clinical management and public health. Klebsiella pneumoniae carbapenemase (KPC) and NDM represent two major families of carbapenemases, which are able to confer resistance to almost all β-lactams, including carbapenems, in the Enterobacteriaceae. Although isolates producing either KPC or NDM enzymes have been reported worldwide, isolates producing both KPC and NDM enzymes rarely have been detected in the same strain before (1–3). The coproduction of two major carbapenemases in a single strain may be significant, as it may confer higher-level resistance to carbapenems. During the ongoing surveillance of CRE in our hospital, an Enterobacter cloacae isolate was found to simultaneously produce a KPC and an NDM carbapenemase and is reported here.
Strain WCHECl-14653 was recovered from a blood sample from a 44-year-old male patient with severe pancreatitis in the intensive care unit (ICU) of West China Hospital, Chengdu, China, in April 2014. The patient had no travel history in the year prior to his admission to the ICU. The strain was identified as E. cloacae using the Vitek II system (bioMérieux, Durham, NC, USA), and species identification was confirmed by partially sequencing the gyrB gene, as described previously (4). In vitro antimicrobial susceptibility tests were performed using the Vitek II system, and the MICs of amikacin, ceftazidime, ciprofloxacin, colistin, imipenem, meropenem, and tigecycline were also determined using the microdilution broth method, according to recommendations of the Clinical and Laboratory Standards Institute (CLSI) (5). Breakpoints defined by CLSI were applied, except for tigecycline and colistin, for which those defined by EUCAST were used. WCHECl-14653 was resistant to ceftazidime (MIC, >512 μg/ml), ciprofloxacin (64 μg/ml), imipenem (128 μg/ml), meropenem (128 μg/ml), and tigecycline (8 μg/ml) but susceptible to amikacin (16 μg/ml) and colistin (2 μg/ml). WCHECl-14653 was also resistant to ampicillin-sulbactam, aztreonam, cefepime, ertapenem, gentamicin, levofloxacin, nitrofurantoin, piperacillin-tazobactam, tobramycin, and trimethoprim-sulfamethoxazole, as determined by the Vitek II system.
Acquired carbapenemase-encoding genes blaGES, blaIMP, blaKPC, blaNDM, blaOXA-48, and blaVIM were screened using PCR, as described previously (6–9). Both blaNDM and blaKPC were detected in WCHECl-14653. The complete coding sequences of blaNDM and blaKPC were obtained with additional primers (6, 10), and sequencing revealed the presence of blaNDM-1 and blaKPC-2.
Conjugation experiments were performed using azide-resistant Escherichia coli strain J53 as the recipient. Potential transconjugants were selected on medium containing 2 μg/ml meropenem and 150 μg/ml sodium azide. Additionally, plasmid DNA from WCHECl-14653 was prepared by alkaline lysis and then electroporated into E. coli strain DH5α. Potential transformants were selected on medium containing 2 μg/ml meropenem for selection. Transconjugants and transformants were screened for blaNDM-1 and blaKPC-2 by PCR. blaNDM-1 was transferred by conjugation, but blaKPC-2 was not. In contrast, transformants containing blaNDM-1 or blaKPC-2 or both genes were obtained via electroporation. The self-transmissible plasmid carrying blaNDM-1 was designated pNDM1_EC14653, while the non-self-transmissible plasmid carrying blaKPC-2 was designated pKPC2_EC14653. The MICs of ceftazidime, imipenem, and meropenem for DH5α, DH5α containing pNDM1_EC14653, DH5α containing pKPC2_EC14653, and DH5α containing both plasmids were determined using the CLSI microdilution method (5). The presence of two carbapenemase genes in a single strain conferred slightly increased resistance to carbapenems compared to that in the presence of either carbapenemase gene individually; a 2- to 4-fold increase in MIC was observed for imipenem and meropenem (Table 1). This might provide an advantage for the host strain to survive carbapenem exposure during treatment. The coexistence of two carbapenemase genes on separate plasmids may allow new platforms to be generated for mediating the spread of antimicrobial resistance genes.
TABLE 1.
MICs of carbapenems and ceftazidime against DH5α containing one or two carbapenemase gene-carrying plasmids
| Strain | MIC (μg/ml) of: |
||
|---|---|---|---|
| Ceftazidime | Imipenem | Meropenem | |
| E. coli DH5α | 0.5 | 0.06 | 0.12 |
| E. coli DH5α:pNDM1_EC14653 | >512 | 16 | 32 |
| E. coli DH5α:pKPC2_EC14653 | 64 | 16 | 16 |
| E. coli DH5α:pNDM1_EC14653:pKPC2_EC14653 | >512 | 32 | 64 |
| E. cloacae WCHECl-14653 | >512 | 128 | 128 |
Whole-genome sequencing of WCHECl-14653 was performed using the HiSeq 2500 sequencer (Illumina, San Diego, CA), with 100× coverage. Using the SPAdes program, reads were assembled into 151 contigs, of which 93 were ≥500 bp in length (N50, 236,790 bp) (11). Strain WCHECl-14653 was of the sequence type 177 (ST177), as determined using the genomic sequence to query the multilocus sequence typing database of E. cloacae (http://pubmlst.org/ecloacae/). ST177 is a single-allele variant of ST93, which comprises the E. cloacae strain ECNIH2 carrying blaKPC-2, recovered from a sink at the National Institutes of Health Clinical Center in 2012 (12). The sequences of pNDM1_EC14653 and pKPC2_EC14653 were completely circularized, with intervals between contigs being filled by PCR and Sanger sequencing.
The 109-kb pNDM1_EC14653 plasmid contained two IncF replicons, FIB (type 36) and FIIY (type 4). The backbone of pNDM1_EC14653 is identical to two FIB36:FIIY4 plasmids, pRJF866 (GenBank accession no. KF732966) from a K. pneumoniae from Shanghai, and pKOX_NDM1 (GenBank accession no. JQ314407) from a Klebsiella oxytoca isolate from Taiwan (13). The genetic context of blaNDM-1 on pNDM1_EC14653 is also almost identical to that on pRJF866 and pKOX_NDM1 (Fig. 1). In addition, the partially sequenced plasmid pKP-NCGM18-1 (GenBank accession no. AB824738) from a K. pneumoniae isolate from Nepal (14) is likely to be similar to pRJF866 and pKOX_NDM1, based on the genetic context of blaNDM-1 and its available sequence (13.4 kb), with only three nucleotide differences (Fig. 1). On these plasmids, blaNDM-1 and with commonly found downstream genes, i.e., ble-trpF-dsbC-cutA1-groL, are bracketed by two copies of a 256-bp miniature inverted-repeat transposable element (MITE) (small nonautonomous mobile element containing repeated sequences) (15). The MITE found here is AT rich (AT content, 57.8%), has a 39-bp imperfect inverted repeat (IR) (33 of the 39 bp matched), and contains no open reading frames. It has been suggested that the mobilization of blaNDM-1 onto the FIB36:FIIY4 plasmid was mediated by the transposition of this MITE (13). Compared to pRJF866 and pKOX_NDM1, which have three copies of the MITE, pNDM1_EC14653 has two copies. pRJF866 and pKOX_NDM1contain a large 11.5-kb region upstream of blaNDM-1, containing a 16S rRNA methylase gene, rmtC (conferring high-level resistance to aminoglycosides), an ISCR14-like element, tni-like transposase/integrase genes, sul1 (conferring resistance to sulfonamides), and a type II restriction-modification system that is not present on pNDM1_EC14653. The presence or absence of this 11.5-kb region on different plasmids is likely due to homologous recombination between two copies of the MITE. The same plasmid backbone and highly similar regions containing blaNDM-1 of pNDM1_EC14653, pRJF866, and pKOX_NDM1 suggests that the three plasmids evolved from a common ancestor and diverged recently.
FIG 1.
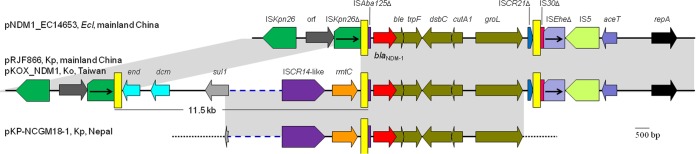
Genetic contexts of blaNDM-1 on pNDM1_ECl14563 and related plasmids. The plasmid names, host species (Ecl, Kp, and Ko stand for E. cloacae, K. pneumoniae, and K. oxytoca, respectively), and locations for each structure are listed. The GenBank accession numbers are KP868647 for pNDM1_EC14653, KF732966 for pRJF866, JQ314407 for pKOX_NDM1, and AB824738 for pKP-NCGM18-1. The identical regions are highlighted in gray. The 256-bp MITEs are indicated as yellow bars. Broken lines in blue, which are not scaled, represent structures that are not shown. Dotted lines in black represent sequences that are not available. end and dcm form a type II restriction-modification system and encode an endonuclease III and a DNA methyltransferase, respectively.
The 88-kb plasmid pKPC2_EC14653 had only a single FIIY replicon, which was classified as a new type, type 5, by a plasmid multilocus sequence typing (MLST) scheme (16). Although both pNDM1_EC14653 and pKPC2_EC14653 contain an FIIY replicon, it has been documented that two plasmids containing FII-like replicons could be compatible in the one isolate (17). FIIY replicons have been thought to originate from Yersinia spp., but it is evident now that they are also present in the Enterobacteriaceae. The FIIY5 replicon is also present on the blaKPC-2-carrying plasmid pKPC-727 of K. oxytoca strain KONIH1 (GenBank accession no. CP008791) (12) and the plasmid pNJST258C1, which does not carry blaKPC-2, of K. pneumoniae strain 30684/NJST258_2 (GenBank accession no. CP006922) (18). Based on available sequences, it appears that blaKPC-2 is commonly flanked by Tn3 upstream and by Tn1722 downstream, as the genetic context of blaKPC-2 on pKPC2_EC14653 is similar to several known plasmids found in isolates from China (Fig. 2). However, the Tn3-like transposon on pKPC2_EC14653 was interrupted by the insertion of ISKpn27 and was also truncated at its blaTEM end. The Tn3-blaKPC-2-Tn1722 region is flanked by various genes on different plasmids (Fig. 2), suggesting that the region has been mobilized onto different plasmids, although the mechanism mediating this event has not been determined. As Tn3 transposons are widespread among the Enterobacteriaceae, we speculate that the mobilization of the Tn3-blaKPC-2-Tn1722 region might be due to homologous recombination between different Tn3 transposons.
FIG 2.
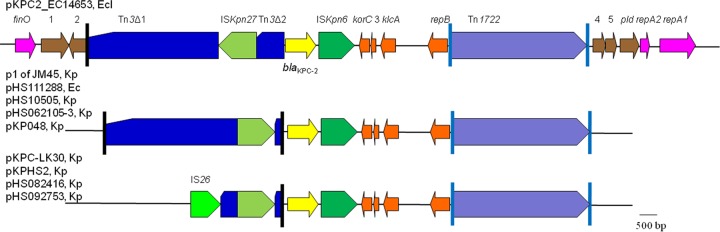
Genetic contexts of blaKPC-2 on pKPC1_ECl14653 and related plasmids. The plasmid names, host species (Ec, Ecl, and Kp stand for E. coli, E. cloacae, and K. pneumoniae, respectively) and locations of each structure are listed. The GenBank accession numbers are KP868647 for pKPC1_EC14653, CP006657 for plasmid p1 of K. pneumoniae strain JM45, KF826291 for pHS111288, KF826292 for pHS10505, KF623109 for pHS062105-3, FJ628167 for pKP048, KC405622 for pKPC-LK30, CP003224 for pKPHS2, KF724507 for pHS082416, and KF826293 for pHS092753. All of the plasmids were found in isolates from mainland China, except pKPC-LK30, which was from an isolate from Taiwan. The vertical lines are the IR of Tn3 (black) and Tn1722 (cyan). Open reading frames (ORFs) of unknown function are indicated by numbers 1 to 5.
The coexistence of blaNDM-1 and blaKPC-2 has been identified in three isolates of the Enterobacteriaceae (Table 2). In these isolates, blaNDM-1 was carried by IncA/C or IncP plasmids, which were generally self-transmissible, while blaKPC-2 was located either on non-self-transmissible plasmids or on a chromosome (Table 2). On an IncN plasmid of Enterobacter hormaechei strain CCBH14397, blaKPC-2 was carried by a Tn4401 transposon (1), which is different from the Tn3-blaKPC-2-Tn1722 structure on pKPC2_EC14653. Otherwise, the genetic contexts of blaNDM-1 and blaKPC-2 are unknown in the three isolates.
TABLE 2.
Strains carrying blaNDM-1 and blaKPC-2
| Strain | Species | Source | Yr | Country | Location of blaNDM-1 | Location of blaKPC-2 | Reference |
|---|---|---|---|---|---|---|---|
| IR98 | K. pneumoniae | Urine | 2010 | India | 160-kb self-transmissible IncA/C plasmid | 70-kb non-transmissible nontypeable plasmid | 3 |
| CCBH14397 | E. hormaechei | Rectal swab | 2013 | Brazil | 160-kb IncA/C plasmida | 50-kb IncN plasmida | 1 |
| E134 | E. cloacae | Blood | 2013 | Brazil | 30-kb IncP self-transmissible plasmid | Chromosome | 2 |
| WCHECl-14653 | E. cloacae | Blood | 2014 | China | 109-kb IncF self-transmissible plasmid | 88-kb IncF non-transmissible plasmid | This study |
Whether the plasmid was self-transmissible was not determined.
In conclusion, the study characterizes a clinical E. cloacae strain carrying both blaKPC-2 and blaNDM-1. The coexistence of two carbapenemase genes confers slightly increased resistance to carbapenems, and such a coexistence may generate new platforms to carry both genes and mediate their further spread.
Nucleotide sequence accession numbers.
The complete sequences of pKPC2_EC14653 and pNDM1_EC14653 have been deposited into GenBank under the accession numbers KP868646 and KP868647, respectively.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Natural Science Foundation of China (project no. 81222025), the New Century Excellent Talents Program, Ministry of Education, China (project no. NCET-13-0399), and a grant from the Sichuan Bureau of Science, China (project no. 2013JQ0042).
We thank Björn Espedido, University of Western Sydney, Australia, for his critical reading and thoughtful comments.
REFERENCES
- 1.Pereira PS, Borghi M, Albano RM, Lopes JC, Silveira MC, Marques EA, Oliveira JC, Asensi MD, Carvalho-Assef AP. 2015. Coproduction of NDM-1 and KPC-2 in Enterobacter hormaechei from Brazil. Microb Drug Resist 21:234–236. doi: 10.1089/mdr.2014.0171. [DOI] [PubMed] [Google Scholar]
- 2.Quiles MG, Rocchetti TT, Fehlberg LC, Kusano EJU, Chebabo A, Pereira RMG, Gales AC, Pignatari ACC. 2015. Unusual association of NDM-1 with KPC-2 and armA among Brazilian Enterobacteriaceae isolates. Braz J Med Biol Res 48:174–177. doi: 10.1590/1414-431X20144154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumarasamy K, Kalyanasundaram A. 2012. Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J Antimicrob Chemother 67:243–244. doi: 10.1093/jac/dkr431. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto S, Harayama S. 1995. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol 61:1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Zong Z, Zhang X. 2013. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J Antimicrob Chemother 68:1007–1010. doi: 10.1093/jac/dks505. [DOI] [PubMed] [Google Scholar]
- 7.Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, Pignatari AC, Tufik S. 2007. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol 45:544–547. doi: 10.1128/JCM.01728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother 44:622–632. doi: 10.1128/AAC.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin Infect Dis 39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Lu X, Zong Z. 2012. Enterobacteriaceae producing the KPC-2 carbapenemase from hospital sewage. Diagn Microbiol Infect Dis 73:204–206. doi: 10.1016/j.diagmicrobio.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Program NCS, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang TW, Wang JT, Lauderdale TL, Liao TL, Lai JF, Tan MC, Lin AC, Chen YT, Tsai SF, Chang SC. 2013. Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan. Antimicrob Agents Chemother 57:4072–4076. doi: 10.1128/AAC.02266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tada T, Miyoshi-Akiyama T, Dahal RK, Mishra SK, Ohara H, Shimada K, Kirikae T, Pokhrel BM. 2013. Dissemination of multidrug-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (NDM-1 and OXA-72) and 16S rRNA methylases (ArmA, RmtC and RmtF) in Nepal. Int J Antimicrob Agents 42:372–374. doi: 10.1016/j.ijantimicag.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Delihas N. 2008. Small mobile sequences in bacteria display diverse structure/function motifs. Mol Microbiol 67:475–481. doi: 10.1111/j.1365-2958.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 17.Froehlich B, Parkhill J, Sanders M, Quail MA, Scott JR. 2005. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J Bacteriol 187:6509–6516. doi: 10.1128/JB.187.18.6509-6516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]


