Abstract
Previous studies of antibiotic resistance dissemination by travel have, by targeting only a select number of cultivable bacterial species, omitted most of the human microbiome. Here, we used explorative shotgun metagenomic sequencing to address the abundance of >300 antibiotic resistance genes in fecal specimens from 35 Swedish students taken before and after exchange programs on the Indian peninsula or in Central Africa. All specimens were additionally cultured for extended-spectrum beta-lactamase (ESBL)-producing enterobacteria, and the isolates obtained were genome sequenced. The overall taxonomic diversity and composition of the gut microbiome remained stable before and after travel, but there was an increasing abundance of Proteobacteria in 25/35 students. The relative abundance of antibiotic resistance genes increased, most prominently for genes encoding resistance to sulfonamide (2.6-fold increase), trimethoprim (7.7-fold), and beta-lactams (2.6-fold). Importantly, the increase observed occurred without any antibiotic intake. Of 18 students visiting the Indian peninsula, 12 acquired ESBL-producing Escherichia coli, while none returning from Africa were positive. Despite deep sequencing efforts, the sensitivity of metagenomics was not sufficient to detect acquisition of the low-abundant genes responsible for the observed ESBL phenotype. In conclusion, metagenomic sequencing of the intestinal microbiome of Swedish students returning from exchange programs in Central Africa or the Indian peninsula showed increased abundance of genes encoding resistance to widely used antibiotics.
INTRODUCTION
The increasing prevalence of antibiotic resistance in clinically relevant pathogens has emerged as a major health crisis around the world (1). To mitigate the resistance problem, it is important to identify sources and dissemination routes of resistant bacteria and antibiotic resistance genes (2). Several human actions accelerate the global emergence and spread of antibiotic resistance, including inappropriate use of antimicrobial drugs, poor infection prevention and control within health care systems, poor control of antibiotic pollution of the environment, and international food trade and travel (3–7). Travel has been known for years to change antibiotic resistance patterns of bacteria residing in the human gut, in particular for species of the Enterobacteriaceae (8). In recent years, acquisition of Enterobacteriaceae strains resistant to cephalosporin antibiotics by production of extended-spectrum beta-lactamases (ESBLs) have been quantified in travelers from the Netherlands (9, 10), France (11), Australia (12), Sweden (13–15), Germany (16), and Finland (17). Certain risk factors, such as intake of antibiotics and a travel destination on the Indian peninsula, appear to increase the risk of acquisition (9, 10, 12–15, 17). Consequently, there is general concern over the contribution of travel to the global dispersal of antibiotic-resistant bacteria.
While the principal role of traveling in the dissemination of resistance genes is established, the research performed thus far has employed either a culturing approach, a PCR-based strategy, or a combination of the two. The studies have largely been focused on a select subset of Enterobacteriaceae species (18–21) or a small number of genes or antibiotics (10, 15, 22). Although many species of the Enterobacteriaceae are important human pathogens, it has repeatedly been shown that these bacteria usually constitute less than 1% of the human gut microbiota (23, 24). As the total bacterial community in the gut is considered a reservoir for antibiotic resistance genes (25, 26), a much larger fraction of the community may contribute to an antibiotic resistance gene catalogue associated with travel. The fast decrease in costs for DNA sequencing has allowed for large-scale investigations of the total DNA in bacterial communities, including in the gut—so-called metagenomics. This provides a tool to investigate, in a single experiment, virtually all known antibiotic resistance genes present in sufficient abundance (27–29). This enables detection of changes in the abundance of any resistance gene regardless of whether they occur in cultivable species or not and without the need of designing specific primers for every gene and gene variant.
We have applied explorative shotgun metagenomic sequencing to human feces from 35 students before and after travel from Sweden to the Indian peninsula or to Central Africa. Our study population of travelers was exposed to a variety of health care settings at each travel destination and environments where the prevalence of antibiotic-resistant bacteria is especially high, but the study population was not exposed to antibiotic treatments (30). Here, we present alterations in the taxonomic composition of the gut microbiota of study subjects and the abundance of >300 antibiotic resistance genes.
MATERIALS AND METHODS
For a more detailed materials and methods section, see the supplemental material.
Subject selection and specimen collection.
The study subjects were recruited among participants in a larger study in which Swedish students from the universities in Umeå, Stockholm, and Gothenburg traveling for international exchange studies were invited to participate from April 2010 through January 2014. The study protocol included responding to questionnaire data and self-submission of separate fecal specimens for metagenomic analysis and culture screening for ESBL-producing Enterobacteriaceae before and after travel. For the present study, only health care students (medical, nursing, and dentistry) traveling to the Indian peninsula or to Central Africa and having submitted a full set of fecal specimens qualified for inclusion. None of the subjects included were allowed to have taken antibiotics within 6 months prior to fecal sampling. The inclusion was consecutive with a target of 15 to 20 study subjects traveling to each of the two destinations. DNA was extracted from the fecal specimens and sequenced using Illumina HiSeq 2000 technology. The study was approved by the regional ethical review board in Umeå, Sweden (2011-357-32M).
Culture of ESBL-producing bacteria.
Screening for ESBL-producing Enterobacteriaceae was done using chromogenic culture media. All positive isolates were analyzed through culture-based methods according to EUCAST guidelines. Antibiotic susceptibility testing was done by disc diffusion on Mueller-Hinton agar. E-tests were used to test for the presence of the ESBLA phenotype (detecting the presence of CTX-M, SHV, and TEM enzymes). ESBL were defined as suggested by Giske et al. (31). Carbapenemase screening was performed with susceptibility testing for meropenem as well as temocillin or piperacillin-tazobactam as proposed by Huang et al. (32). Phenotypic species identification of ESBL-positive Enterobacteriaceae isolates was done using an API identification system. DNA was extracted from ESBL-positive isolates and sequenced using the Illumina MiSeq instrument (250-bp paired-end reads).
Bioinformatic analysis.
Reads were quality filtered using Trim Galore! version 0.2.8 (www.bioinformatics.babraham.ac.uk/projects/trim_galore/), and host sequences were subsequently removed from the metagenomic data by aligning the trimmed sequences to the hg19 human genome reference assembly using Bowtie2 (33). Isolate genomes were assembled using SPAdes (34), and resistance genes were identified using the Resqu database version 1.1 (http://www.1928diagnostics.com/resdb). Metagenomic reads were scanned for antibiotic resistance genes using Vmatch, allowing two mismatched amino acids per translated read (over at least 20 amino acids), with the Resqu database as a reference. They were also searched for small-subunit (SSU) rRNA sequences using Metaxa2 (35). Resistance gene abundances were normalized for gene length and number of bacterial 16S rRNA sequences in each library. Each sample was de novo assembled using Ray Méta (36). Open reading frames (ORFs) were predicted using Prodigal (37) and mapped to the Resqu database. Reads were mapped against the Human Microbiome Project (HMP) gastrointestinal tract reference genome collection using BWA (38) and mapped to the complete set of assembled contigs using Bowtie2.
Statistical analysis.
Significant changes in average abundances between before and after specimens were assessed using paired Student's t tests on log10-transformed values. The Wilcoxon signed-rank test was used as a complement to find genes with changed median abundance. All P values were corrected for multiple testing using a Benjamini-Hochberg false discovery rate (FDR), and genes with an FDR of <0.05 were considered significant (39). The same procedure was adapted for resistance gene categories and taxonomic groups. Correlations between resistance gene abundances and other factors (gender, age, length of visit, time between return from trip and sample delivery, health care work, sickness during travel, diarrhea during travel, use of malaria prophylaxis, and culture recovery of ESBL-producing bacteria) were assessed using linear regression in the R statistical program. The P values for each coefficient were corrected for multiple testing, and tests with an FDR of <0.05 were considered significant.
Nucleotide sequence accession number.
Sequence data have been submitted to the European Nucleotide Archive under accession number PRJEB7369.
RESULTS
Subject description.
In total 35 subjects were included and provided fecal specimens between April 2010 and May 2013. Seventeen traveled to Central Africa and 18 to the Indian peninsula (see Table S1 in the supplemental material). Subjects were 23 to 34 years old, with a gender distribution of 74% females (26/35). The median travel duration was 34 days (range 14 to 150). The fecal specimens were collected at a median of 4 days before departure (range 0 to 23) and 22 days (range 2 to 120) after return to Sweden. Two-thirds (23/35) participated in hospital-based patient work during their exchange. As many as 69% of subjects (12/17 in the Central Africa group and 12/18 in the Indian peninsula group) had travelers' diarrhea during their trip for a median of 4 days. Four out of 35 needed to seek medical care during the stay abroad, and one of them was hospitalized for 5 days due to a traffic accident. All travelers to Central Africa and 44% (8/18) of travelers to the Indian peninsula used antimalarial chemoprophylaxis. More detailed information is provided in Table S1 of the supplemental material.
Abundance of resistance genes in the gut metagenome.
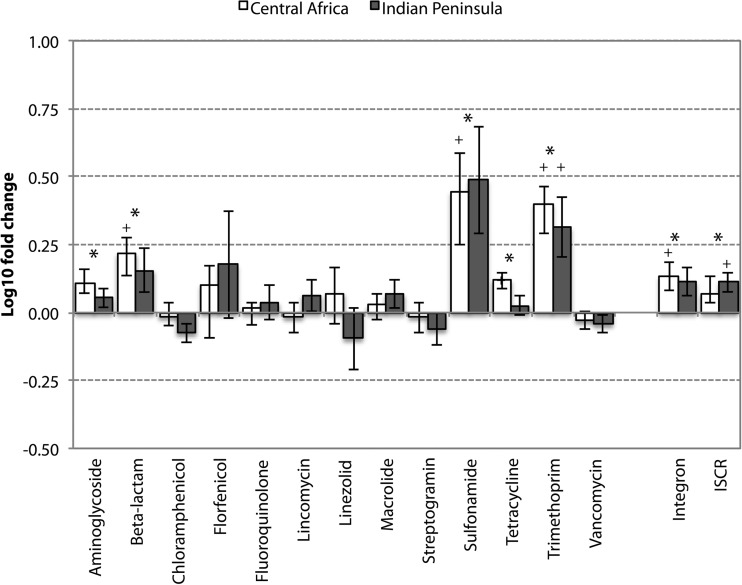
Seventy sequencing libraries in the total size range of 57,708,298 to 463,791,736 read pairs (11.5 to 92.8 Gbp) were generated from before and after specimens of the 35 individuals. After quality filtering, an average of 97,788,318 read pairs (40,538,343 to 178,004,607) remained in each library (8.1 to 35.6 Gbp) (see Table S2 in the supplemental material). The libraries were assembled separately, resulting in a total of 19,988,368 contigs, corresponding to 17.2 assembled Gbp (see Table S3 in the supplemental material). In total, 178 different resistance gene types (referred to hereafter as “resistance genes”) were detected across all specimens. Twenty-three resistance genes were found in all and 35 in ≥90% of the libraries (see Fig. S1a in the supplemental material). The overall fold change increase of resistance genes after international travel across all categories of antibiotics was a modest 1.06. Travel was, however, associated with significantly increased resistance gene abundances of five different categories encompassing genes conferring resistance to tetracyclines, aminoglycosides, beta-lactams, sulfonamides, and trimethoprim. Four additional categories showed increased abundance and another four categories decreased abundances, but none of these potential changes were statistically significant (Fig. 1). The largest significant fold changes in resistance gene abundance were the 2.6-fold increase for sulfonamide, 7.7-fold increase for trimethoprim, and 2.6-fold increase for beta-lactams (Table 1). Tetracycline resistance genes showed the largest significant increase in absolute resistance gene counts per 16S rRNA (0.2306 to 0.2980 resistance genes per 16S rRNA), followed by aminoglycoside resistance genes (0.0085 to 0.0129) and beta-lactam resistance genes (0.0012 to 0.0031). In addition, we observed significant increases of integrases and insertion sequence common region elements (ISCRs). Similar changes were found for the two destinations when assessed according to antibiotic categories. There were no genes with significant differences between the two travel destinations (Central Africa versus the Indian peninsula, all adjusted P values were >0.5). In analyses of travelers to Central Africa separately, the majority of the gene categories with significant changes across the entire cohort was significantly elevated. In analyses of travelers to the Indian peninsula separately, only the abundances of trimethoprim resistance genes and ISCR elements were significantly elevated (Fig. 1).
FIG 1.
Average fold change of resistance gene categories after travel (log10 scale). Changes in the entire cohort significant after correction for multiple testing are indicated with an asterisk. Significance within the Indian peninsula or the Central Africa group is indicated with a plus sign.
TABLE 1.
Abundance of resistance genes and mobile genetic elements before and after travel
| Resistance gene or mobile genetic element | Fold change | Abundance (resistance gene copies per bacterial 16S rRNA) |
||
|---|---|---|---|---|
| Before | After | Difference | ||
| Overall | 1.06 | 0.3207 | 0.3408 | 0.0201a |
| Aminoglycoside | 1.52 | 0.0085 | 0.0129 | 0.0044a |
| Beta-lactam | 2.62 | 0.0012 | 0.0031 | 0.0019a |
| Chloramphenicol | 0.68 | 0.0061 | 0.0042 | −0.0020 |
| Florfenicol | 26.41 | 0.0000 | 0.0001 | 0.0001 |
| Fluoroquinolone | 141.51 | 0.0000 | 0.0004 | 0.0004 |
| Lincomycin | 1.65 | 0.0034 | 0.0056 | 0.0022 |
| Linezolid | 0.89 | 0.0000 | 0.0000 | 0.0000 |
| Macrolide | 1.05 | 0.0322 | 0.0339 | 0.0017 |
| Streptogramin | 0.88 | 0.0011 | 0.0010 | −0.0001 |
| Sulfonamide | 2.61 | 0.0009 | 0.0023 | 0.0014a |
| Tetracycline | 1.04 | 0.2306 | 0.2398 | 0.0091a |
| Trimethoprim | 7.66 | 0.0001 | 0.0011 | 0.0010a |
| Vancomycin | 0.98 | 0.0361 | 0.0354 | −0.0007 |
| Integron | 3.20 | 0.0003 | 0.0008 | 0.0006a |
| ISCR | 6.44 | 0.0000 | 0.0002 | 0.0002a |
Changes after travel that were significant after correction for multiple testing.
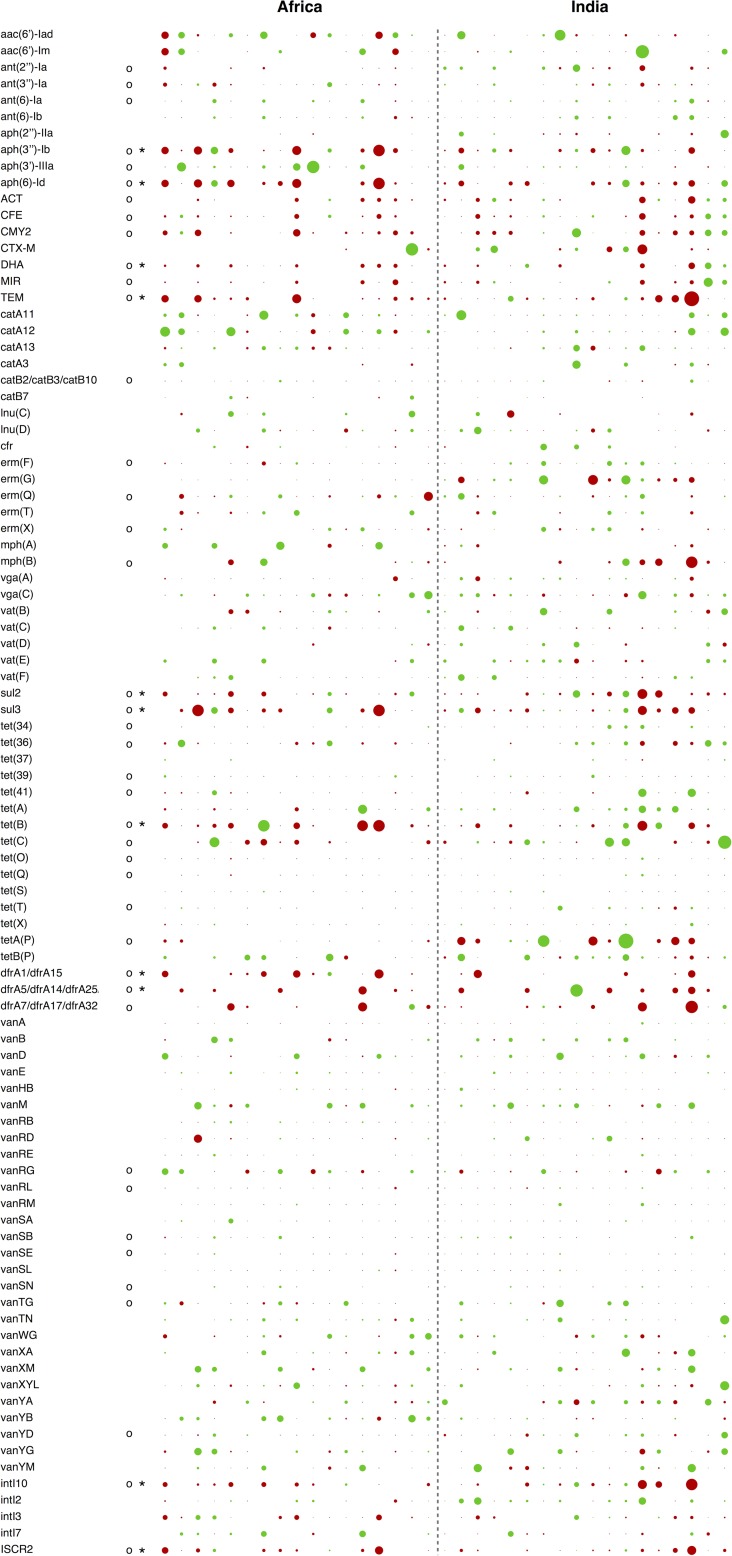
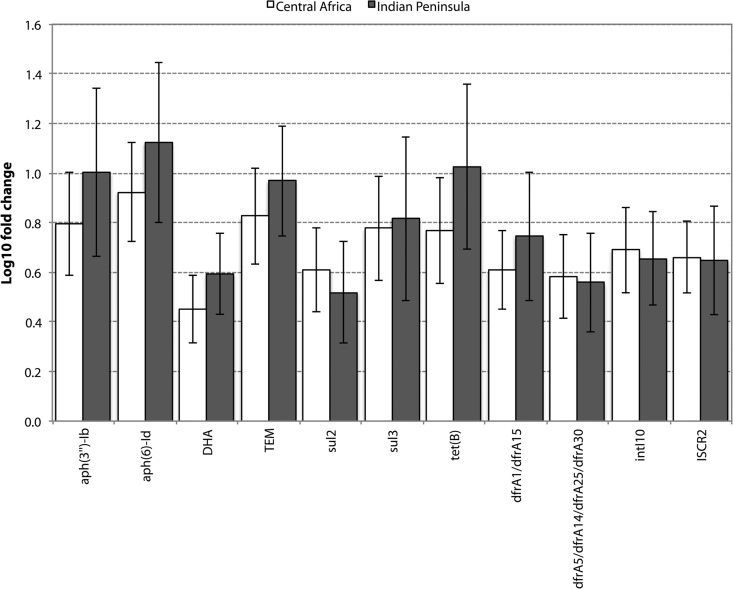
Among the 178 resistance genes detected across the full sample set, 93 were detected in at least 10 individuals and 9 of them exhibited a significant change in abundance (Fig. 2). These nine genes were all rare in absolute counts compared to the genes constituting the “core resistome” (see Fig. S1b in the supplemental material). In contrast, the most common resistance genes of the core resistome, e.g., vancomycin and tetracycline resistance genes, showed no systematic changes, with the exception of tet(B) (Fig. 2 and 3). Notably, the genes with significant increases included genes conferring resistance to clinically important antibiotics, such as beta-lactams (DHA and TEM beta-lactamases), trimethoprim (dfrA variants), and tetracyclines [tet(B)]. In addition, the commonly co-occurring resistance genes against streptomycin [aph(3″)-Ib and aph(6)-Id] and sulfonamides (sul2) were significantly increased after travel, along with intI10 integrases and ISCR2 elements (Fig. 3). A set of putatively increased resistance genes were significantly changed only under the Wilcoxon signed-rank test (Fig. 2). These may be enriched but to a lesser extent than the genes identified as significant using the two testing procedures. We did not detect any significant differences in resistance gene abundances between study subjects reporting traveler's diarrhea and those who did not (P value of 0.96 for total resistance gene abundance and adjusted P values for individual genes of >0.5). We controlled the resistance gene data for significant correlations to a number of other factors (previous international travel and/or previously living abroad, health care work at destination, illness during travel, use of malaria prophylaxis, gender, age, length of visit, and time passed between return to Sweden and post-travel sampling) but found no significant link between changes in resistance gene abundance and any of those factors after correction for multiple testing.
FIG 2.
Changes of resistance genes detected in at least 10 individuals. The diameter of each dot represents the magnitude of change in that individual (log10 scale). Green indicates decreases, and red indicates increases. Changes significant after correction for multiple testing are indicated with asterisks, while changes only significant using the Wilcoxon signed-rank test (putatively changed genes) are indicated by circles.
FIG 3.
Fold changes of resistance genes significantly changed after correction for multiple testing. All shown genes were significantly more abundant after travel, using correction for multiple testing when the two destinations were combined. Error bars represent standard error of the mean.
ESBL genes in enterobacteria detected by culture compared to metagenomic sequencing.
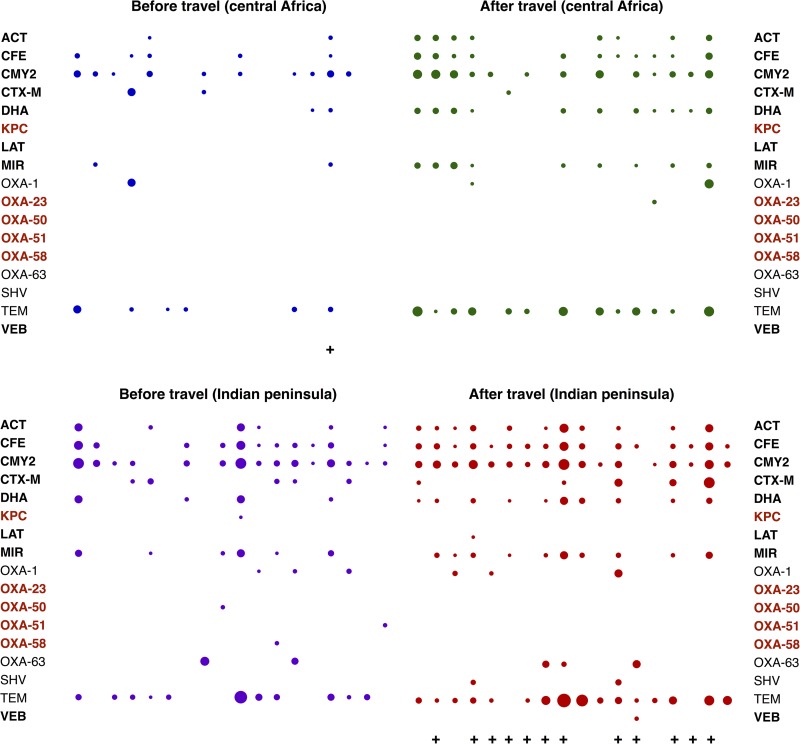
Despite increases of resistance genes in the metagenome being more consistent in the Central Africa group, no ESBL-positive strains were isolated among travelers returning from Africa. In the India group, however, 12/18 (67%) travelers carried ESBLA-positive isolates (all Escherichia coli) after their return (Fig. 4). Notably, no person in the India group had ESBL-positive isolates before travel. An ESBL-producing Escherichia coli isolate was detected in a specimen taken before travel from one of the subjects traveling to Central Africa but not after return. Whole-genome sequencing of the E. coli isolates with ESBL production revealed that all carried the CTX-M-15 gene. In addition, the majority of isolates carried additional beta-lactamases, most commonly OXA-1 and TEM (see Table S4 in the supplemental material). In all study subjects that acquired ESBL-producing E. coli, beta-lactamases associated with ESBL resistance were detected in the corresponding metagenomes, and all but one subject had acquired at least one ESBL gene that was not detected in the before specimens. Notably, the genes detected in the metagenomes were not the same as those present in the sequenced isolates, and in the majority of cases, metagenomics failed to detect any presence of the CTX-M gene. ESBL genes were frequently present in subjects without an ESBL-producing isolate, meaning that culturing for ESBL-producing Enterobacteriacae does not unambiguously predict the total resistance gene content and, conversely, detection of ESBL genes using metagenomics does not necessary imply identification of ESBL-resistant cultures. We found no evidence for acquisition of genes encoding carbapenemases by culturing or by metagenomic sequencing approaches.
FIG 4.
Abundance of beta-lactam resistance genes in all specimens (before and after). Specimens with ESBL-positive isolates are indicated by a plus sign. ESBL resistance gene names are shown in bold, while carbapenemase gene names are indicated in red. The diameter of each dot represents the relative abundance of that gene in that specimen (log10 scale).
Colocated resistance genes.
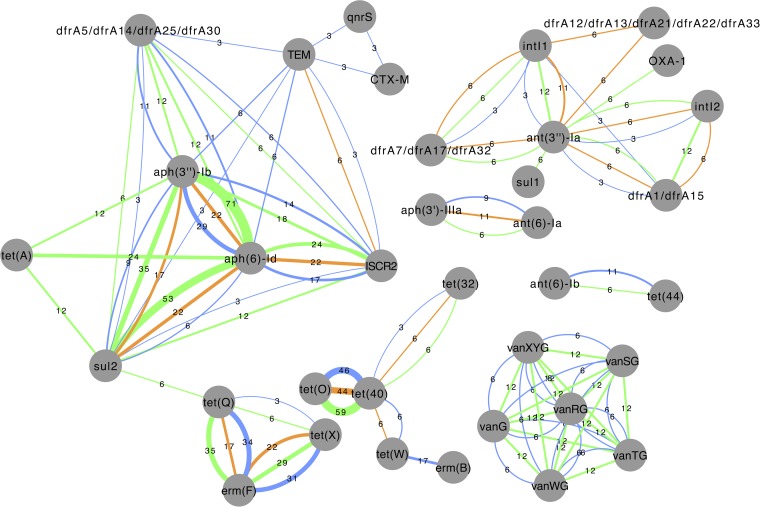
By performing de novo assemblies of the sequence reads from each specimen, we found that many of the significantly increased resistance genes were physically connected on the same assembled contigs. Such examples include the aph(3″)-Ib and aph(6)-Id genes, which were located together on contigs from 26 specimens (10 before and 16 after travel), many of which also included sul2 and the mobile element ISCR2, particularly after return (Fig. 5). Colocalization network analysis showed that the erm(F) gene often occurred together with either tet(Q) or tet(X) (21 and 20 occurrences, respectively). The two tetracycline resistance genes, however, were only rarely observed together. Furthermore, the two tetracycline resistance genes tet(40) and tet(O) were strongly connected to each other before and after travel, being detected on the same contig in 49% of the libraries. By colocalization network analysis, a cluster of vancomycin resistance genes was detected in two individuals both before their departure to Africa and after their return. The colocalization of the six vancomycin resistance genes is consistent with the fact that those genes constitute an operon, verifying that the assembly approach was feasible.
FIG 5.
Resistance genes colocalized on the same assembled contig. Blue edges represent contigs from before specimens, green edges correspond to Central Africa after specimens, and orange edges indicate contigs from the Indian peninsula after specimens. Numbers show the percentage of individuals where the co-occurring genes were detected.
The abundance of Proteobacteria was increased after travel.
The overall taxonomic composition of the gut bacterial community was highly variable between individuals but seemed to be only moderately affected by travel, regardless of destination (see Fig. S2a in the supplemental material). However, despite the fact that the Proteobacteria represented a minor fraction of the total human gut microbiota (on average, less than 4%) (see Fig. S2a in the supplemental material), the phylum showed a significant increase after return from travel (see Fig. S2b in the supplemental material). These increases were not significantly correlated with changes of resistance gene abundances. At the family and genus levels, taxonomic changes were relatively minor and, importantly, in most cases nonsystematic (see Fig. S3 and S4 in the supplemental material). The human gut enterotypes (23), driven by the Prevotella and Bacteroides genera, remained stable during the travel period for 29 of the 35 study subjects. There were no statistically significant changes in the abundances of specific bacterial genera, but the subjects returning from Africa had higher abundances of families within the Proteobacteria phylum (see Fig. S3 and S4 in the supplemental material). For example, the enterobacteria were increased after travel in the African group, particularly members of the Escherichia genus. This finding was further supported by the mapping of all reads to the gastrointestinal genomes from the Human Microbiome Project, which also showed elevated abundances of Escherichia coli and other Escherichia species, although none of those changes were statistically significant after correction for multiple testing.
DISCUSSION
In this study, we found an increase of antibiotic resistance gene abundance in Swedish students after completion of exchange programs in India or Central Africa by explorative metagenomic sequencing of fecal specimens. The most prominent increases were observed in less abundant genes encoding resistance to sulfonamide (2.6-fold), trimethoprim (7.7-fold), and beta-lactams (2.6-fold). The metagenomic results offer a new and broadened perspective on the changes of the antibiotic resistance potential of the human gut microbiome after travel, providing an independent complement to previous studies targeting a limited number of bacterial species or antibiotic resistance genes (11, 13–17, 21).
To avoid detection of large numbers of false-positive resistance gene matches, we used a highly stringent approach with regard to the definition of resistance genes. Our approach included the use of a database containing only resistance genes conferring a verified resistance phenotype and allowing only two mismatching amino acids between a sequence read and its best reference match (to account for sequencing errors). Such stringent criteria may introduce a risk of excluding resistance gene variants that are not yet described, but because resistance genes may show high sequence similarity to genes not conferring resistance, less stringent criteria would improperly inflate the number of predicted resistance genes. Our approach is in line with recent proposals made by Martínez et al. (40) and Bengtsson-Palme and Larsson (41) for the evaluation of risks associated with the presence of antibiotic resistance genes in bacterial metagenomes and includes addressing sequence context and the mobility potential of detected resistance genes.
To study the possible interrelationship between the metagenomic detection of resistance gene abundances on one hand and the demonstration of bacteria with specific resistance phenotypes on the other hand, we used culture to identify Enterobacteriaceae carrying genes encoding ESBLs. These genes, acquired by some students, were not always detected by metagenomic sequencing. Specifically, the CTX-M-15 genes of cultured E. coli were often missed despite using sequencing able to detect a resistance gene present in one out of ∼100,000 bacterial cells. Thus, the sensitivity of metagenomic sequencing may be insufficient for assessing low-abundant genes demonstrable by a specific assay used in routine bacteriological culture. An alternative approach using quantitative PCR (qPCR) assays or amplicon sequencing of specific gene families may have detected the rare resistance genes responsible for the ESBL phenotypes, but such approaches have important limitations in capturing the entire diversity of the human gut resistome because of their dependency on primer sequences targeting specific resistance genes.
Overall, a stable fecal microbiome across a time period of 15 to 150 days on the Indian peninsula or in Central Africa was found in our Swedish students. This is in line with results of previous studies of the microbiota of the human intestine, showing a remarkable stability over several months or even up to 5 years (42, 43). The present results strengthen the concept of long-term stability by showing a stable microbiome in Swedes spending a relatively long period in India or Africa.
We found that several bacterial families belonging to the Proteobacteria phylum exhibited larger abundance alterations than families within the Actinobacteria, Bacteroidetes, and Firmicutes phyla. An increase of Proteobacteria was previously reported to be associated with inflammatory conditions of the large intestine, including ulcerative colitis and Clostridium difficile enteritis, and in chronic HIV infection (44–47). Diarrhea may possibly have led to sufficient inflammation driving similar changes, but we found no significant relation to whether the subjects reported that they experienced travelers' diarrhea or not. Importantly, additional factors at the travel destination may be involved, such as exposure to the bacterial communities present in the destination environments and diet changes contributing to changes of the microbiome (48, 49).
The metagenomic data analysis suggested that there was a core resistome of high abundance genes that remained stable and a fraction of low abundance genes that increased after the completion of exchange programs. The lowly abundant genes conferred resistance toward sulfonamides, trimethoprim, and beta-lactams that are all well known for being part of the worldwide acceleration of the antibiotic resistance problem. The observation of increasing abundance of resistance genes in study subjects traveling from Sweden, a country with a low background of antimicrobial resistance, is in line with a previous study showing that the core resistomes at the population level are increasing over time since the introduction of different antibiotics and with antibiotic consumption levels in different countries (28). Some genes observed to increase in the present study have previously been associated with travel, such as the DHA beta-lactamase (15), while others—to our best knowledge—have not [e.g., tet(B), aph(3″)-Ib, aph(6)-Id, dfrA, sul2, and sul3].
The mechanisms behind the increased levels of resistance genes are unknown and are not likely to be explained solely by increased abundance of Proteobacteria carrying these genes. The absence of correlation between changes in Proteobacteria and resistance genes does not rule out taxonomic changes as a driver but suggests a more general enrichment of the resistome. The intake of antibiotics is also not a reason, since none of the subjects took antibiotics before or during travel. A caveat is the possible antibacterial effect of antimalaria agents, such as mefloquine, used by a majority of the students. Mefloquine has antibacterial effects on some Gram-positive bacteria, while proguanil, which was used by some of the study subjects, at least theoretically may have antibacterial action although with no known cross-resistance with true antibacterial agents (50, 51). Atovaquone is considered to have little antibacterial effect (50). We found no link between increased resistance abundance and the use of antimalaria agents but acknowledge that our study might be too small for detecting such an effect. Another reason for enrichment of resistance genes may be ingestion of resistant bacteria through food (52) or contaminated water (53–55) or by close contact with an environment containing antibiotic-resistant bacteria. The culture results provide support for the conclusion that bacteria carrying resistance genes were indeed taken up during travel, as ESBL-producing bacteria were detected by culture in only 1 of 35 individuals before travel while 12 of 18 students carried E. coli with the CTX-M-15 gene after their return from the Indian peninsula. Nine of the 12 detected CTX-M-15 genes in E. coli were found on DNA sequences that were identical or very similar to known plasmid sequences. The network analysis of resistance genes in the metagenomic data showed frequent colocalization with an integrase or an insertion sequence, giving indirect support for the conclusion that the changing part of the resistome often has potential for mobility.
In conclusion, the use of metagenomic shotgun sequencing provides a novel means for studies of the geographic spread of antibiotic resistance, revealing a previously unseen diversity of resistance genes being enriched in the gut microbiome of Swedish exchange students after visiting the Indian peninsula or Central Africa.
Supplementary Material
ACKNOWLEDGMENTS
D.G.J.L. acknowledges financial support from the Swedish Research Council (VR), the Swedish Research Council for Environment, Agriculture, and Spatial Planning (FORMAS), and the Swedish Foundation for Strategic Environmental Research (MISTRA). A.J. acknowledges financial support from Umeå University and Västerbotten County Council. M.H. and S.K. were supported by a grant from the Knut and Alice Wallenberg Foundation to the Wallenberg Advanced Bioinformatics Infrastructure. M.A. acknowledges financial support provided by Västerbotten County Council and the scholarship fund Stiftelsen JC Kempes Minnes Stipendiefond. J.B.-P. acknowledges financial support from the Adlerbertska Research Foundation. E.K. acknowledges financial support from the Swedish Research Council (VR) and the Life Science Area of Advance at Chalmers University of Technology.
We thank Birgitta Evengård, Margareta Granlund, Joakim Forsell, Maria Casserdahl, and Helén Edebro in their work with organizing the fecal specimen collection and Elin Nilsson for performing the Illumina sequencing of E. coli isolates. We thank Arne Tärnvik and the anonymous reviewers for critical comments on the manuscript, and we acknowledge support from the Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure.
We declare no competing financial interests.
A.J. and H.P. conceived the study. A.J. and D.G.J.L. designed the study. M.A., H.P., and A.J. arranged the sampling and questionnaires. M.A. performed the laboratory experiments. J.B.-P., E.K., M.H., and S.K. performed the bioinformatic and statistical analyses. J.B.-P., M.A., D.G.J.L., and A.J. interpreted the results and wrote the manuscript with contribution from all other authors. All authors have read and approved the final manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00933-15.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Ashbolt NJ, Amézquita A, Backhaus T, Borriello P, Brandt KK, Collignon P, Coors A, Finley R, Gaze WH, Heberer T, Lawrence JR, Larsson DGJ, McEwen SA, Ryan JJ, Schönfeld J, Silley P, Snape JR, Van den Eede C, Topp E. 2013. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ Health Perspect 121:993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernando GA, Collignon PJ, Bell JM. 2010. A risk for returned travellers: the “post-antibiotic era”. Med J Aust 193:59. [DOI] [PubMed] [Google Scholar]
- 4.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 5.Mutreja A. 2012. Bacterial frequent flyers. Nat Rev Microbiol 10:734. doi: 10.1038/nrmicro2899. [DOI] [PubMed] [Google Scholar]
- 6.Molton JS, Tambyah PA, Ang BS, Ling ML, Fisher DA. 2013. The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clin Infect Dis 56:1310–1318. doi: 10.1093/cid/cit020. [DOI] [PubMed] [Google Scholar]
- 7.Woerther P-L, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaarslev K, Stenderup J. 1985. Changes during travel in the composition and antibiotic resistance pattern of the intestinal Enterobacteriaceae flora: results from a study of mecillinam prophylaxis against travellers' diarrhoea. Curr Med Res Opin 9:384–387. doi: 10.1185/03007998509109608. [DOI] [PubMed] [Google Scholar]
- 9.Paltansing S, Vlot JA, Kraakman MEM, Mesman R, Bruijning ML, Bernards AT, Visser LG, Veldkamp KE. 2013. Extended-spectrum β-lactamase-producing Enterobacteriaceae among travelers from the Netherlands. Emerging Infect Dis 19:1206–1213. doi: 10.3201/eid1908.130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Wintersdorff CJ, Penders J, Stobberingh EE, Oude Lashof AML, Hoebe CJPA, Savelkoul PHM, Wolffs PFG. 2014. High rates of antimicrobial drug resistance gene acquisition after international travel, The Netherlands. Emerging Infect Dis 20:649–657. doi: 10.3201/eid2004.131718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruppé E, Armand-Lefèvre L, Estellat C, El-Mniai A, Boussadia Y, Consigny PH, Girard PM, Vittecoq D, Bouchaud O, Pialoux G, Esposito-Farèse M, Coignard B, Lucet JC, Andremont A, Matheron S. 2014. Acquisition of carbapenemase-producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Euro Surveill 19(14):1 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20768. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy K, Collignon P. 2010. Colonisation with Escherichia coli resistant to “critically important” antibiotics: a high risk for international travellers. Eur J Clin Microbiol Infect Dis 29:1501–1506. doi: 10.1007/s10096-010-1031-y. [DOI] [PubMed] [Google Scholar]
- 13.Tängdén T, Cars O, Melhus A, Löwdin E. 2010. Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum beta-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother 54:3564–3568. doi: 10.1128/AAC.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tham J, Odenholt I, Walder M, Brolund A, Ahl J, Melander E. 2010. Extended-spectrum beta-lactamase-producing Escherichia coli in patients with travellers' diarrhoea. Scand J Infect Dis 42:275–280. doi: 10.3109/00365540903493715. [DOI] [PubMed] [Google Scholar]
- 15.Östholm-Balkhed Å, Tärnberg M, Nilsson M, Nilsson LE, Hanberger H, Hällgren A, Travel Study Group of Southeast Sweden. 2013. Travel-associated faecal colonization with ESBL-producing Enterobacteriaceae: incidence and risk factors. J Antimicrob Chemother 68:2144–2153. doi: 10.1093/jac/dkt167. [DOI] [PubMed] [Google Scholar]
- 16.Lübbert C, Straube L, Stein C, Makarewicz O, Schubert S, Mössner J, Pletz MW, Rodloff AC. 2015. Colonization with extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacteriaceae in international travelers returning to Germany. Int J Med Microbiol 305:148–156. doi: 10.1016/j.ijmm.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Kantele A, Lääveri T, Mero S, Vilkman K, Pakkanen SH, Ollgren J, Antikainen J, Kirveskari J. 2015. Antimicrobials increase travelers' risk of colonization by extended-spectrum betalactamase-producing Enterobacteriaceae. Clin Infect Dis 60:837–846. doi: 10.1093/cid/ciu957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collard J-M, Place S, Denis O, Rodriguez-Villalobos H, Vrints M, Weill F-X, Baucheron S, Cloeckaert A, Struelens M, Bertrand S. 2007. Travel-acquired salmonellosis due to Salmonella Kentucky resistant to ciprofloxacin, ceftriaxone and co-trimoxazole and associated with treatment failure. J Antimicrob Chemother 60:190–192. doi: 10.1093/jac/dkm114. [DOI] [PubMed] [Google Scholar]
- 19.Al-Mashhadani M, Hewson R, Vivancos R, Keenan A, Beeching NJ, Wain J, Parry CM. 2011. Foreign travel and decreased ciprofloxacin susceptibility in Salmonella enterica infections. Emerging Infect Dis 17:123–125. doi: 10.3201/eid1701.100999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, Livermore DM, Woodford N. 2011. Variation in the genetic environments of bla(CTX-M-15) in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 66:1005–1012. doi: 10.1093/jac/dkr041. [DOI] [PubMed] [Google Scholar]
- 21.van der Bij AK, Pitout JDD. 2012. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother 67:2090–2100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 22.Patel U, Dasgupta P, Amoroso P, Challacombe B, Pilcher J, Kirby R. 2012. Infection after transrectal ultrasonography-guided prostate biopsy: increased relative risks after recent international travel or antibiotic use. BJU Int 109:1781–1785. doi: 10.1111/j.1464-410X.2011.10561.x. [DOI] [PubMed] [Google Scholar]
- 23.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sommer MO, Dantas G, Church GM. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommer MO, Church GM, Dantas G. 2010. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence 1:299–303. doi: 10.4161/viru.1.4.12010. [DOI] [PubMed] [Google Scholar]
- 27.Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegård B, Söderström H, Larsson DGJ. 2011. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One 6:e17038. doi: 10.1371/journal.pone.0017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forslund K, Sunagawa S, Kultima JR, Mende DR, Arumugam M, Typas A, Bork P. 2013. Country-specific antibiotic use practices impact the human gut resistome. Genome Res 23:1163–1169. doi: 10.1101/gr.155465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ. 2014. Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front Microbiol 5:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modi SR, Collins JJ, Relman DA. 2014. Antibiotics and the gut microbiota. J Clin Invest 124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giske CG, Sundsfjord AS, Kahlmeter G, Woodford N, Nordmann P, Paterson DL, Cantón R, Walsh TR. 2009. Redefining extended-spectrum beta-lactamases: balancing science and clinical need. J Antimicrob Chemother 63:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang T-D, Poirel L, Bogaerts P, Berhin C, Nordmann P, Glupczynski Y. 2014. Temocillin and piperacillin/tazobactam resistance by disc diffusion as antimicrobial surrogate markers for the detection of carbapenemase-producing Enterobacteriaceae in geographical areas with a high prevalence of OXA-48 producers. J Antimicrob Chemother 69:445–450. doi: 10.1093/jac/dkt367. [DOI] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH. 23 March 2015. Metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Res doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 36.Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. 2012. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol 13:R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc 57:289–300. [Google Scholar]
- 40.Martínez JL, Coque TM, Baquero F. 2015. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol 13:116–123. [DOI] [PubMed] [Google Scholar]
- 41.Bengtsson-Palme J, Larsson DGJ. 2015. Antibiotic resistance genes in the environment: prioritizing risks. Nat Rev Microbiol 13:396. doi: 10.1038/nrmicro3399-c1. [DOI] [PubMed] [Google Scholar]
- 42.Zoetendal EG, Akkermans AD, De Vos WM. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 64:3854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, Wanke CA, Ward HD. 2015. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS, Young VB. 2014. Recovery of the gut microbiome following fecal microbiota transplantation. mBio 5:e00893–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walujkar SA, Dhotre DP, Marathe NP, Lawate PS, Bharadwaj RS, Shouche YS. 2014. Characterization of bacterial community shift in human ulcerative colitis patients revealed by Illumina based 16S rRNA gene amplicon sequencing. Gut Pathog 6:22. doi: 10.1186/1757-4749-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winter SE, Bäumler AJ. 2014. Why related bacterial species bloom simultaneously in the gut: principles underlying the “like will to like” concept. Cell Microbiol 16:179–184. doi: 10.1111/cmi.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2013. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunin CM, Ellis WY. 2000. Antimicrobial activities of mefloquine and a series of related compounds. Antimicrob Agents Chemother 44:848–852. doi: 10.1128/AAC.44.4.848-852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grey D, Hamilton-Miller JM. 1977. Trimethoprim-resistant bacteria: cross-resistance patterns. Microbios 19:45–54. [PubMed] [Google Scholar]
- 52.Ruimy R, Brisabois A, Bernede C, Skurnik D, Barnat S, Arlet G, Momcilovic S, Elbaz S, Moury F, Vibet MA, Courvalin P, Guillemot D, Andremont A. 2010. Organic and conventional fruits and vegetables contain equivalent counts of Gram-negative bacteria expressing resistance to antibacterial agents. Environ Microbiol 12:608–615. doi: 10.1111/j.1462-2920.2009.02100.x. [DOI] [PubMed] [Google Scholar]
- 53.Amaya E, Reyes D, Paniagua M, Calderón S, Rashid MU, Colque P, Kühn I, Möllby R, Weintraub A, Nord CE. 2012. Antibiotic resistance patterns of Escherichia coli isolates from different aquatic environmental sources in León, Nicaragua. Clin Microbiol Infect 18:E347–E354. doi: 10.1111/j.1469-0691.2012.03930.x. [DOI] [PubMed] [Google Scholar]
- 54.De Boeck H, Miwanda B, Lunguya-Metila O, Muyembe-Tamfum JJ, Stobberingh E, Glupczynski Y, Jacobs J. 2012. ESBL-positive enterobacteria isolates in drinking water. Emerging Infect Dis 18:1019–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, Endtz HP, Islam MA. 2013. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One 8:e61090. doi: 10.1371/journal.pone.0061090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.