Abstract
A Salmonella enterica serovar Corvallis strain was isolated from a wild bird in Germany. This strain carried the IncA/C2 pRH-1238 plasmid. Complete sequencing of the plasmid was performed, identifying the blaNDM-1, blaCMY-16, fosA3, sul1, sul2, strA, strB, aac(6′)-Ib, aadA5, aphA6, tetA(A), mphA, floR, dfrA7, and merA genes, which confer clinically relevant resistance to most of the antimicrobial classes, including β-lactams with carbapenems, fosfomycin, aminoglycosides, co-trimoxazole, tetracyclines, and macrolides. The strain likely originated from the Asiatic region and was transferred to Germany through the Milvus migrans migratory route.
TEXT
The New Delhi metallo-β-lactamase (NDM) is one of the most widespread carbapenemases. NDM-producing strains have been identified worldwide in Enterobacteriaceae but also in Acinetobacter spp. and Pseudomonas aeruginosa from clinical cases and colonized patients (1). However, one of the most intriguing aspects of the wide diffusion of NDM is the emergence of positive carriers in livestock and in companion and wild animals, denouncing contamination by wildlife and the environment (2, 3). The blaNDM-1 gene has been identified in different genetic environments but is always associated with remnant portions of the Tn125 transposon (4). The blaNDM-1-carrying genetic determinants have been localized on a variety of rare and frequent plasmid types (5). Here, the complete sequence of the blaNDM-1-carrying IncA/C plasmid (pRH-1238), identified in a Salmonella enterica serovar Corvallis strain isolated from a wild bird (Milvus migrans) in Germany (6), was determined.
The NDM-carrying plasmid was transferred by conjugation from the S. Corvallis 12-1738 isolate to the sodium azide-resistant Escherichia coli K-12 strain J53, selecting transconjugants on cefoxitin (10 μg/ml) plus azide (100 μg/ml; MICs determined for donor and transconjugant strains are reported in Table S1 in the supplemental material). The transferred NDM-positive plasmid was classified as IncA/C by a PCR-based replicon typing method (7). The complete DNA sequence of the pRH-1238 plasmid was obtained using a 454-FLX genome sequencer (Roche Diagnostics, Monza, Milan, Italy) on a library obtained using plasmid DNA purified by the Invitrogen PureLink HiPure plasmid filter midiprep kit (Invitrogen, Milan, Italy), according to the manufacturer's protocol. Assembly of DNA sequences was done using the GS-FLX gsAssembler software, followed by PCR-based gap closure of contigs. Homology and phylogenetic trees were obtained by aligning the IncA/C DNA plasmid sequences downloaded from GenBank, using the DNAman phylogenetic analysis software (Lynnon BioSoft, Vaudreuil, Quebec, Canada) for quick alignment. Unrooted phylogenetic trees were generated by the maximum-likelihood method.
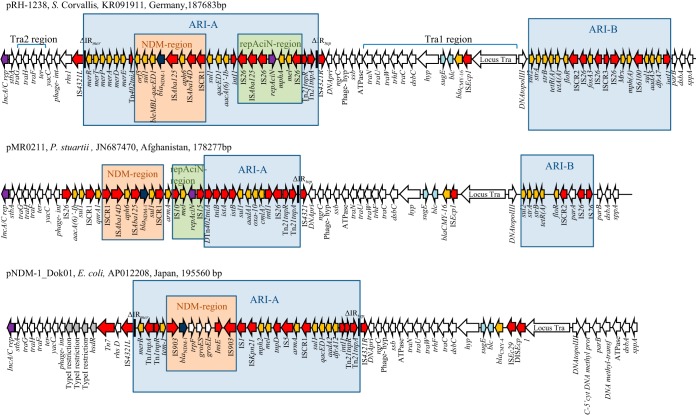
The pRH-1238 plasmid (GenBank accession no. KR091911) is 187,683 bp in size, with a G+C content of 51.7%, and it contains 173 predicted coding sequences (CDSs). pRH-1238 belongs to type 1 A/C2, as defined by the presence of the entire rhs gene in the ARI-A region (type 1 A/C2 reference plasmid pR148). pRH-1238 shows the two typical resistance islands, designated ARI-A and ARI-B, and the A/C2 replicon (8). Homology and phylogenetic analysis performed on the entire plasmid DNA sequence revealed high relatedness and 82% nucleotide identity with the plasmid pMR0211 (GenBank accession no. JN687470) identified in a Providencia stuartii strain isolated from a patient in Afghanistan (9). These two plasmids share an origin, which is different from the other NDM-IncA/C2 plasmids identified in the United States, Canada, Kenya, Australia, and Oman (Fig. 1 and 2). The best homology between the two plasmids is observed in the replicon, conjugation (Tra1 and Tra2 transfer regions), and ARI-B regions; both pRH-1238 and pMR0211 carried the RepAciN and NDM regions, localized in the ARI-A region (Fig. 2).
FIG 1.
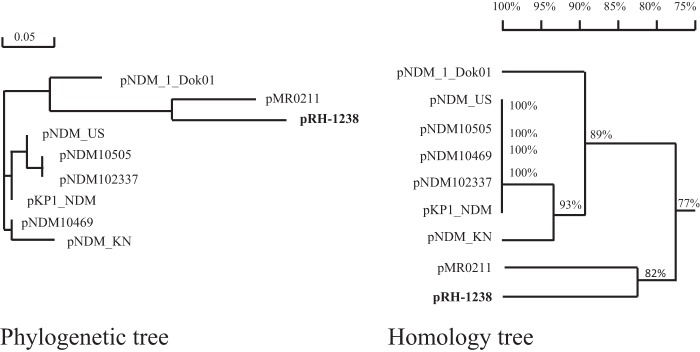
Phylogenetic and homology trees of IncA/C plasmids. The phylogenetic and homology analyses were performed by aligning sequences of the following NDM-1-IncA/C plasmids (GenBank accession numbers): pNDM-1_Dok01 (AP012208), pMR0211 (JN687470), pKP1-NDM-1 (KF992018), pNDM-US (CP006661), pNDM102337 (JF714412), pNDM10469 (JN861072), pNDM-KN (JN157804), and pNDM10505 (JF503991). The phylogenetic (left) and homology (right) trees were obtained using the DNAman software (Lynnon BioSoft, Vaudreuil, Quebec, Canada) for quick alignment. Unrooted phylogenetic trees were generated by the maximum-likelihood method. The bar corresponds to the scale of sequence divergence.
FIG 2.
Major structural features of pRH-1238 plasmid compared with pMR0211 and pNDM-1_Dok01 plasmids identified in P. stuartii and E. coli, respectively. Orange arrows indicate antibiotic resistance genes, and red arrows indicate transposon-related genes (tnpA and tnpR) or insertion sequences. The blaNDM-1 gene is indicated by blue arrows. The repAciN gene is indicated by violet arrows. The white arrows indicate plasmid scaffold regions in common among IncA/C plasmids. The black bars indicate inverted repeat (IR) sites.
pRH-1238 carried the ISEcp1-blaCMY-16 element located in the Tra1 region, while the blaNDM-1 gene was located within ARI-A. This resistance island contains a complete Tn21 transposon carrying the entire mer21 locus (Fig. 2). Most of the previously fully sequenced IncA/C plasmids (pMR0211, pKP1-NDM-1, pNDM-US, pNDM102337, pNDM10469, pNDM-KN and pNDM10505) carried truncated Tn21 transposons with deleted mer21 loci. The IRtnp and IRmer inverted repeats of the Tn21 transposon were interrupted by IS4321L and IS4321R elements in the opposite orientation (Fig. 2). These insertion sequences show target site specificity and have been found to interrupt the terminal ends of members of the Tn21-like subgroup transposons (10). The blaNDM-1 gene environment shows the ISCR1 and the Tn402 tniA transposase genes in a complex class 1 integron carrying the aac(6′)-Ib gene cassette (Fig. 2). Immediately upstream of the blaNDM-1 gene, the partial sequence of the remnant Tn125 with the ISAba125 element, the aphA6 gene, and a partial ISAba14 were identified, as previously described in pMR0211. Downstream of the blaNDM-1 gene, the bleMBL gene, encoding bleomycin resistance, fused with qacEΔ1, and the sul1 and orf5 genes were found. A similar blaNDM-1 genetic environment was previously identified in the chromosome of P. aeruginosa MMA83 strain (a comparison among the two structures is shown in Fig. 3) (11).
FIG 3.
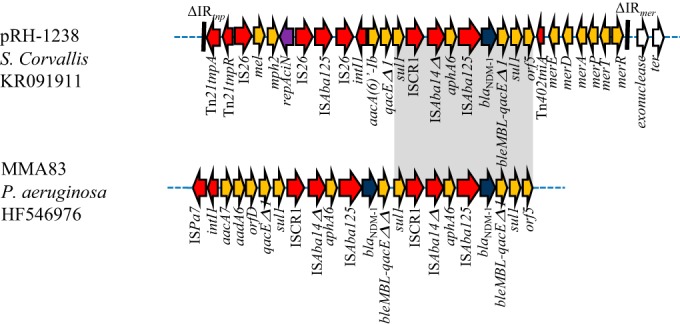
Comparison of the blaNDM-1 gene environments between pRH-1238 plasmid and the genetic context of the genome of the P. aeruginosa MMA83. Orange arrows indicate antibiotic resistance genes, and red arrows indicate transposon-related genes (tnpA and tnpR) or insertion sequences. The blaNDM-1 gene is indicated by blue arrows. The repAciN gene is indicated by the violet arrow. The white arrows indicate plasmid scaffold regions that are in common among IncA/C plasmids. The black bars indicate IR sites.
pRH-1238 contains the macrolide resistance determinant mphA-mel-repAciN flanked by two IS26 elements integrated in the Tn21 transposon of the ARI-A region (Fig. 2). The mphA-mel-repAciN module was previously described in IncA/C plasmids pVC1447 from Vibrio cholerae (GenBank accession no. KM083064), pPG010208 from E. coli (GenBank accession no. HQ023861), and pTR2 from Klebsiella pneumoniae (GenBank accession no. KJ187752), but it was located in the ARI-B region.
The ARI-B region of the pRH-1238 plasmid contains the sul2 (sulfonamide resistance), strA and strB (streptomycin resistance), tetR and tetA(A) (tetracycline resistance), floR (chloramphenicol/florfenicol resistance), and fosA3 (fosfomycin resistance) genes and a class 1 integron carrying the dfrA7 (trimethoprim resistance) and aadA5 (streptomycin resistance) gene cassettes (Fig. 2).
The fosA3-orf1-orf2-orf3 element, conferring resistance to fosfomycin, was identified on this plasmid, flanked by two IS26 elements, as previously described (12). FosA3 is rarely detected in Europe, while it is emerging among E. coli and in K. pneumoniae CTX-M-producing strains from animals and patients in China, Japan, and South Korea. It has been suggested that the increasing prevalence of fosA3 is due to dissemination of the IncI1 and IncN plasmids rather than clonal expansion of specific strains (13, 14). Recent reports from China described fosA3 on IncFII plasmids in S. enterica serovars Derby and Enteritidis from chickens purchased at the market in Hong Kong (15) and the copresence of both blaNDM-1 and fosA3 genes in a clinical E. coli isolate (16). Furthermore, the colocalization of the blaNDM-1 and fosA3 genes on IncA/C plasmids was demonstrated in E. coli and Citrobacter freundii clinical isolates in a report denouncing the high incidence and endemic spread of NDM-1-positive Enterobacteriaceae in the Henan Province of China (17).
pRH-1238 is the first completely sequenced blaNDM-1-fosA3-IncA/C plasmid and is of particular interest because it was identified in Salmonella from a wild animal in Germany (6). The report of this broad-host-range plasmid, which confers multidrug resistance in Salmonella identified in wildlife in Europe, is worrisome. This strain has been found thanks to the German Salmonella routine monitoring system, but it is possible that plasmids of this type are highly represented in other commensal bacteria that are not under surveillance. The structure of the plasmid, the presence of the fosA3 gene that is still so rare in Europe but frequent in China, and the differences observed with the other previously fully sequenced NDM-1-IncA/C2 plasmids identified in bacteria from Western countries are all clues supporting a possible origin of this plasmid in the Asiatic region. This strain probably traveled in the gut of the wild bird M. migrans, which is a migratory diurnal raptor, and its route of migration includes Europe and northern Asia during the summer and wintering in sub-Saharan Africa and southern Asia (18). The report in Germany of this M. migrans bird carrying the S. Corvallis strain with the NDM-1-fosA3-IncA/C plasmid suggests that new routes of environmental diffusion of multidrug-resistant bugs from Eastern to Western countries are possible, for instance, according to wild bird migratory paths.
Nucleotide sequence accession number.
The complete sequence of the pRH-1238 plasmid was deposited in GenBank under accession no. KR091911.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Pfeifer, Robert Koch Institut, Wernigerode, Germany, who kindly provided the fosfomycin MICs for our strains.
The project was supported by the Italian Flagship “InterOmics” project (PB.P05) funded by MIUR and coordinated by the Italian Council of National Research (CNR) (A.C.), the Federal Institute for Risk Assessment (grant 46-001), and the Ecology From Farm to Fork (EFFORT) project funded by the European Community’s Seventh Framework Programme (FP7/2007-2013; grant agreement 613754 to B.G.). This study is part of “The genomics of antimicrobial resistance in bacterial pathogens of relevance for human health” project (A.C. and Z.Z.), which is part of the Italy-China Significant Bilateral Projects sponsored by the Italian and Chinese Ministers of Foreign Affairs.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00944-15.
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerra B, Fischer J, Helmuth R. 2014. An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet Microbiol 171:290–297. doi: 10.1016/j.vetmic.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Woodford N, Wareham DW, Guerra B, Teale C. 2014. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother 69:287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 4.Bonnin RA, Poirel L, Nordmann P. 2014. New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol 9:33–41. doi: 10.2217/fmb.13.69. [DOI] [PubMed] [Google Scholar]
- 5.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Fischer J, Schmoger S, Jahn S, Helmuth R, Guerra B. 2013. NDM-1 carbapenemase-producing Salmonella enterica subsp. enterica serovar Corvallis isolated from a wild bird in Germany. J Antimicrob Chemother 68:2954–2956. doi: 10.1093/jac/dkt260. [DOI] [PubMed] [Google Scholar]
- 7.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Harmer CJ, Hall RM. 2014. pRMH760, a precursor of A/C2 plasmids carrying blaCMY and blaNDM genes. Microb Drug Resist 20:416–423. doi: 10.1089/mdr.2014.0012. [DOI] [PubMed] [Google Scholar]
- 9.McGann P, Hang J, Clifford RJ, Yang Y, Kwak YI, Kuschner RA, Lesho EP, Waterman PE. 2012. Complete sequence of a novel 178-kilobase plasmid carrying blaNDM-1 in a Providencia stuartii strain isolated in Afghanistan. Antimicrob Agents Chemother 56:1673–1679. doi: 10.1128/AAC.05604-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Partridge SR, Hall RM. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J Bacteriol 185:6371–6384. doi: 10.1128/JB.185.21.6371-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jovčić B, Lepsanović Z, Begović J, Rakonjac B, Perovanović J, Topisirović L, Kojić M. 2013. The clinical isolate Pseudomonas aeruginosa MMA83 carries two copies of the blaNDM-1 gene in a novel genetic context. Antimicrob Agents Chemother 57:3405–3407. doi: 10.1128/AAC.02312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. 2010. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother 54:3061–3064. doi: 10.1128/AAC.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SY, Park YJ, Yu JK, Jung S, Kim Y, Jeong SH, Arakawa Y. 2012. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J Antimicrob Chemother 67:2843–2847. doi: 10.1093/jac/dks319. [DOI] [PubMed] [Google Scholar]
- 14.Ho PL, Chan J, Lo WU, Law PY, Li Z, Lai EL, Chow KH. 2013. Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant Escherichia coli from livestock and other animals. J Appl Microbiol 114:695–702. doi: 10.1111/jam.12099. [DOI] [PubMed] [Google Scholar]
- 15.Lin D, Chen S. 2015. First detection of conjugative plasmid-borne fosfomycin resistance gene fosA3 in Salmonella isolates of food origin. Antimicrob Agents Chemother 59:1381–1383. doi: 10.1128/AAC.04750-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao JY, YQ Zhu, Li YN, Mu XD, You LP, Xu C, Qin P, Ma JL. 2015. Coexistence of SFO-1 and NDM-1 β-lactamase genes and fosfomycin resistance gene fosA3 in an Escherichia coli clinical isolate. FEMS Microbiol Lett 362:1–7. doi: 10.1093/femsle/fnu018. [DOI] [PubMed] [Google Scholar]
- 17.Qin S, Fu Y, Zhang Q, Qi H, Wen JG, Xu H, Xu L, Zeng L, Tian H, Rong L, Li Y, Shan L, Xu H, Yu Y, Feng X, Liu HM. 2014. High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob Agents Chemother 58:4275–4282. doi: 10.1128/AAC.02813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.BirdLife International. 2015. Species factsheet: Milvus migrans. BirdLife International, Cambridge, United Kingdom: http://www.birdlife.org/datazone/speciesfactsheet.php?id=3355. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.