Abstract
Healthy carriage of CTX-M-type extended-spectrum β-lactamase (ESBL)-producing Escherichia coli was examined by thrice collecting fecal samples from the same 199 healthy Vietnamese subjects every 6 months. Using pulsed-field gel electrophoresis (PFGE), identical PFGE patterns throughout the three samplings were not observed, although prevalence of E. coli in the subjects was around 50% in the three samplings. Our results suggested a short carriage period of the CTX-M-type ESBL-producing E. coli in healthy Vietnamese subjects.
TEXT
A high prevalence of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae has been reported in Southeast and South Asian countries (1–5). Southeast and South Asian countries are regarded as one of the epicenters of antibiotic-resistant bacteria (6). The emerging ESBL have been classified by their amino acid sequences into more than 10 families, including CTX-M, SHV, and TEM types (7, 8). Antibiotic resistance genes, such as blaCTX-M, are horizontally transferred among bacterial cells via conjugative antibiotic-resistant plasmids, even beyond bacterial species (9). Nosocomial and community-acquired infections of CTX-M-type ESBL-producing Enterobacteriaceae have been distributed worldwide (9, 10).
The pathway of the ESBL-producing Enterobacteriaceae distribution is consistent with environmental water and soil (sewage), wild animals, livestock, and human communities, including hospitals and nursing homes (10, 11). Epidemiological studies have been performed to determine its prevalence in various populations, and the distribution of antibiotic-resistant bacteria has been analyzed (10, 11). However, the nature and role of healthy individuals in the transfer and distribution of ESBL-producing Enterobacteriaceae are not well understood.
The study setting and population in the Bavi district of Hanoi, Vietnam was described previously (12). Stool specimens from the study population were collected three times every 6 months from June 2013 to June 2014. Vietnamese individuals who were under clinical treatments within a week before each sampling began were excluded from this study. Finally, 199 healthy Vietnamese individuals were enrolled, and a total of 597 stool specimens were collected. The mean age of the participants at the last sampling was 40.5 years (standard deviation [SD] = 23.1), and the age range was between 1 and 92 years. The study population consisted of 93 men (46.7%) and 106 women (53.3%). Informed consent was obtained from all participants. The study protocol was reviewed and approved by the participating institutes. Statistical significance was evaluated with the χ2 test using IBM SPSS Statistics software version 20 (International Business Machines Corporation, Armonk, NY).
The collected specimens were subjected to microbiological experiments to isolate ESBL-producing Escherichia coli within a sampling day. ESBL-producing E. coli screening, confirmation of ESBL phenotype, and bacterial species determination were performed as described previously (12). The prevalence of ESBL-producing E. coli in the study population of each sampling was 46.7% in June 2013, 52.8% in November 2013, and 46.2% in June 2014. There was no significant difference among the samplings. Antibiotic resistance profiles of the ESBL-producing E. coli in this study were observed by a standard disc diffusion test described previously (12). Overall, resistance against ampicillin (AMP) (97.9%), streptomycin (STR) (63.0%), tetracycline (TET) (74.1%), trimethoprim-sulfamethoxazole (SXT) (74.6%), and nalidixic acid (NAL) (42.4%) were evident. The ESBL-producing E. coli in this study was completely susceptible to meropenem (MEM).
Phylogenetic groups of the ESBL-producing E. coli were observed by conventional phylogenetic grouping protocol (13). Although phylogenetic group A was prevalent in the June 2013 (37.6%) and June 2014 (40.2%) samplings, phylogenetic group D was the predominant type (53.3%) in November 2013, and its detection rate was significantly different from the detection rate of phylogenetic group D in the other samplings. In this study population, phylogenetic group B2 was a minority (15.1% in June 2013, 4.8% in November 2013, and 12.0% in June 2014).
Then, genotyping of blaCTX-M was performed with TaKaRa EX Taq DNA polymerase (TaKaRa Bio Inc., Shiga, Japan) and primers specific to the blaCTX-M-1 group (5′-TAWTTCGTMTCTTCCAGAATAAGG-3′ and 5′-ATGAGTTTCCCCATTCCGTTTCC-3′), blaCTX-M-2 group (5′-ATGATGACTCAGAGCATTCGC-3′ and 5′-TCAGAAACCGTGGGTTACGAT-3′), blaCTX-M-9 group (5′-TGTAACACGGATTGACCGTAT-3′ and (5′-ACTCAGCAAAAGTTCGATTTATTC-3′), and blaCTX-M-25 group (5′-ATGATGAGAAAAAGCGTAAGG-3′ and 5′-TTAATAACCGTCGGTGACAAT-3′). PCR conditions, except for the blaCTX-M-9 group, were 5 min of initial denaturation at 98°C followed by 35 cycles at 98°C for 10 s, 55°C for 30 s, and 72°C for 1 min. PCR conditions for the blaCTX-M-9 group were the same as the above PCR conditions except for the annealing temperature, which was 58°C. Consequently, the blaCTX-M-9 group was the predominant type in the population throughout the sampling period, and the detection rates of the blaCTX-M-9 group in November 2013 (86.7%) and June 2014 (85.9%) were significantly higher than that in June 2013 (71.0%). The predominance of the blaCTX-M-9 group was also observed in healthy Thai individuals in our previous study (1). Therefore, endemic ESBL-producing E. coli in Southeast Asian countries might predominantly possess blaCTX-M-9 group.
At the individual level, ESBL-producing E. coli, regardless of blaCTX-M group, was detected at least one time in 175 (87.9%) of the 199 participants during the sampling period. The number of participants who harbored ESBL-producing E. coli possessing any kind of blaCTX-M throughout the sampling period was 29 (14.6%). Among those, only three participants (1.5%), who belonged to different household, carried ESBL-producing E. coli isolates that belonged to the same phylogenetic group in addition to the commonly detected blaCTX-M-9 throughout the sampling period. Carriage numbers and their ratios of blaCTX-M groups and phylogenetic groups (Table 1) of the ESBL-producing E. coli are summarized by the sampling periods. The ability to properly classify E. coli isolates by their phylogenetic group has been evaluated using multilocus sequence typing classification in our previous study (12), and the results of our blaCTX-M grouping was completely consistent with the sequence analysis interpretation (T. K. N. Bui, unpublished data). Therefore, the frequent alteration in the healthy carriage of ESBL-producing E. coli was not simply due to the inaccuracy of genetic classifications.
TABLE 1.
Genotyping of E. coli isolates possessing blaCTX-M among the three samplingsa
| Detected sampling(s) of E. coli possessing blaCTX-M |
Detected blaCTX-M group |
Detected phylogenetic group |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| June 2013 | November 2013 | June 2014 | Any CTX-M |
CTX-M-1 |
CTX-M-2 |
CTX-M-9 |
CTX-M-25 |
A |
B1 |
B2 |
D |
|||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |||
| + | 25 | 12.6 | 28 | 14.1 | 1 | 0.5 | 27 | 13.6 | 2 | 1.0 | 25 | 12.6 | 17 | 8.5 | 12 | 6.0 | 17 | 8.5 | ||
| + | 36 | 18.1 | 14 | 7.0 | 0 | 0.0 | 35 | 17.6 | 0 | 0.0 | 29 | 14.6 | 12 | 6.0 | 3 | 1.5 | 45 | 22.6 | ||
| + | 28 | 14.1 | 14 | 7.0 | 0 | 0.0 | 30 | 15.1 | 1 | 0.5 | 29 | 14.6 | 19 | 9.5 | 11 | 5.5 | 14 | 7.0 | ||
| + | + | 22 | 11.1 | 4 | 2.0 | 0 | 0.0 | 15 | 7.5 | 0 | 0.0 | 2 | 1.0 | 0 | 0.0 | 2 | 1.0 | 4 | 2.0 | |
| + | + | 17 | 8.5 | 2 | 1.0 | 0 | 0.0 | 8 | 4.0 | 0 | 0.0 | 7 | 3.5 | 2 | 1.0 | 0 | 0.0 | 2 | 1.0 | |
| + | + | 18 | 9.0 | 0 | 0.0 | 0 | 0.0 | 25 | 12.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 5 | 2.5 | |
| + | + | + | 29 | 14.6 | 0 | 0.0 | 0 | 0.0 | 16 | 8.0 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 2 | 1.0 |
Detection rates of blaCTX-M and phylogenetic group are indicated as percentages of the 199 participants.
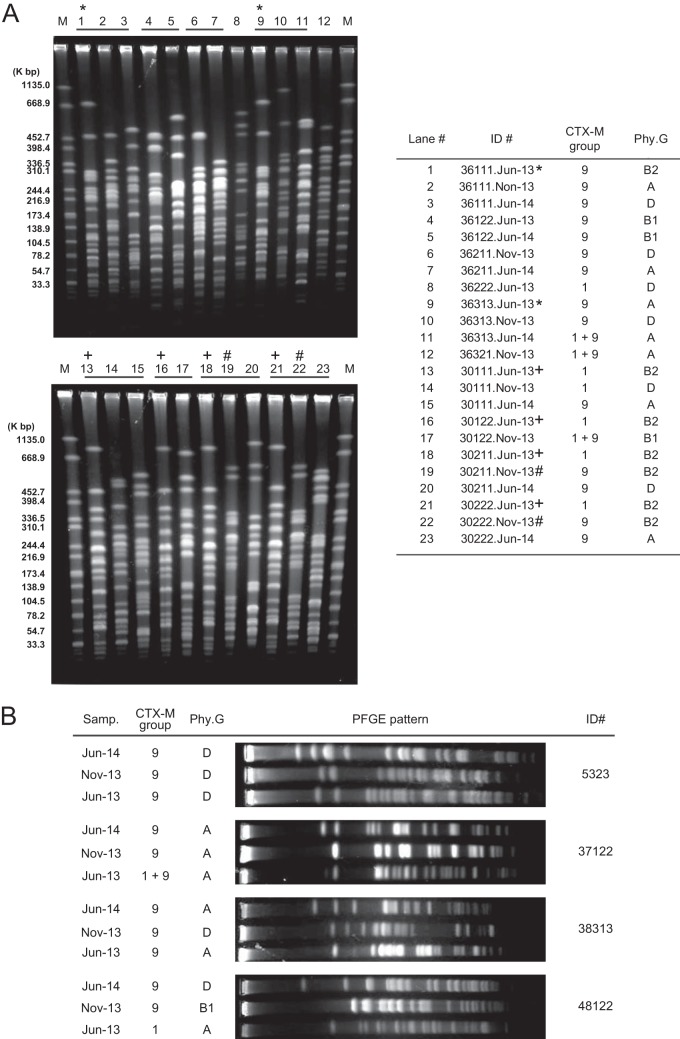
To explain genetic relatedness among the CTX-M-type ESBL-producing E. coli, pulsed-field gel electrophoresis (PFGE) was performed as described in our previous study (14). In the case that number of the CTX-M-type ESBL-producing E. coli isolates detected in each household among the three samplings was at least four, the households' isolates were examined. In this study, 261 of the 290 CTX-M-type ESBL-producing E. coli isolates, which belonged to 36 of the 47 households, were analyzed (Fig. 1A). Various PFGE patterns were observed from the examined CTX-M-type ESBL-producing E. coli isolates. Discrepancy between phylogenetic groups of certain CTX-M-type ESBL-producing E. coli isolates and similarity of their PFGE patterns (e.g., lanes 1 and 9 in Fig. 1A) were observed in 12 (4.6%) of the 261 examined E. coli isolates. In this study, we used PCR-based phylogenetic grouping (13). Generally, mutation in a primer target sequence gives rise to recognition error of the primer. Therefore, the discrepancy in our study might be a consequence of certain mutations on one of the three genetic markers for determination of the phylogenetic grouping. We compared the PFGE patterns obtained from the examined E. coli isolates, and the case of exactly identical PFGE patterns, suggesting clonal distribution, was observed in at least two E. coli isolates. There were two types of clonal distribution, i.e., intrahousehold and interhousehold distributions. Eighteen intrahousehold clonal distributions were observed in 16 of the 36 households (44.4%), and one or two PFGE patterns were observed in two to four CTX-M-type ESBL-producing E. coli isolates obtained from the same household. As interhousehold clonal distributions, seven cases were confirmed. The interhousehold distributions were detected in 15 of the 36 households (41.7%), and two to six CTX-M-type ESBL-producing E. coli isolates shared one of the seven PFGE patterns. However, an identical PFGE pattern was not observed from the CTX-M-type ESBL-producing E. coli isolates in plural sampling periods. Taken together, these results suggested that transient clonal intrafamily and interfamily clonal distributions of certain CTX-M-type ESBL-producing E. coli clones occurred at each sampling period. At the individual level, observed PFGE patterns again varied among the three samplings (Fig. 1B), implying shorter carriage period (less than 6 months) of the CTX-M-type ESBL-producing E. coli in the study subjects. Considering the E. coli in intestinal microflora, commensal E. coli colonizes over longer periods, such as months or years. In contrast, extraintestinal E. coli, such as pathogenic E. coli, does not colonize for longer periods; rather, it colonizes transiently in the gastrointestinal tract (15). Escobar-Paramo et al. reported that the phylogenetic group ratio of commensal E. coli in humans was different from that of other mammals and that the phylogenetic group ratio of other mammals altered with the sampling year, habitat, diet, and climate (16). Therefore, infection of certain extraintestinal E. coli isolates possessing certain types of blaCTX-M might result in the shorter carriage period and the frequent alteration of the participants' carriage of the CTX-M-type ESBL-producing E. coli. In addition to clonal distribution of bacteria, blaCTX-M is horizontally transferred through antibiotic-resistant plasmids among Enterobacteriaceae (17), and this in vivo horizontal transfer has been observed in humans (18, 19). Although there was no supporting data obtained in this study, we did not exclude this possibility.
FIG 1.
Pulsed-field gel electrophoresis profiles of the E. coli possessing blaCTX-M in Vietnamese healthy individuals. Representative PFGE results of the CTX-M-type ESBL-producing E. coli isolates at household level (A) and individual level (B) are shown. The E. coli isolates indicating identical PFGE patterns are indicated by *, +, and #. CTX-M, detected group(s) of blaCTX-M; Phy.G, detected phylogenetic group of the E. coli isolates.
Although our results indicate that the CTX-M-type ESBL-producing E. coli was carried less than 6 month in healthy individuals living in a rural area of Vietnam, many aspects of the epidemiology remain to be explained. Those include the source of certain extraintestinal ESBL-producing Enterobacteriaceae, the reason why regional difference in phylogenetic group and blaCTX-M group are observed, and the different distribution from clinic-oriented isolates such as the E. coli O25b-B2-ST131 clone. Further studies are needed to identify factors contributing to the behavior of the CTX-M-type ESBL-producing E. coli and to determine how to contain this ESBL-producing bacteria.
ACKNOWLEDGMENTS
This work was supported by the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA) as part of the Science and Technology Research Partnership for Sustainable Development (SATREPS).
We sincerely thank N. T. A. Tuyet, H. T. T. Van, P. T. T. Ha, and the National Institute of Nutrition staff for their excellent technical assistance.
REFERENCES
- 1.Luvsansharav UO, Hirai I, Nakata A, Imura K, Yamauchi K, Niki M, Komalamisra C, Kusolsuk T, Yamamoto Y. 2012. Prevalence of and risk factors associated with faecal carriage of CTX-M β-lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob Chemother 67:1769–1774. doi: 10.1093/jac/dks118. [DOI] [PubMed] [Google Scholar]
- 2.Luvsansharav UO, Hirai I, Niki M, Sasaki T, Makimoto K, Komalamisra C, Maipanich W, Kusolsuk T, Sa-Nguankiat S, Pubampen S, Yamamoto Y. 2011. Analysis of risk factors for a high prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in asymptomatic individuals in rural Thailand. J Med Microbiol 60:619–624. doi: 10.1099/jmm.0.026955-0. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama T, Ueda S, Huong BT, Tuyen le D, Komalamisra C, Kusolsuk T, Hirai I, Yamamoto Y. 2015. Wide dissemination of extended-spectrum β-lactamase-producing Escherichia coli in community residents in the Indochinese peninsula. Infect Drug Resist 8:1–5. doi: 10.2147/IDR.S74934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasaki T, Hirai I, Niki M, Nakamura T, Komalamisra C, Maipanich W, Kusolsuk T, Sa-Nguankiat S, Pubampen S, Yamamoto Y. 2010. High prevalence of CTX-M beta-lactamase-producing Enterobacteriaceae in stool specimens obtained from healthy individuals in Thailand. J Antimicrob Chemother 65:666–668. doi: 10.1093/jac/dkq008. [DOI] [PubMed] [Google Scholar]
- 5.Severin JA, Lestari ES, Kloezen W, Lemmens-den Toom N, Mertaniasih NM, Kuntaman K, Purwanta M, Duerink DO, Hadi U, van Belkum A, Verbrugh HA, Goessens WH, Antimicrobial Resistance in Indonesia Prevalence and Prevention (AMRIN) Study Group. 2012. Faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae among humans in Java, Indonesia, in 2001-2002. Trop Med Int Health 17:455–461. doi: 10.1111/j.1365-3156.2011.02949.x. [DOI] [PubMed] [Google Scholar]
- 6.Jean SS, Hsueh PR. 2011. High burden of antimicrobial resistance in Asia. Int J Antimicrob Agents 37:291–295. doi: 10.1016/j.ijantimicag.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Bush K. 2013. Proliferation and significance of clinically relevant β-lactamases. Ann N Y Acad Sci 1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic support and diversity of acquired extended-spectrum β-lactamases in Gram-negative rods. Infect Genet Evol 12:883–893. doi: 10.1016/j.meegid.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 9.D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. 2013. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol 303:305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueda S, Ngan BT, Huong BT, Hirai I, Tuyen le D, Yamamoto Y. 2015. Limited transmission of blaCTX-M-9-type-positive Escherichia coli between humans and poultry in Vietnam. Antimicrob Agents Chemother 59:3574–3577. doi: 10.1128/AAC.00517-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirai I, Fukui N, Taguchi M, Yamauchi K, Nakamura T, Okano S, Yamamoto Y. 2013. Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int J Antimicrob Agents 42:500–506. doi: 10.1016/j.ijantimicag.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Leimbach A, Hacker J, Dobrindt U. 2013. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol 358:3–32. [DOI] [PubMed] [Google Scholar]
- 16.Escobar-Paramo P, Le Menac'h A, Le Gall T, Amorin C, Gouriou S, Picard B, Skurnik D, Denamur E. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ Microbiol 8:1975–1984. doi: 10.1111/j.1462-2920.2006.01077.x. [DOI] [PubMed] [Google Scholar]
- 17.Pfeifer Y, Cullik A, Witte W. 2010. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol 300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Mata C, Miro E, Mirelis B, Garcillan-Barcia MP, de la Cruz F, Coll P, Navarro F. 2010. In vivo transmission of a plasmid coharbouring bla and qnrB genes between Escherichia coli and Serratia marcescens. FEMS Microbiol Lett 308:24–28. doi: 10.1111/j.1574-6968.2010.01980.x. [DOI] [PubMed] [Google Scholar]
- 19.Neuwirth C, Siebor E, Pechinot A, Duez JM, Pruneaux M, Garel F, Kazmierczak A, Labia R. 2001. Evidence of in vivo transfer of a plasmid encoding the extended-spectrum beta-lactamase TEM-24 and other resistance factors among different members of the family Enterobacteriaceae. J Clin Microbiol 39:1985–1988. doi: 10.1128/JCM.39.5.1985-1988.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]