Abstract
Mutation-dependent overproduction of intrinsic β-lactamase AmpC is considered the main cause of resistance of clinical strains of Pseudomonas aeruginosa to antipseudomonal penicillins and cephalosporins. Analysis of 31 AmpC-overproducing clinical isolates exhibiting a greater resistance to ceftazidime than to piperacillin-tazobactam revealed the presence of 17 mutations in the β-lactamase, combined with various polymorphic amino acid substitutions. When overexpressed in AmpC-deficient P. aeruginosa 4098, the genes coding for 20/23 of these AmpC variants were found to confer a higher (2-fold to >64-fold) resistance to ceftazidime and ceftolozane-tazobactam than did the gene from reference strain PAO1. The mutations had variable effects on the MICs of ticarcillin, piperacillin-tazobactam, aztreonam, and cefepime. Depending on their location in the AmpC structure and their impact on β-lactam MICs, they could be assigned to 4 distinct groups. Most of the mutations affecting the omega loop, the R2 domain, and the C-terminal end of the protein were shared with extended-spectrum AmpCs (ESACs) from other Gram-negative species. Interestingly, two new mutations (F121L and P154L) were predicted to enlarge the substrate binding pocket by disrupting the stacking between residues F121 and P154. We also found that the reported ESACs emerged locally in a variety of clones, some of which are epidemic and did not require hypermutability. Taken together, our results show that P. aeruginosa is able to adapt to efficacious β-lactams, including the newer cephalosporin ceftolozane, through a variety of mutations affecting its intrinsic β-lactamase, AmpC. Data suggest that the rates of ESAC-producing mutants are ≥1.5% in the clinical setting.
INTRODUCTION
Pseudomonas aeruginosa is a well-known cause of acute and chronic infections in fragile patients. One of the most remarkable traits of this opportunistic pathogen is its ability to evolve and become resistant to many antibiotics through a variety of mutational and transferable mechanisms (reviewed in reference 1). Some mechanisms tend to prevent the interaction of drugs with their cognate cellular targets, while others result in drug inactivation (1). Like several other Gram-negative species, P. aeruginosa harbors a chromosomal drug-inducible gene, blaAmpC, encoding a wide-spectrum class C β-lactamase (2). This enzyme contributes to the natural resistance of the microorganism toward labile and inducing molecules, such as aminopenicillins, first- and second-generation cephalosporins (3). More importantly, when overproduced as a result of mutations altering the peptidoglycan recycling process, AmpC becomes a major cause of resistance to widely used antipseudomonal penicillins (ticarcillin and piperacillin), monobactams (aztreonam), and third-generation (ceftazidime) and fourth-generation (cefepime) cephalosporins (4–7). The so-called “derepressed mutants” are common in the clinical setting and account for a large proportion of strains resistant to ceftazidime and cefepime in various studies (8–11). This worrisome situation has called for the development of new β-lactams such as ceftolozane and novel AmpC inhibitors such as avibactam that show promising activities against this type of mutants (12, 13).
Class C β-lactamase variants showing unusual substrate specificities (named ESACs for extended-spectrum AmpCs) have been characterized in several Gram-negative pathogens (14). However, because of the high sequence polymorphism of AmpC in P. aeruginosa (affecting more than 22% of its amino acid residues), the occurrence of ESACs in this species has remained largely unexplored (15). This study demonstrates that such variants have potential therapeutic implications as they are responsible for a higher resistance to ceftazidime and ceftolozane-tazobactam than to piperacillin-tazobactam in clinical strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Thirty-five (31 ESAC and 4 non-ESAC) strains of P. aeruginosa isolated between 2009 and 2014 in 21 French hospitals and health care facilities were used in this study as sources of different AmpC variants, named PDCs for Pseudomonas-derived cephalosporinases. These strains were referred to the French National Reference Center (NRC) for Antibiotic Resistance (University Hospital, Besançon, France) for analysis of their resistance mechanisms to β-lactams and were found to produce AmpCs differing from that of reference strain PAO1 (16) by at least one amino acid residue. Strain PA14 (17) was used as the source of PDC-34. The resistance phenotypes conferred by the cloned blaAmpC genes were assessed in strain 4098, an AmpC-deficient mutant from PAO1 (18), while the impact of enzymes PDC-1 and PDC-3 on carbapenem susceptibility was investigated more specifically in mutant PAO1ΔoprD (19), which lacks the carbapenem-specific uptake porin OprD. The PBP4-deficient mutant PAO1ΔdacB (6), which constitutively overproduces PDC-1, was used as a control in MIC experiments. In addition, 44 nonredundant, antibiotic-susceptible strains of P. aeruginosa, including 39 isolates from the University Hospital of Besançon and 5 isolates from surface waters of eastern France, were selected from the laboratory collection to investigate the sequence polymorphism of AmpC in wild-type P. aeruginosa. Molecular biology experiments were performed with Escherichia coli DH5α (Life Technologies) as the recipient strain. The blaAmpC genes were cloned into the broad-host-range plasmid vector pUCP24, kindly supplied by Herbert Schweizer (Colorado University) (20). Bacteria were cultivated at 35 ± 1°C on Mueller-Hinton (MH) agar (Bio-Rad) supplemented with gentamicin (5 μg/ml for E. coli and 50 μg/ml for P. aeruginosa) to maintain pUCP24 and its derivatives where needed.
Drug susceptibility testing.
MICs (shown as ≤ susceptible [S] breakpoint/≥ resistant [R] breakpoint) of ticarcillin (16/128 μg/ml), piperacillin-tazobactam (16-4/128-4 μg/ml), ceftazidime (8/32 μg/ml), cefepime (8/32 μg/ml), ceftolozane-tazobactam (breakpoints pending), aztreonam (8/32 μg/ml), imipenem (2/8 μg/ml), and meropenem (2/8 μg/ml) were determined with and without 250 or 1,000 μg/ml cloxacillin using customized Sensititre plates, according to the instructions of the manufacturer (Thermo Fisher Scientific). All plates were incubated at 35 ± 1°C for 18 ± 2 h with inocula of 105 CFU/ml. The strains were considered “susceptible” (S), “intermediate” (I), or “resistant” (R) to the tested drugs in reference to the Clinical and Laboratory Standards Laboratory (CLSI) breakpoints (21).
Molecular typing of the strains.
The epidemiological relatedness of the clinical P. aeruginosa strains was studied by multilocus sequence typing (MLST) based on the allelic variations of 7 housekeeping genes, namely, acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE (22). The isolates were assigned a sequence type (ST) number according to the allelic profiles available in the MLST Database (http://pubmlst.org/paeruginosa).
Analysis and cloning of blaAmpC genes.
Genomic DNA was extracted and purified from the clinical strains by using QIAamp DNA minikit (Qiagen). For sequencing purposes, the blaAmpC genes were amplified by PCR with primers SeqAmpC-PA14-Fw1 (5′-TGGGGTCGAACCAATCTCTA-3′), SeqAmpC-PA14-Fw2 (5′-CAGATCCGCGACTACTACCG-3′), SeqAmpC-PA14-Rv1 (5′-ACGTCGAGGTGGGTCTGTT-3′), and SeqAmpC-PA14 Rv2 (5′-CTCATGGCACCATCATAGCC-3′). The amplicons were sequenced using the BigDye Terminator chemistry on an automated ABI 3730 sequencer (Applied Biosystems), and the resulting data were edited using BioEdit 7.1.9 software (Tom Hall, North Carolina State University, Raleigh, NC). For cloning purposes, the blaAmpC genes were PCR amplified and cloned into plasmid vector pUCP24, as already described (23). After a DNA sequencing step ensuring that no mutations had been introduced during PCR amplification, the resulting pPDC plasmids were transferred by electroporation (Bio-Rad MicroPulser) into P. aeruginosa 4098. Transformants were selected on MH agar supplemented with gentamicin, the selection marker for pUCP24, and characterized as to their resistance levels to selected β-lactams. Plasmids pPDC-1 and pPDC-3 were further electroporated into porin OprD-deficient mutant 4098ΔoprD.
RT-qPCR experiments.
For the reverse transcription-quantitative PCR (RT-qPCR) experiments, the bacterial strains were cultivated aerobically to the mid-log phase in drug-free Mueller-Hinton broth. Their total RNA content was extracted and reverse transcribed as described by Dumas et al. (24). The expression of gene blaAmpC and housekeeping gene uvrD was then assessed in a Rotor Gene RG6000 instrument (Qiagen, Courtaboeuf, France) by using the intercalating dye Rotor-Gene SYBR green (Qiagen) and primer pairs AmpC1/AmpC2 and UvrD1/UvrD2, respectively (24, 25). Sequence alignment analysis confirmed the absence of base mismatch between the primers and the target genes, ensuring efficient PCR amplification. The mRNA levels of blaAmpC were normalized with those of uvrD for each strain and expressed as a ratio (fold change) to that of wild-type strain PAO1, used as the reference. Mean gene expression values were calculated from two independent bacterial cultures, each assayed in duplicate.
Structure-function analysis of AmpCs.
The AmpC sequence, corresponding to the genomic locations 4594029 to 4595222 on strain PAO1, was extracted from the Pseudomonas Genome Database (http://www.pseudomonas.com/). A BLAST search was performed, choosing the Protein Data Bank (PDB) as the search database (http://blast.ncbi.nlm.nih.gov/), resulting in 43 PDB structures presenting an E value lower than 10−100 and 11 with an E value between 10−11 and 10−4. Among them, those presenting a β-lactamase in complex with a molecule in its active site were selected and superposed on the structure of PAO1 AmpC (PDB code 2WZZ). In order to evaluate the possible effect of the different observed mutations, all of the corresponding positions were mutated using PyMOL on the 2WZZ structure. All the structural figures were drawn using Pymol software (PyMOL Molecular Graphics system [http://pymol.sourceforge.net/]).
Accession numbers.
The sequences of blaAmpC genes corresponding to the following PDCs have been deposited in GenBank under the accession numbers shown in parentheses: PDC-73 (KR057742), PDC-74 (KR057743), PDC-75 (KR057744), PDC-76 (KR057745), PDC-77 (KR057746), PDC-78 (KR057747), PDC-79 (KR057748), PDC-80 (KR057749), PDC-81 (KR057750), PDC-82 (KR057751), PDC-83 (KR057752), PDC-84 (KR057753), PDC-85 (KR057754), PDC-86 (KR057755), PDC-87 (KR057756), PDC-88 (KR057757), PDC-89 (KR057758), PDC-90 (KR057759), PDC-91 (KR057760), PDC-92 (KR057761), and PDC-93 (KR057762).
RESULTS AND DISCUSSION
Sequence polymorphism of P. aeruginosa AmpC.
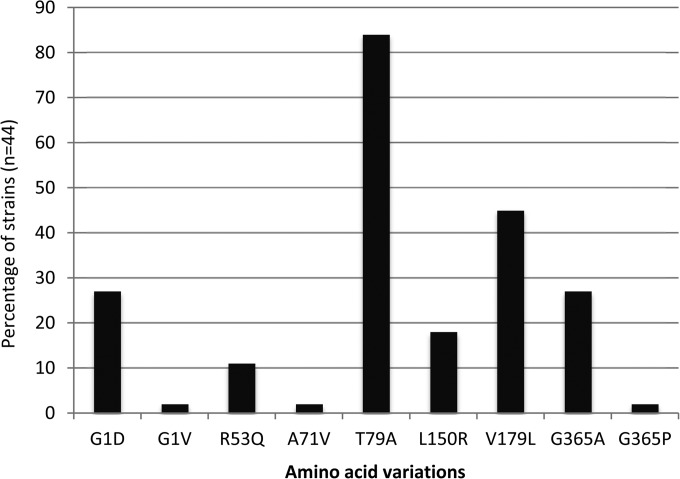
Amino acid substitutions G1D/V, A29T, R53Q, A71V, T79A, K82E, Q129R, L150R, V179L, V330I, and G365A have been reported to occur in AmpC enzymes produced by β-lactam-resistant strains of P. aeruginosa, compared with the mature (i.e., after cleavage of the 26-amino-acid-long signal peptide) protein from reference strain PAO1 (26, 27). In this study, analysis of the coding sequence of blaAmpC genes from 44 antibiotic-susceptible strains isolated at the hospital (n = 39) or in natural environments (n = 5) showed the same amino acid variations described above at positions 1, 53, 71, 79, 150, 179, and 365 (Fig. 1). Given that most of these substitutions are present in the enzyme of wild-type reference strain PA14 (Table 1), collectively, these results support the notion that such a polymorphism has little or no impact on the enzymatic activity of AmpC (i.e., as it is found in both bacteria susceptible to and bacteria resistant to β-lactams). Consistent with this, the interchangeable amino acids appeared to be distant from the active site, with residues at positions 1, 79, 129, and 365 being located on the opposite surface and those at positions 29 and 179 being part of secondary structure elements close to the surface that can easily adapt the local mutations. Referring to the initial classification of AmpC variants (from PDC-1 to PDC-10 for Pseudomonas-derived cephalosporinases 1 to 10) established by Rodriguez-Martinez et al. (26), variant PDC-1 (identical to that of PAO1 by definition) was found to be produced by 7 of the 44 wild-type strains analyzed, PDC-3 by 6 strains, PDC-5 by 4 strains, PDC-7 by 2 strains, and PDC-8 by 8 strains, while the 17 remaining isolates harbored various allelic combinations not reported in this classification scheme. The K82E and V330I changes reported previously were not identified in the present collection (26).
FIG 1.
Amino acid sequence polymorphism of β-lactamase AmpC among nonredundant clinical (n = 39) and environmental (n = 5) antibiotic-susceptible strains of P. aeruginosa.
TABLE 1.
Amino acid variations in the sequences of PDC enzymes
| Strain(s) | PDC | Residue at amino acid position showna |
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 19 | 29 | 71 | 79 | 100 | 121 | 129 | 150 | 154 | 175 | 179 | 213 | 216 | 221 | 223 | 290 | 291 | 292 | 293 | 294 | 295 | 330 | 347 | 365 | ||
| Wild-type AmpC producers | ||||||||||||||||||||||||||
| PAO1b | PDC-1 | G | V | A | A | T | R | F | Q | L | P | M | V | V | G | E | Y | T | P | M | A | L | Q | V | N | G |
| 14.2028 | PDC-3 | A | ||||||||||||||||||||||||
| 13.1642 | PDC-8 | A | R | |||||||||||||||||||||||
| 13.1781 | PDC-24 | A | A | |||||||||||||||||||||||
| PA14b | PDC-34 | D | T | A | R | L | A | |||||||||||||||||||
| 11.773 | PDC-35 | D | V | A | L | A | ||||||||||||||||||||
| ESAC producers | ||||||||||||||||||||||||||
| 12.1227 and 12.1255 | PDC-44 | A | P | |||||||||||||||||||||||
| 10.257 | PDC-50 | A | A | |||||||||||||||||||||||
| 12.1285 | PDC-73 | A | L | |||||||||||||||||||||||
| 12.1111 and 13.1716 | PDC-74 | A | R | |||||||||||||||||||||||
| 12.1129 and 13.1727 | PDC-75 | A | R | I | ||||||||||||||||||||||
| 13.1415 | PDC-76 | A | I | |||||||||||||||||||||||
| 13.1404, 13.1696, and 13.1760 | PDC-77 | R | ||||||||||||||||||||||||
| 11.571 | PDC-78 | H | R | |||||||||||||||||||||||
| 11.516 | PDC-79 | K | ||||||||||||||||||||||||
| 13.1737 | PDC-80 | G | ||||||||||||||||||||||||
| 13.1514 | PDC-81 | A | L | A | ||||||||||||||||||||||
| 13.1770 | PDC-82 | A | L | L | A | |||||||||||||||||||||
| 4836 | PDC-83 | A | A | A | A | |||||||||||||||||||||
| 13.1482 | PDC-84 | A | A | A | I | A | ||||||||||||||||||||
| 12.961 | PDC-85 | A | H | A | ||||||||||||||||||||||
| 13.1755 and 14.1999 | PDC-86 | D | A | L | K | I | A | |||||||||||||||||||
| 11.698 and 13.1775 | PDC-87 | D | A | L | I | I | A | |||||||||||||||||||
| 13.1601 | PDC-88 | D | A | L | Δ | Δ | I | A | ||||||||||||||||||
| 09.236 | PDC-89 | D | A | L | Δ | Δ | Δ | A | ||||||||||||||||||
| 13.1562 | PDC-90 | D | A | L | Δ | Δ | Δ | I | A | |||||||||||||||||
| 11.813 | PDC-91 | D | A | L | Δ | Δ | Δ | Δ | I | A | ||||||||||||||||
| 12.1121 and 13.1389 | PDC-92 | D | A | L | Δ | Δ | A | |||||||||||||||||||
| 14.2036 | PDC-93 | D | A | L | P | A | ||||||||||||||||||||
The numbering of the amino acids refers to the mature protein from strain PAO1, after cleavage of the 26 N-terminal amino acid residues of the signal peptide. Variations highlighted in boldface are considered common polymorphisms. The role of V19A has not been investigated specifically in this study.
Reference strain of P. aeruginosa (http://v2.pseudomonas.com).
Clinical strains with unusual resistance profiles.
From 2009 to 2014, 2,040 clinical isolates of P. aeruginosa were referred to the French National Reference Center for Antibiotic Resistance to characterize their resistance mechanisms to antibiotics, mainly β-lactams. Among these strains, 506 (24.8%) and 471 (23.1%) were found to produce extended-spectrum β-lactamases (ESBLs) and carbapenemases, respectively, by phenotypic and molecular biology techniques (unpublished data). In addition, 810 (37.7%) strains recovering their susceptibility to penicillins and cephalosporins when tested by the disk diffusion method on Mueller-Hinton agar supplemented with class C inhibitor cloxacillin (2) at 1,000 μg/ml were considered AmpC-overproducing mutants. Unlike these typical AmpC overproducers (as illustrated in Table S1 in the supplemental material), 31 (1.5%) isolates that showed rather uncommon resistance profiles characterized by a higher resistance to ceftazidime than to piperacillin-tazobactam were investigated more extensively. None of them appeared to produce transferable β-lactamases (data not shown).
According to the current CLSI breakpoints, 12.9% (n = 4) and 87.1% (n = 27) of these strains were intermediate and resistant to ceftazidime, respectively, with MICs ranging from 16 to >512 μg/ml. Using the same breakpoints (8 μg/ml ≤ S and R ≥ 32 μg/ml), ceftolozane combined with 4 μg/ml tazobactam performed better, with 58.1% (n = 18), 12.9% (n = 4), and 29% (n = 9) of strains being susceptible, intermediate, and resistant to the drug, respectively (MICs from 0.5 to >64 μg/ml). Categorization of bacteria as S, I, and R for the other β-lactams tested was as follows: ticarcillin, 6.4%, 9.7%, and 83.9%; piperacillin plus 4 μg/ml tazobactam, 48.4%, 35.5%, and 16.1%; aztreonam, 16.1%, 12.9%, and 71%; cefepime, 12.9%, 25.8%, and 61.3%; imipenem, 22.6%, 9.7%, and 67.7%; and meropenem, 19.4%, 29%, and 51.6%. In contrast to in vitro mutant PAO1ΔdacB, only 4 of the 31 strains recovered their susceptibility to ceftazidime (MIC, ≤8 μg/ml) in the presence of cloxacillin (see Table S1 in the supplemental material).
Identification of new AmpC variants.
Sequencing of gene blaAmpC revealed the occurrence of 23 different AmpC variants (dubbed PDCs in the current nomenclature) among the selected bacteria, including 21 new variants with respect to the 72 PDCs listed previously (15, 26) (Table 1). Seventeen so far uncharacterized mutations at 15 amino acid positions were combined with 0 to 5 variations presumed to be common polymorphisms (described above): R100H, F121L, P154L, M175L, V213A, G216R, E221K/G, Y223H, ΔT290-ΔP291, ΔT290-ΔM292, ΔT290-ΔA293, ΔL294-ΔQ295, L294P, and N347I (Table 1). Of note, AmpC variant PDC-77 was produced by 3 strains, and PDC-44, PDC-74, PDC-75, PDC-86, PDC-87, and PDC-92 were each expressed by 2 isolates, while the 16 remaining variants were found in single isolates. RT-qPCR experiments showed that all of the selected strains constitutively overexpressed the blaAmpC gene, with relative values ranging from 4.4- to 2,895-fold (median 686-fold) compared with wild-type strain PAO1. As noted previously (27, 28), no correlation could be found between these expression levels and β-lactam MICs, suggesting a posttranscriptional regulation of AmpC production or the influence of undetermined resistance mechanisms in individual strains. Interestingly, the blaAmpC transcripts were quantitatively lower (median, 162-fold) in slow-growing bacteria isolated from cystic fibrosis (CF) patients than in bacteria from chronic obstructive pulmonary disease (COPD) patients (median 1,031-fold) or non-CF, non-COPD patients (median, 906-fold).
Impact of PDCs on β-lactam MICs.
The blaAmpC genes of 20 PDC enzymes harboring the various mutations were amplified by PCR, cloned into the broad-host-range vector pUCP24, and overexpressed in AmpC-deficient P. aeruginosa mutant 4098. The same approach was used to clone the blaAmpC genes from PAO1 (PDC-1), PA14 (PDC-34), and 4 clinical strains presumed to overproduce wild-type AmpCs (PDC-3, PDC-8, PDC-24, and PDC-35) because of their greater resistance to piperacillin-tazobactam than to ceftazidime (Table 1; see Table S1 in the supplemental material). As expected, the resistance levels to β-lactams conferred by these latter AmpC variants and by PDC-34 were nearly identical to that provided by PDC-1, thus confirming the absence of impact of the polymorphic variations G1D, A29T, A71V, T79A, Q129R, L150R, V179L, and G365A individually or collectively on the enzyme activity (PDCs of group 0 in Table 2). Since T79A (T105A in the nonprocessed peptide) had previously been found to be prevalent in carbapenem-nonsusceptible clinical strains (29) and to weakly increase the catalytic activity of AmpC in vitro on carbapenems (26), we overexpressed enzymes PDC-1 (T79) and PDC-3 (A79) in mutant 4098ΔoprD, reasoning that the slow penetration of carbapenems into this porin OprD-negative strain would potentiate the AmpC action and thus reveal slight differences in resistance due to T79A. In agreement with other results (28), the variations at position 79 did not affect the resistance of 4098ΔoprD to carbapenems (MICs of imipenem and meropenem were equal to 4 and 2 μg/ml, respectively, with both PDCs), clearly demonstrating that T79A does not broaden the substrate spectrum of AmpC at least in vivo.
TABLE 2.
Susceptibility profiles of strain 4098 complemented with blaAmpC genes
| PDC-producing strain | MIC (μg/ml)a |
Mutation(s)b | |||||
|---|---|---|---|---|---|---|---|
| TIC | TZP | ATM | CAZ | FEP | CZ/T | ||
| 4098(pUCP24) | 16 (8) | 4 (≤2) | 4 (2) | 1 (1) | 1 (≤0.5) | 0.5 (≤0.25) | |
| 4098(pPDC-1)c | 64 (16) | 64 (≤2) | 16 (2) | 16 (1) | 8 (1) | 1 (0.5) | |
| Group 0 | |||||||
| 4098(pPDC-3) | 64 (16) | 64 (4) | 16 (2) | 16 (2) | 8 (1) | 1 (0.5) | |
| 4098(pPDC-8) | 64 (16) | 32 (4) | 16 (2) | 8 (1) | 4 (1) | 1 (0.5) | |
| 4098(pPDC-24) | 64 (8) | 64 (≤2) | 16 (2) | 16 (1) | 8 (1) | 1 (0.5) | |
| 4098(pPDC-34) | 64 (16) | 64 (≤2) | 16 (2) | 16 (1) | 8 (1) | 1 (0.5) | |
| 4098(pPDC-35) | 64 (16) | 64 (4) | 16 (2) | 8 (1) | 8 (1) | 1 (0.5) | |
| Group I | |||||||
| 4098(pPDC-50) | 128 (16) | 64 (4) | 32 (4) | 64 (2) | 8 (1) | 4 (0.5) | V213A |
| 4098(pPDC-74) | 256 (128) | 8 (4) | 128 (32) | >64 (32) | 8 (4) | 8 (4) | G216R |
| 4098(pPDC-75) | 256 (128) | 16 (8) | 128 (32) | >64 (64) | 8 (4) | 8 (4) | G216R |
| 4098(pPDC-78) | >512 (512) | 32 (16) | >128 (128) | >64 (>64) | 16 (16) | 16 (16) | G216R, R100H |
| 4098(pPDC-79) | 256 (128) | 16 (16) | 32 (16) | >64 (64) | 16 (8) | 64 (32) | E221K |
| 4098(pPDC-80) | 128 (64) | 32 (16) | 16 (8) | 64 (16) | 4 (2) | 32 (8) | E221G |
| 4098(pPDC-85) | 256 (64) | 16 (4) | 32 (8) | 32 (8) | 8 (2) | 4 (4) | Y223H |
| 4098(pPDC-86) | 256 (128) | 16 (16) | 32 (32) | >64 (>64) | 16 (8) | >64 (64) | E221K |
| Group II | |||||||
| 4098(pPDC-44) | 32 (16) | 16 (4) | 8 (2) | 64 (4) | >64 (4) | 2 (0.5) | L294P |
| 4098(pPDC-88) | 64 (16) | 64 (4) | 16 (2) | 64 (2) | >64 (4) | 4 (0.5) | Δ(T290-P291) |
| 4098(pPDC-89) | 32 (16) | 32 (8) | 16 (4) | >64 (16) | >64 (32) | 8 (2) | Δ(T290-M292) |
| 4098(pPDC-90) | 16 (16) | 16 (4) | 8 (2) | >64 (8) | >64 (16) | 4 (1) | Δ(T290-M292) |
| 4098(pPDC-91) | 32 (16) | 64 (8) | 32 (8) | >64 (16) | >64 (64) | 16 (2) | Δ(T290-A293) |
| 4098(pPDC-92) | 32 (16) | 32 (4) | 16 (4) | 64 (8) | >64 (16) | 4 (1) | Δ(L294-Q295) |
| Group III | |||||||
| 4098(pPDC-73) | 128 (16) | 64 (≤2) | 32 (2) | 64 (2) | 16 (1) | 8 (1) | P154L |
| 4098(pPDC-81) | 128 (16) | 64 (≤2) | 16 (2) | 64 (2) | 16 (2) | 8 (1) | P154L |
| 4098(pPDC-82) | 128 (32) | 32 (8) | 16 (4) | 64 (8) | 8 (2) | 16 (8) | F121L, M175L |
| Group IV | |||||||
| 4098(pPDC-76) | 32 (16) | 16 (4) | 16 (4) | >64 (16) | 16 (2) | 4 (1) | N347I |
| 4098(pPDC-87) | 64 (16) | 32 (8) | 32 (4) | >64 (32) | 32 (8) | 8 (2) | N347I |
| Groups I + IV | |||||||
| 4098(pPDC-84) | 32 (16) | 16 (4) | 64 (16) | >64 (64) | 32 (8) | 16 (4) | V213A, N347I |
Abbreviations: TIC, ticarcillin; TZP, piperacillin plus tazobactam at a fixed concentration of 4 μg/ml; ATM, aztreonam; CAZ, ceftazidime; FEP, cefepime; CZ/T, ceftolozane plus tazobactam at a fixed concentration of 4 μg/ml. Shown are values from at least two independent experiments. Values in parentheses correspond to the MICs for bacteria grown in the presence of 1,000 μg/ml cloxacillin. MIC values in boldface are at least 4-fold higher and those in italics are at least 4-fold lower than those for 4098(pPDC-1). MIC values of imipenem and meropenem were equal to 0.5 and 0.25 μg/ml, respectively, for all strains tested.
Shown are mutations different from common polymorphisms G1D, A29T, A71V, T79A, Q129R, L150R, V179L, V330I, and G365A.
blaAmpC genes cloned on broad-spectrum plasmid pUCP24.
Based on their location in the three-dimensional (3D) structure of AmpC and their effects on β-lactam MICs, the other mutations found in this study could be assigned to 4 distinct groups, designated I to IV.
Group I PDCs.
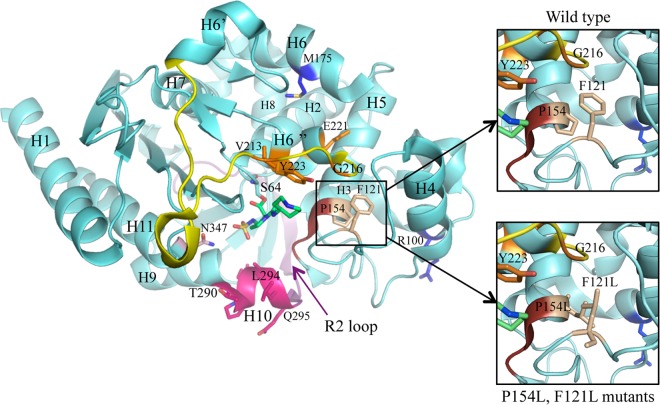
Group I mutations (V213A, G216R, E221K, E221G, and Y223H) are located in the C-terminal region of the Ω loop and in the following α-helix (Fig. 2). Compared with PDC-1, they increased the MICs of ticarcillin, aztreonam (except for PDC-80, which contains E221G), ceftazidime, and ceftolozane-tazobactam, with variable effects on resistance levels to piperacillin-tazobactam and cefepime (Table 2). Remarkably, amino acid changes at position E221 (K/G) had the greatest impact on the ceftolozane-tazobactam MIC (≥32 μg/ml ceftolozane). Reminiscent of these results, AmpC-overexpressing, ceftolozane-tazobactam-resistant clones exhibiting the E221K mutation together with F121L, Q131R, and/or V330I were selected in vitro from a ΔmutS hypermutator PAO1 mutant exposed to increasing drug concentrations (23). The Ω-loop region, which is part of the active site of the enzyme and which accommodates the R1 side chain of the β-lactam nucleus, is a well-known hot spot for mutations able to extend the substrate specificity of chromosome- or plasmid-encoded class C β-lactamases (14). Interestingly, most of the mutated amino acid residues found in P. aeruginosa AmpC mapped at the same position or in the vicinity of residues previously reported as cause of ESACs in other organisms, such as E. coli, Enterobacter sp., Serratia marcescens, and Acinetobacter baumannii (see details in Table S2 in the supplemental material). It should be noted here that the R100H change, which coexists with G216R in PDC-78, slightly enhanced the enzyme activity against all the β-lactams tested (compare the MICs conferred by PDC-74, -75, and -78). No predictable explanation could be found by structural analysis as this residue is situated at the surface of the protein on the back side compared with the binding pocket (Fig. 2). With the exception of PDC-50 which carries the V213A mutation, group I PDCs (PDC-74, -75, -78, -79, -80, -84, -85, and -86) turned out to be poorly antagonized by cloxacillin at 1,000 μg/ml. This observation is of practical importance as tests using this AmpC inhibitor even at high doses may fail to recognize clinical strains overproducing such variants.
FIG 2.
Representation of the AmpC β-lactamase structure from the P. aeruginosa PAO1 strain (light blue) in complex with an inhibitor (green) (PDB code 2WZZ) (41). The different structural regions lining the binding site are colored as follows: omega loop, yellow; helix H-10, pink; R2 loop, purple; and YSN, brick. The amino acids highlighted in this study as playing an important role in ESAC emergence are colored in terms of the function of their pertaining group (see the text): group I, orange; group II, pink; group III, wheat; and group IV, light pink. The two other mutants (M175L and R100H) found in combination with some from the other groups are colored in blue.
Group II PDCs.
Group II relates to mutations, mainly deletions, that are located in the R2 domain of AmpC (ΔT290-ΔQ295, L294P), at or near positions already known to increase the hydrolytic activity of class C β-lactamases toward third- to fourth-generation cephalosporins (see Table S2 in the supplemental material). Indeed, alterations occurring in this region, which corresponds to H-10, the N terminus of the R2 loop, are able to widen the active site, making the enzyme more accessible to β-lactam molecules carrying a bulky R2 side chain (14). Consistent with this notion and with respect to PDC-1, group II PDCs (PDC-44, -88, -89, -90, -91, and -92) conferred higher resistance to ceftazidime, cefepime, and ceftolozane-tazobactam, with no or a negative impact on the MICs of penicillins and aztreonam (Table 2). Among all of the ESACs identified in this study, the group II AmpC variants were associated with the highest resistance to cefepime (>64 μg/ml).
Group III PDCs.
P154 and F121, which are situated near the conserved YSN loop (at the extremity of H-5 and in loop α3-α4, respectively) are predicted to interact by making a stacking involving their aromatic side chains. According to this model, mutations P154L and F121L that occur in PDC-73, -81, or -82 are expected to abrogate this interaction, forcing loop α3-α4 to move away, enlarging the substrate binding pocket, with consequent modification of AmpC substrate specificity (Fig. 2). Supporting this hypothesis, both mutations resulted in similar effects on β-lactam MICs, with a strong increase in resistance of strain 4098 to ceftazidime and ceftolozane-tazobactam relative to PDC-1 (Table 2). As mentioned above, F121L was already found to coexist with group I mutation E221K in a ceftolozane-tazobactam-resistant mutant selected in vitro; however, its contribution to the whole resistance of the mutant was not investigated further (23). M175L, concurrent with F121L in PDC-82, is localized in H-6 sided to H-6″ involved in the substrate-binding site at the C terminus of the Ω loop. Its mutation into leucine would require a small rearrangement of the surrounding region, but without clear knowledge on the necessary movements. To our knowledge, mutations at positions corresponding to P154 and F121 in P. aeruginosa AmpC have not been identified in other class C β-lactamases.
Group IV PDCs.
Group IV is characterized by the N347I substitution as found in PDC-76, -84 (in combination with V213A), and -87. Reminiscent of the phenotype associated with these AmpC variants, N347I was reported to increase the activity of plasmid-determined enzyme CMY-2 and Enterobacter AmpC on cephalosporins ceftazidime and cefepime (30, 31) (see Table S2 in the supplemental material). According to the results of site-directed mutagenesis experiments and structural studies, the highly conserved N347 (346 in E. coli AmpC) is part of the C3/C4 carboxylate recognition region that directly contributes to substrate positioning in the catalytic site of class C enzymes through hydrogen bonding (32). How N347I affects this positioning and accounts for the phenotype associated with group IV PDCs is unclear so far.
Because of the structural plasticity of class C β-lactamases, enabling them to accommodate new substrates when mutated in specific regions, one could assume that coexistence of mutations in several of these regions would further increase the activity of AmpC enzymes toward newer cephalosporins and/or carbapenems. Actually, this was not verified here, as the occurrence of two ESAC-associated mutations in the same enzyme (PDC-84), namely, V213A (group I) and N347I (group IV), did not result in significant additive effects on the levels of resistance to cephalosporins and did not impact carbapenem MICs (Table 2).
Finally, the data presented above confirmed that the common polymorphism, which also includes V330I in P. aeruginosa, has no or negligible impact on AmpC activity when combined with ESAC-associated mutations (compare PDC-74 and PDC-75, PDC-79 and PDC-86, PDC-89 and PDC-90, PDC-73 and PDC-81, and PDC-76 and PDC-87 in Table 2).
Strain genotyping.
The clonal relatedness of the 31 ESAC-producing P. aeruginosa strains was investigated by MLST. The bacteria could be grouped into 14 different sequence types (STs), some of which correspond to widespread clonal complexes or clones, such as ST175 (n = 5 isolates), ST235 (n = 3), ST260 (n = 1), ST298 (n = 1), ST308 (n = 4), ST309 (n = 1), and ST313 (n = 2) (33) (see Table S1 in the supplemental material). Interestingly, most of the strains isolated in the context of cystic fibrosis appeared to belong to clones already found associated with this pathology, such as ST170 (n = 1), ST242 (n = 2), ST282 (n = 2), and ST274 (n = 4) (34–36). Our genotypic analysis also revealed the occurrence of clonally related isolates producing PDC-44 (n = 2), PDC-75 (n = 2), PDC-87 (n = 2), and PDC-92 (n = 2) in distinct institutions. With the exception of PDC-77 found in ST175 isolates from 3 distant cities (i.e., which suggests a geographical diffusion of a single clone), our results provide evidence that the ESACs emerged locally in genotypically distinct and rather prevalent P. aeruginosa clones (see Table S1). Since 15 (48.4%) ESAC-producing isolates came from chronically infected patients with cystic fibrosis (n = 10) or chronic obstructive pulmonary disease (COPD [n = 5]), we asked whether hypermutability, which is common in these diseases (37), was required for the emergence of ESAC-associated mutations, as suggested elsewhere (23, 38). Actually, only 1 (CF isolate 12.1111) out of 10 randomly selected ESAC strains (the other 9 strains including 5 CF, 1 COPD, and 3 non-CF, non-COPD isolates) turned out to give rise to rifampin-resistant mutants at rates significantly higher (≥20-fold) than those of reference strain PAO1 (10−8) and to fit with the definition of hypermutator (39; data not shown).
Conclusions.
Despite differences in their amino acid sequences, class C β-lactamases share the same general structure and conserved sequence motifs near the active-site serine (14). So, it is not surprising per se that mutations occurring in specific regions of the substrate binding pocket, such as the Ω loop, the R2 domain, or the C-terminal domain, result in conformational changes prone to enhance the catalytic efficiencies of these enzymes toward poor substrates. Like in other Gram-negative species, these mutations increase the hydrolytic activity of P. aeruginosa AmpC on cephalosporins (ceftazidime, cefepime, and/or ceftolozane), with mitigated effects on penicillins and/or aztreonam (14). Our assumption that P154L and F121L (which apparently belong to a new group of mutations) prevent the interaction between P154 and F121, with consequent remodeling of the substrate cavity, needs to be substantiated by crystallography experiments. In this work, none of the ESACs studied could increase the resistance of P. aeruginosa to carbapenems. Furthermore, confirming other data (28), we could not demonstrate any effect of the T79A substitution on carbapenem MICs even in the low-outer-membrane-permeability mutant 4098ΔoprD, consistent with T79 being distant from the active site, at the surface of the protein, in a hydrophobic environment showing no evidence of negative consequence of replacing a threonine by an alanine.
The prevalence of ESAC-producing strains among clinical P. aeruginosa isolates is still largely unknown. In the collection of the French NRC, mainly composed of β-lactam-resistant bacteria, 31 of 2,040 isolates (1.5%) were characterized as ESAC producers. However, sequencing of gene blaAmpC was not systematic and only applied to those strains exhibiting uncommon phenotypes (higher resistance to ceftazidime than to piperacillin-tazobactam) and devoid of secondary β-lactamases (ESBLs or carbapenemases), so we cannot rule out the possibility that the use of specific phenotypic criteria to screen for ESACs may miss bacteria in which these variants are phenotypically masked by other resistance mechanisms (e.g., active efflux pumps, penicillin-binding protein [PBP] alterations). Alternatively, some ESACs may well be responsible for phenotypes that are different from those reported here. Interestingly, we examined the recently released AmpC sequences of 531 P. aeruginosa isolates collected in 25 different countries and associated with multiple clinical situations (15). This search revealed the presence of 9 strains (1.7%) harboring several of the mutations described in this study (including 7 strains with V231A, 1 with L294P, and 1 with N347I). Therefore, if one considers that still unreported ESAC-associated mutations are likely to occur in the clinical setting, the prevalence of AmpC variants should be higher than 2%. That AmpC variants conferring high resistance to ceftolozane (in combination with tazobactam) preexist the introduction of this AmpC-stable molecule into clinical practice is of particular interest (40). Although clinical data were not available in this study, one can assume that older antipseudomonal cephalosporins (ceftazidime and cefepime) cross-selected resistance to this new antibiotic. Therefore, in the future it will be necessary to pay a special attention to the emergence of ceftolozane-resistant mutants under treatment since AmpC does not necessarily require hypermutability to give rise to ESACs in P. aeruginosa. The use of cloxacillin as an AmpC inhibitor may fail to characterize these variants, which as shown here, can potentially emerge in epidemic clones.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the following clinical microbiologists for their collaboration with the NRC: T. Hadou (Nancy), B. Alves Pereira (Wissous), O. Belmonte (Saint Denis de la Réunion), H. Boulestreau and N. Milpied (Bordeaux), J. Caillon (Nantes), D. Chrisment (Pessac), B. Compan and H. Jean-Pierre (Montpellier), F. Gouriet (Marseille), T. Lambert (Paris), V. Nègre (Martigues), E. Petat (Bellegarde), E. Pluquet and H. Guillon (Amiens), C. Recule (Grenoble), C. Segonds (Toulouse), G. Suermondt (Bourg Saint Maurice), A. Tachet (Pau), D. Tandé (Brest), B. Vaché (Gap), and N. Van Der Mee (Tours). We also thank Barbara Dehecq, Emeline Gilliot, Amélie Mille, and Pauline Chatelain for excellent technical assistance.
Ceftolozane-tazobactam was provided by Cubist Pharmaceuticals. VYNE was supported by the cystic fibrosis associations Vaincre la Mucoviscidose and Association Grégory Lemarchal. The French NRC for Antibiotic Resistance is funded by the French Ministry of Health through the InVS agency.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00825-15.
REFERENCES
- 1.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabath LD, Jago M, Abraham EP. 1965. Cephalosporinase and penicillinase activities of a β-lactamase from Pseudomonas pyocyanea. Biochem J 96:739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masuda N, Gotoh N, Ishii C, Sakagawa E, Ohya S, Nishino T. 1999. Interplay between chromosomal β-lactamase and the MexAB-OprM efflux system in intrinsic resistance to β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livermore DM, Williams RJ, Lindridge MA, Slack RC, Williams JD. 1982. Pseudomonas aeruginosa isolates with modified beta-lactamase inducibility: effects on beta-lactam sensitivity. Lancet i:1466–1467. [DOI] [PubMed] [Google Scholar]
- 5.Fung-Tomc J, Dougherty TJ, DeOrio FJ, Simich-Jacobson V, Kessler RE. 1989. Activity of cefepime against ceftazidime- and cefotaxime-resistant Gram-negative bacteria and its relationship to beta-lactamase levels. Antimicrob Agents Chemother 33:498–502. doi: 10.1128/AAC.33.4.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moya B, Dötsch A, Juan C, Blazquez J, Zamorano L, Haussler S, Oliver A. 2009. β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabot G, Ocampo-Sosa AA, Dominguez MA, Gago JF, Juan C, Tubau F, Rodriguez C, Moyà B, Pena C, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castanheira M, Mills JC, Farrell DJ, Jones RN. 2014. Mutation-driven β-lactam resistance mechanisms among contemporary ceftazidime-nonsusceptible Pseudomonas aeruginosa isolates from U.S. hospitals. Antimicrob Agents Chemother 58:6844–6850. doi: 10.1128/AAC.03681-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob Agents Chemother 51:4062–4070. doi: 10.1128/AAC.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabot G, Ocampo-Sosa AA, Tubau F, Macia MD, Rodriguez C, Moya B, Zamorano L, Suarez C, Pena C, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother 55:1906–1911. doi: 10.1128/AAC.01645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hocquet D, Berthelot P, Roussel-Delvallez M, Favre R, Jeannot K, Bajolet O, Marty N, Grattard F, Mariani-Kurkdjian P, Bingen E, Husson M-O, Couedic G, Plésiat P. 2007. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob Agents Chemother 51:3531–3536. doi: 10.1128/AAC.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moya B, Zamorano L, Juan C, Perez JL, Ge Y, Oliver A. 2010. Activity of a new cephalosporin, CXA-101 (FR264205), against beta-lactam-resistant Pseudomonas aeruginosa mutants selected in vitro and after antipseudomonal treatment of intensive care unit patients. Antimicrob Agents Chemother 54:1213–1217. doi: 10.1128/AAC.01104-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mushtaq S, Warner M, Livermore DM. 2010. In vitro activity of ceftazidime+NXL104 against Pseudomonas aeruginosa and other non-fermenters. J Antimicrob Chemother 65:2376–2381. doi: 10.1093/jac/dkq306. [DOI] [PubMed] [Google Scholar]
- 14.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahiri SD, Johnstone MR, Ross PL, McLaughlin RE, Olivier NB, Alm RA. 2014. Avibactam and class C β-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 58:5704–5713. doi: 10.1128/AAC.03057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 17.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tomkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 18.Bryan LE, Kwan S, Godfrey AJ. 1984. Resistance of Pseudomonas aeruginosa mutants with altered control of chromosomal β-lactamase to piperacillin, ceftazidime, and cefsulodin. Antimicrob Agents Chemother 25:382–384. doi: 10.1128/AAC.25.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardot C, Plésiat P, Fournier D, Monlezun L, Broutin I, Llanes C. 2015. Carbapenem resistance in cystic fibrosis strains of Pseudomonas aeruginosa as a result of amino acid substitutions in porin OprD. Int J Antimicrob Agents 45:529–532. doi: 10.1016/j.ijantimicag.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 20.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 21.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. M100-S24 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 22.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabot G, Bruchmann S, Mulet X, Zamorano L, Moya B, Juan C, Haussler S, Oliver A. 2014. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 58:3091–3099. doi: 10.1128/AAC.02462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumas J-L, vanDelden C, Perron K, Köhler T. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol Lett 254:217–225. doi: 10.1111/j.1574-6968.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 25.Jo JTH, Brinkman FSL, Hancock REW. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob Agents Chemother 47:1101–1111. doi: 10.1128/AAC.47.3.1101-1111.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Martinez JM, Poirel L, Nordmann P. 2009. Extended-spectrum cephalosporinases in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:1766–1771. doi: 10.1128/AAC.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam VH, Schilling AN, LaRocco MT, Gentry LO, Lolans K, Quinn JP, Garey KW. 2007. Prevalence of AmpC over-expression in bloodstream isolates of Pseudomonas aeruginosa. Clin Microbiol Infect 13:413–418. doi: 10.1111/j.1469-0691.2006.01674.x. [DOI] [PubMed] [Google Scholar]
- 28.Zamorano L, Moya B, Juan C, Oliver A. 2010. Differential β-lactam resistance response driven by ampD or dacB (PBP4) inactivation in genetically diverse Pseudomonas aeruginosa strains. J Antimicrob Chemother 65:1540–1542. doi: 10.1093/jac/dkq142. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Martinez JM, Poirel L, Nordmann P. 2009. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahyot S, Broutin I, de Champs C, Guillon H, Mammeri H. 2013. Contribution of asparagine 346 residue to the carbapenemase activity of CMY-2. FEMS Microbiol Lett 345:147–153. doi: 10.1111/1574-6968.12199. [DOI] [PubMed] [Google Scholar]
- 31.Livermore DM, Mushtaq S, Barker K, Hope R, Warner M, Woodford N. 2012. Characterization of beta-lactamase and porin mutants of Enterobacteriaceae selected with ceftaroline + avibactam (NXL104). J Antimicrob Chemother 67:1354–1358. doi: 10.1093/jac/dks079. [DOI] [PubMed] [Google Scholar]
- 32.Drawz SM, Taracila M, Caselli E, Prati F, Bonomo RA. 2011. Exploring sequence requirements for C3/C4 carboxylate recognition in the Pseudomonas aeruginosa cephalosporinase: insights into plasticity of the AmpC β-lactamase. Protein Sci 20:941–958. doi: 10.1002/pro.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-Olmos A, Garcia-Castillo M, Alba JM, Morosini MI, Lamas A, Romero B, Galan JC, del Campo R, Canton R. 2013. Population structure and antimicrobial susceptibility of both nonpersistent and persistent Pseudomonas aeruginosa isolates recovered from cystic fibrosis patients. J Clin Microbiol 51:2761–2765. doi: 10.1128/JCM.00802-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Castillo M, Maiz L, Morosini MI, Rodriguez-Banos M, Suarez L, Fernandez-Olmos A, Baquero F, Canton R, del Campo R. 2012. Emergence of a mutL mutation causing multilocus sequence typing-pulsed-field gel electrophoresis discrepancy among Pseudomonas aeruginosa isolates from a cystic fibrosis patient. J Clin Microbiol 50:1777–1778. doi: 10.1128/JCM.05478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren AE, Boulianne-Larsen CM, Chandler CB, Chiotti K, Kroll E, Miller SR, Taddei F, Sermet-Gaudelus I, Ferroni A, McInnerney K, Franklin MJ, Rosenzweig F. 2011. Genotypic and phenotypic variation in Pseudomonas aeruginosa reveals signatures of secondary infection and mutator activity in certain cystic fibrosis patients with chronic lung infections. Infect Immun 79:4802–4818. doi: 10.1128/IAI.05282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver A, Mena A. 2010. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect 16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- 38.Crémet L, Caroff N, Giraudeau C, Reynaud A, Caillon J, Corvec S. 2013. Detection of clonally related Escherichia coli isolates producing different CMY β-lactamases from a cystic fibrosis patient. J Antimicrob Chemother 68:1032–1035. doi: 10.1093/jac/dks520. [DOI] [PubMed] [Google Scholar]
- 39.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 40.Zhanel GG, Chung P, Adam H, Zelenitsky S, Denisuik A, Schweizer F, Lagace-Wiens PR, Rubinstein E, Gin AS, Walkty A, Hoban DJ, Lynch JP, Karlowsky JA. 2014. Ceftolozane/tazobactam: a novel cephalosporin/beta-lactamase inhibitor combination with activity against multidrug-resistant Gram-negative bacilli. Drugs 74:31–51. doi: 10.1007/s40265-013-0168-2. [DOI] [PubMed] [Google Scholar]
- 41.Blizzard TA, Chen H, Kim S, Wu J, Young K, Park YW, Ogawa A, Raghoobar S, Painter RE, Hairston N, Lee SH, Misura A, Felcetto T, Fitzgerald P, Sharma N, Lu J, Ha S, Hickey E, Hermes J, Hammond ML. 2010. Side chain SAR of bicyclic beta-lactamase inhibitors (BLIs). 1. Discovery of a class C BLI for combination with imipinem. Bioorg Med Chem Lett 20:918–921. doi: 10.1016/j.bmcl.2009.12.069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.