Abstract
Acquisition of blaNDM-1 in bacterial species, such as Proteus mirabilis that is intrinsically resistant to tetracycline, tigecycline and colistin, will make clinical treatment extremely difficult. Here, we characterized an NDM-1-producing clinical isolate of P. mirabilis (PM58) that displayed an extensively drug-resistant (XDR) phenotype, susceptible only to aztreonam. Molecular analysis revealed that PM58 harbored both a conjugative NDM-1 plasmid and a novel Salmonella genomic island 1 variant on chromosome.
TEXT
Global dissemination of New Delhi metallo-β-lactamase 1 (NDM-1), an Ambler class B metallo-β-lactamase (MBL) able to hydrolyze all β-lactams except monobactams, in Gram-negative bacteria represents a great threat for public health (1). Transmission of blaNDM-1 is of especial concern with bacterial species of intrinsic antibiotic resistance, such as Proteus mirabilis that is intrinsically resistant to tetracycline, tigecycline and colistin, which will make clinical treatment extremely difficult.
Salmonella genomic island 1 (SGI1), consisting of a conserved backbone structure and a multidrug resistance (MDR) region, is integrated into the chromosome specifically at the last 18 bp of the 3′-end of the thdF gene and was initially reported in Salmonella enterica serovar Typhimurium DT104 (2). To date, multiple variants of SGI1 have been discovered, and they are classified from SGI1-A to SGI1-Y, corresponding to various structures of the MDR region. Since several novel variants of SGI1, such as SGI1-L and SGI1-U to SGI1-Y, have been identified in P. mirabilis, this species is considered an acceptor of SGI independent of Salmonella spp. (3–5). Recently, the blaNDM-1 gene was reportedly integrated into a novel SGI1 variant, named PGI1 (Proteus genomic island 1), in a P. mirabilis clinical isolate (6). However, the blaNDM-1 carrying PGI1 was nontransferable in the presence of helper plasmid pR55, and this PGI1-harboring isolate remained susceptible to multiple antibiotics, including meropenem, ertapenem, gentamicin, and fosfomycin (6). Here, we describe a clinical extensively drug resistant (XDR) P. mirabilis isolate that harbors both a conjugative NDM-1 plasmid and a novel SGI1 variant on chromosome.
A 3-year-old girl was admitted to the emergency department of The First Affiliated Hospital of Zhengzhou University on August 10, 2013, with a diagnosis of spinal cord injury (SCI) caused by a knife injury. After the surgery operation on August 11, intravenous cefathiamidine was given empirically (1 g administered 2 times/day) for 1 week to prevent bacterial infections. After receiving treatment, the patient was in a stable condition and was transferred to the rehabilitation department for restorative treatment. Unfortunately, the patient developed symptoms of frequent and urgent micturition and urodynia on December 10, 2013, and was treated with gentamicin bladder irrigations via catheter (20 mg administered 2 times/day). On December 15, an XDR P. mirabilis isolate named PM58 in this study was obtained from the urine culture. According to the drug resistance pattern of PM58, antibiotic treatment was switched to aztreonam (30 mg/kg administered as an intravenous drip 2 times/day). After 1 week of treatment, the girl recovered, and the urine was negative for P. mirabilis.
Antimicrobial susceptibilities were tested by the microdilution method and the agar dilution method (for fosfomycin) following the CLSI guidelines, and the MIC results were interpreted according to the CLSI breakpoints (7). PM58 was resistant to all the tested antibiotics used for treatment of infections caused by Enterobacteriaceae, with the exception of aztreonam. (Table 1). The positive result for the imipenem-ethylenediaminetetraacetic acid (EDTA) double-disk synergy test indicated that PM58 produced metallo-β-lactamase (MBL). PCR and sequencing analysis identified the MBL and β-lactamase genes blaNDM-1 and blaTEM-1, as well as other genes conferring resistance to aminoglycoside (rmtB), quinolones (qnrA1), tetracyclines (tet[C], tet[J]), chloramphenicol (floR), and fosfomycin (fosA3). Primers used for detecting these resistance genes were described in previous studies (8–11). S1-pulsed field gel electrophoresis and Southern blotting demonstrated that the blaNDM-1 gene, together with rmtB and fosA3 genes, was located on an ∼85-kb plasmid (see Fig. S1 in the supplemental material), designated pNDM-PM58. Conjugative assay was performed using an established method to evaluate the transferability of pNDM-PM58 (8). The result showed that this plasmid was successfully transferred to Escherichia coli J53 at a frequency of 9.4 × 10−7 per donor cell. In addition, all of the resistance determinants detected in PM58 except blaTEM-1 were cotransferred with the blaNDM-1 gene to E. coli J53 by the mating experiment. This was confirmed by using PCR and sequencing.
TABLE 1.
Antibiotic susceptibilities of P. mirabilis PM58, its transconjugant PM58-J53, and recipient strain E. coli J53
| Antimicrobial category | Antimicrobial agent | MIC (μg/ml) |
||
|---|---|---|---|---|
| PM58 | PM58-J53 | E. coli J53 | ||
| Penicillins | Ampicillin | >256 | >256 | 8 |
| Penicillins + β-lactamase inhibitors | Ampicillin-sulbactam | >256 | >256 | 8 |
| Antipseudomonal penicillins + β-lactamase inhibitors | Piperacillin-tazobactam | >256 | >256 | 2 |
| Non-extended-spectrum cephalosporins | Cefazolin | >256 | >256 | 2 |
| Extended-spectrum cephalosporins | Ceftazidime | >256 | >256 | 0.5 |
| Cefepime | >256 | >256 | 0.25 | |
| Cephamycins | Cefotetan | >256 | >256 | 1 |
| Carbapenems | Imipenem | >64 | >64 | <0.125 |
| Meropenem | 8 | 8 | <0.125 | |
| Monobactams | Aztreonam | 1 | 1 | <0.125 |
| Fluoroquinolones | Levofloxacin | 32 | 8 | <0.125 |
| Aminoglycosides | Gentamicin | >64 | >64 | 1 |
| Amikacin | >64 | >64 | 1 | |
| Folate pathway inhibitors | Trimethoprim-sulfamethoxazole | >32 | >32 | 1 |
| Phenicols | Chloroamphenicol | >64 | >64 | 4 |
| Phosphonic acids | Fosfomycin | >1,024 | >1,024 | 2 |
| Tetracyclines | Doxycycline | 128 | 64 | 2 |
| Tetracycline | NDa | >64 | 2 | |
| Glycylcyclines | Tigecycline | NDa | 0.5 | 0.25 |
| Polymyxins | Colistin | NDa | 1 | 0.5 |
ND, not determined: P. mirabilis is intrinsically resistant to tetracycline, tigecycline and colistin.
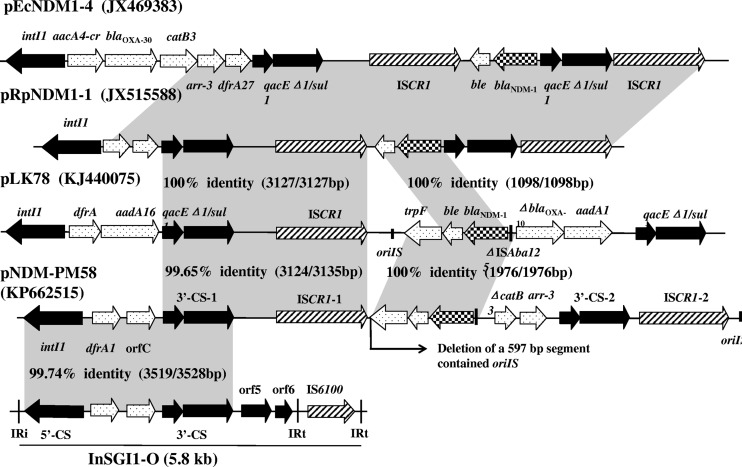
The surrounding genetic environment of the blaNDM-1 gene on pNDM-PM58 was sequenced by a modified random primer walking strategy (12), yielding a 12,146-bp fragment (GenBank accession number KP662515; Fig. 1). The blaNDM-1 gene was embedded in a complex class 1 integron, flanked by two copies of ISCR1 elements (ISCR1-1 and ISCR1-2) located in the same orientation. Similar structures were reported in E. coli and Raoultella planticola isolates from China in a very recent study (13). However, the resistance gene cassettes and genetic arrangement within the complex class 1 integron in PM58 were different from those of pEcNDM1-4, a 58-kb plasmid carrying blaNDM-1 in E. coli, and pRpNDM1-1, a 280-kb plasmid carrying blaNDM-1 in R. planticola (13) (Fig. 1). In PM58, a classical class 1 integron carrying a dfrA1-orfC cassette array was located upstream of the ISCR1-1 (immediately downstream of the trpF gene) and exhibited 99.74% (3,519 of 3,528 bp) nucleotide sequence identity to a corresponding segment of InSGI1-O, an In4 type integron that has been previously identified in P. mirabilis in China (14). Notably, the qacEΔ1/sul1-ISCR1-1-trpF-bleMBL-blaNDM-1-ΔISAba125 cassette array is quite similar to that of plasmid pLK78 (Genbank accession number KJ440075) (15), which differs by the deletion of a 597-bp segment-contained oriIS site immediately downstream of the ISCR1-1 element (Fig. 1). It has been documented that the ISCR1-like elements might be responsible for the mobilization of blaNDM-1 in Enterobacteriaceae and non-Enterobacteriaceae species via the rolling circle replication process (13, 15, 16). The acquisition of the blaNDM-1-carrying segment in the complex class 1 integron in PM58 may be related to ISCR1-2, since the oriIS site was identified downstream of this element.
FIG 1.
Genetic environment of the blaNDM-1 gene in the pNDM-PM58 of P. mirabilis 58 and structural comparison with Klebsiella pneumoniae plasmid pLK78 (Genbank accession number KJ440075), E. coli plasmid pEcNDM1-4 (Genbank accession number JX469383), pRpNDM1-1 (Genbank accession number JX515588) from Raoultella planticola, and the MDR region of SGI1-O from P. mirabilis. The boxed arrows indicate the positions and directions of transcription of the genes. The gray-shaded areas represent regions sharing >99% DNA identity.
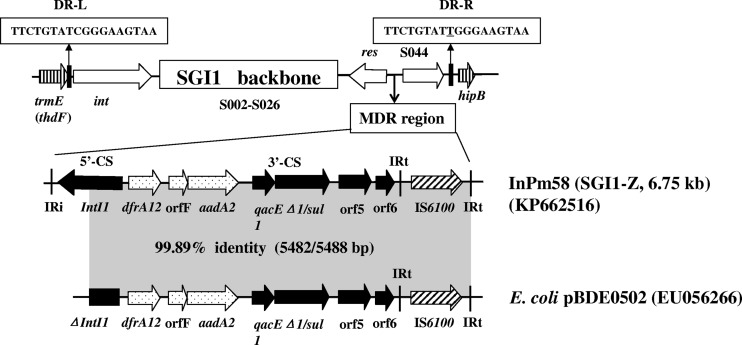
The presence of an InSGI1-O-like segment on pNDM-PM58 prompted us to investigate the possible presence of an SGI1-like element in PM58. A 35-kb chromosomal DNA fragment of PM58 was obtained by using PCR mapping and DNA sequencing, as described previously (5). It contained a novel SGI1 variant of 34.1 kb, named SGI1-Z in this study. The complete structure of SGI1-Z (backbone and MDR region) is shown in Fig. 2. Sequence analysis revealed that SGI1-Z was integrated between the chromosomal genes thdF and hipB, and the 18-bp sequences of the chromosomal attB site (direct repeat right [DR-R]) and the SGI1 attP site (direct repeat left [DR-L]) were confirmed. In addition, the DR-R located 109 bp downstream of open reading frame (ORF) S044 diverged in 1 bp from the DR-L (Fig. 2). Compared with SGI1 backbone of S. Typhimurium DT104, only seven single-base changes were observed. The MDR region of SGI1-Z belonging to the In4-type integron was integrated between the SGI1 backbone genes res (S027) and S044. The structure of the MDR region was further confirmed by long-range PCR (LR-PCR) and DNA sequencing using forward primer LR-F and reverse primer LR-R (located in the conserved regions of the S026 and hipB genes, respectively), which yielded an 8,645-bp amplicon that contained the MDR region of SGI1-Z. InSGI1-Z (6,755 bp) contained a dfrA12-orfF-aadA2 cassette array conferring resistance to trimethoprim, spectinomycin, and streptomycin. Comparative analysis of DNA sequence revealed InSGI1-Z was nearly identical to an In4-like backbone structure located on plasmid pBDE0502 from a clinical isolate of E. coli (GenBank accession number EU056266) (17). Given that the SGI1 is considered a hot spot for acquiring complex In4-type integrons (18), we speculate that InSGI1-Z might be derived from pBDE0502-like plasmids. The observations of an InSGI1-O-like structure on pNDM-PM58 and InSGI1-Z corresponding to an In4-type integron on pBDE0502 suggest the exchange of gene cassettes between plasmid and chromosome via integron-mediated recombination.
FIG 2.
Schematic view of SGI1-Z integrated into the chromosome of P. mirabilis 58, and comparison of the MDR region of SGI1-Z (InPm58) with an In4-like backbone structure on E. coli plasmid pBDE0502 (EU056266). The boxed arrows indicate the positions and directions of transcription of the genes. The gray-shaded areas represent regions sharing >99% DNA identity. DR-L and DR-R represent the 18-bp direct repeats at the ends of SGI1. CS, conserved segment; IR, inverted repeat.
In conclusion, our study represents the first report of coexistence of a self-transferable NDM-1 plasmid and a novel chromosomal SGI1 variant in an XDR P. mirabilis isolate that is susceptible only to aztreonam. It is unclear how this XDR isolate originated in the patient. It could have originated from the treatment process due to overuse or inappropriate use of antibiotic therapy. In this case, the patient received intravenous cefathiamidine for a week to prevent bacterial infection, but this strategy is non-evidence based and not plausible, which might have served as a selection pressure for the emergence of the XDR isolate. Alternatively, the patient could have been infected by the XDR P. mirabilis isolate from other sources. Regardless, rational and evidence-based use of antibiotics should be emphasized in clinical therapy. A recent review recommended a combination of two or even three antibiotics among aminoglycoside, fosfomycin, high-dose tigecycline, and colistin for the treatment of infections caused by carbapenem-resistant Enterobacteriaceae with MICs for carbapenems higher than 8 μg/ml (19). Since P. mirabilis is intrinsically resistant to tigecycline and colistin, the emergence and spread of XDR P. mirabilis carrying plasmid-mediated resistance to carbapenems (blaNDM-1), fosfomycin (fosA3), and aminoglycosides (rmtB) will severely compromise the utility of combination regimens in treating infections attributed to P. mirabilis. Thus, detection and surveillance of XDR P. mirabilis and its resistance elements are urgently warranted to control their further spread.
Nucleotide sequence accession number.
The sequence of the novel complex class 1 integron carrying blaNDM-1 and the complete SGI1-Z sequence were submitted to GenBank under accession numbers KP662515 and KP662516, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant 81172937 from the National Natural Science Foundation of China, grant 112102310165 from the Scientific and Technologic Development Program of Henan Province, and grant 201303046 from the Medical Science and Technique Foundation of Henan Province.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00292-15.
REFERENCES
- 1.Dortet L, Poirel L, Nordmann P. 2014. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55:1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 3.Bi S, Yan H, Chen M, Zhang Z, Shi L, Wang H. 2011. New variant Salmonella genomic island 1-U in Proteus mirabilis clinical and food isolates from South China. J Antimicrob Chemother 66:1178–1179. doi: 10.1093/jac/dkr030. [DOI] [PubMed] [Google Scholar]
- 4.Lei CW, Zhang AY, Liu BH, Wang HN, Guan ZB, Xu CW, Xia QQ, Cheng H, Zhang DD. 2014. Molecular characteristics of Salmonella genomic island 1 in Proteus mirabilis isolates from poultry farms in China. Antimicrob Agents Chemother 58:7570–7572. doi: 10.1128/AAC.03992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siebor E, Neuwirth C. 2013. Emergence of Salmonella genomic island 1 (SGI1) among Proteus mirabilis clinical isolates in Dijon, France. J Antimicrob Chemother 68:1750–1756. doi: 10.1093/jac/dkt100. [DOI] [PubMed] [Google Scholar]
- 6.Girlich D, Dortet L, Poirel L, Nordmann P. 2015. Integration of the blaNDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-PmPEL) in a Proteus mirabilis clinical isolate. J Antimicrob Chemother 70:98–102. doi: 10.1093/jac/dku371. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI M100-S24 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Qin S, Fu Y, Zhang Q, Qi H, Wen JG, Xu H, Xu L, Zeng L, Tian H, Rong L, Li Y, Shan L, Yu Y, Feng X, Liu HM. 2014. High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob Agents Chemother 58:4275–4282. doi: 10.1128/AAC.02813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 10.Fan W, Hamilton T, Webster-Sesay S, Nikolich MP, Lindler LE. 2007. Multiplex real-time SYBR green I PCR assay for detection of tetracycline efflux genes of Gram-negative bacteria. Mol Cell Probes 21:245–256. doi: 10.1016/j.mcp.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Kadlec K, Kehrenberg C, Schwarz S. 2007. Efflux-mediated resistance to florfenicol and/or chloramphenicol in Bordetella bronchiseptica: identification of a novel chloramphenicol exporter. J Antimicrob Chemother 59:191–196. doi: 10.1093/jac/dkl498. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Wu C, Zhang Q, Qi J, Liu H, Wang Y, He T, Ma L, Lai J, Shen Z, Liu Y, Shen. 2012. Identification of New Delhi metallo-β-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One 7:e37152. doi: 10.1371/journal.pone.0037152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Lan R, Xiong Y, Ye C, Yuan M, Liu X, Chen X, Yu D, Liu B, Lin W, Bai X, Wang Y, Sun Q, Zhao H, Meng Q, Chen Q, Zhao A, Xu J. 2014. Sequential isolation in a patient of Raoultella planticola and Escherichia coli bearing a novel ISCR1 element carrying blaNDM-1. PLoS One 9:e89893. doi: 10.1371/journal.pone.0089893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd DA, Shi X, Hu QH, Ng LK, Doublet B, Cloeckaert A, Mulvey MR. 2008. Salmonella genomic island 1 (SGI1), variant SGI1-I, and new variant SGI1-O in Proteus mirabilis clinical and food isolates from China. Antimicrob Agents Chemother 52:340–344. doi: 10.1128/AAC.00902-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CJ, Wu TL, Lu PL, Chen YT, Fung CP, Chuang YC, Lin JC, Siu LK. 2014. Closely related NDM-1-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. PLoS One 9:e104899. doi: 10.1371/journal.pone.0104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janvier F, Jeannot K, Tesse S, Robert-Nicoud M, Delacour H, Rapp C, Merens A. 2013. Molecular characterization of blaNDM-1 in a sequence type 235 Pseudomonas aeruginosa isolate from France. Antimicrob Agents Chemother 57:3408–3411. doi: 10.1128/AAC.02334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae IK, Lee YH, Jeong HJ, Hong SG, Lee SH, Jeong SH. 2008. A novel bla(CTX-M-14) gene-harboring complex class 1 integron with an In4-like backbone structure from a clinical isolate of Escherichia coli. Diagn Microbiol Infect Dis 62:340–342. doi: 10.1016/j.diagmicrobio.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Doublet B, Praud K, Weill FX, Cloeckaert A. 2009. Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J Antimicrob Chemother 63:282–289. doi: 10.1093/jac/dkn500. [DOI] [PubMed] [Google Scholar]
- 19.Rafailidis PI, Falagas ME. 2014. Options for treating carbapenem-resistant Enterobacteriaceae. Curr Opin Infect Dis 27:479–483. doi: 10.1097/QCO.0000000000000109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.